From August to December 2011, the National Institutes of Health Clinical Center experienced an outbreak of carbapenem-resistant Klebsiella pneumoniae infections; interventions implemented in an attempt to contain spread, as well as the challenges and pitfalls encountered with this outbreak, are described.
Keywords: carbapenem-resistant enterobacteriaceae, whole genome sequencing, isolation, transmission, multi-drug resistance
Abstract
In 2011, the National Institutes of Health Clinical Center experienced a cluster of infection and colonization caused by carbapenem-resistant Klebsiella pneumoniae among profoundly immunocompromised inpatients. This manuscript describes the approach and interventions that were implemented in an attempt to curtail the cluster. Interventions employed included engagement of all stakeholders involved in care of at-risk patients; detailed and frequent communication with hospital staff about issues relating to the outbreak; aggressive microbial surveillance; use of techniques that facilitate rapid identification of resistant organisms; rapid characterization of resistance mechanisms; whole-genome sequencing of outbreak isolates to characterize the spread and to investigate mechanisms of healthcare-associated spread; implementation of enhanced contact precautions for all infected or colonized patients; geographic and personnel cohorting; daily chlorhexidine gluconate baths; dedicating equipment to be used solely for cohorted patients and aggressive decontamination of equipment that had to be reused on uncohorted patients; monitoring adherence to infection control precautions, including unwavering attention to adherence to appropriate hand hygiene procedures; and attention to the details of environmental decontamination. In addition, the manuscript discusses some of the challenges associated with managing such an event, as well as a few of the unanticipated consequences associated with the aftermath of the case cluster.
Hospitals are increasingly plagued by resistant gram-negative pathogens. Although not the most numerous healthcare-associated infections [1], these organisms are an enormous challenge for healthcare facilities. They are easily transmissible from asymptomatic carriers and survive in the environment to varying degrees [2–4], facilitating spread in healthcare institutions. Their incidence in healthcare facilities has quadrupled over the past decade [5]. Guidelines from the Centers for Disease Control and Prevention (CDC) recommend stringent measures to control transmission of carbapenem-resistant Enterobacteriaceae (CRE) [6].
From August to December 2011, the National Institutes of Health (NIH) Clinical Center experienced an outbreak of carbapenem-resistant Klebsiella pneumoniae infections and colonization among profoundly immunocompromised inpatients that resulted in a high attributable mortality rate. The cluster began with the transfer of a patient from a New York City facility where carbapenem-resistant Enterobacteriaceae species are endemic. Despite isolation measures implemented at the beginning of hospitalization, silent transmission spawned a cluster that led to Klebsiella pneumoniae carbapenemase–producing Klebsiella pneumoniae (KPC-Klebsiella pneumoniae) infections in 8 patients, 6 of whom died of infection, and KPC-Klebsiella colonization in 9 others. Seven months after the putative end of the outbreak, 1 additional patient acquired the bacteria through nosocomial spread and died from KPC-Klebsiella infection. The lessons learned during the cluster led to a prolonged, high-intensity effort to control and contain resistant organisms in our hospital. This manuscript describes our approach and the interventions that were implemented in an attempt to contain spread, as well as the challenges and pitfalls encountered.
SURVEILLANCE
Identifying colonized patients is key to limiting risk for transmission [6–9]. Patients colonized with CRE serve as asymptomatic reservoirs for transmission; ultimately, as was the case with other outbreaks [10, 11], detection and careful management of these patients in isolation was temporally associated with cluster termination.
During and since our CRE cluster, because of the highly vulnerable nature of our patient population, we obtain surveillance cultures frequently. Although stool and perirectal swabs have the highest yield for CRE [12], our routine surveillance procedure samples other sites as well, primarily to detect colonization with other multidrug-resistant gram-negative bacteria, such as Acinetobacter. In the intensive care unit (ICU), perirectal, throat, and inguinal swabs are collected by nurses at admission and twice weekly. In the medical ward in closest proximity to the CRE-cohorted area, the same 3 cultures are collected twice weekly. Once each month, perirectal swabs are ordered for hospitalized patients of all ages, excluding those on locked behavioral health units. Given the ubiquity of health care-associated CRE [5, 13, 14], we decided to begin collecting perirectal surveillance swabs from all medical–surgical patients at admission.
Swabs are plated on KPC CHROMagar (CHROMagar, Paris, France) to identify carbapenem-resistant organisms. Groin and throat swabs are inoculated together on a single plate to limit consumption of costly plates. Within 18 hours of inoculation, the chromogenic agar labels colonies with vivid colors warning of possible carbapenem resistance; organisms from these colonies are identified within minutes using matrix-assisted laser desorption/ionization time of flight (MALDI-TOF) mass spectrometry [15]. Polymerase chain reaction (PCR) testing for the KPC gene is performed on isolates suspected of being carbapenemase producers, typically within hours; modified Hodge testing is used when PCR is not immediately available. Automated antibiotic susceptibility testing is performed (Phoenix, BD, Sparks, Maryland) and, when appropriate, testing for susceptibility to colistin, tigecycline, and other antibiotics is performed using the Etest method.
The results of surveillance cultures obtained during and after the outbreak are summarized in Figure 1.
Figure 1.
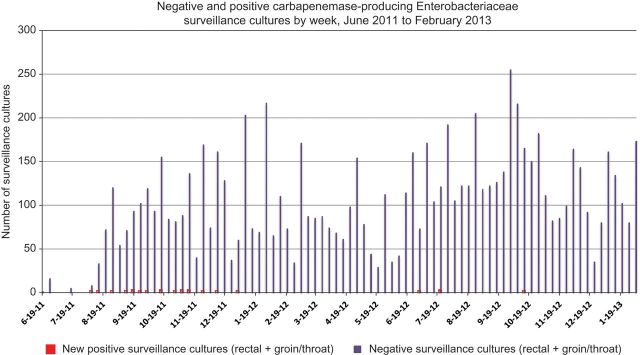
Surveillance cultures obtained from June 2011 to February 2013.
In mid-September 2011 when the initial results of whole-genome sequencing of the first 5 isolates were pooled with epidemiological data, our genomics collaborators deciphered the pattern of transmission. We immediately recognized the complexity of transmission as well as the counterintuitive transmission sequence. These data prompted us routinely to conduct monthly whole-house surveillance of all medical–surgical inpatients. Whereas 2 of the first 4 patients were identified as a result of clinical cultures, the remaining 15 patients who acquired the outbreak strain were detected with surveillance cultures.
PREVENTING TRANSMISSION FROM COLONIZED PATIENTS
Isolation Precautions
Identifying CRE colonization or infection mandates an effort to contain its spread. Barrier precautions and geographic and staff cohorting are practices supported by national guidelines and tradition in healthcare epidemiology. Little controversy surrounds use of private rooms, gowns, and gloves; however, compliance with these measures is far from uniform [16–18].
We implemented a modified isolation category (“enhanced contact precautions”) for managing CRE patients. Patients in enhanced contact isolation can leave their rooms only for medically necessary reasons, wear gowns and gloves when leaving the room, and are accompanied by staff members. Their visitors wear gowns and gloves at all times in patient rooms. Patients receive meals on disposable trays, using disposable dishes and utensils. Staff cannot touch pagers, cell phones, or other personal equipment in these patients' rooms. Equipment is either dedicated for single-patient use (eg, keyboards, stethoscopes) or decontaminated whenever possible (eg, computers, ultrasound machines) with hydrogen peroxide vapor before use on another patient (discussed below).
Cohorting
The goal of geographic cohorting, or placing patients colonized with similar or identical pathogens in shared space, is to avoid cross-transmission by segregating colonized from uncolonized patients. In its most effective form, geographic cohorting is accompanied by staff cohorting, in which a cadre of healthcare personnel is assigned only to care for cohorted patients during a shift. During our outbreak, nurses, respiratory therapists, housekeeping personnel, physical therapists, and staff from other allied fields were cohorted. Several challenges made ideal staff cohorting difficult. We lack sufficient numbers of physicians to provide around-the-clock duplicate staffing. The cohorting of nurses placed significant strain on the nursing department's ability to staff other parts of the hospital, particularly when a staff member who provided coverage for a short time in the cohorted area could not then be reassigned to other (uncohorted) patients. Morale among cohorted staff flagged at times due to the strain of dealing with sustained high-acuity illness, relative isolation from colleagues, and the frustrations of family members. Medical emergencies, such as cardiovascular or respiratory decompensation, elicited a flood of staff members either from or to the cohorted area, often with an urgency that overrode infection control precautions. Finally, the use of precautions and cohorting for months may have given way to “precaution fatigue,” or a waning of the level of vigilance toward the details of infection control. To this day, CRE-colonized patients who are inpatients or outpatients receive cohorted nursing care.
Hand Hygiene
Hand hygiene played a major role in the campaign to control CRE transmission. Meticulous compliance with hand hygiene was encouraged and enforced by adherence monitors. The manufacturer's instructions for our alcohol-based hand gel do not recommend a specific volume. Based on experimental data on the volume of hand-gel required for optimal bacterial killing on hands [19–23], and based on the volume dispensed with each pump of our hand-gel bottles, we recommended—and later mandated—that staff double hand-gel use to 2 pumps at every hand hygiene opportunity and rub the gel into their hands for 20 seconds. The motto “Two pumps, 20 seconds” was used widely in our hand hygiene campaign.
At the time the outbreak began, hand hygiene adherence rates were similar to recent historical trends at our institution (ie, 80%–85% adherence, as directly documented by trained observers). During the 3-month period when transmission was brought largely under control, hand hygiene compliance was virtually 100%, a reflection of the high level of attention that staff were devoting to infection prevention. Outside monitored areas, spot checks revealed that few staff complied regularly with the “2-pump” requirement. In the 6 months following the last nosocomial transmission, hand hygiene adherence fell to previous rates, effectively demonstrating a regression to the mean and emphasizing the difficulty in maintaining very high levels of hand hygiene adherence.
Chlorhexidine Gluconate Baths
Daily antiseptic baths for patients using 2% chlorhexidine gluconate had been introduced into the ICU 6 months before the first KPC-colonized patient arrived. Adherence to the bathing schedule had been <70%, largely because some physicians declined to have patients receive chlorhexidine baths. Starting in August 2011 when nosocomial transmission of CRE became evident, the ICU and infection control staff made efforts to improve compliance, and all physicians agreed to have patients receive the baths, ultimately resulting in a >90% compliance rate.
Adherence Monitoring
As soon as nosocomial transmission was recognized, we implemented around-the-clock adherence monitoring. Previously, we used this strategy during outbreak settings to assure adherence to CDC-recommended infection control measures [24]. In a 2009 Acinetobacter baumannii outbreak, monitors collected nearly 4900 observations of staff entering and exiting patient rooms, and determined that 2.6% were noncompliant, despite monitor intervention; physicians were responsible for more violations than others. In a setting with highly immunocompromised patients, even a handful of failures can result in sustained transmission.
At peak outbreak activity, monitors simultaneously staffed 3 positions—1 in each cohorted area and 1 in the ICU. Monitors were patient care technicians whose exclusive assignments were to ensure that staff and visitors adhere to appropriate hand hygiene, barrier precautions, and environmental disinfection techniques. Monitors were trained to handle friction from monitored individuals, but they encountered occasional conflicts, particularly when patient volume and acuity were high. Monitors were instructed to notify infection prevention staff when someone was oppositional or persistently noncompliant; in such instances, intervention resolved the problem in some cases, and fueled conflict in others. Monitors more skilled in diplomacy tended to be more effective; those who were ineffective or could not maintain amicable staff and visitor relationships were removed from the monitor workforce and reassigned to standard duties.
In monitored areas, the only documented breaches of precautions were rare missed hand hygiene opportunities and breaches occurring during medical emergencies, such as cardiovascular or respiratory decompensation.
PREVENTING TRANSMISSION FROM THE ENVIRONMENT
Several factors led us to pursue aggressive environmental decontamination: cultures from a ventilator that had been used on a CRE-colonized patient grew KPC-Klebsiella, despite 3 prior rounds of manual cleaning and disinfection; a bed was suspected as the source of contamination in 1 transmission; and cultures obtained from 4 sink drains within, and outside, rooms of known colonized patients grew KPCs. Other researchers have raised similar concerns about environmental risks associated with these organisms [2–4]. Double disinfection of all high-touch surfaces with sodium hypochlorite (bleach)–impregnated wipes was added to routine cleaning, to try to increase effectiveness and to signal to housekeepers and other staff the importance of environmental cleaning in an outbreak setting. A company (Bioquell, Philadelphia, Pennsylvania) was hired to perform hydrogen peroxide vapor decontamination of patient rooms and equipment [25]. Although only 7 of 260 environmental cultures performed during the outbreak grew KPCs, concern that small inocula might transmit lethal infection to immunosuppressed patients and concern that inadequate environmental cleaning might foster transmission prompted continued use of hydrogen peroxide vapor decontamination. Hydrogen peroxide vapor decontamination can reduce environmental bioburdens [26–28] and has been shown in 1 study to be associated with reduced risks for acquisition of multidrug-resistant organisms [3], though its efficacy remains to be demonstrated in this specific setting.
When a KPC-Klebsiella–colonized or infected patient vacated a patient room, the room was double cleaned, disinfected with bleach, and decontaminated with hydrogen peroxide vapor before use by a patient not previously identified as harboring KPC-Klebsiella. On several occasions, large sections of the ICU and other cohorted areas were similarly decontaminated to attempt to decrease the ICU bioburden.
Several outbreak reports cite contaminated sink drains as possible sources of transmission of resistant pathogens [2, 29–32], although we had no definitive evidence that splashback onto hands of healthcare personnel was a source of transmission. Hydrogen peroxide vapor does not penetrate sink drain biofilm (M. Hodgson, Bioquell, personal communication, October 2011). Lacking an evidence-based method for reliably and permanently decontaminating sink drains, we managed them with the goal of reducing the bioburden. For ongoing management, sink drains were removed and cleaned thoroughly, and bleach is sprayed daily down drains on affected wards to attempt to suppress resistant pathogens; drains are recultured periodically to monitor the effectiveness of these measures.
Communication
Weekly multidisciplinary “all-hands” meetings were convened to discuss new developments, interventions, and investigative findings. Ensuring that clinical stakeholders are completely informed about the outbreak and about the rationale for the selected interventions and assuring their alignment to interventions are keys to successful outbreak control [24]. These well-attended meetings involved physicians, nurses, infection preventionists, respiratory therapists, housekeepers, nutritionists, pharmacists, hospital administration, patient and environmental safety, and other staff. These meetings provided opportunities to air concerns and suggestions, as well as to educate staff. Smaller daily staff meetings were designed to manage details of outbreak investigation and control. Hospital epidemiologists and infection preventionists made dozens of presentations at departmental meetings, rounds, and teaching conferences to attempt to reach all staff who had patient contact with medical and surgical patients. Frequent questions from staff pertained to the occupational risk of caring for colonized patients and to the risk that they or their family members could become ill from KPC-Klebsiella. The most thoughtful inquiries often required deliberation and simulation (eg, infection preventionists practiced actions required to minimize the potential to contaminate paperwork requiring signature by a KPC-Klebsiella–colonized patient before recommending steps to staff). Multiple email notifications providing status updates and infection control reminders were distributed to all clinical staff, both whenever new information became available, and routinely every few weeks.
During the cluster, information regarding enhanced contact isolation and perirectal surveillance cultures was made available to patients and staff. An information sheet describing the risk of nosocomial multidrug-resistant organisms was developed and was included in patients' admissions materials.
WHOLE-GENOME SEQUENCING
Details of the methodology used, and rationale for the use of, whole-genome sequencing of KPC isolates in this setting have been published previously [33]. The standard typing techniques did not have sufficient resolution to differentiate isolates that shared the most common strain type (sequence type 258), and therefore could not discern whether the early cases represented separate introductions or nosocomial transmission. As new isolates were detected through active surveillance and clinical cultures, they were added to the queue for sequencing. In mid-September when the results of sequencing demonstrated the pattern of transmission among the first few patients, the complexity of transmission and the counterintuitive sequence of transmission led us to conduct whole-house surveillance in the hospital, as described above. In this outbreak, the sequencing data informed outbreak management but did not come in time to make targeted interventions based on specific sequencing data. In the future, at institutions that are equipped with genomic sequencing equipment and expertise, real-time whole-genome sequencing could enable the application of measures trained at the most likely sources or modes of spread in recent instances of transmission, rather than large-scale blanket strategies that carry a high cost in finances and other resources. A limitation of sequencing is that it explains whence a nosocomial transmission came, but not the modality of spread; we can only still surmise that the most common modes of spread, transmission on the hands of healthcare personnel and contaminated surfaces, were also applicable in this cluster.
WHAT WORKED?
When a set of interventions is implemented simultaneously under the pressure of time, morbidity, and mortality, institutions do not have the luxury of evaluating which strategic elements are most and least effective, and which are key to the success of the multifaceted approach. We can only infer from the temporal association that our strategy, and the associated improvement in hand hygiene and adherence to infection control precautions, led to conclusion of the outbreak. In reflection, we believe that meticulously implementing infection control precautions that have been recommended by CDC [6], such as hand hygiene, cohorting, and active surveillance, most likely contributed the most, and remediation of environmental contamination the least, to our success, based on the paucity of positive environmental cultures and the lack of epidemiological data suggesting environmental spread.
UNINTENDED CONSEQUENCES OF PUBLICATION
When the paper reporting the use of whole-genome sequencing was published, it received considerable attention from both the science press and the lay press. The science press focused on the use of genomics to tackle a clinical problem; the broader media focused initially on the “genomic detective work,” then quickly turned to the aspects that might be better understood (or misunderstood) by their lay audience. The idea that bacteria transmitted in a hospital could lead to death in >75% of those who became infected was very easily taken out of context and used to stir fear that the general public was unknowingly at risk. The fact that the infections were limited to critically ill, highly vulnerable patients in the hospital was not included in media reports to mitigate the concern of the public. Local community residents and even some NIH employees from nonclinical parts of the campus became incensed that they had not been informed about the outbreak. Community anxiety led to a significant effort at local, state, and federal levels to reassure and educate the public on nosocomial infections and multidrug-resistant organisms.
We emphasize that much of what we did in managing this outbreak is not evidence-based (a common problem for the discipline of healthcare epidemiology [34, 35]). We attempted to mitigate literally every risk we could identify and took the most conservative approach that was feasible in virtually every instance. We are not suggesting that our use of any of these interventions implies efficacy and underscore that we implemented all of them virtually simultaneously, so we have no way of knowing which of them were effective. We also recognize that our “kitchen sink” approach may not be practical for implementation in settings other than our own.
The problem of multidrug-resistant organisms will be present for the foreseeable future. Several categories of carbapenemase-producing organisms are threatening healthcare, among them KPC-, New Delhi metallo-β-lactamase (NDM-1)–, Verona integron-encoded metallo-β-lactamase (VIM-1)–, IMP β-lactamase–, and OXA-β-lactamase–producing isolates [36–40]. To combat these organisms, we need strategies for detecting the organisms, enhanced techniques for tracking them in healthcare institutions, and novel interventions to prevent spread, and we desperately need new classes of antimicrobials to manage infections caused by these resistant pathogens.
Note
Potential conflicts of interest. Both authors: No reported conflicts.
Both authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.Sievert DM, Ricks P, Edwards JR, et al. Antimicrobial-resistant pathogens associated with healthcare-associated infections: summary of data reported to the National Healthcare Safety Network at the Centers for Disease Control and Prevention, 2009-2010. Infect Control Hosp Epidemiol. 2013;34:1–14. doi: 10.1086/668770. [DOI] [PubMed] [Google Scholar]
- 2.Kotsanas D, Wijesooriya WR, Korman TM, et al. Down the drain”: carbapenem-resistant bacteria in intensive care unit patients and handwashing sinks. Med J Australia. 2013;198:267–9. doi: 10.5694/mja12.11757. [DOI] [PubMed] [Google Scholar]
- 3.Landman D, Babu E, Shah N, et al. Transmission of carbapenem-resistant pathogens in New York City hospitals: progress and frustration. J Antimicrob Chemother. 2012;67:1427–31. doi: 10.1093/jac/dks063. [DOI] [PubMed] [Google Scholar]
- 4.Lerner A, Adler A, Abu-Hanna J, Meitus I, Navon-Venezia S, Carmeli Y. Environmental contamination by carbapenem-resistant Enterobacteriaceae. J Clin Microbiol. 2013;51:177–81. doi: 10.1128/JCM.01992-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Centers for Disease Control and Prevention. Vital signs: carbapenem-resistant Enterobacteriaceae. MMWR Morbid Mortal Wkly Rep. 2013;62:165–70. [PMC free article] [PubMed] [Google Scholar]
- 6.Centers for Disease Control and Prevention. CRE toolkit–guidance for control of carbapenem-resistant Enterobacteriaceae (CRE) 2012. Available at: http://www.cdc.gov/hai/organisms/cre/cre-toolkit/index.html. Accessed 21 February 2013.
- 7.Ben-David D, Maor Y, Keller N, et al. Potential role of active surveillance in the control of a hospital-wide outbreak of carbapenem-resistant Klebsiella pneumoniae infection. Infect Control Hosp Epidemiol. 2010;31:620–6. doi: 10.1086/652528. [DOI] [PubMed] [Google Scholar]
- 8.Calfee D, Jenkins SG. Use of active surveillance cultures to detect asymptomatic colonization with carbapenem-resistant Klebsiella pneumoniae in intensive care unit patients. Infect Control Hosp Epidemiol. 2008;29:966–8. doi: 10.1086/590661. [DOI] [PubMed] [Google Scholar]
- 9.Kochar S, Sheard T, Sharma R, et al. Success of an infection control program to reduce the spread of carbapenem-resistant Klebsiella pneumoniae. Infect Control Hosp Epidemiol. 2009;30:447–52. doi: 10.1086/596734. [DOI] [PubMed] [Google Scholar]
- 10.Munoz-Price LS, De La Cuesta C, Adams S, et al. Successful eradication of a monoclonal strain of Klebsiella pneumoniae during a K. pneumoniae carbapenemase-producing K. pneumoniae outbreak in a surgical intensive care unit in Miami, Florida. Infect Control Hosp Epidemiol. 2010;31:1074–7. doi: 10.1086/656243. [DOI] [PubMed] [Google Scholar]
- 11.Munoz-Price LS, Hayden MK, Lolans K, et al. Successful control of an outbreak of Klebsiella pneumoniae carbapenemase-producing K. pneumoniae at a long-term acute care hospital. Infect Control Hosp Epidemiol. 2010;31:341–7. doi: 10.1086/651097. [DOI] [PubMed] [Google Scholar]
- 12.Chen IL, Lee CH, Su LH, Tang YF, Chang SJ, Liu JW. Antibiotic consumption and healthcare-associated infections caused by multidrug-resistant gram-negative bacilli at a large medical center in Taiwan from 2002 to 2009: implicating the importance of antibiotic stewardship. PLoS One. 2013;8:e65621. doi: 10.1371/journal.pone.0065621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gupta N, Limbago BM, Patel JB, Kallen AJ. Carbapenem-resistant Enterobacteriaceae: epidemiology and prevention. Clin Infect Dis. 2011;53:60–7. doi: 10.1093/cid/cir202. [DOI] [PubMed] [Google Scholar]
- 14.Patel G, Bonomo RA. Stormy waters ahead”: global emergence of carbapenemases. Front Microbiol. 2013;4:48. doi: 10.3389/fmicb.2013.00048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Seng P, Drancourt M, Gouriet F, et al. Ongoing revolution in bacteriology: routine identification of bacteria by matrix-assisted laser desorption ionization time-of-flight mass spectrometry. Clin Infect Dis. 2009;49:543–51. doi: 10.1086/600885. [DOI] [PubMed] [Google Scholar]
- 16.Clock SA, Cohen B, Behta M, Ross B, Larson EL. Contact precautions for multidrug-resistant organisms: current recommendations and actual practice. Am J Infect Control. 2010;38:105–11. doi: 10.1016/j.ajic.2009.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Manian FA, Griesenauer S, Senkel D, et al. Isolation of Acinetobacter baumannii complex and methicillin-resistant Staphylococcus aureus from hospital rooms following terminal cleaning and disinfection: can we do better? Infect Control Hosp Epidemiol. 2011;32:667–72. doi: 10.1086/660357. [DOI] [PubMed] [Google Scholar]
- 18.Morgan DJ, Day HR, Harris AD, Furuno JP, Perencevich EN. The impact of contact isolation on the quality of inpatient hospital care. PLoS One. 2011;6:e22190. doi: 10.1371/journal.pone.0022190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goroncy-Bermes P, Koburger T, Meyer B. Impact of the amount of hand rub applied in hygienic hand disinfection on the reduction of microbial counts on hands. J Hosp Infect. 2010;74:212–8. doi: 10.1016/j.jhin.2009.09.018. [DOI] [PubMed] [Google Scholar]
- 20.Larson E. Draft guideline for use of topical antimicrobial agents. Am J Infect Control. 1987;15:25A–36A. doi: 10.1016/0196-6553(87)90133-7. [DOI] [PubMed] [Google Scholar]
- 21.Macinga DR, Beausoleil CM, Campbell E, et al. Quest for a realistic in vivo test method for antimicrobial hand-rub agents: introduction of a low-volume hand contamination procedure. Appl Environ Microbiol. 2011;77:8588–94. doi: 10.1128/AEM.06134-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mackintosh CA, Hoffman PN. An extended model for transfer of micro-organisms via the hands: differences between organisms and the effect of alcohol disinfection. J Hyg (Lond) 1984;92:345–55. doi: 10.1017/s0022172400064561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sickbert-Bennett EE, Weber DJ, Gergen-Teague MF, Rutala WA. The effects of test variables on the efficacy of hand hygiene agents. Am J Infect Control. 2004;32:69–83. doi: 10.1016/j.ajic.2003.09.003. [DOI] [PubMed] [Google Scholar]
- 24.Palmore TN, Michelin AV, Bordner M, et al. Use of adherence monitors as part of a team approach to control clonal spread of multidrug-resistant Acinetobacter baumannii in a research hospital. Infect Control Hosp Epidemiol. 2011;32:1166–72. doi: 10.1086/662710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Otter JA, Yezli S, Schouten MA, van Zanten AR, Houmes-Zielman G, Nohlmans-Paulssen MK. Hydrogen peroxide vapor decontamination of an intensive care unit to remove environmental reservoirs of multidrug-resistant gram-negative rods during an outbreak. Am J Infect Control. 2010;38:754–6. doi: 10.1016/j.ajic.2010.03.010. [DOI] [PubMed] [Google Scholar]
- 26.Fu TY, Gent P, Kumar V. Efficacy, efficiency and safety aspects of hydrogen peroxide vapour and aerosolized hydrogen peroxide room disinfection systems. J Hosp Infect. 2012;80:199–205. doi: 10.1016/j.jhin.2011.11.019. [DOI] [PubMed] [Google Scholar]
- 27.Otter JA, French GL. Survival of nosocomial bacteria and spores on surfaces and inactivation by hydrogen peroxide vapor. J Clin Microbiol. 2009;47:205–7. doi: 10.1128/JCM.02004-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Otter JA, Yezli S, French GL. The role played by contaminated surfaces in the transmission of nosocomial pathogens. Infect Control Hosp Epidemiol. 2011;32:687–99. doi: 10.1086/660363. [DOI] [PubMed] [Google Scholar]
- 29.Hota S, Hirji Z, Stockton K, et al. Outbreak of multidrug-resistant Pseudomonas aeruginosa colonization and infection secondary to imperfect intensive care unit room design. Infect Control Hosp Epidemiol. 2009;30:25–33. doi: 10.1086/592700. [DOI] [PubMed] [Google Scholar]
- 30.Lowe C, Willey B, O'Shaughnessy A, et al. Outbreak of extended-spectrum beta-lactamase-producing Klebsiella oxytoca infections associated with contaminated handwashing sinks. Emerg Infect Dis. 2012;18:1242–7. doi: 10.3201/eid1808.111268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Starlander G, Melhus A. Minor outbreak of extended-spectrum beta-lactamase-producing Klebsiella pneumoniae in an intensive care unit due to a contaminated sink. J Hosp Infect. 2012;82:122–4. doi: 10.1016/j.jhin.2012.07.004. [DOI] [PubMed] [Google Scholar]
- 32.Van Saene HK, Van Putte JC, Van Saene JJ, Van de Gronde TW, Van Warmerdam EG. Sink flora in a long-stay hospital is determined by the patients’ oral and rectal flora. Epidemiol Infect. 1989;102:231–8. doi: 10.1017/s0950268800029903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Snitkin ES, Zelazny AM, Thomas PJ, et al. Tracking a hospital outbreak of carbapenem-resistant Klebsiella pneumoniae with whole-genome sequencing. Sci Transl Med. 2012;4:148ra16. doi: 10.1126/scitranslmed.3004129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Henderson DK, Palmore TN. Critical gaps in knowledge of the epidemiology and pathophysiology of healthcare-associated infections. Infect Control Hosp Epidemiol. 2010;31(suppl 1):S4–6. doi: 10.1086/655984. [DOI] [PubMed] [Google Scholar]
- 35.Weinstein RA, Henderson DK. A double-edged sword and a golden opportunity for healthcare epidemiology. Infect Control Hosp Epidemiol. 2009;30:1–3. doi: 10.1086/596559. [DOI] [PubMed] [Google Scholar]
- 36.Castanheira M, Deshpande LM, Farrell SE, Shetye S, Shah N, Jones RN. Update on the prevalence and genetic characterization of NDM-1–producing Enterobacteriaceae in Indian hospitals during 2010. Diagn Microbiol Infect Dis. 2013;75:210–3. doi: 10.1016/j.diagmicrobio.2012.10.017. [DOI] [PubMed] [Google Scholar]
- 37.Centers for Disease Control and Prevention. Notes from the field: hospital outbreak of carbapenem-resistant Klebsiella pneumoniae producing New Delhi metallo-beta-lactamase—Denver, Colorado, 2012. MMWR Morbid Mortal Wkly Rep. 2013;62:108. [PMC free article] [PubMed] [Google Scholar]
- 38.Kim Y, Cunningham MA, Mire J, Tesar C, Sacchettini J, Joachimiak A. NDM-1, the ultimate promiscuous enzyme: substrate recognition and catalytic mechanism. FASEB J. 2013;27:1917–27. doi: 10.1096/fj.12-224014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kumarasamy K, Kalyanasundaram A. Emergence of Klebsiella pneumoniae isolate co-producing NDM-1 with KPC-2 from India. J Antimicrob Chemother. 2012;67:243–4. doi: 10.1093/jac/dkr431. [DOI] [PubMed] [Google Scholar]
- 40.Lowe CF, Kus JV, Salt N, et al. Nosocomial transmission of New Delhi metallo-beta-lactamase-1-producing Klebsiella pneumoniae in Toronto, Canada. Infect Control Hosp Epidemiol. 2013;34:49–55. doi: 10.1086/668778. [DOI] [PubMed] [Google Scholar]


