Key Points
This study has identified a novel capture mechanism for host-derived vesicles within the spleen and lymph node.
This pathway modulates the immune response to circulating particulate antigens.
Abstract
Exosomes are lipid nanovesicles released following fusion of the endosoma limiting membrane with the plasma membrane; however, their fate in lymphoid organs after their release remains controversial. We determined that sialoadhesin (CD169; Siglec-1) is required for the capture of B cell-derived exosomes via their surface-expressed α2,3-linked sialic acids. Exosome-capturing macrophages were present in the marginal zone of the spleen and in the subcapsular sinus of the lymph node. In vitro assays performed on spleen and lymph node sections confirmed that exosome binding to CD169 was not solely due to preferential fluid flow to these areas. Although the circulation half-life of exosomes in blood of wild-type and CD169−/− mice was similar, exosomes displayed altered distribution in CD169−/− mice, with exosomes freely accessing the outer marginal zone rim of SIGN-R1+ macrophages and F4/80+ red pulp macrophages. In the lymph node, exosomes were not retained in the subcapsular sinus of CD169−/− mice but penetrated deeper into the paracortex. Interestingly, CD169−/− mice demonstrated an enhanced response to antigen-pulsed exosomes. This is the first report of a role for CD169 in the capture of exosomes and its potential to mediate the immune response to exosomal antigen.
Introduction
Exosomes are vesicles released from multivesicular endosomes following fusion with the plasma membrane. Exosomes are a potential source of self-antigen for modulating the immune response against self-tissues, including tumors.1,2 Several cellular and molecular interactions direct the binding of exosomes to populations of leukocytes or stromal cells.3-10 In lymphoid organs, antigen presenting cells (APCs) in the marginal zone (MZ) of the spleen11 and follicular dendritic cells (DCs) in the B-cell areas of lymph node (LN)12 have been suggested to interact with exosomes, although the presence of a specific exosome receptor has yet to be demonstrated.
Sialic acid binding immunoglobulin lectins (Siglec) are sialic acid binding molecules expressed on a variety of leukocytes and stromal cells. CD169 (Sialoadhesin), the first Siglec family member identified, contains 17 immunoglobulin-like domains with the sialic acid binding site within the V-set terminal immunoglobulin domain. The short cytoplasmic tail of CD169 lacks signal transduction and endocytosis motifs, although recent data have implicated CD169 in endocytosis.13 Sialic acids decorate the surface of all cells and most secreted proteins14; however, due to the low (millimolar) affinity of CD169 for sialic acid, only heavily sialylated, multimeric structures bind strongly to CD169+ macrophages.13 CD169−/− mice do not display overt immune response defects but have depressed immunoglobulin M (IgM) levels and subtle alterations in the proportions of T- and B-cell subsets.15 Interestingly, CD169−/− mice show lower levels of autoreactive T-cell activation in mouse models of multiple sclerosis and uveoretinitis autoimmune mice, likely due to altered regulatory T-cell activity.13,16
CD169 is strongly expressed on the subcapsular sinus (SCS) and medullary macrophages in LN and on marginal metallophilic macrophages in the MZ of the spleen.13,17 CD169+ macrophages sample a wide variety of antigens and participate in generation of immunity to tumors and viruses but may also down-regulate immune responses to self-tissue.13 CD169+ macrophages directly present captured antigen to T cells or natural killer (NK) T cells17 and are adept at transferring antigen to CD8α+ DC and B cells.17 LN CD169+ macrophages transfer their own membrane material to closely associated T cells and NK cells.18
Exosomes express carbohydrate modifications, such as complex N-linked glycans, high mannose, polylactosamine, and sialic acids.19-21 We report that the preferred ligand of CD169, α2,3-linked sialic acid,22 is enriched on B cell–derived exosomes, allowing their capture by CD169+ macrophages in both spleen and LN. In the absence of this pathway, exosome access to the lymphoid system is dysregulated, resulting in aberrant trafficking of exosomes into the splenic red pulp or LN cortex. In addition, CD169−/− mice demonstrate enhanced cytotoxic T-cell responses to exosomal antigen. This suggests that CD169 controls the access of exosomes to lymphoid organs, possibly to minimize immune responses to self-antigen.
Materials and methods
Mice
C57BL/6 (wild-type) mice and ovalbumin peptide-specific OT-I and OT-II transgenic T-cell ovalbumin were obtained from Jackson Laboratories. OT-I and OT-II mice were crossed with CD45.1+ B6.SJL-PtprcaPep3b/BoyJArc (Animal Resources Centre) to generate F1 CD45.1+ OT-I or OT-II progeny. CD169−/− mice15 were from the University of Dundee. All mice were bred in specific pathogen free conditions at the University of Otago Hercus Taieri Resource Unit as described.23 All intravenous injections (100 µL phosphated-buffered saline [PBS]) were in the lateral tail vein. Subcutaneous injections were in the forelimb with 50 µL PBS. Animal studies were approved by the regional Animal Ethics Committee.
Exosome purification and labeling
Exosomes were isolated from anti-CD40 (FGK-45; 5 µg/mL) and interleukin-4 (50 ng/mL; R&D Systems, Auckland, NZ) stimulated C57BL/6 splenocytes cultured at 2 × 106/mL for 3 days in R10 (RPMI-1640; Gibco #31800-022) supplemented with 10% vesicle-depleted fetal calf serum (FCS; PAA Laboratories), 100 U/mL penicillin (Gibco #15140-122), 100 µg/mL streptomycin (Gibco #15140-122), 55 µM β-mercaptoethanol (Gibco #21985-023), and 2 g/L NaHCO3.24 In brief, culture supernatant was centrifuged at 450g for 5 minutes and then 2000g for 20 minutes (4°C) to deplete cells and debris, respectively. The supernatant was 0.2 µm filtered, and exosomes were pelleted by ultracentrifugation at 120 000g for 1 hour at 4°C. Pellets were washed twice in PBS. Where stated, exosomes were resuspended in 12 mL of PBS, overlaid onto 4 mL of 30% sucrose/200 mM Tris/D2O cushion, and ultracentrifuged at 100 000g for 75 minutes at 4°C. Exosomes were located 1 mL above to 2 mL below the interface; these fractions were pooled (supplemental Figure 1 on the Blood website). Pellets were resuspended in the final 0.5 mL. Sucrose was removed by washing twice in PBS by ultracentrifugation. Protein content of exosome preparations was performed using the Bradford assay, and exosome quality was routinely controlled by flow cytometry and electron microscopy.24 For in vivo capture experiments and modified Stamper-Woodruff25 assays, exosomes were biotinylated (Exo-bio) for 10 minutes at 4°C using 1 mg/mL sulfo-NHS-LC-biotin (Pierce #21335) in PBS (Gibco #21600-010), quenched with 100 mM glycine/PBS (pH 7.4), and washed twice with 30 mL PBS by ultracentrifugation. Alternatively, where stated, exosomes were labeled with 1 mg/mL sulfo-NHS-LC-fluorescein (Pierce #46410) following the same procedure as described for biotin.
Peptide and protein loading
DCs were generated from C57BL/6 bone marrow cells as previously described,26 matured overnight at day 6 with 200 ng/mL lipopolysaccharide (Salmonella Typhimurium; Sigma #L6511), and harvested on day 7. For peptide experiments, pelleted exosomes, parental B cells, or DCs were pulsed for 4 hours at 37°C with 1 µM ovalbumin (OVA) peptides OVA257-264 and/or OVA323-339 (Genscript; Exo257/323, B cell257/323 and DC257/323, respectively). Alternatively, B cells or DCs were incubated for 2 days with 200 µg/mL OVA protein (Sigma #A5503). B cell–derived exosomes (Exo-pro) were then isolated from OVA-pulsed B-cell supernatants. Exosomes were washed twice in 30 mL PBS by ultracentrifugation. Exo257/323 and Exo-pro were subsequently sucrose cushion purified. Peptide- and protein-pulsed B cells and DCs were washed twice in PBS.
Sialidase treatment of exosomes
Biotinylated exosomes were treated with 0.1 U/mL Vibrio cholerae–derived sialidase (SIAL-V; Roche #11-080-725-001) in digestion buffer (14.7 mg CaCl2-dihydrate, 580 mg NaCl to 100 mL 0.1 M Na-acetate, pH 5.5) for 30 minutes at 37°C. Exosomes were washed twice in 0.1% bovine serum albumin (BSA; Gibco #30063-572)/PBS by ultracentrifugation. Where stated, exosomes were sucrose cushion purified. Untreated or SIAL-V–treated exosomes were bound to 4 µm aldehyde-sulfate microspheres24 (Molecular Probes #A8244A); BSA-conjugated microspheres were used as a negative control. Exosomes were analyzed for α2,3- and α2,6-linked sialic acid expression with biotinylated lectins 5 µg/mL MAL-II (Vector #B-1265) or 0.6 µg/mL SNA-I (Vector #B-1305), or for CD9, CD19, CD21, CD24, MHC-II, and immunoglobulin expression (see supplemental Methods) before flow cytometric analysis (BD LSRFortessa; FlowJo).
Modified Stamper-Woodruff assay
Frozen naïve C57BL/6 and CD169−/− mouse spleen or LN (axillary, brachial, inguinal, and mesenteric) tissue was cryosectioned as previously described.26 Sections were blocked with 1% BSA/PBS for 10 minutes, and 50 µg/mL Exo-bio (±sialidase treatment) diluted in 0.1% BSA/Iscove's modified Dulbecco's media (Gibco #12440-053) was added for 2 hours at 37°C in a humid box. Sections were rinsed with PBS, fixed with 1% paraformaldehyde/PBS, quenched with 100 mM glycine/PBS, and blocked with 1% goat serum (GS)/PBS each for 10 minutes. Sections were incubated for 1 hour with primary antibodies from cell supernatant prepared from MOMA-1 (anti-CD169), F4/80, and ER-TR9 (anti–SIGN-R1) hybridomas obtained from Professor Georg Kraal (VU Medical Centre, Amsterdam, Netherlands). Primary antibodies were detected with 10 µg/mL anti-rat IgG-Alexa-488 or -Alexa-594 (Invitrogen #A21208 and Invitrogen #A21209, respectively) in 1% GS/PBS. Biotin was detected with 5 µg/mL streptavidin-Alexa-488 or -Alexa-594 (Invitrogen #S11223 and Invitrogen #S11227, respectively), and nuclei were counterstained with 25 ng/mL 4′,6 diamidino-2-phenylindole (DAPI; Invitrogen #D3571). Sections were mounted in Prolong Gold anti-fade (Invitrogen #P36930). For blocking experiments, sections were first incubated with 10 µg/mL neutralizing antibody anti-CD169 (clone SER-4) or negative control antibody anti–interleukin-4 (clone 11B11; purified in house) diluted in 1% BSA/PBS for 1 hour. Biotinylated exosomes (50 µg/mL) were then added directly to sections (without removal of blocking antibody) for 2 hours at 37°C. Sections were viewed with an Olympus BX-51 upright fluorescent microscope with UPLAN FL lenses (FN26.5) with identical exposure conditions and images analyzed using DP Manager software.
Fluorescence microscopy of in vivo captured exosomes and beads
C57BL/6 or CD169−/− mice were injected intravenously or subcutaneously with 100 or 50 µg of Exo-bio, respectively, and 2 × 1011 or 1 × 1011 100 nm fluorescent microspheres (ex-488 nm; Polysciences #24061), respectively. For intravenous and subcutaneous routes, respectively, mice were killed at 5, 60, or 120 minutes and spleen and liver were harvested or at 30 minutes and draining LN (axillary and brachial) was harvested. Organs were cryosectioned, fixed, and quenched before being blocked with 1% GS/PBS and labeled for CD169, F4/80, or SIGN-R1. Exosomes were detected as detailed above.
Colocalization analysis
Colocalization between exosome signal (red, Alexa-594) or 100 nm microspheres (green, Alexa-488) and macrophage marker signal (green, Alexa-488; red, Alexa-594) was determined from intravenously injected C57BL/6 or CD169−/− mice killed at 5 minutes. Colocalization was calculated from 10 individual photos (×20 objective lens) per mouse, using the Manders’ coefficient (fraction of exosome or bead signal overlapping with macrophage signal) with the software ImageJ and JaCoP plugin using autothresholding.27
In vivo T-cell proliferation
CD45.1+ splenocytes and LN cells (5 × 107 cells/mL) from OT-I or OT-II mice were labeled with differing dyes; 2.5 µM carboxyfluorescein diacetate succinimidyl ester (CFSE; Invitrogen #C34554) or 2.5 µM cell proliferation dye (CPD) V450 (BD #562158) for 7 minutes at 20°C and quenched with 5 mL FCS. Cells were washed once in 10% FCS/PBS and twice in PBS. Labeled cells were intravenously injected into recipient mice (OT-I and OT-II cells; 107 each were pooled and cotransferred). For LN or splenic responses, C57BL/6 or CD169−/− mice were immunized subcutaneously or intravenously with PBS, 50 or 100 µg sucrose cushion purified Exo257/323, respectively, 50 µg sucrose cushion purified Exo-pro, or 105 peptide- or protein-pulsed B cells or DCs. As a control, the pellet from Exo257/323 sucrose cushion purification was washed and resuspended identically to Exo257/323 fractions, and mice were immunized subcutaneously with equivalent volumes. Five days later, draining LN or spleen was removed, and T cells were labeled for CD4, CD8, and CD45.1 and analyzed by flow cytometry (supplemental Methods).
In vivo cytotoxicity assays
C57BL/6 or CD169−/− mice were immunized intravenously or subcutaneously with PBS, Exo257 or Exo-pro (50 µg sucrose cushion purified or 100 µg by ultracentrifugation only), 105 peptide, or protein pulsed B cells or DCs. As a control, the pellet from Exo-pro sucrose cushion purification was washed and resuspended identically to Exo-pro fractions, and mice were immunized intravenously with equivalent volumes. Where stated, mice were adoptively transferred intravenously 1 day prior to immunization with 107 OT-I splenocytes. Seven days after immunization, targets were prepared as follows: naïve C57BL/6 splenocytes (2 × 107 cells/mL) were unpulsed (R10) or pulsed with 1 µM OVA257 in R10 for 1 hour at 37°C and then washed in 0.1% BSA/PBS/2 mM EDTA. Unpulsed and pulsed cells were stained with 0.2 and 2 µM CFSE, respectively, for 7 minutes at 20°C, quenched, and washed as described above. Equal numbers of unpulsed and pulsed targets were pooled (1.5 × 107 total) for intravenous injection. Mice were killed 18 hours later, and spleens were analyzed by flow cytometry.
Kinetics of exosome clearance from blood
C57BL/6 and CD169−/− mice were anesthetized with ketamine and medetomidine as previously described23 and maintained on a warming tray at 37°C. Biotinylated B cell–derived exosomes (100 µg exosomal protein/mouse) were intravenously injected. Total mouse blood volume was estimated to be 5.5% of body weight. Blood was removed from the opposite lateral tail vein to that of exosome administration. MHC-II+ exosomes were detected by enzyme-linked immunosorbent assay. In brief, Nunc Maxisorp plates were coated with 1 µg/mL purified streptavidin (Jackson #016-000-084) in PBS overnight at 4°C. Plates were washed with 0.02% Tween-20/PBS and blocked for 10 minutes at 20°C with 0.1% caseinate (Arotech)/PBS. Exosome concentration in plasma was determined using a standard of biotinylated exosomes of known protein concentration. EDTA plasma samples were diluted 1/20 with PBS and added overnight at 4°C. After washing, 1 µg/mL fluorescein isothiocyanate (FITC)-M5/114 (anti-mouse I-Ab,d,q/I-Ed,k; purified and FITC-conjugated in-house) was added for 1 hour at 37°C to detect bound MHC-II+ exosomes.28 Plates were washed and then incubated with anti–FITC-horseradish peroxidase (1/5000; Roche #1426346) for 1 hour at 37°C. After washing, the plate was developed in tetramethyl benzidine (Zymed #00-2023), and the reaction was stopped with 2 N H2SO4. Optical density at 450 nm was determined using a Tecan Infinite M200 microplate reader.
Statistical analysis
All statistical analyses were performed with GraphPad Prism 6 by 1-way (or 2-way in Figure 3) ANOVA with a Bonferroni postcorrection test.
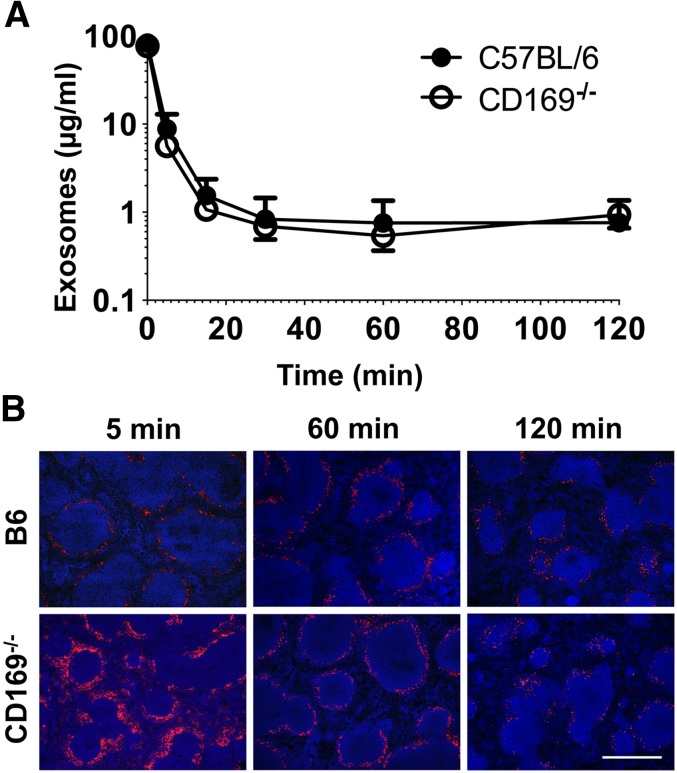
Figure 3.
Exosome clearance and distribution in vivo. (A) C57BL/6 or CD169−/− mice were anesthetized and then IV injected with 100 µg Exo-bio (purified by ultracentrifugation). Mice were tail bled at the indicated time points. MHC-II+ exosome concentration was analyzed by enzyme-linked immunosorbent assay from plasma samples; Exo-bio spiked plasma was used as a standard. Closed circles, C57BL/6 mice; open circles, CD169−/− mice. (B) C57BL/6 or CD169−/− mice were IV injected with 100 µg Exo-bio. Mice were killed at the indicated time points, and spleens were harvested. Exo-bio was detected with streptavidin-Alexa-594 and nuclei counterstained with DAPI. (B) Original magnification, ×100. Bar represents 500 µm. Results representative of (A,B) 6 mice (120 minutes) and (B) 3 to 6 mice (5 and 60 minutes).
Results
Identification of spleen, liver, and LN as the main exosome targets
To determine the in vivo target of exosomes, primary B cell–derived exosomes were isolated. Quality controls demonstrated effective enrichment of MHC-II+ exosomes using ultracentrifugation alone (supplemental Figure 1).24 Vesicles were then biotinylated and injected intravenously or subcutaneously into mice. Results showed distinct MZ or SCS distribution of exosomes in the spleen or LN, respectively (Figure 1). Further analysis showed a distinct colocalization of exosome and CD169 labeling (Figure 1A), suggesting a role for CD169+ macrophages in exosome capture. To investigate the potential for CD169+ macrophage-mediated binding of exosomes, we used CD169−/− mice.15 Compared with B6 mice, CD169−/− mice exhibited altered distribution of intravenously transferred exosomes, with exosomes penetrating the splenic red pulp and outer MZ sinus, the signal overlapping with SIGN-R1+ and F4/80+ macrophages (Figure 1B). To a lesser extent, liver macrophages (Kupffer cells) also bound intravenously transferred exosomes (Figure 1C). Kupffer cells also express CD169, but at levels severalfold below that of splenic or LN macrophages.29 Interestingly, binding of exosomes to Kupffer cells was not altered in CD169−/− mice (Figure 1C).
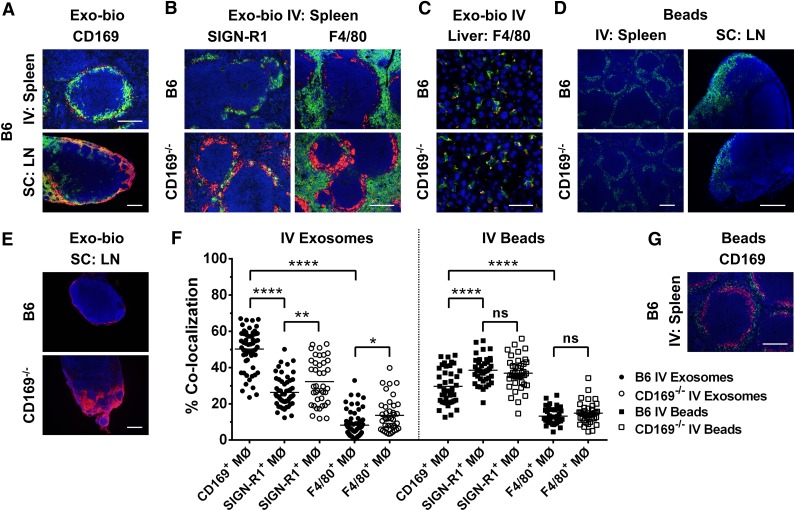
Figure 1.
Aberrant distribution of exosomes in lymphoid tissues of CD169−/− in vivo. C57BL/6 or CD169−/− mice were (A [spleen],B,C,F) intravenously (IV) or (A [LN],E) subcutaneously (SC) injected in the forelimb with 100 or 50 µg of biotinylated B cell–derived exosomes (Exo-bio; purified by ultracentrifugation), respectively. (D,F,G) Alternatively, C57BL/6 or CD169−/− mice were IV or SC injected with 2 × 1011 or 1 × 1011 100 nm fluorescent microspheres, respectively (green). For IV or SC routes, mice were killed at 5 minutes with spleens and livers harvested or at 30 min and draining LN harvested, respectively. (A-C,E) Exo-bio was detected with streptavidin-Alexa-594 (red). Sections were colabeled for (A,G) marginal metallophilic or subcapsular sinus macrophages with anti-CD169 (MOMA-1), (B) MZ or red pulp macrophages with anti–SIGN-R1 (ER-TR9) or anti-F4/80, respectively, or (C) Kupffer cells with anti-F4/80. Primary antibodies were detected with (A-C) anti-rat IgG-Alexa-488 (green) or (G) anti-rat IgG-Alexa-594 (red), and nuclei were counterstained with DAPI (blue). Original magnification, (A [LN],D[spleen],E) ×100 , (A [spleen],B,D [LN],G) ×200, and (C) ×400. Bar represents (A [spleen],B,D [LN],G) 200 µm, (A [LN],D [spleen],E) 250 µm, and (C) 50 µm. All results are representative of ≤4 mice per group. (F) Percent colocalization was calculated from fluorescent microscopy photos of (A [spleen],B,G) spleen sections. Ten individual photographs per mouse (original magnification, ×200) were analyzed for colocalization of green (Alexa-488) and red (Alexa-594) signal using the Manders’ coefficient with ImageJ. Each point represents an individual photograph; line indicates mean. Circles, exosomes; squares, beads; closed symbols, C57BL/6 mice; open symbols, CD169−/− mice. One-way ANOVA with Bonferroni postcorrection test was performed: ns, not significant; *P < .05; **P < .01; ****P < .0001.
Although CD169−/− mice retain populations of marginal metallophilic macrophages in the MZ of the spleen and SCS macrophages in LN,30 it is possible that these macrophages display altered barrier properties to particulate antigens. However, intravenous or subcutaneous administration of inert 100 nm microspheres into wild-type and CD169−/− mice demonstrated no strain difference in distribution of microspheres into the splenic MZ or SCS of the LN, respectively (Figure 1D), suggesting no loss of MZ or SCS barrier function in CD169−/− mice.
Subcutaneous administration of exosomes resulted in localization of exosomes to CD169+ macrophages within the SCS (Figure 1E). However, in CD169−/− mice, exosome binding to SCS macrophages was reduced, and exosomes penetrated deeper areas of the LN cortex (Figure 1E). Exosome binding to splenic CD169+ macrophages in wild-type mice was significantly greater than to other macrophage subsets (Figure 1F). In contrast, inert 100 nm beads preferentially colocalized with SIGN-R1+ macrophages (Figure 1F-G; supplemental Figure 1). In the absence of CD169, exosomes showed greater access to SIGN-R1+ and F4/80+ macrophages (Figure 1B,F).
Exosome binding to splenic and lymph node macrophages in vitro
It could be argued that colocalization of exosomes to CD169+ macrophages was simply due to anatomical constraints, because the primary site of fluid entry into the spleen and LN is the MZ and SCS, respectively. We developed an in vitro assay (modified Stamper-Woodruff assay25; Figure 2A-B) using Exo-bio (Figure 2C) applied to tissue sections to determine if specific exosome receptors were present in the spleen and LN. Interestingly, the distribution pattern of exosome binding matched that of exosome capture in vivo (cf. Figure 1A with Figure 2A-B). These results clearly demonstrated that exosomes bound CD169+ macrophages in both wild-type spleen and LN but not CD169−/− mice (Figure 2B,D). BSA-biotin failed to bind to tissue sections, ruling out nonspecific binding effects due to amine-linked biotinylation (supplemental Figure 3). In addition, labeling of exosomes with an alternate fluorochrome confirmed these results (supplemental Figure 3), and cold (nonbiotinylated) exosomes effectively inhibited exosome binding to tissue sections (supplemental Figure 3). Blocking experiments with an anti-CD169 neutralizing antibody (SER-4)31 further confirmed that binding of both B cell–derived (Figure 2D) and DC-derived (supplemental Figure 4) exosomes was CD169 dependent.
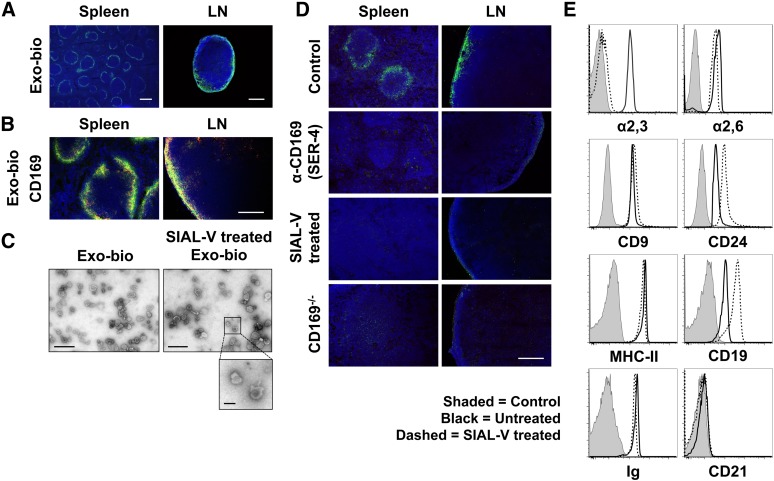
Figure 2.
Exosomes are bound by CD169+ macrophages in the spleen and LN in the absence of blood or lymph flow. Exo-bio was applied to naïve C57BL/6 or CD169−/− spleen and LN sections using a modified Stamper-Woodruff assay. Biotin was detected with (A) streptavidin-Alexa-488 (green) or (B) -Alexa-594 (red) and marginal metallophilic or subcapsular sinus macrophages stained with anti-CD169 (MOMA-1). Primary antibody was detected with (B) anti-rat IgG-Alexa-488 (green) and nuclei counterstained with DAPI (blue). (C) Exo-bio ± treatment with Vibrio cholerae–derived sialidase (α2,3-linked sialic acid preferential cleavage [SIAL-V]) were purified using a sucrose cushion, negatively stained and visualized by transmission electron microscopy. No apparent differences in morphology were observed between samples; diameter range was 70 to 120 nm. Photographs are representative of the preparations as a whole. (D) Exo-bio was bound to naïve C57BL/6 sections in the presence of negative control antibody or CD169 neutralizing antibody (SER-4). Alternatively, Exo-bio or SIAL-V–treated Exo-bio was applied to naïve C57BL/6 or CD169−/− sections, respectively. Exosomes and nuclei were detected as described in A. (E) Exosomes ± SIAL-V treatment was bound to aldehyde-sulfate microspheres and analyzed by flow cytometry for α2,3- and α2,6-linked sialic acid expression using biotinylated MAL-II and SNA lectins, respectively. In addition, CD9, CD24, MHC-II, CD19, immunoglobulin, and CD21 expression was measured. Shaded peak, negative control (BSA-conjugated aldehyde-sulfate microspheres); black line, untreated exosomes; dashed line, SIAL-V–treated exosomes. Results representative of ≥3 separate experiments and/or exosome preparations, with (D:LN) LN sections from ≥4 anatomically distinct locations per experiment. Original magnification, (A [spleen]) ×40, (A [LN]) ×100, (B,D) ×200, (C [Exo-bio]) ×24 500, and (C [SIAL-V–treated Exo-bio]) ×17 500. Bar represents (A [spleen]) 500 µm, (A [LN]) 250 µm, (B,D) 200 µm, (C) 500 nm, and (C, inset) 100 nm.
Exosomes were shown to display high levels of α2,3-linked sialic acids—the preferred ligand of CD169 (Figure 2E).29,32 Sialidase-treated exosomes (Figure 2C) failed to bind the SCS or splenic MZ (Figure 2D), further confirming that exosome binding was CD169 dependent. Sialidase treatment did not alter exosome bouyant density on the sucrose cushion (supplemental Figure 1), exosome morphology (Figure 2C), α2,6-linked sialic acid levels (Figure 2E), or the detection of surface marker expression. Note, sialidase treatment caused a modest increase in antibody binding to the CD19 and CD24 glycoproteins (Figure 2E).
Plasma clearance rates of exosomes in wild-type and CD169−/− mice
Surprisingly, despite our observation of aberrant exosome trafficking in lymphoid organs of CD169−/− mice, MHC-II+ exosomes were cleared from the blood of wild-type and CD169−/− mice at similar rates, with a half-life of ∼2 minutes (Figure 3A). Interestingly, after 120 minutes, exosomes were still detectable in the spleen, indicating that longer-lived reservoirs of exosomes may persist after the majority of exosomes have been cleared from circulation (Figure 3B).
Enhanced responses of CD169−/− mice to protein and peptide-loaded exosomes
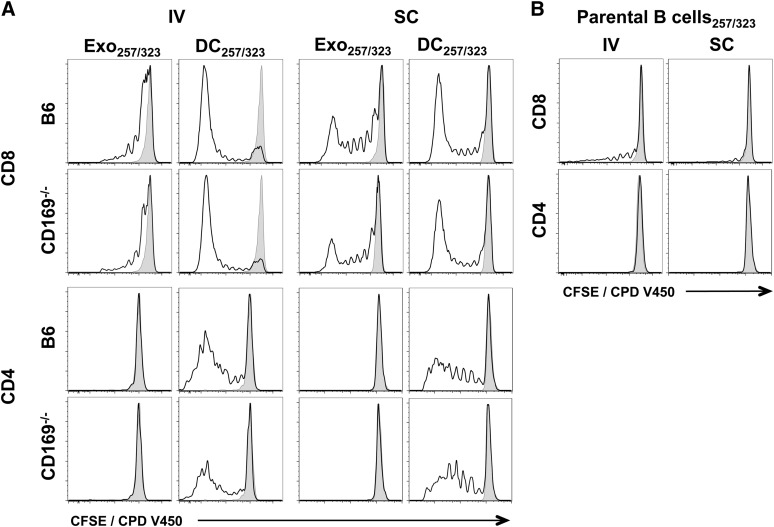
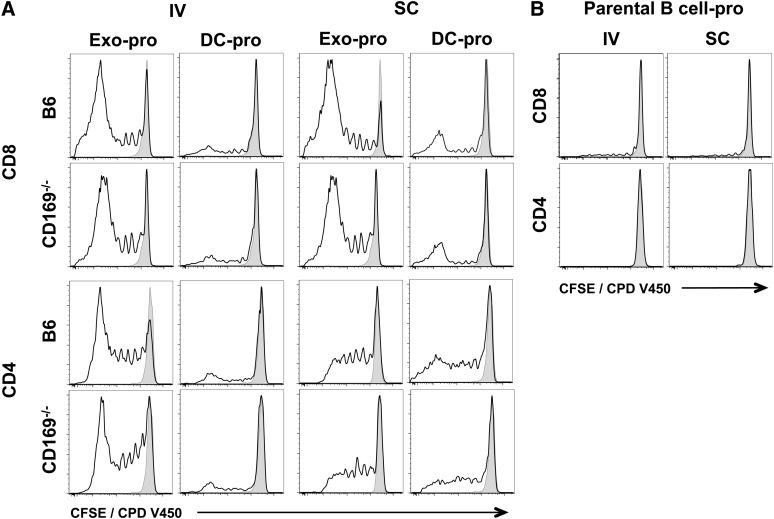
Because B cell–derived exosomes express both MHC-I and MHC-II,24 we next compared their ability to induce an immune response in wild-type and CD169−/− mice. Interestingly, although protein-pulsed exosomes induced both CD4+ and CD8+ T-cell proliferation, peptide-pulsed exosomes did not reproducibly induce CD4+ T-cell proliferation (Figures 4 and 5; supplemental Figures 5 and 6). No differences in OT-I or OT-II proliferation were noted between wild-type and CD169−/− mice by either the intravenous or subcutaneous route for both peptide and protein (Figures 4 and 5; supplemental Figures 5 and 6). Surprisingly, peptide- or protein-pulsed parental B cells did not, or only weakly induced, CD4+ or CD8+ T-cell proliferation, or cytotoxicity, by both subcutaneous and intravenous routes (Figures 4-7).
Figure 4.
In vivo T-cell proliferation in response to exosomal-peptide antigen. C57BL/6 or CD169−/− mice were immunized IV or SC in the forelimb with (A) PBS, 100 µg sucrose cushion purified Exo257/323, and 105 DC257/323 or (B) 105 parental B cell257/323. Exosomes and cells were all pulsed simultaneously with 1 μM ovalbumin peptides OVA257-264 and OVA323-339. T-cell proliferation of adoptively cotransferred OT-I (CD8) and OT-II (CD4) cells (CFSE or CPD V450) were analyzed 5 days after immunization by flow cytometry. Black line, test group; shaded peak, PBS-immunized mice. Results representative of ≥6 mice per group.
Figure 5.
T-cell proliferation in response to exosomal-protein antigen. C57BL/6 or CD169−/− mice were immunized IV or SC in the forelimb with (A) PBS, 50 µg sucrose cushion purified exosomes derived from B cells cultured with 200 µg/mL ovalbumin protein for 2 days (Exo-pro), and 105 DC-pro or (B) 105 parental B cell-pro. DC and B cells were cultured with 200 µg/mL ovalbumin protein for 2 days. T-cell proliferation of adoptively cotransferred OT-I (CD8) and OT-II (CD4) cells (CFSE or CPD V450) were analyzed 5 days after immunization by flow cytometry. Black line, test group; shaded peak, PBS-immunized mice. Results representative of ≥6 mice per group.
Figure 7.
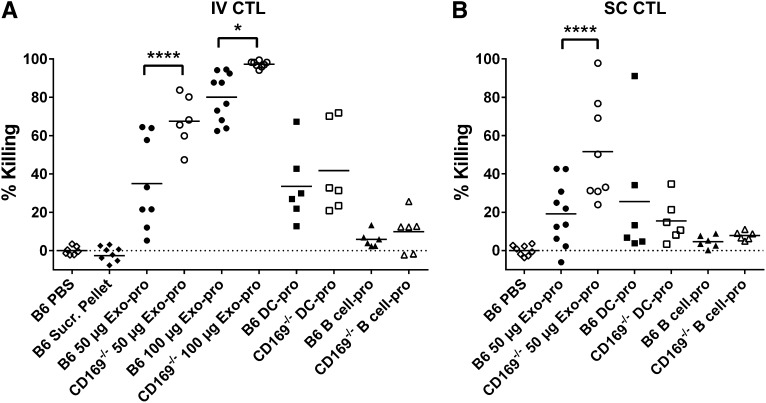
Enhanced cytotoxic responses to exosomal-protein in CD169−/− mice. C57BL/6 or CD169−/− mice were immunized (A) IV or (B) SC with PBS, pellet from exosome sucrose cushion purification (IV: B6 Sucr. Pellet), 50 μg sucrose cushion purified Exo-pro or 100 µg Exo-pro (purified by ultracentrifugation), 105 DC-pro, or 105 parental B cell-pro. Seven days after immunization, mice were adoptively transferred with unpulsed (CFSE low) or OVA257-264-pulsed (CFSE high) target cells. In vivo killing was analyzed 18 hours later by flow cytometry. Results representative of ≥6 mice per group. One-way ANOVA with Bonferroni postcorrection was performed: *P < .05; ****P < .0001.
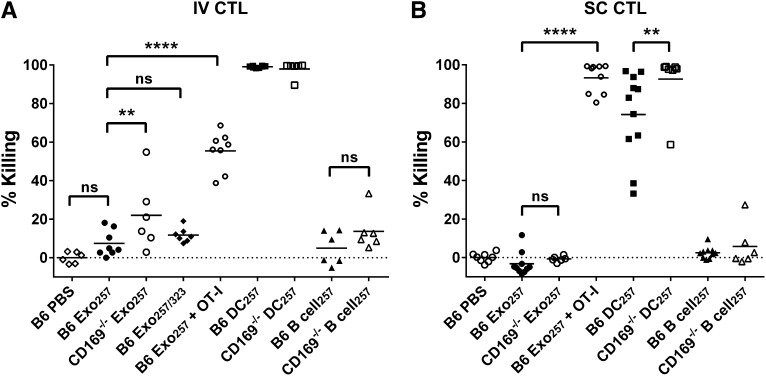
Consistent with a recent report on DC-derived exosomes,10 peptide-pulsed exosomes induced relatively weak cytotoxic T-lymphocyte (CTL) responses. This response was not enhanced by inclusion of the T-cell helper epitope peptide OVA323-339 (Figure 6). However, supplementation of naïve OT-I T cells resulted in a significantly enhanced CTL response to Exo257, demonstrating that increasing CTL precursor levels significantly enhanced the cytotoxic response induced by peptide-pulsed exosomes. Interestingly, the level of cytotoxicity was dependent on antigen type, with significantly higher levels of cytotoxicity noted in mice immunized with protein-pulsed compared with peptide-pulsed exosomes (Figures 6 and 7; P < .0001 for both B6 or CD169−/− mouse strains). Control experiments demonstrated that particulate antigenic material (either protein or peptide) was not responsible for the observed immune responses (Figure 7; supplemental Figure 5). Strikingly, compared with wild-type mice, CD169−/− mice demonstrated significantly enhanced CTL responses to peptide-pulsed (intravenous only; Figure 6) and protein-pulsed exosomes (intravenous and subcutaneous; Figure 7). This difference was maintained using differing amounts (50 and 100 µg) of protein-pulsed exosomes via intravenous immunization (Figure 7). A small but significant increase in the CTL response of CD169−/− mice to subcutaneously injected peptide-pulsed, but not protein pulsed, DCs was also noted (Figure 6B).
Figure 6.
Enhanced cytotoxic responses to intravenous exosomal-peptide in CD169−/− mice. C57BL/6 or CD169−/− mice were immunized (A) IV or (B) SC with PBS and 100 or 50 µg Exo257, respectively, 100 μg Exo257/323 (IV), 105 DC257, or 105 parental B cell257. Where stated, mice were supplemented IV with 107 OT-I splenocytes prior to immunization. Seven days after immunization, mice were adoptively transferred with unpulsed (CFSE low) or OVA257-264-pulsed (CFSE high) target cells. In vivo killing was analyzed 18 hours later by flow cytometry. Results representative of ≥6 mice per group using exosomes purified by ultracentrifugation. Line, mean. One-way ANOVA with Bonferroni postcorrection was performed: ns, not significant; **P < .01; ****P < .0001.
Discussion
We identified CD169 as a specific binding-partner for exosomes in lymphoid tissue. Its restriction to subpopulations of macrophages dedicated to antigen capture and direct presentation and/or transfer of antigen to other APCs17 suggests an important role of CD169-mediated capture in the immune response to exosomal antigen. CD169+ macrophages are strategically situated at antigen entry points into the spleen and LN and are involved in capture of sialylated pathogens13,30; however, a role for CD169 in capturing exosomes has not previously been demonstrated.
Despite the low affinity (Kd ∼ 1.4 mM) of CD169 for α2,3-linked sialic acid,33 we observed abundant binding of exosomes to this receptor. This may be a result of high level expression of α2,3-linked sialic acid expression on exosomes, as linear increases in CD169 ligand availability results in logarithmic increases in avidity.13 Moreover, due to their size, exosomes may suffer less shear stress, therefore gaining greater access to receptors, compared with larger cells or particles.
In the spleen, McGaha et al noted a similar role for MZ macrophages in the exclusion of particulate antigen from the red pulp following depletion of MZ macrophages by clodronate.34 Thus, the MZ macrophage barrier acts together with the MZ MADCAM-1+ sinus-lining cells to limit antigen entry into the red pulp.35 Such partitioning of antigen may optimize antigen availability for the initiation of appropriate immune responses at the MZ and white pulp.35 Interestingly, McGaha et al observed enhanced immune responses to antigen associated with apoptotic cells in mice depleted of MZ macrophages.34 It has previously been demonstrated that exosomes entering the venous circulation localize to the splenic MZ.11 In addition, there are several reports of MZ macrophages being involved in immune tolerance to apoptotic cells, either through modulation of cytokine secretion or by the induction of the immunosuppressive enzyme indoleamine 2,3-dioxygenase.34,36,37
DC-derived exosomes abundantly express the ligand milk-fat globule lactadherin, a potential ligand for the αvβ3/β5 integrins.38 Others have proposed that a number of molecules, including CD9, CD11a, CD81, CD91, intercellular adhesion molecule-1, and phosphatidyl serine, are involved in exosome capture by DCs.3,6,8,11 Stromal interactions of exosomes to collagen or fibronectin may be facilitated in humans by the β1 and β2 interactions.4,7 Additionally, C3b deposition on exosomes enhances antigen presentation and splenic uptake.10,39 There is 1 description of the rat-restricted Galectin 5 mediating the binding of erythrocyte exosomes to macrophages,40 but to our knowledge, ours is the first report of a macrophage-specific exosome receptor expressed in lymphoid tissue.
Similar exosome clearance rates were noted in the circulation of wild-type and CD169−/− mice, suggesting that CD169-independent exosome clearance mechanisms are active in CD169−/− mice. Removal of exosomes by complement-mediated destruction and uptake by phagocytes,10,39 or uptake by fenestrated endothelium in the liver sieve,41 may mask the contribution of a selectively expressed receptor present only in a subpopulation of LN and splenic macrophages. In contrast to our findings, liposomes displaying high-affinity glycan ligands for CD169 exhibited delayed clearance from circulation in CD169−/− mice.42 However, the combination of multivalent, high-affinity glycan ligands expressed on these liposomes together with enhanced stealth properties due to polyethylene glycol modification may explain this difference.42 A key distinction between exosomes and glycan-expressing liposomes is that the latter does not present any ligands for host receptors, apart from sialic acid. In contrast, exosomes possess many potential host ligands including adhesion molecules and complement receptors.4,6-8,10,11,38-40 As this is the first report on exosome circulation clearance rates, we are unable to compare our findings. However, similar rapid rates of reticulocyte microvesicle uptake and destruction were reported by Willekens et al.43
Interestingly, protein- but not peptide-pulsed exosomes induced strong endogenous CTL responses in agreement with another study.10 This enhancement was reportedly due to cooperation of helper T cells with antigen-specific B cells.10,44 This is an important finding as it distinguishes exosomes from cellular APCs namely, DCs, which have been reported to be 50-fold more efficient at presenting OVA peptides compared with equimolar concentrations of whole OVA.45 Given the inherent inefficiency of processing proteins for cross-priming, it is surprising that protein immunization resulted in high-level CTL activity by exosomes.
Our results are the first report of exosome capture by CD169 and of enhanced cytotoxicity to protein-pulsed exosomes in CD169−/− mice. Similarly, it was shown that CD169+ macrophages promote an inhibitory effect in response to apoptotic cells,36 although CD169 may alternatively promote autoimmunity and inflammatory responses to sialylated pathogens.13,30 It is possible that the outcome of antigen targeted to CD169+ macrophages is greatly influenced by the antigenic context, such as the presence of microbial products or inflammation.13,17 Our current focus is the role of APC subsets mediating the enhanced immune response observed in CD169−/− mice.
Supplementary Material
Acknowledgments
The authors thank Tri Phan and Robert Brink (Garvan Institute) for critical input into this study, Vernon Ward for support, and Lane Black for performing initial bead-localization experiments. The authors acknowledge the scientific assistance from the Otago Centre for Electron Microscopy (OCEM; University of Otago).
This work was supported by the Marsden Fund (08-UOO-106), a University of Otago Research grant, a bequest from the Otago School of Medical Sciences, and a scholarship from New Zealand Lottery Health Research. P.R.C. was funded by Wellcome Trust Senior Research Fellowship WT081882.
Footnotes
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: S.C.S. performed the research presented, analyzed the data, prepared the figures, and aided in design of the study and manuscript preparation; A.C.D. provided technical assistance and aided in manuscript preparation; P.R.C. provided antibodies and the CD169−/− mice; and A.D.M. designed the study, assisted with laboratory work, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Alexander McLellan, Department of Microbiology and Immunology, Otago School of Medical Sciences, University of Otago, PO Box 56, Dunedin, Otago, New Zealand; e-mail: alex.mclellan@otago.ac.nz; and Sarah Saunderson, Department of Microbiology and Immunology, Otago School of Medical Sciences, University of Otago, PO Box 56, Dunedin, Otago, New Zealand; e-mail: sarah.saunderson@otago.ac.nz.
References
- 1.Zitvogel L, Regnault A, Lozier A, et al. Eradication of established murine tumors using a novel cell-free vaccine: dendritic cell-derived exosomes. Nat Med. 1998;4(5):594–600. doi: 10.1038/nm0598-594. [DOI] [PubMed] [Google Scholar]
- 2.André F, Chaput N, Schartz NE, et al. Exosomes as potent cell-free peptide-based vaccine. I. Dendritic cell-derived exosomes transfer functional MHC class I/peptide complexes to dendritic cells. J Immunol. 2004;172(4):2126–2136. doi: 10.4049/jimmunol.172.4.2126. [DOI] [PubMed] [Google Scholar]
- 3.Nolte-’t Hoen EN, Buschow SI, Anderton SM, Stoorvogel W, Wauben MH. Activated T cells recruit exosomes secreted by dendritic cells via LFA-1. Blood. 2009;113(9):1977–1981. doi: 10.1182/blood-2008-08-174094. [DOI] [PubMed] [Google Scholar]
- 4.Rieu S, Geminard C, Rabesandratana H, Sainte-Marie J, Vidal M. Exosomes released during reticulocyte maturation bind to fibronectin via integrin alpha4beta1. Eur J Biochem. 2000;267(2):583-590. [DOI] [PubMed]
- 5.Wubbolts R, Leckie RS, Veenhuizen PT, et al. Proteomic and biochemical analyses of human B cell-derived exosomes. Potential implications for their function and multivesicular body formation. J Biol Chem. 2003;278(13):10963–10972. doi: 10.1074/jbc.M207550200. [DOI] [PubMed] [Google Scholar]
- 6.Skokos D, Botros HG, Demeure C, et al. Mast cell-derived exosomes induce phenotypic and functional maturation of dendritic cells and elicit specific immune responses in vivo. J Immunol. 2003;170(6):3037–3045. doi: 10.4049/jimmunol.170.6.3037. [DOI] [PubMed] [Google Scholar]
- 7.Clayton A, Turkes A, Dewitt S, Steadman R, Mason MD, Hallett MB. Adhesion and signaling by B cell-derived exosomes: the role of integrins. FASEB J. 2004;18(9):977-979. [DOI] [PubMed]
- 8.Segura E, Nicco C, Lombard B, et al. ICAM-1 on exosomes from mature dendritic cells is critical for efficient naive T-cell priming. Blood. 2005;106(1):216–223. doi: 10.1182/blood-2005-01-0220. [DOI] [PubMed] [Google Scholar]
- 9.Segura E, Guérin C, Hogg N, Amigorena S, Théry C. CD8+ dendritic cells use LFA-1 to capture MHC-peptide complexes from exosomes in vivo. J Immunol. 2007;179(3):1489–1496. doi: 10.4049/jimmunol.179.3.1489. [DOI] [PubMed] [Google Scholar]
- 10.Qazi KR, Gehrmann U, Domange Jordö E, Karlsson MC, Gabrielsson S. Antigen-loaded exosomes alone induce Th1-type memory through a B-cell-dependent mechanism. Blood. 2009;113(12):2673–2683. doi: 10.1182/blood-2008-04-153536. [DOI] [PubMed] [Google Scholar]
- 11.Morelli AE, Larregina AT, Shufesky WJ, et al. Endocytosis, intracellular sorting, and processing of exosomes by dendritic cells. Blood. 2004;104(10):3257–3266. doi: 10.1182/blood-2004-03-0824. [DOI] [PubMed] [Google Scholar]
- 12.Denzer K, van Eijk M, Kleijmeer MJ, Jakobson E, de Groot C, Geuze HJ. Follicular dendritic cells carry MHC class II-expressing microvesicles at their surface. J Immunol. 2000;165(3):1259–1265. doi: 10.4049/jimmunol.165.3.1259. [DOI] [PubMed] [Google Scholar]
- 13.Klaas M, Crocker PR. Sialoadhesin in recognition of self and non-self. Semin Immunopathol. 2012;34(3):353–364. doi: 10.1007/s00281-012-0310-3. [DOI] [PubMed] [Google Scholar]
- 14.Varki A. Sialic acids in human health and disease. Trends Mol Med. 2008;14(8):351–360. doi: 10.1016/j.molmed.2008.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Oetke C, Vinson MC, Jones C, Crocker PR. Sialoadhesin-deficient mice exhibit subtle changes in B- and T-cell populations and reduced immunoglobulin M levels. Mol Cell Biol. 2006;26(4):1549–1557. doi: 10.1128/MCB.26.4.1549-1557.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Crocker PR, Paulson JC, Varki A. Siglecs and their roles in the immune system. Nat Rev Immunol. 2007;7(4):255–266. doi: 10.1038/nri2056. [DOI] [PubMed] [Google Scholar]
- 17.Martinez-Pomares L, Gordon S. CD169+ macrophages at the crossroads of antigen presentation. Trends Immunol. 2012;33(2):66–70. doi: 10.1016/j.it.2011.11.001. [DOI] [PubMed] [Google Scholar]
- 18.Gray EE, Friend S, Suzuki K, Phan TG, Cyster JG. Subcapsular sinus macrophage fragmentation and CD169+ bleb acquisition by closely associated IL-17-committed innate-like lymphocytes. PLoS ONE. 2012;7(6):e38258. doi: 10.1371/journal.pone.0038258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Colino J, Snapper CM. Dendritic cell-derived exosomes express a Streptococcus pneumoniae capsular polysaccharide type 14 cross-reactive antigen that induces protective immunoglobulin responses against pneumococcal infection in mice. Infect Immun. 2007;75(1):220–230. doi: 10.1128/IAI.01217-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Batista BS, Eng WS, Pilobello KT, Hendricks-Muñoz KD, Mahal LK. Identification of a conserved glycan signature for microvesicles. J Proteome Res. 2011;10(10):4624–4633. doi: 10.1021/pr200434y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Escrevente C, Keller S, Altevogt P, Costa J. Interaction and uptake of exosomes by ovarian cancer cells. BMC Cancer. 2011;11:108. doi: 10.1186/1471-2407-11-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vinson M, van der Merwe PA, Kelm S, May A, Jones EY, Crocker PR. Characterization of the sialic acid-binding site in sialoadhesin by site-directed mutagenesis. J Biol Chem. 1996;271(16):9267–9272. doi: 10.1074/jbc.271.16.9267. [DOI] [PubMed] [Google Scholar]
- 23.Baker NJ, Schofield JC, Caswell MD, McLellan AD. Effects of early atipamezole reversal of medetomidine-ketamine anesthesia in mice. J Am Assoc Lab Anim Sci. 2011;50(6):916–920. [PMC free article] [PubMed] [Google Scholar]
- 24.Saunderson SC, Schuberth PC, Dunn AC, et al. Induction of exosome release in primary B cells stimulated via CD40 and the IL-4 receptor. J Immunol. 2008;180(12):8146–8152. doi: 10.4049/jimmunol.180.12.8146. [DOI] [PubMed] [Google Scholar]
- 25.Stamper HB, Jr, Woodruff JJ. Lymphocyte homing into lymph nodes: in vitro demonstration of the selective affinity of recirculating lymphocytes for high-endothelial venules. J Exp Med. 1976;144(3):828–833. doi: 10.1084/jem.144.3.828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McLellan AD, Terbeck G, Mengling T, et al. Differential susceptibility to CD95 (Apo-1/Fas) and MHC class II-induced apoptosis during murine dendritic cell development. Cell Death Differ. 2000;7(10):933–938. doi: 10.1038/sj.cdd.4400734. [DOI] [PubMed] [Google Scholar]
- 27.Bolte S, Cordelières FP. A guided tour into subcellular colocalization analysis in light microscopy. J Microsc. 2006;224(Pt 3):213–232. doi: 10.1111/j.1365-2818.2006.01706.x. [DOI] [PubMed] [Google Scholar]
- 28.MacKay PA, Leibundgut-Landmann S, Koch N, Dunn AC, Reith W, Jack RW, McLellan AD. Circulating, soluble forms of major histocompatibility complex antigens are not exosome-associated. Eur J Immunol. 2006;36(11):2875–2884. doi: 10.1002/eji.200636041. [DOI] [PubMed] [Google Scholar]
- 29.Crocker PR, Kelm S, Dubois C, et al. Purification and properties of sialoadhesin, a sialic acid-binding receptor of murine tissue macrophages. EMBO J. 1991;10(7):1661–1669. doi: 10.1002/j.1460-2075.1991.tb07689.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Klaas M, Oetke C, Lewis LE, et al. Sialoadhesin promotes rapid proinflammatory and type I IFN responses to a sialylated pathogen, Campylobacter jejuni. J Immunol. 2012;189(5):2414–2422. doi: 10.4049/jimmunol.1200776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Crocker PR, Gordon S. Mouse macrophage hemagglutinin (sheep erythrocyte receptor) with specificity for sialylated glycoconjugates characterized by a monoclonal antibody. J Exp Med. 1989;169(4):1333–1346. doi: 10.1084/jem.169.4.1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.May AP, Robinson RC, Vinson M, Crocker PR, Jones EY. Crystal structure of the N-terminal domain of sialoadhesin in complex with 3′ sialyllactose at 1.85 A resolution. Mol Cell. 1998;1(5):719–728. doi: 10.1016/s1097-2765(00)80071-4. [DOI] [PubMed] [Google Scholar]
- 33.Crocker PR, Vinson M, Kelm S, Drickamer K. Molecular analysis of sialoside binding to sialoadhesin by NMR and site-directed mutagenesis. Biochem J. 1999;341(Pt 2):355–361. [PMC free article] [PubMed] [Google Scholar]
- 34.McGaha TL, Chen Y, Ravishankar B, van Rooijen N, Karlsson MC. Marginal zone macrophages suppress innate and adaptive immunity to apoptotic cells in the spleen. Blood. 2011;117(20):5403–5412. doi: 10.1182/blood-2010-11-320028. [DOI] [PubMed] [Google Scholar]
- 35.Mebius RE, Kraal G. Structure and function of the spleen. Nat Rev Immunol. 2005;5(8):606–616. doi: 10.1038/nri1669. [DOI] [PubMed] [Google Scholar]
- 36.Miyake Y, Asano K, Kaise H, Uemura M, Nakayama M, Tanaka M. Critical role of macrophages in the marginal zone in the suppression of immune responses to apoptotic cell-associated antigens. J Clin Invest. 2007;117(8):2268–2278. doi: 10.1172/JCI31990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ravishankar B, Liu H, Shinde R, et al. Tolerance to apoptotic cells is regulated by indoleamine 2,3-dioxygenase. Proc Natl Acad Sci USA. 2012;109(10):3909–3914. doi: 10.1073/pnas.1117736109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Théry C, Boussac M, Véron P, Ricciardi-Castagnoli P, Raposo G, Garin J, Amigorena S. Proteomic analysis of dendritic cell-derived exosomes: a secreted subcellular compartment distinct from apoptotic vesicles. J Immunol. 2001;166(12):7309–7318. doi: 10.4049/jimmunol.166.12.7309. [DOI] [PubMed] [Google Scholar]
- 39.Papp K, Végh P, Prechl J, et al. B lymphocytes and macrophages release cell membrane deposited C3-fragments on exosomes with T cell response-enhancing capacity. Mol Immunol. 2008;45(8):2343–2351. doi: 10.1016/j.molimm.2007.11.021. [DOI] [PubMed] [Google Scholar]
- 40.Barrès C, Blanc L, Bette-Bobillo P, André S, Mamoun R, Gabius HJ, Vidal M. Galectin-5 is bound onto the surface of rat reticulocyte exosomes and modulates vesicle uptake by macrophages. Blood. 2010;115(3):696–705. doi: 10.1182/blood-2009-07-231449. [DOI] [PubMed] [Google Scholar]
- 41.Fraser R, Cogger VC, Dobbs B, Jamieson H, Warren A, Hilmer SN, Le Couteur DG. The liver sieve and atherosclerosis. Pathology. 2012;44(3):181–186. doi: 10.1097/PAT.0b013e328351bcc8. [DOI] [PubMed] [Google Scholar]
- 42.Chen WC, Completo GC, Sigal DS, Crocker PR, Saven A, Paulson JC. In vivo targeting of B-cell lymphoma with glycan ligands of CD22. Blood. 2010;115(23):4778–4786. doi: 10.1182/blood-2009-12-257386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Willekens FL, Werre JM, Kruijt JK, et al. Liver Kupffer cells rapidly remove red blood cell-derived vesicles from the circulation by scavenger receptors. Blood. 2005;105(5):2141–2145. doi: 10.1182/blood-2004-04-1578. [DOI] [PubMed] [Google Scholar]
- 44.Näslund TI, Gehrmann U, Qazi KR, Karlsson MC, Gabrielsson S. Dendritic cell-derived exosomes need to activate both T and B cells to induce antitumor immunity. J Immunol. 2013;190(6):2712–2719. doi: 10.4049/jimmunol.1203082. [DOI] [PubMed] [Google Scholar]
- 45.Met O, Buus S, Claesson MH. Peptide-loaded dendritic cells prime and activate MHC-class I-restricted T cells more efficiently than protein-loaded cross-presenting DC. Cell Immunol. 2003;222(2):126–133. doi: 10.1016/s0008-8749(03)00128-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.