Abstract
Previous findings have demonstrated that variants in nicotinic receptor genes are associated with nicotine, alcohol and cocaine dependence. Because of the substantial comorbidity, it has often been unclear whether a variant is associated with multiple substances or whether the association is actually with a single substance. To investigate the possible contribution of rare variants to the development of substance dependencies other than nicotine dependence, specifically alcohol and cocaine dependence, we undertook pooled sequencing of the coding regions and flanking sequence of CHRNA5, CHRNA3, CHRNB4, CHRNA6 and CHRNB3 in 287 African American and 1028 European American individuals from the Collaborative Study of the Genetics of Alcoholism (COGA). All members of families for whom any individual was sequenced (2504 African Americans and 7318 European Americans) were then genotyped for all variants identified by sequencing. For each gene, we then tested for association using FamSKAT. For European Americans, we find increased DSM-IV cocaine dependence symptoms (FamSKAT P = 2 × 10−4) and increased DSM-IV alcohol dependence symptoms (FamSKAT P = 5 × 10−4) among carriers of missense variants in CHRNB3. Additionally, one variant (rs149775276; H329Y) shows association with both cocaine dependence symptoms (P = 7.4 × 10−5, β = 2.04) and alcohol dependence symptoms (P = 2.6 × 10−4, β = 2.04). For African Americans, we find decreased cocaine dependence symptoms among carriers of missense variants in CHRNA3 (FamSKAT P = 0.005). Replication in an independent sample supports the role of rare variants in CHRNB3 and alcohol dependence (P = 0.006). These are the first results to implicate rare variants in CHRNB3 or CHRNA3 in risk for alcohol dependence or cocaine dependence.
INTRODUCTION
Twin and adoption studies have suggested that in addition to genetic variants that contribute to the risk of becoming dependent on a specific drug, there exist genetic factors underlying a generalized risk for becoming dependent on multiple substances [ (1) for review, (2,3)]. Epidemiological studies have shown high levels of comorbidity among alcohol, cocaine and nicotine dependence (4,5). Because of the substantial comorbidity, it is often unclear whether a variant is associated with multiple substances or whether the association is actually with a single substance but the extensive comorbidity leads to apparent association with other substances. Numerous studies have found that variants altering the function or expression of neuronal cholinergic nicotinic receptors (CHRNs) alter risk for becoming nicotine dependent and several have shown effects of variants in nicotinic receptors on risk for both alcohol and cocaine dependence [ (6) for review]. Common single nucleotide polymorphisms (SNPs) in CHRNA5 and CHRNB3 are associated with nicotine dependence or cigarette consumption (7,8). The most strongly associated SNP in several genome-wide association studies (GWAS) of nicotine dependence, a non-synonymous change (rs16969968/D398N) in CHRNA5, has also been shown to mitigate risk for cocaine dependence (9,10). Additionally, a highly correlated group of variants near CHRNA5, previously shown to alter CHRNA5 mRNA expression in vivo, have been reported to alter risk for alcohol dependence (11) in addition to nicotine dependence (12). GWAS of cigarette consumption has also implicated a group of common SNPs near the CHRNB3-CHRNA6 gene cluster (13) that have been suggested to be involved in alcohol consumption, although not with cocaine-related behaviors (14).
In addition to mediating the effects of nicotine, neuronal nicotinic receptors have also been shown to alter the cellular and behavioral effects of alcohol and cocaine independently of nicotine (15–37). Evidence from in vitro studies indicates altered response to ethanol upon perturbation of nicotinic receptors in neurons (18,29–31,38). Studies in rats have shown that partial agonists of α4β2* or α3β4* nicotinic receptors affect alcohol consumption in vivo (15,18–21,24–28). These results were confirmed and expanded using transgenic and knockout mice for various nicotinic receptor genes (16,17,32–37). The physiological and behavioral effects of cocaine are also dependent on nicotinic receptors. Studies in mice, rats and non-human primates have suggested that cocaine administration alters nicotinic receptor expression and in so doing leads to altered response to cocaine (39–42). A recent human trial also found that the α4β2 subunit partial agonist Varenicline reduced alcohol consumption among smokers (43). Together these results suggest that altered nicotinic receptor expression and function likely has an effect on alcohol and cocaine consumption independent of its effect on nicotine consumption. However, despite the promising evidence of synergy and cross-substance influences, the combined effects of discovered common variants have explained only a small proportion of the variance (∼5%) in nicotine dependence and even less of the variance in alcohol or cocaine dependence (44). Rare variants may explain much of the unexplained heritability of these complex substance use disorders.
As a class, rare variants constitute the majority of genetic variation. A recent survey of >4000 exomes from individuals of European and African descent showed that as much as 86% of coding variants have a frequency <0.05% and that these rare variants are four times as likely to be deleterious (45). Many examples now exist of rare variants contributing to common human diseases [(46) for review], both in genes previously reported to cause Mendelian forms of disease and in genes harboring common SNP associations. (47–49). Several studies have demonstrated associations between rare variation in nicotinic receptor genes and nicotine dependence (50–52). An efficient method of determining the full spectrum of genetic variation in a region and thus the relative contribution of rare genetic variation to a trait of interest is to sequence pools of DNA from multiple individuals and to use statistical methods to determine which sites are polymorphic and at what frequency. We have employed this approach to determine the contribution of rare non-synonymous variants in the CHRNA5, CHRNA3, CHRNB4, CHRNA6 and CHRNB3 genes to alcohol and cocaine dependence.
RESULTS
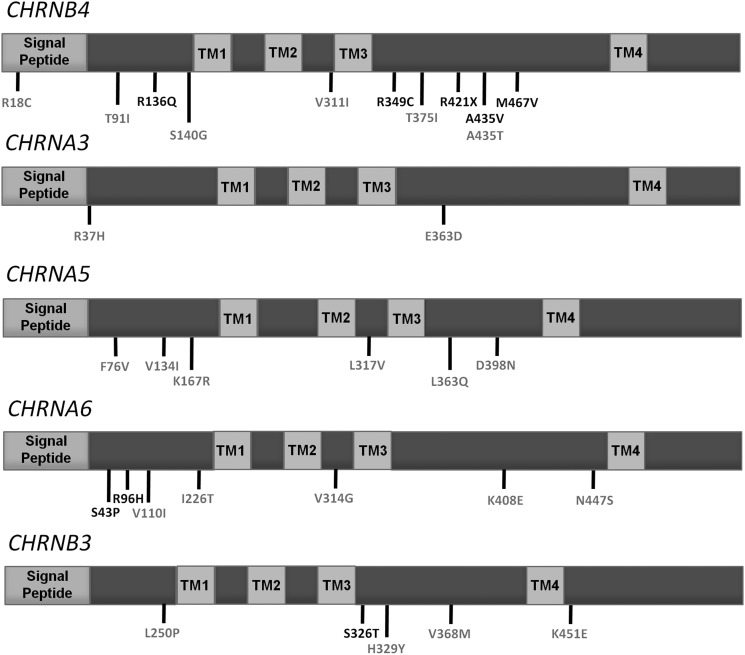
We sequenced the protein coding regions of the five human cholinergic nicotinic receptor subunit genes constituting the gene clusters previously reported to harbor common variants associated with nicotine dependence (CHRNA5-CHRNA3-CHRNB4 and CHRNA6-CHRNB3) in 287 unrelated African Americans (147 DSM-IV alcohol-dependent cases and 140 controls) and 1028 unrelated European Americans (480 DSM-IV alcohol-dependent cases and 548 controls) as part of the Collaborative Study of the Genetics of Alcoholism (COGA). DNA pools were sequenced on an Illumina GAIIx and analyzed using the SPLINTER algorithm (53). In total, we validated 31 non-synonymous variants (Fig. 1 and Table 1). Of these, all but rs16969968 were rare (<5% MAF), 9 (29%) were novel and 15 (48%) were present in only one sequenced individual. We only attempted to validate non-synonymous variants as these are more likely to have a functional impact than non-coding or synonymous coding changes. As the SPLINTER algorithm produces a P-value for the probability that a predicted variant is a true positive, we also genotyped 19 variants below our cut-off for significance in order to determine the positive predictive value of the algorithm. Only 1 of these 19 variants was validated while 31 of 40 variants predicted to be true positives were validated. All variants included as part of the amplified positive control vector were found upon achieving greater >30-fold coverage at mutated sites (Sensitivity = 100%) and only 63 sites in the 1908 bp negative control vector were predicted to be polymorphic (Specificity = ∼97%).
Figure 1.
Schematics of the CHRN genes studied and the locations of missense variants identified in sequenced individuals from the COGA study. Each protein consists of a signal peptide, two extracellular domains, four transmembrane domains (TM1-4) and three intracellular domains. Conserved sites (phyloP44 Vertebrate score >2) are shown in gray, while all others are shown in black.
Table 1.
Characteristics and frequencies of variants observed in the COGA data set
| Gene | Amino acid position | CHR | hg19 position | A1 | A2 | PhyloP score | SIFT | Polyphen | Frequency in COGA EAs | Frequency in COGA AAs | Frequency in COGEND EAs | Frequency in COGEND AAs |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CHRNB3 | L250P | 8 | 42587199 | C | T | 5.117 | + | ++ | 0.0021 | 0 | 0 | 0 |
| CHRNB3 | S326T | 8 | 42587426 | T | A | 0.672 | + | 0 | 0.0028 | 0 | 0.0022 | |
| CHRNB3 | H329Y | 8 | 42587435 | C | T | 3.982 | + | ++ | 0.0019 | 0.0120 | 0.0020 | 0 |
| CHRNB3 | V368M | 8 | 42587552 | A | G | 4.767 | 0.0188 | 0 | 0 | 0 | ||
| CHRNB3 | K451E | 8 | 42591735 | G | A | 2.350 | 0.0096 | 0.0423 | 0.0007 | 0.0515 | ||
| CHRNA6 | N447S | 8 | 42611002 | T | C | 3.502 | + | ++ | 0.0007 | 0 | 0 | 0.0006 |
| CHRNA6 | K408E | 8 | 42611120 | C | T | 3.527 | 0.0028 | 0 | 0 | 0 | ||
| CHRNA6 | V314G | 8 | 42611401 | C | A | 7.129 | + | ++ | 0.0087 | 0.0005 | 0 | 0 |
| CHRNA6 | I226T | 8 | 42611665 | C | T | 5.214 | + | 0.0069 | 0 | 0 | 0 | |
| CHRNA6 | V110I | 8 | 42612117 | C | T | 2.128 | 0.0035 | 0 | 0 | 0 | ||
| CHRNA6 | R96H | 8 | 42612158 | C | T | 0.939 | + | + | 0.0007 | 0.0002 | 0 | 0.0010 |
| CHRNA6 | S43P | 8 | 42620300 | A | G | 1.234 | + | 0.0034 | 0.0110 | 0.0003 | 0.0145 | |
| CHRNA5 | F76V | 15 | 78873272 | T | G | 4.712 | ++ | 0.0021 | 0 | 0 | 0 | |
| CHRNA5 | V134I | 15 | 78880752 | A | G | 6.721 | + | ++ | 0.0175 | 0.0042 | 0.0147 | 0.0011 |
| CHRNA5 | K167R | 15 | 78882233 | A | G | 5.403 | + | ++ | 0.0007 | 0.0158 | 0.0003 | 0.0157 |
| CHRNA5 | L317V | 15 | 78882682 | G | C | 2.262 | + | ++ | 0.0038 | 0.0014 | 0 | 0 |
| CHRNA5 | L363Q | 15 | 78882821 | T | A | 4.767 | + | + | 0.0034 | 0.0433 | 0.0005 | 0.0576 |
| CHRNA5 | D398N | 15 | 78882925 | A | G | 3.766 | 0.2920 | 0.0520 | 0.3494 | 0.0478 | ||
| CHRNA3 | E363D | 15 | 78893895 | G | C | 3.285 | 0.0042 | 0 | 0 | 0 | ||
| CHRNA3 | R37H | 15 | 78911230 | C | T | 2.671 | + | ++ | 0.0458 | 0.0116 | 0.0521 | 0.0088 |
| CHRNB4 | M467V | 15 | 78917573 | T | C | 1.584 | 0.0192 | 0.0637 | 0 | 0.0759 | ||
| CHRNB4 | A435V | 15 | 78921343 | A | G | −0.549 | + | ++ | 0.0145 | 0.0007 | 0 | 0 |
| CHRNB4 | A435T | 15 | 78921344 | T | C | 3.053 | + | + | 0 | 0.0011 | 0 | 0 |
| CHRNB4 | R421X | 15 | 78921386 | A | G | 1.653 | + | + | 0.0014 | 0 | 0 | 0 |
| CHRNB4 | T375I | 15 | 78921523 | G | A | 2.262 | + | 0 | 0.0168 | 0 | 0.0169 | |
| CHRNB4 | R349C | 15 | 78921602 | A | G | 1.960 | + | ++ | 0.0842 | 0.0030 | 0.0072 | 0.0006 |
| CHRNB4 | V311I | 15 | 78921716 | T | C | 2.402 | + | 0.0062 | 0 | 0 | 0 | |
| CHRNB4 | S140G | 15 | 78922229 | C | T | 2.269 | + | 0.0555 | 0.0410 | 0.0040 | 0.0434 | |
| CHRNB4 | R136Q | 15 | 78922240 | C | T | 0.410 | 0.1243 | 0.0024 | 0.0114 | 0.0016 | ||
| CHRNB4 | T91I | 15 | 78923505 | A | G | 2.161 | 0.3911 | 0.0074 | 0.0457 | 0.0067 | ||
| CHRNB4 | R18C | 15 | 78933424 | A | G | 5.625 | 0.0014 | 0 | 0 | 0 |
For SIFT predictions, variants predicted to be damaging are depicted with +. For polyphen predictions, variants predicted to be ‘possibly damaging’ are depicted with + and variants predicted to be ‘probably damaging’ are depicted with ++.
We genotyped each missense variant validated from sequencing in every individual from the COGA study for whom DNA existed, totaling 2504 African Americans and 7318 European Americans. We then tested for association between non-synonymous variant carrier status at each sequenced gene and either alcohol or cocaine dependence symptom count using the family-based Sequence Kernel Association Test (FamSKAT), which incorporates relatedness into the model using a kinship matrix. Age, sex and the number of cigarettes smoked per day (CPD) were included in the model as covariates. To increase the chances of novel findings, we did not include the SNP rs16969968 in analyses of European Americans or African Americans as it might overshadow the effects of other variants in the CHRNA5 gene and because it is common in European Americans (Frequency = 0.35).
Analysis of European Americans
For European Americans, we find increased DSM-IV alcohol dependence symptoms (0.97 ± 2.2 versus. 0.65 ± 1.8, famSKAT P = 6 × 10−4) as well as increased DSM-IV cocaine dependence symptoms (2.37 ± 2.6 versus. 2.07 ± 2.4, famSKAT P = 4 × 10−4) DSM-IV cocaine dependence symptoms (2.37 ± 2.6 versus. 2.07 ± 2.4, famSKAT P = 4 × 10−4) among carriers of missense variants in the CHRNB3-A6 gene cluster. We observed increased DSM-IV alcohol dependence symptoms (1.3 ± 2.5 versus. 0.65 ± 1.8, famSKAT P = 5 × 10−4) and increased DSM-IV cocaine dependence symptoms (2.27 ± 2.7 versus. 2.07 ± 2.4, famSKAT P = 2 × 10−4) among carriers of missense variants in CHRNB3 but not for variants in CHRNA6 (Table 2, Table 3 and Fig. 2). Further, among the SNPs comprising the CHRNB3 carrier genotype (S326T, H329Y, K451E and V368M), one variant (rs149775276; H329Y) was significantly associated in a linear regression model with increases in both alcohol dependence symptoms (P = 2.6 × 10− each gene for association with CPD using famSKAT and found no significant associations between CPD and rare missense variants at any of the genes sequenced in this data set, suggesting that this is a direct effect of these variants on risk for alcohol and cocaine dependence phenotypes (Table 2).
Table 2.
Collapsed frequencies and famSKAT results for each sequenced gene, sequenced gene cluster or all genes combined
| European Americans |
African Americans |
|||||||
|---|---|---|---|---|---|---|---|---|
| cMAF | Alcohol P-value | Cocaine P-value | Nicotine P-value | cMAF | Alcohol P-value | Cocaine P-value | Nicotine P-value | |
| CHRNA5 | 0.0037 | 0.764 | 0.696 | 0.868 | 0.0130 | 0.405 | 0.405 | 0.219 |
| CHRNA6 | 0.0003 | 0.265 | 0.937 | 0.762 | 0.0017 | 0.510 | 0.646 | 0.295 |
| CHRNB3 | 0.0010 | 5.5 × 10−4 | 2.4 × 10−4 | 0.202 | 0.0125 | 0.604 | 0.754 | 0.565 |
| CHRNB4 | 0.0065 | 0.265 | 0.376 | 0.820 | 0.0133 | 0.087 | 0.065 | 0.898 |
| CHRNA3 | 0.0234 | 0.417 | 0.360 | 0.854 | 0.0077 | 0.021 | 0.005 | 0.566 |
| CHRNA5-A3-B4 | 0.0076 | 0.437 | 0.521 | 0.493 | 0.0128 | 0.05 | 0.014 | 0.684 |
| CHRNB3-A6 | 0.0006 | 5.9 × 10−4 | 4.0 × 10−4 | 0.396 | 0.0063 | 0.734 | 0.734 | 0.499 |
| All | 0.0047 | 0.121 | 0.138 | 0.496 | 0.0101 | 0.122 | 0.047 | 0.721 |
For alcohol, DSMIV alcohol dependence symptom count was used as the phenotype and age, sex and CPD were used as covariates. For cocaine, DSMIV cocaine dependence symptom count was used as the phenotype and age, sex and CPD were used as covariates. For nicotine, CPD was used as the phenotype and age and sex were used as covariates.
Table 3.
Demographic and phenotypic characteristics of COGA and COGEND African American and European American samples
| CHRNB3 |
CHRNA3 |
|||||
|---|---|---|---|---|---|---|
| Non-carriers | Carriers | P-value | Non-carriers | Carriers | P-value | |
| COGA European Americans | ||||||
| No. of individuals | 4124 | 44 | – | 3765 | 398 | – |
| Age (years) | 37 ± 16 | 37 ± 15 | 0.95 | 39 ± 15 | 40 ± 15 | 0.37 |
| Women/men | 2076/2048 | 25/19 | 0.51 | 1878/1887 | 185/213 | 0.21 |
| Cigarettes per day | 21 ± 13 | 19 ± 12 | 0.6 | 20 ± 13 | 20 ± 13 | 0.86 |
| Alcohol-dependent cases–controls | 1796/2328 | 22/22 | 0.01 | 1634/2128 | 177/221 | 0.71 |
| Alcohol symptom count | 2.07 ± 2.4 | 2.27 ± 2.7 | 5.6 × 10−4 | 2.9 ± 2.51 | 3.04 ± 2.48 | 0.28 |
| Cocaine-dependent cases–controls | 677/3447 | 12/32 | 0.005 | 601/3158 | 77/320 | 0.1 |
| Cocaine symptom count | 0.65 ± 1.8 | 1.3 ± 2.5 | 2.4 × 10−4 | 1.03 ± 2.21 | 1.16 ± 2.3 | 0.26 |
| COGA African Americans | ||||||
| No. of individuals | 1121 | 143 | – | 1237 | 27 | – |
| Age (years) | 36 ± 13 | 37 ± 13 | 0.34 | 36 ± 12 | 33 ± 11 | 0.19 |
| Women/men | 503/618 | 64/79 | 1 | 558/679 | 9/18 | 0.24 |
| Cigarettes per day | 14 ± 11 | 14 ± 10 | 0.82 | 14 ± 10 | 16 ± 11 | 0.25 |
| Alcohol-dependent cases–controls | 530/590 | 74/69 | 0.32 | 595/641 | 9/18 | 0.17 |
| Alcohol symptom count | 2.99 ± 2.51 | 3.32 ± 2.56 | 0.13 | 3.04 ± 2.52 | 2.66 ± 2.18 | 0.445 |
| Cocaine-dependent cases–controls | 344/768 | 51/92 | 0.25 | 388/840 | 7/20 | 0.67 |
| Cocaine symptom count | 2.02 ± 2.91 | 2.30 ± 3.02 | 0.29 | 1.14 ± 2.4 | 0.81 ± 1.8 | 0.006 |
| COGEND European Americans | ||||||
| No. of individuals | 2036 | 11 | – | 1869 | 178 | – |
| Age (years) | 36 ± 5 | 36 ± 5 | 0.96 | 37 ± 5 | 36 ± 6 | 0.14 |
| Women/men | 1247/789 | 41489 | 0.54 | 1142/727 | 113/65 | 0.57 |
| Cigarettes per day | 18 ± 17 | 18 ± 19 | 0.99 | 19 ± 17 | 17 ± 17 | 0.14 |
| Alcohol-dependent cases–controls | 464/1530 | 4/7 | 0.3 | 434/1395 | 34/142 | 0.22 |
| Alcohol symptom count | 1.72 ± 1.66 | 1.81 ± 1.72 | 0.852 | 1.74 ± 1.67 | 1.50 ± 1.55 | 0.06 |
| Cocaine-dependent cases–controls | 131/1905 | 1/10 | 0.52 | 126/1743 | 6/172 | 0.11 |
| Cocaine symptom count | 0.43 ± 1.44 | 0.55 ± 1.81 | 0.8 | 0.45 ± 1.48 | 0.26 ± 1.12 | 0.08 |
| COGEND African Americans | ||||||
| No. of individuals | 641 | 69 | – | 700 | 10 | – |
| Age (years) | 37 ± 6 | 33 ± 5 | 0.93 | 37 ± 5 | 38 ± 6 | 0.14 |
| Women/men | 408/233 | 41/28 | 0.55 | 1142/727 | 113/65 | 0.57 |
| Cigarettes per day | 16 ± 13 | 16 ± 12 | 0.63 | 16 ± 13 | 12 ± 10 | 0.26 |
| Alcohol-dependent cases–controls | 66/575 | 13/56 | 0.06 | 78/622 | 1/9 | 1 |
| Alcohol symptom count | 1.35 ± 1.72 | 2.00 ± 2.34 | 0.004 | 1.43 ± 1.80 | 1.43 ± 2.6 | 0.99 |
| Cocaine-dependent cases–controls | 43/598 | 6/63 | 0.46 | 49/651 | 0/10 | 1 |
| Cocaine symptom count | 0.71 ± 1.93 | 0.79 ± 2.10 | 0.77 | 0.42 ± 1.54 | 0 ± 0 | 0.39 |
CPD, age, alcohol symptom count and cocaine symptom count values are mean ± SD. P-values for sex and age were calculated with a Fisher's exact test and Wilcoxon Rank-Sum test, respectively. In COGA, P-values for primary phenotypes were calculated using FamSKAT in R using a kinship matrix with age, sex and CPD as covariates. In COGEND, P-values for primary phenotypes were calculated using a Wilcoxon Rank-Sum test.
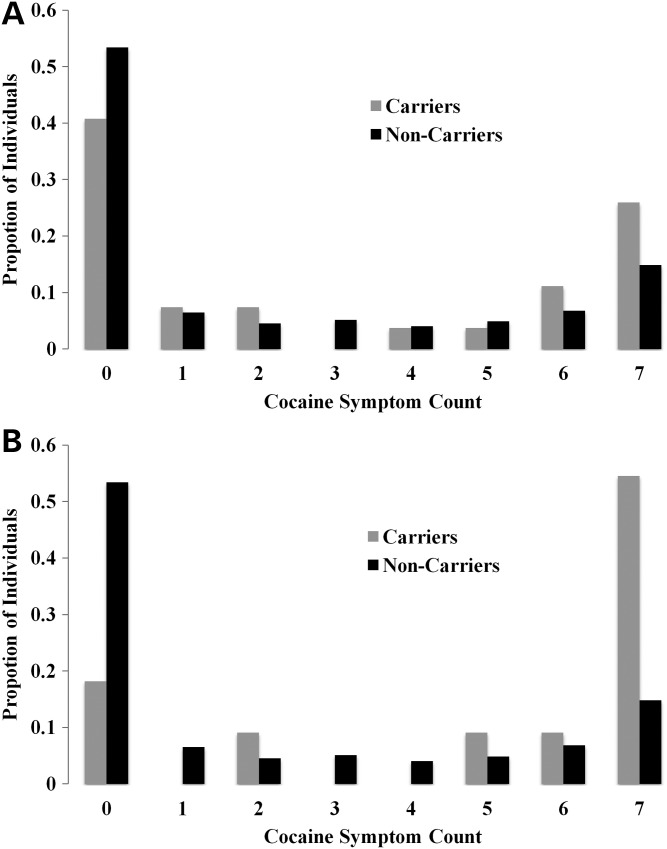
Figure 2.
Proportion of individuals among cocaine symptom counts in carriers and non-carriers of (A) any CHRNB3 variant or (B) CHRNB3 H329Y.
To determine whether these findings were the result of a correlation between alcohol or cocaine symptom count and smoking, which had not been accounted for using CPD as a covariate, we tested whether these associations remained when only heavy smokers (CPD ≥20) were analyzed, as there is no significant correlation between alcohol symptom count or cocaine symptom count and CPD among these heavy smoking individuals. Using only heavy smokers (n = 2509), the association between CHRNB3 and alcohol symptom count (famSKAT P = 0.0064) and cocaine dependence symptom count (famSKAT P = 0.002) remained significant, although reduced. No associations between carrier status at any sequenced gene and alcohol or cocaine symptom count were observed in non-smokers (n = 3177). Despite large numbers of non-smokers, very few non-smokers are alcohol dependent (n = 387; 12%) or cocaine dependent (n = 73; 2%), greatly reducing power to detect associations with these phenotypes in this subset of individuals. Overall, these analyses suggest that rare CHRNB3 variants are associated with cocaine dependence symptoms and alcohol dependence symptoms and that these associations cannot be fully explained by comorbid smoking behavior.
As expected, there are high rates of comorbidity between cocaine dependence and alcohol dependence in our sample. For example, only 3% of European cocaine-dependent cases and ∼5% of African-Americans cocaine-dependent cases in our sample have no symptoms of alcohol dependence. To determine whether the observed association in European Americans between CHRNB3 variants and either cocaine dependence symptoms or alcohol dependence symptoms could be explained by a correlation between the two phenotypes, we included alcohol dependence symptom count as a covariate in the analysis of cocaine dependence and vice versa. The association between cocaine dependence symptoms and both CHRNB3 (famSKAT P = 0.009) and rs149775276 (linear regression P = 0.002) remained significant when alcohol dependence was used as a covariate. Using cocaine dependence symptom count as a covariate, however, CHRNB3 carrier status did not remain significantly associated with alcohol dependence symptoms (famSKAT P = 0.66). These findings suggest that variants in CHRNB3 increase risk for cocaine dependence, but only increase risk for alcohol dependence indirectly by increasing risk for cocaine dependence symptoms or that the structure of the data set is such that one cannot correct for cocaine dependence symptoms without disrupting all associations with alcohol dependence symptoms.
One group of common SNPs within the CHRNB3-CHRNA6 gene cluster (tagged in our data set by rs6474412) was previously shown to affect levels of nicotine consumption (8,13). To determine whether our findings in CHRNB3 could be due to linkage disequilibrium with this variant, we tested for association between rs6474412 and both alcohol and cocaine dependence symptom count, but in neither case was a significant association observed (alcohol symptom count P = 0.49, β = −0.03; cocaine symptom count: P = 0.91, β = −0.01). This suggests that our observed associations are not due to a correlation between rare variants in CHRNB3 and the previously described common variant.
To ensure that our findings within the European American portion of the COGA sample are not due to population stratification, we calculated principal components using EIGENSTRAT (54). We then tested for association between CHRNB3 missense variants and both alcohol and cocaine dependence symptom count including the first two principal components (PC1 and PC2) as covariates. CHRNB3 carrier status remained associated with both alcohol dependence symptom count (famSKAT P = 5.6 × 10−4) and cocaine dependence symptom count in European Americans (famSKAT P = 2.3 × 10−4). These findings suggest that the observed association is not due to population stratification.
Analysis of African Americans
For African Americans, we found decreased DSM-IV cocaine dependence symptoms (famSKAT P = 0.01) and decreased DSM-IV alcohol dependence symptoms (famSKAT P = 0.05) among carriers of missense variants in the CHRNA5-B4-A3 gene cluster. This association was driven by decreased DSM-IV cocaine dependence symptoms (famSKAT P = 0.005) and decreased DSM-IV alcohol dependence symptoms (famSKAT P = 0.02) among carriers of missense variants in CHRNA3 (Tables 2 and 3). As for European Americans, age, sex and CPD were included as covariates in the analysis. As there were only two missense variants observed in CHRNA3 in African Americans, this association was due largely to the association between cocaine dependence symptoms and one variant (rs8192475; R37H). This variant was previously shown to decrease risk for nicotine dependence and increase cellular response to nicotine in vitro (52). The association between CHRNA3 carrier status and cocaine dependence symptom count remained significant even after alcohol dependence symptoms was included as a covariate (famSKAT P = 0.04).
To determine whether our findings in African Americans could be explained by subtle population stratification, we calculated principal components using EIGENSTRAT (54) and included the first two principal components (PC1 and PC2) as covariates in the linear regression. CHRNA3 carrier status remained associated with cocaine dependence symptom count in African Americans (famSKAT P = 0.01). No sequenced gene other than CHRNA3 was significant before or after addition of the first two principal components as covariates in the analysis. Additionally, we calculated local admixture using LAMP (55). CHRNA3 carrier status remained significantly associated with cocaine dependence symptoms when we performed linear regression including local admixture estimates from the 100 kb encompassing the CHRNA5-CHRNA3-CHRNB4 gene cluster as a covariate (famSKAT P = 0.03). No sequenced gene other than CHRNA3 was significant before or after addition of estimated local ancestry as a covariate in the analysis. These findings suggest that the observed association is not due to population stratification, either on a genome or a local scale.
Replication in COGEND
Previous findings from the Collaborative Genetic Study of Nicotine Dependence (COGEND) have suggested that rare variants in CHRNB4 reduce risk of nicotine dependence (52). Using previously obtained sequencing data from COGEND (51), we aimed to determine whether we could replicate the findings from COGA. We tested for association between alcohol dependence symptom count and cocaine dependence symptom count and variants in the CHRNB3 and CHRNA3 genes in the COGEND sample as these genes were associated with these phenotypes here in the COGA data set. We used both linear regression and a non-parametric Wilcoxon Rank-Sum test as appropriate for this sample of unrelated subjects. Linear regression using age, sex, PC1, PC2 and CPD as covariates revealed that, among African Americans, variants in CHRNB3 are associated with increased alcohol dependence symptom count (P = 0.0038, β = 0.48). This was then confirmed using a Wilcoxon Rank-Sum test comparing the distribution of age, sex, PC1, PC2 and CPD corrected alcohol symptom count in CHRNB3 carriers versus non-carriers (P = 0.005). There was no association between either alcohol or cocaine symptom count and CHRNA3 variants among African Americans in the COGEND sample. More than 60% (n = 1685) of individuals in the COGEND sample have ≥1 alcohol dependence symptom, but there is a severe lack of power to interrogate cocaine dependence symptoms in COGEND because only ∼10% (n = 290) of individuals in this data set have non-zero cocaine dependence symptoms.
DISCUSSION
We find that rare missense variants in CHRNB3 are associated with increased risk for cocaine dependence and alcohol dependence among European Americans and that rare missense variants in CHRNA3 are associated with decreased risk of cocaine dependence among African Americans. Having tested five genes and two primary phenotypes, alcohol and cocaine dependence symptom count, the association between CHRNB3 variants and both alcohol and cocaine dependence symptom count among European Americans in the COGA data set are significant after multiple test correction for 10 tests (P < 0.005) and the association between CHRNA3 variants and cocaine dependence symptom count among African Americans in COGA nearly passes that significance threshold (P = 0.005). We note that the two primary phenotypes are correlated in the COGA data set (EA r2 = 0.22; AA r2 = 0.36) and thus do not represent two fully independent tests. These 10 tests of association were performed in both European Americans and African Americans within the COGA sample. The observed associations in COGA were then tested for association in the COGEND data set for replication. We were able to observe an association between CHRNB3 variants and alcohol dependence in the African-American portion of COGEND but were unable to replicate the association between CHRNA3 variants and either cocaine or alcohol dependence symptom count in COGEND.
One possible explanation for the observed association between rare variants in these genes and both alcohol and cocaine dependence symptom count is the high rate of comorbidity for these traits in the COGA sample. We attempted to determine which of these two phenotypes was driving the associations by including each as a covariate when analyzing the other. Our findings suggest that the observed association with cocaine dependence symptom count is not due to correlations between co-morbid phenotypes, either alcohol or smoking, but that the association between variants in CHRNB3 and alcohol dependence symptoms among European Americans may reflect the correlation of this trait with cocaine dependence symptoms in the COGA data set. We find no evidence that the observed associations are due to co-morbidity with smoking as we observe no association between rare variants in these genes with CPD and including CPD as a covariate in the analyses did not alter the results.
The association between CHRNB3 and both alcohol and cocaine dependence was largely due to one variant (rs149775276; H329Y). This variant was rare in European Americans, but absent in African Americans, partially explaining the lack of association at this gene among African Americans in the COGA data set. We were able to replicate the association between CHRNB3 and alcohol dependence in the COGEND African-American sample. It is likely that we were able to observe an association between CHRNB3 variants and alcohol dependence symptoms but not cocaine dependence symptoms in COGEND because alcohol dependence symptoms appear more often among COGEND individuals (∼60%) than do cocaine dependence symptoms (∼10%). Additionally, this association was only observed among African-Americans individuals in COGEND. The frequency of CHRNB3 carriers among COGEND European Americans was exceedingly small (0.6% of individuals) compared with CHRNB3 carriers among African Americans (∼4%).
The association between CHRNA3 and cocaine dependence among African Americans was due almost entirely to the effect of one variant (R37H; rs8192475). This variant was previously shown to decrease risk for nicotine dependence and increase cellular response to nicotine in vitro (52). Because CHRNA3 R37H is rarer in African Americans (∼0.1%) than European Americans (∼4%), the absence of association among European Americans is not likely to be due to lack of power. However, there may be an effect of differing genetic background between the two ethnic groups that cannot be accounted for in our analyses. Additionally, controlling for both global and local admixture in the African-American cohort does not affect the association observed between CHRNA3 and cocaine dependence symptom counts, suggesting that this association is not due to population stratification, however subtle.
For the other genes studied (CHRNB4, CHRNA6 and CHRNA5), no associations were observed between rare variants and either alcohol or cocaine dependence in European Americans or African Americans in COGA. This may have been due to insufficient power resulting from low minor allele frequencies even after collapsing genotypes within genes. Analysis of all variants within each gene cluster (CHRNB3-A6 and CHRNA5-A3-B4) revealed that each cluster was associated only when one of the constituent genes was significantly associated by itself. This suggests that the observed associations, between rare variants across gene clusters, are merely a reflection of the single-gene associations reported here.
There are several limitations to this study. First, we were not able to fully account for the possible confounding of nicotine dependence on the observed associations. We included CPD as a covariate in our analyses, but ideally we would perform the association in never-smokers or use Fagerstrom Test of Nicotine Dependence (FTND) score as a covariate. Unfortunately, very few non-smokers are alcohol dependent (n = 387; 12%) or cocaine dependent (n = 73; 2%) and FTND score had not been assessed in much of the COGA data set and therefore could not be included as a covariate in the analyses. Another limitation is the lack of a comparable replication data set. The COGEND data set was assessed for both alcohol and cocaine dependence phenotypes, but lacks power due to limited sample size and limited numbers of cocaine-dependent cases. Furthermore, the COGEND sample is a case–control series, whereas COGA sample includes extended families. If a rare variant is detected in family, we were able to examine segregation with drug dependence phenotypes in the rest of the family, which is not possible in a case–control series. Lastly, rare variant associations are more dependent on chance observation, namely the sampling of rare allele carriers from a population, than studies of common genetic variants. It is possible that our observed associations may be due to chance fluctuations in rare variant alleles.
There is a debate in the scientific community regarding the relative role of common versus rare genetic variants in determining an individual's risk of developing common diseases. Whether or not the majority of variability in any given trait will eventually be attributed to common or rare variants is yet to be determined. However, this and other studies continue to demonstrate the power of rare variant association testing in the identification and validation of genes contributing to common disease. Moreover, rare variant associations have the ability to identify specific genes and variants, rather than large genomic regions. This suggests that in the era of whole-genome sequencing, genome-wide gene-based rare variant associations may be more powerful in identifying novel susceptibility loci. We were not able to replicate our previously reported association between rare missense variants in CHRNB4 and nicotine dependence in the COGA cohort. One explanation for this is that the COGA sample is not well suited for analysis of smoking-related traits. The sample is a family data set ascertained to study alcohol dependence and as such does not have the same power as the COGEND sample to investigate smoking-related traits. Secondly, not all of the subjects in the COGA sample have been assessed for nicotine dependence.
In conclusion, we find an association between missense variants in CHRNB3 and CHRNA3 and alcohol and cocaine dependence symptom counts. Variants in CHRNB3 are associated with increased risk for cocaine dependence and alcohol dependence, while variants in CHRNA3 are associated with decreased risk for cocaine dependence. Both CHRNB3 and CHRNA3 are associated with cocaine dependence independently of smoking quantity as measured by CPD, a fact that is not surprising given the role of CHRN genes in the physiological response to multiple substances and the comorbidity of nicotine with both alcohol and cocaine use in human populations. We hypothesize that these variants act either by altering generalized reward pathways, by impacting impulsivity and related personality characteristics or by altering the effect of alcohol or cocaine on nicotinic receptors directly. Further sequencing of individuals for whom detailed dependence-related information has been obtained is necessary to confirm and elaborate on these findings and functional studies will be necessary to elucidate the role of these genes on the physiological effects of cocaine.
MATERIALS AND METHODS
COGA sample
DNA samples were collected as part of the COGA. All members of the COGA sample underwent a semi-structured interview, the SSAGA, which assessed alcohol, cocaine and nicotine use as well as comorbid psychiatric conditions. The COGA sample utilized in this study consisted of 2504 African Americans and 7318 European Americans. Maximum lifetime CPD was used as a covariate in all analyses. CPD was calculated by comparing reports across multiple interviews for an individual and selecting the value of CPD that reflected the largest number smoked during the time when the subject was smoking the most. Only individuals who had at one point in their life smoked have CPD values. Controls consisted of individuals who had tried a substance but never became dependent. A total of 287 African American (147 cases and 140 controls) and 1028 European American (480 cases and 548 controls) individuals were sequenced. For African Americans, pools ranging in size from 44 to 96 individuals were used (two case pools and two control pools). For European Americans, pools ranging in size from 68 to 96 individuals were used (five case pools and six control pools). Follow-up genotyping of SNPs identified and validated in the sequenced individuals was done in the remaining members of each family represented in the sequenced set of individuals (2217 African Americans and 6290 European Americans).
COGEND sample
DNA samples were collected as part of the Collaborative Genetic Study of Nicotine Dependence (COGEND). All members of the COGEND sample underwent a semi-structured interview, which assessed smoking behavior, other substance use and comorbid psychiatric conditions. The COGEND sample includes 710 African Americans and 2055 European Americans. The coding regions of the CHRNA3, CHRNB4, CHRNA5, CHRNB3 and CHRNA6 genes were sequenced in a total of 352 African Americans (176 ND cases and 176 smoking controls) and 400 European Americans (200 ND cases and 200 smoking controls) (51). Of these sequenced individuals, 120 (34%) of the African Americans had non-zero DSM-IV alcohol dependence symptom counts, 300 (75%) of the European Americans had non-zero DSM-IV alcohol dependence symptom counts, 23 (6.5%) of the African Americans had non-zero DSM-IV cocaine dependence symptom counts and 50 (12.5%) of the European Americans had non-zero DSM-IV cocaine dependence symptom counts. All missense variants observed in these sequenced genes were genotyped in all members of COGEND using Sequenom.
Pooled sequencing
Pooled DNA sequencing was performed as previously described (52,56). The concentrations of individual DNA samples were first measured using Quant-iT™ PicoGreen reagent and pooled in equimolar amounts. Each pooled DNA sample was then used as the template for the amplification of each protein coding exon of the CHRNA3, CHRNA5, CHRNA6, CHRNB3 and CHRNB4 genes. Primers for the amplification of each exon were designed using Primer3 and reference sequences were taken from the human genome reference assembly build 37 (hg19). To ensure complete coverage of each desired exon, a minimum of 50 bp of a flanking sequence on each side was required for each amplicon. We used Pfu high fidelity DNA polymerase in all PCR reactions to reduce the identification of SNPs generated as a result of the PCR (false-positive SNPs). After PCR amplification of desired genomic regions, PCR products were cleaned using QIAquick PCR purification kits, quantified using Quant-iT PicoGreen reagent and ligated in equimolar amounts using T4 Ligase and T4 Polynucleotide Kinase. At this stage, positive and negative control vectors were amplified and added to each pool to serve as internal quality standards and to be used in data analysis. After ligation, concatenated PCR products were randomly sheared by sonication and prepared for Illumina sequencing on an Illumina Genome Analyzer IIx (GAIIx) according to the manufacturer's specifications. As previously shown by Vallania et al. (53) an average coverage of 30-fold per allele per pool was shown to correlate to optimal positive predictive value for the SNP-calling algorithm and was, therefore, the target level of coverage in this study.
Sequencing analysis
For analysis, sequencing reads (36 bp reads) were aligned using an alignment algorithm developed by Vallania et al. (53) which aligns sequences allowing for two mismatches or indels of up to 4 bp. To quantify the specificity and sensitivity of this method, positive and negative control DNA were introduced as PCR products in the pooled sequencing protocol outlined above. As a positive control to estimate sensitivity for variant calling, a pool of 10 plasmids with a 72 bp insert was generated. One plasmid acts as the ‘wild-type’ insert, while the remaining nine plasmids contain one or two synthetically engineered mutations. All 10 plasmids were combined such that each the allele frequency of each known mutation would mimic either the allele frequency of a single allelic variant or 10 allelic variants in our human DNA pool. Following mixing of the vectors, PCR amplification across the insert sequence was performed and the PCR product was added to the normalized pool of human target PCR reactions during sequencing library preparation. We then obtained more coverage for each amplicon than that which was required to detect all variants in this amplified positive control vector pool in order to ensure a minimal SNP detection false-negative rate. As a negative control and to model the sequencing error rate, 1908 bp of the pCMV6-XL5 plasmid was also amplified and included. One half of the pCMV6-XL5 plasmid sequence was used to train the SNP finding algorithm and the other to test the error model. Finally, to identify variants, in the pooled sequence data we used the SPLINTER algorithm (53). The SPLINTER algorithm produces a P-value for the probability that a predicted variant is a true positive. This probability is produced using large deviation theory to quantify the difference between observed allele frequencies within the sequence data to the background error rate for the same type of sequence changes seen in the amplified pCMV6-XL5 plasmid. A P-value cut-off value for each lane of Illumina sequencing was defined as the value at which all positive controls were identified. Only those variants falling below this cut-off value were considered ‘predicted’ by SPLINTER. All protein coding or splice site variants predicted by SPLINTER were then validated by individual Sequenom genotyping in each person from the source DNA pools. SNPs validated in the sequenced individuals were then genotyped in all members of the COGA sample. All individual genotyping was performed using the Sequenom platform as described previously (11).
Association analysis
Single missense variants were tested for association in African Americans and European Americans from the COGA study using linear regression as implemented in the GWAF package in R using age, sex and CPD as covariates. Each sequenced gene was tested as a whole for association using famSKAT using age, sex and CPD as covariates. The GWAF and famSKAT programs were used as the COGA study is family-based and requires programs that can integrate family information. Associations in COGEND between alcohol dependence symptom count or cocaine dependence symptom count and variants in the CHRNB3 or CHRNA3 genes were performed using linear regression in PLINK using age, sex and CPD as covariates and in R comparing the residuals of alcohol or cocaine dependence symptom count after correcting for age, sex and CPD in CHRNB3 missense variant carriers versus. non-carriers and CHRNA3 missense variant carriers versus. non-carriers using a Wilcoxon Rank-Sum Test. Linear regression and Wilcoxon Rank-Sum tests were used as COGEND is a sample of unrelated cases and controls.
FUNDING
COGA is a national collaborative study supported by NIH Grant U10AA008401 from the National Institute on Alcohol Abuse and Alcoholism (NIAAA) and the National Institute on Drug Abuse (NIDA). COGEND is a collaborative research group and part of the National Institute on Drug Abuse (NIDA) Genetics Consortium. Subject collection was supported by NIH grant P01 CA89392 (PI - L Bierut) from the NCI. Phenotypic and genotypic data are stored in the NIDA Center for Genetic Studies (NCGS) at http://zork.wustl.edu/ under NIDA Contract HHSN271200477451C (PIs J Tischfield and J Rice).
ACKNOWLEDGEMENTS
The Collaborative Study on the Genetics of Alcoholism (COGA): COGA, Principal Investigators B. Porjesz, V. Hesselbrock, H. Edenberg, L. Bierut includes 10 different centers: University of Connecticut (V. Hesselbrock); Indiana University (H.J. Edenberg, J. Nurnberger Jr, T. Foroud); University of Iowa (S. Kuperman, J. Kramer); SUNY Downstate (B. Porjesz); Washington University in Saint Louis (L. Bierut, A. Goate, J. Rice, K. Bucholz); University of California at San Diego (M. Schuckit); Rutgers University (J. Tischfield); Southwest Foundation (L. Almasy), Howard University (R. Taylor) and Virginia Commonwealth University (D. Dick). A. Parsian and M. Reilly are the NIAAA Staff Collaborators. We continue to be inspired by our memories of Henri Begleiter and Theodore Reich, founding PI and Co-PI of COGA, and also owe a debt of gratitude to other past organizers of COGA, including Ting-Kai Li, currently a consultant with COGA, P. Michael Conneally, Raymond Crowe and Wendy Reich, for their critical contributions. In memory of Theodore Reich, founding Principal Investigator of COGEND, we are indebted to his leadership in the establishment and nurturing of COGEND and acknowledge with great admiration his seminal scientific contributions to the field. Lead investigators directing data collection for COGEND are Laura Bierut, Naomi Breslau, Dorothy Hatsukami and Eric Johnson. The authors thank Heidi Kromrei and Tracey Richmond for their assistance in COGEND data collection.
Conflict of Interest statement. L.J.B. and A.M.G. are listed as inventors on the patent ‘Markers for Addiction’ (US 20070258898) covering the use of certain SNPs in determining the diagnosis, prognosis and treatment of addiction. N.S. is the spouse of Dr S. Saccone, who is also listed as an inventor on the above patent.
REFERENCES
- 1.Wang J.C., Kapoor M., Goate A.M. The genetics of substance dependence. Ann. Rev. Genom. Human Genet. 2012;13:241–261. doi: 10.1146/annurev-genom-090711-163844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kendler K.S., Schmitt E., Aggen S.H., Prescott C.A. Genetic and environmental influences on alcohol, caffeine, cannabis, and nicotine use from early adolescence to middle adulthood. Arch. Gen. Psychiatry. 2008;65:674–682. doi: 10.1001/archpsyc.65.6.674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rhee S.H., Hewitt J.K., Young S.E., Corley R.P., Crowley T.J., Stallings M.C. Genetic and environmental influences on substance initiation, use, and problem use in adolescents. Arch. Gen. Psychiatry. 2003;60:1256–1264. doi: 10.1001/archpsyc.60.12.1256. [DOI] [PubMed] [Google Scholar]
- 4.Bierut L.J., Strickland J.R., Thompson J.R., Afful S.E., Cottler L.B. Drug use and dependence in cocaine dependent subjects, community-based individuals, and their siblings. Drug Alcohol Depend. 2008;95:14–22. doi: 10.1016/j.drugalcdep.2007.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stinson F.S., Grant B.F., Dawson D.A., Ruan W.J., Huang B., Saha T. Comorbidity between DSM-IV alcohol and specific drug use disorders in the United States: results from the National Epidemiologic Survey on Alcohol and Related Conditions. Drug Alcohol Depend. 2005;80:105–116. doi: 10.1016/j.drugalcdep.2005.03.009. [DOI] [PubMed] [Google Scholar]
- 6.Bierut L.J. Genetic vulnerability and susceptibility to substance dependence. Neuron. 2011;69:618–627. doi: 10.1016/j.neuron.2011.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thorgeirsson T.E., Gudbjartsson D.F., Surakka I., Vink J.M., Amin N., Geller F., Sulem P., Rafnar T., Esko T., Walter S., et al. Sequence variants at CHRNB3-CHRNA6 and CYP2A6 affect smoking behavior. Nat. Genet. 2010;42:448–453. doi: 10.1038/ng.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Saccone S.F., Hinrichs A.L., Saccone N.L., Chase G.A., Konvicka K., Madden P.A., Breslau N., Johnson E.O., Hatsukami D., Pomerleau O., et al. Cholinergic nicotinic receptor genes implicated in a nicotine dependence association study targeting 348 candidate genes with 3713 SNPs. Hum. Mol. Genet. 2007;16:36–49. doi: 10.1093/hmg/ddl438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grucza R.A., Wang J.C., Stitzel J.A., Hinrichs A.L., Saccone S.F., Saccone N.L., Bucholz K.K., Cloninger C.R., Neuman R.J., Budde J.P., et al. A risk allele for nicotine dependence in CHRNA5 is a protective allele for cocaine dependence. Biol. Psychiatry. 2008;64:922–929. doi: 10.1016/j.biopsych.2008.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sherva R., Kranzler H.R., Yu Y., Logue M.W., Poling J., Arias A.J., Anton R.F., Oslin D., Farrer L.A., Gelernter J. Variation in nicotinic acetylcholine receptor genes is associated with multiple substance dependence phenotypes. Neuropsychopharmacology. 2010;35:1921–1931. doi: 10.1038/npp.2010.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang J.C., Grucza R., Cruchaga C., Hinrichs A.L., Bertelsen S., Budde J.P., Fox L., Goldstein E., Reyes O., Saccone N., et al. Genetic variation in the CHRNA5 gene affects mRNA levels and is associated with risk for alcohol dependence. Mol. Psychiatry. 2009;14:501–510. doi: 10.1038/mp.2008.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang J.C., Cruchaga C., Saccone N.L., Bertelsen S., Liu P., Budde J.P., Duan W., Fox L., Grucza R.A., Kern J., et al. Risk for nicotine dependence and lung cancer is conferred by mRNA expression levels and amino acid change in CHRNA5. Hum. Mol. Genet. 2009;18:3125–3135. doi: 10.1093/hmg/ddp231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thorgeirsson T.E., Gudbjartsson D.F., Surakka I., Vink J.M., Amin N., Geller F., Sulem P., Rafnar T., Esko T., Walter S., et al. Sequence variants at CHRNB3-CHRNA6 and CYP2A6 affect smoking behavior. Nat. Genet. 2010;42:448–453. doi: 10.1038/ng.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hoft N.R., Corley R.P., McQueen M.B., Huizinga D., Menard S., Ehringer M.A. SNPs in CHRNA6 and CHRNB3 are associated with alcohol consumption in a nationally representative sample. Genes Brain Behav. 2009;8:631–637. doi: 10.1111/j.1601-183X.2009.00495.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wouda J.A., Riga D., De Vries W., Stegeman M., van Mourik Y., Schetters D., Schoffelmeer A.N., Pattij T., De Vries T.J. Varenicline attenuates cue-induced relapse to alcohol, but not nicotine seeking, while reducing inhibitory response control. Psychopharmacology. 2011;216:267–277. doi: 10.1007/s00213-011-2213-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Farook J.M., Lewis B., Gaddis J.G., Littleton J.M., Barron S. Lobeline, a nicotinic partial agonist attenuates alcohol consumption and preference in male C57BL/6J mice. Physiol. Behav. 2009;97:503–506. doi: 10.1016/j.physbeh.2009.02.031. [DOI] [PubMed] [Google Scholar]
- 17.Hendrickson L.M., Zhao-Shea R., Pang X., Gardner P.D., Tapper A.R. Activation of alpha4* nAChRs is necessary and sufficient for varenicline-induced reduction of alcohol consumption. J. Neurosci. 2010;30:10169–10176. doi: 10.1523/JNEUROSCI.2601-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Proctor W.R., Dobelis P., Moritz A.T., Wu P.H. Chronic nicotine treatment differentially modifies acute nicotine and alcohol actions on GABA(A) and glutamate receptors in hippocampal brain slices. Br. J. Pharmacol. 2011;162:1351–1363. doi: 10.1111/j.1476-5381.2010.01141.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rezvani A.H., Slade S., Wells C., Petro A., Lumeng L., Li T.K., Xiao Y., Brown M.L., Paige M.A., McDowell B.E., et al. Effects of sazetidine-A, a selective alpha4beta2 nicotinic acetylcholine receptor desensitizing agent on alcohol and nicotine self-administration in selectively bred alcohol-preferring (P) rats. Psychopharmacology. 2010;211:161–174. doi: 10.1007/s00213-010-1878-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lof E., Olausson P., deBejczy A., Stomberg R., McIntosh J.M., Taylor J.R., Soderpalm B. Nicotinic acetylcholine receptors in the ventral tegmental area mediate the dopamine activating and reinforcing properties of ethanol cues. Psychopharmacology. 2007;195:333–343. doi: 10.1007/s00213-007-0899-4. [DOI] [PubMed] [Google Scholar]
- 21.Kuzmin A., Jerlhag E., Liljequist S., Engel J. Effects of subunit selective nACh receptors on operant ethanol self-administration and relapse-like ethanol-drinking behavior. Psychopharmacology. 2009;203:99–108. doi: 10.1007/s00213-008-1375-5. [DOI] [PubMed] [Google Scholar]
- 22.Bowers B.J., McClure-Begley T.D., Keller J.J., Paylor R., Collins A.C., Wehner J.M. Deletion of the alpha7 nicotinic receptor subunit gene results in increased sensitivity to several behavioral effects produced by alcohol. Alcohol. Clin. Exp. Res. 2005;29:295–302. doi: 10.1097/01.alc.0000156116.40817.a2. [DOI] [PubMed] [Google Scholar]
- 23.Bell R.L., Eiler B.J., 2nd, Cook J.B., Rahman S. Nicotinic receptor ligands reduce ethanol intake by high alcohol-drinking HAD-2 rats. Alcohol. 2009;43:581–592. doi: 10.1016/j.alcohol.2009.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Steensland P., Simms J.A., Holgate J., Richards J.K., Bartlett S.E. Varenicline, an alpha4beta2 nicotinic acetylcholine receptor partial agonist, selectively decreases ethanol consumption and seeking. Proc. Natl Acad. Sci. USA. 2007;104:12518–12523. doi: 10.1073/pnas.0705368104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Robles N., Sabria J. Effects of moderate chronic ethanol consumption on hippocampal nicotinic receptors and associative learning. Neurobiol. Learn. Mem. 2008;89:497–503. doi: 10.1016/j.nlm.2008.01.006. [DOI] [PubMed] [Google Scholar]
- 26.Chatterjee S., Steensland P., Simms J.A., Holgate J., Coe J.W., Hurst R.S., Shaffer C.L., Lowe J., Rollema H., Bartlett S.E. Partial agonists of the alpha3beta4* neuronal nicotinic acetylcholine receptor reduce ethanol consumption and seeking in rats. Neuropsychopharmacology. 2011;36:603–615. doi: 10.1038/npp.2010.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bito-Onon J.J., Simms J.A., Chatterjee S., Holgate J., Bartlett S.E. Varenicline, a partial agonist at neuronal nicotinic acetylcholine receptors, reduces nicotine-induced increases in 20% ethanol operant self-administration in Sprague-Dawley rats. Addict. Biol. 2011;16:440–449. doi: 10.1111/j.1369-1600.2010.00309.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zarrindast M.R., Meshkani J., Rezayof A., Beigzadeh R., Rostami P. Nicotinic acetylcholine receptors of the dorsal hippocampus and the basolateral amygdala are involved in ethanol-induced conditioned place preference. Neuroscience. 2010;168:505–513. doi: 10.1016/j.neuroscience.2010.03.019. [DOI] [PubMed] [Google Scholar]
- 29.Marszalec W., Aistrup G.L., Narahashi T. Ethanol-nicotine interactions at alpha-bungarotoxin-insensitive nicotinic acetylcholine receptors in rat cortical neurons. Alcohol. Clin. Exp. Res. 1999;23:439–445. [PubMed] [Google Scholar]
- 30.Frohlich R., Patzelt C., Illes P. Inhibition by ethanol of excitatory amino acid receptors and nicotinic acetylcholine receptors at rat locus coeruleus neurons. Naunyn-Schmiedebergs Arch. Pharmacol. 1994;350:626–631. doi: 10.1007/BF00169367. [DOI] [PubMed] [Google Scholar]
- 31.Aistrup G.L., Marszalec W., Narahashi T. Ethanol modulation of nicotinic acetylcholine receptor currents in cultured cortical neurons. Mol. Pharmacol. 1999;55:39–49. doi: 10.1124/mol.55.1.39. [DOI] [PubMed] [Google Scholar]
- 32.Symons M.N., Weng J., Diehl E., Heo E., Kleiber M.L., Singh S.M. Delineation of the role of nicotinic acetylcholine receptor genes in alcohol preference in mice. Behav. Genet. 2010;40:660–671. doi: 10.1007/s10519-010-9366-9. [DOI] [PubMed] [Google Scholar]
- 33.Kamens H.M., McKinnon C.S., Li N., Helms M.L., Belknap J.K., Phillips T.J. The alpha 3 subunit gene of the nicotinic acetylcholine receptor is a candidate gene for ethanol stimulation. Genes Brain Behav. 2009;8:600–609. doi: 10.1111/j.1601-183X.2008.00444.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kamens H.M., Andersen J., Picciotto M.R. The nicotinic acetylcholine receptor partial agonist varenicline increases the ataxic and sedative-hypnotic effects of acute ethanol administration in C57BL/6J mice. Alcohol. Clin. Exp. Res. 2010;34:2053–2060. doi: 10.1111/j.1530-0277.2010.01301.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hendrickson L.M., Gardner P., Tapper A.R. Nicotinic acetylcholine receptors containing the alpha4 subunit are critical for the nicotine-induced reduction of acute voluntary ethanol consumption. Channels (Austin) 2011;5:124–127. doi: 10.4161/chan.5.2.14409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Taslim N., Soderstrom K., Dar M.S. Role of mouse cerebellar nicotinic acetylcholine receptor (nAChR) alpha(4)beta(2)- and alpha(7) subtypes in the behavioral cross-tolerance between nicotine and ethanol-induced ataxia. Behav. Brain Res. 2011;217:282–292. doi: 10.1016/j.bbr.2010.10.026. [DOI] [PubMed] [Google Scholar]
- 37.Sajja R.K., Dwivedi C., Rahman S. Nicotinic ligands modulate ethanol-induced dopamine function in mice. Pharmacology. 2010;86:168–173. doi: 10.1159/000317063. [DOI] [PubMed] [Google Scholar]
- 38.Adermark L., Clarke R.B., Soderpalm B., Ericson M. Ethanol-induced modulation of synaptic output from the dorsolateral striatum in rat is regulated by cholinergic interneurons. Neurochem. Int. 2011;58:693–699. doi: 10.1016/j.neuint.2011.02.009. [DOI] [PubMed] [Google Scholar]
- 39.Gould R.W., Garg P.K., Garg S., Nader M.A. Effects of nicotinic acetylcholine receptor agonists on cognition in rhesus monkeys with a chronic cocaine self-administration history. Neuropharmacology. 2013;64:479–488. doi: 10.1016/j.neuropharm.2012.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Metaxas A., Keyworth H., Yoo J., Chen Y., Kitchen I., Bailey A. The stereotypy-inducing and OCD-like effects of chronic ‘binge’ cocaine are modulated by distinct subtypes of nicotinic acetylcholine receptors. Br. J. Pharmacol. 2012;167:450–464. doi: 10.1111/j.1476-5381.2012.02023.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xie X., Arguello A.A., Reittinger A.M., Wells A.M., Fuchs R.A. Role of nicotinic acetylcholine receptors in the effects of cocaine-paired contextual stimuli on impulsive decision making in rats. Psychopharmacology. 2012;223:271–279. doi: 10.1007/s00213-012-2715-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McGranahan T.M., Patzlaff N.E., Grady S.R., Heinemann S.F., Booker T.K. Alpha4beta2 nicotinic acetylcholine receptors on dopaminergic neurons mediate nicotine reward and anxiety relief. J. Neurosci. 2011;31:10891–10902. doi: 10.1523/JNEUROSCI.0937-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mitchell J.M., Teague C.H., Kayser A.S., Bartlett S.E., Fields H.L. Varenicline decreases alcohol consumption in heavy-drinking smokers. Psychopharmacology. 2012;223:299–306. doi: 10.1007/s00213-012-2717-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Saccone N.L., Culverhouse R.C., Schwantes-An T.H., Cannon D.S., Chen X., Cichon S., Giegling I., Han S., Han Y., Keskitalo-Vuokko K., et al. Multiple independent loci at chromosome 15q25.1 affect smoking quantity: a meta-analysis and comparison with lung cancer and COPD. PLoS Genet. 2010;6:e1001053. doi: 10.1371/journal.pgen.1001053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tennessen J.A., Bigham A.W., O'Connor T.D., Fu W., Kenny E.E., Gravel S., McGee S., Do R., Liu X., Jun G., et al. Evolution and functional impact of rare coding variation from deep sequencing of human exomes. Science. 2012;337:64–69. doi: 10.1126/science.1219240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schork N.J., Murray S.S., Frazer K.A., Topol E.J. Common versus rare allele hypotheses for complex diseases. Curr. Opin. Genet. Dev. 2009;19:212–219. doi: 10.1016/j.gde.2009.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Johansen C.T., Wang J., Lanktree M.B., Cao H., McIntyre A.D., Ban M.R., Martins R.A., Kennedy B.A., Hassell R.G., Visser M.E., et al. Excess of rare variants in genes identified by genome-wide association study of hypertriglyceridemia. Nat. Genet. 2010;42:684–687. doi: 10.1038/ng.628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Haller G., Torgerson D.G., Ober C., Thompson E.E. Sequencing the IL4 locus in African Americans implicates rare noncoding variants in asthma susceptibility. J. Allergy Clin. Immunol. 2009;124:1204–1209.e9. doi: 10.1016/j.jaci.2009.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Momozawa Y., Mni M., Nakamura K., Coppieters W., Almer S., Amininejad L., Cleynen I., Colombel J.F., de Rijk P., Dewit O., et al. Resequencing of positional candidates identifies low frequency IL23R coding variants protecting against inflammatory bowel disease. Nat. Genet. 2011;43:43–47. doi: 10.1038/ng.733. [DOI] [PubMed] [Google Scholar]
- 50.Wessel J., McDonald S.M., Hinds D.A., Stokowski R.P., Javitz H.S., Kennemer M., Krasnow R., Dirks W., Hardin J., Pitts S.J., et al. Resequencing of nicotinic acetylcholine receptor genes and association of common and rare variants with the Fagerstrom test for nicotine dependence. Neuropsychopharmacology. 2010;35:2392–2402. doi: 10.1038/npp.2010.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xie P., Kranzler H.R., Krauthammer M., Cosgrove K.P., Oslin D., Anton R.F., Farrer L.A., Picciotto M.R., Krystal J.H., Zhao H., et al. Rare nonsynonymous variants in alpha-4 nicotinic acetylcholine receptor gene protect against nicotine dependence. Biol. Psychiatry. 2011;70:528–536. doi: 10.1016/j.biopsych.2011.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Haller G., Druley T., Vallania F.L., Mitra R.D., Li P., Akk G., Steinbach J.H., Breslau N., Johnson E., Hatsukami D., et al. Rare missense variants in CHRNB4 are associated with reduced risk of nicotine dependence. Hum. Mol. Genet. 2012;21:647–655. doi: 10.1093/hmg/ddr498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vallania F.L., Druley T.E., Ramos E., Wang J., Borecki I., Province M., Mitra R.D. High-throughput discovery of rare insertions and deletions in large cohorts. Genome Res. 2010;20:1711–1718. doi: 10.1101/gr.109157.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Price A.L., Patterson N.J., Plenge R.M., Weinblatt M.E., Shadick N.A., Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 55.Sankararaman S., Sridhar S., Kimmel G., Halperin E. Estimating local ancestry in admixed populations. Am. J. Hum. Genet. 2008;82:290–303. doi: 10.1016/j.ajhg.2007.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Druley T.E., Vallania F.L., Wegner D.J., Varley K.E., Knowles O.L., Bonds J.A., Robison S.W., Doniger S.W., Hamvas A., Cole F.S., et al. Quantification of rare allelic variants from pooled genomic DNA. Nat. Methods. 2009;6:263–265. doi: 10.1038/nmeth.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]