Abstract
Current understanding of the population genetics of free-living unicellular eukaryotes is limited, and the amount of genetic variability in these organisms is still a matter of debate. We characterized—reproductively and genetically—worldwide samples of multiple Paramecium species belonging to a cryptic species complex, Paramecium aurelia, whose species have been shown to be reproductively isolated. We found that levels of genetic diversity both in the nucleus and in the mitochondrion are substantial within groups of reproductively compatible P. aurelia strains but drop considerably when strains are partitioned according to their phylogenetic groupings. Our study reveals the existence of discrepancies between the mating behavior of a number of P. aurelia strains and their multilocus genetic profile, a controversial finding that has major consequences for both the current methods of species assignment and the species problem in the P. aurelia complex.
Keywords: Paramecium, genetic diversity, effective population size, speciation, cryptic species, mating group switching
Introduction
Protists are among the most abundant and diverse eukaryotic groups (Patterson 1999), yet current knowledge of these organisms is surprisingly sparse, compared with that for animals, plants, and fungi. Two aspects of microbial eukaryotes that still remain controversial are the level of species diversity and their geographic distribution (Finlay and Fenchel 1999; Foissner 1999). One school of thought maintains that protist species are relatively few, cosmopolitan, and ubiquitous (Fenchel et al. 1997); another supports essentially the opposite view, that is, protist species are numerous and tend to be endemic and isolated (Foissner 2006). The currently limited understanding of the protist world (Weisse 2008) makes it difficult at this time to support one hypothesis or the other, in part due to the paucity of population-genetic studies that could shed light on the global (effective) population sizes of free-living species.
In a species, the genetic effective population size (Ne) (Wright 1931)—which modulates the efficiency of natural selection via its effects on random genetic drift (Ohta 1992)—reflects the number of breeding individuals as well as aspects of the breeding system, population structure, and degree of linkage, and 4Neμ (where μ is the neutral mutation rate) is the expected neutral genetic variation (average nucleotide heterozygosity per site) of a diploid population at mutation–drift equilibrium. As estimates of μ are available for a number of species (Lynch 2006a), measures of neutral genetic variation (e.g., diversity at synonymous sites in protein-coding genes) allow Ne to be inferred indirectly from estimates of variation at silent sites in natural populations.
Although large-scale polymorphism studies of protist pathogens (e.g., Grigg et al. 2001; Mu et al. 2002) have provided measures of neutral genetic variation (Lynch and Conery 2003; Lynch 2006b), the neutral genetic variation of pathogenic organisms is expected to be biased downward and not directly applicable to free-living taxa. On the other hand, a number of small-scale polymorphism surveys of protein-coding loci in free-living microbes, freshwater ciliates in particular, have produced conflicting estimates of intraspecific variation (Gerber et al. 2002; Barth et al. 2006; Katz et al. 2006; Snoke et al. 2006), thus impeding wider conclusions.
Not only can polymorphism surveys based on single or a few genes produce estimates of 4Neμ that are potentially misleading, but poorly defined species boundaries also make the estimation of intraspecific variation problematic. The latter issue may well represent a confounding factor in the computation of genetic diversity for microbial organisms (Daubin and Moran 2004), as the process of “species” identification is a difficult task, given the frequent existence of cryptic species (Waugh 2007), and may require a combination of approaches that include molecular, morphological, and ecological studies (Weisse 2008).
In this sense, the free-living Paramecium is a very suitable system, as the “species problem” in this ciliate group, and in P. aurelia in particular, has been intensely investigated. Paramecium aurelia is a complex that contains 15 species (Sonneborn 1975; Aufderheide et al. 1983). Despite being morphologically identical, these species are genetically isolated and, to quote Sonneborn (1975), “the distinguishing characters of each of the [then 14] species include its mating reactions, breeding relations, and mode of inheritance of mating type.” Specifically, each of the P. aurelia species contains two mating types—E (for “even”) and O (for “odd”)—that determine intraspecific sexual reproduction by conjugation (Sonneborn 1937). When two clones that have either the same mating type or belong to different species are mixed, no conjugation occurs. Although weak cross-reactions between distinct mating types of different species may be observed (e.g., between Paramecium tetraurelia and Paramecium octaurelia or between Paramecium primaurelia and Paramecium pentaurelia) (Beale and Preer 2008), these reactions result in the death of the conjugating cells or in the sterility of their F1s. The reproductive cohesion of P. aurelia species (Sonneborn 1975), the available macronuclear genome sequence of P. tetraurelia (Aury et al. 2006), and a variety of worldwide samples collected during both the past century and in very recent years, make this species complex an appealing candidate for a population-genetic study.
In the attempt to shed light on extant controversial results (Katz et al. 2006; Snoke et al. 2006) and to expand our knowledge on the levels of genetic diversity in free-living unicellular eukaryotes, we performed a polymorphism survey of multiple nuclear and mitochondrial loci in a large set of P. aurelia strains. Our analysis aims to overcome potential limitations of previous studies on free-living unicellular eukaryotes, such as limited sample sizes and both restricted number of genes and sampled locations. Along with a number of insights to the population genetics of multiple species of the P. aurelia complex, we found a surprising incongruence between mating behavior and strains’ genetic profiles, which complicates definitive conclusions on intraspecific levels of genetic variability.
Material and Methods
Paramecium Strains
We examined a total of 80 Paramecium strains, 79 members of the P. aurelia species complex (Sonneborn 1975; Aufderheide et al. 1983) (table 1), and 1 Paramecium multimicronucleatum strain. A total of 26 strains were kindly provided by J. Cohen, H. Schmidt, M. Simon, T. Berendonk, and K. Aufderheide, whereas the remaining 54 strains are maintained in the Collection of Paramecium of the Laboratory of Protozoan Karyology, St Petersburg State University. All strains are fully homozygous, following multiple rounds of autogamy.
Table 1.
List of Paramecium Strains/Species Surveyed
| Strain | Species | Origin |
| 16 | Paramecium primaurelia | Woodstock, MD, United States |
| 168 | Japan | |
| 33 | Unknown | |
| 60 | United States | |
| 61 | Unknown | |
| 90 | Pennsylvania, United States | |
| AZ15-8 | Astrakhan Nature Reserve, Russia | |
| AZ9-3 | Astrakhan Nature Reserve, Russia | |
| Ir4-1 | The Irtysh river, Omsk, Russia | |
| KK2-7 | Kaliningrad region, Russia | |
| TB10 | Rejkjavik, Iceland | |
| V7-6 | Volgograd region, Russia | |
| 223-7 | The Volga river, Tver’ region, Russia | |
| 112-1 | Vologda Region, Russia | |
| 227 | Unknown | |
| 1038s | Paramecium biaurelia | Unknown |
| 114 | Bloomington, IN, United States (uncertain) | |
| AB7-16 | Boston, United States | |
| AZ25-2 | Astrakhan Nature Reserve, Russia | |
| F2 | Unknown | |
| GA62-1 | Altai Mountains, Russia | |
| Kr133-2 | Krasnoyarsk, Russia | |
| P2 | Unknown | |
| PK2 | Kraków, Poland | |
| RR2-1 | Saint-Petersburg region, Russia | |
| TB15 | Teneriffa, Canary Islands, Spain | |
| UM2 | Unknown | |
| V1-4 | Volgograd region, Russia | |
| W7 | Unknown | |
| 152alpha | Paramecium triaurelia | New Haven, United States |
| EP_2/3 | Natural reserve Complex Volga-Ahtuba, Russia | |
| kr128 | Krasnoyarsk, Russia | |
| Kr149-1 | Krasnoyarsk, Russia | |
| TB17 | Greece | |
| TB18 | Santuhalm near Deva, Romania | |
| V10-7 | Volgograd region, Russia | |
| 172 | Paramecium tetraurelia | Perú |
| 298s | Unknown | |
| 32 | Unknown | |
| 51s | Indiana, United States | |
| d4-2 | — | |
| nr7-1 | Novorossiysk, Russia | |
| PF15 | Paris, France | |
| S | Sydney, Australia | |
| ST | Tatry, Slovakia | |
| BGD19 | Unknown | |
| AZ6-24 | Paramecium pentaurelia | Astrakhan Nature Reserve, Russia |
| GA1-9 | Altai Mountains, Russia | |
| Nr1-10 | Novorossiysk, Russia | |
| RA81-8 | Altai Forelands, Russia | |
| V2-7 | Volgograd region, Russia | |
| 159 | Paramecium sexaurelia | Unknown |
| AZ8-4 | Astrakhan Nature Reserve, Russia | |
| CB16-2 | Beijing, China | |
| AZ5-2 | Paramecium septaurelia | Astrakhan Nature Reserve, Russia |
| AZ8-3 | Astrakhan Nature Reserve, Russia | |
| GFg-1 | Freiburg, Germany | |
| V5-13 | Volgograd region, Russia | |
| 138 | Paramecium octaurelia | Florida, United States |
| 565s | Unknown | |
| IEE | Ein Efek, Israel | |
| K8 | Unknown | |
| K9 | Unknown | |
| AB8-22 | Paramecium novaurelia | Boston, United States |
| Iz16 | Izmail, Ukraine | |
| V9-6 | Volgograd region, Russia | |
| VL4-8 | Vladimir, Russia | |
| 223 | Paramecium decaurelia | Florida, United States |
| 93-21 | Yaroslavl region, Russia | |
| GA1-12 | Altai Mountains, Russia | |
| JN | Nara, Japan | |
| 219 | Paramecium undecaurelia | United States |
| 246 | Paramecium dodecaurelia | Mississippi, United States |
| 256-2 | Yaroslavl region, Russia | |
| UV1-3 | Vinnitsa, Ukraine | |
| 209 | Paramecium tredecaurelia | Paris, France |
| 328 | Paramecium quadecaurelia | Australia |
| AN1 | Namibia | |
| Paramecium sonneborni | Texas, United States | |
| Paramecium multimicronucleatum | Krasnoyarsk, Russia |
Culturing and Identification of Paramecium Strains
Paramecia were cultured in a medium made of dried lettuce (or Yeast extract, Cerophyl, Na2HPO4, and Stigmasterol) and distilled water inoculated with Enterobacter aerogenes and identified according to the methods of Sonneborn (1970). Clones matured for conjugation were mated with the reactive complementary mating types of standard strains of all species of the P. aurelia complex.
In intrastrain and interstrain crosses, the F1 generation was obtained by conjugation and F2 by autogamy (using the method of daily isolation lines). The occurrence of the desired stage of autogamy (specimens at the stage of two macronuclear anlagen) was examined on preparations stained with aceto-carmine. Survival of clones in both generations was estimated as percentages. According to Chen (1956), clones can be considered as surviving after passing six to seven fissions during 72 h after separation of partners of conjugation or postautogamous caryonides. The methods are described in detail in Przyboś (1975).
DNA Extraction and Gene Sequencing
Genomic DNA extraction was performed by incubating Paramecium cells for 20 min at 90 °C with 5% Chelex 100 Resin. We surveyed genetic variability at 10 nuclear and 5 mitochondrial protein-coding loci (table 2). In addition, we sequenced three rDNA fragments: a portion of the nuclear small rDNA subunit (SSU), the two internal transcribed spacers (ITS1 and ITS2), and a mitochondrial fragment encompassing the 5.8S and large rDNA subunit (LSU) (table 2).
Table 2.
Paramecium Genes Surveyed and Primers Used for PCR Amplification
| Nuclear Loci | Molecular Function | PCR Primers (5′–3′) |
| GSPATG00000151001 | DNA mismatch repair system Msh2 | gagagacatctaaactgtgcgtttt |
| tggcattttgatccatcttc | ||
| GSPATG00000223001 | Calcium binding protein | ttccctgaccgaataggattt |
| ggccataagcatccaagatt | ||
| GSPATG00000363001 | tRNA methyltransferase | gcagcagctgttggtcataa |
| tccagtgtgtccataagcttaattt | ||
| GSPATG00000454001 | GTP-binding protein RAB2 homolog | tgaacatgatgcaactattggag |
| gtggatttgcaggtttgcta | ||
| GSPATG00000546001 | GrpE protein homolog | tcccttgatgaatctcacga |
| tgttcctggttctttttcagg | ||
| GSPATG00027725001 | 3-Oxoadipate enol-lactonase | ttggcaggatggtctttagg |
| tggcatatgaccaacacctg | ||
| GSPATG00028095001 | Serine carboxypeptidase | ttacatcattagtgtttgtcaggtg |
| cctgcataactctccccaaa | ||
| GSPATG00028110001 | Hypothetical protein | agttggcgataatggaggac |
| aaccatcagctccatcaacc | ||
| GSPATG00022332001 | Ubiquitin-like 1 activating enzyme E1B | tggcattggataatgcagaa |
| gtagaggcaatggcgtgaat | ||
| GSPATG00020758001 | Hypothetical protein | aagccacctccagattttca |
| tttcccagatcgaacctttg | ||
| Mitochondrial loci | ||
| NADH 3 | tgggtagtatgacattgcttttc | |
| ggccagcgagttctggac | ||
| ORF189-2 & yegR | gccctctcagagctcaacat | |
| acaccgggtggacgtaga | ||
| COX I | ccaaaattggggttacgatg | |
| attgcgtttttgaggacgac | ||
| COX II | gcatagaagtagtcatcagcaacc | |
| ttttgtgaggttgtatttccaca | ||
| rDNA loci | ||
| SSUa (nuclear) | gcaagtctggtgccagcagcc | |
| cttccgtcaattcctttaag | ||
| ITS–5.8S rDNA–ITSa (nuclear) | tcctccgcttattgatatgc | |
| ggaagtaaaagtcgtaacaagg | ||
| 5.8S-LSU (mitochondrial) | ttggttgagggcgtaaatct | |
| ctaccccgcacaaaagaaaa | ||
Primer sequences match NS3, NS4, ITS4, and ITS5 in White et al. (1990).
We used the P. tetraurelia macronuclear (http://paramecium.cgm.cnrs-gif.fr/) (Aury et al. 2006) and mitochondrial (Pritchard et al. 1990) genome sequences and the freely available online software “Primer3” (Rozen and Skaletsky 2000) to design DNA oligo pairs to coding regions for polymerase chain reaction (PCR) amplifications (table 2 and supplementary table 3 in Supplementary Material online). More than a single pair of oligos was often required to amplify a locus across most or all strains surveyed. The nuclear loci studied were random single-copy genes in the P. tetraurelia macronuclear genome. We amplified the gene regions using the following PCR conditions: 3 min at 94 °C, 40 cycles of 40 s at 94 °C, 40 s at 55 °C, and 1 min at 72 °C. Amplification products were subsequently checked on 1% agarose gels, and reactions giving unique products were sequenced in both directions. We verified the quality of the sequenced gene fragments both automatically, using the software Codon Code Aligner, and visually. We used ClustalW (Thompson et al. 1994), as implemented in the software Bioedit (Hall 1999), to align the consensus sequences.
Data Analysis
Sequences were exported to the software MEGA 4.0 (Nei and Kumar 2000) for data analysis. For each protein-coding locus, we measured intraspecies and interspecies diversity at both silent sites and replacement sites, following the Kumar method (Nei and Kumar 2000). Diversity values were multiplied by n/(n − 1), where n is the appropriate number of strains for each species, to correct for small sample size, and the average values were weighted by the number of base pairs sequenced for each locus. The Maximum Composite Likelihood model implemented in MEGA 4.0 and bootstrap statistics (1,000 replicates) were used to build Neighbor-Joining (NJ) trees using all the sites of the regions examined. We also measured diversity in single or concatenated intronic regions (the latter when genes contained more than one intron) contained in each of the surveyed loci, after removing the conserved GT–AG ending sites. The package GENECONV (Sawyer 1999) was employed to estimate both the number and the location of possible recombinant alleles.
All the sequences generated in this study were submitted to GenBank, accession numbers FJ002921–FJ004151.
Results and Discussion
High Levels of Silent Genetic Variation within Reproductively Compatible P. aurelia Strains
Levels of intraspecific diversity at nuclear synonymous sites (πs) in the P. aurelia species complex average 0.1493 (SEM = 0.0382) (table 3 and Supplementary Material online). Diversity at silent sites is ∼13-fold higher than at nonsynonymous sites (πa = 0.0117 [SEM = 0.0031]) but is comparable with that calculated for intronic sites (πi = 0.0944 [SEM = 0.0246]). No intron presence or absence polymorphisms were detected across species, and levels of diversity in coding and intronic regions show significant positive correlations when estimates for single loci are examined (πa vs. πi, Spearman's rho = 0.643, P < 0.01; πs vs. πi, Spearman's rho = 0.540, P < 0.01). In the mitochondria, loci also showed a substantial degree of variation (πs = 0.3257 [SEM = 0.0674] and πa = 0.0173 [SEM = 0.0057]), with silent sites having on average ∼2.2 times the nucleotide diversity of nuclear genes (table 4 and Supplementary Material online). Levels of net interspecific divergence (i.e., in excess of within-species divergence) at silent sites average 0.3307 (SEM = 0.0209) for the nuclear loci (supplementary table 1 in Supplementary Material online) and 0.5214 (SEM = 0.0184) for mitochondrial loci (supplementary table 2 in Supplementary Material online). At amino acid replacement sites, divergence drops to 0.0239 (SEM = 0.0012) and 0.0175 (SEM = 0.0175) for nuclear and mitochondrial loci, respectively. The ratio of mitochondrial to nuclear divergence at (neutrally evolving) silent sites reflects the ratio of the mutation rates in the two cellular compartments and averages 4.0223 (SEM = 2.6470) when all species are examined but decreases to 1.2504 (SEM = 0.2451) after the removal of Paramecium sexaurelia, a species previously reported to show unusually divergent genotypes across different geographic locations and where F2 generations of interstrain crosses show low rates of survival (Stoeck et al. 1998; Przybos, Rautian, et al. 2007).
Table 3.
Intraspecific Genetic Diversities at Nonsilent Sites (πa), Silent Sites (πs), and Intronic Sites (πi) with Corresponding Standard Errors (SE), Estimated for All Strains across 10 Nuclear Loci
| Species | All Strains |
Only Nondiscordant Strains |
||||||||
| Average No. of Strains | πa (SE) | πs (SE) | πa/πs | πi (SE) | Average No. of Strains | πa (SE) | πs (SE) | πa/πs | πi (SE) | |
| Paramecium primaurelia | 12 | 0.0011 (0.0003) | 0.0248 (0.0058) | 0.0444 | 0.0249 (0.0142) | 11 | 0.0010 (0.0003) | 0.0228 (0.0055) | 0.0439 | 0.0240 (0.0144) |
| Paramecium biaurelia | 10 | 0.0131 (0.0047) | 0.1683 (0.0695) | 0.0778 | 0.1302 (0.1010) | 10 | 0.0004 (0.0003) | 0.0038 (0.0015) | 0.1053 | 0.0075 (0.0095) |
| Paramecium triaurelia | 7 | 0.0029 (0.0007) | 0.0344 (0.0069) | 0.0843 | 0.0167 (0.0083) | 6 | 0.0003 (0.0003) | 0.0066 (0.0025) | 0.0455 | 0.0010 (0.0013) |
| Paramecium tetraurelia | 10 | 0.0076 (0.0013) | 0.0908 (0.0116) | 0.0837 | 0.0446 (0.0130) | 9 | 0.0007 (0.0004) | 0.0079 (0.0028) | 0.0886 | 0.0040 (0.0034) |
| Paramecium pentaurelia | 4 | 0.0108 (0.0056) | 0.1208 (0.0555) | 0.0891 | 0.0416 (0.0351) | 4 | 0.0009 (0.0012) | 0.0017 (0.0021) | 0.5294 | 0.0036 (0.0046) |
| Paramecium sexaurelia | 3 | 0.0003 (0.0006) | 0.0038 (0.0079) | 0.0789 | 0.0058 (0.0166) | 3 | 0.0003 (0.0006) | 0.0038 (0.0079) | 0.0789 | 0.0057 (0.0163) |
| Paramecium septaurelia | 4 | 0.0197 (0.0040) | 0.2748 (0.0470) | 0.0717 | 0.1514 (0.0505) | 3 | 0.0004 (0.0003) | 0.0034 (0.0024) | 0.1176 | 0.0053 (0.0060) |
| Paramecium octaurelia | 5 | 0.0057 (0.0014) | 0.0743 (0.0081) | 0.0767 | 0.0498 (0.0211) | 4 | 0.0003 (0.0002) | 0.0036 (0.0016) | 0.0833 | 0.0085 (0.0069) |
| Paramecium novaurelia | 3 | 0.0166 (0.0037) | 0.2561 (0.0336) | 0.0648 | 0.2012 (0.1128) | 2 | 0 | 0.0091 (0.0130) | 0.0000 | 0 |
| Paramecium decaurelia | 3 | 0.0294 (0.0094) | 0.2940 (0.0711) | 0.1000 | 0.2451 (0.1141) | 2 | 0.0023 (0.0020) | 0.0104 (0.0068) | 0.2212 | 0.0109 (0.0184) |
| Paramecium dodecaurealia | 3 | 0.0310 (0.0064) | 0.4154 (0.0740) | 0.0746 | 0.1971 (0.0849) | 2 | 0.0008 (0.0009) | 0.0078 (0.0061) | 0.1026 | 0.0109 (0.0158) |
| Paramecium quadecaurelia | 2 | 0.0019 (0.0040) | 0.0345 (0.0115) | 0.0551 | 0.0241 (0.0811) | 2 | 0.0019 (0.0040) | 0.0345 (0.0115) | 0.0551 | 0.0245 (0.0823) |
| 1 | ||||||||||
| Average | 0.0117 (0.0031) | 0.1493 (0.0382) | 0.0751 (0.0043) | 0.0944 (0.0246) | 0.0008 (0.0002) | 0.0096 (0.0028) | 0.1226 (0.0401) | 0.0088 (0.0023) | ||
Table 4.
Intraspecific Genetic Diversities at Nonsilent (πa) and Silent Sites (πs), with Corresponding SE, Estimated for All Strains across Five Mitochondrial Loci
| Species | All Strains |
Only Nondiscordant Strains |
||||||
| Average No. of Strains | πa (SE) | πs (SE) | πa/πs | Average No of Strains | πa (SE) | πs (SE) | πa/πs | |
| Paramecium primaurelia | 15 | 0.0022 (0.0005) | 0.1074 (0.0279) | 0.0205 | 14 | 0.0020 (0.0005) | 0.0986 (0.0257) | 0.0203 |
| Paramecium biaurelia | 13 | 0.0123 (0.0057) | 0.2207 (0.0417) | 0.0557 | 11 | 0.0002 (0.0001) | 0.0043 (0.0013) | 0.0465 |
| Paramecium triaurelia | 7 | 0.0039 (0.0016) | 0.2124 (0.0378) | 0.0184 | 6 | 0 | 0 | |
| Paramecium tetraurelia | 10 | 0.0100 (0.0052) | 0.1255 (0.0309) | 0.0797 | 9 | 0.0006 (0.0006) | 0.0157 (0.0046) | 0.0382 |
| Paramecium pentaurelia | 5 | 0.0123 (0.0075) | 0.1688 (0.0833) | 0.0729 | 4 | 0 | 0 | |
| Paramecium sexaurelia | 2 | 0 | 0.1563 (0.1260) | 0 | 2 | 0 | 0.1563 (0.1260) | 0 |
| Paramecium septaurelia | 4 | 0.0225 (0.0096) | 0.6567 (0.1080) | 0.0343 | 3 | 0 | 0.0052 (0.0041) | 0 |
| Paramecium octaurelia | 5 | 0.0112 (0.0079) | 0.2904 (0.0477) | 0.0386 | 4 | 0 | 0.0115 (0.0040) | 0 |
| Paramecium novaureliaa | 7 | 0.0308 (0.0164) | 0.5744 (0.0848) | 0.0536 | 5 | 0.0016 (0.0018) | 0.1100 (0.1118) | 0.0145 |
| Paramecium decaurelia | 3 | 0.0695 (0.0530) | 0.7804 (0.1740) | 0.0891 | 1 | 0 | 0 | |
| Paramecium dodecaurealia | 2 | 0.0314 (0.0307) | 0.4729 (0.2252) | 0.0664 | 2 | 0 | 0.0080 (0.0129) | 0 |
| Paramecium quadecaurelia | 2 | 0.0016 (0.0080) | 0.1420 (0.0599) | 0.0113 | 1.7 | 0.0016 (0.0080) | 0.1420 (0.0599) | 0.0113 |
| Average | 0.0173 (0.0057) | 0.3257 (0.0674) | 0.0450 (0.0083) | 0.0005 (0.0002) | 0.0460 (0.0177) | 0.0145 (0.0059) | ||
COX I sequences of 21 P. novaurelia strains (Przybos, Tarcz, and Skoblo 2007; Tarcz S., unpublished data) were retrieved from NCBI and analyzed along with the strains surveyed in this study.
Low Levels of Genetic Variation within Monophyletic Taxa
The analysis of the gene genealogies across all nuclear and mitochondrial loci revealed that 13 “discordant” strains consistently group with species that are different from that to which they were originally assigned by mating tests (table 5 and supplementary fig. 1 in Supplementary Material online). We verified this unexpected finding by further studying the macronuclear SSU and ITSs and the mitochondrial 5.8 and LSU sequences (Supplementary Material online) and reperforming mating experiments on this set of suspect strains. We conclude that although these strains contain a multilocus genetic profile that is highly similar to that of a given P. aurelia species, they preferentially mate with strains belonging to a different species within the species complex. As it is legitimate to question whether or not these discordant strains ought to be included in the calculation of intraspecific genetic variability, we also calculated levels of polymorphism after their exclusion. We found that this leads to a considerable decrease in within-species genetic variability, with an average πs of 0.0096 (SEM = 0.0028) and 0.0460 (SEM = 0.0177) in the nucleus and in the mitochondrion, respectively (tables 3 and 4 and Supplementary Material online). The net interspecific average divergence at silent sites increases to 0.4034 (SEM = 0.0198) and to 0.6717 (SEM = 0.0203) at nuclear and mitochondrial loci, respectively (supplementary tables 1 and 2 in Supplementary Material online), but it remains similar at nonsynonymous sites (0.0293 [SEM = 0.0012] and 0.0269 [SEM = 0.0016]). In addition, levels of diversity at intronic sites and at amino acid replacement sites are no longer significantly correlated (πa vs. πi, Spearman's rho = 0.032, P = 0.772; πs vs. πi, Spearman's rho = 0.292, P < 0.01). The ratio of mitochondrial-to-nuclear divergence at silent sites averages 3.8049 (SEM = 2.5900) and decreases to 1.1597 (SEM = 0.4212) after the removal of P. sexaurelia.
Table 5.
List of Discordant Strains
| Strain | Species According to Mating Tests | Species According to mt rRNAs and Nuclear ITS | Species According to Multiple Nuclear and mt Loci |
| 61 | Paramecium primaurelia | P. primaurelia/Paramecium pentaurelia | P. pentaurelia |
| PK2 | Paramecium biaurelia | Paramecium septaurelia/Paramecium octaurelia | P. septaurelia/P. octaurelia |
| GA62-1 | P. biaurelia | Paramecium triaurelia | P. triaurelia |
| TB18 | P. triaurelia | P. primaurelia | P. primaurelia |
| ST | Paramecium tetraurelia | P. primaurelia | P. primaurelia |
| GA1-9 | P. pentaurelia | Unclear | Unclear |
| AZ8-3 | P. septaurelia | P. primaurelia | P. primaurelia |
| IEE | P. octaurelia | P. tetraurelia | P. tetraurelia |
| AB8-22 | Paramecium novaurelia | P. primaurelia | P. primaurelia |
| V9-6 | P. novaurelia | P. primaurelia | P. primaurelia |
| JN | Paramecium decaurelia | P. primaurelia | P. primaurelia |
| 223 | P. decaurelia | Unclear | Unclear |
| 246 | Paramecium dodecaurelia | P. septaurelia/P. octaurelia | P. septaurelia/P. octaurelia |
Discordant and Phylogenetically Distinct P. aurelia Lines
The existence of distinct and reproductively isolated species (or “varieties” or “syngens” as originally reported [Sonneborn 1937, 1957]) within the P. aurelia complex has been convincingly described (Sonneborn 1975) and correlated with molecular makers by many workers (e.g., Stoeck and Schmidt 1998). Yet, as reported above, our polymorphism survey reveals that a number of P. aurelia species are not monophyletic. Allele sharing between P. aurelia species has been detected by previous studies (Coleman 2005; Hori et al. 2006; Snoke et al. 2006; Tarcz et al. 2006; Barth et al. 2008), but little significance has typically been given to this finding. The limited number of species, strains, or loci in these earlier investigations leads to a number of nonmutually exclusive interpretations of the data, including 1) the occurrence of recent speciation events; 2) a large Ne, whereby strains still contain segregating alleles that predate species divergence (i.e., transspecies polymorphisms); and 3) incomplete reproductive isolation.
Our analysis crucially shows that allele sharing between strains with different mating behavior is not sporadic but rather extends across all macronuclear and mitochondrial loci surveyed. Also, allele sharing does not involve the whole set of strains for a species but often concerns all nondiscordant strains for one species and only a single strain for a second species. Finally, distinct P. aurelia species tend to form separate clades—after the exclusion of the discordant strains—with very short internal branches.
Consistent with a number of previous findings (e.g., Haggard 1974), our observations seem to rule out the possibility of recurrent gene flow among P. aurelia species, as the genetic profile of the surveyed strains is never discontinuous, that is, we do not find evidence of introgressive hybridization or episodes of cross-species recombination. The hypothesis of transspecies polymorphisms due to a large Ne in Paramecium also seems unlikely, given the modest level of intraspecific genetic variability (= 4Neμ) calculated after the removal of the discordant strains. Finally, the hypothesis of interspecific allele sharing due to recent speciation could well explain the intimate phylogenetic relationship between Paramecium septaurelia andParamecium octaurelia, whose strains tend to consistently group together across all gene genealogies but hardly explains the allele sharing between multiple strains of one species and an isolated strain of a second species.
A more parsimonious explanation for our observations is that the locus (loci) that is responsible for mating behavior in Paramecium—whose identity is still unknown—has undergone changes in the discordant strains that switch their mating preference. These changes would only alter the mating preference of these strains, which would otherwise normally react with genetically similar conspecific strains, leaving intact the overall genetic profile. It is notable that all 13 discordant strains show apparently normal viability in their matings to genetically distinct species (supplementary table 4 in Supplementary Material online).
Although not reported for any other ciliate or, to our knowledge, eukaryotic species, the incongruence we observe between mating behavior and genetic profile may not be limited to the P. aurelia species complex, and “species switches” might at least extend to other ciliates with cryptic species (e.g., P. multimicronucleatum, Tetrahymena pyriformis). Under this hypothesis, it would be informative to study the genetic profile of species other than P. aurelia and examine whether conspecific strains (following successful mating reactions) carry alleles that tend to be highly divergent at multiple loci while similar to alleles of a related species, an analysis that is currently hampered in ciliates by the dearth of available molecular markers and/or large-scale population-genetic surveys. If the discordance between mating preferences and genetic profile is confirmed to be caused by an alteration of the mating locus (loci), then mating locus switches could be considered a novel mechanism of speciation that, for its effects, could be comparable with other known mechanisms, such as, for example, cytoplasmic incompatibility (Bordenstein et al. 2001).
If the above hypothesis is correct, the genetic and ecological effects of two distinct genetic profiles, combined in one nucleus, remains an issue to be addressed in future investigations. Future studies will determine whether the F1 generation obtained from such a cross is fertile (they do survive autogamy) and less or more vigorous than the parental strains. In addition, it is currently not known if the pronounced interspecific sequence divergence would disfavor homologous recombination—thus gene flow and introgression—as has been observed for other organisms (Datta et al. 1997; Lukacsovich and Waldman 1999; Opperman et al. 2004), and/or if the set of genes of one of the two conjugating strains might be invariably discarded (gynogenesis or hybridogenesis) (Macgregor and Uzzell 1964; Schultz 1969; Mantovani and Scali 1992; Ragghianti et al. 2007). The latter event emerges as a possibility from earlier studies, where a cross between two P. aurelia species, P. tetraurelia, and P. octaurelia was reported to produce survivor clones that are not true hybrids and behave as one or the other species (the true F1 hybrids were sterile) (Levine 1953; Haggard 1974). Also, unless gynogenesis or hybridogenesis occurs, these discordant species would lead to introgression (which we have not observed) if they generated fertile progeny.
Finally, it is worth noting that even after the exclusion of discordant strains, P. aurelia species do not always form monophyletic groups. This can be seen, for example, in the case of P. primaurelia, where strain 227 branches off alone, making this taxon paraphyletic. Levels of sequence divergence between this P. primaurelia strain and all other conspecific strains are higher, both for macronuclear and mitochondrial loci, than the divergence estimated between P. primaurelia and P. pentaurelia. Strain 227 also diverges considerably from its other conspecific strains at rRNA loci, where it tends to cluster with Paramecium triaurelia strains (data not shown). It is tempting to conjecture that this P. primaurelia strain—of unknown geographic origin—may represent an example of incipient speciation in the P. aurelia complex.
Identification of Paralogous Loci
Paramecium tetraurelia has undergone at least three whole genome duplications (WGDs), the most recent of which has been suggested to coincide with the burst of speciation events that led to the emergence of the P. aurelia species complex (Aury et al. 2006). An erroneous sampling of WGD-associated paralogs in our study could have inflated the true genetic variation at macronuclear loci (but not at mitochondrial loci), affecting both intraspecific and interspecific estimates of diversity. The strain we used to design PCR primers (d4-2, P. tetraurelia) does not contain macronuclear paralogs for the loci we have sequenced. Nevertheless, we cannot rule out the possibility that either P. tetraurelia strains other than d4-2 or the remaining P. aurelia species are not paralog-free. Also, divergent resolution of paralogous genes following polyploidization and speciation (Lynch and Conery 2000; Lynch and Force 2000b) might facilitate such an erroneous sampling, but whether or not such a model applies to the P. aurelia complex is yet unknown.
We found two cases in which we PCR amplified paralogous loci and we removed these cases from our analyses. At locus GSPATG00022332001—the only nuclear intronless gene we surveyed—DNA sequencing revealed the existence of multiple double peaks in single-band PCR products obtained for the P. biaurelia species. We scored double peaks for all but five of the P. biaurelia strains, and we only included these five strains in the final analyses. As single-gene duplications appear to be uncommon in Paramecium (Aury et al. 2006) and no other double-peaked DNA sequences were observed for other species at this locus, this additional gene copy in P. biaurelia may be in the process of being lost from the genome. Alternatively, this gene could be a young duplicate that arose in P. biaurelia. GSPATG00022332001 is also the only gene for which we could not obtain any PCR product in P. primaurelia (notably, strain 227 was the only one producing a band in P. primaurelia). Overall, our observations suggest that GSPATG00022332001, which codes for a protein that is involved in the ubiquitin cycle, is an evolutionarily labile gene in P. aurelia.
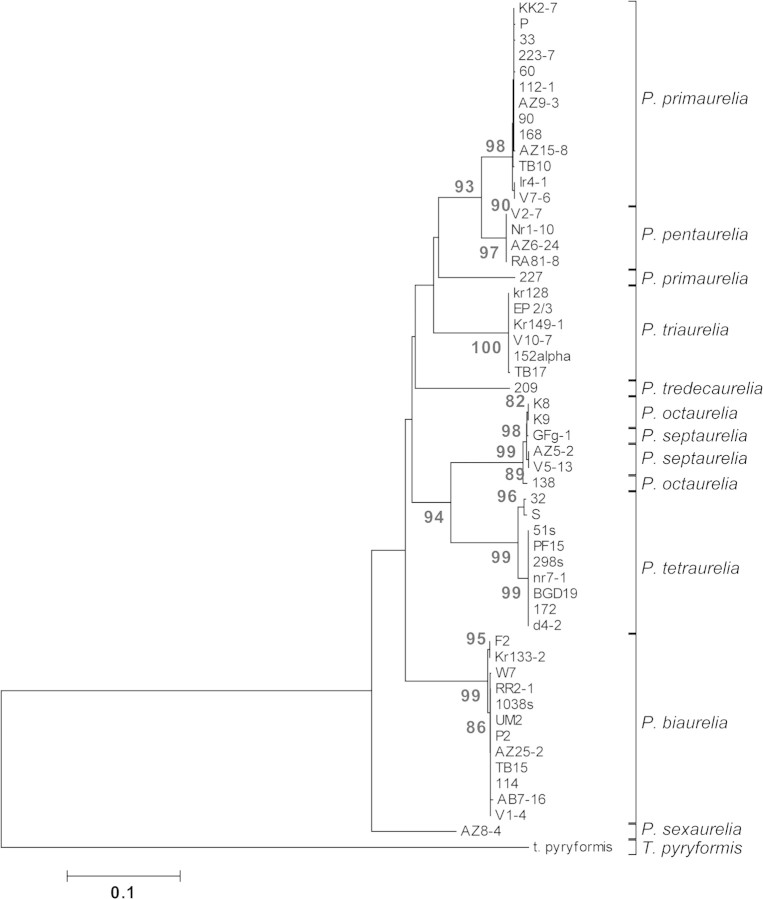
We detected the second case of paralogy after inspecting possible inconsistencies among gene genealogies. In particular, a phylogenetic tree constructed with five concatenated mitochondrial protein-coding loci strongly supports a close relatedness between P. primaurelia and P. pentaurelia and this relationship was supported by a tree constructed from three concatenated nuclear loci (fig. 2) and is observed for all nuclear loci but one, namely, GSPATG00028095001. At this locus, strains belonging to P. pentaurelia still cluster together (they share identical sequences) but are isolated from all other species in the tree. The level of divergence at silent sites between P. primaurelia and P. pentaurelia is as high as 80% at GSPATG00028095001 but drops to an across-loci average of <7% when this locus is removed from the analysis. We observed no PCR band at this locus for P. pentaurelia when we used an additional PCR primer that is external to the former pair. Although further experiments are needed to unequivocally show that the orthologous locus to the P. tetraurelia‘s GSPATG00028095001 is not present in P. pentaurelia, our observations are consistent with a differential retention of paralogous loci between the two P. aurelia species at this locus.
FIG. 2.—
Unrooted NJ tree built by concatenating the nuclear loci GSPATG00000363001, GSPATG00027725001, and GSPATG00000546001 (1,233 bp). Only nondiscordant strains are included in the analysis. Bootstrap values equal to or higher than 75 are shown.
Molecular Phylogeny, Mating Features, and Speciation in the P. aurelia Species Complex
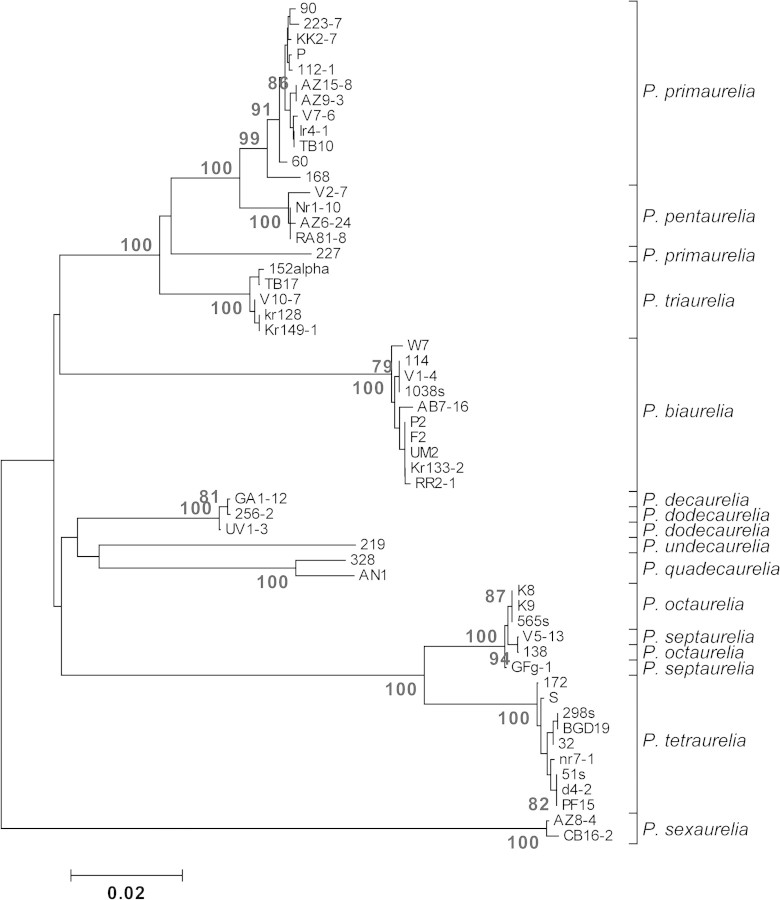
We obtained well-supported phylogenetic relationships only for the P. aurelia species that have diverged most recently (figs. 1 and 2). In these cases, the groupings broadly reflect the tendency that species have to cross-react during mating experiments (Sonneborn 1975). In particular, P. primaurelia and P. pentaurelia as well as P. tetraurelia and P. octaurelia, which pair closely in all gene genealogies, are known to cross-react (the latter pair more strongly than the former) and conjugate, without however giving rise to viable or fertile hybrids (Sonneborn 1974; Beale and Preer 2008). For either combination of mating types, 40% of cells will form true conjugants in a mixed culture of P. primaurelia and P. pentaurelia species, and the frequency rises to 90% when mating type E of P. tetraurelia is mixed with mating type O of P. octaurelia. Paramecium primaurelia mating type E also weakly reacts with type O of P. triaurelia. One would also expect a tendency for a cross-reaction between the closely related P. septaurelia and P. octaurelia or between P. septaurelia and P. tetraurelia. In fact, no such reactions are reported in the literature, but both P. septaurelia and P. octaurelia can—depending on their mating type—weakly react with P. primaurelia (Sonneborn 1975). It is worth noting that the genetic profile of most of the detected discordant strains corresponds to that of P. primaurelia, which can cross-react with more species within the P. aurelia complex than any other, an observation that remains unexplained.
FIG. 1.—
NJ tree built by concatenating three mitochondrial protein-coding loci, NADH dehydrogenase subunit 3 (NADH3), cytochrome c oxidase subunit I (COXI) and II (COXII) (1,057 bp). Only nondiscordant strains are included in the analysis. Bootstrap values equal to or higher than 75 are shown. Tetrahymena pyriformis used as outgroup.
We consistently find that the internal nodes of the molecular phylogeny of the P. aurelia species complex are only supported by low bootstrap values. A similar poor phylogenetic resolution has also been observed by previous studies (e.g., Coleman 2005; Hori et al. 2006; Barth et al. 2008). The virtual lack of phylogenetic resolution does not depend on the type of marker examined, as neither nuclear nor mitochondrial, and neither coding nor noncoding markers appear to be sufficiently informative. A plausible explanation for the latter observation is that speciation in P. aurelia may not have always proceeded as a branching process, followed by genetic isolation, but may have been the result of an initial burst of speciation events, perhaps accompanied by extensive hybridization. As the average divergence at silent sites between recent paralogs in P. tetraurelia is equal to 2.17 (Aury et al. 2006)—>2 times higher than the maximum species divergence per silent site, we estimate in this study (0.9804, between P. tetraurelia and P. sonneborni)—our observations indicate that the P. aurelia radiation postdates the most recent WGD and that speciation events may be associated with differential retain of gene copies across isolated populations (Lynch and Force 2000a).
Species Diversity and Geographic Distribution
Although our results do not suggest that huge global populations of the P. aurelia species exist, we find the same alleles in strains isolated from multiple continents, consistent with regular (or recent) global dispersal of most of the species. These observations intriguingly suggest that P. aurelia species might both have a limited (effective) population size but also be cosmopolitan. In an attempt to provide an explanation for this counterintuitive conclusion, we suggest that individual Paramecia inhabiting relatively few and durable habitats, which provide the long-term persistence of the species, would be dispersed to multiple geographically separated (and often ephemeral) environments, generating clonal populations whose life span is limited and depends on the features of the newly colonized habitats. The proposed scenario suggests a possibly cyclical and rapid transition from limited geographic distribution to an effectively, temporary, and cosmopolitan distribution of Paramecium species, a dynamic that is similar to that proposed by source–sink or source–pseudosink ecological models (Pulliam 1988; Watkinson and Sutherland 1995) and that may lead to a decrease in Ne (Whitlock and Barton 1997).
Additional and nonmutually exclusive explanations for allele sharing between strains inhabiting widely separated geographical areas would be strong selection against mutations at silent sites and/or an exceptionally low mutation rate in Paramecium. These factors would influence the accumulations of nucleotide changes after geographical separation of conspecific strains, and the extensive interspecific sequence divergence we observe would indicate fairly old events of speciation in the P. aurelia species complex. Whereas some support for selection against mutation at silent sites has been already provided (Salim et al. 2008), our ongoing study on the rate of spontaneous mutations in the genome of P. tetraurelia will determine the likelihood of the low-mutation-rate hypothesis.
Conclusions
The average estimate of neutral genetic variability (πs) in a species is expected to reflect the global effective population size of the species (Lynch 2006a). We found that within the P. aurelia species complex, estimates of πs are considerably higher when strains are grouped according to their reproductive compatibility but decrease greatly when the grouping criterion is the similarity of the strains’ genetic profiles. This effect is due to the exclusion from the analysis of a small set of strains with apparently aberrant mating behavior.
Overall, our study reveals that the average level of nuclear genetic diversity in the free-living ciliate Paramecium may not be as exceptionally high as previously reported (Snoke et al. 2006) but is, on average, not as low as the degree of neutral genetic diversity reported for another ciliate, Tetrahymena thermophila (∼0.003) (Katz et al. 2006). Depending on the criteria that we use to tell true species apart, values of macronuclear πs within P. aurelia may be either comparable with or higher than the average diversity reported for a set of mostly pathogenic unicellular eukaryotes (0.0573 [SD = 0.0777]) (Lynch 2006b) and for the unicellular green alga Chlamydomonas reinhardtii (0.032) (Smith and Lee 2008). Similar conclusions apply to the average genetic diversity observed at mitochondrial loci, where the πs within the P. aurelia complex could be one of the highest values ever reported, together with P. multimicronucleatum (πs = 0.308), a close outgroup of the P. aurelia complex (Snoke et al. 2006) or comparable with the πs of five other unicellular (mostly pathogenic) eukaryotes (0.011 [SEM = 0.004]) and C. reinhardtii (0.0085) (Smith and Lee 2008), after the removal of discordant strains.
Although this study cannot provide a definitive answer to the amount of genetic variability in Paramecium, our and previous observations (Katz et al. 2006) seem to suggest that levels of genetic diversity in free-living species may be relatively low and comparable to those of pathogenic taxa. Our findings support the proposal that speciation in Paramecium may not have always followed a cladistic process and highlight how the species problem in P. aurelia, an issue extensively addressed by Sonneborn during the last century (Sonneborn 1937, 1957, 1974, 1975), may prove even more complex than anticipated.
Supplementary Material
Supplementary tables 1-4, supplementary figure 1, and supplementary material are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Acknowledgments
We thank T.G. Doak and C.L. McGrath for insightful comments and suggestions for the improvement of the manuscript. We are grateful to E. Choi and V. Cloud for the maintenance of the Paramecium strains and to J.R. Preer Jr for his patience and for sharing with us his own experience and knowledge of Paramecium. We thank the Russian Ministry of Education and Science (project RNP 2.2.3.1.4148) for supporting the maintenance of Collection of Paramecium strains of the Laboratory of Protozoan Karyology, St Petersburg State University, from which originate most of the strains examined in this study. This work was supported by the MetaCyte funding from the Lilly Foundation to Indiana University.
References
- Aufderheide KJ, Daggett PM, Nerad TA. Paramecium-sonneborni N-Sp, a new member of the Paramecium-aurelia species-complex. J Protozool. 1983;30:128–131. [Google Scholar]
- Aury JM, Jaillon O, Duret L, et al. (42 co-authors) Global trends of whole-genome duplications revealed by the ciliate Paramecium tetraurelia. Nature. 2006;444:171–178. doi: 10.1038/nature05230. [DOI] [PubMed] [Google Scholar]
- Barth D, Krenek S, Fokin SI, Berendonk TU. Intraspecific genetic variation in Paramecium revealed by mitochondrial cytochrome C oxidase I sequences. J Eukaryot Microbiol. 2006;53:20–25. doi: 10.1111/j.1550-7408.2005.00068.x. [DOI] [PubMed] [Google Scholar]
- Barth D, Przybos E, Fokin SI, Schlegel M, Berendonk TU. Cytochrome b sequence data suggest rapid speciation within the Paramecium aurelia species complex. Mol Phylogenet Evol. 2008;49:669–673. doi: 10.1016/j.ympev.2008.08.007. [DOI] [PubMed] [Google Scholar]
- Beale G, Preer JR. Paramecium: genetics and epigenetics. Boca Raton (FL): CRC Press; 2008. [Google Scholar]
- Bordenstein SR, O'Hara FP, Werren JH. Wolbachia-induced incompatibility precedes other hybrid incompatibilities in Nasonia. Nature. 2001;409:707–710. doi: 10.1038/35055543. [DOI] [PubMed] [Google Scholar]
- Chen TT. Varieties and mating types in Paramecium Bursaria. 2. Variety and mating types found in China. J Exp Zool. 1956;132:255–268. [Google Scholar]
- Coleman AW. Paramecium aurelia revisited. J Eukaryot Microbiol. 2005;52:68–77. doi: 10.1111/j.1550-7408.2005.3327r.x. [DOI] [PubMed] [Google Scholar]
- Datta A, Hendrix M, Lipsitch M, Jinks-Robertson S. Dual roles for DNA sequence identity and the mismatch repair system in the regulation of mitotic crossing-over in yeast. Proc Natl Acad Sci USA. 1997;94:9757–9762. doi: 10.1073/pnas.94.18.9757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daubin V, Moran NA. Comment on “The origins of genome complexity”. Science. 2004;306:978. doi: 10.1126/science.1100559. author reply 978. [DOI] [PubMed] [Google Scholar]
- Fenchel T, Esteban GF, Finlay BJ. Local versus global diversity of microorganisms: cryptic diversity of ciliated protozoa. Oikos. 1997;80:220–225. [Google Scholar]
- Finlay BJ, Fenchel T. Divergent perspectives on protist species richness. Protist. 1999;150:229–233. doi: 10.1016/S1434-4610(99)70025-8. [DOI] [PubMed] [Google Scholar]
- Foissner W. Protist diversity: estimates of the near-imponderable. Protist. 1999;150:363–368. doi: 10.1016/S1434-4610(99)70037-4. [DOI] [PubMed] [Google Scholar]
- Foissner W. Biogeography and dispersal of micro-organisms: a review emphasizing protists. Acta Protozool. 2006;45:111–136. [Google Scholar]
- Gerber CA, Lopez AB, Shook SJ, Doerder FP. Polymorphism and selection at the SerH immobilization antigen locus in natural populations of Tetrahymena thermophila. Genetics. 2002;160:1469–1479. doi: 10.1093/genetics/160.4.1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigg ME, Bonnefoy S, Hehl AB, Suzuki Y, Boothroyd JC. Success and virulence in Toxoplasma as the result of sexual recombination between two distinct ancestries. Science. 2001;294:161–165. doi: 10.1126/science.1061888. [DOI] [PubMed] [Google Scholar]
- Haggard BW. Interspecies crosses in Paramecium aurelia (syngen 4 by syngen 8) J Protozool. 1974;21:152–159. doi: 10.1111/j.1550-7408.1974.tb03630.x. [DOI] [PubMed] [Google Scholar]
- Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser. 1999;41:95–98. [Google Scholar]
- Hori M, Tomikawa I, Przybos E, Fujishima M. Comparison of the evolutionary distances among syngens and sibling species of Paramecium. Mol Phylogenet Evol. 2006;38:697–704. doi: 10.1016/j.ympev.2005.09.006. [DOI] [PubMed] [Google Scholar]
- Katz LA, Snoeyenbos-West O, Doerder FP. Patterns of protein evolution in Tetrahymena thermophila: implications for estimates of effective population size. Mol Biol Evol. 2006;23:608–614. doi: 10.1093/molbev/msj067. [DOI] [PubMed] [Google Scholar]
- Levine M. The interactions of nucleus and cytoplasm in the isolation and evolution of species of Paramecium. Evolution. 1953;7:366–385. [Google Scholar]
- Lukacsovich T, Waldman AS. Suppression of intrachromosomal gene conversion in mammalian cells by small degrees of sequence divergence. Genetics. 1999;151:1559–1568. doi: 10.1093/genetics/151.4.1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M. Streamlining and simplification of microbial genome architecture. Annu Rev Microbiol. 2006;60:327–349. doi: 10.1146/annurev.micro.60.080805.142300. [DOI] [PubMed] [Google Scholar]
- Lynch M. The origins of eukaryotic gene structure. Mol Biol Evol. 2006;23:450–468. doi: 10.1093/molbev/msj050. [DOI] [PubMed] [Google Scholar]
- Lynch M, Conery JS. The evolutionary fate and consequences of duplicate genes. Science. 2000;290:1151–1155. doi: 10.1126/science.290.5494.1151. [DOI] [PubMed] [Google Scholar]
- Lynch M, Conery JS. The origins of genome complexity. Science. 2003;302:1401–1404. doi: 10.1126/science.1089370. [DOI] [PubMed] [Google Scholar]
- Lynch M, Force A. Gene duplication and the origin of interspecific genomic incompatibility. Am Nat. 2000;156:590–605. doi: 10.1086/316992. [DOI] [PubMed] [Google Scholar]
- Lynch M, Force A. The probability of duplicate gene preservation by subfunctionalization. Genetics. 2000;154:459–473. doi: 10.1093/genetics/154.1.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macgregor HC, Uzzell TM., Jr Gynogenesis in salamanders related to Ambystoma Jeffersonianum. Science. 1964;143:1043–1045. doi: 10.1126/science.143.3610.1043. [DOI] [PubMed] [Google Scholar]
- Mantovani B, Scali V. Hybridogenesis and androgenesis in the stick-insect Bacillus-Rossius Grandii-Benazzii (Insecta, Phasmatodea) Evolution. 1992;46:783–796. doi: 10.1111/j.1558-5646.1992.tb02084.x. [DOI] [PubMed] [Google Scholar]
- Mu J, Duan J, Makova KD, Joy DA, Huynh CQ, Branch OH, Li WH, Su XZ. Chromosome-wide SNPs reveal an ancient origin for Plasmodium falciparum. Nature. 2002;418:323–326. doi: 10.1038/nature00836. [DOI] [PubMed] [Google Scholar]
- Nei M, Kumar S. Molecular evolution and phylogenetics. New York: Oxford University Press; 2000. [Google Scholar]
- Ohta T. The nearly neutral theory of molecular evolution. Annu Rev Ecol Syst. 1992;23:263–286. [Google Scholar]
- Opperman R, Emmanuel E, Levy AA. The effect of sequence divergence on recombination between direct repeats in Arabidopsis. Genetics. 2004;168:2207–2215. doi: 10.1534/genetics.104.032896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson DJ. The diversity of eukaryotes. Am Nat. 1999;154:S96–S124. doi: 10.1086/303287. [DOI] [PubMed] [Google Scholar]
- Pritchard AE, Seilhamer JJ, Mahalingam R, Sable CL, Venuti SE, Cummings DJ. Nucleotide sequence of the mitochondrial genome of Paramecium. Nucleic Acids Res. 1990;18:173–180. doi: 10.1093/nar/18.1.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Przybos E. Genetic studies of Paramecium jenningsi strains (Diller, Earl, 1958) Folia Biol (Krakow) 1975;23:425–471. [PubMed] [Google Scholar]
- Przybos E, Rautian M, Greczek-Stachura M, Potekhin A. Polymorphism within Paramecium sexaurelia (Ciliophora, Oligohymenophorea) and description of a new stand of the species in China. Folia Biol (Krakow) 2007;55:121–125. doi: 10.3409/173491607781492632. [DOI] [PubMed] [Google Scholar]
- Przybos E, Tarcz S, Skoblo I. First American stand of Paramecium novaurelia and intra-specific differentiation of the species. Folia Biologica-Krakow. 2007;55:53–63. doi: 10.3409/173491607780006371. [DOI] [PubMed] [Google Scholar]
- Pulliam HR. Sources, sinks, and population regulation. Am Nat. 1988;132:652–661. [Google Scholar]
- Ragghianti M, Bucci S, Marracci S, Casola C, Mancino G, Hotz H, Guex GD, Plotner J, Uzzell T. Gametogenesis of intergroup hybrids of hemiclonal frogs. Genet Res. 2007;89:39–45. doi: 10.1017/S0016672307008610. [DOI] [PubMed] [Google Scholar]
- Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol. 2000;132:365–386. doi: 10.1385/1-59259-192-2:365. [DOI] [PubMed] [Google Scholar]
- Salim HMW, Ring KL, Cavalcanti ARO. Patterns of codon usage in two ciliates that reassign the genetic code: tetrahymena thermophila and Paramecium tetraurelia. Protist. 2008;159:283–298. doi: 10.1016/j.protis.2007.11.003. [DOI] [PubMed] [Google Scholar]
- Sawyer SA. GENECONV: a computer package for the statistical detection of gene conversion. 1999. Distributed by the author, Department of Mathematics, Washington University in St. Louis, available at http://www.math.wustl.edu/∼sawyer. [Google Scholar]
- Schultz RJ. Hybridization, unisexuality, and polyploidy in teleost Poeciliopsis (Poeciliidae) and other vertebrates. Am Nat. 1969;103:605–619. [Google Scholar]
- Smith DR, Lee RW. Nucleotide diversity in the mitochondrial and nuclear compartments of Chlamydomonas reinhardtii: investigating the origins of genome architecture. BMC Evol Biol. 2008;8:156. doi: 10.1186/1471-2148-8-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snoke MS, Berendonk TU, Barth D, Lynch M. Large global effective population sizes in Paramecium. Mol Biol Evol. 2006;23:2474–2479. doi: 10.1093/molbev/msl128. [DOI] [PubMed] [Google Scholar]
- Sonneborn TM. Sex, sex inheritance and sex determination in Paramecium Aurelia. Proc Natl Acad Sci USA. 1937;23:378–385. doi: 10.1073/pnas.23.7.378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonneborn TM. Breeding systems, reproductive methods, and species problems in protozoa. In: Mayr E, editor. The species problem. Washington DC: AAAS; 1957. [Google Scholar]
- Sonneborn TM. Methods in cell physiology. New York, London: DMPA Press; 1970. Methods in Paramecium research. p. 241–339. [Google Scholar]
- Sonneborn TM. Handbook of genetics. New York: R.C.K.P. Press; 1974. Paramecium aurelia. p. 469–594. [Google Scholar]
- Sonneborn TM. The Paramecium-aurelia complex of 14 sibling species. Trans Am Microsc Soc. 1975;94:155–178. [Google Scholar]
- Stoeck T, Przybos E, Schmidt HJ. A combination of genetics with inter- and intra-strain crosses and RAPD-fingerprints reveals different population structures within the Paramecium aurelia species complex. Eur J Protistol. 1998;34:348–355. [Google Scholar]
- Stoeck T, Schmidt HJ. Fast and accurate identification of European species of the Paramecium aurelia complex by RAPD-fingerprints. Microb Ecol. 1998;35:311–317. doi: 10.1007/s002489900086. [DOI] [PubMed] [Google Scholar]
- Tarcz S, Przybos E, Prajer M, Greczek-Stachura M. Intraspecific variation of diagnostic rDNA genes in Paramecium dodecaurelia, P. tredecaurelia and P. quadecaurelia (Ciliophora: oligohymenophorea) Acta Protozool. 2006;45:255–263. [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkinson AR, Sutherland WJ. Sources, sinks and pseudo-sinks. J Anim Ecol. 1995;64:126–130. [Google Scholar]
- Waugh J. DNA barcoding in animal species: progress, potential and pitfalls. Bioessays. 2007;29:188–197. doi: 10.1002/bies.20529. [DOI] [PubMed] [Google Scholar]
- Weisse T. Distribution and diversity of aquatic protists: an evolutionary and ecological perspective. Biodivers Conserv. 2008;17:243–259. [Google Scholar]
- White TJ, Bruns T, Lee S, Taylor J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenies. In: Innis MA, Gelford DH, Sninsky JJ, White TJ, editors. CR protocols: a guide to methods and applications. New York: Academic Press; 1990. pp. 315–322. [Google Scholar]
- Whitlock MC, Barton NH. The effective size of a subdivided population. Genetics. 1997;146:427–441. doi: 10.1093/genetics/146.1.427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright S. Evolution in Mendelian populations. Genetics. 1931;16:97–159. doi: 10.1093/genetics/16.2.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.