Abstract
Zika virus (ZIKV) is a mosquito-borne flavivirus first isolated in Uganda in 1947. Although entomological and virologic surveillance have reported ZIKV enzootic activity in diverse countries of Africa and Asia, few human cases were reported until 2007, when a Zika fever epidemic took place in Micronesia. In the context of West Africa, the WHO Collaborating Centre for Arboviruses and Hemorrhagic Fever at Institut Pasteur of Dakar (http://www.pasteur.fr/recherche/banques/CRORA/) reports the periodic circulation of ZIKV since 1968. Despite several reports on ZIKV, the genetic relationships among viral strains from West Africa remain poorly understood. To evaluate the viral spread and its molecular epidemiology, we investigated 37 ZIKV isolates collected from 1968 to 2002 in six localities in Senegal and Côte d'Ivoire. In addition, we included strains from six other countries. Our results suggested that these two countries in West Africa experienced at least two independent introductions of ZIKV during the 20th century, and that apparently these viral lineages were not restricted by mosquito vector species. Moreover, we present evidence that ZIKV has possibly undergone recombination in nature and that a loss of the N154 glycosylation site in the envelope protein was a possible adaptive response to the Aedes dalzieli vector.
Author Summary
Zika fever is a mosquito-borne illness caused by a flavivirus. Human infections with Zika virus (ZIKV) could cause fever, malaise and cutaneous rash. Despite several ZIKV reports since 1947 when it was first isolated at Zika forest in Uganda, molecular evolution of ZIKV as an emerging agent remains poorly understood. Moreover, despite several ZIKV reports from Africa and Asia, few human cases were notified until 2007 when an epidemic took place in Micronesia. In West Africa, surveillance programs have reported periodic circulation of the virus since 1968. To help fill the gap in understanding ZIKV evolution, 43 ZIKV samples were analyzed. We focused on: (i) adaptive genetic changes including protein glycosylation patterns, (ii) phylogenetic relationship among isolates and their spatiotemporal patterns of spread across Africa and Asia and, (iii) dispersion among vertebrate reservoirs and invertebrate vector species. Our results indicated that ZIKV may have experienced recombination in nature and that, after it emerged from Uganda in the early of the 20th century, it moved to West Africa and Asia in the first half of the century, without any clear preference for host and vector species.
Introduction
Zika virus (ZIKV) is a mosquito-borne flavivirus, a member of the Spondweni serocomplex, whose natural transmission cycle involves mainly vectors from the Aedes genus (A. furcifer, A. taylori, A. luteocephalus and A. africanus) and monkeys [1], while humans are occasional hosts. Clinical pictures range from asymptomatic cases to an influenza-like syndrome associated to fever, headache, malaise and cutaneous rash [2], [3]. Likewise, direct contact is also considered a potential route of transmission among humans, probably during sexual intercourse [4]. The first isolation of ZIKV was in 1947 from the blood of a sentinel Rhesus monkey No. 766, stationed in the Zika forest, near the Lake Victoria in Uganda, and in 1948 ZIKV was also isolated in the same forest from a pool of A. africanus mosquitoes [5]. Thereafter, serological and entomological data indicated ZIKV infections in the African continent in Nigeria in 1971 and 1975 [6], Sierra Leone in 1972 [7], Gabon in 1975 [8], Uganda in 1969 and 1970 [9], Central African Republic in 1979 [10], Senegal from 1988 to 1991 [11] and Côte d'Ivoire in 1999 [12]. Recently, ZIKV was detected in Senegal in 2011 and 2012 (unpublished data). In addition, ZIKV infections in Asia were reported in Pakistan [13], Malaysia [14], Indonesia in 1977 and 1978 [15], Micronesia in 2007 [16], [17] and Cambodia in 2010 [18]. Although ZIKV was repeatedly isolated, only 14 human cases were reported before April 2007, when a Zika fever (ZF) epidemic occurred in Yap island in Micronesia, where 49 confirmed cases and 73% of the residents older than 3 years provided serologic evidence for recent ZIKV infection [16]. This outbreak showcased the potential of ZF as an emerging disease, which could be misdiagnosed as dengue fever, as happened during the beginning of the Micronesian outbreak [16], [17].
The ZIKV genome consists of a single-stranded positive sense RNA molecule with 10794 kb of length with 2 flanking non-coding regions (5′ and 3′ NCR) and a single long open reading frame encoding a polyprotein: 5′-C-prM-E-NS1-NS2A-NS2B-NS3-NS4A-NS4B-NS5-3′, that is cleaved into capsid (C), precursor of membrane (prM), envelope (E) and seven non-structural proteins (NS) [19], [20]. The E protein (≈53 kDa) is the major virion surface protein. E is involved in various aspects of the viral cycle, mediating binding and membrane fusion [21]. The NS5 protein (≈103 kDa) is the largest viral protein whose C-terminal portion has RNA-dependent RNA polymerase (RdRP) activity and the N-terminus is involved in RNA capping by virtue of its processing due to methyl transferase activity [21]. The 3′NCR of the ZIKV genome contains about 428 nucleotides, including 27 folding patterns [20] that may be involved in the recognition by cellular or viral factors, translation, genome stabilization, RNA packaging, or cyclization [21]. Although diverse studies have contributed greatly to our understanding of the evolutionary biology of flaviviruses in general [22]–[25], few studies have addressed ZIKV evolutionary biology [17], [26]. Those studies report three main ZIKV lineages, one from Asia and two from Africa. Aiming to fill this gap and gain better insights ZIKV molecular evolution in the 20th century, we investigated 43 ZIKV strains, sampled from 1947 to 2007 in Africa and Asia, to describe phylogenetic relationships, selective influences, recombination events, phylodynamics, phylogeography, host correlations with viral lineages and glycosylation patterns.
Methods
Ethical statements
Samples used in this study are part of the Institute Pasteur in Dakar collection (WHO Collaborating Centre for Arboviruses and/or Hemorrhagic Fever Reference and Research). Monkey and human strains (AnD 30332 and HD 78788) were obtained respectively in 1979 and 1991 in Senegal during routine surveillance. None of the data was directly derived from human or animal samples but rather from cell culture supernatant. Therefore all the samples were anonymous and only reference numbers were used during the analysis that originated this study.
Virus isolates
ZIKV strains were provided by CRORA at the Institute Pasteur of Dakar. The strains were obtained from mosquitoes, humans and other mammals isolated in Burkina Faso, Central African Republic, Côte d'Ivoire and Senegal in West Africa (Table S1). Viral stocks were prepared by inoculating viral strains into Aedes pseudoscutellaris clone 61 monolayer in Leibovitz 15 (L-15) growth medium (GibcoBRL, Grand Island, NY, USA) supplemented with 5% fetal bovine serum (FBS) (GibcoBRL, Grand Island, NY, USA), 10% Tryptose Phosphate and antibiotics (Sigma, Gmbh, Germany). Viral infection was confirmed after seven days of propagation by an indirect immunofluorescence assay (IFA) using specific hyper-immune mouse ascitic fluid, as described previously [27]. Cultures supernatants were collected for virus RNA isolation.
RNA extraction
RNA was extracted from ZIKV stocks using the QIAamp RNA Viral Kit (Qiagen, Hilden, Germany) according to the manufacturer's recommendations. RNA was eluted in 50 µl of AVE buffer and stored at −80°C until use.
RT-PCR amplification
For cDNA synthesis, 10 µl of viral RNA was mixed with 1 µl of each of a reverse primer (2 pmol), 1 µl of deoxynucleotide triphospahte (dNTP) (10 mM each dNTP and the mixture was heated at 65°C for 5 min. Reverse transcription was performed in 20 µl mixture containing mixed of 2.5 U RNasin (Promega, Madison, USA) 5 U of Superscript III reverse transcriptase (Invitrogen, Carlsbad, USA) and incubated at 55°C for 50 min, followed by 70°C for 15 min. PCR products were generated independently using the primers Unifor/Unirev, FD3/FU1 and VD8/EMFI to amplify partial E, NS5 and NS5/3′NC region respectively [28]. Five microliters of cDNA were mixed with 10× buffer, 5 µl of each primer, 5 µl of dNTPs 10 mM, 3 µl of MgCl2, and 0.5 µl of Taq polymerase (Promega, Madison, USA).
Nucleotide sequencing
PCR products of the expected size were purified from agarose gels with the QiaQuick Gel Extraction Kit (Qiagen, Hilden, Germany) as specified by the manufacturer. Both strands of each PCR product were sequenced directly with the ABI Prism BigDye Terminator Cycle Sequencing Ready Reaction Kit V3.1 on an Applied Biosystems 3100 DNA Analyzer (Applied Bisoystem, Foster City, CA, USA) at the Laboratory of Molecular Evolution and Bioinformatics, Biomedical Sciences Institute, University of Sao Paulo, Brazil. We deposited thirty two 753 bp-long sequences from the E gene (Accession numbers: KF383015-KF383046), thirty one of NS5 (708 bp) (Accession numbers: KF38304-KF383114), thirty seven of 3′NCR (537 bp) (Accession numbers: KF383047-KF383083) and six genomes (10274 bp) (Accession numbers: KF383115–KF383120) in GenBank (www.ncbi.nlm.nih.gov/genbank/) from thirty eight viral strains (Table S1). Additional sequences representing strains from Kedougou in Senegal, Nigeria, Malaysia, the Ugandan prototype MR766, the strain related to Micronesian outbreak in 2007 and the Spondweni virus were obtained from GenBank, with the following accession numbers, respectively: HQ234501, HQ234500, HQ234499, NC_012532, EU545988 and DQ859064.1 (Table S1).
Recombination detection
Prior to the analyses, all sequences were aligned with MUSCLE v3.7 [29] and manually edited with SeaView v4.3.3 [30]. To prevent potential biases during phylogenetic inference due to recombination, we first analyzed the sequences of available ZIKV genomes with RDP v4.4.8 program [31] that incorporates RDP [32], GENECONV [33], Chimaera [34], MaxChi [35], Bootscan [36], SiScan [37] and 3Seq [38] methods to uncover evidence for recombination events. Only events with p-values≤0.01 that were confirmed by four or more methods were considered, using the Bonferroni correction to prevent false positive results [39], as implemented in the RDP program [31]. In addition, the occurrence of recombination in genomes was also investigated with the Rec-HMM program that estimates breakpoints based on the Phylo-HMM approach, which models tree topology changes over the columns of a multiple alignment [40]. Moreover, potential intra-gene recombination was also inspected with RDP using individual gene datasets, and the incompatibility among phylogenies inferred from genes (NS5 and E) was also investigated with GiRaF v1.01 [41] that searches incompatible clades among posterior set of trees (PST) obtained from different genomic regions with threshold of 70% for incompatible clades. The PST was obtained during Monte Carlo Markov chain (MCMC) stationarity using four chains, one ‘cold’ and three ‘heated’, after 20 million of generations, sampling every 5000 generations using MrBayes v3.2.1 [42].
Phylogenetic analyses
The phylogenetic signal content of the sequence datasets to phylogenetic reconstruction was investigated by Likelihood mapping method [43], implemented in TREE-PUZZLE v5.2 [44]. The concordance among gene (E and NS5) datasets without recombinant sequences was further assessed using a permutation test with significance level (α) of 0.05 after 10000 permutations, implemented in the Congruence among Distance Matrices (CADM) package [45]. Phylogenetic trees were generated by Maximum Likelihood (ML) criterion using GARLI v2.0 [46] that uses a stochastic algorithm to estimate simultaneously the best topology, branch lengths and substitution model parameters that maximize the log Likelihood (lnL). The confidence of ML trees was assessed by the convergence of lnL scores from ten independent replicates. We used a substitution model based on general time reversible (GTR) model with gamma-distributed rate variation (Γ) and a proportion of invariant sites (I). Support for the topology was obtained after 1000 non-parametric bootstrap replicates with GARLI. Then, we summarized the bootstrap trees into one consensus tree to access bootstrap values, using Dendropy v3.10.1 [47].
Selection analyses
To infer the selection pressures acting on each gene of ZIKV, we estimated the difference between the non-synonymous (dN) and synonymous (dS) rates per codon site using the single likelihood ancestor counting (SLAC) algorithm available in HyPhy v0.99 [48], assuming a significance level of 1% (α = 0.01). In the HyPhy output, values of ω are expressed as ω = dN - dS. Therefore, ω smaller than zero (ω<0) indicate purifying, negative selection.
Prediction of glycosylation sites
Potential glycosylation sites that may have adaptive value were previously described in ZIKV proteins [17], [20], [26]. Thus, we investigated partial E sequences to detect potential glycosylation sites using NetCGlyc v1.0 [49], NetOGlyc v3.1 [50], YinOYang v1.2 [51] and NetNGlyc v1.0 [51], [52] methods that employ algorithms based in neural networks to predict, respectively, C-mannosylated, mucin-type O-linked, N-acetylglucosamine (GlcNAc) and N-linked glycosylation sites. To infer the structural position of the predicted glycosylation sites, we modeled the tridimensional structures of E regions of viral polyprotein of the Micronesian strain (GenBank accession number ACD75819). We used the homologous sequences from Japanese Encephalitis virus (PDB code 3p54), West Nile virus (PDB code 2i69) and Dengue virus type 3 (PDB code 1uzg). The amino acids sequences were aligned using MUSCLE v3.7 [29], a total of 1000 independent models were generated and optimized using Modeller v9.10 [53], and the best models were validated with PROCHECK v3.5.4 [54].
Phylodynamic analysis
Maximum Clade Credibility (MCC) trees were inferred using a Markov Chain Monte Carlo (MCMC) Bayesian approach implemented on the program BEAST v1.6.2 [55] under GTR + Γ + I and a relaxed (uncorrelated lognormal) molecular clock [56]. MCMC convergence was obtained for four independent runs with 50 million generations, which were sufficient to obtain a proper sample for the posterior at MCMC stationarity, assessed by effective sample sizes (ESS) above 200 inspected using Tracer v1.5 (http://tree.bio.ed.ac.uk/software/tracer/). Furthermore, using the concatenated sequences of E and NS5 genes, we employed a discrete model attributing state characters representing isolation locality, animal source, recombination and N- linked glycosylation on E protein of each of the strains with the Bayesian Stochastic Search Variable (BSSVS) algorithm [57], implemented in BEAST. This method estimates the most probable state at each node in the MCC trees, allowing us to reconstruct plausible ancestral states on these nodes. Moreover, we represented the viral migration in Google Earth (http://www.google.com/earth/), using the SPREAD v1.0.3 program [58]. We evaluated the correlation among viral states and inferred phylogenies from PST by the parsimony score (PS), association index (AI) and monophyletic clade size (MC), with BaTS v1.0 [59] after 10000 null replications. In addition, we investigated the occurrence of correlated evolutionary change among ZIKV phenotypes (glycosylation pattern and vector host) along PST, employing a ML approach to test the fit of the two evolutionary models, one where the two traits evolve independently on the phylogenetic tree (independent model), and one where they evolve in a correlated way (dependent model) [60], using BayesTraits program (http://www.evolution.rdg.ac.uk/). To evaluate model suitability to ZIKV data, we estimated the marginal likelihoods for both models after 1000 bootstrap replications and compared Bayes factors (BF) between models [61], using Tracer v1.5.
Results/Discussion
Recombination among ZIKV strains
The primary analyses with RDP suggested 13 recombination events in ZIKV complete genomes (Table S2), Rec-HMM also detected genomic breakpoints with confidence in the following alignment positions: 1044 to 1095, 5181 to 5238, 9007 to 9132 and 9580 to 9631 (Figure S1). Since the results obtained by both methods revealed breakpoints in the E and NS5 genomic regions, we investigated these evidences with RDP on partial gene sequences. We found a single event in E sequences with estimated breakpoints near to the 414th and 1065th site of E gene reaching nine viral strains: ArA986, HD78788, ArA27101, ArA27290, ArA27096, ArA27443, ArA27407, ArA27106 and ArA982. These results were found by Bootscan, Maxchi, Chimaera, SiSscan and 3Seq methods and supported by significant p-values of 1.31E-5, 2.85E-3, 1.59E-3, 1.79E-29 and 6.85E-19, respectively. Likewise, only one recombination event was detected in NS5 sequences with estimated breakpoints near sites 1581 and 2152 of the NS5 gene from strains ArD158084, ArB1362 and ArD157995. These findings were confirmed by Bootscan, Maxchi, Chimaera, SiSscan and 3Seq methods and supported by significant p-values of 9.93E-9, 3.32E-7, 3.32E-7, 5.27E-28 and 7.65E-24, respectively. These potential recombinant sequences were excluded from further analyses to avoid inferential biases [62], [63]. To perform the phylogenetic analysis we concatenated E and NS5 sequences and replaced inferred recombinant fragments with missing data. This is in line with the use of Maximum Likelihood approaches, which is fairly robust to the introduction of gaps [64], [65]. In addition, we found incompatibilities between E and NS5 phylogenies using GiRaF. The three discordant strains (ArD128000, ArA1465 and ArD142623) were excluded, and we used 40 (31 from E and 36 from NS5) concatenated sequences for phylogenetic analysis. Moreover, we also found that the two remaining datasets for E and NS5 have no conflicting phylogenetic signal, as estimated by a CADM test (p-value = 9.99E-5 and α = 0.05). Given the limited sampling that we investigated, these results indicate that ZIKV may be experiencing recombination in the field, which is uncommon among flaviviruses [66]. These findings remain to be properly evaluated and assessed related to their effects on viral spread, zoonotic maintenance and epidemiologic potential. The possibility that our findings could be a consequence of cross contamination among isolates seems highly improbable given the extreme precautions that were taken. RNA extraction and reverse transcription were done separately for each isolate under BSL-II cabinets, sequenced several times leading to identical sequences, even when processed in different laboratories in Sao Paulo, Brazil, and Dakar, Senegal.
Phylogenetic analysis
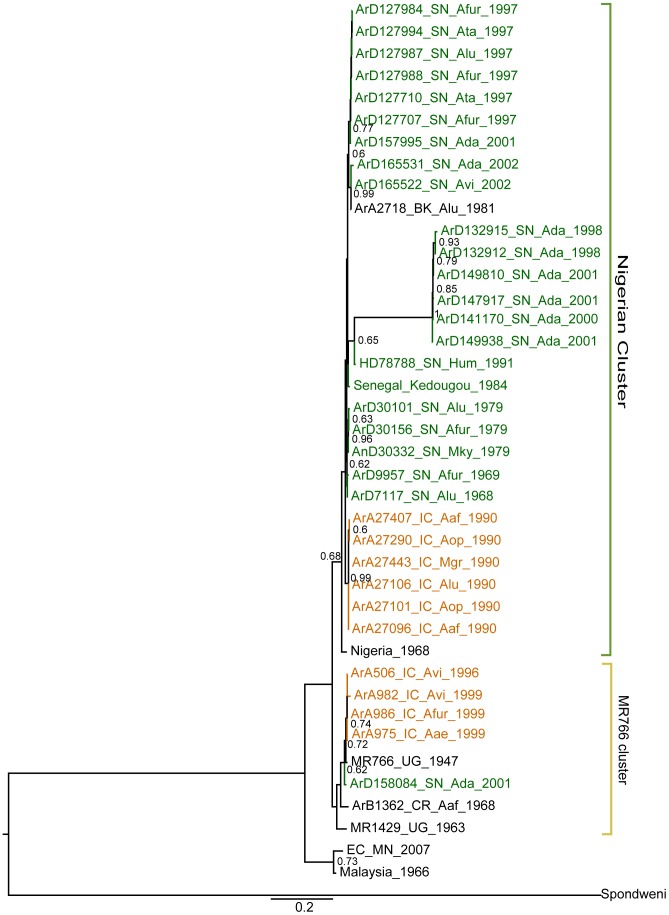
We first investigated the phylogenetic signal content in our data by reconstructing 50000 quartets for each gene segment using the likelihood mapping method (see methods section). Our results indicated that NS5 and E datasets had high phylogenetic signal content given their lower percentage of unresolved quartets (3.2% and 3.4%, respectively), while 3′NCR showed less signal (16.4% of unresolved quartets) and was not considered. The ML trees for E (data not shown), NS5 (data not shown) and the two concatenated genes (Figure 1) reinforced that ZIKV strains could be classified in three major clusters [17]. Accordingly, the African strains were arranged into two groups: the MR766 prototype strain cluster (yellow sector on Figure 1) and the Nigerian cluster (green sector on Figure 1); and the Micronesian and Malaysian strains clustered together forming the Asian clade (Figure 1), in agreement with [26]. For West Africa, the strains from Côte d'Ivoire and Senegal were found in both African clusters, suggesting that at least two distinct lineages of ZIKV circulated in these countries. Interestingly, we found that the position of the Senegalese cluster, comprising viruses isolated from 1998 to 2001 associated with A. dalzieli, branching as a sister group of HD78788 isolated in Senegal in 1991, was not simply explained by recombination (with both Giraf and RDP) or poor rooting of the tree, since it did not depend on the inclusion (Figure 1) or exclusion (Figure S2) of the Spondweni, which is a bonafide outgroup. It was observed 65% of the time during a highly stringent maximum likelihood (ML) analysis with GARLI, not taking into account dates of isolation, but crucially it had a posterior probability of one during Bayesian Inference (BI) that do take into account dates of isolation. Although we cannot rule out systematic topological errors, BI was certainly better informed than ML, since RNA viruses evolve fast, making their times of isolation important parameters for phylogenetic inference. Moreover, since we did not find compositional or codon usage biases in those sequences and in agreement with the consistent BI results, we could not rule out that the long branch length observed was not due to a detected increase of almost 10 fold increase in the rate of change along that lineage, which was not caused by detectable positive selection, as evaluated using HyPhy.
Figure 1. Maximum likelihood phylogenetic tree inferred for concatenated of sequences from Envelope and NS5 genes of Zika virus.
Consensus tree summarized after 1000 non-parametric bootstrap replicates, with support values greater than 60% shown in the nodes. The cluster the Ugandan MR766 prototype strain was highlighted by the yellow sector and the Nigerian cluster was highlighted by the green sector. The strains from Senegal and Côte d'Ivoire are shown in green and orange, respectively. The tree has been rooted with the Spondweni lineage isolated in South Africa was used as outgroup to root the tree.
Selection analyses
Selection analyses of E and NS5 genes uncovered several sites under strong negative selection indicated by ω<0. This suggests frequent purging of deleterious polymorphisms in functionally important genes. Likewise, the lack of positively selected sites, indicated by ω>0, is typical of highly adapted phenotypes and shows no detectable directional change on the available data. Our findings were expected, as the infection and transmission modes of ZIKV allow the accumulation of synonymous mutations and negatively selected sites [67]. The alternation between arthropod vector and mammal hosts may impose several barriers to non-synonymous mutations in important genes [68].
Phylodynamic analyses
The μ and the highest posterior densities (HPD with 95% of confidence interval) estimated with Beast for E, NS5 and 3′NCR genomic regions were, respectively, 2.135E-3 (2.04E-3 to 2.33221E-3), 7.1789E-4 (6.9466E-4 to 7.417E-4) and 1.1285E-3 (2.708E-4 to 2.504E-3) substitutions per site per year, which are similar to μ estimated other flaviviruses [69]. As evolutionary rates are the result of spontaneous mutations followed by selection, differences per gene are expected and in accordance with their biological role, given that the NS5 is a polymerase and the E is a surface protein. In addition, the root date estimates and 95% HPDs of the phylogenetic trees for E, NS5 and 3′NCR genomic regions were, respectively, 1900 (1851 to 1937), 1927 (1887 to 1940) and 1923 (1874 to 1959). These dates suggest a recent origin for the ZIKV strains (included in this study) near to the beginning of the 20th century.
Movement of ZIKV
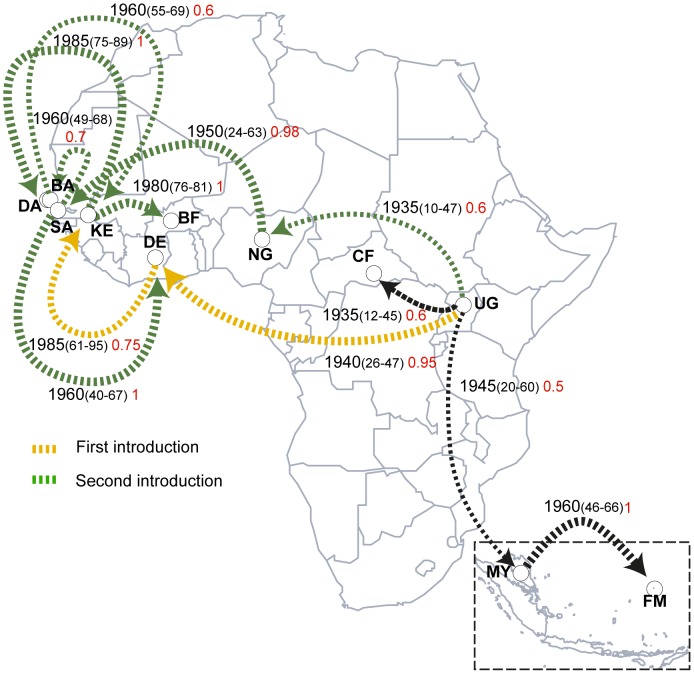
Based on our samples we inferred the most likely geographical pathway connecting ZIKV lineages. These results indicated that ZIKV emerged in Uganda around 1920, most probably between 1892 and 1943. This inference is in line with the first known ZIKV isolation in Uganda in 1947 [5]. We found two independent ZIKV introductions into West Africa from the Eastern portion of the continent (Figures 2 and S2A, and kml file in Dataset S1). The first viral introduction into Côte d'Ivoire (CI) and Senegal (SN) was related to the MR766 cluster (yellow lines in Figure 2), which possibly moved from Uganda around 1940 into Dezidougou (CI). From there, this lineage probably spread to Kedougou in Senegal (SN) around 1985 and to Sokala-Sobara (CI) around 1995. The second introduction was related to a Nigerian cluster (green lines in Figure 2), when ZIKV probably moved from Uganda to the Central African Republic and Nigeria around 1935. From Nigeria, the virus probably spread to Saboya (SN) around 1950 and from there to Dezidougou (CI) and Bandia (SN) around 1960. From Bandia, ZIKV was introduced into Kedougou (SN) around 1965 and from there to Burkina Faso around 1980 and to Dakar (SN) around 1985. Moreover, an additional ZIKV lineage from Uganda probably spread to Malaysia around 1945 and from there, the virus reached Micronesia around 1960, forming the Asian cluster [26]. The correlation between viral location (coded as character states) and phylogenies was strongly supported by significant AI and PS values, p-values≤1.00 E-4 (Dataset S2). Thus, assuming an origin of ZIKV in Uganda, our findings revealed at least two independent exits from East Africa in agreement with the two previously proposed African clades [17] and also pointed to a viral migratory flow from Eastern Africa to Asia. Although our sampling was the most comprehensive to this date, our conclusions about ZIKV movement are informed conjectures at best on the most plausible hypotheses on ZIKV spreading patterns, which are limited by the inherent biases of this type of analyses.
Figure 2. Geographic spread of ZIKV in Africa and Asia.
The directed lines connect the most probable sources and target localities of viral lineages (shown by arrows), with widths proportional to the posterior probabilities and values shown in red. Only plausible routes with probabilities above 50% are shown. The distinct introductions into Senegal and Côte d'Ivoire were represented by different colors. The estimated time to the most recent common ancestor of strains from different countries are shown with 95% posterior time intervals in parenthesis and could be interpreted as the oldest possible year of introduction of that lineage at that locality.
Animal sources of ZIKV
The association of the animal sources with viral lineages (Figure S2B) suggested that ZIKV dispersed widely among distinct animal species without a clear pattern of preference, maybe associated to the enzootic cycle of ZIKV in Africa, whose natural cycle allows a broad range of hosts [70]. Nevertheless, we found significant MC (p-value≈1.00 E-4, Dataset S2) for ZIKV strains isolated from A. dalzieli, suggesting a possible important role of this zoophilic vector [71] in West Africa. This association was found to be robust to the exclusion of vertebrate host from the analysis. The plausibility of the putative recombination events we detected (Table S2), could in part be explained by mosquitoes taking sequential blood meals from distinct animal species harboring distinct ZIKV lineages, which is in line with ours and others host range findings [70]. Also, when analyzing the increase of ZIKV activity in Kedougou, (where most of the viruses analyzed herein were collected), we noticed that ZIKV activity is much more frequent, with an interval of 1–2 years, compared to the 5 to 8 years cycle of dengue and yellow fever virus. Hence from 1972 to 2002, ZIKV emerged in 1973, 1976, 1979, 1980 and 1981. Such frequent activity can also be an opportunity of co-circulation and mixing of multiple genotypes present in the forest and that may favor recombination among them.
A phylodynamic context for recombination events
The occurrence of recombination among ZIKV strains in time-scaled phylogenetic trees suggested that some ZIKV lineages sampled in Dezidougou (CI) in 1990 (ArA27101, ArA27290, ArA27096, ArA27443, ArA27407 and ArA27106) with recombinant E (Figure S2C) shared a common ancestor around 1962 (ranging from 1951 to 1967 HPD with 95% of confidence interval). Likewise, the strain ArA982 was also isolated at Dezidougou in 1999 and its sister-group ArA986, which shared a common ancestor with the former around 1992 (ranging from 1981 to 1996 HPD with 95% of confidence interval), was sampled in the neighbor province Sokala-Sobara (CI) in 1999. Together these results indicated that recombination in envelope protein could be an important trend among the enzootic cycle of ZIKV at this region in Côte d'Ivoire, as ZIKV lineages did not show a clear pattern of host preference and recombination requires the infection of the same host by more than one viral strain. Besides, the other E recombinant strain (HD78788), isolated from a human case at Dakar (SN) in 1991, shared a common ancestor around 1984 (ranging from 1976 to 1988 HPD with 95% of confidence interval) with ZIKV strains from Kedougou (SN). Conversely, the three NS5 recombinants did not cluster along phylogenetic trees (Figure S2C), although two of them were isolated in Kedougou from A. dalzieli mosquitoes in 2001 (ArD157995 and ArD158084) and the other (ArB1362) was isolated in Bouboui, Central African Republic, from A. africanus mosquitoes in 1968. The preferential distribution of recombinant strains along phylogenies was supported by significant p-values of AI and PS ≤2.00E-4 (Dataset S2) and the adjacency patterns of E and NS5 recombinants were also confirmed by MC statistics (Dataset S2).
Glycosylation patterns in ZIKV envelope protein
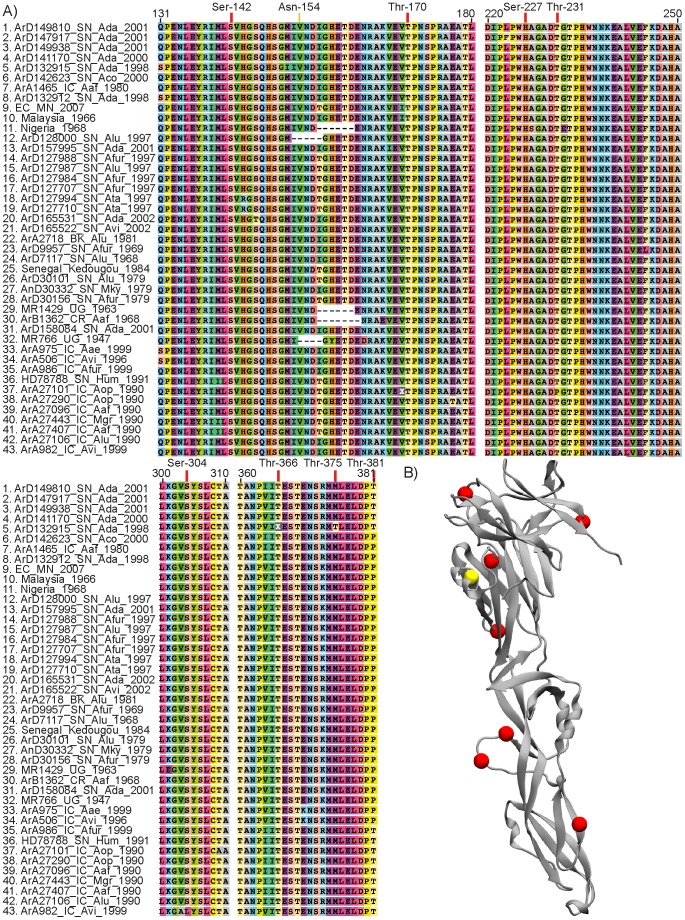
Our analyses predicted several glycosylation sites in the E protein (Figure 3). We detected a probable motif (Asn-X-Thr) among E sequences from several ZIKV strains (Figure 3A), which suggests a N-linked glycosylation site in the residue Asn-154, in agreement with [17], [26]. This residue is located on an α-helix in the E protein structure (yellow arrow in Figure 3A and yellow bead in Figure 3B). Our results also pointed several O-linked glycosylation sites in the E protein (red arrows in Figure 3A and red beads Figure 3B) but no C-mannosylated site. We found a probable mucin-type O-linked glycosylated site at residue Thr-170 in E protein from all ZIKV strains, and other mucin sites at residues Thr-245 and Thr-381 in some isolates (Figure 3A). In addition, we also uncovered probable O-GlcNAc attachment sites at residues Ser-142, Ser-227, Thr-231, Ser-304, Thr-366 and Thr-381 in E from some strains (Figure 3A).
Figure 3. Mapping of predicted glycosylation sites on envelope protein of ZIKV.
A) Alignment of E protein showing predicted glycosylation sites. Red arrows point to O-linked glycosylation sites (Ser or Thr residues) and the yellow arrow points to the N-linked glycosylation site (Asn-X-Thr motif). B) Tridimensional structure of E protein. Red beads indicate O-linked glycosylation sites and the yellow bead indicates the unique N-linked glycosylation site.
Given the importance of the N-linked glycosylation site around position 154 of the E protein for infectivity and assembly of flaviviruses [72]–[74] and the fact that we observed polymorphisms in this motif (deletions and substitutions 156 Thr/Iso), we investigated the correlation between the conservation of this motif (Asn-X-Thr) and phylogenies for ZIKV strains. Our results suggested that the acquisition of this glycosylation site is a recurrent event in the history of ZIKV, given the observed changes from Isoleucine to Threonine and vice-versa more than once in the MCC tree (Figure S2D), supported by p-values for AI and PS ≤7.00E-4 (Dataset S2). However, our conclusions are limited due to serial passages of the former ZIKV strains (Figure S2D) in mouse brain [26], which could result in the loss of this glycosylation site, as observed in West Nile virus [75].
Correlated evolutionary change along ZIKV phylogenies
Since it was demonstrated that the absence of an N-linked glycosylation site on the E protein enhances viral infectivity for C6/36 mosquito cells [72], [73] and E protein of ZIKV strains from A. dalzieli, which was the unique vector source with significant MC–showed an absence of this glycosylation site, we investigated the correlation between this mosquito-source and N-linked glycosylation patterns of E protein along PST. Our results indicated the changes in glycosylation patterns (presence or absence) and vector (A. dalzieli or not) were correlated during ZIKV emergence, which was supported by BF for dependent model (BF≈47.004) greater than for them to independent model. These findings could be related to the enzootic cycle of ZIKV in West Africa and the zoophilic behavior of A. dalzieli [71], whose females take blood meals from a broad range of vertebrates, which provides additional evidence for the absence of host preference (as described in Animal sources of ZIKV). Hence, further studies are necessary to understand the consequences of our results to ZIKV transmission cycle in nature.
Biological correlates of our findings
Our analyses indicated that ZIKV may have experienced several recombination events, which is uncommon among flaviviruses [66]. The recurrent loss and gain of the N-linked glycosylation site in the E protein could be related to mosquito-cell infectivity [73] and the correlated loss of this glycosylation site in ZIKV strains from A. dalzieli also provides indirect evidence for the enzootic cycle, since this vector has a zoophilic behavior [71] that may spread ZIKV among several hosts. Crucially, our results corroborated the notion that at least three distinct ZIKV clusters shared a common ancestor possibly with Ugandan lineages around 1920, followed by two events of East to West Africa spread (Figure 2): (i) one related to the MR766 cluster introduction to Côte d'Ivoire and posterior spread to Senegal and; (ii) other related to the Nigerian cluster introduction in Senegal and posterior dispersion to Côte d'Ivoire and Burkina Faso.
Supporting Information
Spread of ZIKV strains in Africa and Asia. A kml file to picture the history of ZIKV movement into Africa and Asia during the time, it is executable in Google Earth program (http://www.google.com/earth/).
(KML)
Significance of the correlation among phylogenies and attributes of ZIKV lineages.
(DOC)
Recombination analysis using Rec-HMM along ZIKV genomes. The dashed green lines indicate estimated breakpoints in the genomes.
(TIF)
Maximum clade credibility (MCC) trees for concatenated sequences summarizing lineage states along a time-scaled tree, with posterior probability values shown near the nodes. (A) Most probable geographical location coded according to map (Figure 2): Uganda (UG), Central African Republic (CF), Dezidougou in Côte d'Ivoire (DE), Sokala-Sobara in Côte d'Ivoire (SS), Kedougou in Senegal (KE), Saboya in Senegal (SA), Bandia in Senegal (BA), Dakar in Senegal (DA), Burkina Faso (BF), Nigeria (NG), Malaysia (MY) and Yap Island in the Federated States of Micronesia (FM); (B) most probable animal source; (C) recombination events per region; and (D) glycosylation polymorphisms.
(TIF)
Source, country and year of isolation from ZIKV strains used in this study.
(DOC)
Detection of recombination events in ZIKV genomes.
(DOC)
Acknowledgments
We thank Dr. Scott Weaver for helpful comments and critical reading of the manuscript.
Funding Statement
This study was made possible by the Institute Pasteur of Dakar in Senegal, FAPESP (Fundação de Amparo a Pesquisa do Estado de Sao Paulo, Brazil) projects #00/04205-6 (VGDN program), #08/17013-6 and #10/19341-4. CCMF has a CAPES studentship, AI (project #12/04818-5) and JVCdO received FAPESP scholarships and PMAZ holds a CNPq-PQ grant. Funding was also provided by NIH grant R01-AI069145. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Hayes EB (2009) Zika virus outside Africa. Emerg Infect Dis 15: 1347–1350 doi:10.3201/eid1509.090442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Simpson DI (1964) Zika virus infection in man. Trans R Soc Trop Med Hyg 58: 335–338. [PubMed] [Google Scholar]
- 3. Bearcroft WG (1956) Zika virus infection experimentally induced in a human volunteer. Trans R Soc Trop Med Hyg 50: 442–448. [PubMed] [Google Scholar]
- 4. Foy BD, Kobylinski KC, Chilson Foy JL, Blitvich BJ, Travassos da Rosa A, et al. (2011) Probable non-vector-borne transmission of Zika virus, Colorado, USA. Emerg Infect Dis 17: 880–882 doi:10.3201/eid1705.101939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Dick GWA, Kitchen SF, Haddow AJ (1952) Zika virus. I. Isolations and serological specificity. Trans R Soc Trop Med Hyg 46: 509–520. [DOI] [PubMed] [Google Scholar]
- 6. Fagbami AH (1979) Zika virus infections in Nigeria: virological and seroepidemiological investigations in Oyo State. J Hyg (Lond) 83: 213–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Robin Y, Mouchet J (1978) Serological and entomological study on yellow fever in Sierra Leone. Bull Soc Pathol Exot Filiales 68: 249–258. [PubMed] [Google Scholar]
- 8. Jan C, Languillat G, Renaudet J, Robin Y (1978) A serological survey of arboviruses in Gabon. Bull Soc Pathol Exot Filiales 71: 140–146. [PubMed] [Google Scholar]
- 9. McCrae AW, Kirya BG (1982) Yellow fever and Zika virus epizootics and enzootics in Uganda. Trans R Soc Trop Med Hyg 76: 552–562. [DOI] [PubMed] [Google Scholar]
- 10. Saluzzo JF, Gonzalez JP, Hervé JP, Georges AJ (1981) Serological survey for the prevalence of certain arboviruses in the human population of the south-east area of Central African Republic. Bull Soc Pathol Exot Filiales 74: 490–499. [PubMed] [Google Scholar]
- 11. Monlun E, Zeller H, Le Guenno B, Traoré-Lamizana M, Hervy JP, et al. (1993) Surveillance of the circulation of arbovirus of medical interest in the region of eastern Senegal. Bull Soc Pathol Exot 86: 21–28. [PubMed] [Google Scholar]
- 12. Akoua-Koffi C, Diarrassouba S, Bénié VB, Ngbichi JM, Bozoua T, et al. (2001) Investigation surrounding a fatal case of yellow fever in Côte d'Ivoire in 1999. Bull Soc Pathol Exot 94: 227–230. [PubMed] [Google Scholar]
- 13. Darwish MA, Hoogstraal H, Roberts TJ, Ahmed IP, Omar F (1983) A sero-epidemiological survey for certain arboviruses (Togaviridae) in Pakistan. Trans R Soc Trop Med Hyg 77: 442–445. [DOI] [PubMed] [Google Scholar]
- 14. Marchette NJ, Garcia R, Rudnick A (1969) Isolation of Zika virus from Aedes aegypti mosquitoes in Malaysia. Am J Trop Med Hyg 18: 411–415. [DOI] [PubMed] [Google Scholar]
- 15. Olson JG, Ksiazek TG, Suhandiman, Triwibowo (1981) Zika virus, a cause of fever in Central Java, Indonesia. Trans R Soc Trop Med Hyg 75: 389–393. [DOI] [PubMed] [Google Scholar]
- 16. Duffy MR, Chen T-H, Hancock WT, Powers AM, Kool JL, et al. (2009) Zika virus outbreak on Yap Island, Federated States of Micronesia. N Engl J Med 360: 2536–2543 doi:10.1056/NEJMoa0805715 [DOI] [PubMed] [Google Scholar]
- 17. Lanciotti RS, Kosoy OL, Laven JJ, Velez JO, Lambert AJ, et al. (2008) Genetic and serologic properties of Zika virus associated with an epidemic, Yap State, Micronesia, 2007. Emerg Infect Dis 14: 1232–1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Heang V, Yasuda CY, Sovann L, Haddow AD, Travassos da Rosa AP, et al. (2012) Zika virus infection, cambodia, 2010. Emerg Infect Dis 18: 349–351 doi:10.3201/eid1802.111224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Chambers TJ, Hahn CS, Galler R, Rice CM (1990) Flavivirus genome organization, expression, and replication. Annu Rev Microbiol 44: 649–688 doi:10.1146/annurev.mi.44.100190.003245 [DOI] [PubMed] [Google Scholar]
- 20. Kuno G, Chang G-JJ (2007) Full-length sequencing and genomic characterization of Bagaza, Kedougou, and Zika viruses. Arch Virol 152: 687–696 doi:10.1007/s00705-006-0903-z [DOI] [PubMed] [Google Scholar]
- 21. Lindenbach BD, Rice CM (2003) Molecular biology of flaviviruses. Adv Virus Res 59: 23–61. [DOI] [PubMed] [Google Scholar]
- 22. Kuno G, Chang GJ, Tsuchiya KR, Karabatsos N, Cropp CB (1998) Phylogeny of the genus Flavivirus. J Virol 72: 73–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gould EA, de Lamballerie X, Zanotto PM, Holmes EC (2003) Origins, evolution, and vector/host coadaptations within the genus Flavivirus. Adv Virus Res 59: 277–314. [DOI] [PubMed] [Google Scholar]
- 24. Gaunt MW, Sall AA, de Lamballerie X, Falconar AK, Dzhivanian TI, et al. (2001) Phylogenetic relationships of flaviviruses correlate with their epidemiology, disease association and biogeography. J Gen Virol 82: 1867–1876. [DOI] [PubMed] [Google Scholar]
- 25. Zanotto PM, Gould EA, Gao GF, Harvey PH, Holmes EC (1996) Population dynamics of flaviviruses revealed by molecular phylogenies. Proc Natl Acad Sci U S A 93: 548–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Haddow AD, Schuh AJ, Yasuda CY, Kasper MR, Heang V, et al. (2012) Genetic Characterization of Zika Virus Strains: Geographic Expansion of the Asian Lineage. PLoS Negl Trop Dis 6: e1477 doi:10.1371/journal.pntd.0001477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Digoutte JP, Calvo-Wilson MA, Mondo M, Traore-Lamizana M, Adam F (1992) Continuous cell lines and immune ascitic fluid pools in arbovirus detection. Res Virol 143: 417–422. [DOI] [PubMed] [Google Scholar]
- 28. Faye O, Faye O, Dupressoir A, Weidmann M, Ndiaye M, et al. (2008) One-step RT-PCR for detection of Zika virus. J Clin Virol 43: 96–101 doi:10.1016/j.jcv.2008.05.005 [DOI] [PubMed] [Google Scholar]
- 29. Edgar RC (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32: 1792–1797 doi:10.1093/nar/gkh340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Gouy M, Guindon S, Gascuel O (2010) SeaView version 4: A multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol Biol Evol 27: 221–224 doi:10.1093/molbev/msp259 [DOI] [PubMed] [Google Scholar]
- 31. Martin DP, Lemey P, Lott M, Moulton V, Posada D, et al. (2010) RDP3: a flexible and fast computer program for analyzing recombination. Bioinformatics 26: 2462–2463 doi:10.1093/bioinformatics/btq467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Martin D, Rybicki E (2000) RDP: detection of recombination amongst aligned sequences. Bioinformatics 16: 562–563. [DOI] [PubMed] [Google Scholar]
- 33. Padidam M, Sawyer S, Fauquet CM (1999) Possible emergence of new geminiviruses by frequent recombination. Virology 265: 218–225 doi:10.1006/viro.1999.0056 [DOI] [PubMed] [Google Scholar]
- 34. Posada D, Crandall KA (2001) Evaluation of methods for detecting recombination from DNA sequences: computer simulations. Proc Natl Acad Sci U S A 98: 13757–13762 doi:10.1073/pnas.241370698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Smith JM (1992) Analyzing the mosaic structure of genes. J Mol Evol 34: 126–129. [DOI] [PubMed] [Google Scholar]
- 36. Martin DP, Posada D, Crandall KA, Williamson C (2005) A modified bootscan algorithm for automated identification of recombinant sequences and recombination breakpoints. AIDS Res Hum Retroviruses 21: 98–102 doi:10.1089/aid.2005.21.98 [DOI] [PubMed] [Google Scholar]
- 37. Gibbs MJ, Armstrong JS, Gibbs AJ (2000) Sister-scanning: a Monte Carlo procedure for assessing signals in recombinant sequences. Bioinformatics 16: 573–582. [DOI] [PubMed] [Google Scholar]
- 38. Boni MF, Posada D, Feldman MW (2006) An Exact Nonparametric Method for Inferring Mosaic Structure in Sequence Triplets. Genetics 176: 1035–1047 doi:10.1534/genetics.106.068874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Bland JM, Altman DG (1995) Multiple significance tests: the Bonferroni method. BMJ 310: 170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Westesson O, Holmes I (2009) Accurate detection of recombinant breakpoints in whole-genome alignments. PLoS Comput Biol 5: e1000318 doi:10.1371/journal.pcbi.1000318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Nagarajan N, Kingsford C (2011) GiRaF: robust, computational identification of influenza reassortments via graph mining. Nucleic Acids Res 39: e34 doi:10.1093/nar/gkq1232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, et al. (2012) MrBayes 3.2: Efficient Bayesian Phylogenetic Inference and Model Choice Across a Large Model Space. Syst Biol 61: 539–42 doi:10.1093/sysbio/sys029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Strimmer K, von Haeseler A (1997) Likelihood-mapping: a simple method to visualize phylogenetic content of a sequence alignment. Proc Natl Acad Sci U S A 94: 6815–6819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Schmidt H a, Strimmer K, Vingron M, von Haeseler A (2002) TREE-PUZZLE: maximum likelihood phylogenetic analysis using quartets and parallel computing. Bioinformatics 18: 502–504 doi:10.1093/bioinformatics/18.3.502 [DOI] [PubMed] [Google Scholar]
- 45. Campbell V, Legendre P, Lapointe F-J (2011) The performance of the Congruence Among Distance Matrices (CADM) test in phylogenetic analysis. BMC Evol Biol 11: 64 doi:10.1186/1471-2148-11-64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zwickl D (2006) Genetic algorithm approaches for the phylogenetic analysis of large biological sequence datasets under the maximum likelihood criterion. The University of Texas at Austin.
- 47. Sukumaran J, Holder MT (2010) DendroPy: a Python library for phylogenetic computing. Bioinformatics 26: 1569–1571 doi:10.1093/bioinformatics/btq228 [DOI] [PubMed] [Google Scholar]
- 48. Pond SLK, Frost SDW, Muse S V (2005) HyPhy: hypothesis testing using phylogenies. Bioinformatics 21: 676–679 doi:10.1093/bioinformatics/bti079 [DOI] [PubMed] [Google Scholar]
- 49. Julenius K (2007) NetCGlyc 1.0: prediction of mammalian C-mannosylation sites. Glycobiology 17: 868–876 doi:10.1093/glycob/cwm050 [DOI] [PubMed] [Google Scholar]
- 50. Julenius K, Mølgaard A, Gupta R, Brunak S (2005) Prediction, conservation analysis, and structural characterization of mammalian mucin-type O-glycosylation sites. Glycobiology 15: 153–164 doi:10.1093/glycob/cwh151 [DOI] [PubMed] [Google Scholar]
- 51. Gupta R, Brunak S (2002) Prediction of glycosylation across the human proteome and the correlation to protein function. Pac Symp Biocomput 310–322. [PubMed] [Google Scholar]
- 52. Blom N, Sicheritz-Pontén T, Gupta R, Gammeltoft S, Brunak S (2004) Prediction of post-translational glycosylation and phosphorylation of proteins from the amino acid sequence. Proteomics 4: 1633–1649 doi:10.1002/pmic.200300771 [DOI] [PubMed] [Google Scholar]
- 53. Sali A, Blundell TL (1993) Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol 234: 779–815 doi:10.1006/jmbi.1993.1626 [DOI] [PubMed] [Google Scholar]
- 54. Laskowski RA, MacArthur MW, Moss DS, Thornton JM (1993) PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Crystallogr 26: 283–291 doi:10.1107/S0021889892009944 [Google Scholar]
- 55. Drummond AJ, Rambaut A (2007) BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol 7: 214 doi:10.1186/1471-2148-7-214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Drummond AJ, Ho SYW, Phillips MJ, Rambaut A (2006) Relaxed phylogenetics and dating with confidence. PLoS Biol 4: e88 doi:10.1371/journal.pbio.0040088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Lemey P, Rambaut A, Drummond AJ, Suchard MA (2009) Bayesian phylogeography finds its roots. PLoS Comput Biol 5: e1000520 doi:10.1371/journal.pcbi.1000520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Bielejec F, Rambaut A, Suchard MA, Lemey P (2011) SPREAD: spatial phylogenetic reconstruction of evolutionary dynamics. Bioinformatics 27: 2910–2912 doi:10.1093/bioinformatics/btr481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Parker J, Rambaut A, Pybus OG (2008) Correlating viral phenotypes with phylogeny: accounting for phylogenetic uncertainty. Infect Genet Evol 8: 239–246. [DOI] [PubMed] [Google Scholar]
- 60. Barker D, Pagel M (2005) Predicting functional gene links from phylogenetic-statistical analyses of whole genomes. PLoS Comput Biol 1: e3 doi:10.1371/journal.pcbi.0010003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Suchard MA, Weiss RE, Sinsheimer JS (2001) Bayesian Selection of Continuous-Time Markov Chain Evolutionary Models. Mol Biol Evol 18: 1001–1013 doi:10.1093/oxfordjournals.molbev.a003872 [DOI] [PubMed] [Google Scholar]
- 62. Posada D, Crandall K a (2002) The effect of recombination on the accuracy of phylogeny estimation. J Mol Evol 54: 396–402 doi:10.1007/s00239-001-0034-9 [DOI] [PubMed] [Google Scholar]
- 63. Schierup MH, Hein J (2000) Consequences of recombination on traditional phylogenetic analysis. Genetics 156: 879–891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Wiens JJ (2003) Missing Data, Incomplete Taxa, and Phylogenetic Accuracy. Syst Biol 52: 528–538 doi:10.1080/10635150390218330 [DOI] [PubMed] [Google Scholar]
- 65. Wiens JJ (2006) Missing data and the design of phylogenetic analyses. J Biomed Inform 39: 34–42 doi:10.1016/j.jbi.2005.04.001 [DOI] [PubMed] [Google Scholar]
- 66. Simon-Loriere E, Holmes EC (2011) Why do RNA viruses recombine? Nat Rev Microbiol 9: 617–626 doi:10.1038/nrmicro2614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Hanada K, Suzuki Y, Gojobori T (2004) A large variation in the rates of synonymous substitution for RNA viruses and its relationship to a diversity of viral infection and transmission modes. Mol Biol Evol 21: 1074–1080 doi:10.1093/molbev/msh109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Holmes EC (2003) Patterns of intra- and interhost nonsynonymous variation reveal strong purifying selection in dengue virus. J Virol 77: 11296–11298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Twiddy SS, Holmes EC, Rambaut a (2003) Inferring the Rate and Time-Scale of Dengue Virus Evolution. Mol Biol Evol 20: 122–129 doi:10.1093/molbev/msg010 [DOI] [PubMed] [Google Scholar]
- 70. Kuno G, Chang G-JJ (2005) Biological transmission of arboviruses: reexamination of and new insights into components, mechanisms, and unique traits as well as their evolutionary trends. Clin Microbiol Rev 18: 608–637 doi:10.1128/CMR.18.4.608-637.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Diallo M, Thonnon J, Traore-Lamizana M, Fontenille D (1999) Vectors of Chikungunya virus in Senegal: current data and transmission cycles. Am J Trop Med Hyg 60: 281–286. [DOI] [PubMed] [Google Scholar]
- 72. Hanna SL, Pierson TC, Sanchez MD, Ahmed AA, Murtadha MM, et al. (2005) N-linked glycosylation of west nile virus envelope proteins influences particle assembly and infectivity. J Virol 79: 13262–13274 doi:10.1128/JVI.79.21.13262-13274.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Lee E, Leang SK, Davidson A, Lobigs M (2010) Both E protein glycans adversely affect dengue virus infectivity but are beneficial for virion release. J Virol 84: 5171–5180 doi:10.1128/JVI.01900-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Mondotte JA, Lozach P-Y, Amara A, Gamarnik A V (2007) Essential role of dengue virus envelope protein N glycosylation at asparagine-67 during viral propagation. J Virol 81: 7136–7148 doi:10.1128/JVI.00116-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Chambers TJ, Halevy M, Nestorowicz A, Rice CM, Lustig S (1998) West Nile virus envelope proteins: nucleotide sequence analysis of strains differing in mouse neuroinvasiveness. J Gen Virol 79 (Pt 10) 2375–2380. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Spread of ZIKV strains in Africa and Asia. A kml file to picture the history of ZIKV movement into Africa and Asia during the time, it is executable in Google Earth program (http://www.google.com/earth/).
(KML)
Significance of the correlation among phylogenies and attributes of ZIKV lineages.
(DOC)
Recombination analysis using Rec-HMM along ZIKV genomes. The dashed green lines indicate estimated breakpoints in the genomes.
(TIF)
Maximum clade credibility (MCC) trees for concatenated sequences summarizing lineage states along a time-scaled tree, with posterior probability values shown near the nodes. (A) Most probable geographical location coded according to map (Figure 2): Uganda (UG), Central African Republic (CF), Dezidougou in Côte d'Ivoire (DE), Sokala-Sobara in Côte d'Ivoire (SS), Kedougou in Senegal (KE), Saboya in Senegal (SA), Bandia in Senegal (BA), Dakar in Senegal (DA), Burkina Faso (BF), Nigeria (NG), Malaysia (MY) and Yap Island in the Federated States of Micronesia (FM); (B) most probable animal source; (C) recombination events per region; and (D) glycosylation polymorphisms.
(TIF)
Source, country and year of isolation from ZIKV strains used in this study.
(DOC)
Detection of recombination events in ZIKV genomes.
(DOC)