Abstract
Background
The phytohormone auxin is involved in a wide range of developmental processes and auxin signaling is known to modulate the expression of target genes via two types of transcriptional regulators, namely, Aux/IAA and Auxin Response Factors (ARF). ARFs play a major role in transcriptional activation or repression through direct binding to the promoter of auxin-responsive genes. The present study aims at gaining better insight on distinctive structural and functional features among ARF proteins.
Results
Building on the most updated tomato (Solanum lycopersicon) reference genome sequence, a comprehensive set of ARF genes was identified, extending the total number of family members to 22. Upon correction of structural annotation inconsistencies, renaming the tomato ARF family members provided a consensus nomenclature for all ARF genes across plant species. In silico search predicted the presence of putative target site for small interfering RNAs within twelve Sl-ARFs while sequence analysis of the 5′-leader sequences revealed the presence of potential small uORF regulatory elements. Functional characterization carried out by transactivation assay partitioned tomato ARFs into repressors and activators of auxin-dependent gene transcription. Expression studies identified tomato ARFs potentially involved in the fruit set process. Genome-wide expression profiling using RNA-seq revealed that at least one third of the gene family members display alternative splicing mode of regulation during the flower to fruit transition. Moreover, the regulation of several tomato ARF genes by both ethylene and auxin, suggests their potential contribution to the convergence mechanism between the signaling pathways of these two hormones.
Conclusion
All together, the data bring new insight on the complexity of the expression control of Sl-ARF genes at the transcriptional and post-transcriptional levels supporting the hypothesis that these transcriptional mediators might represent one of the main components that enable auxin to regulate a wide range of physiological processes in a highly specific and coordinated manner.
Introduction
The plant hormone auxin, indole-3-acetic acid (IAA), is a simple signaling molecule that plays a critical role in plant development and growth. This phytohormone regulates cell division and cell elongation and exerts pleiotropic effects on a wide range of developmental processes including organ differentiation, embryogenesis, lateral root initiation, apical dominance, gravitropism and phototropism, leaf elongation, shoot architecture and fruit development [1], [2], [3], [4], [5]. A critical move towards understanding the mechanisms underlying auxin action [6] happened when it was shown that the hormone coordinates plant development essentially through transcriptional regulation of genes such as Aux/IAA, Gretchen Hagen3 (GH3), Small Auxin Up RNA (SAUR) and Auxin Response Factor (ARF). It was subsequently found that these so-called early auxin-responsive genes contain in their promoter one or more copies of a conserved motif, TGTCTC or its variants, known as the auxin-responsive element (AuxRE) [7]. Experimental evidences were then provided showing that transcription factors from the ARF type specifically bind to this AuxRE to mediate the transcription of auxin responsive genes [8]. The components of the pathway linking auxin perception to gene expression are now well established indicating that ubiquitination of Aux/IAA proteins by the TIR1/AFB subunit of the SCFTIR1/AFB ubiquitin ligase leads to their degradation by the 26S proteasome thus releasing the Aux/IAA-mediated inhibition of ARFs which allows these transcription factors to modulate the expression of their target genes [9].
Three types of transcriptional regulators are required for the control of auxin-responsive genes, Auxin Response Factors (ARFs), Aux/IAAs and Topless (TPLs) [10], [11]. Members of the Aux/IAA and TPL families have been reported to function as repressors of auxin-induced gene expression [10], [12], [13], [14]. An increasing number of studies demonstrate the critical role of ARFs in a variety of developmental processes, such as embryo patterning [15], [16], leaf expansion and senescence [17], [18], [19], lateral root growth [18], [20], [21], floral organ abscission and petal growth [19], [22], fruit set and development [23], [24], [25], [26], apical hook formation [27], and various responses to environmental stimuli [28]. In addition, ARF genes are involved in the cross-talk between auxin and other hormones like gibberellins [29], ethylene [30], ABA [31] and brassinosteroid signaling [32]. A typical ARF protein consists of a conserved N-terminal B3-type DNA Binding Domain (DBD) that regulates the expression of early auxin response genes, a variable middle region (MR) that function as a transcriptional activation or repression domain (AD or RD), and a conserved C-terminal dimerization domain (CTD) that contributes to the formation of either ARF/ARF homo- and hetero-dimers or ARF/Aux-IAA hetero-dimers [8], [33], [34]. The amino acid composition of MRs, located between the DBD and CTD, showed that AD types are rich in glutamine(Q), serine (S), and leucine (L) residues while RD types are rich in proline (P), serine (S), threonine (T), and glycine (G) residues [33], [35].
Since the cloning of the first AtARF1 from Arabidopsis, 22 members of this family, distributed over all five chromosomes, have been identified [33]. The functional characterization of AtARF genes was revealed by mutant analysis approach. For instance, arf1 and arf2 T-DNA insertion mutations indicated that ARF2 regulates leaf senescence [17] and floral organ abscission [19]. The arf7/arf19 double mutant had stronger auxin resistance than the single mutant and displayed phenotypes not seen in the single mutant [30]. ARF8 was reported to regulate fertilization and fruit development, and arf8-4 mutation results in the uncoupling of fruit development from pollination and fertilization giving rise to parthenocarpic fruit [23], while flowers in arf6/arf8 double mutant are arrested as infertile closed buds with short petals, short stamen filaments, undehiscent anthers and immature gynoecia [36]. In tomato, recent studies have shown the involvement of ARF genes in fruit set, development, ripening and fruit quality [3], [4], [5], [24], [25], [26], [37]. Because of these findings, members of this gene family are becoming one of the main targets towards improving fruit traits in tomato and more broadly in fleshy fruits.
Studies using different species have indicated a total of 25 ARF genes in rice (Oryza sativa), 39 ARF genes in Populus trichocarpa, 24 ARF genes in sorghum (Sorghum vulgare) and 31 ARF genes in maize [38], [39], [40], [41]. Though 21 ARF genes have been previously identified in the tomato (Solanum lycopersicum), yet, the list was incomplete and some the family members were either misannotated or suffered structural inconsistency due to the lack at that time of a high quality assembled tomato genome sequence [42], [43]. The present study, while comprehensively revising the entire ARF gene family in tomato, brings new insight on the complexity of their expression control at the post-transcriptional level. The distinctive spatio-temporal pattern of expression of tomato ARF genes and their differential responsiveness to auxin and ethylene lay the foundation for a deeper functional characterization of these transcriptional mediators.
Results
Genome-wide search for tomato ARF genes
Comprehensive identification of the ARF gene family members in the tomato was achieved using all ARF proteins previously reported from Arabidopsis and other plant species in BLAST queries on the recently published tomato genome sequence (SL2.40 genome sequence and iTAG2.30 whole protein sequences). Twenty four significant hits corresponding to non-redundant putative Sl-ARF genes were identified. PCR amplification of full length coding sequences (CDS) revealed two structural annotation inconsistencies reducing the total number of ARFs in the tomato genome to 22 (Table 1). Indeed, four sequences previously annotated as distinct ARF genes in iTAG2.30 corresponded to C-terminal or N-terminal parts of two ARF proteins (Solyc12g006340/Solyc00g196060; Solyc11g013480/Solyc11g013470). The mapping of tomato RNA-Seq data allowed to further improve the annotation of tomato ARFs by identifying the 3′ and/or 5′ UTR regions for 13 Sl-ARF genes (Table 1). All Sl-ARF proteins were found to contain a typical DBD domain (Figure 1A) as revealed by the Pfam analysis tool (http://pfam.sanger.ac.uk/). The molecular weight of the deduced Sl-ARF proteins showed a large variation ranging from 68 to 126 kDa (Table 1). Of particular note, Sl-ARF6B contains a premature stop codon in the region corresponding to the DBD domain and is therefore likely to be a pseudo-gene whose expression at the protein level is not expected (data not shown). Using cNLS Mapper, nuclear localisation signals (NLS) were also identified in all of Sl-ARFs (data not shown).
Table 1. Sl-ARF gene family in tomato.
| Generic namea | Aliasb | CDS lengthc | Str | Chr | Domainsd | Name in Wu et al.e | improvementf | New locationg |
| Sl-ARF1 | Solyc01g103050 | 1965 | + | 1 | B3,ARF,AUX/IAA,SPL-Rich RD | SlARF1(HM061154.1) | - | |
| Sl-ARF2A | Solyc03g118290 | 2541 | + | 3 | B3,ARF,AUX/IAA,SPL-Rich RD | SlARF2(DQ340255.1) | - | |
| Sl-ARF2B | Solyc12g042070 | 2490 | - | 12 | B3,ARF,AUX/IAA,SPL-Rich RD | SlARF11(HM143940.1) | 5′, 3′ UTR | 42538600..42544937 |
| Sl-ARF3 | Solyc02g077560 | 2244 | + | 2 | B3,ARF,SL/G-Rich RD | SlARF3(DQ340254.1) | - | |
| Sl-ARF4 | Solyc11g069190 | 2436 | - | 11 | B3,ARF,AUX/IAA,SPL-Rich RD | SlARF4(DQ340259.1) | 5′, 3′ UTR | 50900912..50910023 |
| Sl-ARF5 | Solyc04g081240 | 2793 | - | 4 | B3,ARF,AUX/IAA,QSL-Rich AD | SlARF5(HM195248.1) | - | |
| Sl-ARF6A | Solyc12g006340(Nter);Solyc00g196060(Cter) | 2643 | −/+ | 12/0 | B3,ARF,AUX/IAA,QSL-Rich AD | SlARF6(HM594684.1) | 5′ UTR, CDS | 857256..859656(Nter) |
| Sl-ARF6B | Solyc07g043620 | 2673 | - | 7 | B3,ARF,QSL-Rich AD | SlARF6-1(NM_001247611.1) | 5′UTR | 54884781..54890560 |
| Sl-ARF7A | Solyc07g016180 | 3339 | - | 7 | B3,ARF,AUX/IAA,QSL-Rich AD | SlARF19(HM130544.1) | - | |
| Sl-ARF7B | Solyc05g047460 | 3294 | - | 5 | B3,ARF,AUX/IAA,QSL-Rich AD | SlARF19-1(HM565130.1) | 5′UTR | 58050744..58057040 |
| Sl-ARF8A | Solyc03g031970 | 2535 | + | 3 | B3,ARF,AUX/IAA,QSL-Rich AD | SlARF8-1(HM560979.1) | 5′UTR | 8739535..8747501 |
| Sl-ARF8B | Solyc02g037530 | 2529 | + | 2 | B3,ARF,AUX/IAA,QSL-Rich AD | SlARF8(EF66734F2.1) | 5′UTR | 21756022..21766699 |
| Sl-ARF9A | Solyc08g082630 | 1977 | + | 8 | B3,ARF,AUX/IAA,SPL-Rich RD | SlARF9(HM037250.1) | 5′UTR | 62527409..62531812 |
| Sl-ARF9B | Solyc08g008380 | 2052 | + | 8 | B3,ARF,AUX/IAA,SPL-Rich RD | SlARF12(HM565127.1) | 5′UTR | 2807931..2812983 |
| Sl-ARF10A | Solyc11g069500 | 2100 | - | 11 | B3,ARF,AUX/IAA,SL/G-Rich RD | SlARF10(HM143941.1) | 5′, 3′ UTR | 51188434..51192539 |
| Sl-ARF10B | Solyc06g075150 | 2016 | + | 6 | B3,ARF,AUX/IAA,SL/G-Rich RD | SlARF16(HM195247.1) | 3′ UTR | 43020594..43023604 |
| Sl-ARF24 | Solyc05g056040 | 1953 | - | 5 | B3,ARF,SPL-Rich RD | SlARF13(HM565128.1); SlARF13-1(HM565129.1) | - | |
| Sl-ARF16A | Solyc09g007810 | 2085 | - | 9 | B3,ARF,AUX/IAA,SL/G-Rich RD | No | 3′ UTR | 1332230..1335760 |
| Sl-ARF16B | Solyc10g086130 | 1896 | - | 10 | B3,ARF,SL/G-Rich RD | SlARF16(NM_001247861.1) | - | |
| Sl-ARF17 | Solyc11g013480(Nter);Solyc11g013470(Cter) | 1869 | - | 11 | B3,ARF,SL/G-Rich RD | SlARF17(HQ456923) | 3′ UTR, CDS | 6495469..6511349 |
| Sl-ARF18 | Solyc01g096070 | 2058 | + | 1 | ARF,AUX/IAA,SPL-Rich RD | No | 5′UTR | 78941268..78946012 |
| Sl-ARF19 | Solyc07g042260 | 3357 | - | 7 | B3,ARF,AUX/IAA,QSL-Rich AD | SlARF7(EF121545.1) | - |
a Sl-ARF gene names
b the alias of each ARF gene in iTAG2.30 genome annotation
c Length of the corresponding Coding Sequence (CDS) in base pairs.
d Conserved Domains found in PFAM database: B3 means DNA binding domain, ARF means Auxin response Factor conserved domain, AUX/IAA means AUX/IAA dimerization domain, AD means transcriptional activation domain, RD means transcriptional repression domain.
e Corresponding names in Wu et al.; accession numbers are in the parentheses.
f Gene Model modification type: UTR means that the UTR sequence have been identified and annotated, CDS means that the the coding sequence have been corrected.
g New locations in the tomato genome version Sl2.40 taking into account the manual curation of the previous gene annotation in iTAG2.30
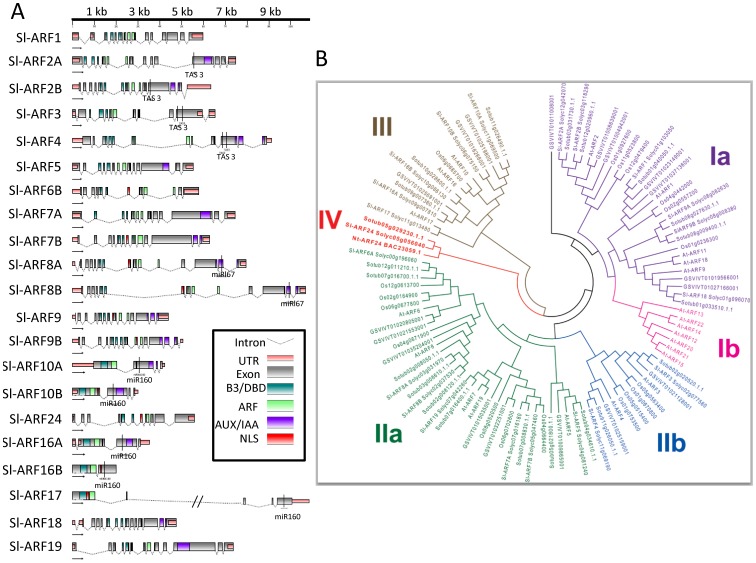
Figure 1. The ARFfamily structures in tomato and phylogenetic relationship between rice, potato, tomato, grape and Arabidopsis.
(A) The generic structures of Sl-ARF family except Sl-ARF6A. The gene size (kb) is indicated in the upper panel. The domain of Sl-ARF gene is indicated by different colours. The marker in Sl-ARF family showsSl-ARF2A, 2B, 3 and 4genes are spliced by TAS 3, Sl-ARF8A and 8B spliced by miRl67, and Sl-ARF10A, 10B, 16A, 16B and 17 spliced by miR160.(B) The unrooted tree was generated using MEGA4 program by neighbor-joining method. Bootstrap values (above 50%) from 1000 replicates are indicated at each branch. All Sl-ARFs contain a DBD (brown). Most of the Sl-ARF proteins except Sl-ARF3, 10, 24, 16 and 17 contain a carboxy-terminal domain related to the domains III and IV found in the Aux/IAA proteins (blue).Sl-ARF5, 6A, 7, 8A, 8B, 19 contains a middle region that corresponds to the predicted activation domain (green) found in some AtARFs. The remaining Sl-ARFs contains a predicted repression domain (red). Sl-ARF-6B and AtARF23 contain only a truncated DBD (B3 domain).
Building on the available tomato genome assembly sequence, the mapping of Sl-ARF genes revealed that Sl-ARF family members are distributed among the 12 tomato chromosomes. Chromosome 7 and 11 are found to harbor three ARFgenes each; chromosome 1, 2, 3, 5, 8 and 12 bear two ARFs, while each of chromosome 4, 6, 9 and 10 contains only a single ARF gene (Figure S1 in File S1). Unlike the situation prevailing in Arabidopsis, there is no evidence for tandem or segmental duplication events involving members of the tomato ARF family.
Phylogenetic relationship and consensus nomenclature for Sl-ARFs
To explore phylogenetic relationship among ARF proteins in largely distributed land plant species, a phylogenetic tree (Figure 1B) was constructed that included ARF family members from tomato, Arabidopsis, potato, grape and rice. The phylogenetic distribution revealed that ARF genes group into four major classes named Class I, II, III and IV. ARFs predicted to function as transcriptional activators, based on the presence the Q-rich activation domain in their middle region, belong to sub-class IIa (Sl-ARF5, Sl-ARF6A, Sl-ARF7A, Sl-ARF7B, Sl-ARF8A, Sl-ARF8B and Sl-ARF19) while ARFs from the remaining classes (Ia, IIb, III and IV) all harbor a repression domain in the middle region and are consequently predicted to function as transcriptional repressors.
Compared to Arabidopsis which contains 23 members, the size of the tomato Sl-ARF gene family is slightly contracted to 22 members. In order to reach a consensual nomenclature for ARF genes across species, the tomato members of this gene family were renamed, based on phylogenetic relationship and according to the numbering of the closest Arabidopsis homolog. While complying with the most complete classification available in Arabidopsis [33], the proposed nomenclature better clarifies the correspondence between ARF subclasses in plant species. Noteworthy, sub-class Ib which has no representative in the tomato, contains 7 members in Arabidopsis that are likely to derive from multiple duplications of At-ARF13 which has no ortholog in any of the plant species tested in the present study. A distinctive feature of the tomato ARF family is the expanded size of the activators' sub-class (IIa) which represents 36.5% of the ARF genes whereas the activators only account for 21.7% of Arabidopsis ARFs. Another specific feature of the tomato ARF family is the presence of Sl-ARF24 (sub-class IV) that is not found out of the Solanaceae family. Interestingly, this presumably Solanaceae-specific gene encodes an ARF protein that lacks domain III and IV involved in protein/protein interactions and required for the binding to Aux/IAA proteins. Likewise, Sl-ARF3, Sl-ARF16B and Sl-ARF17 are also deprived of domain III and IV necessary for interaction with Aux/IAAs (Figure 1A and Figure S2 in File S1). It is therefore likely that these Sl-ARFs escape the classical mechanism underlying auxin signaling which implies the sequestration of ARF proteins through interaction with Aux/IAAs.
Predicted siRNA-mediated degradation and multiple upstream ORFs in the 5′ leader sequences of tomato ARF transcripts
ARF genes have been already reported to undergo post-transcriptional regulation involving small interfering RNAs. In silico analysis at the RNA level predicted that 12 out of the 22 tomato Sl-ARFs have a putative target site for small interfering RNAs (Figure 1A). That is, Sl-ARF2A, Sl-ARF2B, Sl-ARF3 and Sl-ARF4 are predicted to be potentially targeted by TAS3; Sl-ARF6A, Sl-ARF8A and Sl-ARF8B by miR167; and Sl-ARF10A, Sl-ARF10B, Sl-ARF16A, Sl-ARF16B and Sl-ARF17 by miR160.
The uORFs are elements found in the 5′-leader sequences of specific mRNAs that modulate the translation of downstream ORFs by ribosomal stalling and inefficient re-initiation or by affecting transcript accumulation through nonsense-mediated mRNA decay pathway. Among the 19 Sl-ARFs for which the 5′ leader sequences are available in iTAG2.30 (8 members) or identified in this study (11 members), uORFs were predicted for 17 genes, ranging from 1 to 52 amino acids in size with four genes (Sl-ARF2A, Sl-ARF5, Sl-ARF10A and Sl-ARF16A) having five or more uORFs (Table S1 in File S1).The average number of uORF per Sl-ARF gene is similar in tomato (2.8/leader) and Arabidopsis (3.3/leader), indicating that tomato ARFs are suitable candidates to be regulated through translational uORFs depending mechanism
Transcriptional activation and repression activities of tomato ARFs
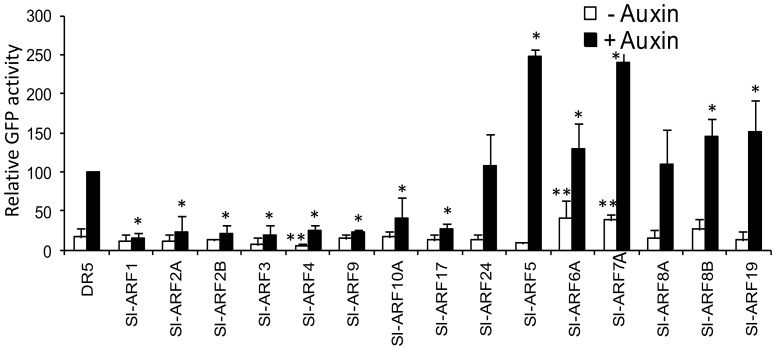
To characterize the capacity of tomato ARF proteins to in vivo activate or repress gene transcription, tobacco cells were co-transfected with an effector construct expressing the full-length coding sequence of Sl-ARFs and a reporter construct carrying the auxin-responsive DR5 promoter fused to GFP coding sequence [44]. DR5 is a synthetic auxin-responsive promoter made of 9 inverted repeats of the conserved Auxin-Responsive Element, the so-called TGTCTC box, fused to a CaMV35S minimal promoter. The DR5-driven GFP chimeric gene showed low basal activity which was induced up to 5-fold by exogenous auxin treatment (Figure 2). Co-transfection of tobacco protoplasts with the DR5::GFP reporter construct and effector plasmids expressing either Sl-ARF1, Sl-ARF2A, Sl-ARF2B, Sl-ARF3, Sl-ARF4, Sl-ARF9A, Sl-ARF10A orSl-ARF17 coding sequences, resulted in repression of the auxin-induced expression of the reporter gene (Figure 2). By contrast, co-transfection with effector constructs expressing Sl-ARF5, Sl-ARF6A, Sl-ARF7, Sl-ARF8B or Sl-ARF19 enhanced slightly the auxin-induced expression of the reporter gene. Noteworthy, with the exception of Sl-ARF6A and Sl-ARF7, these activator ARFs were unable to enhance the basal activity of the DR5 promoter in the absence of auxin treatment (Figure 2) suggesting that most ARFs require the input of an active auxin signalling for transcriptional activation of target genes.
Figure 2. Sl-ARF factors differentially regulate the expression of reporter genes driven by synthetic and native auxin-responsive promoters.
Sl-ARF factors were challenged with a synthetic auxin-responsive promoter called DR5, consisting of seven tandem copies of the AuxREtgtctc element. A transient expression using a single cell system was performed to measure the reporter gene activity. The fluorescence was measured by flux cytometry. Because of the very low basal activity of the DR5 promoter without auxin treatment, the auxin inducible fluorescence obtained by co-transformation with the promoter fused to the reporter gene and with the empty vector was standardized to 100 and taken as reference. Biological triplicates were averaged and analysed statistically using Student's t-test at (P<0.05). (*) indicates significant changes corresponding to co-transformation with effector Sl-ARF and reporter DR5-GFP constructs compared to basal activity of DR5 promoter in the absence of auxin treatment. (**) indicates significant changes for the same experiment carried out in the presence of auxin Bars indicate the SEM.
Expression of Sl-ARF genes in different tomato tissues
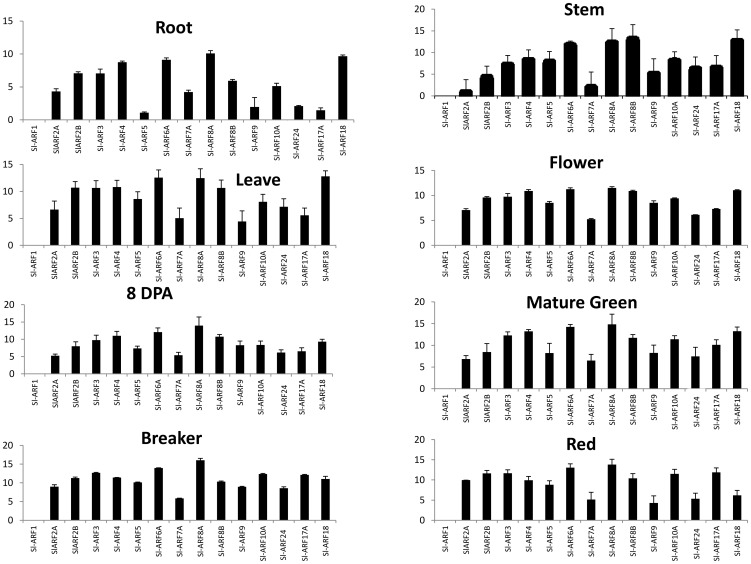
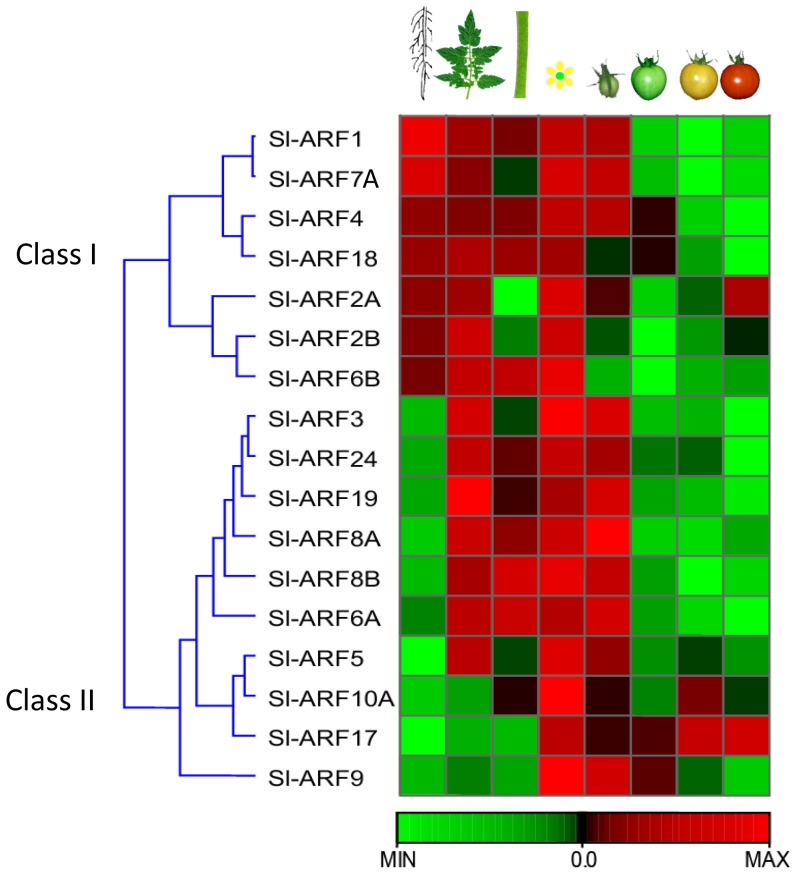
To gain clues on the physiological function of tomato ARFs, the spatio-temporal expression of individual members of the gene family was examined at the transcriptional level using qRT-PCR. Transcript accumulation could be assessed for 15 ARF genes in different tissues including root, stem, leaves, flower and fruit at various developmental stages. For the remaining 7 tomato ARF genes, transcript detection was unsuccessful in any of the samples tested suggesting their extremely low expression in these tissues. The data indicate that the expression of ARF genes is ubiquitous in all tissues with most genes being expressed in reproductive tissues suggesting their putative role in flower and fruit development (Figure 3). Heatmap representation (Figure 4) allowed the clustering of tomato ARFs into two main groups based on their expression pattern: group I (Sl-ARF1, Sl-ARF2A, Sl-ARF2B, Sl-ARF4, Sl-ARF7A, Sl-ARF6B and Sl-ARF18) are genes preferentially expressed in roots and group II Sl-ARFs in the areal part of the plant.
Figure 3. Real-time PCR expression profiles of individual Sl-ARF genes.
Total of 15 Sl-ARFgenes were performed in different tomato organs (root, stem, leaf, flower, 8DPA, Mature Green, Breaker and Red). X-axis represents different Sl-ARF genes, while Y-axis represents three relative expressions of those genes. 8DPA: 8 days after pollination, Mature Green, Breaker and Red represent different stage of the fruit development.
Figure 4. Heatmap showing Sl-ARF gene expression in different tomato tissues.
Changes in RNA accumulation in different tomato tissues (Roots, Leaves, Stems, Flowers, Early Immature Green (8 DPA), Mature Green, Breaker, Red (Breaker + 7 days) as schematically depicted above the displayed array data, are shown relative to the RNA accumulation levels in roots. Levels of down expression (green) or up expression (red) are shown on a log2 scale from the high to the low expression of each Sl-ARF gene.
Sl-ARF6B displays a very low expression in all tomato tissues analyzed and the corresponding CT values showed high variability among repeats making these Sl-ARF6B expression data not meaningful. Therefore, they were not included in Figure 3 but retained for the heat map (Figure 4) in despite of the variability between the repeats in order to give a general idea about its expression in different tissues.
Auxin and ethylene regulation of Sl-ARF genes
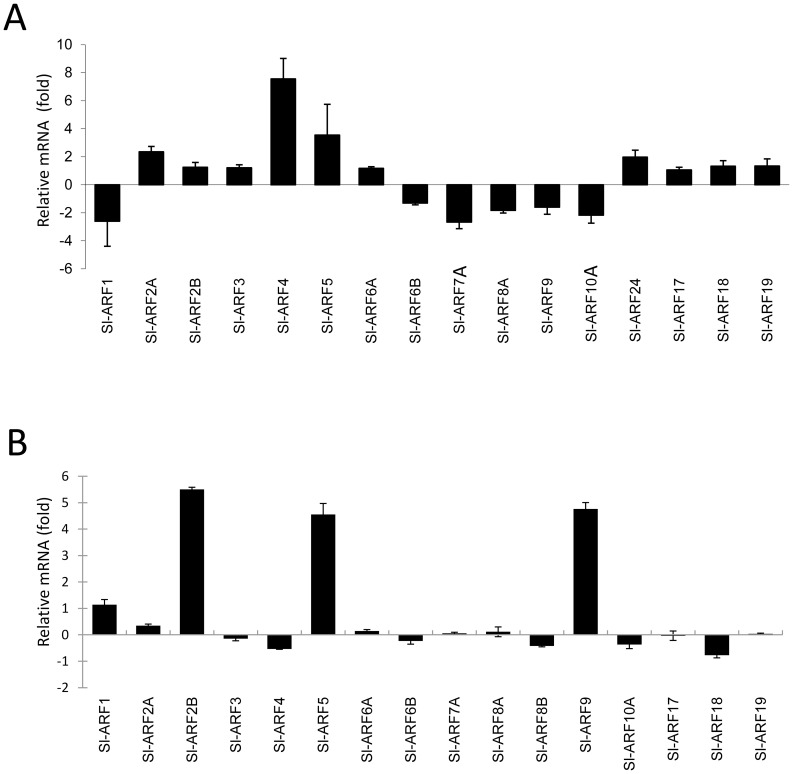
Screening for cis-acting elements corresponding to Auxin Response Elements (AuxRE) within the promoter regions using the Place database (http://www.dna.affrc.go.jp/PLACE/signalscan.html) identified conserved (TGTCTC) and degenerate (TGTCCC) motifs in most tomato ARF promoters. In addition to these AuxRE, Sl-ARF promoters contain conserved Ethylene-Response motifs, the so-called ERELEE4 motif found in the promoter of tomato E4 gene (AWTTCAAA) (Table S2 in File S1). The presence of these cis-regulatory elements suggests a potential regulation of ARF genes by both auxin and ethylene. To test the responsiveness of tomato ARF genes to both hormones, transcript accumulation was assessed by qRT-PCR in seedlings treated with auxin or ethylene. All Sl-ARFs were found to be auxin-responsive after 2 hour treatment (Figure 5A), with Sl-ARF4, Sl-ARF5 and Sl-ARF2A showing the highest up-regulation whereas Sl-ARF1, Sl-ARF7 Sl-ARF10 displayed the most significant down-regulation. On the other hand, the expression of Sl-ARF2B, Sl-ARF5 and Sl-ARF9A showed strong up-regulation (more than four folds increase) when treated 5 hours with ethylene (Figure 5B). Of particular interest, Sl-ARF5 is strongly up-regulated by both hormones and may therefore be involved in mediating responses to both hormones.
Figure 5. The expression of Sl-ARF family genes in response to auxin and ethylene.
(A) Auxin induction of Sl-ARF genes on light grown seedlings. Quantitative RT-PCR of Sl-ARF transcripts in RNA samples extracted from 12-day-old tomato seedlings soaked in liquid MS medium with 10 µM IAA for 2 hours. ΔΔCT refers to the fold of difference in Sl-ARF expression to the untreated seedlings. The SAUR gene was used as control to validate the auxin treatment.(B) Ethylene regulation of Sl-ARF genes on dark grown seedlings. Quantitative RT-PCR of Sl-ARF transcripts in RNA samples extracted from5-days dark-grown tomato seedlings treated 5 hours with ethylene (50 µL/L). ΔΔCT refers to fold differences in Sl-ARF expression relative to untreated seedlings. The E4 gene was used as control for efficient ethylene treatment.
Expression of Sl-ARF genes during tomato fruit set
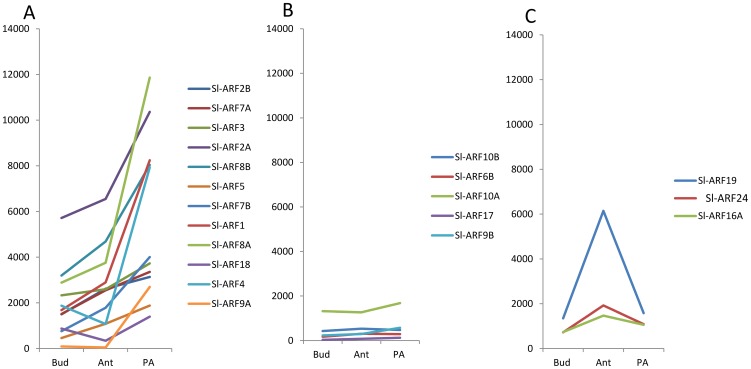
The expression of a high number of Sl-ARFs in reproductive tissues (Figure 3 and 4) along with the previously reported role of auxin in controlling the fruit set process, prompted us to investigate the expression of Sl-ARF genes during the flower-to-fruit transition. To determine the expression dynamics throughout the fruit set process, transcript accumulation of tomato ARFs was monitored by RNA-seq approach at flower buds, anthesis and pos-anthesis stages (young fruit at 4 DPA). For each stage, RNA libraries were generated from three independent biological replicates and subjected to Illumina mRNA-Seq technology sequencing (Data desposited at NCBI SRA database under accession number SRP029978). Reads were then mapped on the tomato genome sequence and read counts were determined as described in Maza et al. 2013 [45]. The data indicate that most Sl-ARFs undergo a strong change in their expression associated with the flower-to-fruit transition (Figure 6). Three groups could be discriminated based on RNA counts distribution during the fruit set process. Group 1 corresponds to Sl-ARFs whose expression increased following pollination, Group 2 to ARFs with unchanged expression and Group 3 to Sl-ARFs displaying decreased expression following pollination (Figure 6).
Figure 6. The expression profile of Sl-ARF family genes in tomato fruit set.
(A)12 Sl-ARF genes are over-expressed after pollination and fertilization (4DPA), which are Sl-ARF9A, 4, 18, 8A, 1, 7B, 5, 8B, 2A, 3, 7A and 2B genes in turn according to the log change of P/A (Post-anthesis/Anthesis). (B) 5 Sl-ARF genes keep stable expression from flower bud to post-anthesis, includingSl-ARF10A, 10B, 6B, 9B, 17 genes.(C) 3 Sl-ARF genes are up-regulated from flower bud to anthesis and down-regulated after pollination and fertilization (4DPA), including Sl-ARF24, 19, and 16A genes. The expression values are taken from RNA-sequencing data and the colors represent different Sl-ARF genes.
Sl-ARF transcripts undergo intense alternative splicing during tomato fruit set
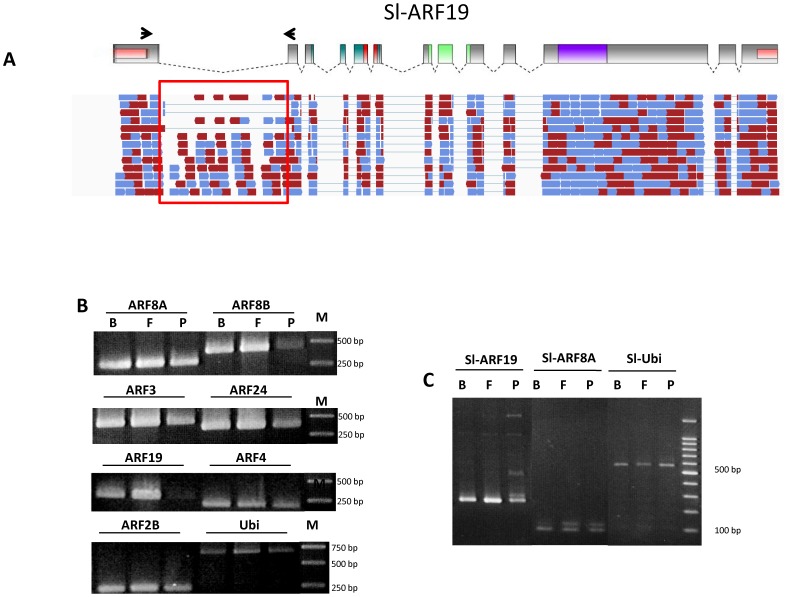
Closer analysis of the mapping of RNA-seq data on the gene models revealed possible alternative splicing regulation during fruit set for 30% of Sl-ARF genes. Sl-ARF2B and ARF19 shows one possible alternative splicing occurring at intron 11 and intron 1, respectively (Figure 6A and Figures S3.1 in File S1). Sl-ARF3 and Sl-ARF4 could putatively give rise to two alternative splicing events at introns 7 and 9, and at introns 6 and 10, respectively. Three possible alternative splicings were found at introns 3, 6 and 10 in Sl-ARF8A and at introns 9, 11 and 13 in Sl-ARF8B. Finally, Sl-ARF24 offers up to four alternative splicing possibilities at introns 1, 3, 6 and 10 (Figures S3.1–6 in File S1). In all cases, the detected Sl-ARF splice variants resulted in a frame shift within the coding region that generates a premature stop codon. To further validate the occurrence of the alternative splicing forms and assess the relative levels of the various splice variants, a semi quantitative PCR approach was conducted. To this purpose, two pairs of primers were designed, one aiming to specifically amplify the retained intron fragment while the second pair was designed in the margins of the two exons framing the retained intron. A PCR product with the expect size was detected for all genes confirming the presence of the splice variant in each RNA extraction (Figure 7B). Interestingly, the data indicate that the abundance of the Sl-ARF8B_int11 transcript variant decreases dramatically in young fruits whereas the global expression of the corresponding Sl-ARF8B gene increases significantly. This finding suggests that the down-regulation of the Sl-ARF8B_int11 transcript variant may potentially play a role in the regulation of the flower to fruit transition. By contrast, increased accumulation of the Sl-ARF19_int1 was observed concomitant to the transition from flower to fruit. Taking together, these data uncover a potential role for alternative splicing in regulating the expression of tomato ARFs during the fruit set process.
Figure 7. The ARF family genes showed alternative spilcing mode of regulation in tomato fruit set.
(A) RNA-seq reads generated during the fruit-set and mapped on Sl-ARF19 gene structure showing one alternative spicing that can be generated in the Intron 1. Reads are represented by red and blue rod arrows (B) The RT-PCR was carried out using pairs of primers designed within the introns of 7 Sl-ARF genes highlighted in Figures S3.1 to S3.6 in File S1, such as Sl-ARF8A_Intron 6, Sl-ARF8B_Intron 11, Sl-ARF3_Intron 9, Sl-ARF24_Intron 3, Sl-ARF19_Intron 1, Sl-ARF4_Intron 6 and Sl-ARF2B_Intron 11. The ubiquitin gene was used as the reference. (C) The RT-PCR was performed using pairs of primers nested in the two exons encompassing the intron of target Sl-ARF genes, such as Exon1-Exon2 in Sl-ARF19 and Exon6-Exon7 in Sl-ARF8A. The cDNAs generated from flower bud (B), flower at anthesis (F) and young fruit 4 days post-pollination (P) tissues were used as the template. The ubiquitin was used as the reference.
Discussion
Being down-stream components of auxin signalling pathway, ARFs likely contribute to the specificity of the hormone responses. Hence, the functional characterization of these transcriptional mediators is essential towards understanding the mechanisms by which auxin triggers appropriate growth and developmental responses in a timely and tissue-specific manner. To better define the role of ARFs in mediating specific auxin responses, the present study brings a complete picture on the main structural features of the tomato ARF gene family. Identification of tomato ARFs has been already described but this attempt built on a draft tomato genome sequence and ESTs and could therefore not be comprehensive [42], [43]. The present work takes advantage of the most updated tomato reference genome sequence [46] to isolate the complete ARF family members and perform functional analysis and expression profiling of these transcriptional regulators. Using these extended resources, the list of tomato ARFs has been enlarged to 22 members and manual annotation based on deep RNA-Seq data, allowed the curation of some structural annotation inconsistencies as well as the identification of the 3′ and 5′ UTR regions for more than 50% of the Sl-ARF gene family. The tomato members of the ARF family were renamed according to the numbering of the closest Arabidopsis homolog, which provides a consensus nomenclature for ARF genes across plant species. In this way, the proposed nomenclature better clarifies the correspondence between ARF subclasses in various plant species. The phyllogenetic approach applied on a well distributed set of plant ARFs allowed to identify a specific sub-class (sub-class IV) that is absent out of the Solanaceae family. Interestingly, this sub-class contains a specific gene, Sl-ARF24, encoding a putative ARF protein that lacks the two protein/protein interaction domains, known as domain III and IV and required for the binding to Aux/IAA proteins. It is therefore likely that Sl-ARF24 escapes the classical mechanism underlying auxin signaling which implies the sequestration of ARF proteins through interaction with Aux/IAAs.
As a preliminary approach towards functional characterization of members of the tomato ARF family, the present study describes their expression pattern, their post-transcriptional regulation and their ability to activate or repress transcriptional activity on synthetic or native auxin-responsive promoters. Transactivation assays revealed that 36% of tomato ARFs are strong repressors of transcriptional activity while only 22% are transcriptional activators. The repressor/activator ratio among ARFs is more than twice higher in tomato (3.6) compared to Arabidopsis (1.7), yet, it remains to be elucidated whether this feature may account for differences in developmental and growth behaviour between the two species. In contrast to repressor ARFs, most activator Sl-ARFs promote transcription of target genes only upon exogenous auxin treatment thus suggesting that activator ARFs require some input from a highly activated auxin signalling pathway in order to potentiate transcriptional activity. It is conceivable that when the auxin level is low, the amount of Aux/IAA proteins available is sufficient to block ARFs at the protein level thus preventing these latter from activating the transcription of the target genes. In this perspective, it has to be postulated that Aux/IAAs are present in excess in the cell when the tissue is not subjected to auxin treatment.
The spatio-temporal pattern of expression indicated that all Sl-ARF genes are expressed in flower and fruit suggesting a putative important role in reproductive tissue development. The shift from the static flower ovary to fast-growing young fruit is a phenomenon known as fruit set and auxin has been shown to play a crucial role in controlling this developmental process [47], [48] representing an important step in the development of all sexually reproducing higher plants. Adding to the primary role of Aux/IAAs in triggering the fruit set process previously reported [3], [49], the present study reveals the potential active role of a number of Sl-ARFs during this process based on genome-wide transcriptomic profiling of the flower to fruit transition. The expression of 12 members of the gene family sharply increases upon pollination/fertilization, while the expression of a fewer number of Sl-ARF genes peaks at anthesis and then dramatically declines at post-pollination stage. Given the role of auxin signaling in the fruit set process [48], [50], the dynamics of the expression pattern of these Sl-ARFs is indicative of their putative involvement in mediating auxin responses during the flower-to-fruit transition. This is consistent with the prominent role reported for Sl-ARF8A and Sl-ARF7 (referred as Sl-ARF19 in the present study) during fruit set and parthenocarpy in Arabidopsis and tomato, respectively [24], [26]. Of particular interest, Sl-ARF8A shows the most dramatic rise in expression at post-anthesis stage which may designate this ARF among all family members as the main actor of the fruit set process.
The data indicate that tomato ARFs are subject to multi-levels post-transcriptional regulation of their expression. In line with Arabidopsis ARFs [51], [52], [53], it is shown here that 11 out of the 22 tomato ARF genes are potentially regulated by siRNAs. Moreover, the direct evidence for active alternative splicing described here uncover a new layer of complexity in the post-transcriptional regulation of ARF genes in the tomato. This mode of regulation may account for a significant part of the control of ARF expression in developmental processes such as fruit set in the tomato as indicated by the abundance of some transcript splice variants concomitant to the flower to fruit transition. An additional mean towards controlling ARF expression in the tomato may also take place at the translational level via upstream ORFs (uORFs) that have been predicted in most members of ARF genes. This mode of regulation has been first suggested in Arabidopsis where in silico search revealed an enrichment of uORFs in the ARF 5′-leader sequences that is not seen in other auxin-related genes such Aux/IAA, YUCCA, TIR1 auxin receptors homologs and PIN family of auxin transporters [54]. Subsequently, translational control of AtARFs by upstream ORF (uORFs) has been proposed as a regulatory mechanism required in modulating auxin responses during plant development [55]. Though direct experimental evidence is still lacking, tomato ARFs may also undergo the same mode of regulation.
In addition of being auxin-responsive, the expression of some Sl-ARFs was found to be regulated by ethylene. The presence of auxin and ethylene cis-regulatory elements in the promoter region of a number of Sl-ARFs, supports the potential regulation of ARF genes by both auxin and ethylene and suggests that these transcription factors have the ability to mediate both auxin and ethylene responses. In support to this hypothesis, Arabidopsis ARF19 has been shown to be inducible by ethylene and has been reported to contribute to ethylene sensitivity through a cross-talk between auxin and ethylene signalling [27], [30]. Also, ARF2 has been shown to regulate the hook curvature of etiolated Arabidopsis seedlings, a typical ethylene response [27]. Taking together, these data suggest that ARFs may act at the crossroads of auxin and ethylene signaling.
Altogether, the data provide molecular clues on how ARFs can contribute to the specificity and selectivity of auxin responses through (i) structural features, (ii) differential expression of family members at the tissue and organ levels and, (iii) ability to negatively or positively impact transcriptional activity of target genes. The auxin and ethylene regulation of some ARF members suggest their specific role in the multi-hormonal cross-talks. The regulation of the expression of ARFs by alternative splicing during fruit set provides new insight into the complexity of regulation of these genes at the post-transcriptional level.
Materials and Methods
Plant material and growth conditions
Tomato seeds (Solanumlycopersicum cv MicroTom or Ailsa Craig) were sterilized, rinsed in sterile water and sown in recipient Magenta vessels containing 50 mL of 50% Murashige and Skoog (MS) culture medium added with R3 vitamin (0.5 mg L−1 thiamine, 0.25 mg L−1 nicotinic acid and 0.5 mg L−1pyridoxine), 1.5% (w/v) sucrose and 0.8% (w/v) agar, pH 5.9. Plants were grown under standard greenhouse conditions. The culture chamber rooms are set as follows: 14-h-day/10-h-night cycle, 25/20°C day/night temperature, 80% hygrometry, 250 µmol m−2s−1 intense luminosity.
In silico Identification of the tomato ARFs
All the ARF gene sequences (ITAG2.3_gene_models.gff3) are download from the Sol Genomics Network (http://solgenomics.net/), and analyzed in Notepad++ software. The NLS location was searched using cNLS Mapper (http://nls-mapper.iab.keio.ac.jp/cgi-bin/NLS_Mapper_form.cgi). All the obtained sequences were sorted for the unique sequences and these were further used for B3, AUX_RESP, and Aux/IAA domain search using InterProScan Sequence Search (http://www.ebi.ac.uk/Tools/pfa/iprscan/). The UTR of Sl-ARFs were found by two steps, first, the whole tomato genome and Sl-ARF gene structures (ITAG2.3_gene_models.gff3) were loaded into the Java, and then, the complete cDNA sequences from RNA-Seq data including three stages (flower bud, anthesis and post-anthesis) were blast with Sl-ARF gene structures to identify the final 5′ or 3′ UTRs in Sl-ARFs. The miRNA location on the Sl-ARFs were searched depend on the GBF data (http://tata.toulouse.inra.fr/gbf/blast/blast.html) and SGN Blast tools. Taken together, all of the Sl-ARF family structures were drawn by Fancy Gene v1.4 (http://host13.bioinfo3.ifom-ieo-campus.it/fancygene/) with manual correction.
Transient Expression Using a Single Cell System
Protoplasts were obtained from suspension-cultured tobacco (Nicotianatabacum) BY-2 cells and transfected by a modified polyethylene glycol method as described by Abel and Theologis [56]. For nuclear localization of the selected ARF fusion proteins, the coding sequence of genes were cloned as a C-terminal fusion in frame with GFP under the control of the 35S CaMV, a cauliflower mosaic virus promoter. Transfected protoplasts were incubated for 16 h at 25°C and analysed for GFP fluorescence by confocal microscopy. For co-transfection assays, aliquots of protoplasts (0.5×106) were transformed either with 10 µg of the reporter vector alone containing the promoter fused to the GFP reporter gene or in combination with 10 µg of ARF contructs as the effector plasmid. Transformation assays were performed in three independent replicates. After 16 h, GFP expression was analyzed and quantified by flow cytometry (FACS Calibur II instrument, BD Biosciences, San Jose, CA) on the flow cytometry platform, IRF31, Inserm, Toulouse and and cell sorting platform, INSERM UPS UMR 1048, Toulouse RIO imaging platform. Data were analyzed using Cell Quest software. For each sample, 100 to 1000 protoplasts were gated on forward light scatter and the GFP fluorescence per population of cells corresponds to the average fluorescence intensity of the population of cells after subtraction of autofluorescence determined with non transformed BY-2 protoplasts. The data are normalised using an experiment, in presence of 50 µM 2.4 D, with protoplasts transformed with the reporter vector in combination with the vector used as the effector plasmid but lacking Sl-ARF coding region.
RNA isolation and Quantitative RT-PCR
Total RNA from fruit was extracted according to the method of Hamilton et al. [57]. Total RNA from leaves and seedlings was extracted using a Plant RNeasy Mini kit (Qiagen) according to the manufacturer's instruction. Total RNA was treated by DNase I to remove any genomic DNA contamination. First strand cDNA was reverse transcribed from 2 µg of total RNA using Omniscript kit (Qiagen) according to the manufacturer's instruction.The qRT-PCR analysis was performed as previously described [3]. The sequences of primers are listed in Table S3 in File S1. Relative fold differences were calculated based on the comparative Ct method using the Sl-Actin as an internal standard. To determine relative fold differences for each sample in each experiment, the Ct value of genes was normalized to the Ct value for Sl-Actin-51 (accession number Q96483/Solyc11g005330) and was calculated relative to a calibrator using the formula 2−ΔΔCt. At least two to three independent RNA isolations were used for cDNA synthesis and each cDNA sample was subjected to real-time PCR analysis in triplicate. Heat map representation was performed using centring and normalized ΔCt value, with Cluster 3.0 software and Java Tree view to visualize dendogram.
Hormone treatment
For auxin treatment on light grown seedlings, 12-day-old tomato seedlings (30 seedlings) were soaked in liquid MS medium with or without (mock treatment) 10 µM IAA for 2 hours. The efficiency of the treatment was checked by measuring the induction of the tomato early auxin-responsive SAUR gene. For ethylene treatment on dark grown seedlings, 5-days-old MicroTom seedlings (100 seedlings) were treated with air or ethylene gas (50 µL/L) for 5 hours. The efficiency of the treatment was checked by measuring the induction of the tomato ethylene-responsive E4 gene. Experiment was repeated for 3 biological times.
RNA-Sequencing and RT-PCR
Total RNA was extracted from bud, flower and post-flower (4DPA) for three biological repeats using a TRIZOL Reagent (invitrogen) according to the manufacturer's instruction. Total RNA was treated by DNase I to remove any genomic DNA contamination and checked by RNA gel and Agilent RNA 6000 Nano Assay, which the RIN value above 7 was determined to be qualified. After that, the best RNA were sent out for deep RNA sequencing using Illumina Hiseq2000 and the reads generated were mapped to the tomato genome sequence SL2.40. The data are desposited at NCBI SRA database under the accession number SRP029978 The gene expression was calculated for each annotated tomato gene (iTAG2.30). For continuous validation, first strand cDNA was synthesized as previously described and PCR was performed using primers designed from the intron and exon of 7 Sl-ARF genes. The primer sequences are listed in Table S4 in File S1. An aliquot of 1 ul of the product was used as a template. The PCR amplification cycle was as follows: 95°C for 30 s, 56–60°C for 40 s, 72°C for 30 s-2.5 min. Samples were taken after 25, 30 or 35 cycles and 10 ul of the PCR product was visualized on a 2–2.5% agarose gel. All PCRs were carried out in a Mastercycler (Eppendorf, Hamburg, Germany). DNA was stained with ethidium bromide in the gel. Sl-Ubi3 expression was used as an internal control.
Supporting Information
Supporting tables and figures. Table S1. uORF prediction in the 5′UTR leader sequences of Sl-ARFs. Table S2. In silico analysis of Sl-ARF gene promoters. Table S3. Quantitative RT-PCR primers of Sl-ARF genes. Table S4. PCR primers for identifying the alternative splicing expressed forms in Sl-ARF genes. Figure S1. Sl-ARF genes genomic distribution on the tomato chromosomes. The arrows next to gene names show the direction of transcription. The number near to each Sl-ARF designates the position megabases (Mb) of the first ATG in the tomato chromosome pseudomolecules (tomato genome version SL2.40). The chromosome numbers and their corresponding size are indicated at the top and bottom of each bar. Figure S2. Phylogenetic relationship between tomato Sl-ARF genes. The unrooted tree was generated using MEGA4 program by neighbor-joining method. Bootstrap values (above 50%) from 1000 replicates are indicated at each branch. Sl-ARFs with a star (*) are deprived of domain III and IV necessary for interaction with Aux/IAAs. Figure S3.1-6. Predicted alternative splicing in six Sl-ARFs (Figure S3.1 to Figure S3.6). RNA-seq reads generated during the fruit-set and mapped on the corresponding Sl-ARF gene sequence (Sl-ARF2B, 3, 4, 8A, 8B, and 24) showing predicted alternative splicing events. RNA-seq reads are represented by red and blue rod arrows.
(PDF)
Acknowledgments
This work benefited from the networking activities within the European funded COST ACTION FA1106 “Qualityfruit”. The authors thank C. Pecher and A. Zakaroff-Girard for their technical assistance and expertise in flow cytometry (Cytometry and cell sorting platform, INSERM UPS UMR 1048, Toulouse RIO imaging platform). The authors also thank O. Boucher and N. Marsaud for their technical assistance and expertise in RNA-seq data production (plateforme genotoul, GeT).
Funding Statement
This work was supported by the Laboratoire d'Excellence (LABEX, TULIP ANR-10-LABX-41). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Quint M, Gray WM (2006) Auxin signaling. Curr Opin Plant Biol 9: 448–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Teale WD, Ditengou FA, Dovzhenko AD, Li X, Molendijk AM, et al. (2008) Auxin as a model for the integration of hormonal signal processing and transduction. Mol Plant 1: 229–237. [DOI] [PubMed] [Google Scholar]
- 3. Wang H, Schauer N, Usadel B, Frasse P, Zouine M, et al. (2009) Regulatory features underlying pollination-dependent and -independent tomato fruit set revealed by transcript and primary metabolite profiling. Plant Cell 21: 1428–1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Sagar M, Chervin C, Mila I, Hao Y, Roustan JP, et al. (2013) SlARF4, an auxin response factor involved in the control of sugar metabolism during tomato fruit development. Plant Physiol 161: 1362–1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sagar M, Chervin C, Bouzayen M, Zouine M (2013) Under-expression of the Auxin Response Factor Sl-ARF4 improves post-harvest behavior of tomato fruits. Plant Signal Behav 8.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Theologis A, Huynh TV, Davis RW (1985) Rapid induction of specific mRNAs by auxin in pea epicotyl tissue. J Mol Biol 183: 53–68. [DOI] [PubMed] [Google Scholar]
- 7. Guilfoyle T, Hagen G, Ulmasov T, Murfett J (1998) How does auxin turn on genes? Plant Physiol 118: 341–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Tiwari SB, Hagen G, Guilfoyle T (2003) The roles of auxin response factor domains in auxin-responsive transcription. Plant Cell 15: 533–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hayashi K (2012) The interaction and integration of auxin signaling components. Plant Cell Physiol 53: 965–975. [DOI] [PubMed] [Google Scholar]
- 10. Szemenyei H, Hannon M, Long JA (2008) TOPLESS mediates auxin-dependent transcriptional repression during Arabidopsis embryogenesis. Science 319: 1384–1386. [DOI] [PubMed] [Google Scholar]
- 11. Causier B, Lloyd J, Stevens L, Davies B (2012) TOPLESS co-repressor interactions and their evolutionary conservation in plants. Plant Signal Behav 7: 325–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ulmasov T, Murfett J, Hagen G, Guilfoyle TJ (1997) Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell 9: 1963–1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wang L, Kim J, Somers DE (2013) Transcriptional corepressor TOPLESS complexes with pseudoresponse regulator proteins and histone deacetylases to regulate circadian transcription. Proc Natl Acad Sci U S A 110: 761–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Audran-Delalande C, Bassa C, Mila I, Regad F, Zouine M, et al. (2012) Genome-wide identification, functional analysis and expression profiling of the Aux/IAA gene family in tomato. Plant Cell Physiol 53: 659–672. [DOI] [PubMed] [Google Scholar]
- 15. Rademacher EH, Lokerse AS, Schlereth A, Llavata-Peris CI, Bayer M, et al. (2012) Different auxin response machineries control distinct cell fates in the early plant embryo. Dev Cell 22: 211–222. [DOI] [PubMed] [Google Scholar]
- 16. Cole M, Chandler J, Weijers D, Jacobs B, Comelli P, et al. (2009) DORNROSCHEN is a direct target of the auxin response factor MONOPTEROS in the Arabidopsis embryo. Development 136: 1643–1651. [DOI] [PubMed] [Google Scholar]
- 17. Lim PO, Lee IC, Kim J, Kim HJ, Ryu JS, et al. (2010) Auxin response factor 2 (ARF2) plays a major role in regulating auxin-mediated leaf longevity. J Exp Bot 61: 1419–1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wilmoth JC, Wang S, Tiwari SB, Joshi AD, Hagen G, et al. (2005) NPH4/ARF7 and ARF19 promote leaf expansion and auxin-induced lateral root formation. Plant J 43: 118–130. [DOI] [PubMed] [Google Scholar]
- 19. Ellis CM, Nagpal P, Young JC, Hagen G, Guilfoyle TJ, et al. (2005) AUXIN RESPONSE FACTOR1 and AUXIN RESPONSE FACTOR2 regulate senescence and floral organ abscission in Arabidopsis thaliana. Development 132: 4563–4574. [DOI] [PubMed] [Google Scholar]
- 20. Marin E, Jouannet V, Herz A, Lokerse AS, Weijers D, et al. (2010) miR390, Arabidopsis TAS3 tasiRNAs, and their AUXIN RESPONSE FACTOR targets define an autoregulatory network quantitatively regulating lateral root growth. Plant Cell 22: 1104–1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Yoon EK, Yang JH, Lee WS (2010) Auxin and Abscisic Acid Responses of Auxin Response Factor 3 in Arabidopsis Lateral Root Development. Journal of Plant Biology 53: 150–154. [Google Scholar]
- 22. Varaud E, Brioudes F, Szecsi J, Leroux J, Brown S, et al. (2011) AUXIN RESPONSE FACTOR8 regulates Arabidopsis petal growth by interacting with the bHLH transcription factor BIGPETALp. Plant Cell 23: 973–983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Goetz M, Vivian-Smith A, Johnson SD, Koltunow AM (2006) AUXIN RESPONSE FACTOR8 is a negative regulator of fruit initiation in Arabidopsis. Plant Cell 18: 1873–1886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Goetz M, Hooper LC, Johnson SD, Rodrigues JC, Vivian-Smith A, et al. (2007) Expression of aberrant forms of AUXIN RESPONSE FACTOR8 stimulates parthenocarpy in Arabidopsis and tomato. Plant Physiol 145: 351–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Guillon F, Philippe S, Bouchet B, Devaux MF, Frasse P, et al. (2008) Down-regulation of an Auxin Response Factor in the tomato induces modification of fine pectin structure and tissue architecture. J Exp Bot 59: 273–288. [DOI] [PubMed] [Google Scholar]
- 26. de Jong M, Wolters-Arts M, Feron R, Mariani C, Vriezen WH (2009) The Solanum lycopersicum auxin response factor 7 (SlARF7) regulates auxin signaling during tomato fruit set and development. Plant J 57: 160–170. [DOI] [PubMed] [Google Scholar]
- 27. Li H, Johnson P, Stepanova A, Alonso JM, Ecker JR (2004) Convergence of signaling pathways in the control of differential cell growth in Arabidopsis. Dev Cell 7: 193–204. [DOI] [PubMed] [Google Scholar]
- 28. Jain M, Khurana JP (2009) Transcript profiling reveals diverse roles of auxin-responsive genes during reproductive development and abiotic stress in rice. Febs Journal 276: 3148–3162. [DOI] [PubMed] [Google Scholar]
- 29. de Jong M, Wolters-Arts M, Garcia-Martinez JL, Mariani C, Vriezen WH (2011) The Solanum lycopersicum AUXIN RESPONSE FACTOR 7 (SlARF7) mediates cross-talk between auxin and gibberellin signalling during tomato fruit set and development. J Exp Bot 62: 617–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Li J, Dai X, Zhao Y (2006) A role for auxin response factor 19 in auxin and ethylene signaling in Arabidopsis. Plant Physiol 140: 899–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Liu ZH, Yu YC, Xiang FN (2011) [Auxin response factors and plant growth and development]. Yi Chuan 33: 1335–1346. [DOI] [PubMed] [Google Scholar]
- 32. Vert G, Walcher CL, Chory J, Nemhauser JL (2008) Integration of auxin and brassinosteroid pathways by Auxin Response Factor 2. Proc Natl Acad Sci U S A 105: 9829–9834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Guilfoyle TJ, Hagen G (2007) Auxin response factors. Curr Opin Plant Biol 10: 453–460. [DOI] [PubMed] [Google Scholar]
- 34. Guilfoyle TJ, Hagen G (2001) Auxin response factors. Journal of Plant Growth Regulation 20: 281–291. [Google Scholar]
- 35. Ulmasov T, Hagen G, Guilfoyle TJ (1999) Activation and repression of transcription by auxin-response factors. Proceedings of the National Academy of Sciences of the United States of America 96: 5844–5849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Nagpal P, Ellis CM, Weber H, Ploense SE, Barkawi LS, et al. (2005) Auxin response factors ARF6 and ARF8 promote jasmonic acid production and flower maturation. Development 132: 4107–4118. [DOI] [PubMed] [Google Scholar]
- 37. Jones B, Frasse P, Olmos E, Zegzouti H, Li ZG, et al. (2002) Down-regulation of DR12, an auxin-response-factor homolog, in the tomato results in a pleiotropic phenotype including dark green and blotchy ripening fruit. Plant Journal 32: 603–613. [DOI] [PubMed] [Google Scholar]
- 38. Wang DK, Pei KM, Fu YP, Sun ZX, Li SJ, et al. (2007) Genome-wide analysis of the auxin response factors (ARF) gene family in rice (Oryza sativa). Gene 394: 13–24. [DOI] [PubMed] [Google Scholar]
- 39. Kalluri UC, Difazio SP, Brunner AM, Tuskan GA (2007) Genome-wide analysis of Aux/IAA and ARF gene families in Populus trichocarpa. BMC Plant Biol 7: 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Paterson AH, Bowers JE, Bruggmann R, Dubchak I, Grimwood J, et al. (2009) The Sorghum bicolor genome and the diversification of grasses. Nature 457: 551–556. [DOI] [PubMed] [Google Scholar]
- 41. Xing H, Pudake RN, Guo G, Xing G, Hu Z, et al. (2011) Genome-wide identification and expression profiling of auxin response factor (ARF) gene family in maize. BMC Genomics 12: 178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kumar R, Tyagi AK, Sharma AK (2011) Genome-wide analysis of auxin response factor (ARF) gene family from tomato and analysis of their role in flower and fruit development. Molecular Genetics and Genomics 285: 245–260. [DOI] [PubMed] [Google Scholar]
- 43. Wu J, Wang F, Cheng L, Kong F, Peng Z, et al. (2011) Identification, isolation and expression analysis of auxin response factor (ARF) genes in Solanum lycopersicum. Plant Cell Reports 30: 2059–2073. [DOI] [PubMed] [Google Scholar]
- 44. Ottenschlager I, Wolff P, Wolverton C, Bhalerao RP, Sandberg G, et al. (2003) Gravity-regulated differential auxin transport from columella to lateral root cap cells. Proc Natl Acad Sci U S A 100: 2987–2991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Maza E, Frasse P, Senin P, Bouzayen M, Zouine M (2013) Comparison of normalization methods for differential gene expression analysis in RNA-Seq experiments: A matter of relative size of studied transcriptomes. Communicative & Integrative Biology 6.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. The tomato genome sequence provides insights into fleshy fruit evolution. Nature 485: 635–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Serrani JC, Ruiz-Rivero O, Fos M, Garcia-Martinez JL (2008) Auxin-induced fruit-set in tomato is mediated in part by gibberellins. Plant Journal 56: 922–934. [DOI] [PubMed] [Google Scholar]
- 48. de Jong M, Mariani C, Vriezen WH (2009) The role of auxin and gibberellin in tomato fruit set. Journal of Experimental Botany 60: 1523–1532. [DOI] [PubMed] [Google Scholar]
- 49. Wang H, Jones B, Li Z, Frasse P, Delalande C, et al. (2005) The tomato Aux/IAA transcription factor IAA9 is involved in fruit development and leaf morphogenesis. Plant Cell 17: 2676–2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Devoghalaere F, Doucen T, Guitton B, Keeling J, Payne W, et al. (2012) A genomics approach to understanding the role of auxin in apple (Malus x domestica) fruit size control. BMC Plant Biol 12: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Wang JW, Wang LJ, Mao YB, Cai WJ, Xue HW, et al. (2005) Control of root cap formation by MicroRNA-targeted auxin response factors in Arabidopsis. Plant Cell 17: 2204–2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Williams L, Carles CC, Osmont KS, Fletcher JC (2005) A database analysis method identifies an endogenous trans-acting short-interfering RNA that targets the Arabidopsis ARF2, ARF3, and ARF4 genes. Proc Natl Acad Sci U S A 102: 9703–9708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Wu MF, Tian Q, Reed JW (2006) Arabidopsis microRNA167 controls patterns of ARF6 and ARF8 expression, and regulates both female and male reproduction. Development 133: 4211–4218. [DOI] [PubMed] [Google Scholar]
- 54. Zhou F, Roy B, von Arnim AG (2010) Translation reinitiation and development are compromised in similar ways by mutations in translation initiation factor eIF3h and the ribosomal protein RPL24. BMC Plant Biol 10: 193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Rosado A, Li R, van de Ven W, Hsu E, Raikhel NV (2012) Arabidopsis ribosomal proteins control developmental programs through translational regulation of auxin response factors. Proc Natl Acad Sci U S A 109: 19537–19544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Abel S, Theologis A (1994) Transient transformation of Arabidopsis leaf protoplasts: a versatile experimental system to study gene expression. Plant J 5: 421–427. [DOI] [PubMed] [Google Scholar]
- 57. Hamilton AJ, Lycett GW, Grierson D (1990) Antisense Gene That Inhibits Synthesis of the Hormone Ethylene in Transgenic Plants. Nature 346: 284–287. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting tables and figures. Table S1. uORF prediction in the 5′UTR leader sequences of Sl-ARFs. Table S2. In silico analysis of Sl-ARF gene promoters. Table S3. Quantitative RT-PCR primers of Sl-ARF genes. Table S4. PCR primers for identifying the alternative splicing expressed forms in Sl-ARF genes. Figure S1. Sl-ARF genes genomic distribution on the tomato chromosomes. The arrows next to gene names show the direction of transcription. The number near to each Sl-ARF designates the position megabases (Mb) of the first ATG in the tomato chromosome pseudomolecules (tomato genome version SL2.40). The chromosome numbers and their corresponding size are indicated at the top and bottom of each bar. Figure S2. Phylogenetic relationship between tomato Sl-ARF genes. The unrooted tree was generated using MEGA4 program by neighbor-joining method. Bootstrap values (above 50%) from 1000 replicates are indicated at each branch. Sl-ARFs with a star (*) are deprived of domain III and IV necessary for interaction with Aux/IAAs. Figure S3.1-6. Predicted alternative splicing in six Sl-ARFs (Figure S3.1 to Figure S3.6). RNA-seq reads generated during the fruit-set and mapped on the corresponding Sl-ARF gene sequence (Sl-ARF2B, 3, 4, 8A, 8B, and 24) showing predicted alternative splicing events. RNA-seq reads are represented by red and blue rod arrows.
(PDF)