Abstract
Toxoplasmosis has been recognized as parasitic zoonosis with the highest human incidence. The human infection by the parasite can lead to severe clinical manifestations in congenital toxoplasmosis and immunocompromised patients. Contamination occurs mainly by foodborne ways especially consumption of raw or undercooked meat. In contrast to other foodborne infections, toxoplasmosis is a chronic infection which would make its economic and social impact much higher than even previously anticipated. Ovine meat was advanced as a major risk factor, so we investigated its parasite survey, under natural conditions. Serological MAT technique and touchdown PCR approaches were used for prevalence determination of the parasite in slaughtered sheep intended to human consumption in Tunis City. The genotyping was carried by SNPs analysis of SAG3 marker. Anti-Toxoplasma antibodies were present in 38.2% of young sheep and in 73.6% of adult sheep. Molecular detection revealed the contamination of 50% of ewes’ tissue. Sequencing and SNPs analysis enabled unambiguous typing of meat isolates and revealed the presence of mixed strains as those previously identified from clinical samples in the same area. Our findings conclude that slaughtered sheep are highly infected, suggesting them as a major risk factor of Toxoplasma gondii transmission by meat consumption. Special aware should target consequently this factor when recommendations have to be established by the health care commanders.
Introduction
Toxoplasma gondii (T.gondii) is a cosmopolite parasitic protozoon. It infects all warm-blooded vertebrate species, including humans. Human infection can leads to drastic consequences during congenital transmission [1] and in immunocompromised patients [2]. Transmission occurs mainly by foodborne way through the ingestion of contaminated vegetable /water with oocysts, as well as the ingestion of contaminated raw / undercooked meat with tissue cysts [3]. The European Food Safety Authority (EFSA) has recognized toxoplasmosis as parasitic zoonosis with the highest human incidence [4]. Researchers considered the consumption of undercooked infected meat as the biggest risk and suspected ovine meat, to be a major risk factor for human infection [5]. In our country, Tunisia (North of Africa), data still fragmentary about the survey of this parasite in slaughtered ovine. No previous study investigated relevantly the detection the parasite in the meat and its genotyping for epidemiological correlation with clinical data [6]. This is what we propose to cover by our present work to check if ovine meat could or not be a risk factor for human contamination in Tunis City, the capital of Tunisia.
Materials and Methods
Sheep sera
Blood samples were collected, in slaughterhouse upon the individual owners request, from 217 lambs (3–11 months old) and 125 ewes (≥ 1-year-old) intending for human consumption in the neighborhood of Tunis City. No endangered or protected species have been included in our study. The blood was drawn via the jugular vein from each animal at the time of slaughter by exsanguinations as approved by the Tunisian National Ethics Committee. Sera were separated after centrifugation 2300 x g for 10 min and stored at –20°C until analysis.
Serum samples were initially screened at 1∶20 and 1∶200 dilutions with the modified agglutination test (MAT) using whole formalin-preserved tachyzoites as antigen [7]. Seropositive sera were then end-titrated using 2-fold dilutions.
Sheep tissues
We purchased 72 ewes heart from different butchers in Tunis City. Hearts were collected in individual plastic bags and kept refrigerated (+4°C). Intra-cardiac blood or meat juices were collected for serological examination by MAT technique. When the serological sampling wasn't possible, tissue specimen were grouped in pools which will be, like seropositive hearts, subjected to enzymatic treatment for the molecular identification.
Briefly, heart tissue was ground in 2 volumes (w/v) of 0.9% NaCl and digested 2:3 (v/v) with 2.5% fresh trypsin solution in PBS. Lysis was achieved after incubation during 1h30/150 rpm at 37°C. The product was then filtrated twice, pelted by cold centrifugation 10 min at 2500 rpm, washed and resuspended in saline containing Penicillin G/Streptomycin.
DNA extraction
700 µl of the treated pellet was subjected to DNA extraction protocol. The same volume of aqueous lysis buffer (100 mM Tris HCl pH 8, 1.4 M NaCl, 20 mM EDTA pH8, 2% CTAB) containing fresh 70 µl of Proteinase K (10 mg/ml) was added for overnight incubation in 65°C water bath. Lysate extraction was done by chloroform (v/v). After the cold centrifugation at maximum speed, clear aqueous phase must be recovered otherwise chloroform treatment should be repeated for the upper phase. DNA was precipitated by the addition of Sodium acetate at a final concentration of 0.3 M and 0.6 volume of isopropanol, followed by 15 min incubation at –80°C. After cold centrifugation, wash step by cold 70% ethanol, the pellet was dried and resuspended in 70 µl warm TE buffer.
PCR reaction
T.gondii diagnostic was carried out by B1 PCR using the primers B22: AACGGGCGAGTAGCACCTGAGGAGA and B23: TGGGTCTACGTCGATGGCATGACAAC. DNA extract was tested in duplicate with the use of an internal amplification control (0.1pg of reference DNA) to enable the identification of false negative PCR results. Amplification was performed in a final volume of 25 µl with 10X PCR buffer, 2.5 mM MgCl2, 200 µM dNTP each, 0.6 mg BSA, 10 pmol of primers, 1U of Gold Taq Polymerase (Applied Biosystems) and 5 µl of sample DNA.
Amplification program started with preliminary steps of 2min at 50°C then of 6min at 95°C and followed by 40 cycles of 30sec at 94°C, 30sec at 57°C and 1min at 72°C adding 1sec/cycle. The final elongation step was achieved after 7min at 72°C. Reaction products were resolved on 3% agarose gel.
Genotyping
Single nucleotide polymorphism (SNPs) was investigated for the positive tissues at the marker SAG3 as described elsewhere [8]. Sample profiles were compared to profiles of references strains RH, PRU and NED kindly provides by Toxoplasma BRC (Limoges-France). Phylogenetic network analysis was performed using the software SplitsTree 4 [9].
Statistical test
The difference in the seroprevalence between the age categories was analyzed by the chi-square test [10].
Results
Antibodies to T. gondii (Table 1) were found in 83 of 217 lambs (38.2%) in titers of 20 (25 lambs), 40 (23 lambs), 80 (12 lambs), 160 (9 lambs), 320 (3 lambs) and ≥ 640 (11 lambs). In ewes, positives sera were 92 of 125 (73.6%) with 56 samples within 1∶20 dilution, 11 within 1∶40, 3 within 1∶80, within 1∶160, one sample within 1∶320 and 11 within ≥1∶640 dilution. Statistical analysis showed that difference in prevalence observed between ages was significant (p <0.001).
Table 1. Summary of antibodies prevalence.
| Age | Ni | N+ | N- | % | |
| Lambs | 3–11 months | 217 | 83 | 134 | 38.2 |
| Ewes | ≥ 1-year-old | 125 | 92 | 33 | 73.6 |
Ni: Initial number of tested serums; N+: Number of sera with positive reaction; N–: Number of sera with negative reaction; %: percentage of seroprevalence.
In an attempt to identify T.gondii from sheep tissue, we weren't able to do serological sampling for 18 hearts; consequently tissues were grouped in 3 pools: ShrTgTnP1, ShrTgTnP2 and ShrTgTnP3. For the remaining 54 tissues, we detected Toxoplasma antibodies in 34 (63%) hearts from ewes slaughtered in Tunis City within dilutions 1∶20 in 5 samples, 1∶40 in 5 samples, 1∶80 in 6 samples, 1∶160 in 14 samples, 1∶320 in 3 samples and titter ≥ 1∶640 in 1 ewe.
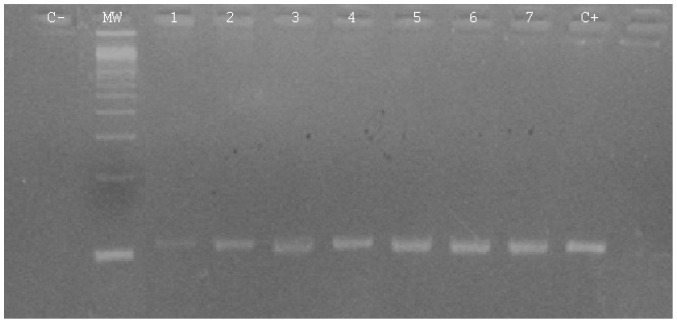
With the new extraction protocol, inhibition reaction wasn’t observed, however best visualization of the result was observed after optimization of DNA dilution [data not shown]. Parasite DNA amplification was obtained in 17 hearts (50%) and in 1 of the 3 tissue pools by visualizing the 114 bp band after agarose gel electrophoresis (Figure 1). T.gondii was detected mainly from tissues with high antibodies titer (Table 2).
Figure 1. Electrophoretic pattern of the direct PCR on sheep hearts.
C- corresponds to negative control of PCR; MW: 100bp ladder; 1–9: amplification of different sheep isolates and C+ corresponds to the positive control of the reaction.
Table 2. Data about positive sheep heart samples.
| Strains | MAT titer | SAG3 type |
| ShrTgTn P1 | ND | II/III |
| ShrTgTn 1 | 160 | III |
| ShrTgTn 2 | 40 | I |
| ShrTgTn 8 | 160 | II |
| ShrTgTn 10 | 320 | III |
| ShrTgTn 17 | 20 | III |
| ShrTgTn 20 | 40 | II/III |
| ShrTgTn 24 | 160 | II/III |
| ShrTgTn 30 | 160 | III |
| ShrTgTn 31 | 160 | II/III |
| ShrTgTn 34 | 160 | III |
| ShrTgTn 35 | 160 | III |
| ShrTgTn 37 | 320 | III |
| ShrTgTn 42 | 20 | III |
| ShrTgTn 44 | 80 | II |
| ShrTgTn 49 | 160 | III |
| ShrTgTn 53 | 40 | III |
| ShrTgTn 54 | 20 | I |
ND: not done.
Amplification was retained for all the 18 samples by SAG3 marker. The 225bp products were purified and subjected to NciI restriction reaction. Comparison between samples and reference strains profile’s showed: profiles identical to strain I profile within two samples, two other samples were typed as strain II and ten samples revealed genotype III. The remaining 4 samples, advanced complex profiles suggesting the presence of type II and III simultaneously (Table 2). Mixed infection was confirmed by sequencing (Table 3) and well demonstrated by the intermediate position of isolates within the phylogenetic network (Figure 2). Sequences analysis revealed also homology with clinical isolates identified in our previous work with the clustering of Tunisian clinical and environmental samples in the phylogenetic network (Figure 2).
Table 3. Summary of strains polymorphism at SAG3 marker.
| SNPs polymorphism positions at the marker SAG3 | |||||||||
| Strains/Positions | 64 | 84 | 88 | 120 | 127 | 129 | 136 | 144 | 159 |
| RH Type I | C | G | A | T | G | G | A | G | A |
| PRU Type II | T | A | G | T | A | G | C | C | G |
| NED Type III | C | G | A | C | G | A | C | C | A |
| ShrTgTn 24 | Y | R | R | Y | R | R | C | C | R |
Figure 2. SplitsTree network of toxoplasmic strains at the marker SAG3.
RH, PRU and CEP represent reference strains of type I, II and III respectively. TgUgCh: strains isolated form free-ranging chicken in Uganda. LA, PL, LCR: Tunisian clinical strains. ShrTgTn: strains isolated from sheep in Tunisia. CSe and CSd: atypical strains.
Discussion
Previous studies showed that selected areas for which seroprevalence was high in sheep corresponded to those for which seroprevalence was also high in humans [11]. The direct link between sheep infection and human contamination at the area level is difficult to assess because meat is not always consumed in the same area where animals are slaughtered. Nevertheless, the high seroprevalence of the parasite in several species within the same areas could be related to an intense circulation of the parasite, perhaps due to specific environmental conditions (felids densities, climatic specificities, etc.). Those areas may be considered as “risky areas” for toxoplasmosis infection and specific investigations should be conducted [12]. In Tunisia, Tunis City (located in the north), is the largest city and the densest human agglomeration. Within it, Toxoplasma prevalence was estimated to 47.7% among pregnant women [13]. Regarding prevalence in sheep, two published studies are reported. The first work published in 1970 [14] advanced 75% dye-test sero-prevalence in slaughtered sheep, regardless animal age. Recently, another analysis investigated the prevalence of the parasite within age, sex and locality differentiation [15]. Low prevalences were reported in northern cities: 1.8% by ELISA and 12.7% by nPCR, with insignificant difference between young and old animals. Consequently our present work is, to our knowledge, the first survey in Africa of T.gondii prevalence in ovine livestock using combined serological analysis, molecular detection and genotypic characterization.
For the serological test, we opted for MAT technique as its relative sensitivity is the highest when performed on cardiac fluids [16]. Contradictory to what was published recently in our country, seroprevalence among adult sheep was significantly higher than among young sheep. This confirms previous reports indicating a higher risk of exposure as age increases in animals [17], [18] and humans. The seroprevalence in lambs cannot be related to a transfer of maternal antibodies through colostrums, which occurs within 3 months after birth [19]. Risk factors for infection with T.gondii in sheep are within the age of 7 months or older, suggesting that most of the animals acquired the infection progressively after birth [12].
In our study, the advanced prevalence of antibodies in Tunisian ovine was higher to those observed as average in African countries: 5.6% in South Africa [20], 6.7% in Nigeria [21], 22.9% in Ethiopia [22] and 32.2% in Ghana [23]. Moreover, unexpectedly seroprevalence in Morocco, a close country, is about 27.6% [24] and 43.7% in Egypt [25], other geographically close country. However similar results are observed in some localities from: Turkey 66.66% [26], France 65.5% [27], Switzerland 61.6% [28] and from Suriname 67% [29]. The studied areas are known to be warm, moist tending to favour the survival of oocysts. Indeed, Tunis City is located in humid zone with an average annual rainfall over 400 mm. Its climatic characteristics increase the chance of viability of the parasite and as result, confer generally high prevalence. In the present study, the fact that 16.8% of the tested sheep had a high antibodies titer (≥160) is an indication of frequent exposure to the parasite in farms.
To explore whether this high prevalence could be a risk factor or not to human contamination, we investigated to study the prevalence of the parasite in the ovine meat. The most sensitive tissues of the sheep to T.gondii are the brain and the heart. Since adult-sheep hearts are more frequently consumed in Tunisia, we focused our study on the analysis of ewes hearts purchased from butchers so directly intended to human consumption. We tested antibodies in meat juice since it was described as: reasonable alternative to serum when testing carcasses and relevant matrix for toxoplasmosis survey in meat [30], [16]. The serological result was in concordance with seroprevalence study showing high rate of anti-T.gondii antibodies in adult animals. The positive tissues were digested by trypsin to liberate the eventual toxoplasmic cysts present in the meat. We had chosen to use the CTAB in the lysis step of our DNA extraction protocol. This reagent, frequently used in plant extraction protocols [31], is described with high power of binding and purifying DNA from other tissues components. Even without the use of phenol which lead predict to higher protein contamination, the efficiency of CTAB was well demonstrated by obtaining a clear upper phase well separated from the proteins ring and the organic phase. The extracted DNA was enough pure to not induce any inhibition during PCR diagnosis reaction.
For the parasite detection we used the primers B22/B23 as they were described the most sensitive ones even in tissues [32]. T.gondii was detected in the half of the seropositives tissues which indicate the high rate of meat contamination in contrast to what was reported previously in our country [15]. The distribution of T.gondii parasites within same tissue is random, and parasite density may be low. Therefore, a negative result has to be interpreted carefully because it is possible that the parasite could be present in unexamined parts of the target tissue. The Gharbi et al team analysed the apex of the heart for Toxoplasma detection, however our approach investigated the detection in the whole animal heart which can explain the higher prevalence that we obtained. The same rate was also observed in Netherland where molecular detection advanced the contamination of 50% of seropositive hearts [33]. However the used technique was combination of magnetic capture and PCR, much more complex than the touchdown PCR we used.
All the positive samples were able to be amplified by the marker SAG3 for genotyping [34]. SAG3 is one of the most sensitive described markers with detection level up to 5 parasites /sample. It is also a frequently used RFLP marker in the literature, offering thus abundant data about worldwide T.gondii genotyped isolates. The observed genotypes within our meat isolates concord with previous ones identified from human clinical cases in the same area [6]. The presence of mixed genotypes could be explained by the presence of a pool of tissues in one case, but its presence in three single samples underlines more that mixed infections are frequents on African content [35], [6], [1]. The phylogentic analyses demonstrate well the close relation between our isolates and African ones (TgUgCH) as it demonstrates the clustering between Tunisian clinical and sheep isolates.
Conclusions
According to Tunisian traditions, pregnant women are encouraged to consume undercooked meat (ovine, free ranging chicken and horse meat) to have “healthier baby”. In previous work, we investigated the prevalence of the parasite among horses; however the low observed prevalence disqualify it from risk factors list [36]. Within our current study, considering the presence of T.gondii in slaughtered sheep and the clustering of the identified strains with those described from Tunisian clinical samples, ovine meat is suggesting as a major risk factor of human contamination in Tunis City. Special aware must be taken, especially with the identification of mixed genotypes which are presumed to be more virulent than clonal types I, II and III [1].
Funding Statement
This study was supported by the Ministry of Higher Education Research and Technology in Tunisia and carried out within the framework of the Research Lab "Parasitoses emergentes" LR 05SP03. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Boughattas S, Ben-Abdallah R, Siala E, Souissi O, Maatoug R, et al. (2011) Case of fatal congenital toxoplasmosis associated with I/III recombinant genotype. Trop Biomed 28: 615–619. [PubMed] [Google Scholar]
- 2. Cenci-Goga BT, Rossitto PV, Sechi P, McCrindle CM, Cullor JS (2011) Toxoplasma in animals, food, and humans: an old parasite of new concern. Foodborne Pathog Dis 8: 751–762. [DOI] [PubMed] [Google Scholar]
- 3. Gajadhar AA, Scandrett WB, Forbes LB (2006) Overview of food- and water-borne zoonotic parasites at the farm level. Rev Sci Tech 25: 595–606. [PubMed] [Google Scholar]
- 4. EFSA (2007) Surveillance and monitoring of Toxoplasma in humans, food and animals: Scientific Opinion of the Panel on Biological Hazards. The EFSA Journal 583: 1–64. [Google Scholar]
- 5. Tenter AM (2009) Toxoplasma gondii in animals used for human consumption. Mem Inst Oswaldo Cruz 104: 364–369. [DOI] [PubMed] [Google Scholar]
- 6. Boughattas S, Ben-Abdallah R, Siala E, Souissi O, Aoun K, et al. (2010) Direct genotypic characterization of Toxoplasma gondii strains associated with congenital toxoplasmosis in Tunisia (North Africa). Am J Trop Med Hyg 82: 1041–1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Dubey JP, Desmonts G (1987) Serological responses of equids fed Toxoplasma gondii oocysts. Equine Vet J 19: 337–339. [DOI] [PubMed] [Google Scholar]
- 8. Grigg ME, Ganatra J, Boothroyd JC, Margolis TP (2001) Unusual abundance of atypical strains associated with human ocular toxoplasmosis. J Infect Dis 184: 633–639. [DOI] [PubMed] [Google Scholar]
- 9. Huson DH, Bryant D (2006) Application of Phylogenetic Networks in Evolutionary Studies. Mol Biol Evol 23: 254–267. [DOI] [PubMed] [Google Scholar]
- 10.Preacher KJ (2001) Calculation for the chi-square test: An interactive calculation tool for chi-square tests of goodness of fit and independence [Computer software]. Available from http://quantpsy.org.
- 11.Berger F, Goulet V, Le Strat Y, De Valk H, Désenclos JC (2007) La toxoplasmose en France chez la femme enceinte en 2003: séroprévalence et facteurs associés. Available : http://www.invs.sante.fr/pmb/invs/(id)/PMB_3908
- 12. Halos L, Thébault A, Aubert D, Thomas M, Perret C, et al. (2010) An innovative survey underlining the significant level of contamination by Toxoplasma gondii of ovine meat consumed in France. Int J Parasitol 40: 193–200. [DOI] [PubMed] [Google Scholar]
- 13. Fakhfakh N, Kallel K, Ennigro S, Kaouech E, Belhadj S, et al. (2013) Facteurs de risque pour Toxoplasma gondii et statut immunitaire des femmes parturientes : relation de cause à effet ? Tunis Med 91: 188–190. [PubMed] [Google Scholar]
- 14. Ben Rachid MS, Blaha R (1970) Human and animal toxoplasmosis in Tunisia. Tunis Med 48: 101–110. [PubMed] [Google Scholar]
- 15. Gharbi M, Zribi L, Jedidi M, Chakkhari H, Hamdi S, et al. (2013) Prévalence d’infection des ovins par Toxoplasma gondii en Tunisie. Bull Soc Pathol Exot 10.1007/s13149-013-0290-4 [DOI] [PubMed] [Google Scholar]
- 16. Villena I, Durand B, Aubert D, Blaga R, Geers R, et al. (2012) New strategy for the survey of Toxoplasma gondii in meat for human consumption. Vet Parasitol 183: 203–208. [DOI] [PubMed] [Google Scholar]
- 17. Alvarado-Esquivel C, García-Machado C, Alvarado-Esquivel D, Vitela-Corrales J, Villena I, et al. (2012) Seroprevalence of Toxoplasma gondii infection in domestic sheep in Durango State, Mexico. J Parasitol 98: 271–273. [DOI] [PubMed] [Google Scholar]
- 18.Hecker YP, Moore DP, Manazza JA, Unzaga JM, Späth EJ, et al. (2013) First report of seroprevalence of Toxoplasma gondii and Neospora caninum in dairy sheep from Humid Pampa, Argentina. Trop Anim Health Prod. [DOI] [PubMed]
- 19. Waldeland H (1976) Toxoplasmosis in sheep. The prevalence of Toxoplasma antibodies in lambs and mature sheep from different parts of Norway. Acta Vet Scand 17: 432–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Samra NA, McCrindle CM, Penzhorn BL, Cenci-Goga B (2007) Seroprevalence of toxoplasmosis in sheep in South Africa. J S Afr Vet Assoc 78: 116–120. [DOI] [PubMed] [Google Scholar]
- 21. Kamani J, Mani AU, Egwu GO (2010) Seroprevalence of Toxoplasma gondii infection in domestic sheep and goats in Borno state, Nigeria. Trop Anim Health Prod 42: 793–797. [DOI] [PubMed] [Google Scholar]
- 22. Bekele T, Kasali OB (1989) Toxoplasmosis in sheep, goats and cattle in central Ethiopia. Vet Res Commun 13: 371–375. [DOI] [PubMed] [Google Scholar]
- 23. van der Puije WN, Bosompem KM, Canacoo EA, Wastling JM, Akanmori BD (2000) The prevalence of anti-Toxoplasma gondii antibodies in Ghanaian sheep and goats. Acta Trop 76: 21–26. [DOI] [PubMed] [Google Scholar]
- 24. Sawadogo P, Hafid J, Bellete B, Sung RT, Chakdi M, et al. (2005) Seroprevalence of T. gondii in sheep from Marrakech, Morocco. Vet Parasitol 130: 89–92. [DOI] [PubMed] [Google Scholar]
- 25. Shaapan RM, El-Nawawi FA, Tawfik MA (2008) Sensitivity and specificity of various serological tests for the detection of Toxoplasma gondii infection in naturally infected sheep. Vet Parasitol 153: 359–362. [DOI] [PubMed] [Google Scholar]
- 26. Oncel T, Vural G, Babür C, Kiliç S (2005) Detection of Toxoplasma gondii seropositivity in sheep in Yalova by Sabin Feldman Dye Test and Latex Agglutination Test. Turkiye Parazitol Derg 29: 10–12. [PubMed] [Google Scholar]
- 27. Dumètre A, Ajzenberg D, Rozette L, Mercier A, Dardé ML (2006) Toxoplasma gondii infection in sheep from Haute-Vienne, France: seroprevalence and isolate genotyping by microsatellite analysis. Vet Parasitol 142: 376–379. [DOI] [PubMed] [Google Scholar]
- 28. Berger-Schoch AE, Bernet D, Doherr MG, Gottstein B, Frey CF (2011) Toxoplasma gondii in Switzerland: A Serosurvey Based on Meat Juice Analysis of Slaughtered Pigs, Wild Boar, Sheep and Cattle. Zoonoses Public Health 58: 472–478. [DOI] [PubMed] [Google Scholar]
- 29. Bastiaensen P, Dorny P (1999) Ovine toxoplasmosis in Suriname and its possible impact on human infection. Tropicultura 16–17: 18–20. [Google Scholar]
- 30. Glor SB, Edelhofer R, Grimm F, Deplazes P, Basso W (2013) Evaluation of a commercial ELISA kit for detection of antibodies against Toxoplasma gondii in serum, plasma and meat juice from experimentally and naturally infected sheep. Parasites & Vectors 6: 85 10.1186/1756-3305-6-85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Rogers SO, Bendich AJ (1985) Extraction of DNA from milligram amounts of fresh, herbarium and mummified plant tissues. Plant Molecular Biology 5: 69–76. [DOI] [PubMed] [Google Scholar]
- 32. Chabbert E, Lachaud L, Crobu L, Bastien P (2004) Comparison of two widely used PCR primer systems for detection of toxoplasma in amniotic fluid, blood, and tissues. J Clin Microbiol 42: 1719–1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Opsteegh M, Langelaar M, Sprong H, den Hartog L, De Craeye S, et al. (2010) Direct detection and genotyping of Toxoplasma gondii in meat samples using magnetic capture and PCR. Int J Food Microbiol 139: 193–201. [DOI] [PubMed] [Google Scholar]
- 34. Khan A, Su C, German M, Storch GA, Clifford DB, et al. (2005) Genotyping of Toxoplasma gondii strains from immunocompromised patients reveals high prevalence of type I strains. J Clin Microbiol 43: 5881–5887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lindström I, Sundar N, Lindh J, Kironde F, Kabasa JD, et al. (2008) Isolation and genotyping of Toxoplasma gondii from Ugandan chickens reveals frequent multiple infections. Parasitology 135: 39–45. [DOI] [PubMed] [Google Scholar]
- 36. Boughattas S, Bergaoui R, Essid R, Aoun K, Bouratbine A (2011) Seroprevalence of Toxoplasma gondii infection among horses in Tunisia. Parasites & Vectors 4: 218 10.1186/1756-3305-4-218 [DOI] [PMC free article] [PubMed] [Google Scholar]