Summary
The human fungal pathogen Candida albicans can switch between two phenotypic cell types, termed “white” and “opaque.” Both cell types are heritable for many generations, and the switch between the two types occurs epigenetically, that is, without a change in the primary DNA sequence of the genome. Previous work identified six key transcriptional regulators important for white-opaque switching: Wor1, Wor2, Wor3, Czf1, Efg1, and Ahr1. In this work, we describe the structure of the transcriptional network that specifies the white and opaque cell types and governs the ability to switch between them. In particular, we use a combination of genome-wide chromatin immunoprecipitation, gene expression profiling, and microfluidics-based DNA binding experiments to determine the direct and indirect regulatory interactions that form the switch network. The six regulators are arranged together in a complex, interlocking network with many seemingly redundant and overlapping connections. We propose that the structure (or topology) of this network is responsible for the epigenetic maintenance of the white and opaque states, the switching between them, and the specialized properties of each state.
Keywords: White-opaque switching, Transcriptional Regulation, Transcription Networks, Transcriptional circuits, Candida albicans
Introduction
Candida albicans has a long history of causing disease in humans. It can colonize and infect many environmental niches of its host, such as the gastrointestinal tract, the oral cavity, the bloodstream, and the vagina. These environments are diverse, and C. albicans has evolved many mechanisms to modulate cell morphology, nutrient uptake, pH homeostasis, and oxygen use (to name a few regulated systems) in response to these differences in environmental conditions. Here, we address a mechanism that epitomizes the ability of C. albicans to generate different cell types from the same genome, the white-opaque switch.
The two types of cells generated by white-opaque switching were first recognized because colonies of white cells are shiny and dome-shaped, while those of opaque cells are dull and flat (Slutsky et al., 1987; for reviews, see Soll et al., 1993; Bennett et al., 2003; Johnson, 2003; Lohse and Johnson, 2009; Soll, 2009; Morschhäuser, 2010). Both cell types are heritable for many generations, and because the switch between the two types occurs without any change in the DNA sequence of the genome, it will be referred to as epigenetic. Although genetically identical, these cell types differ in many ways, including the size and shape of the individual cells (Slutsky et al., 1987), their metabolic preferences (Lan et al., 2002), their ability to mate (Miller and Johnson, 2002), their pathogenic properties (Kvaal et al., 1997; Kvaal et al., 1999; Geiger et al., 2004; Lohse and Johnson, 2008), and the expression of nearly 15% of the transcriptome (Lan et al., 2002; Tsong et al., 2003; Tuch et al., 2010). Switching from the white cell type (in essence, the default cell type) to the “differentiated” opaque cell type occurs stochastically in approximately one out of every 10,000 cell divisions (Rikkerink et al., 1988). The reverse switch (opaque to white) occurs at approximately the same frequency at 25°C under a standard set of laboratory conditions, but occurs at much higher frequencies (100%) when the temperature is shifted to 37°C (Rikkerink et al., 1988). Other conditions (oxygen, carbon dioxide, and nutrient levels, for example) can also affect the forward and reverse switching frequencies. White-opaque switching appears to be limited to C. albicans and the closely related pathogenic species Candida dubliniensis (Pujol et al., 2004) and Candida tropicalis (Porman et al., 2011).
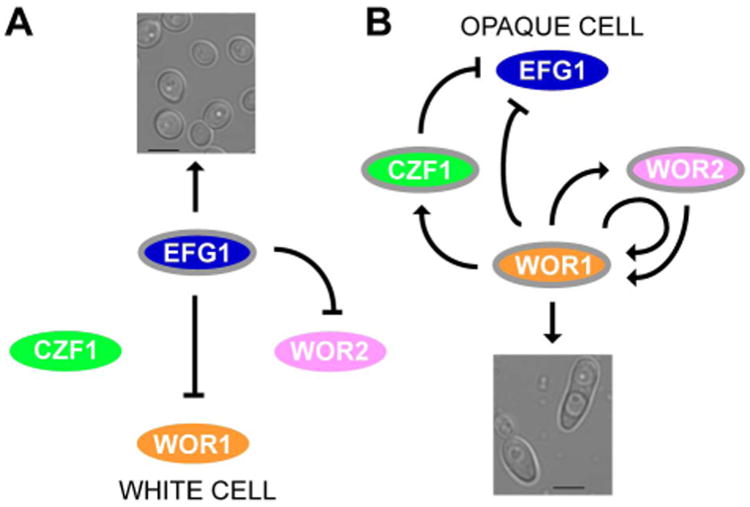
Previous analysis of the white-opaque switch identified a core circuit of four transcriptional regulators – Wor1, Wor2, Czf1 and Efg1 – that control switching via a network of negative and positive feedback loops (Zordan et al., 2006; Zordan et al., 2007) (Figs. 1A and 1B). Wor1, the “master regulator” of opaque cell formation, is a transcriptional regulator that is highly upregulated in opaque cells; it is required for both the transition to, and heritable maintenance of, the opaque cell type. High-level expression of WOR1 in opaque cells is sustained – in part – by a positive auto-regulatory loop, which is believed to form the basis of the heritable maintenance of opaque cells. Consequently, ectopic expression of WOR1 can drive ∼100% of cells from the white to the opaque form (Huang et al., 2006; Srikantha et al., 2006; Zordan et al., 2006). Wor2 is an opaque-enriched transcriptional regulator that is required for the heritable maintenance of the opaque cell type. Czf1 is a third opaque-enriched transcriptional regulator that plays an important role in modulating the white-to-opaque switch frequency, but is not required for the heritable maintenance of the opaque cell type. The white-enriched regulator Efg1 is a negative regulator of the white-to-opaque transition which, when expressed ectopically in opaque cells, can drive switching to the white cell type. Efg1 represses the opaque transcriptional program by binding directly to the upstream region of WOR1 (Sriram et al., 2009; Lassak et al., 2011). More recently, Wang et al. (Wang et al., 2011) identified the regulator Ahr1 (Zcf37/Orf19.7381) as another repressor of the white-to-opaque transition that also drives switching to the white cell type in an Efg1-dependent manner. We recently identified and characterized a sixth regulator of white-opaque switching, Wor3 (Orf19.467), which is highly upregulated in the opaque cell type; moreover, ectopic expression of WOR3 in white cells causes mass switching to the opaque cell type (Lohse et al., 2013).
Figure 1.
Working model of the white-opaque regulatory circuit and its activity in the two cell types. (A) In white cells, Efg1 represses WOR1 directly as well as indirectly through WOR2 to maintain white cell identity. (B) In opaque cells, Wor1, Wor2, and Czf1 establish a series of positive feedback loops, maintaining opaque cell identity and repressing EFG1. Upregulated genes are bordered in gray and active relationships are indicated in black. Downregulated genes lack a border. Arrows and bars represent activation and repression respectively.
Through a series of genetic epistasis experiments, Zordan et al. determined a limited regulatory hierarchy for white-opaque switching (Zordan et al., 2006; Zordan et al., 2007). In this paper, we fully describe the C. albicans white-opaque circuit based on the six known regulators of white-opaque switching; in particular we identify a large set of target genes bound by these regulators. We perform and analyze ChIP-chip experiments with the known members of the transcriptional regulatory network. We combine the existing high resolution ChIP-chip analysis of Wor1 (Zordan et al., 2007) and Wor3 (Lohse et al., 2013) with newly generated ChIP-chip analyses of the other four regulators. We also carry out genome-wide transcriptional profiling of wild-type and transcriptional regulator deletion strains, to identify the direct and indirect regulatory interdependencies of this network. Finally, we use a microfluidic-based approach to examine the inherent DNA-binding specificities of the regulators, and we also incorporate this information into the circuit. Our principle results are as follows: in white cells we observed a compact circuit where only three of the six regulators, Czf1, Ahr1, and Efg1, are active; they bind to and control expression of a relatively small number of target genes with a limited degree of overlap. In contrast, the opaque regulatory circuit is comprised of all six regulators in a large and tightly interwoven arrangement, involving 748 target genes, numerous interactions between regulators and target genes, and multiple interactions among the regulators themselves. Overall, the integrated network controlling white-opaque switching that we describe provides a detailed, comprehensive view of a circuit that orchestrates an epigenetic change, and we believe this circuit can serve as a model for other types of cell differentiation that use complex transcriptional circuits as a basis for generating heritable cell types.
Results
Determining transcriptional relationships among regulators of white-opaque switching
To identify genes directly bound by the six regulators of white-opaque switching (Wor1, Wor2, Efg1, Czf1, Ahr1, and Wor3), we performed ChIP-chip experiments to map the positions across the genome to which each of the six regulators are bound; we carried out these experiments in both white and opaque cell types. Our methods and analyses are very similar to those recently described in Nobile et al. (Nobile et al., 2012), and are reiterated in detail in the Supplemental Experimental Procedures. We first discuss the ChIP-chip results for the relatively simple circuit that is found in white cells and, later, for the more complex opaque cell circuit. We focus on the highlights of these analyses below; the full datasets and further analyses are included in Supplementary Information (File S1, Supplementary Information). In the following discussion, we will typically refer to regulators bound to a given number of intergenic regions. If any of these regions lie between divergently transcribed genes we will assume that the bound regulator is in position to potentially regulate two genes. Thus, the number of genes potentially regulated will almost always be higher than the number of intergenic regions bound.
White cell circuit
ChIP-chip experiments with Wor1, Wor2, and Wor3 in white cells did not produce detectable enrichment at any genomic location, consistent with the fact that these regulators are expressed at much lower levels in white cells compared to opaque cells. In white cells, the remaining three regulators, Czf1, Ahr1, and Efg1, bind to a total of 131 discrete intergenic regions (corresponding to 179 genes), with Czf1 in position to control 55 genes, Ahr1, 93, and Efg1, 73. There are 23 intergenic regions that are bound by at least two regulators, and five that are bound by all three regulators (Fig. 2A, Table S1A). Efg1 binds to its own upstream region and to those of WOR1 and WOR2; Czf1 binds to its own upstream region and to those for EFG1 and WOR2; and Ahr1 binds to its own upstream region and to those of EFG1 and WOR2 (Fig. 3, Tables S1A and S1B, File S2).
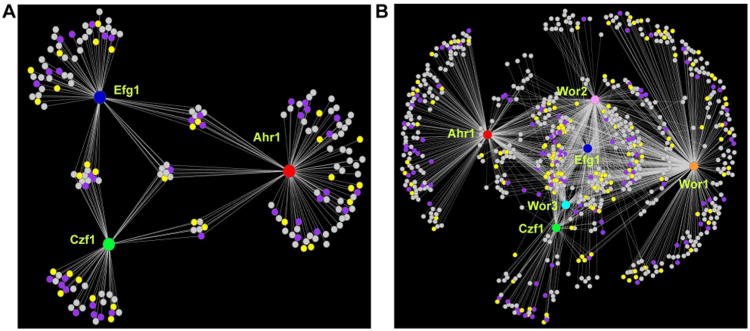
Figure 2.
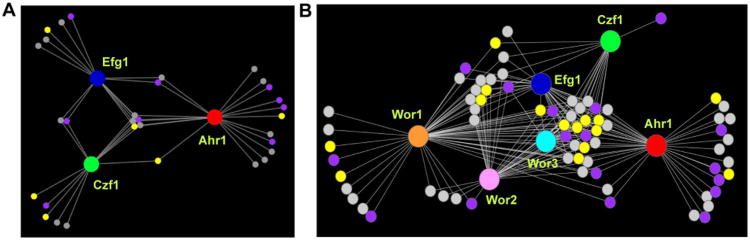
The white and opaque cell regulatory networks. (A) The white cell network consists of three core regulators (Ahr1, red; Czf1, green; Efg1, blue) represented by the large circular hubs. (B) The opaque cell network consists of six core regulators (Wor1, orange; Wor2, pink; Wor3, light blue; Ahr1, red; Czf1, green; Efg1, blue) represented by the large circular hubs. For both (A) and (B), smaller circles represent target genes, which are connected to their respective regulators by white lines, indicative of a direct binding interaction assessed by ChIP-chip. Genes differentially regulated as determined by RNA-seq from Tuch et al. (Tuch et al., 2010) in opaque compared to white cells are shown in yellow for genes upregulated in opaque cells, in light purple for genes downregulated in opaque cells, and in grey for genes with no change.
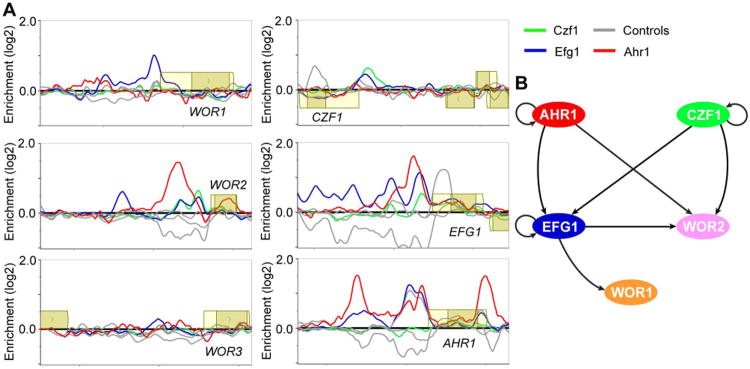
Figure 3.
ChIP-chip of the regulators of white-opaque switching and core regulatory network established in white cells. (A) ChIP-chip data for Ahr1 (red), Czf1 (green), Efg1 (blue) and their respective controls (grey) at the upstream regions for the six core regulators of white-opaque switching in white cells. Open reading frames are represented by yellow boxes, lighter yellow represents the untranslated region, genes above the bold line are transcribed in the sense direction and genes below the bold line are transcribed in the antisense direction. The x-axis represents ORF chromosomal locations and the y-axis represents the ChIP-chip enrichment value (log2). (B) Regulatory network in white cells, based on our ChIP-chip data. Arrows represent direct binding interactions for the various transcriptional regulators.
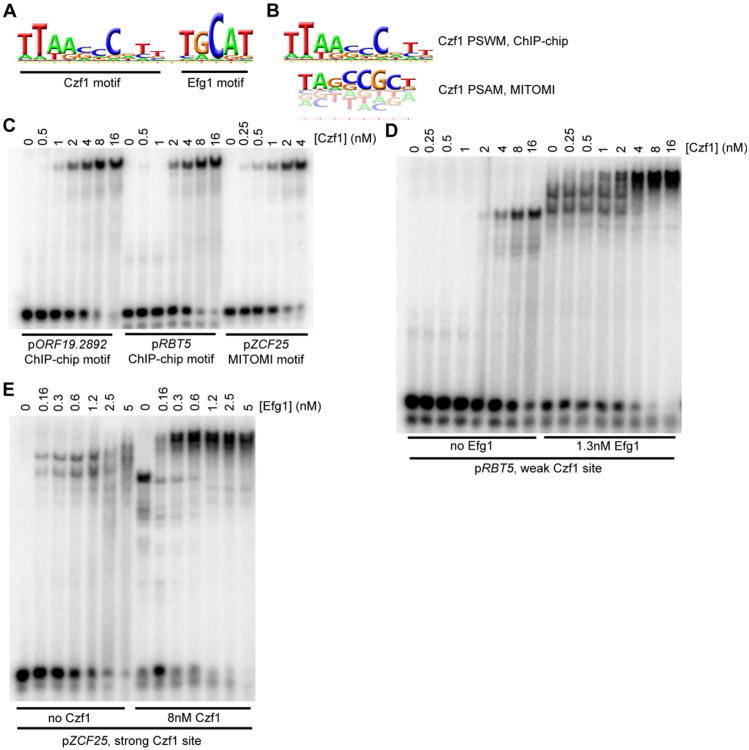
The binding data for all three regulators was further processed using MochiView (Homann and Johnson, 2010) to identify regions of maximum peak enrichment, as previously described (Cain et al., 2012; Nobile et al., 2012). These regions were then examined for cis-regulatory DNA sequences (binding motifs) corresponding to bound Czf1, Ahr1, and Efg1. The Ahr1 binding sites were enriched for the presence of the motif “TCGNYWAWWSTTGCC” (Fig. 4A, File S3), which is similar to that reported by Askew et al. (Askew et al., 2011) and can explain roughly 80% of Ahr1 binding events with a less than 10% false positive rate. Efg1 binding sites were enriched for the presence of the sequence “TGCAT” (Fig. 4B, File S3), which matches both the previously reported Efg1 binding sequence in C. albicans (Nobile et al., 2012) as well as those for its orthologs Sok2 and Phd1 in S. cerevisiae (Harbison et al., 2004; Badis et al., 2008; Zhu et al., 2009). Czf1 binding sites were enriched for the sequence “TTWRSCGCCG” (Fig. 4C, File S3) as well as for the Efg1 motif; these two sequences were consistently separated by a fixed distance (Fig. 5A), suggesting that the two proteins may act together, a hypothesis tested directly as discussed below.
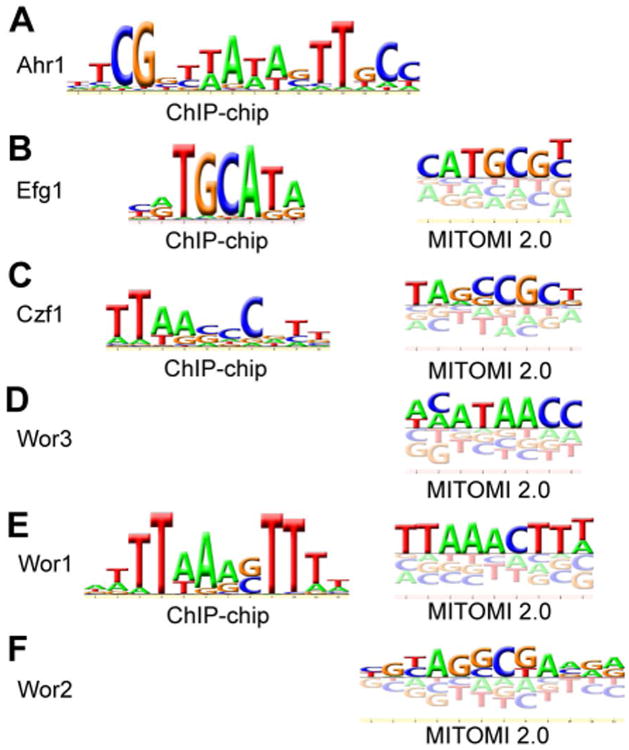
Figure 4.
DNA motifs recognized by the six regulators of white-opaque switching. Motifs developed from ChIP-chip binding sites and MITOMI 2.0 analyses are shown. (A) Ahr1, ChIP-chip only; (B) Efg1, ChIP-chip (left) and MITOMI 2.0 (right); (C) Czf1, ChIP-chip (left) and MITOMI 2.0 (right); (D) Wor3, MITOMI 2.0 only; (E) Wor1, ChIP-chip (left) and MITOMI 2.0 (right); and (F) Wor2, MITOMI 2.0 only.
Figure 5.
Further characterization of Czf1 and Efg1 binding sites. (A) Full PSWM for Czf1 developed from the white cell ChIP-chip data, including the Efg1 site (positions 12-16). (B) Comparison of ChIP-chip developed PSWM (top) and MITOMI 2.0 developed PSAM (bottom) for Czf1. (C) EMSAs for DNA fragments containing instances of the ChIP-chip motif (pORF19.2892, pRBT5) or the MITOMI 2.0 motif (pZCF25), performed using a 6×His-Czf1 construct. (D-E) EMSAs examining cooperative interactions between Czf1 and Efg1 at binding sites taken from either pRBT5 (D) or pZCF25 (E). Levels of one protein were increased stepwise (Czf1 in panel D, Efg1 in panel E) while the other protein was either absent or held at a fixed concentration (Efg1 in panel D, Czf1 in panel E).
As an independent validation of the cis-regulatory motifs generated from the ChIP-chip data for Efg1 and Czf1, we measured sequence binding preferences for both proteins in vitro using a recently developed mechanically induced trapping of molecular interactions (MITOMI 2.0) technique (Maerkl and Quake, 2007; Fordyce et al., 2010). In this approach, an in vitro translated transcriptional regulator is presented with all possible 8-mer DNA sequences and its binding to each is quantitatively monitored in vitro. This approach therefore examines, in an unbiased way, the inherent DNA-binding specificity of a regulator. While the Efg1 motif produced by this method was similar to the motif developed from the ChIP-chip data (Fig. 4B, Files S3 and S4), we observed subtle differences between the Czf1 motifs derived from ChIP-chip and from MITOMI 2.0 (Figs. 4C and 5B, Files S3 and S4). By examining Czf1 binding sites more closely, we observed that the ChIP-chip derived Czf1 motif frequently co-occurred with a consensus Efg1 motif, while the MITOMI 2.0-derived Czf1 motif was more common when a “non-optimal” Efg1 motif was present (Table S1C). We hypothesized that the ChIP-chip-derived motif was weighted towards lower affinity Czf1 sites (given the presence of Efg1 adjacent to these sites) and that the MITOMI 2.0 motif represented the optimal binding site.
To test this hypothesis, we expressed and purified full length Czf1 and performed electrophoretic mobility shift assays (EMSAs) with DNA fragments containing the two motifs. The observed dissociation constant (Kd) for binding was four to ten times tighter for Czf1 binding to the MITOMI 2.0-derived motif than to the ChIP-chip-derived motif (Fig. 5C), indicating that the MITOMI 2.0-derived motif represents the optimal Czf1 recognition sequence. Interactions between Czf1 and Efg1 in yeast two-hybrid experiments have been previously reported (Giusani et al., 2002; Vinces et al., 2006; Noffz et al., 2008; Petrovska and Kumamoto, 2012), and to test whether cooperative binding with Efg1 could account for the differences in affinities of the two Czf1 binding sites, we expressed and purified full length Efg1 and performed additional EMSAs using both proteins. We observed greater occupancy on DNA for both Efg1 and Czf1 when fixed amounts of the other regulator were included in the binding reactions, confirming that the two proteins bind to DNA cooperatively (Figs. 5D and 5E), and that they do so in the absence of other proteins. This cooperative interaction between Czf1 and Efg1 explains the non-optimal nature of the Czf1 sequence found at a majority of Czf1 binding sites and resolves the difference between the ChIP-chip and MITOMI 2.0-derived motifs (Table S1C).
The logic of the white cell circuit
In white cells, the “core regulatory circuit” consists of Efg1 binding to its own upstream region and to those of WOR1 and WOR2, Ahr1 binding to its own upstream region and to those of EFG1 and WOR2, and Czf1 binding to its own upstream region and to those of WOR2 and EFG1 (Fig. 3). These observations, combined with previous results (Zordan et al., 2007; Sriram et al., 2009; Askew et al., 2011), lead to a model whereby Efg1 directly represses WOR1 and both Efg1 and Ahr1 directly repress WOR2 to stabilize the white cell type and reduce switching to the opaque cell type. Czf1, on the other hand, both represses EFG1 and acts as a positive regulator of WOR2, thereby promoting the establishment of the opaque cell type.
To test this model and to identify target genes controlled by each regulator, we performed genome-wide transcriptional profiling of strains that carry homozygous deletions of EFG1, CZF1, or AHR1 (Figs. S1A-S1C, File S1). Deletion of EFG1 in white cells leads to changes in 171 transcripts, 96 of which are formally repressed by Efg1; these include 59 opaque-enriched transcripts, indicating that Efg1 directly or indirectly represses approximately one-fourth of all opaque-enriched transcripts in white cells. Our ChIP-chip data, combined with the transcriptional profiling data, reveal that Efg1 directly activates eight white-enriched transcripts, indicating that Efg1 is responsible not only for repressing the white-to-opaque transition, but also for activating white-specific transcripts (Fig. S1A). Deletion of Ahr1 in white cells results in the differential expression of 84 transcripts, 54 of which are formally repressed (including eighteen opaque-enriched genes). Only twelve of the 84 regulated transcripts are directly associated with Ahr1 binding, and ten of these are downregulated in the Ahr1 deletion. These results suggest that Ahr1 functions, like Efg1, as both an activator of the white cell program and a repressor of the opaque cell program, although fewer genes are affected (Fig. S1B). Deletion of Czf1 in white cells affects expression of an even smaller number of genes, seven (Fig. S1C). Considering the white circuit as a whole, the upstream regions of 26 additional transcriptional regulators are bound by Ahr1, Efg1 or Czf1 in white cells, three of which (RFG1, CRZ2, AAF1) are bound by all three regulators (Fig. 6A, Table S1D). This arrangement likely accounts for the large number of genes regulated in the efg1 deletion mutant strain that are not directly regulated by Efg1.
Figure 6.
Transcriptional regulators bound by the white and opaque cell networks. (A) The white cell network consists of three core regulators (Ahr1, red; Czf1, green; Efg1, blue) represented by the large circular hubs. (B) The opaque cell network consists of six core regulators (Wor1, orange; Wor2, pink; Wor3, light blue; Ahr1, red; Czf1, green; Efg1, blue) represented by the large circular hubs. For both (A) and (B), smaller circles represent other transcriptional regulators that are bound by the core network regulators. These downstream regulators are connected to their respective core regulators by white lines, indicative of a direct binding interaction assessed by ChIP-chip. Downstream regulators that are differentially regulated as determined by RNA-seq from Tuch et al. (Tuch et al., 2010) in opaque compared to white cells are shown in yellow for those upregulated in opaque cells, in light purple for those downregulated in opaque cells, and in grey for those with no change.
We conclude from this analysis that Efg1 is the major controller of the white state – its expression blocks the opaque program by repressing WOR1 and WOR2, and promotes the expression of the white-specific program, in part by activating additional “downstream” transcriptional regulators. Ahr1 formally carries out the same role (a repressor of the opaque state and an activator of the white state), although it controls fewer genes than Efg1. Finally, Czf1 is needed to control only a small number of white-specific genes; its main role is modulating the switching rate by repressing EFG1 and activating WOR2. When Czf1 is deleted, white-to-opaque switching decreases tenfold (Vinces and Kumamoto, 2007; Zordan et al., 2007).
Opaque cell circuit
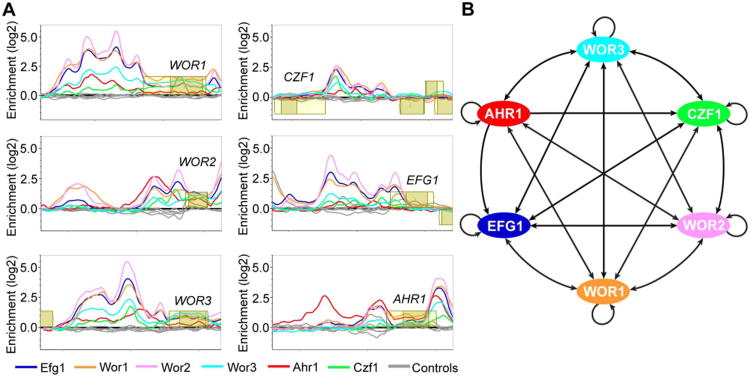
We used ChIP-chip to identify the transcriptional regulatory circuit defined by Efg1, Czf1, Wor2, and Ahr1 in the opaque cell type and combined this with the previously published ChIP-chip data generated by this lab for Wor1 (Zordan et al., 2007) and for Wor3 (Lohse et al., 2013) (Fig. 7A, Files S1, S5, and S6). Even the “core circuit” (that is, the circuit composed of only the six regulators and the interactions between them) is significantly more complex in opaque cells than in white cells, with the WOR1, WOR2, WOR3, CZF1, and EFG1 upstream regions bound by all six of the switch regulators, and the AHR1 upstream region bound by Wor1, Wor2, Wor3, and itself (Fig. 7B). Overall, the entire opaque regulatory circuit encompasses 546 intergenic regions (corresponding to 748 genes) which are bound by one or more of the six white-opaque regulators; of these, the majority are associated with Wor1 (68%), while Wor2 (42%), Ahr1 (38%), Efg1 (30%), Wor3 (15%), and Czf1 (13%) make up the rest (Fig. 2B, Files S5 and S6). This network is highly interwoven, with 45% of the 546 intergenic regions bound by at least two of the six regulators, and 32% bound by three or more.
Figure 7.
ChIP-chip of the regulators of white-opaque switching and core regulatory network established in opaque cells. (A) ChIP-chip data for Ahr1 (red), Czf1 (green), Efg1 (blue), Wor1 (orange), Wor2 (pink), Wor3 (light blue), and their respective controls (grey) at the upstream regions for the six core regulators of white-opaque switching in opaque cells. Open reading frames are represented by yellow boxes, lighter yellow represents the untranslated region, genes above the bold line are transcribed in the sense direction and genes below the bold line are transcribed in the antisense direction. The x-axis represents ORF chromosomal locations and the y-axis represents the ChIP-chip enrichment value (log2). (B) Regulatory network in opaque cells, based on our ChIP-chip data as well as that from Zordan et al. (Zordan et al., 2007) and Lohse et al. (Lohse et al., 2013). Arrows represent direct binding interactions for the various transcriptional regulators.
To explore the overlapping nature of this network in more detail, we classified all binding events for the six core regulators based on the combination of regulators that were present at given intergenic regions in the opaque cell ChIP-chip experiments (see Supplemental Experimental Procedures for details). The results of this analysis are as follows: there are large numbers of sites where, of the six core regulators, only Wor1 (137) or Ahr1 (102) are bound, but many fewer sites where only Czf1 (18), Efg1 (7), Wor2 (19), or Wor3 (16) are bound. Of the 57 theoretical combinations of two or more regulators, a subset of only eight combinations comprises roughly 80% of observed binding events (Tables S1E and S1F). Specifically, the combinations of Wor1+Wor2 and Wor1+Ahr1 represent the bulk of two-regulator binding events, the Wor1+Wor2+Efg1 and Wor1+Wor2+Ahr1 combinations the bulk of three-regulator events, and the Wor1+Wor2+Efg1+Wor3 and Ahr1+Wor1+Wor2+Efg1 combinations the bulk of four-regulator events. The Ahr1+Wor1+Wor2+Wor3+Efg1 combination forms the majority of the five-regulator events. In addition, there are a number of intergenic regions where all six regulators are bound (Table S1E). The observed combinations are suggestive of a combinatorial hierarchy where Wor1 is the most important regulator for binding in opaque cells, followed by Wor2, Efg1, Wor3, and then Czf1; indeed the regulators may well be recruited to the intergenic regions in this order. In contrast, Ahr1 appears capable of joining at any point in this hierarchy, and, as described above, it is also observed at a significant number of upstream regions in the absence of the other five core regulators. The trends noted above are greater than would be expected by chance (Table S2A). We also note that Wor1, Wor2, Wor3, and Efg1 ChIP-chip enrichment levels are considerably higher at locations bound by four or more regulators than at those bound by three or fewer (Figs. S2A and S2B).
Opaque cell binding motif analysis
As described above in the white cell analysis section, the cis-regulatory motifs recognized by Efg1, Czf1, and Ahr1 were determined; here we turn to the opaque-specific transcriptional regulators Wor1, Wor2, and Wor3. Based on MITOMI 2.0 experiments, the optimal sequence recognized by Wor3 is “KGGTTAYK” (Lohse et al., 2013) (Fig. 4D, Files S3 and S4), that recognized by Wor1 is “WTTWARSTTT” (Fig. 4E, Files S3 and S4), and that recognized by Wor2 is “TARSCKA” (Fig. 4F, File S3 and S4). The Wor1 motif derived from the MITOMI 2.0 experiments was similar to that derived by EMSAs using purified protein and DNA (Lohse et al., 2010; Cain et al., 2012) and the MITOMI 2.0-derived Wor2 motif was confirmed using purified protein in EMSAs (Fig. S3). We then examined whether these motifs, plus the previously developed motifs for Wor3, Efg1, Ahr1, and Czf1, could explain the ChIP-chip data generated from opaque cells.
Of the six core transcriptional regulators, the Ahr1, Wor1, Wor2, Czf1, and Efg1 motifs show a high correlation with “singleton” binding events, i.e. sites where only one of the six regulators is bound (Table S2B). The correlation between a given motif and a bound regulator becomes weaker as more proteins are bound to the intergenic regions. However, there are some clear trends within this generalization. The following sets of binding events all have very strong Wor1 motifs: Wor1+Wor2, Wor1+Wor2+Efg1, Wor1+Wor2+Efg1+Wor3, Wor1+Wor2+Efg1+Wor3+Ahr1 sites and sites with all six regulators bound. In contrast, other prominent combinations (Ahr1+Wor1+Wor2, Ahr1+Wor1+Wor2+Efg1) are correlated strongly with Ahr1 sites. Finally, the Wor1+Ahr1 dual binding events have strong correlations with both the Ahr1 and Wor1 motifs (Table S2B). We note that further attempts to combine motifs from multiple regulators did not provide a significant improvement in predictive power over those noted above.
From this analysis, we conclude that the Wor1 and Ahr1 motifs are the most important determinants of higher order binding with the Wor3 and Efg1 motifs making contributions to only a portion of the sites bound by four or more regulators. Further work will be required to rigorously define the detailed “rules” of binding that would provide a truly deterministic model; we note, nonetheless, that the simple analysis described above provides significant predictive value.
Distinct properties of the opaque cell circuit
The most noticeable difference in the regulatory circuit of opaque cells compared to that of white cells is the large increase in the number of intergenic regions bound by the regulators (546 compared to 131). Moreover, nearly half (247 of 546, 45%) of intergenic regions bound in opaque cells are bound by multiple transcriptional regulators, compared to less than a fifth of intergenic regions bound in white cells (23 of 131, 18%). Even the white cell regulators undergo an expansion in the number of targets they bind in opaque cells. Although levels of Efg1 decrease four-fold in opaque cells relative to white cells, the protein is found at more than three times as many genomic locations (169 compared to 49). Likewise, levels of Ahr1 do not change, but it is found at three times as many genomic locations in opaque cells as in white cells (212 compared to 71). Finally, the number of genomic locations bound by Czf1 roughly doubles relative to white cells (72 compared to 39) (Tables S1A and S1E). We note that increases in binding events in the opaque circuit relative to the white circuit are largely additive; that is, the white cell circuit expands. However, although additive in structures, these circuits are not additive in their transcriptional outputs. For example, there are approximately 168 white-enriched target genes and each gene is downregulated when cells switch to the opaque form.
Higher order binding events correlate with white or opaque cell type-specific regulation
To explain how the output of the circuit (gene expression) relates to its structure (binding events), we next determined whether intergenic regions bound by multiple regulators correspond to genes differentially regulated between the white and opaque cell types (Supplemental Experimental Procedures). Intergenic regions bound by at least four regulators were correlated with cell type-specific expression relative to an equivalently sized control set of intergenic regions (hyper-geometric test, P<1E-6, Table S2C). Lesser, although still statistically significant, increases were also found for regions bound by at least three or at least two regulators (P<1E-3, Table S2C). We also found that the intergenic regions bound by a given regulator in opaque cells were roughly twice as likely (relative to an equivalently sized control set of intergenic regions) to be adjacent to genes that were mis-regulated when that regulator was deleted. These increases were statistically significant for genes mis-regulated in wor2, ahr1, czf1, and efg1 deleted opaque strains (hyper-geometric test, P<0.01, Table S2D). Thus, there is a strong correlation between regulator binding and changes in expression between white and opaque cell types. We note that our analyses (full genome ChIP-chip and transcriptional profiling) were carried out in only one condition (rich medium) and there is no reason to expect the correlation to be perfect; based on other circuits in fungi, we predict that different environmental conditions will alter the spectrum of white- and opaque-enriched genes without changing the binding pattern of the core regulators. For example, the Saccharomyces cerevisiae regulatory protein Gal4 remains bound to its control regions in both non-inducing (raffinose) and inducing (galactose) medium, whereas the expression of genes controlled by Gal4 various enormously between these two conditions (Ren et al., 2000; for review, see Traven et al., 2006).
Opaque cell logic
Our analysis also shows that the opaque cell network includes 65 additional transcriptional regulators whose intergenic regions are bound by at least one of the core switch regulators (Fig. 6B). Roughly half of these 65 regulators are differentially regulated between white and opaque cells (Table S1G). Of these 65, the upstream regions of six are bound by all six members of the circuit in opaque cells; these genes are BRG1, RFG1, NRG1, CRZ2, AAF1, and ORF19.6713 as well as the core regulators WOR1, WOR2, WOR3, CZF1, and EFG1. These results indicate that many of the features that distinguish white cells from opaque cells are specified by transcriptional regulators that lie “downstream” of the core switch regulators. We believe this observation explains why many of the white-opaque enriched genes are not bound directly by the core regulators we have described. We have constructed strains deleted for three of these other regulators (BRG1, CRZ2, and AAF1) and tested them for effects on white-opaque switching frequencies. Unlike the six “core” regulators, none of these three significantly affect white to opaque switching rates when deleted (Table S2E). This observation supports our view that these regulators function primarily to specify features of the opaque state and lie downstream of the actual switching circuit. A full examination of the transcriptional changes in opaque cells following deletion of WOR2, CZF1, EFG1, and AHR1 can be found in Supplemental Information (Figs. S1D-S1G).
Discussion
Using a combination of approaches including full genome ChIP-chip, full genome transcriptional analysis, MITOMI 2.0, and biochemical experiments with purified proteins, we have described a circuit of six transcription regulators and 748 target genes that is responsible for the white-opaque switch in C. albicans. The switch from the white to the opaque cell type involves a major rewiring of the cell circuitry, leading to the differential expression of one in every six genes in the genome. We have shown that the core of this network in opaque cells is made up of many interactions among the six white-opaque switch regulators. For example, in opaque cells, Wor1 binds to its own regulatory region and those of WOR2, EFG1, CZF1, AHR1, and WOR3 (Fig. 7); the other five regulators show similar patterns. This network appears resistant to many perturbations. For example, deletion of AHR1, CZF1, EFG1 or WOR3 affects switching rates and results in the improper expression of portions of the white or opaque cell transcriptional programs, but does not prevent the formation of either cell type or even the proper expression of a majority of the cell type-specific programs. Even Wor2, necessary for the heritable transmission of the opaque state from mother to daughter, is dispensable when WOR1 is ectopically overexpressed. Our previous work (Lohse and Johnson, 2010) indicated that the circuit was very slow to respond temporally to major changes in the levels of the core regulators. Consistent with the idea that the network is stable to perturbations, the white and opaque states themselves are very stable with switching rates of approximately 1 in 104 generations at 25°C. We believe that this stability to perturbation results from the large number of interactions between the regulators including multiple positive feedback loops that are predicted to maintain a consistent output (opaque or white cell type) of the circuit. On a broader level, the interconnected properties of the white-opaque switch network may allow the two cell types to be stable under the wide variety of different environmental conditions characteristic of the diverse host niches that C. albicans can occupy.
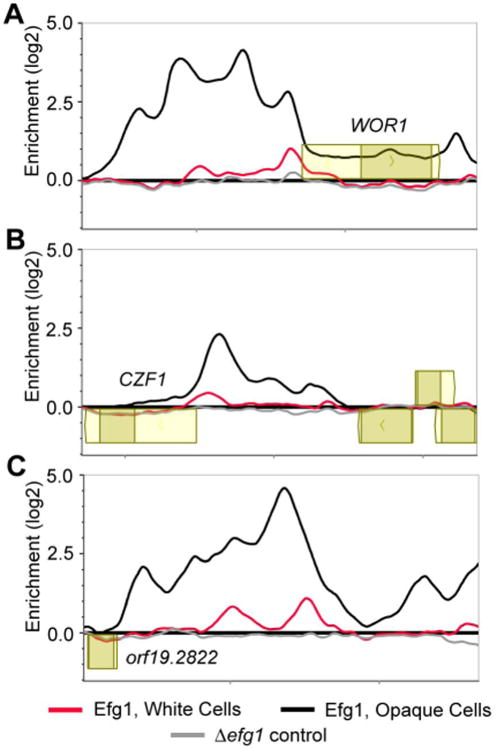
Our analysis shows that, although switching between the white and opaque cell type is “all-or-none,” the binding patterns of the regulators in white and opaque cells are not mutually exclusive, as might have been predicted. For example, in the bacteriophage lambda switch, the occupancies of the right operator by the lambda repressor and cro are mutually exclusive (Ptashne, 2004). In contrast, the white circuit (which is relatively small) is incorporated into the much larger opaque circuit. For example, Efg1, whose expression level is reduced four-fold in opaque cells, remains bound to most of its white-cell targets, but roughly triples its number of bound upstream regions in opaque cells. Even the nature of the binding by these six regulators differs between the two cell types. In white cells, we observed narrow, tapered peaks while in opaque cells we observed broad peaks that extended across entire intergenic regions, some several kilobases in length (Fig. 8). The narrow peaks observed for white cells from our ChIP-chip data are typical of ChIP-chip binding events we have observed for other C. albicans regulators (See, for example Nobile et al., 2009; Nobile et al., 2012), whereas the broad peaks we reproducibly observed for opaque cells appear unusual compared to those of other systems.
Figure 8.
Efg1 binding changes between the white and opaque cell types. (A-C) ChIP-chip for Efg1 in white cells (red), opaque cells (black), and ΔΔefg1 (grey, control) strains at the (A) WOR1, (B) CZF1, or (C) ORF19.2822 upstream regions. Open reading frames are represented by yellow boxes, lighter yellow boxes indicate untranslated regions, genes above the bold line are transcribed in the sense direction and ones below the bold line are transcribed in the antisense direction. The x-axis represents chromosomal locations and the y-axis represents the ChIP-chip enrichment value (log2).
Our data, taken as a whole, indicates that the increase in the size of the network, and much of the higher order binding (that is, the binding of multiple regulators to the same intergenic region) is due to large assemblies of regulators on target genes, nucleated largely by Wor1, which is 50-fold upregulated in opaque cells. The Wor1 motif is the most important determinant of higher order binding with the Ahr1, Wor3, and Efg1 motifs making contributions to only a portion of the sites bound by four or more regulators. Their assembly on control regions in opaque cells is a complex function of motif strengths, motif numbers, and the spacing between motifs. We propose that this assembly contributes to the all-or-none nature of the switch and to the stability of the opaque cell type through many cycles of DNA replication.
Although we do not yet know how these assemblies form on intergenic regions in opaque cells, we can offer a speculation. Sequences containing polyglutamine stretches have been implicated in stabilizing protein-protein interactions and in some cases, protein aggregation (Schaefer et al., 2012). We note that five of the switch regulators (all except Ahr1) contain one or more polyglutamine stretches, and our working hypothesis is that these sequences promote aggregation of these proteins and DNA, nucleated largely by Wor1 binding.
A remarkable feature of the white-opaque circuitry is its topological similarity to other complex circuits. These include the C. albicans biofilm regulatory network (Nobile et al., 2012) and the network regulating pseudohyphal growth in S. cerevisiae (Borneman et al., 2006; Borneman et al., 2007; Cain et al., 2012). Each of these networks contain at least six “core” regulators, and they regulate each other in addition to hundreds of target genes including dozens of “downstream” transcriptional regulators. This same basic type of network structure has also been observed in formation of induced pluripotent stem cells from differentiated tissues (Hochedlinger and Plath, 2009; Scheper and Copray, 2009) and in the hematopoietic and embryonic stem cell differentiation pathways (Wilson et al., 2010; Young, 2011). All these networks share a group of “master” or “core” regulators, that appear to regulate each other in a complex, almost circular (as opposed to linear) pathway. The upstream regions of target genes in these networks are typically bound by multiple master regulators, resulting in a highly connected network that contains hundreds of feedback and feed-forward loops. These complex fungal and mammalian networks do not appear to have evolved from a common ancestral network; for example, the fungal networks utilize several regulators that are not present in metazoans and even the target genes of the different fungal networks are not highly conserved across the fungal kingdom. Thus, it seems likely that each of the fungal and mammalian networks discussed above independently evolved a highly interconnected, very similar network structure.
The S. cerevisiae pseudohyphal growth circuit controls cell elongation and cell division, the C. albicans biofilm network orchestrates biofilm development, and the C. albicans white-opaque switching network allows two different cell types to form, each of which is stably transmitted to subsequent generations. Of these three circuits, only white-opaque switching is known to exhibit cell memory. We do not presently know which collection of parameters in the white-opaque switch network is responsible for the “memory” of the two cell states. However, the network structure contains so many feed-forward loops (3,225 for the opaque network and 36 for the white network) that it seems likely that “cell memory” could easily be generated from such a circuit: as cells divide, daughters will inherit molecules of the “master” regulators, which can then, through the numerous positive feedback loops, re-excite the circuit. Indeed, modeling of an earlier, much simpler version of the white-opaque circuit illustrates that such a circuit can, in principle, produce cell memory (Sriram et al., 2009). We propose that this type of circuit structure can account for other cases where cells “remember” who they are through many cell divisions. However, we know that cell memory (at least over many cell divisions) is not an inevitable consequence of this type of network structure, as the biofilm and pseudohyphal growth programs are readily reversible.
The white-opaque network is central to C. albicans biology and is believed to enable C. albicans to occupy different niches in its mammalian host. It is remarkable that the network structure controlling this process resembles those found in a wide variety of other species and that these network structures apparently arose convergently. Despite their similar overall structures, the outputs of these networks vary enormously, as they depend on connections between regulators and specific target genes. We further propose that individual parameters of such a network determine whether a transcriptional state can be stably passed on to descendent cells, as it is in white-opaque switching.
Experimental Procedures
All methods are briefly described below. For additional experimental details, please see Supplemental Experimental Procedures.
Growth Conditions
All cells were grown on synthetic complete media supplemented with 100 μg mL−1 uridine and 2% glucose (synthetic dextrose + amino acids + uridine) unless otherwise indicated. Growth was at room temperature (25°C) unless otherwise noted.
In order to confirm that homogenous cell populations were present before microarrays and ChIP-chip experiments were performed, white and opaque cell populations were assessed by microscopy.
Strain construction
A list of strains used in this study can be found in Table S3A. Deletion strains were generated as described previously (Hernday et al., 2010). The WOR1, WOR2, WOR3, CZF1, and EFG1 deletion strains have been previously reported (Zordan et al., 2006; Lohse et al., 2013). C-terminal myc-tagged transcription factor strains were generated using plasmid pADH34 (Nobile et al., 2009) and PCR-directed genomic integration as described previously (Hernday et al., 2010).
Ectopic WOR1 expression strains were generated by cloning the open reading frame behind the pMET3 upstream region in pADH33 (Lohse et al., 2013). Following digestion with NcoI, the SAT1-marked ectopic expression construct containing WOR1 was integrated at the RP10 locus and confirmed by colony PCR.
Plasmid construction
A list of plasmids used in this study can be found in Table S3B. A list of oligonucleotides used in this study can be found in Table S3C. Full details of plasmid construction can be found in the Supplemental Experimental Procedures .
Protein Expression and Purification
Proteins were expressed for 4 hours at 25°C with 0.1mM (Wor2) or 0.4mM (Czf1, Efg1) IPTG in 2×YT media with 0.15% glucose and 1mM MgSO4 (Wor2) or LB (Czf1 and Efg1). Protein purification of 6-His MBP-Wor2 231-343aa and 6-His Efg1 and 6-His Czf1 were performed as previously described (Lohse et al., 2010; Cain et al., 2012). In brief, cell pellets were resuspended, cells lysed by sonication, centrifuged, purified using Ni-NTA Agarose (Qiagen), concentrated, and moved into storage buffer (10 mM Tris, pH 7.4, 100 mM NaCl, 5 mM DTT, and 50% glycerol). Protein concentrations were estimated based on comparison to a dilution series of known BSA amounts on a SDS-PAGE gel.
Electrophoretic mobility shift assays
EMSAs were performed using previously described conditions (Lohse et al., 2010), with the omission of Poly(dI-dC) and a reduction of NaCl concentration to 50mM. 21bp oligonucleotides were used for Wor2 EMSAs, 36bp oligonucleotides were used for Czf1 and Efg1 EMSAs. The motif, motifs, or mutated motifs were located at the center of the oligonucleotides.
MITOMI 2.0 random library experiments
MITOMI 2.0 experiments for de novo identification of transcription factor binding sites using a pseudorandom library of DNA sequences were performed as described previously (Fordyce et al., 2010), with the following modifications. First, we used a new pseudorandom 8mer DNA library that included all possible 8mer DNA sequences within 740 oligonucleotides (Table S3C). This library was based on a previously published algorithm (Mintseris and Eisen, 2006). Second, we designed a new, smaller version of the microfluidic devices with 1568 chambers arrayed in 28 channels with 56 chambers per channel. Third, we modified the protocol for both mold and device fabrication (as described below). Finally, we printed 2 arrays per 2″×3″ SuperChip EpoxySilane glass slide (ThermoFisher Scientific). MITOMI 2.0 molding master and device fabrications were performed as previously described (Lohse et al., 2013). Position Specific Affinity Matrices (PSAMs) developed for Wor1, Wor2, Efg1, and Czf1 using MITOMI 2.0 are included in File S3.
MITOMI 2.0 binding curve experiments
Experiments assessing concentration-dependent binding to oligonucleotides containing systematic mutations of candidate “consensus” transcription factor target sites were performed largely as described previously (Fordyce et al., 2012), with final concentrations of DNA before printing set to be 10 μM, 6.7 μM, 4 μM, 3 μM, 2 μM, 1.3 μM, 0.9 μM, and 0.4 μM. The oligonucleotide sequences used for binding curves are provided in Table S3C. Single-site binding model fits shown are from globally fitting all binding curves simultaneously for Czf1, Wor1, and Wor2 transcription factors. Experiments assessing concentration-dependent binding for Efg1 protein did not adequately capture both the linear and saturated binding regimes; therefore, we chose an arbitrary but reasonable constant maximum value for measured fluorescence intensity ratios and determined relative Ka values from individual single-site binding model fits. The “consensus” binding sites were developed based on a combination of existing ChIP-chip and random library MITOMI 2.0 experimentally developed sites. Binding curves for Wor1, Wor2, Efg1, and Czf1 in these experiments are included in File S4, and the related PSAMs are included in File S3.
Microarrays
Cultures for gene expression microarray analysis were harvested during mid-log phase by centrifugation and pellets were snap-frozen in liquid nitrogen prior to RNA extraction. See Supplemental Experimental Procedures for details. Equal amounts of white vs. opaque or wild-type vs. mutant cDNA were hybridized to custom-designed Agilent 8×15k microarrays (AMADID #020166), and scanned using a Genepix 4000B scanner (Axon/Molecular Devices). Data was extracted using GenePix Pro version 5.1 and normalized by Global Lowess normalization using the Goulphar script (Lemoine et al., 2006) for R (The R foundation for Statistical Computing). Transformations (i.e. white vs. opaque or wild-type vs. deletion strain) were performed prior to the extraction of median differential expression values for each ORF. Differential expression was determined by using a 2-fold cutoff. Two biological replicates were performed for each condition. Raw gene expression array data are available at the Gene Expression Omnibus (www.ncbi.nlm.nih.gov/geo, accession #GSE42134). The analyzed gene expression array data are included in File S1.
ChIP-chip
Cultures for chromatin immunoprecipitation were harvested during mid-log phase by centrifugation. Chromatin immunoprecipitation, strand-displacement amplification, dye-coupling and hybridization to a custom 1×244k Agilent tiling microarray (AMADID #016350) was performed as described previously (Hernday et al., 2010). Chromatin shearing was consistent between samples for white and opaque strains. See Supplemental Experimental Procedures for details. Arrays were scanned using a Genepix 4000B scanner (Axon/Molecular Devices) and processed as described previously (Nobile et al., 2012) using the thresholds described in Supplemental Experimental Procedures . ChIP enrichment values for each target ORF were selected from the most highly-enriched peak within each bound intergenic region. Raw ChIP-chip data are available at the Gene Expression Omnibus (www.ncbi.nlm.nih.gov/geo, accession # GSE42837). The analyzed ChIP-chip data are included in File S1. Plots of the areas of peak enrichment for each regulator are included in Files S2, S5, and S6.
Motif finding
Motif finding from ChIP-chip peak calls was performed as previously described (Nobile et al., 2012). In brief, we used the “Motif Finder” utility in MochiView v1.45 to find over-represented sequences in the 500bp maximum enrichment location sets for Ahr1, Czf1, and Efg1 in white cells. We used the same technique to find over-represented sequences in the previously reported 500bp maximum enrichment location set for Wor1 in opaque cells (Zordan et al., 2007; Lohse et al., 2013). The Position Specific Weight Matrices (PSWMs) developed for these regulators are included in File S3.
Analysis of genome-wide data
Unless otherwise noted, all of the analysis described was performed in MochiView following the procedures described in Supplemental Experimental Procedures.
We used both the previously published RNA-seq data (Tuch et al., 2010) as well as the newly generated microarray data for comparing binding to white-opaque regulation (Lohse et al., 2013). Both of these datasets are included in File S1.
Motif enrichment at binding sites
Examination of the ability of the various regulator motifs to explain the in vivo binding sites used a previously described procedure (Lohse et al., 2013). The analysis was performed using the ChIP-chip developed PSWM motif for Ahr1 and MITOMI 2.0 PSAM motifs for the other regulators (File S3). See Supplemental Experimental Procedures for details.
Supplementary Material
Acknowledgments
We are grateful to Oliver Homann for developing MochiView, Weihan Li for help on the binding overlap among the six regulators, Victor Hanson-Smith for advice on bioinformatics and statistics, Trevor Sorrells for help with the network motif analysis of the white and opaque cell circuits, and Richard Bennett and Liron Noiman for critically reading the manuscript. We thank Sudarsi Desta, Jeanselle Dea, and Jorge Mendoza for technical assistance. This study was supported by NIH grants R01AI049187 (A.D.J.), F32AI071433 (A.D.H.), and K99AI100896 (C.J.N.), and the Howard Hughes Medical Institute (J.L.D., P.M.F.). The content is the sole responsibility of the authors and does not necessarily represent the views of the NIH or HHMI.
Abbreviations
- EMSAs
Electrophoretic Mobility Shift Assays
- ChIP-chip
Genome-wide Chromatin Immunoprecipitation
- MITOMI 2.0
Mechanically Induced Trapping of Molecular Interactions
- PSAMs
Position Specific Affinity Matrices
- PSWMs
Position Specific Weight Matrices
References
- Askew C, Sellam A, Epp E, Mallick J, Hogues H, Mullick A, et al. The zinc cluster transcription factor Ahr1p directs Mcm1p regulation of Candida albicans adhesion. Mol Microbiol. 2011;79:940–953. doi: 10.1111/j.1365-2958.2010.07504.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badis G, Chan ET, van Bakel H, Pena-Castillo L, Tillo D, Tsui K, et al. A library of yeast transcription factor motifs reveals a widespread function for Rsc3 in targeting nucleosome exclusion at promoters. Mol Cell. 2008;32:878–887. doi: 10.1016/j.molcel.2008.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett RJ, Uhl MA, Miller MG, Johnson AD. Identification and characterization of a Candida albicans mating pheromone. Mol Cell Biol. 2003;23:8189–8201. doi: 10.1128/MCB.23.22.8189-8201.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borneman AR, Leigh-Bell JA, Yu H, Bertone P, Gerstein M, Snyder M. Target hub proteins serve as master regulators of development in yeast. Genes Dev. 2006;20:435–438. doi: 10.1101/gad.1389306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borneman AR, Zhang ZD, Rozowsky J, Seringhaus MR, Gerstein M, Snyder M. Transcription factor binding site identification in yeast: a comparison of high-density oligonucleotide and PCR-based microarray platforms. Funct Integr Genomics. 2007;7:335–345. doi: 10.1007/s10142-007-0054-7. [DOI] [PubMed] [Google Scholar]
- Cain CW, Lohse MB, Homann OR, Sil A, Johnson AD. A conserved transcriptional regulator governs fungal morphology in widely diverged species. Genetics. 2012;190:511–521. doi: 10.1534/genetics.111.134080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fordyce PM, Gerber D, Tran D, Zheng J, Li H, DeRisi JL, Quake SR. De novo identification and biophysical characterization of transcription-factor binding sites with microfluidic affinity analysis. Nat Biotechnol. 2010;28:970–975. doi: 10.1038/nbt.1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fordyce PM, Pincus D, Kimmig P, Nelson CS, El-Samad H, Walter P, Derisi JL. Basic leucine zipper transcription factor Hac1 binds DNA in two distinct modes as revealed by microfluidic analyses. Proc Natl Acad Sci USA. 2012;109:E3084–3093. doi: 10.1073/pnas.1212457109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiger J, Wessels D, Lockhart SR, Soll DR. Release of a potent polymorphonuclear leukocyte chemoattractant is regulated by white-opaque switching in Candida albicans. Infect Immun. 2004;72:667–677. doi: 10.1128/IAI.72.2.667-677.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giusani AD, Vinces M, Kumamoto CA. Invasive filamentous growth of Candida albicans is promoted by Czf1p-dependent relief of Efg1p-mediated repression. Genetics. 2002;160:1749–1753. doi: 10.1093/genetics/160.4.1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harbison CT, Gordon DB, Lee TI, Rinaldi NJ, Macisaac KD, Danford TW, et al. Transcriptional regulatory code of a eukaryotic genome. Nature. 2004;431:99–104. doi: 10.1038/nature02800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernday AD, Noble SM, Mitrovich QM, Johnson AD. Genetics and molecular biology in Candida albicans. Methods Enzymol. 2010;470:737–758. doi: 10.1016/S0076-6879(10)70031-8. [DOI] [PubMed] [Google Scholar]
- Hochedlinger K, Plath K. Epigenetic reprogramming and induced pluripotency. Development. 2009;136:509–523. doi: 10.1242/dev.020867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homann OR, Johnson AD. MochiView: versatile software for genome browsing and DNA motif analysis. BMC Biol. 2010;8:49. doi: 10.1186/1741-7007-8-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang G, Wang H, Chou S, Nie X, Chen J, Liu H. Bistable expression of WOR1, a master regulator of white-opaque switching in Candida albicans. Proc Natl Acad Sci USA. 2006;103:12813–12818. doi: 10.1073/pnas.0605270103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson A. The biology of mating in Candida albicans. Nat Rev Microbiol. 2003;1:106–116. doi: 10.1038/nrmicro752. [DOI] [PubMed] [Google Scholar]
- Kvaal C, Lachke SA, Srikantha T, Daniels K, McCoy J, Soll DR. Misexpression of the opaque-phase-specific gene PEP1 (SAP1) in the white phase of Candida albicans confers increased virulence in a mouse model of cutaneous infection. Infect Immun. 1999;67:6652–6662. doi: 10.1128/iai.67.12.6652-6662.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kvaal CA, Srikantha T, Soll DR. Misexpression of the white-phase-specific gene WH11 in the opaque phase of Candida albicans affects switching and virulence. Infect Immun. 1997;65:4468–4475. doi: 10.1128/iai.65.11.4468-4475.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan CY, Newport G, Murillo LA, Jones T, Scherer S, Davis RW, Agabian N. Metabolic specialization associated with phenotypic switching in Candida albicans. Proc Natl Acad Sci USA. 2002;99:14907–14912. doi: 10.1073/pnas.232566499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lassak T, Schneider E, Bussmann M, Kurtz D, Manak JR, Srikantha T, et al. Target specificity of the Candida albicans Efg1 regulator. Mol Microbiol. 2011;82:602–618. doi: 10.1111/j.1365-2958.2011.07837.x. [DOI] [PubMed] [Google Scholar]
- Lemoine S, Combes F, Servant N, Le Crom S. Goulphar: rapid access and expertise for standard two-color microarray normalization methods. BMC Bioinformatics. 2006;7:467. doi: 10.1186/1471-2105-7-467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohse MB, Hernday AD, Fordyce PM, Noiman L, Sorrells TR, Hanson-Smith V, et al. Identification and characterization of a previously undescribed family of sequence-specific DNA-binding domains. Proc Natl Acad Sci USA. 2013;110:7660–7665. doi: 10.1073/pnas.1221734110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohse MB, Johnson AD. Differential phagocytosis of white versus opaque Candida albicans by Drosophila and mouse phagocytes. PLoS One. 2008;3:e1473. doi: 10.1371/journal.pone.0001473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohse MB, Johnson AD. White-opaque switching in Candida albicans. Curr Opin Microbiol. 2009;12:650–654. doi: 10.1016/j.mib.2009.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohse MB, Johnson AD. Temporal anatomy of an epigenetic switch in cell programming: the white-opaque transition of C. albicans. Mol Microbiol. 2010;78:331–343. doi: 10.1111/j.1365-2958.2010.07331.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohse MB, Zordan RE, Cain CW, Johnson AD. Distinct class of DNA-binding domains is exemplified by a master regulator of phenotypic switching in Candida albicans. Proc Natl Acad Sci USA. 2010;107:14105–14110. doi: 10.1073/pnas.1005911107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maerkl SJ, Quake SR. A systems approach to measuring the binding energy landscapes of transcription factors. Science. 2007;315:233–237. doi: 10.1126/science.1131007. [DOI] [PubMed] [Google Scholar]
- Miller MG, Johnson AD. White-opaque switching in Candida albicans is controlled by mating-type locus homeodomain proteins and allows efficient mating. Cell. 2002;110:293–302. doi: 10.1016/s0092-8674(02)00837-1. [DOI] [PubMed] [Google Scholar]
- Mintseris J, Eisen MB. Design of a combinatorial DNA microarray for protein-DNA interaction studies. BMC Bioinformatics. 2006;7:429. doi: 10.1186/1471-2105-7-429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morschhäuser J. Regulation of white-opaque switching in Candida albicans. Med Microbiol Immunol. 2010;199:165–172. doi: 10.1007/s00430-010-0147-0. [DOI] [PubMed] [Google Scholar]
- Nobile CJ, Fox EP, Nett JE, Sorrells TR, Mitrovich QM, Hernday AD, et al. A recently evolved transcriptional network controls biofilm development in Candida albicans. Cell. 2012;148:126–138. doi: 10.1016/j.cell.2011.10.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobile CJ, Nett JE, Hernday AD, Homann OR, Deneault JS, Nantel A, et al. Biofilm matrix regulation by Candida albicans Zap1. PLoS Biol. 2009;7:e1000133. doi: 10.1371/journal.pbio.1000133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noffz CS, Liedschulte V, Lengeler K, Ernst JF. Functional mapping of the Candida albicans Efg1 regulator. Eukaryot Cell. 2008;7:881–893. doi: 10.1128/EC.00033-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrovska I, Kumamoto CA. Functional Importance of the DNA Binding Activity of Candida albicans Czf1p. PLoS One. 2012;7:e39624. doi: 10.1371/journal.pone.0039624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porman AM, Alby K, Hirakawa MP, Bennett RJ. Discovery of a phenotypic switch regulating sexual mating in the opportunistic fungal pathogen Candida tropicalis. Proc Natl Acad Sci USA. 2011;108:21158–21163. doi: 10.1073/pnas.1112076109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ptashne M. A Genetic Switch: Phage Lambda Revisited. Cold Spring Harbor Laboratory Press; Cold Spring Harbor: 2004. p. 164. [Google Scholar]
- Pujol C, Daniels KJ, Lockhart SR, Srikantha T, Radke JB, Geiger J, Soll DR. The closely related species Candida albicans and Candida dubliniensis can mate. Eukaryot Cell. 2004;3:1015–1027. doi: 10.1128/EC.3.4.1015-1027.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren B, Robert F, Wyrick JJ, Aparicio O, Jennings EG, Simon I, et al. Genome-wide location and function of DNA binding proteins. Science. 2000;290:2306–2309. doi: 10.1126/science.290.5500.2306. [DOI] [PubMed] [Google Scholar]
- Rikkerink EH, Magee BB, Magee PT. Opaque-white phenotype transition: a programmed morphological transition in Candida albicans. J Bacteriol. 1988;170:895–899. doi: 10.1128/jb.170.2.895-899.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer MH, Wanker EE, Andrade-Navarro MA. Evolution and function of CAG/polyglutamine repeats in protein-protein interaction networks. Nucleic Acids Res. 2012;40:4273–4287. doi: 10.1093/nar/gks011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheper W, Copray S. The Molecular Mechanism of Induced Pluripotency: A Two-Stage Switch. Stem Cell Rev. 2009;5:204–223. doi: 10.1007/s12015-009-9077-x. [DOI] [PubMed] [Google Scholar]
- Slutsky B, Staebell M, Anderson J, Risen L, Pfaller M, Soll DR. “White-opaque transition”: a second high-frequency switching system in Candida albicans. J Bacteriol. 1987;169:189–197. doi: 10.1128/jb.169.1.189-197.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soll DR. Why does Candida albicans switch? FEMS Yeast Res. 2009;9:973–989. doi: 10.1111/j.1567-1364.2009.00562.x. [DOI] [PubMed] [Google Scholar]
- Soll DR, Morrow B, Srikantha T. High-frequency phenotypic switching in Candida albicans. Trends Genet. 1993;9:61–65. doi: 10.1016/0168-9525(93)90189-O. [DOI] [PubMed] [Google Scholar]
- Srikantha T, Borneman AR, Daniels KJ, Pujol C, Wu W, Seringhaus MR, et al. TOS9 regulates white-opaque switching in Candida albicans. Eukaryot Cell. 2006;5:1674–1687. doi: 10.1128/EC.00252-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sriram K, Soliman S, Fages F. Dynamics of the interlocked positive feedback loops explaining the robust epigenetic switching in Candida albicans. J Theor Biol. 2009;258:71–88. doi: 10.1016/j.jtbi.2009.01.008. [DOI] [PubMed] [Google Scholar]
- Traven A, Jelicic B, Sopta M. Yeast Gal4: a transcriptional paradigm revisited. EMBO Rep. 2006;7:496–499. doi: 10.1038/sj.embor.7400679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsong AE, Miller MG, Raisner RM, Johnson AD. Evolution of a combinatorial transcriptional circuit: a case study in yeasts. Cell. 2003;115:389–399. doi: 10.1016/s0092-8674(03)00885-7. [DOI] [PubMed] [Google Scholar]
- Tuch BB, Mitrovich QM, Homann OR, Hernday AD, Monighetti CK, De La Vega FM, Johnson AD. The Transcriptomes of Two Heritable Cell Types Illuminate the Circuit Governing Their Differentiation. PLoS Genet. 2010;6:e1001070. doi: 10.1371/journal.pgen.1001070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinces MD, Haas C, Kumamoto CA. Expression of the Candida albicans morphogenesis regulator gene CZF1 and its regulation by Efg1p and Czf1p. Eukaryot Cell. 2006;5:825–835. doi: 10.1128/EC.5.5.825-835.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinces MD, Kumamoto CA. The morphogenetic regulator Czf1p is a DNA-binding protein that regulates white opaque switching in Candida albicans. Microbiology. 2007;153:2877–2884. doi: 10.1099/mic.0.2007/005983-0. [DOI] [PubMed] [Google Scholar]
- Wang H, Song W, Huang G, Zhou Z, Ding Y, Chen J. Candida albicans Zcf37, a zinc finger protein, is required for stabilization of the white state. FEBS Letters. 2011;585:797–802. doi: 10.1016/j.febslet.2011.02.005. [DOI] [PubMed] [Google Scholar]
- Wilson NK, Foster SD, Wang X, Knezevic K, Schutte J, Kaimakis P, et al. Combinatorial transcriptional control in blood stem/progenitor cells: genome-wide analysis of ten major transcriptional regulators. Cell Stem Cell. 2010;7:532–544. doi: 10.1016/j.stem.2010.07.016. [DOI] [PubMed] [Google Scholar]
- Young RA. Control of the embryonic stem cell state. Cell. 2011;144:940–954. doi: 10.1016/j.cell.2011.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu C, Byers KJ, McCord RP, Shi Z, Berger MF, Newburger DE, et al. High-resolution DNA-binding specificity analysis of yeast transcription factors. Genome Res. 2009;19:556–566. doi: 10.1101/gr.090233.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zordan RE, Galgoczy DJ, Johnson AD. Epigenetic properties of white-opaque switching in Candida albicans are based on a self-sustaining transcriptional feedback loop. Proc Natl Acad Sci USA. 2006;103:12807–12812. doi: 10.1073/pnas.0605138103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zordan RE, Miller MG, Galgoczy DJ, Tuch BB, Johnson AD. Interlocking transcriptional feedback loops control white-opaque switching in Candida albicans. PLoS Biol. 2007;5:e256. doi: 10.1371/journal.pbio.0050256. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.