Abstract
The ROP1 GTPase-based signaling network controls tip growth in Arabidopsis pollen tubes. Our previous studies imply that ROP1 might be directly activated by RopGEF1, which belongs to a plant-specific family of Rho guanine nucleotide exchange factors (RopGEFs) and in turn may be activated by an unknown factor through releasing RopGEF1’s auto-inhibition. In this study, we found that RopGEF1 forms a complex with ROP1 and AtPRK2, a receptor-like protein kinase previously shown to interact with RopGEFs. AtPRK2 phosphorylated RopGEF1 in vitro and the atprk1,2,5 triple mutant showed defective pollen tube growth, similar to the phenotype of the ropgef1,9,12,14 quadruple mutant. Overexpression of a dominant negative form of AtPRK2 (DN-PRK2) inhibited pollen germination in Arabidopsis and reduced pollen elongation in tobacco. The DN-PRK2-induced pollen germination defect was rescued by overexpressing a constitutively active form of RopGEF1, RopGEF1(90–457), implying that RopGEF1 acts downstream of AtPRK2. Moreover, AtPRK2 increased ROP1 activity at the apical plasma membrane whereas DN-PRK2 reduced ROP1 activity. Finally, two mutations at the C-terminal putative phosphorylation sites of RopGEF1 (RopGEF1S460A and RopGEF1S480A) eliminated the function of RopGEF1 in vivo. Taken together, our results support the hypothesis that AtPRK2 acts as a positive regulator of the ROP1 signaling pathway most likely by activating RopGEF1 through phosphorylation.
Key words: AtPRK2, RopGEF1, ROP GTPase, auto-inhibition, polarity growth.
INTRODUCTION
The ROP (Rho-like small GTPase from plant) GTPase family is an important molecular switch in plant signaling (Yang, 2002; Gu et al., 2003, 2004; Yang and Fu, 2007; Lee and Yang, 2008; Yang, 2008). ROPs control polar cell growth and, as such, play a critical role in many developmental processes, including pollen tube and root hair growth, pavement cell morphogenesis, and plant responses to auxin and ABA (Lin et al., 1996; Lin and Yang, 1997; Li et al., 1999; Fu et al., 2001; Jones et al., 2002; Zheng et al., 2002; Gu et al., 2003; Fu et al., 2005; Gu et al., 2005; Hwang et al., 2005; Chang et al., 2007; Jones et al., 2007; Fu et al., 2009; Xu et al., 2010; Chen et al., 2011).
Pollen tubes are an excellent model to study the regulation of polar growth (Cheung and Wu, 2008; Yang, 2008). Three functionally redundant ROPs (ROP1, ROP3, and ROP5) are required for pollen tube polar growth (Yang, 2002, 2008). ROP1 promotes pollen tube polar growth via two counteracting pathways: the RIC4 pathway, which promotes apical F-actin assembly, and the RIC3 pathway, which promotes Ca2+ signaling at the tip and subsequently increases the disassembling of apical F-actin (Gu et al., 2005). These two counteracting pathways work together to regulate F-actin dynamics and F-actin/Ca2+-dependent exocytosis (Yang and Fu, 2007; Lee and Yang, 2008; Lee et al., 2008; Yang, 2008).
Rho GTPases are mainly regulated by three classes of upstream regulators: guanine exchange factors (GEFs), GTPase activating proteins (GAPs), and guanine nucleotide dissociation inhibitors (GDIs) (Yang, 2002). GEFs activate ROPs by promoting the conversion of Rho GTPases from their GDP-bound inactive form to the GTP-bound active form. GAPs promote GTP hydrolysis and deactivate ROPs. GDIs bind to the GDP-bound form of Rho GTPases and negatively regulate Rho GTPases via preventing both guanine nucleotide exchange and Rho GTPase localization to the PM.
Plants possess a plant-specific RhoGEF family whose members contain a highly conserved novel GEF catalytic domain, the PRONE domain, instead of the conserved PH or CZH domains found in yeast, animal, and human RhoGEFs (Berken et al., 2005; Gu et al., 2006). Fourteen members of the RopGEF family have been found in Arabidopsis thaliana and seven members are specifically or highly expressed in pollen tubes (Gu et al., 2006; Zhang and McCormick, 2007). Gu et al. transiently overexpressed five Arabidopsis RopGEFs in tobacco pollen and found that RopGEF1 overexpression induced the most severely swollen tubes, similarly to the phenotype induced by the expression of a constitutively active ROP1 (CA-ROP1) (Gu et al., 2006). The PRONE domain (amino acid 90–457) of RopGEF1 was found to exhibit high GEF activity on ROP1, while the full-length RopGEF1 protein was inactive in vitro due to the presence of an auto-inhibitory domain in the C-terminal region. Therefore, it was proposed that an in vivo mechanism must exist to release the auto-inhibition of RopGEF1 (Gu et al., 2006).
Recent studies have revealed the function of receptor-like protein kinases (RLKs) in the regulation of pollen development, germination, and pollen tube polar growth (Mu et al., 1994; Muschietti et al., 1998; Kim et al., 2002; Zhang and McCormick, 2007; Zhang et al., 2008). The first Pollen-specific Receptor-like Kinase (PRK1) was reported to play an important role in postmeiotic development of pollen in petunia (Mu et al., 1994). The tomato pollen-specific RLKs LePRK1 and LePRK2 have been proposed to receive signals from LAT52 and LeSTIG1 at different stages to promote pollen germination and pollen tube growth (Muschietti et al., 1998; Tang et al., 2002; Johnson and Preuss, 2003; Wengier et al., 2003; Tang et al., 2004; Zhang et al., 2008). Interestingly, the McCormick group found that the intracellular kinase domain of tomato pollen receptor-like kinases (PRKs) can physically interact with KPP (kinase partner protein) (Kaothien et al., 2005), whose Arabidopsis homologs were later shown to belong to the RopGEF family (Berken et al., 2005; Gu et al., 2006). KPP was found to be phosphorylated in vivo, although the kinase responsible for its phosphorylation is unknown (Kaothien et al., 2005). Importantly, the same group showed that an Arabidopsis homolog of PRKs, AtPRK2a (for simplicity, referred to as AtPRK2 here), can also interact with the pollen-specific RopGEF12 and that a phospho-mimic mutation (S510D) of RopGEF12 enhanced RopGEF12 promotion of growth depolarization (Zhang and McCormick, 2007). Co-expression studies in tobacco pollen suggested AtPRK2 could be involved in the activation of RopGEF12 (Zhang and McCormick, 2007). Similar results were found in tomato (Solanum lycopersicum) (Zhang et al., 2008). These results imply that AtPRK2 may act upstream of RopGEF12 to activate ROP signaling pathways in pollen tubes. Recent work in Arabidopsis root hairs suggests that the FER RLK acts as an upstream regulator of RAC/ROP2 signaling pathway most likely by interacting with RopGEF1 (Duan et al., 2010). Together, these studies suggest RLKs are likely to work as an upstream regulator of ROP signaling via RopGEFs. However, it remains unclear how RopGEFs are activated and how the RLKs regulate ROP activity.
In this study, we present new findings on the mechanisms employed by AtPRK2 to regulate RopGEF1 and ROP1 activities. Specifically, AtPRK2 increased ROP1 activity, and physically forms a complex with RopGEF1 and ROP1. In addition, AtPRK2 directly phosphorylated RopGEF1 and two serine-to-alanine mutations in the RopGEF1 C-terminal region abolished the release of auto-inhibition, suggesting AtPRK2 may activate RopGEF1 through phosphorylation in the C-terminal region. Moreover, a constitutively active form of RopGEF1 rescued the germination deficiency induced by DN-PRK2 overexpression, suggesting RopGEF1 bridges the signaling transduction from AtPRK2 to ROP1 signaling pathways. Based on these observations, we propose that the AtPRK2 phosphorylation of RopGEF1 in its C-terminal region releases its auto-inhibition, thereby activating RopGEF1, which in turn activates ROP1. Therefore, RopGEF1 bridges the signaling transduction from AtPRK2 to ROP1 to control polarized pollen tube growth.
RESULTS
Pollen Receptor-Like Kinases (PRKs) Regulate Pollen Tube Polar Growth
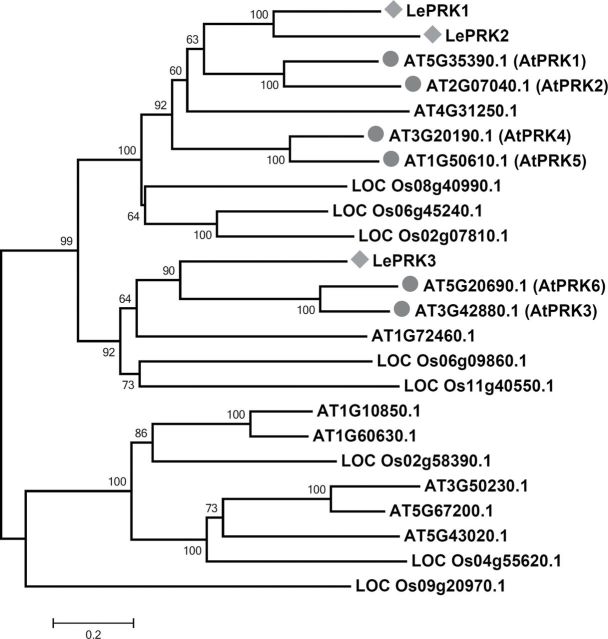
A pollen RLK (AtPRK2a) has been implicated in the regulation of ROP1 signaling through its interaction of RopGEF12 in Arabidopsis (Zhang and McCormick, 2007). To further explore RLK regulation of ROP1 signaling, we surveyed a group of Arabidopsis receptor-like kinases that are homologs of the tomato LePRKs (Figure 1). Using rice genes as outgroups, we found eight Arabidopsis RLKs are closely related to LePRKs. Six out of these eight genes are highly expressed in pollen according to the Genevestigator Expression Database, and were named AtPRK1-6 (Figure 1 and Supplemental Figure 1). According to the phylogenetic tree, AtPRK1 and AtPRK2 are close to LePRK1 and LePRK2, and, although AtPRK4 and AtPRK5 are further apart, they are also derived from the same common ancestor that existed before the divergence of rice and Arabidopsis. AtPRK3 and AtPRK6 are closer to LePRK3, and diverged from the common ancestor of Arabidopsis and rice.
Figure 1.
Phylogenetic Tree of Selected AtPRKs and LePRKs. LePRK1–3 from tomato, six pollen-specific (AtPRK1–6), and several non-pollen-specific AtPRKs from Arabidopsis were included in this tree. Rice genes are included as outgroups.
To test whether these AtPRKs are important for pollen tube polar growth, we transiently overexpressed full-length AtPRK1–5 (cloning AtPRK6 was unsuccessful) driven by the pollen-specific LAT52 promoter (pLAT52::AtPRK) in tobacco pollen, and analyzed the width and length of pollen tubes after germination and growth for 5 h. Overexpressing (OX) AtPRK2 through 5 resulted in all cases in increased width and reduced length of pollen tubes (Table 1) as did ROP1 overexpression (Li et al., 1999), suggesting that these AtPRKs may be involved in the control of pollen tube polar growth, and may regulate ROP1 signaling pathways. AtPRK2 overexpression induced the most severe growth depolarization with the most swollen tips (Table 1). In contrast, AtPRK1 was found to increase pollen tube length (Table 1).
Table 1.
RLKs Induce Depolarized Growth in Pollen Tubes.
| Lengtha | Widtha | Width/length | |
|---|---|---|---|
| GFP | 528±11 | 9.4±0.8 | 0.02 |
| GFP + PRK1 | 861±18 | 10.1±1.3 | 0.01 |
| GFP + PRK2 | 423±22 | 27.2±3.2 | 0.06 |
| GFP + PRK3 | 324±15 | 20.2±2.4 | 0.06 |
| GFP + PRK4 | 307±16 | 13.7±2.1 | 0.04 |
| GFP + PRK5 | 411±21 | 11.3±0.7 | 0.03 |
The length and the maximum tip width of tobacco pollen tubes were measured 6–7 h after bombardment. a Data are the mean ± SD.
Triple prk and Quadruple ropgef Mutants Exhibit Reduced Pollen Germination
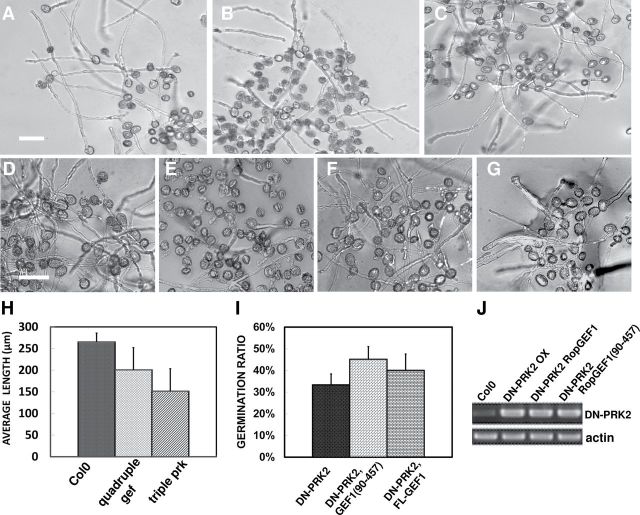
To test the role of AtPRKs in regulating pollen germination and pollen tube growth in Arabidopsis, T-DNA insertion mutant lines (Supplemental Table 1) of AtPRKs were obtained from ABRC (www.arabidopsis.org) and in vitro pollen germination and pollen tube growth were examined. Pollen from each of the prk1-1, prk1-2, prk1-2, prk2-1, and prk5-1 single homozygous mutant lines exhibited normal germination and growth. We generated pollen from the homozygous prk1-2 prk2-1 double mutant, prk2-1 prk5-1 double mutant, and prk1-2 prk2-1 prk5-1 triple mutant. Only pollen from the homozygous prk1 prk2 prk5 triple mutant line showed reduced tube elongation compared to wild-type (Figure 2B and 2H). Given additional AtPRKs are highly expressed in pollen, it is likely that one or more AtPRKs are functionally redundant with AtPRK1, AtPRK2, and AtPRK5. Similarly, none of ropgef1-1, ropgef9-1, ropgef12-1, ropgef14-1, and ropgef14-2 single knockout mutant lines exhibited altered pollen phenotype, while the homozygous ropgef1-1 ropgef9-1 ropgef12-1 ropgef14-2 quadruple mutant line showed reduced pollen tube elongation (Figure 2C and 2H), suggesting high redundancy among these RopGEFs in the regulation of pollen tube growth. The similarity in pollen phenotype between the triple prk mutant and quadruple ropgef mutant is consistent with the notion that AtPRKs and RopGEFs may work in the same signaling pathway to regulate pollen tube growth.
Figure 2.
Pollen Tube Phenotypes of the Quadruple gef Mutant and Triple prk Mutant, and the DN-PRK2 OX/RopGEF1(90–457) Transgenic Plants. (A–C) Pollen tubes of Col0 (A), prk1 prk2 prk5 triple mutant (B), and gef1 gef9 gef12 gef14 quadruple mutant (C), after 5-h incubation in pollen germination medium.(D–G) Pollen germination assay of Col0 (D), DN-PRK2 (E), DNPRK2, RopGEF1 (F), and DNPRK2, RopGEF1(90–457) (G) plants.(H, I) Statistical analysis of pollen tube growth (H) and the germination rate of various genotypes (I).(J) RT–PCR results suggest that the expression level of DNPRK2 does not change in all of the tested lines. Bar = 100 μm. (A–C) same magnification and (D–G) same magnification.
DN-PRK2 Inhibits Pollen Germination in Arabidopsis
Given the weak phenotype of the prk1 prk2 prk5 triple mutant, we reasoned that overexpression of a dominant-negative (DN) form of AtPRK2 (DN-PRK2) would effectively inhibit the function of redundant PRKs. Deletions of the cytoplasmic kinase domain of other RLKs have been shown to have a DN effect on the wild-type endogenous copy (Shpak et al., 2003). Therefore, we generated DN-PRK2 by deleting the kinase domain of AtPRK2 (Supplemental Figure 2) and fusing it to the pollen-specific LAT52 promoter. We introduced LAT52::DN-PRK2 in Arabidopsis and isolated 39 T1 transgenic lines, which carried a Basta resistance selection marker, and analyzed the segregation ratio of the Basta resistant to Basta-sensitive T2 progenies. We anticipated that a distortion of segregation ratios from the Mendelian genetics would suggest an effect of DN-PRK2 on pollen fertility, because the LAT52 promoter is pollen-specific. Among these 39 lines, only seven exhibited a nearly 3:1 ratio, which is consistent with single-locus insertions that did not alter pollen fertility. Twenty-two lines exhibited less than 3:1 segregation ratios for Basta resistance to sensitivity, implicating a defect in pollen fertility in these lines. Three lines showed 7:1 to 10:1 ratio, and seven lines displayed ratios higher than 15:1, suggesting that they contained multiple T-DNA insertions at more than one site (Supplemental Table 2). We generated homozygous transgenic lines from lines showing a segregation ratio < 3:1, and performed the pollen phenotype analysis. Pollen from DN-PRK2 OX lines exhibited abnormally low germination rates (Supplemental Figure 2B), suggesting that AtPRKs are important for pollen germination. Since the same phenotype was observed in multiple mutants affected in ROP1 signaling (Li et al., 1999; Hwang et al., 2008), we propose AtPRKs may participate in ROP1 signaling in the modulation of pollen germination and pollen tube growth.
RopGEF1(90–457) Suppresses the DN-PRK2 OX-Induced Pollen Germination Defect
RopGEF1 overexpression produces a pollen tube phenotype similar to that induced by CA-rop1 (Gu et al., 2006); thus, we sought to test whether RopGEF1 acts downstream of AtPRK2. For this purpose, we developed a constitutively active form of RopGEF1 (CA-RopGEF1). We previously showed that the PRONE domain of RopGEF1, RopGEF1(90–457), exhibits constitutive RopGEF activity in vitro (Gu et al., 2006). Thus, we anticipated that RopGEF1(90–457) would also be constitutively active in vivo, and introduced it in Arabidopsis with a C-terminal GFP tag and driven by the LAT52 promoter. RopGEF1(90–457)–GFP overexpression (OX) induced a much greater growth depolarization in pollen tubes compared to that induced by RopGEF1 OX (data not shown), suggesting that RopGEF1(90–457) acts as an activated form of RopGEF1 in vivo. Therefore, we crossed a RopGEF1(90–457)–GFP OX line with DN-PRK2 OX line 23–12 to determine whether RopGEF1(90–457) could suppress the reduced germination phenotype induced by DN-PRK2. As expected, the pollen germination rates of DN-PRK2 OX lines were increased in the RopGEF1(90–457)–GFP OX background (Figure 2G and 2I). Overexpression of full-length RopGEF1–GFP also showed a similar but weaker effect, compared to RopGEF1(90–457)–GFP (Figure 2F and 2I). We verified that the increased pollen germination rate was not due to altered DN-PRK2 expression levels (Figure 2J). These results imply that the full-length RopGEF1 needs to be activated, probably by AtPRK2.
AtPRK2 and DN-PRK2 Activate and Inhibit ROP1 Activity, Respectively
To test whether AtPRK2 acts as a positive regulator of ROP1, we determined the effect of AtPRK2 expression on ROP1 activity in tobacco pollen tubes. ROP1 activity was monitored using the ROP1 activity marker GFP–RIC4ΔC, which specifically interacts with active ROP1 but does not induce growth depolarization in pollen tubes (Hwang et al., 2005, 2008, 2010). GFP–RIC4ΔC was transiently expressed together with AtPRK2a or DN-PRK2a, and PM-associated GFP–RIC4ΔC fluorescence was measured as previously described (Huang et al., 2005). The average ROP1 activity in the apical PM (Intensity-Ave-PM) was calculated by means of PM GFP–RIC4ΔC fluorescence standardized with the cytosolic GFP–RIC4ΔC signal (Average-cyt) (Huang et al., 2005). Active ROP1 on the PM was presented as (Intensity-Ave-PM/Ave-cyt) X PM distribution. We set the active ROP1 of control pollen tubes to 100%, and calculated the relative change of ROP1 activity caused by AtPRK2 or DN-PRK2 overexpression. ROP1 activity increased by 47% in AtPRK2-overexpressing tubes, but was reduced by 26% in DN-PRK2-overexpressing pollen tubes (Figure 3A–3D). These results further support our hypothesis that AtPRK2 acts as a positive regulator of ROP1 in the control of polar growth of pollen tubes.
Figure 3.
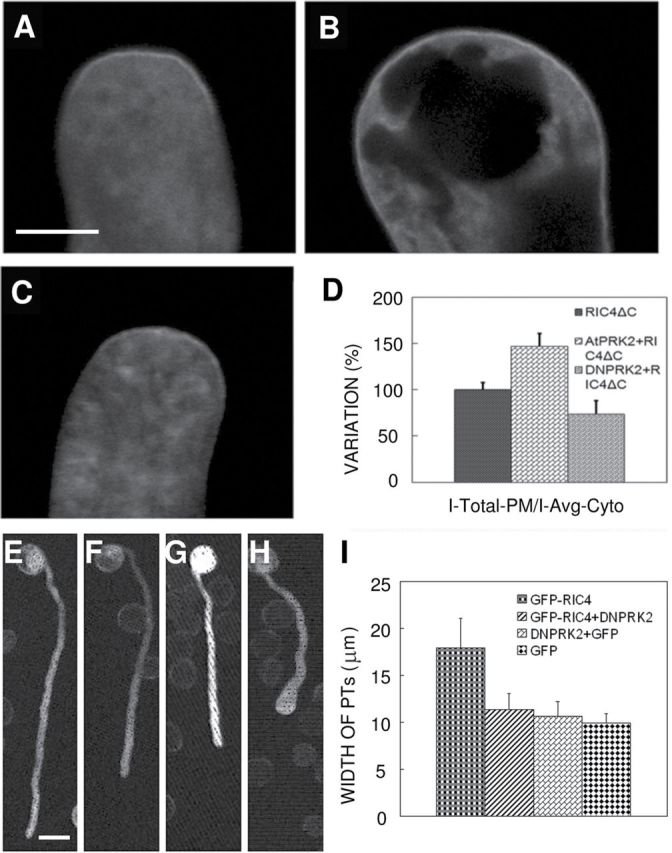
ROP1 Activity Is Positively Regulated by AtPRK2 and Negatively Regulated by DNPRK2 in Pollen Tube Growth.(A–E) ROP1 activity was increased by AtPRK2 and decreased by DN-PRK2. The distribution of the active ROP1 marker, GFP–RIC4 and control (A), AtPRK2 OX (B), and DN-PRK2 OX (C) pollen tubes are shown. Distribution of GFP–RIC4ΔC was enlarged in most AtPRK2 OX pollen tubes and narrowed in most DNPRK2 pollen tubes. (D) Statistical analysis of GFP–RIC4ΔC distribution in each background compared to wild-type pollen tube. Data were collected from three independent experiments.(F–I) DN-PRK2 suppresses RIC4-OX-induced defect in polar pollen tube growth. (E) GFP, (F) DN-PRK2+GFP, (G) DN-PRK2+GFP–RIC4, and (H) GFP–RIC4 were transiently expressed in tobacco pollen by bombardment and germinated for 5 h at RT before visualization. (I) Quantitative data of pollen tube widths from various backgrounds. Bar shows SD in (H) and (I). Scale bar = 5 μm in (A); Scale bar = 25 μm in (E).
To confirm this hypothesis, we co-expressed DN-PRK2 with one of the ROP1 effectors, RIC4, which is directly activated by ROP1 (Wu et al., 2001; Gu et al., 2005). RIC4 overexpression induces severe tip swelling, as ROP1 overexpression does. We reasoned that, if AtPRK2 acts upstream of ROP1, overexpression of the negative form of AtPRK2, DN-RPK2, would suppress the phenotype caused by RIC4-OX, similar to the suppression of RIC4 overexpression phenotype by DN-ROP1 (Gu et al., 2005). Indeed, DN-PRK2 overexpression suppressed the GFP–RIC4-induced depolarization of pollen tube growth. Compared to GFP–RIC4 OX pollen tubes, DN-PRK2 and GFP–RIC4 co-overexpressing pollen tubes revealed reduced width and increased elongation (Figure 3E–3I).
RopGEF1 Interacts with and Is Phosphorylated by AtPRKs
We next investigated the possible mechanisms by which AtPRKs regulate the RopGEF1/ROP1 signaling pathway. To test whether AtPRKs directly associate with RopGEF1, the kinase domains of four AtPRKs (AtPRK1, AtPRK2, AtPRK3, and AtPRK4) were cloned and fused to the C-terminus of GST and an MBP-tagged RopGEF1/RopGEF1(90–457) were expressed in E. coli. The interaction was determined by pull-down assays using GST–AtPRKs-linked glutathione-beads and anti-MBP antibodies. All tested AtPRKs were able to interact with RopGEF1 and its PRONE domain, RopGEF1(90–457), through the kinase domain (Figure 4A), consistent with the genetic results that suggest functional redundancy among these AtPRKs.
Figure 4.
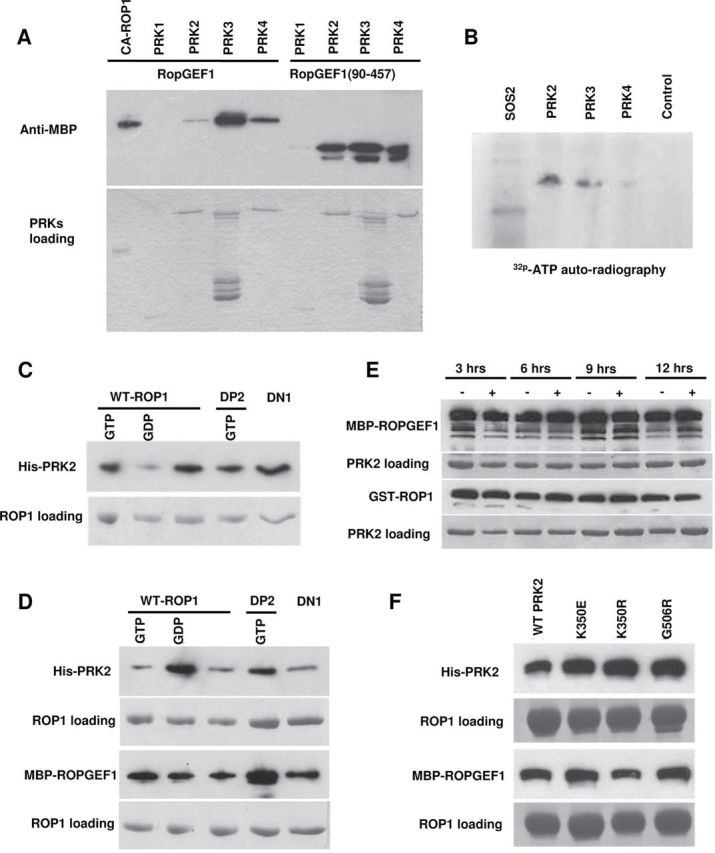
RopGEF1 Is Phosphorylated by AtPRKs and Forms a Complex with AtPRK2 and ROP1.(A) Both full-length RopGEF1 and RopGEF1(90–457) interact with the kinase domains of AtPRK2, AtPRK3, and AtPRK4.(B) RopGEF1 is phosphorylated by AtPRK2, AtPRK3, and possibly AtPRK4. SOS2 was used as a positive control and GST alone was used as the negative control.(C) The kinase domain of AtPRK2 interacts with various forms of ROP1. GDP/GTP-bound form ROP1, GTP–DP2, and DN-ROP1 were used in each reaction as indicated.(D) The HIS-tagged kinase domain of AtPRK2, MBP–RopGEF1, and various forms of ROP1 form complexes in vitro.(E) Interaction assay between the kinase domain of AtPRK2, RopGEF1, and ROP1 after incubation for 3, 6, 9, and 12 h with/without alkaline phosphatase.(F) Pull-down assay to verify the interaction between ROP1, MBP-tagged ROPGEF1, and wild-type or mutated kinase domain of AtPRK2.
The interaction of AtPRKs with RopGEF1 suggested that AtPRKs may directly phosphorylate RopGEF1. This hypothesis was tested using an in vitro kinase assay. As shown in Figure 4B, RopGEF1 was phosphorylated by AtPRK2 and AtPRK3, maybe also AtPRK4, while it was not phosphorylated by the control protein (GST alone was used as a negative control). To investigate whether the kinase activity of AtPRK2 or AtPRK3 was calcium-dependent, Ca2+-free conditions in the kinase assay were achieved by adding excess EGTA (10mM). The phosphorylation of RopGEF1 was not altered by EGTA inclusion (data not shown), suggesting that the phosphorylation of RopGEF1 is calcium-independent.
RopGEF1 Forms a Complex with ROP1 and AtPRK2
The physical and functional interactions between AtRPKs and RopGEF1, and between AtRPK2 and ROP1, suggest that these proteins might form complexes. We chose AtPRK2 for our further analysis because AtPRK2 overexpression induced the most severe depolarization of pollen tubes and strongly phosphorylated RopGEF1 (Table 1 and Figure 4B). To test whether AtPRK2 physically interacts with and with which form of ROP1 in vitro, the kinase domain of AtPRK2 was fused to the C-terminus of HIS tag (HIS–AtPRK2). The GTP-bound GST–ROP1 and GST–DP2 (a constitutively active form ROP1) were used as active form ROP1 (Li et al., 1999; Wu et al., 2001), the GDP-bound GST–ROP1 and GST–DN (Dominant Negative) ROP1 as inactive form (Li et al., 1999; Wu et al., 2001). All these active, inactive, and free forms of GST–ROP1 fusion proteins on glutathione-agarose beads were incubated with eluted HIS–AtPRK2. Purified fusion proteins were pulled down with GST–ROP1 and detected with HIS-antibodies. We found that AtPRK2 strongly interacts with all ROP1, GTP–ROP1, GDP–ROP1, GTP–DP2, and DN-ROP1 (Figure 4C). As it was shown that ROP1 interacts with RopGEF1 and is activated by RopGEF1 (Gu et al., 2006), we proposed that AtPRK2, RopGEF1, and ROP1 may form a complex. To test this, we performed another pull-down experiment with all these forms of GST–ROP1, HIS–AtPRK2, and MBP–RopGEF1. GST–ROP1 proteins on glutathione-agarose beads were incubated with eluted HIS–AtPRK2 and MBP–RopGEF1. After extensive washes, glutathione-agarose beads were subjected to Western blot analysis using HIS or MBP-antibody. As expected, both AtPRK2 and RopGEF1 were detected using all these forms of GST–ROP1 (Figure 4D).
To test whether the phosphorylation status of AtPRK2 may affect the formation of AtPRK2–RopGEF1–ROP1 complex, we treated the complex using alkaline phosphatase and tested their integrity. As shown in Figure 4E, the formation of the AtPRK2–RopGEF1–ROP1 complex was not markedly affected by alkaline phosphatase treatment. We also showed that mutations in the conserved kinase catalytic sites of AtPRK2 did not affect the complex formation (Figure 4F). These results suggest the interaction between AtPRK2, RopGEF1, and ROP1 is phosphorylation-independent.
The C-Terminal Region of RopGEF1 Is Important for Its Interaction with ROP1 and AtPRK2
We next investigated how AtPRK2 regulates the RopGEF1–ROP1 signaling pathway. Our previous results suggest that the C-terminal variable region regulates the GEF activity of RopGEF1 in an auto-inhibitory manner (Gu et al., 2006). Thus, it is reasonable to postulate that the C-terminal region may be regulated by AtPRK2. First, we tested whether the C-terminal region, RopGEF1(458–548), interacts with AtPRK2, using several truncated constructs for RopGEF1 fused to the C-terminal of an MBP tag (Figure 5A). MBP–RopGEF1 was incubated with HIS–PRK2 or the truncated forms of GST–ROP1. As shown in Figure 5B, the MBP-tagged C-terminal region, MBP–RopGEF1(458–548), interacted with both ROP1 and AtPRK2. Furthermore, full-length MBP–RopGEF1 showed a strong interaction with AtPRK2, while MBP–RopGEF1(1–457) exhibited a much weaker interaction with AtPRK2. Similarly, MBP–RopGEF1(365–548) showed a strong interaction with AtPRK2 while MBP–RopGEF1(365–457) exhibited no interaction with AtPRK2 (Figure 5B). These results demonstrate that the C-terminal region of RopGEF1(458–548) is important for RopGEF1 to interact with AtPRK2.
Figure 5.
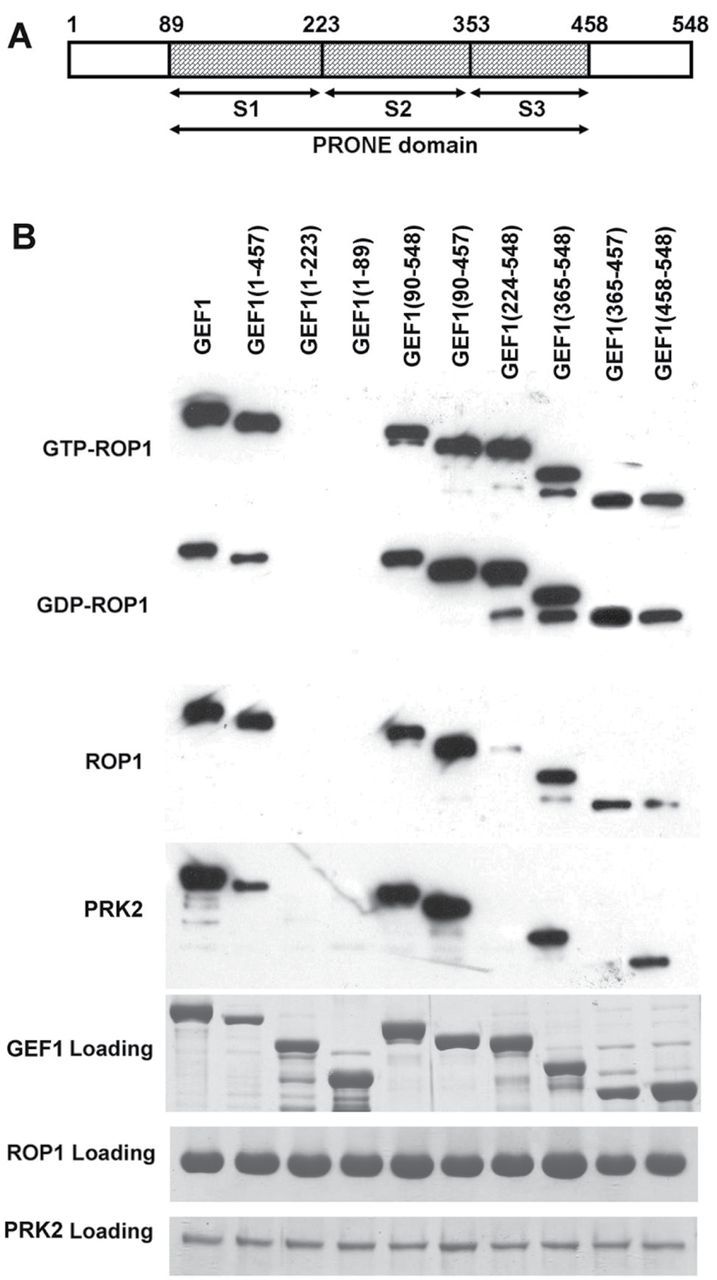
The C-Terminal Region of RopGEF1 Is Important for Its Interaction with AtPRK2 and ROP1.(A) A diagram of RopGEF1 motifs. The PRONE domain, S1-3 subdomains, N-terminal, and C-terminal regions of RopGEF1 are indicated.(B) MBP-tagged truncated RopGEF1 was used in pull-down assays with the His-tagged kinase domain of AtPRK2 (His–PRK2) and various forms of GST-tagged ROP1. His–PRK2 or various GST–ROP1 included in the assay are indicated on the left. Different truncated RopGEF1 forms used in each assay are indicated above and signals were detected by anti-MBP antibodies. Proteins loaded in the assay are also shown.
Phosphorylation of the C-Terminal Region Is Required for RopGEF1 Activity
Full-length RopGEF1 has no in vitro GEF activity towards ROP1. However, overexpression of RopGEF1 induces growth depolarization of pollen tubes in both Arabidopsis and tobacco (Gu et al., 2006). We reasoned that RopGEF1 activity might be regulated by phosphorylation. RopGEF1 has more than 20 predicted serine, threonine, and tyrosine phosphorylation sites (http://myhits.isb-sib.ch/cgi-bin/motif_scan), including six in the C-terminal region. Because the C-terminal region of RopGEF1 is likely to be essential for its auto-inhibitory regulation (Gu et al., 2006), the six C-terminal sites of RopGEF1 were chosen for site-directed mutagenesis to test whether phosphorylation at these sites is critical for the release of auto-inhibition. If the phosphorylation of RopGEF1 is required for its in vivo function, a serine-to-alanine substitution should abolish its ability to induce growth depolarization in pollen tubes.
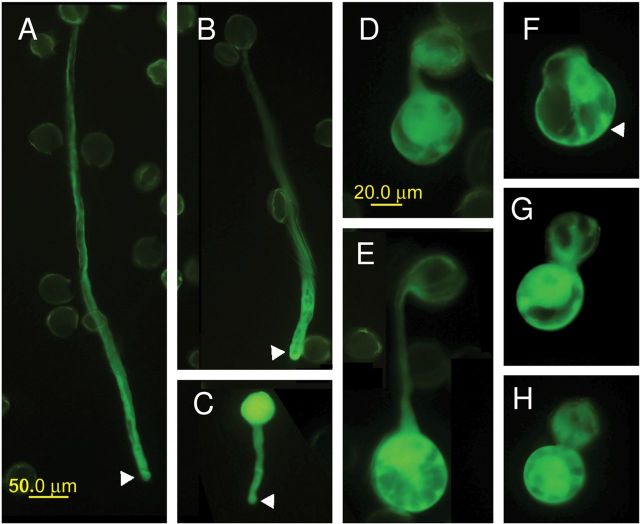
We generated GFP-tagged RopGEF1 constructs with a serine-to-alanine substitution at each of the six sites and transiently overexpressed these constructs in tobacco pollen tubes. Overexpression of the GFP–RopGEF1S460A mutant failed to induce depolarized pollen tube growth (Figure 6B and Supplemental Table 3), suggesting that phosphorylation at the S460 site is required for the functional activity of RopGEF1. The GFP–RopGEF1S480A mutant not only failed to cause balloon-like pollen tips, but also inhibited pollen tube elongation, similar to the effect of the dominant-negative ROP1 (DN-ROP1) mutant (Figure 6C and Supplemental Table 3), suggesting that this mutant may be dominant-negative. Four other mutations did not appear to abolish RopGEF1 activity (Figure 6D, 6E, 6G, 6H, and Supplemental Table 3).
Figure 6.
Phosphorylation in C-Terminal Regions Is Critical for RopGEF1 Activity.Serine-to-alanine mutation of several sites in the RopGEF1 C-terminal region decreases RopGEF1 activity. Wild-type and mutant RopGEF1 were transiently expressed in tobacco pollen for 5 h before observation. Compared to GFP (A), GFP–RopGEF1wt (F) caused severe depolarization of pollen tubes with balloon-like tips. The GFP–S460A mutation (B) and GFP–S480A mutation (C) show greatly reduced RopGEF1-induced depolarization; pollen tubes overexpressing the GFP–S460A mutant are much longer than GFP–RopGEF1wt. Overexpression of GFP–S484A (D), GFP–S488A (E), GFP–S458A (G), and GFP–S501A (H) mutations caused severely depolarized pollen tubes as wild-type RopGEF1 does. Scale bars shown, and images (A–C) with the same magnification and (D–H) with the same magnification. Arrowhead indicates the tip of pollen tubes.
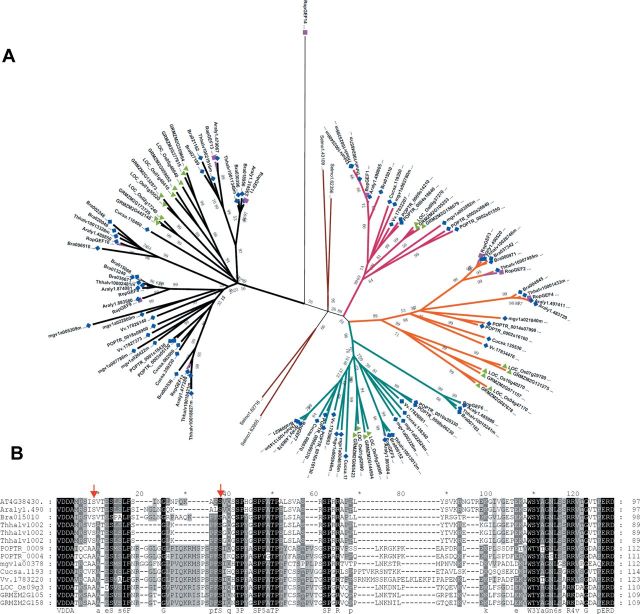
To investigate whether these two serine phosphorylation sites (S460/S480) of RopGEF1 are conserved, we first constructed a phylogenic tree using the conserved PRONE domain in Arabidopsis and other dicots and monocots species (Figure 7A and Supplemental Figure3). An alignment between Arabidopsis RopGEF family members or RopGEF1 orthologs with the full-length amino acids revealed that S460 is shared among Brassicaceae RopGEF1 orthologs, and S480 is conserved among plant RopGEF1 orthologs (Figure 7B), but neither S460 nor S480 is conserved among other Arabidopsis RopGEFs (Supplemental Figure 4).
Figure 7.
The S480 Site of RopGEF1 Is Conserved in RopGEF1 Orthologs and S460 Is Conserved in Brassicaceae.(A) A phylogenetic tree of RopGEF genes in dicots and monocots. Arabidopsis RopGEFs are highlighted in dark pink squares, other dicots genes are labeled by blue rhombuses, and the monocots genes are indicated by green triangles. Selaginella genes were included to root the tree.(B) Alignment of the C-terminal region of RopGEF1 orthologous genes. The S460 and S480 sites are indicated by red arrows.
DISCUSSION
ROPs are central regulators of many important cellular processes such as cell polarity formation, cell morphogenesis, and polar cell growth (Yang, 2002, 2008), and are modulated by a number of regulators including RhoGAPs, GDIs, and RopGEFs (Yang, 2002, 2008), although little is known of how these direct regulators of ROPs are connected to extracellular signals. Recent studies suggest that RLKs directly interact with ROPs and may act upstream of RopGEFs to regulate polar growth in pollen tubes and root hairs (Zhang and McCormick, 2007; Duan et al., 2010). In this study, we provide genetic and biochemical evidence in support of the hypothesis that the Arabidopsis pollen RLK–AtPRK2 phosphorylates and activates RopGEF1 to regulate ROP1-dependent polar growth in pollen tubes.
Based on our findings, we propose a model to explain the regulatory mechanism for pollen tube growth (Figure 8). The GEF activity of ROPGEF1 is inhibited/disrupted by the binding of its C-terminal region to its PRONE catalytic domain. Upon AtPRK2 interaction with RopGEF1 and phosphorylation of the serine sites in the RopGEF1 C-terminal region, a conformational change is induced and the PRONE1 domain is exposed, resulting in an active form of RopGEF1. Active RopGEF1 subsequently catalyzes the conversion of GDP-bound inactive form ROP1 into GTP active form, thereby activating ROP1 signaling pathways and promoting pollen germination and pollen tube growth (Figure 8). This model explains the observed higher activity of ROP1 in AtPRK2 OX pollen tubes and lower ROP1 activity in DN-PRK2 OX pollen tubes.
Figure 8.
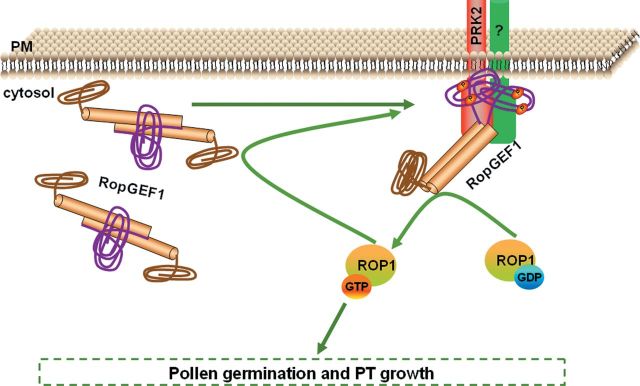
A Model for AtPRK2-Induced Release of RopGEF1’s Auto-Inhibition and Activation of ROP1 Signaling Pathways. RopGEF1 is subjected to auto-inhibitory regulation, which is controlled by its C-terminal region. Auto-inhibition is released through the phosphorylation of the serine amino acids in its C-terminal region upon RopGEF1 interaction with AtPRK2. The phosphorylation-induced conformational change in RopGEF1 exposes the PRONE domain. Active RopGEF1 can then promote the conversion of the GDP-bound inactive form of ROP1 into its GTP-bound active form, thereby enhancing the ROP1 signaling pathways and regulating pollen germination and pollen tube growth. We also propose that the GTP form of ROP1 interacts with both RopGEF1 and AtPRK2 and somehow, through positive feedback, regulates RopGEF1 activity. The PRONE domain of RopGEF1 is shown as brown cylinders, the N-terminal region is shown as a brown curve, and the C-terminal region is shown as a purple curve. Green arrow indicates positive regulation.
Since RopGEF1 OX pollen tubes showed severe depolarization similar to CA-ROP1 OX pollen tubes, we further propose a positive feedback regulation of RopGEF1 activity (Figure 8). In our previous study, the result that the inactive form of RopGEF1 shows the strongest interaction with GTP-bound ROP1 also supports this idea (Gu et al., 2006). We propose that, after the phosphorylation and activation of RopGEF1 by AtPRK2, the active GTP-bound ROP1 interacts with AtPRK2 and/or RopGEF1 and enhances the interaction between AtPRK2 and RopGEF1. This enhancement could be the result of an increased interaction frequency or affinity between AtPRK2 and RopGEF1. Consequently, more RopGEF1s are phosphorylated and active, as supported by the observation that RopGEF1 OX pollen tubes displayed CA-ROP1-like bulbous tips. This would also be similar to the mechanism regulating the Ras GEF Sos in animal cells (Margarit et al., 2003).
RopGEFs Directly Interact with and Are Regulated by RLKs: An Emerging Theme in RLK/ROP Signaling
An increasing number of studies suggest critical roles for Arabidopsis RopGEFs (Berken et al., 2005; Gu et al., 2006) in the regulation of diverse ROP-mediated processes. A recent study shows that RopGEF7 modulates the maintenance of the root stem cell niches by regulating the activity of AtRAC1 (AtROP3) and the expression of PLETHORA1 (PLT1), PLT2, and the auxin efflux protein PIN1 (Chen et al., 2011). Another Arabidopsis RopGEF, PIRF1 (phytochrome-interacting ROP guanine nucleotide exchange factor 1, also referred to as RopGEF11), has been found to interact with phytochromes via its conserved PRONE domain and to activate ROPs in a phytochrome-dependent manner to regulate primary root development (Shin et al., 2010). Studies in rice and legume also implicate RopGEFs in regulating the development of root hairs, cuticular papillae on the leaf surface by regulating the substrate ROP activity (Riely et al., 2011; Yoo et al., 2011; Yamaguchi et al., 2012).
The physiological significance of RopGEFs and ROPs begs the question of how RopGEFs are regulated. This study, together with several recent reports, supports an emerging theme of RLK-based RopGEF regulation at the cell surface. McCormick’s group first demonstrated a physical and functional interaction between AtPRK2 and RopGEF12 and proposed that AtPRK2 functions to dock RopGEF12 to the plasma membrane of pollen tubes (Zhang and McCormick, 2007). Recently, Cheung’s group showed that the RLK FERONIA (FER) interacts with RopGEF1 and is required for ROP activation and ROP-mediated root hair development (Duan et al., 2010). Our data here support the hypothesis that AtPRK2-mediated phosphorylation regulates RopGEF1 activation. This regulation is most different from AtPRK2’s regulation of RopGEF12, because the functional phosphorylation sites in RopGEF1 are not conserved in RopGEF12 (see below). It is unclear why AtPRK2 regulates two different RopGEFs via distinct mechanisms. Nonetheless, these results suggest that the RLK–RopGEF connection may provide a common cell surface signaling mechanism in plants.
Activation of RopGEF by Phosphorylating Its C-Terminal Region and Releasing Auto-Inhibition Could Be a Common Mechanism for RopGEF Regulation
Based on our data that the C-terminal S-to-A mutations inhibited/prevented the release of RopGEF1 auto-inhibition, we propose that the phosphorylation of the serine sites in the C-terminal region of RopGEF1 releases auto-inhibition and activates RopGEF1. In addition to RopGEF1, RopGEF12 also showed C-terminal auto-inhibition in our study, and this inhibition is likely to be released by phosphorylating the C-terminal region, possibly also by AtPRK2 (Zhang and McCormick, 2007). Overexpression of wild-type RopGEF12 slightly affects the polarity of pollen tubes, while a C-terminal truncation shows a much stronger phenotype, suggesting the auto-inhibition of GEF activity is controlled by its C-terminus. Overexpression of C-terminal phosphorylation mimicking the mutation of RopGEF12 resulted in significant tip swelling, strongly suggesting that phosphorylation is likely to be critical for release of the C-terminal inhibition (Zhang and McCormick, 2007). Thus, we propose that the release of auto-inhibition by C-terminal phosphorylation could be a common mechanism for RopGEF regulation. However, the detailed mechanisms for the phosphorylation-mediated regulation may differ among different RopGEFs because different phosphorylation sites are critical for differential RopGEFs. The critical S510 phosphorylation site of RopGEF12 is conserved among several closely related RopGEFs (Zhang and McCormick, 2007) and their orthologs (Supplemental Figure 5), and the essential phosphorylation sites in the C-terminal region of RopGEF1 sites are also conserved among its orthologs (Figure 7B).
N-Terminal Regions May Contribute Differently to GEF Activity in Different RopGEFs
Previous results suggested that the PRONE domain of RopGEF1 is sufficient for its RopGEF activity (Gu et al., 2006), but the same domain of RopGEF12 was not (Zhang and McCormick, 2007). We propose that the function of the various N-terminal regions among RopGEFs might be different. The N-terminal region of RopGEF1 contributed to the auto-inhibition caused mainly by the C-terminal region (Gu et al., 2006) rather than being required for GEF activity. In contrast, the N-terminal region of RopGEF12 was important for the normal GEF activity of RopGEF12; the overexpression of PRONE12 had a mild phenotype on pollen tube while overexpression of PRONE12 along with the N-terminal region of RopGEF12 caused a much stronger phenotype (Zhang and McCormick, 2007). Further studies, such as structural analysis using crystallography of full-length RopGEFs, could uncover the conformational change induced by phosphorylation and provide insights into the common and distinct mechanisms for regulating activity between these two RopGEFs.
Redundancy between AtRopGEF and AtPRK Family Members
Arabidopsis has 14 RopGEF members and seven members are expressed in the pollen according to previous studies (Gu et al., 2006; Zhang and McCormick, 2007). Single mutants showed no defects in pollen germination or pollen tube growth. In contrast, the ropgef1,9,12,14 quadruple mutant exhibited defective pollen tube growth, implying functional redundancy between these genes. Similarly, functional redundancy exists between AtPRK members, as indicated by the pollen tube growth deficiency in the prk1,2,5 triple mutant but not in any of the single or double mutants. It is possible that RopGEF1 may not be the only RopGEF family member that can perceive the signal from AtPRK2 and that is phosphorylated by AtPRK2, and AtPRK2 may not be the only AtPRK that can phosphorylate and activate RopGEF1. This is supported by our in vitro biochemical results that RopGEF1 interacts with and can be phosphorylated by more than one AtPRK.
METHODS
Database Search, Sequence Alignment, and Phylogenetic Analysis
LePRK protein sequences were used to identify Arabidopsis and rice (Oryza sativa ssp. japonica) homology sequences by BLAST against TAIR database (www.arabidopsis.org/Blast) and phytozome database (www.phytozome.org/search.php?show=blast). Protein sequences were initially aligned using MUSCLE 3.6 (Edgar, 2004). Data generated from this alignment were loaded onto MEGA5.05 (Tamura et al., 2011) to reconstruct the rooted Maximum Likelyhood phylogenetic tree using default settings. The supporting values were estimated by using bootstrap analysis (1000 replicates). The creation of the RopGEF phylogenetic tree was performed using the same method.
Site-Directed Mutagenesis
RopGEF1 mutations in designed sites were made using the Fast mutagenesis system following the manufacturer’s protocols (FM111, Transgen, Beijing, CN). The resulting constructs were then verified by sequencing.
Protein Expression and Purification
The full-length and the various truncated forms of RopGEF1 were cloned in pMALC2 fusion vector, and ROP1 was cloned in pGEX–KG fusion vector (Gu et al., 2006). The kinase domains of AtPRKs were constructed in pHIS8 fusion vector. Fusion protein expression in E. coli was carried out as described previously with some modifications (Gu et al., 2006). RopGEF1 and ROP1 fusion proteins were expressed at 30°C for 4h after induction with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG). Cells were harvested by centrifugation at 5000 g for 10 min. AtPRKs fusion proteins were expressed at 16°C for 20 h after induction by 0.2 mM IPTG. For GST-fusion proteins and MBP-fusion proteins, cell pellets were re-suspended in a binding buffer (20 mM Tris, pH 7.4, 200 mM NaCl, 1 mM EDTA) and sonicated using 5-s pulses 20 times. After centrifugation at 16 000 g for 30 min, the supernatant was collected and mixed with glutathione-agarose beads (Sigma) or amylose beads (Biolabs) for GST-fusion proteins and MBP-fusion proteins, respectively. After 2-h incubation, the beads were washed with the binding buffer and the fusion protein was eluted using maltose (10 mM) or glutathione (30 mM), respectively. For His-tagged proteins, cell pellets were re-suspended in a lysis buffer (50 mM NaH2PO4, 300 mM NaCl, 10 mM imidazole, 10% glycerol (V/V), pH 8.0). After sonication and centrifugation as described above, the supernatant was mixed with Ni-NTA beads (Qiagen) for 2 h. The mixture was then washed with the wash buffer (50 mM NaH2PO4, 300 mM NaCl, 20 mM imidazole, 10% glycerol, pH 8.0). His-tagged proteins were eluted using the elution buffer (50 mM NaH2PO4, 300 mM NaCl, 250 mM imidazole, 10% glycerol, pH 8.0).
In Vitro Protein–Protein Interaction Assays
In vitro protein–protein interaction assay was performed as described previously (Gu et al., 2006). To detect whether ROP1, AtPRK2, and RopGEF1 formed a complex, GST–ROP1 was preloaded with GDP or GTP in a nucleotide loading buffer containing 3 mM of the corresponding nucleotide, 25 mM Tris-HCl (pH 7.5), 1 mM DTT, 10 mg ml–1 BSA, and 5 mM EDTA. Approximately 10 µg of GST–ROP1 fusion proteins were bound to glutathione-conjugated agarose beads, and similar amounts of MBP–RopGEF1 and His–AtPRK2 fusion proteins were used in each assay. Beads containing GST–ROP1 fusion proteins were incubated with MBP–RopGEF1 and HIS–AtPRK2 fusion proteins in an interaction buffer containing 20 mM Hepes (pH 7.4), 5 mM MgCl2, 1 mM DTT, 0.1% Triton X-100, and 1 mM EDTA for 2 h. After binding, beads containing GST–ROP1 and bound MBP–RopGEF1 and His–AtPRK2 were washed extensively to remove unbound proteins. The bound proteins were detected using a polyclonal antibody against MBP or HIS (New England Biolabs, Beverly, MA).
In Vitro Kinase Assay
The substrate protein (2 μg) was equilibrated with 1X kinase buffer containing 20 mM Tris, pH 7.4, 5 mM MgCl2, 1 mM CaCl2, 1 mM DTT, and 10 μM ATP. Purified kinase protein and substrate were incubated in 1 kinase buffer containing 5–10 μCi per reaction [32p-ATP] at 30°C for 30 min. Reactions were stopped with SDS–PAGE sample buffer. Proteins were separated by SDS–PAGE and detected using autoradiography.
Analyses of AtPRKs and RopGEF1 Overexpression Phenotype
The full-length AtPRK1–5 driven by the pollen-specific LAT52 promoter (pLAT52::AtPRK) were transiently expressed in tobacco pollen, together with the pLAT52::GFP vector as an indicative marker. The wild-type and point mutation RopGEF1 were cloned in the pLAT52::GFP vector and transiently expressed in tobacco pollen. Approximately 5 h after bombardment, pollen tubes were visualized using an Olympus BX51 microscope equipped with an Olympus DP70 CCD camera (Japan). Widths and lengths of pollen tubes were measured using the AIM software (Zeiss, Germany).
Pollen tubes co-expressing GFP–RIC4ΔC and AtPRK2/DN-PRK2 were analyzed using laser scanning confocal microscopy under a Nikon Axioplan2 microscope (Japan) equipped with a Zeiss LSM 510 META confocal laser scanning device (Germany). 1 μm optical sections were scanned and captured using the AIM software. Confocal laser scanning images were analyzed using the MetaMorph 4.5 software (Universal Imaging, West Chester, PA). The degree of depolarized growth was determined by measuring the diameter of the widest region of the pollen tube tip. Experiments were repeated three times and more than 30 pollen tubes were measured for each sample each time.
Generation of the AtPRK2-Dominant Mutant
To generate AtPRK2–DN construct, the ATPRK2 cDNA sequence without the cytoplasmic kinase domain was cloned into a pC1300LAT52 vector derived from pCAMBIA1300 (CAMBIA). The construct was introduced into Agrobacterium tumefaciens GV3101 by electroporation and transformed into A. thaliana ecotype Columbia. T2 homozygous plants were selected for analysis.
Isolation of T-DNA Insertional Mutants
The gef1-1, gef9-1, gef12-1, gef14-1, gef14-2, prk1-1, prk1-2, prk2-1, and prk5-1 T-DNA insertion mutant lines were screened from the SIGNAL collection (http://signal.salk.edu/cgi-bin/tdnaexpress/) based on a combination of database searches and PCR amplification of T-DNA flanking regions (Supplemental Table 1). For T-DNA lines identified from the SIGNAL collection, seeds were obtained from the Arabidopsis Biological Resource Center (http://abrc.osu.edu/order-stocks). PCR reactions were carried out to identify single plants for the T-DNA insertion with gene-specific primers and LBb1.3 primer (ATTTTGCCGATTTCGGAAC) for SALK lines or GAB-LB1 primer (CCCATTTGGACGTGAATGTAGACAC) for GABI lines.
SUPPLEMENTARY DATA
Supplementary Data are available at Molecular Plant Online.
FUNDING
This work is supported by a MOST 973 project (grant 2007CB108704), National Institute of General Medical Research (GM100130) and DOE (DE-FG02-04ER15555, which supported F.C. and the biochemical experiments described in this work) to Z.Y., and a National Science Foundation of China (31070274) to F.C.
Supplementary Material
ACKNOWLEDGMENTS
We thank the Salk Institute Genomic Analysis Laboratory for providing the sequence-indexed A. thaliana T-DNA insertion mutants, and the Ohio State University Arabidopsis Biological Resources Center for seeds. We thank Dr Ying Fu in China Agricultural University and Mingzhang Wen in NSFC for discussion on the organization of this paper, and Dr Ning Zhang in Fudan University for discussion on the phylogenetic analysis of PRKs and RopGEFs. No conflict of interest declared.
REFERENCES
- Berken A., Thomas C., Wittinghofer A. 2005. A new family of RhoGEFs activates the Rop molecular switch in plants. Nature. 436, 1176–1180 [DOI] [PubMed] [Google Scholar]
- Chang F., Yan A., Zhao L., Wu W., Yang Z. 2007. A putative calcium-permeable Cyclic Nucleotide-Gated Channel, CNGC18, regulates polarized pollen tube growth. J. Int. Plant Biol. 49, 1261–1270 [Google Scholar]
- Chen M., Liu H., Kong J., Yang Y., Zhang N., Li R., Yue J., Huang J., Li C., Cheung A.Y., et al. 2011. RopGEF7 regulates PLETHORA-dependent maintenance of the root stem cell niche in Arabidopsis . Plant Cell. 23, 2880–2894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung A.Y., Wu H.M. 2008. Structural and signaling networks for the polar cell growth machinery in pollen tubes. Annu. Rev. Plant Biol. 59, 547–572 [DOI] [PubMed] [Google Scholar]
- Duan Q.H., Kita D., Li C., Cheung A.Y., Wu H.M. 2010. FERONIA receptor-like kinase regulates RHO GTPase signaling of root hair development. Proc. Natl Acad. Sci. U S A. 107, 17821–17826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R.C. 2004. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics. 5, 113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y., Gu Y., Zheng Z., Wasteneys G., Yang Z. 2005. Arabidopsis interdigitating cell growth requires two antagonistic pathways with opposing action on cell morphogenesis. Cell. 120, 687–700 [DOI] [PubMed] [Google Scholar]
- Fu Y., Wu G., Yang Z. 2001. Rop GTPase-dependent dynamics of tip-localized F-actin controls tip growth in pollen tubes. J. Cell Biol. 152, 1019–1032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y., Xu T., Zhu L., Wen M., Yang Z. 2009. A ROP GTPase signaling pathway controls cortical microtubule ordering and cell expansion in Arabidopsis . Curr. Biol. 19, 1827–1832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Y., Fu Y., Dowd P., Li S.D., Vernoud V., Gilroy S., Yang Z.B. 2005. A Rho family GTPase controls actin dynamics and tip growth via two counteracting downstream pathways in pollen tubes. J. Cell Biol. 169, 127–138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Y., Li S., Lord E.M., Yang Z. 2006. Members of a novel class of Arabidopsis Rho guanine nucleotide exchange factors control Rho GTPase-dependent polar growth. Plant Cell. 18, 366–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Y., Vernoud V., Fu Y., Yang Z. 2003. ROP GTPase regulation of pollen tube growth through the dynamics of tip-localized F-actin. J. Exp. Bot. 54, 93–101 [DOI] [PubMed] [Google Scholar]
- Gu Y., Wang Z., Yang Z. 2004. ROP/RAC GTPase: an old new master regulator for plant signaling. Curr. Opin. Plant Biol. 7, 527–536 [DOI] [PubMed] [Google Scholar]
- Hwang J.U., Gu Y., Lee Y.J., Yang Z. 2005. Oscillatory ROP GTPase activation leads the oscillatory polarized growth of pollen tubes. Mol. Biol. Cell. 16, 5385–5399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang J.U., Vernoud V., Szumlanski A., Nielsen E., Yang Z. 2008. A tip-localized RhoGAP controls cell polarity by globally inhibiting Rho GTPase at the cell apex. Curr. Biol. 18, 1907–1916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang J.U., Wu G., Yan A., Lee Y.J., Grierson C.S., Yang Z. 2010. Pollen-tube tip growth requires a balance of lateral propagation and global inhibition of Rho-family GTPase activity. J. Cell Sci. 123, 340–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson M.A., Preuss D. 2003. On your mark, get set, GROW! LePRK2-LAT52 interactions regulate pollen tube growth. Trends Plant Sci. 8, 97–99 [DOI] [PubMed] [Google Scholar]
- Jones M.A., Raymond M.J., Yang Z., Smirnoff N. 2007. NADPH oxidase-dependent reactive oxygen species formation required for root hair growth depends on ROP GTPase. J. Exp. Bot. 58, 1261–1270 [DOI] [PubMed] [Google Scholar]
- Jones M.A., Shen J.J., Fu Y., Li H., Yang Z., Grierson C.S. 2002. The Arabidopsis Rop2 GTPase is a positive regulator of both root hair initiation and tip growth. Plant Cell. 14, 763–776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaothien P., Ok S.H., Shuai B., Wengier D., Cotter R., Kelley D., Kiriakopolos S., Muschietti J., McCormick S. 2005. Kinase partner protein interacts with the LePRK1 and LePRK2 receptor kinases and plays a role in polarized pollen tube growth. Plant J. 42, 492–503 [DOI] [PubMed] [Google Scholar]
- Kim H.U., Cotter R., Johnson S., Senda M., Dodds P., Kulikauskas R., Tang W.H., Ezcurra I., Herzmark P., McCormick S. 2002. New pollen-specific receptor kinases identified in tomato, maize and Arabidopsis: the tomato kinases show overlapping but distinct localization patterns on pollen tubes. Plant Mol. Biol. 50, 1–16 [DOI] [PubMed] [Google Scholar]
- Lee Y.J., Yang Z. 2008. Tip growth: signaling in the apical dome. Curr. Opin. Plant Biol. 11, 662–671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y.J., Szumlanski A., Nielsen E., Yang Z. 2008. Rho-GTPase-dependent filamentous actin dynamics coordinate vesicle targeting and exocytosis during tip growth. J. Cell Biol. 181, 1155–1168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Lin Y., Heath R.M., Zhu M.X., Yang Z. 1999. Control of pollen tube tip growth by a Rop GTPase-dependent pathway that leads to tip-localized calcium influx. Plant Cell. 11, 1731–1742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y., Yang Z. 1997. Inhibition of pollen tube elongation by microinjected anti-Rop1Ps antibodies suggests a crucial role for Rho-type GTPases in the control of tip growth. Plant Cell. 9, 1647–1659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y., Wang Y., Zhu J.K., Yang Z. 1996. Localization of a Rho GTPase implies a role in tip growth and movement of the generative cell in pollen tubes. Plant Cell. 8, 293–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margarit S.M., Sondermann H., Hall B.E., Nagar B., Hoelz A., Pirruccello M., Bar-Sagi D., Kuriyan J. 2003. Structural evidence for feedback activation by Ras.GTP of the Ras-specific nucleotide exchange factor SOS. Cell. 112, 685–695 [DOI] [PubMed] [Google Scholar]
- Mu J.H., Lee H.S., Kao T.H. 1994. Characterization of a pollen-expressed receptor-like kinase gene of Petunia inflata and the activity of its encoded kinase. Plant Cell. 6, 709–721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muschietti J., Eyal Y., McCormick S. 1998. Pollen tube localization implies a role in pollen-pistil interactions for the tomato receptor-like protein kinases LePRK1 and LePRK2. Plant Cell. 10, 319–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riely B.K., He H., Venkateshwaran M., Sarma B., Schraiber J., Ane J.M., Cook D.R. 2011. Identification of legume RopGEF gene families and characterization of a Medicago truncatula RopGEF mediating polar growth of root hairs. Plant J. 65, 230–243 [DOI] [PubMed] [Google Scholar]
- Shin D.H., Cho M.H., Kim T.L., Yoo J., Kim J. I., Han Y. J., Song P. S., Jeon J. S., Bhoo S. H., Hahn T. R. 2010. A small GTPase activator protein interacts with cytoplasmic phytochromes in regulating root development. J. Biol. Chem. 285, 32151-–32159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shpak E.D., Lakeman M.B., Torii K.U. 2003. Dominant-negative receptor uncovers redundancy in the Arabidopsis ERECTA leucine-rich repeat receptor-like kinase signaling pathway that regulates organ shape. Plant Cell. 15, 1095–1110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Bio. Evol. 28, 2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W., Ezcurra I., Muschietti J., McCormick S. 2002. A cysteine-rich extracellular protein, LAT52, interacts with the extracellular domain of the pollen receptor kinase LePRK2. Plant Cell. 14, 2277–2287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W., Kelley D., Ezcurra I., Cotter R., McCormick S. 2004. LeSTIG1, an extracellular binding partner for the pollen receptor kinases LePRK1 and LePRK2, promotes pollen tube growth in vitro . Plant J. 39, 343–353 [DOI] [PubMed] [Google Scholar]
- Wengier D., Valsecchi I., Cabanas M.L., Tang W.H., McCormick S., Muschietti J. 2003. The receptor kinases LePRK1 and LePRK2 associate in pollen and when expressed in yeast, but dissociate in the presence of style extract. Proc. Natl Acad. Sci. U S A. 100, 6860–6865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G., Gu Y., Li S., Yang Z. 2001. A genome-wide analysis of Arabidopsis ROP-interactive CRIB motif genome-wide analysis of f is of ormic GTPase targets. Plant Cell. 13, 2841ell [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu T., Wen M, Nagava S., Fu Y., Chen J., Wu M., Rechenmann C., Frimi J., Jones A.M., Yang Z. 2010. Cell surface- and rho GTPase-based auxin signaling controls cellular interdigitation in Arabidopsis . Cell. 143, 99–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi K., Imai K., Akamatsu A., Mihashi M., Hayashi N., Shimanoto K., Kawasaki T. 2012. SWAP70 functions as a Rac/Rop guanine nucleotide-exchange factor in rice. Plant J. 70, 389–397 [DOI] [PubMed] [Google Scholar]
- Yang Z. 2002. Small GTPases: versatile signaling switches in plants. Plant Cell. 14 Suppl., S375–S388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. 2008. Cell polarity signaling in Arabidopsis . Annu. Rev. Cell Dev. Biol. 24, 551–575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z., Fu Y. 2007. ROP/RAC GTPase signaling. Curr. Opin. Plant Biol. 10, 490–494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo J.H., Park J.H., Cho S.H., Yoo S.C., Li J., Zhang H., Kim K.S., Koh H.J., Paek N.C. 2011. The rice bright green leaf (bgl) locus encodes OsRopGEF10, which activates the development of small cuticular papillae on leaf surfaces. Plant Mol. Biol. 77, 631–641 [DOI] [PubMed] [Google Scholar]
- Zhang D., Wengier D., Shuai B., Cui C.P., Muschietti J., McCormick S., Tang W.H. 2008. The pollen receptor kinase LePRK2 mediates growth-promoting signals and positively regulates pollen germination and tube growth. Plant Physiol. 148, 1368–1379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., McCormick S. 2007. A distinct mechanism regulating a pollen-specific guanine nucleotide exchange factor for the small GTPase Rop in Arabidopsis thaliana . Proc. Natl Acad. Sci. U S A. 104, 18830–18835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Z.L., Nafisi M., Tam A., Li H., Crowell D.N., Chary S.N., Schroeder J.I., Shen J., Yang Z. 2002. Plasma membrane-associated ROP10 small GTPase is a specific negative regulator of abscisic acid responses in Arabidopsis . Plant Cell. 14, 2787–2797 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.