Abstract
During a blood meal, Lutzomyia intermedia sand flies transmit Leishmania braziliensis, a parasite causing tegumentary leishmaniasis. In experimental leishmaniasis, pre-exposure to saliva of most blood-feeding sand flies results in parasite establishment in absence of any skin damages in mice challenged with dermotropic Leishmania species together with saliva. In contrast, pre-immunization with Lu. intermedia salivary gland sonicate (SGS) results in enhanced skin inflammatory exacerbation upon co-inoculation of Lu. intermedia SGS and L. braziliensis. These data highlight potential unique features of both L. braziliensis and Lu. intermedia. In this study, we investigated the genes modulated by Lu. intermedia SGS immunization to understand their potential impact on the subsequent cutaneous immune response following inoculation of both SGS and L. braziliensis. The cellular recruitment and global gene expression profile was analyzed in mice repeatedly inoculated or not with Lu. intermedia. Microarray gene analysis revealed the upregulation of a distinct set of IFN-inducible genes, an immune signature not seen to the same extent in control animals. Of note this INF-inducible gene set was not induced in SGS pre-immunized mice subsequently co-inoculated with SGS and L. braziliensis. These data suggest the parasite prevented the upregulation of this Lu. intermedia saliva-related immune signature. The presence of these IFN-inducible genes was further analyzed in peripheral blood mononuclear cells (PBMCs) sampled from uninfected human individuals living in a L. braziliensis-endemic region of Brazil thus regularly exposed to Lu. intermedia bites. PBMCs were cultured in presence or absence of Lu. intermedia SGS. Using qRT-PCR we established that the IFN-inducible genes induced in the skin of SGS pre-immunized mice, were also upregulated by SGS in PBMCs from human individuals regularly exposed to Lu. intermedia bites, but not in PBMCs of control subjects. These data demonstrate that repeated exposure to Lu. intermedia SGS induces the expression of potentially host-protective IFN-inducible genes.
Author Summary
Leishmaniasis is a vector-borne parasitic disease of serious public health importance. No efficient vaccine is currently available. Parasites are transmitted to mammalian hosts during sand fly bites. During this process, both parasites and sand fly salivary products are delivered into the skin. Immunization with salivary proteins from most sand fly species can protect mice against cutaneous leishmaniasis; however, immunization with sand fly saliva of Lutzomyia intermedia leads to aggravation of leishmaniasis. We investigated the impact of Lutzomyia intermedia saliva exposure on the development of immune response to Leishmania braziliensis, the major causative agent of tegumentary leishmaniasis in Brazil. To this end, we analyzed in mice the gene expression pattern induced by immunization with salivary gland extracts. Among the genes highly induced were the interferon-inducible genes known to contribute to resistance against parasite infections. These genes were also induced in blood cells of human individuals that were naturally pre-exposed to bites of Lutzomyia intermedia sand flies. Interestingly, subsequent infection with Leishmania braziliensis blocked the induction of these genes in mice. These data show that the induction of potentially protective genes by insect saliva can be altered by the infecting parasite. This should be considered when including salivary components in a vaccine.
Introduction
Leishmania protozoan parasites induce a broad spectrum of disease including cutaneous lesions and visceral leishmaniasis the latter being fatal if not treated. L. braziliensis parasites can be transmitted by Lu. intermedia sand flies in Central and South America where they are the leading cause of American cutaneous and mucocutaneous leishmaniasis. During a blood meal, the host is exposed to a variety of sand fly factors. Sand fly saliva contains many pharmacological agents aimed at obtaining the optimal amount of blood for nutrition, egg development and survival. In addition, the proteophosphoglycan gel which is synthesized by the parasites inside the fly midgut can exacerbate cutaneous leishmaniasis [1], [2]. Individuals living in an endemic region are bitten by both uninfected and infected sand flies, and thus are repeatedly being exposed to sand fly saliva, leading progressively to the induction of an immune response to saliva. In Brazil, Lu. intermedia is the predominant sand fly species harboring L. braziliensis [3], [4] and in Corte de Pedra, Bahia, the endemic area studied in this report, both Lu. intermedia and Lu. whitmani sand fly species exist sympatrically with fluctuations reported for these populations [5].
The role of sand fly salivary factors is also important in the establishment of infection and thus the outcome of disease. Salivary factors include mediators that circumvent the host's hemostatic responses by preventing blood clotting, vasoconstriction and platelet aggregation for optimal feeding [6], [7]. Sand fly saliva is immunogenic and the immune response to salivary antigens modulates the microenvironment at the site of the bite with an impact on the development of disease. Co-inoculation of L. major parasites and sand fly salivary gland sonicate (SGS) from either Phlebotomus papatasi or Lutzomyia longipalpis leads to increased lesion sizes and parasite numbers [8], [9]. In contrast, several studies demonstrated that pre-immunization with P. papatasi SGS, individual components of SGS, or even uninfected sand fly bites followed by infection with L. major resulted in protection characterized by decreased lesion sizes and parasite numbers compared to controls [8], [10], [11]. These studies suggest that the immune response associated with sand fly SGS may be detrimental to the establishment of Leishmania infection and salivary molecules may be included in the design of a vaccine against leishmaniasis.
In contrast, pre-exposure to Lu. intermedia SGS surprisingly leads to enhanced disease development after infection with L. braziliensis. The exacerbated disease in these mice was associated with increased parasite burdens and low IFNγ/IL-4 ratios [12]. Pre-sensitization to Lu. intermedia SGS induced cell recruitment, an anti-SGS antibody response and a cell-mediated immune response [12], [13]. To understand the parameters involved in the increased lesion development at the site of L. braziliensis inoculation in mice pre-exposed to Lu. intermedia SGS, we examined gene expression in the skin after repeated SGS inoculations. We wanted to understand the mechanisms by which Lu. intermedia SGS modulates the microenvironment and how it may enhance susceptibility to L. braziliensis infection. The genes that were most induced in mice were further analyzed in SGS-stimulated PBMCs of healthy individuals naturally pre-exposed to Lu. intermedia sand fly bites.
Materials and Methods
Ethical Statement
For animal studies, all animal protocols were approved by the Swiss Federal Veterinary Office and experiments were performed adhering to ethical guidelines established by this office. Recommendations in the guidelines for the care and use of laboratory animals were obtained from the Department of Security and Environment of the state of Vaud, Switzerland. The protocol has been approved by the Ethics and Veterinary Office of Regulations of the state of Vaud (SAV), Switzerland under the administrative authorization number 1266-5. For human studies, written informed consent was obtained from all enrolled subjects; all procedures were approved by the Ethical Committee of the Federal University of Bahia.
Mice
Female BALB/c mice were purchased from Charles River (Lyon, France), housed under pathogen-free conditions in the BIL Epalinges Center and used for experiments between 6–8 weeks old.
Sand Flies and SGS Preparation
Adult Lu. intermedia female sand flies were captured in Corte de Pedra, Bahia, Brazil. Entomological gathering was done on private land with permission from owners for the study to be conducted on their land and within their residences. Lu. intermedia sand flies were morphologically identified according to the identification key proposed by Young and Duncan [14]. Sand fly salivary glands were dissected and stored in groups of 20 pairs in 20 mL NaCl (150 mM), Hepes buffer (10 mM; pH 7.4) at −70°C. Immediately before use, salivary glands were disrupted by ultrasonication in 1.5 mL conical tubes. Tubes were centrifuged at 10,000×g for 2 min, and the resultant supernatant (SGS) was used for the studies. All SGS batches were below the limit of detection for endotoxin activity (<0.01 EU/µg) using the LAL QCL-1000 assay (Lonza, Portsmouth, NH).
Parasites and Infections
L. braziliensis (MHOM/BR/01/BA788 strain) parasite which does not contain the Leishmania RNA virus [15] was used for experiments. The parasites were maintained in vivo in BALB/c mice and grown in vitro in M199 media (GIBCO, Paisley, UK) supplemented with 10% FCS (PAA Laboratories, Pasching, Austria), 4% HEPES (Amimed) and 2% antibiotics (penicillin, streptomycin, neomycin) (GIBCO). For infections, 1×106 stationary phase promastigotes with or without SGS (equivalent of 1 pair of Lu. intermedia salivary glands) in 10 µL PBS were injected intradermally into the ear.
Sand Fly Saliva Immunizations
Mice were immunized with salivary gland sonicate supernatant (SGS) as previously described [13]. BALB/c mice (at least 3–5 per group) were immunized 3 times with SGS (equivalent to 1 pair of Lu. intermedia salivary glands) or PBS in 10 µL in the right ear at 2-week intervals. After 2 weeks, the opposing left ear was challenged with SGS (equivalent to 1 pair of Lu. intermedia salivary glands) in the presence or absence of 1×106 stationary phase L. braziliensis promastigotes. Ear lesion size was monitored weekly and measured using a caliper. To determine cellular content, ears were digested 2 weeks after challenge in the left ear using 0.2 mg/mL Liberase TL (Roche, Rotkreuz, Switzerland) for 2 h at 37°C followed by FACS analysis [16].
Flow Cytometry
For cell surface molecules, mAb 24G2 was used to block FcRs and cells were stained using α-F4/80-biotin, α-Ly6C-FITC, α-Ly6G-APC/Cy7 (clone 1A8), α-MHCII-Alexa Fluor 700 from BioLegend (San Diego, CA) and α-CD11b-eFluor 450, α-CD11c-PE/Cy5, α-DEC205-APC, α-pan-NK CD49b-PE (clone DX5) and streptavidin-PE/Cy7 from eBioscience (San Diego, CA). All cell events were acquired on an LSRII flow cytometer (BD Biosciences, San Jose, CA) and analyzed using FlowJo (Tree Star, Ashland, OR).
Mouse Ear Pinna Processing for mRNA Isolation and Microarray Analysis
Ears were harvested 2 weeks after challenge, homogenized using a tissue lyser (Qiagen, Hilden, Germany) and mRNA was extracted by the RNeasy Plus Mini kit (Qiagen). For microarray analysis RNA was harvested from ears 2 weeks post inoculation and for each sample condition, three independent sets of 200 ng of total RNA were isolated and used as a template for probe generation using an Ambion WT expression kit (Applied Biosystems, Foster City, CA) and the cDNA was fragmented and labeled with WT DNA terminal labeling kit (Affymetrix, Santa Clara, CA). Biotinylated sense strand fragments were hybridized to Affymetrix Mouse Gene 1.0 ST GeneChips using the Hybridization Control and Hybridization Wash and Stain kits at 45°C for 18 h. The stained array was scanned using an Affymetrix GeneChip Scanner 3000 7G to generate the CEL files. The chip data were imported with Partek Genomics Suite 6.5 (Partek, Inc., St. Louis, MO), normalized and summarized using the RMA (Robust Multiarray Average) algorithm. The relative log expression was examined to ensure that the data were properly corrected by normalization and that there were no outliers. Scatter plots were generated using Matlab 2012a (MathWorks, Natick, MA) and DataGraph 3.0 (Visual Data Tools Inc., Chapel Hill, NC). To identify expression changes between genotypes, a one-way ANOVA with contrast was performed by using the methods-of-moments.
Mouse Ear Pinna Processing for Quantitative Real-Time PCR
Quantitative real-time PCR was carried out using random 9-mers, M-MLV reverse transcriptase RNase H- (Promega, Madison, WI) and SYBR green on a LightCycler 480 system (Roche). The primer sequences are listed in Table S1. Thermal cycle conditions started with a 5 min denaturation at 95°C and 45 cycles at 95°C for 10 sec, 60°C for 10 sec and 72°C for 10 sec. The results were normalized to the housekeeping gene hypoxanthine phosphoribosyl transferase (HPRT) using the comparative threshold cycle method (2ΔΔCT) for relative quantification [16].
Processing of Human Blood Samples for Quantitative Real-Time PCR
Samples used in the present study were obtained from individuals enrolled in an epidemiological survey conducted in Corte de Pedra, Brazil, an endemic region for American cutaneous leishmaniasis, where Lu. intermedia sand flies transmit L. braziliensis [5]. Details of the area and patients are described elsewhere [17]. For the present study, individuals (n = 7) were selected based on a positive ELISA for anti-Lu. intermedia salivary molecules; the cutoff OD values for a positive anti-Lu. intermedia SGS response were established using control individuals [9]. None of control individuals had history of Leishmania infection and all had a negative Leishmania skin test. For the control group, four individuals living in a non-endemic area of Salvador, Bahia were selected based on their lack of exposure to Lu. intermedia SGS as determined by serology using SGS-specific ELISAs.
Following Ficoll-Hypaque gradient centrifugation, peripheral blood mononuclear cells (PBMCs) were resuspended in RPMI-1640 supplemented with 2 mM L-glutamine, penicillin (100 U/mL), streptomycin (100 µg/mL) (all from Invitrogen), and heat inactivated human serum AB Rh+ (Sigma Chemical Co., MO). PBMCs (3×106/mL) were washed two times and resuspended in complete RPMI. Cells were plated in 24-well plates (Corning Incorporated Life Sciences, Lowell, MA) and incubated at 37°C, 5% CO2 in the presence or not of SGS (equivalent to 1.5 pairs of salivary glands) for 72 h. Following stimulation, cells were harvested and total RNA was extracted using Trizol (Life Technologies, Rockville, MD), according to manufacturer's instructions. RNA was eluted in water and used for cDNA synthesis (ImProm-II reverse transcription system-Promega). Real-time PCR was performed on the ABI Prism 7500 (Applied Biosystems). The primer sequences are found in Table S1. Thermal cycle conditions consisted of a two-min initial incubation at 50°C followed by a 10 min denaturation at 95°C and 50 cycles at 95°C for 15 sec and 60°C for one min each. Samples were analyzed in triplicate and the comparative method was used where gene expression cycle threshold (Ct) values were normalized to HPRT expression as determined by the equation ΔCt = Ct (target gene)−Ct (hprt). Fold change was determined by 2−ΔΔCt, where ΔΔCt = ΔCt (SGS)−ΔCt (medium) [18].
Statistics
Statistical analysis was performed using GraphPad Prism 5 software (San Diego, CA). For murine experiments, a two-tailed Student's unpaired t-test was carried out. For human experiments, a nonparametric Mann-Whitney test was applied.
Results
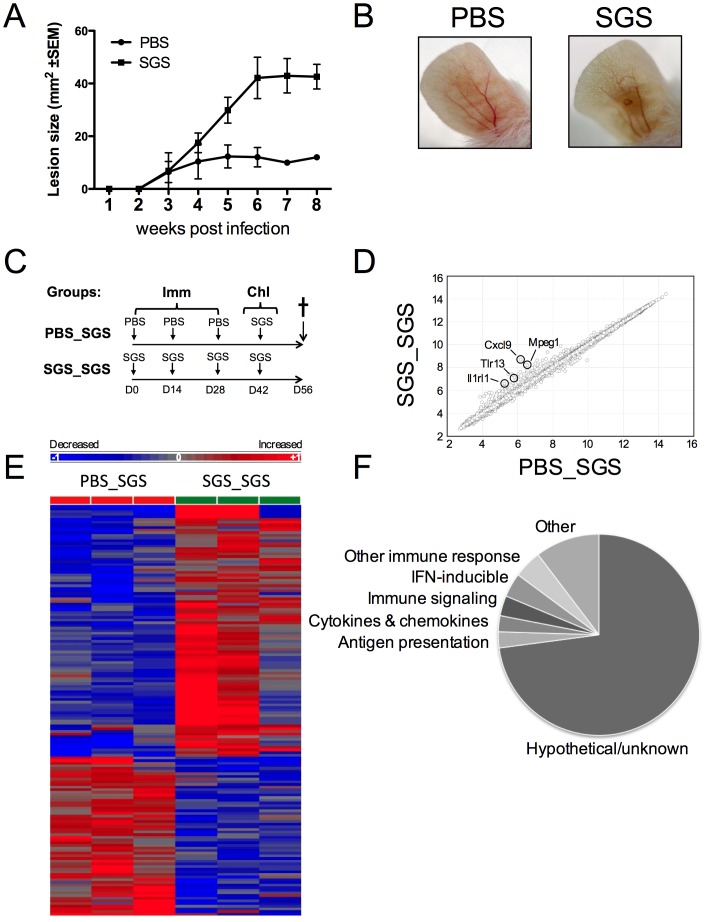
Repeated pre-exposure of BALB/c mice to Lu. intermedia SGS enhances susceptibility to L. braziliensis infection with a lesion beginning at 3 weeks post-infection (Fig. 1A and 1B) in line with previously published results [12]. Thus, we wanted to determine if differences in cellular recruitment due to pre-immunization with Lu. intermedia SGS prior to infection could explain the differences in disease status. Therefore, mice were repeatedly pre-immunized with SGS or inoculated with PBS and both groups were challenged with SGS in the contralateral ear. We examined the cellular infiltrate of the ear two weeks after SGS challenge, when the adaptive immune response is ongoing and the parasite has typically already established infection, despite a lack of detectable differences in lesion size. At this point, no significant differences were observed in the total number of cells, or the numbers of neutrophils, macrophages or DCs in the ears of mice pre-immunized with SGS compared to those inoculated with PBS (Fig. S1).
Figure 1. Gene categories modulated by SGS pre-immunization.
BALB/c mice were inoculated 3 times in the right ear pinna every 2 wks with SGS from 1 pair of Lu. intermedia salivary glands and then challenged 2 wks later with Lu. intermedia SGS plus 1×106 L. braziliensis parasites. (A) Lesion development was monitored weekly. Each point is the mean ±SEM of 5 animals per group. (B) Lesion images of ear pinna at 8 wks p.i. and these data are representative of two independent experiments. (C) BALB/c mice were pre-immunized (Imm) 3 times in the right ear every 2 wks with Lu. intermedia SGS or PBS and then challenged (Chl) in the left ear 2 wks later with Lu. intermedia SGS. The challenged left ears were collected after 2 wks, homogenized and gene expression was determined by microarray analysis. (D) Global significant differences in gene expression shown in log2 were determined comparing mice pre-immunized with SGS to those given PBS (n = 3 mice per group) and hierarchical clustering revealed genes differentially expressed >1.5× with a p value of <0.05 are presented in a heat map (E) and separated based on functional categories in a pie chart (F).
As a result, we hypothesized that alterations in gene expression in response to repeated exposures to Lu. intermedia SGS may be modulating the local skin microenvironment, impacting the innate and adaptive immune responses and thus the outcome of disease. To examine the effect of SGS pre-immunization at the inoculation site, we carried out a microarray analysis in mice that were pre-immunized with SGS or inoculated with PBS and later challenged with SGS in the opposing ear dermis. The ear pinna was processed and analyzed two weeks after the last SGS challenge (Fig. 1C).
Overall, there were few differences in the global gene expression patterns between mice that were repeatedly pre-exposed to SGS and challenged with SGS compared to those inoculated with PBS and challenged with SGS. However, hierarchical clustering analysis revealed that 95 genes were increased and 60 genes were decreased in response to SGS pre-immunization compared to control mice (Fig. 1D and 1E). Of the 155 transcripts modulated by SGS pre-immunization, only 49 transcripts have been annotated, or ascribed to a specific gene, and the rest are classified as hypothetical or unknown. Despite the majority of these genes being classified as hypothetical or unknown, many of the transcripts that were differentially expressed in response to SGS pre-sensitization are known to play a role in immune processes like antigen presentation and signaling as well as transcripts encoding for cytokines, chemokines and their receptors (Fig. 1F).
Of the 49 annotated genes differentially regulated with SGS pre-immunization, the microarray analysis revealed all but one of these annotated genes was increased upon SGS challenge in mice pre-immunized with SGS compared to those inoculated with PBS (Table 1). Of the 49 annotated genes, 4 transcripts had greater than 2-fold expression in SGS pre-exposed mice compared to controls; the gene most highly expressed in SGS pre-immunized mice compared to PBS-inoculated animals was CXCL9. Mpeg1, a transcript indicative of macrophage presence, as well as IL-1rl1 and TLR13 which are members of the toll-like superfamily of receptors, were also significantly elevated in response to SGS challenge in SGS pre-exposed mice compared to controls. The microarray results revealed an especially high frequency (14.3% of the annotated genes) of genes induced in response to SGS pre-immunization to be IFN-inducible genes including immunity-related GTPases (IRGs) and guanylate-binding proteins (GBPs) [19]–[25].
Table 1. Genes differentially expressed in response to SGS pre-immunization.
| Gene Symbol | Description | Fold Change | p-value |
| Antigen Presentation | |||
| CD74 | CD74 antigen (invariant chain) | 1.65 | 0.018 |
| H2-gs10 | MHCI like protein | 1.72 | 0.003 |
| H2Q6 | Histocompatibility 2 | 1.61 | 0.009 |
| Tap1 | Transporter 1 | 1.51 | 0.0006 |
| Chemokine, cytokines and their receptors | |||
| CXCL9 (MIG) | Chemokine ligand 9 | 5.72 | 0.037 |
| IL-1rl1 (ST2) | IL-1 receptor-like 1 | 2.58 | <0.05 |
| IL-7r (CD127) | IL-7 receptor | 1.53 | 0.017 |
| IL-10rα | IL-10 receptor α | 1.69 | 0.016 |
| Immune response signaling | |||
| CD180 | CD180 antigen | 1.99 | 0.034 |
| Sfpi1 (PU.1) | SFFV proviral integration 1 | 1.54 | 0.038 |
| Sla | Src-like adaptor | 1.55 | 0.027 |
| Stat1 | Signal transducer and activator of transcription 1 | 1.51 | 0.017 |
| TLR13 | Toll-like receptor 13 | 2.48 | 0.049 |
| IFN-inducible genes | |||
| Gbp6 | Guanylate-binding protein 6 | 1.74 | 0.031 |
| Gpb8 | Guanylate-binding protein 8 | 1.86 | 0.037 |
| Ifit1 | IFN-induced protein | 1.62 | 0.007 |
| Iigp1 | IFN-inducible GTPase | 3.38 | 0.049 |
| Irgm1 | Immunity-related GTPase | 1.65 | 0.033 |
| Irgm2 | Immunity-related GTPase | 2.16 | 0.027 |
| Other immune response genes | |||
| Aif1 | Allograft inflammatory factor 1 | 1.84 | 0.012 |
| Chi3l1 (Ym1) | Chitinase 3-like 1 | 1.77 | 0.005 |
| Igsf6 | Immunoglobulin superfamily member | 1.98 | 0.044 |
| Klrd1 | Killer cell lectin-like receptor | 1.66 | 0.030 |
| Mpeg1 | Macrophage expressed gene | 3.23 | 0.044 |
| Nkg7 | Natural killer cell group 7 | 1.67 | 0.040 |
| Pdcd1lg | Programmed cell death 1 ligand 2 | 1.96 | 0.035 |
| Other genes | |||
| Apobec1 | Apolipoprotein B | 1.65 | 0.021 |
| Atp8b4 | ATPase 8B | 1.70 | 0.004 |
| Dpep2 | Dipeptidase 2 | 1.52 | 0.040 |
| F10 | Coagulation factor X | 1.88 | 0.038 |
| Fyb | FYN binding protein | 1.94 | 0.010 |
| Havcr2 | Hepatitis A virus cellular receptor | 1.51 | 0.038 |
| Lgals3bp | Lectin, galactoside-binding | 1.50 | 0.042 |
| Mir203 | microRNA 203 | −1.53 | 0.022 |
| Mrgpra9 | MAS-related GPR | 1.54 | 0.031 |
| Ms4a4b | Membrane-spanning 4 domains | 1.99 | 0.020 |
| Myo1f | Myosin 1F | 1.65 | 0.023 |
| Naaa | N-acylethanolamine acid amidase | 1.52 | 0.033 |
| Ptprc | Protein tyrosine phosphatase receptor 1 | 1.91 | 0.046 |
| Samhd1 | SAM domain and HD domain | 1.51 | 0.048 |
| Slfn1 | Schlafen 1 | 1.76 | 0.030 |
| Sp110 | Nuclear body protein | 1.57 | <0.05 |
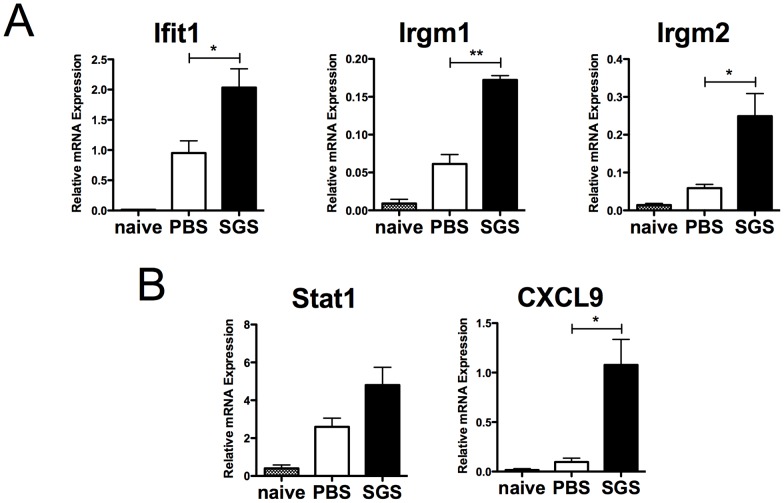
Given the surprisingly large proportion of the modulation of IFN-inducible genes in mice pre-immunized with SGS compared to control mice, we carried out real-time qPCR for IFN-inducible genes as well as genes associated with IFN-induced responses on a biological replicate experiment to confirm the findings of the microarray analysis. Cells from mice pre-sensitized with SGS and challenged with SGS had a higher expression of Ifit1, Irgm1 and Irgm2 compared to mice inoculated with PBS and challenged with SGS. Of note, despite these differences, challenge with SGS in mice pre-immunized or not with SGS had a higher expression of these genes compared to naïve mice, suggesting SGS inoculation alone can already induce this gene family (Fig. 2A). In addition, pre-immunization with SGS also led to an increased expression of Stat1, a signaling molecule responsible for the subsequent expression of IFN-inducible genes. CXCL9, a chemokine involved in T cell migration induced by IFNγ, was also expressed at higher levels in SGS pre-exposed mice compared to control mice (Fig. 2B).
Figure 2. IFN-inducible genes are upregulated in response to SGS pre-immunization.
BALB/c mice were inoculated 3 times in the right ear every 2 wks with Lu. intermedia SGS or PBS and then challenged in the left ear 2 wks later with Lu. intermedia SGS. The left ears were collected 2 wks after SGS challenge, homogenized and the expression of IFN-inducible genes such as (A) Ifit1, Irgm1 and Irgm2, and the expression of IFN-related genes like (B) Stat1 and CXCL9 was determined by real-time quantitative PCR normalized relative to HPRT mRNA levels. Similar analysis of ear pinna of naïve mice that did not receive any injections and were not challenged with SGS is also shown. Data are an independent biological replicate to the microarray analysis; relative mRNA expression normalized to the housekeeping gene HPRT is presented as the mean +SEM with 3–5 mice per group; ** p<0.005, * p<0.05 by Student's t-test.
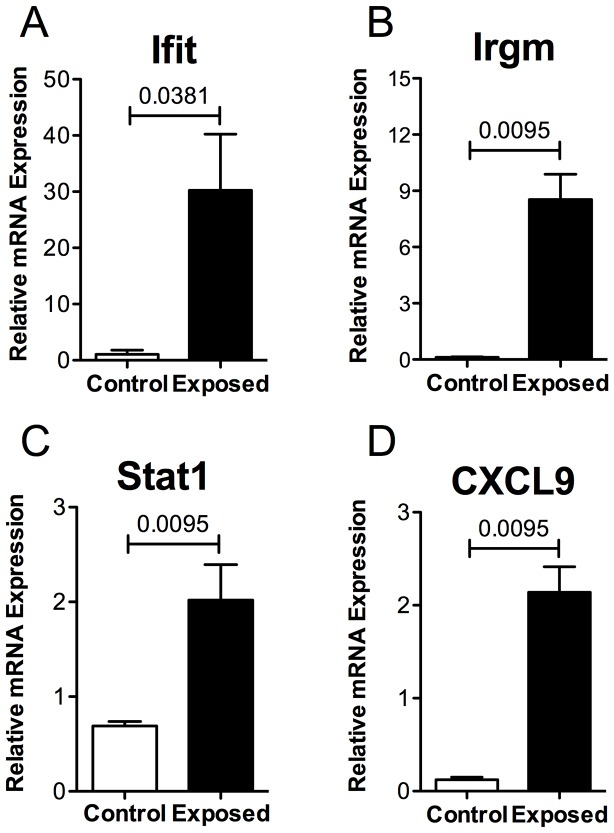
Despite a reduced number of IRG homologues in humans, some of the IFN-inducible genes modulated in the mouse upon SGS pre-immunization have homologues in humans [26]–[30]. In order to determine if the same genes were upregulated in humans naturally exposed to Lu. intermedia saliva, we isolated PBMCs from individuals living in Corte de Pedra, Brazil, an area endemic for L. braziliensis with active Lu. intermedia sand fly transmission [31]. Exposure to Lu. intermedia bites was determined based on a positive serology result for anti-Lu. intermedia SGS antibodies using a mean OD cutoff of 0.2711 (+/− 0.1006 SD) (Carvalho et al., unpublished data). Following PBMC isolation, cells were cultured in the presence or absence of Lu. intermedia SGS followed by mRNA isolation. PBMCs from these exposed individuals exhibited higher levels of Ifit1, Irgm, Stat1 and CXCL9 mRNA in response to SGS compared to PBMCs isolated from people living in a non-endemic area (Fig. 3). Similarly, supernatants from PBMCs of individuals living in an endemic area stimulated with SGS also produced significantly more CXCL9 protein as measured by ELISA compared to controls (data not shown). These data demonstrate that IFN-inducible genes are induced in response to SGS pre-sensitization in both the experimental model and in cells from human individuals pre-exposed to Lu. intermedia bites.
Figure 3. IFN-inducible genes are upregulated in human PBMCs by individuals exposed to sand fly bites.
PBMCs from people exposed to Lu. intermedia bites living in an endemic area who expressed high anti-SGS antibody were isolated and stimulated in vitro with SGS (equivalent to 1.5 pairs of salivary glands) for 72 hours. For the control group, PBMCs were isolated from individuals living in a non-endemic area of Salvador, Bahia, Brazil and stimulated with SGS for 72 hours. The expression of (A) Ifit, (B) Irgm, (C) Stat1, and (D) CXCL9 was determined by real-time quantitative PCR. *** p<0.0001.
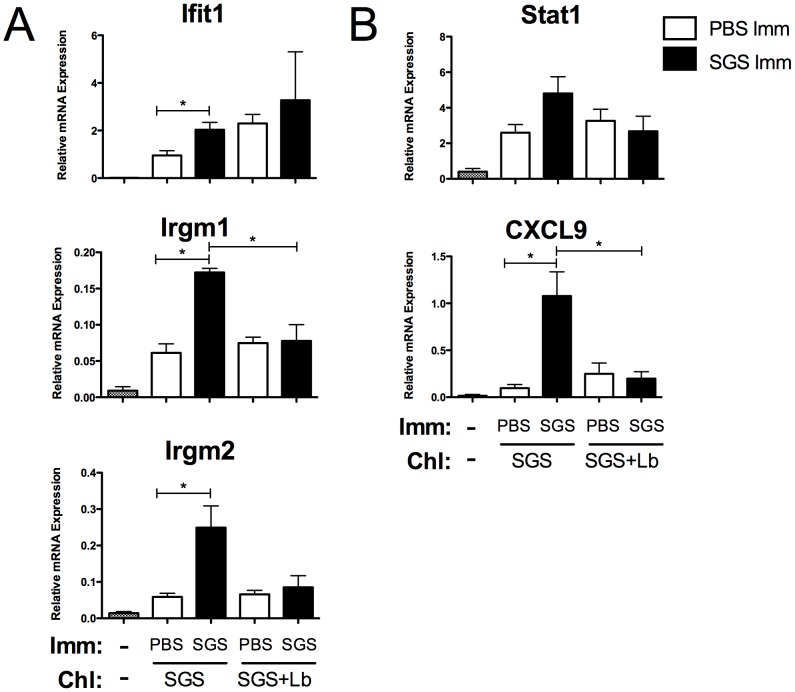
Here, we show that SGS pre-immunization induces the expression of IFN-inducible genes, which are typically associated with a protective response as Irgm1−/− animals are highly susceptible to Leishmania infection (mentioned as data not shown in [21]). However, pre-immunization with Lu. intermedia SGS has been reported to enhance L. braziliensis infection [12]. To evaluate the effect of the parasite on the local immune response induced by pre-immunization with SGS, mice were repeatedly pre-exposed to SGS or PBS and challenged with SGS in the presence or absence of L. braziliensis parasites. Gene expression profiling studies were carried out 2 weeks later. Mice that were pre-sensitized with SGS and challenged with SGS alone significantly upregulated the expression of Ifit1, Irgm1, Irgm2, Stat1 and CXCL9 compared to PBS pre-inoculated mice in line with our microarray data (Fig. 4). However, the mice that were pre-exposed to SGS and challenged with SGS and L. braziliensis did not significantly upregulate the expression of these genes compared to controls (Fig. 4 and Table S2). Taken together, these data suggest that L. braziliensis parasites modulate host gene expression at the site of infection creating a more hospitable environment for parasite establishment which is associated with increased lesion development.
Figure 4. L. braziliensis infection prevents the upregulation of IFN-inducible genes due to SGS pre-immunization.
BALB/c mice were inoculated 3 times in the right ear pinna every 2 wks with Lu. intermedia SGS and then challenged in the left ear 2 wks later with Lu. intermedia SGS plus 1×106 L. braziliensis stationary phase promastigotes. The left ears were collected 2 wks after infection, homogenized and the expression of IFN-inducible genes such as (A) Ifit1, Irgm1 and Irgm2, and the expression of IFN-related genes like (B) Stat1 and CXCL9 was determined by real-time quantitative PCR. Relative mRNA expression normalized to the housekeeping gene HPRT is presented as the mean +SEM (n = 5 mice per group); * p<0.05 by Student's t-test.
Discussion
Many studies have suggested the anti-saliva response against sand fly species such as P. papatasi or L. longipalpis is detrimental for the establishment of Leishmania infection. In contrast, the Lu. intermedia anti-saliva response does not prevent the development of disease, but rather may modulate the outcome of infection. Studies in a mouse experimental model have demonstrated that L. braziliensis infection alone induces a strong Th1 cell immune response with high levels of IFNγ and elevated numbers of IFNγ-producing CD4+ and CD8+ T cells in the dLN [32]–[34]. The strong protective immune response characterized by the presence of IFNγ was thought to correlate with the strong resistance to L. braziliensis infection [34]–[39]. Here, we show that repeated pre-immunizations with Lu. intermedia SGS alters the skin microenvironment and induces the expression of a variety of genes involved in the immune response, especially from the family of IFN-inducible genes. Genomic analysis of the skin of mice pre-immunized with SGS reveals an inflammatory setting with an increase in genes involved in immune responses including antigen presentation and cell signaling. Genes associated with Th1 cell immune responses such a CXCL9, a chemokine typically linked with the recruitment of Th1 cells, exhibited the greatest fold induction at >5 times over control mice. The IL-7R, also known to influence the Th1 cell immune response, was also elevated following SGS pre-exposure (this study and [40]–[42]). Of note, cytokines typically associated with a Th2 cell immune response such as Chi3l1 (Ym1), or regulatory cytokines such as IL-10Rα were also detected at higher levels in SGS pre-immunized mice.
In the periphery IFNγ binds to its receptor and initiates the JAK/STAT signaling pathway leading to the phosphorylation and translocation of STAT1 to the nucleus which induces the transcription of more than 2000 genes including effector molecules that suppress the growth and survival of intracellular pathogens (Phox, iNOS, IDO, NRAMP1, GTPases, Ifits and chemokines) [21]. Remarkably, several IFN-inducible genes as well as the IFN signaling molecule, STAT1, were upregulated at the site Lu. intermedia challenge in mice that were pre-exposed through immunization. For example, p47 GTPases such as Irgm1 (formerly Lrg47) and Irgm2 (Gtpi) and p65 GTPases such as GBP6 and GBP8 were expressed at high levels in pre-immunized mice.
Interestingly, IFN-inducible genes were similarly induced in SGS-stimulated PBMCs isolated from humans living in an area endemic for L. braziliensis with active Lu. intermedia sand fly transmission. It was not possible to perform skin biopsy in the human population studied due to ethical considerations; however, the expression of IFN-inducible genes in SGS-stimulated blood cells of individuals naturally exposed to sand fly bites was similar to that observed at the site of SGS challenge in mice. Collectively, these data demonstrate that the induction of IFN-inducible genes by SGS is also occurring in humans.
To our knowledge this is the first report demonstrating an induction in the expression of IFN-inducible GTPases in response to vector saliva. These products have been well characterized for their role in host defense against viruses but they also contribute to resistance against protozoans. Mice deficient for either Irgm1 or many of the other GTPases are highly susceptible to infection with Toxoplasma gondii, Trypanosoma cruzi and Leishmania major, and many mimic the dramatic susceptibility phenotypes seen in IFNγR-deficient mice [21]. It should be noted that IFN-inducible genes are turned on in response to IFNγ but type I IFNs may also contribute, although to a lesser degree [21]. In our analysis neither IFNγ or type I IFNs were elevated in cells from mice pre-sensitized with SGS but this may be a reflection of the time point analyzed (14 days post inoculation).
In this study mice were immunized with SGS to mimic one of the features of natural transmission of Leishmania where individuals are pre-exposed to several sand fly bites prior to deposition of parasites by the sand fly. A high dose of L. braziliensis promastigotes was co-inoculated in mice with SGS in an attempt to reproduce the cell recruitment rapidly observed at the site of infection after a sand fly bite. However, it is important to note that upon a blood meal, the sand fly is inoculating fewer parasites and also regurgitating many other factors including metacyclic promastigotes embedded in a proteophosphoglycan gel in a blood pool [2]. These factors are not all present during needle inoculation of the parasites and SGS. It is clear that further studies using natural sand fly infection will be required for a better understanding of the transmission dynamics during Leishmania infection.
Repeated exposures to Lu. intermedia SGS followed by challenge with L. braziliensis parasites in the presence of SGS leads to an enhanced disease compared to control mice (Fig. 1A and [12]). Interestingly, in this prior study the SGS pre-immunized mice challenged with L. braziliensis plus SGS, and analyzed two weeks later had a lower parasite load compared to mice not immunized with SGS, suggesting a transient protection conferred by SGS pre-immunization [12]. However, the trend was inversed from 3 weeks on and the SGS-immunized group showed increased lesion size and parasite load [12]. Additionally, the mice pre-immunized with SGS and subsequently infected with L. braziliensis plus SGS had lower levels of IFNγ to IL-4 ratios compared to mice inoculated with PBS and infected with L. braziliensis plus SGS at 2 weeks post infection. Thus, in that study, the highest levels of IFNγ were not associated with the decreased parasite numbers in vivo suggesting IFNγ is not the major factor contributing to parasite killing by macrophages at this time point. In our study, increased expression of IFN-inducible genes and of IFNγ was also not detected in the microarray performed in SGS pre-immunized mice 2 weeks post co-inoculation of L. braziliensis and SGS compared to PBS controls challenged with L. braziliensis plus SGS. In this and the previous study, the levels of IFNγ upon challenge with L. braziliensis and SGS were not elevated following SGS pre-immunization. Thus other factors may be involved in the transient control of parasite load observed by de Moura and colleagues [12]. Furthermore, higher concentrations of IFNγ are required for optimal parasite killing of L. braziliensis compared to L. major suggesting differences in the susceptibilities to IFNγ-mediated killing between different parasite strains [15]. Nevertheless, following L. braziliensis and SGS co-inoculation, both studies showed increased inflammatory lesions in the group pre-immunized with SGS compared to that injected with PBS.
This increase in disease severity to L. braziliensis infection in SGS pre-immunized mice, corresponds to a silencing of many of the genes turned on by SGS pre-sensitization, including IFN-inducible genes. This suggests that the parasite is actively modulating the host's immune response to the SGS. Interestingly, this observation is consistent with previous findings showing a decreased ratio of IFNγ/IL-4 production in the dLNs of mice pre-exposed to SGS and challenged with parasites [12]. In the same line, the same group further reported that challenge with L. braziliensis plus SGS after SGS pre-immunization also silenced CXCL10, another IFN-inducible gene [13]. Despite differences in the methodology used between these studies (air pouch model in the former studies and needle inoculation in the ear pinna in the current study), the outcomes are going in the same direction. In addition, the impact of SGS on the skin microbiome which was shown to influence skin immunity may also contribute to the phenotype observed [43]. We show here that there is an obvious benefit for the parasite to down-modulate the IRG system expressed in response to SGS pre-exposure to allow for parasite establishment. However, modulation of IFN-inducible genes is most likely not the only mechanism for enhancing disease. It is unclear how the parasite is altering the host's response to the SGS in the present study and this will require further investigation.
In conclusion, we have shown that in both humans and mice, an array of IFN-inducible genes were up-regulated in response to Lu. intermedia SGS pre-exposure. Interestingly, these genes were silenced when the parasite was present during the challenge. Given the marked changes in the skin microenvironment resulting from repeated exposures to Lu. intermedia SGS, and the different outcomes to Leishmania infection, understanding the relationship between pathogens and their homologous vectors is essential. Since SGS proteins from different sand fly species can either exacerbate or protect from disease, subsequent studies will aim to understand how the parasite is modulating SGS impact on the microenvironment [44]. This will help determine risk factors for disease development, markers of exposure and defining potential vaccine candidates.
Supporting Information
SGS pre-immunization does not modify cellular recruitment in response to L. braziliensis inoculation. BALB/c mice were inoculated 3 times in the right ear every 2 wks with 1 pair of Lu. intermedia salivary glands and then challenged 2 wks later in the left ear with L. intermedia SGS. Ears were digested 2 wks post inoculation and cellular content was analyzed by FACS. Cell numbers are shown as the mean +SEM with 5 mice per group. Data are results from one experiment and representative of 2 individual experiments.
(TIF)
List of the primers used in this study to analyze gene expression by RT-PCR in mouse and human samples.
(DOC)
List of the genes that were positively (>1.5 fold) or negatively (<1.5 fold) regulated at the site of parasite inoculation in mice immunized with SGS and subsequently infected with L. braziliensis plus SGS, compared to mice pretreated with PBS and infected with L. braziliensis plus SGS. Contralateral ears were isolated for microarray analysis 2 weeks after parasite challenge. The data are presented as the fold change of mice pre-immunized with SGS (3 times every 2 weeks) over mice inoculated with PBS and challenged with L. braziliensis plus SGS. p-values<0.05: statistically significant. The values for IFN-inducible genes (below the IFN-inducible shaded line) are given but they did not vary >1.5 times and were not statistically significant between L. braziliensis samples that were pretreated with SGS or PBS.
(DOC)
Acknowledgments
The authors thank Dr. Floriane Auderset, Steffen Schuster, Manuel Coutaz, Benjamin Hurrell for their discussion and Corentin Pasche for technical support (UNIL, Lausanne, Switzerland). We would like to acknowledge Prof. Edgar Carvalho input (Universidade Federal da Bahia, Salvador, Bahia, Brazil), the personnel at the Health Poste of Corte de Pedra, Brazil and all the field workers involved in this study. We acknowledge the Genomic Platform of the University of Geneva, Switzerland for performing the transcriptomic experiments with special appreciation to Didier Chollet and Olivier Schaad for their assistance with the microarray protocol and analysis, and Dan Beiting of the University of Pennsylvania for computational support.
Funding Statement
This work was supported by the Brazilian-Swiss Research Program [BSJRP 011007 to FTC and AB] and the Swiss National Foundation [3100030-129852 to FTC]. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Andrade BB, Teixeira CR (2012) Biomarkers for exposure to sand flies bites as tools to aid control of leishmaniasis. Front Immunol 3: 121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Rogers ME (2012) The role of leishmania proteophosphoglycans in sand fly transmission and infection of the Mammalian host. Front Microbiol 3: 223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Rangel EF, Azevedo AC, Andrade CA, Souza NA, Wermelinger ED (1990) Studies on sandfly fauna (Diptera: Psychodidae) in a foci of cutaneous leishmaniasis in mesquita, Rio de Janeiro State, Brazil. Mem Inst Oswaldo Cruz 85: 39–45. [DOI] [PubMed] [Google Scholar]
- 4. Rangel EF, Souza NA, Wermelinger ED, Azevedo AC, Barbosa AF, et al. (1986) [Phlebotomus of Vargem Grande, a focus of cutaneous leishmaniasis in the State of Rio de Janeiro]. Mem Inst Oswaldo Cruz 81: 347–349. [DOI] [PubMed] [Google Scholar]
- 5. Miranda JC, Reis E, Schriefer A, Goncalves M, Reis MG, et al. (2002) Frequency of infection of Lutzomyia phlebotomines with Leishmania braziliensis in a Brazilian endemic area as assessed by pinpoint capture and polymerase chain reaction. Mem Inst Oswaldo Cruz 97: 185–188. [DOI] [PubMed] [Google Scholar]
- 6. Andrade BB, de Oliveira CI, Brodskyn CI, Barral A, Barral-Netto M (2007) Role of sand fly saliva in human and experimental leishmaniasis: current insights. Scand J Immunol 66: 122–127. [DOI] [PubMed] [Google Scholar]
- 7. Bates PA (2007) Transmission of Leishmania metacyclic promastigotes by phlebotomine sand flies. Int J Parasitol 37: 1097–1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Belkaid Y, Kamhawi S, Modi G, Valenzuela J, Noben-Trauth N, et al. (1998) Development of a natural model of cutaneous leishmaniasis: powerful effects of vector saliva and saliva preexposure on the long-term outcome of Leishmania major infection in the mouse ear dermis. J Exp Med 188: 1941–1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Titus RG, Ribeiro JM (1988) Salivary gland lysates from the sand fly Lutzomyia longipalpis enhance Leishmania infectivity. Science 239: 1306–1308. [DOI] [PubMed] [Google Scholar]
- 10. Kamhawi S, Belkaid Y, Modi G, Rowton E, Sacks D (2000) Protection against cutaneous leishmaniasis resulting from bites of uninfected sand flies. Science 290: 1351–1354. [DOI] [PubMed] [Google Scholar]
- 11. Valenzuela JG, Belkaid Y, Garfield MK, Mendez S, Kamhawi S, et al. (2001) Toward a defined anti-Leishmania vaccine targeting vector antigens: characterization of a protective salivary protein. J Exp Med 194: 331–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. de Moura TR, Oliveira F, Novais FO, Miranda JC, Clarencio J, et al. (2007) Enhanced Leishmania braziliensis infection following pre-exposure to sandfly saliva. PLoS Negl Trop Dis 1: e84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. de Moura TR, Oliveira F, Rodrigues GC, Carneiro MW, Fukutani KF, et al. (2010) Immunity to Lutzomyia intermedia saliva modulates the inflammatory environment induced by Leishmania braziliensis. PLoS Negl Trop Dis 4: e712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Young DG, Duncan MA (1994) Guide to the idenfication and geographic distribution of Lutzomyia sand flies in Mexico, the West Indies, Central and South America (Diptera: Psychodidae). Gainesville: Memories American Entomological Institute No 54 Asscoiated Publishers. [Google Scholar]
- 15. Weinkopff T, Mariotto A, Simon G, Hauyon-La Torre Y, Auderset F, et al. (2013) Role of Toll-like receptor 9 signaling in experimental Leishmania braziliensis infection. Infect Immun 81: 1575–1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Charmoy M, Brunner-Agten S, Aebischer D, Auderset F, Launois P, et al. (2010) Neutrophil-derived CCL3 is essential for the rapid recruitment of dendritic cells to the site of Leishmania major inoculation in resistant mice. PLoS Pathog 6: e1000755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Schnorr D, Muniz AC, Passos S, Guimaraes LH, Lago EL, et al. (2012) IFN-gamma production to leishmania antigen supplements the leishmania skin test in identifying exposure to L. braziliensis infection. PLoS Negl Trop Dis 6: e1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25: 402–408. [DOI] [PubMed] [Google Scholar]
- 19. Santiago HC, Feng CG, Bafica A, Roffe E, Arantes RM, et al. (2005) Mice deficient in LRG-47 display enhanced susceptibility to Trypanosoma cruzi infection associated with defective hemopoiesis and intracellular control of parasite growth. J Immunol 175: 8165–8172. [DOI] [PubMed] [Google Scholar]
- 20. Collazo CM, Yap GS, Sempowski GD, Lusby KC, Tessarollo L, et al. (2001) Inactivation of LRG-47 and IRG-47 reveals a family of interferon gamma-inducible genes with essential, pathogen-specific roles in resistance to infection. J Exp Med 194: 181–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Taylor GA, Feng CG, Sher A (2004) p47 GTPases: regulators of immunity to intracellular pathogens. Nat Rev Immunol 4: 100–109. [DOI] [PubMed] [Google Scholar]
- 22. Taylor GA (2007) IRG proteins: key mediators of interferon-regulated host resistance to intracellular pathogens. Cell Microbiol 9: 1099–1107. [DOI] [PubMed] [Google Scholar]
- 23. Taylor GA, Feng CG, Sher A (2007) Control of IFN-gamma-mediated host resistance to intracellular pathogens by immunity-related GTPases (p47 GTPases). Microbes Infect 9: 1644–1651. [DOI] [PubMed] [Google Scholar]
- 24. Hunn JP, Feng CG, Sher A, Howard JC (2011) The immunity-related GTPases in mammals: a fast-evolving cell-autonomous resistance system against intracellular pathogens. Mamm Genome 22: 43–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Howard JC, Hunn JP, Steinfeldt T (2011) The IRG protein-based resistance mechanism in mice and its relation to virulence in Toxoplasma gondii. Curr Opin Microbiol 14: 414–421. [DOI] [PubMed] [Google Scholar]
- 26. Bekpen C, Hunn JP, Rohde C, Parvanova I, Guethlein L, et al. (2005) The interferon-inducible p47 (IRG) GTPases in vertebrates: loss of the cell autonomous resistance mechanism in the human lineage. Genome Biol 6: R92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Singh SB, Davis AS, Taylor GA, Deretic V (2006) Human IRGM induces autophagy to eliminate intracellular mycobacteria. Science 313: 1438–1441. [DOI] [PubMed] [Google Scholar]
- 28. Pichlmair A, Lassnig C, Eberle CA, Gorna MW, Baumann CL, et al. (2011) IFIT1 is an antiviral protein that recognizes 5′-triphosphate RNA. Nat Immunol 12: 624–630. [DOI] [PubMed] [Google Scholar]
- 29. Liu L, Okada S, Kong XF, Kreins AY, Cypowyj S, et al. (2011) Gain-of-function human STAT1 mutations impair IL-17 immunity and underlie chronic mucocutaneous candidiasis. J Exp Med 208: 1635–1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lee HH, Farber JM (1996) Localization of the gene for the human MIG cytokine on chromosome 4q21 adjacent to INP10 reveals a chemokine “mini-cluster”. Cytogenet Cell Genet 74: 255–258. [DOI] [PubMed] [Google Scholar]
- 31. Jirmanus L, Glesby MJ, Guimaraes LH, Lago E, Rosa ME, et al. (2012) Epidemiological and clinical changes in American tegumentary leishmaniasis in an area of Leishmania (Viannia) braziliensis transmission over a 20-year period. Am J Trop Med Hyg 86: 426–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. de Souza-Neto SM, Carneiro CM, Vieira LQ, Afonso LC (2004) Leishmania braziliensis: partial control of experimental infection by interleukin-12 p40 deficient mice. Mem Inst Oswaldo Cruz 99: 289–294. [DOI] [PubMed] [Google Scholar]
- 33. Indiani de Oliveira C, Teixeira MJ, Teixeira CR, Ramos de Jesus J, Bomura Rosato A, et al. (2004) Leishmania braziliensis isolates differing at the genome level display distinctive features in BALB/c mice. Microbes Infect 6: 977–984. [DOI] [PubMed] [Google Scholar]
- 34. Rocha FJ, Schleicher U, Mattner J, Alber G, Bogdan C (2007) Cytokines, signaling pathways, and effector molecules required for the control of Leishmania (Viannia) braziliensis in mice. Infect Immun 75: 3823–3832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Childs GE, Lightner LK, McKinney L, Groves MG, Price EE, et al. (1984) Inbred mice as model hosts for cutaneous leishmaniasis. I. Resistance and susceptibility to infection with Leishmania braziliensis, L. mexicana, and L. aethiopica. Ann Trop Med Parasitol 78: 25–34. [DOI] [PubMed] [Google Scholar]
- 36. DeKrey GK, Lima HC, Titus RG (1998) Analysis of the immune responses of mice to infection with Leishmania braziliensis. Infect Immun 66: 827–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Maioli TU, Takane E, Arantes RM, Fietto JL, Afonso LC (2004) Immune response induced by New World Leishmania species in C57BL/6 mice. Parasitol Res 94: 207–212. [DOI] [PubMed] [Google Scholar]
- 38. Neal RA, Hale C (1983) A comparative study of susceptibility of inbred and outbred mouse strains compared with hamsters to infection with New World cutaneous leishmaniases. Parasitology 87 (Pt 1) 7–13. [DOI] [PubMed] [Google Scholar]
- 39. Samuelson J, Lerner E, Tesh R, Titus R (1991) A mouse model of Leishmania braziliensis braziliensis infection produced by coinjection with sand fly saliva. J Exp Med 173: 49–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Colpitts SL, Dalton NM, Scott P (2009) IL-7 receptor expression provides the potential for long-term survival of both CD62Lhigh central memory T cells and Th1 effector cells during Leishmania major infection. J Immunol 182: 5702–5711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lee LF, Axtell R, Tu GH, Logronio K, Dilley J, et al. (2011) IL-7 promotes T(H)1 development and serum IL-7 predicts clinical response to interferon-beta in multiple sclerosis. Sci Transl Med 3: 93ra68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Groom JR, Luster AD (2011) CXCR3 ligands: redundant, collaborative and antagonistic functions. Immunol Cell Biol 89: 207–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Naik S, Bouladoux N, Wilhelm C, Molloy MJ, Salcedo R, et al. (2012) Compartmentalized control of skin immunity by resident commensals. Science 337: 1115–1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Oliveira F, Lawyer PG, Kamhawi S, Valenzuela JG (2008) Immunity to distinct sand fly salivary proteins primes the anti-Leishmania immune response towards protection or exacerbation of disease. PLoS Negl Trop Dis 2: e226. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
SGS pre-immunization does not modify cellular recruitment in response to L. braziliensis inoculation. BALB/c mice were inoculated 3 times in the right ear every 2 wks with 1 pair of Lu. intermedia salivary glands and then challenged 2 wks later in the left ear with L. intermedia SGS. Ears were digested 2 wks post inoculation and cellular content was analyzed by FACS. Cell numbers are shown as the mean +SEM with 5 mice per group. Data are results from one experiment and representative of 2 individual experiments.
(TIF)
List of the primers used in this study to analyze gene expression by RT-PCR in mouse and human samples.
(DOC)
List of the genes that were positively (>1.5 fold) or negatively (<1.5 fold) regulated at the site of parasite inoculation in mice immunized with SGS and subsequently infected with L. braziliensis plus SGS, compared to mice pretreated with PBS and infected with L. braziliensis plus SGS. Contralateral ears were isolated for microarray analysis 2 weeks after parasite challenge. The data are presented as the fold change of mice pre-immunized with SGS (3 times every 2 weeks) over mice inoculated with PBS and challenged with L. braziliensis plus SGS. p-values<0.05: statistically significant. The values for IFN-inducible genes (below the IFN-inducible shaded line) are given but they did not vary >1.5 times and were not statistically significant between L. braziliensis samples that were pretreated with SGS or PBS.
(DOC)