Abstract
Accurate segregation of genetic material in eukaryotes relies on the kinetochore, a multiprotein complex that connects centromeric DNA with microtubules. In yeast and humans, two proteins – Mif2/CENP-C and Chl4/CNEP-N – interact with specialized centromeric nucleosomes and establish distinct but cross-connecting axes of chromatin-microtubule linkage. Proteins recruited by Chl4/CENP-N include a subset that regulates chromsome transmission fidelity. We show that Chl4 and a conserved member of this subset, Iml3, both from S. cerevisiae, form a stable protein complex, which interacts with Mif2 and Sgo1. We have determined the structures of an Iml3 homodimer and an Iml3-Chl4 heterodimer, which suggest a mechanism for regulating assembly of this functional axis of the kinetochore. We propose that at the core centromere, the Chl4-Iml3 complex participates in recruiting factors, such as Sgo1, that influence sister chromatid cohesion and encourage sister kinetochore biorientation.
Introduction
Nucleosomes with an H3-like histone variant, CENP-A, are distinguishing marks of centromeric chromatin. In human cells, two proteins -- CENP-C and CENP-N -- bind specifically to CENP-A nucleosomes (Carroll, 2009; 2010), directing further assembly of the kinetochore components that link centromeric chromatin to spindle microtubules. The budding yeast orthologues of these proteins are Mif2 and Chl4, respectively. An N-terminal segment of CENP-C binds the four-protein Mis12/Mtw1 complex, which connects in turn to the microtubule (MT)-binding Ndc80 complex (Przewloka, 2012; Screpanti, 2012). This organization, virtually universal in eukaryotes, defines one principal axis of kinetochore structure but does not explain the presence of CENP-N and its orthologues. In budding yeast, Chl4 is required for accurate transmission of whole chromosomes during cell division, but we do not yet know how its position in the kinetochore and its contacts with other proteins contribute to this function.
The fidelity of chromosome segregation relies on biorientation of sister kinetochores. To ensure biorientation, centromeres accumulate high local levels of cohesins during both mitosis and meiosis II (Tanaka, 2000; Kiburz, 2007; Ng, 2009). Strains of budding yeast that fail to protect centromeric cohesins from destruction or removal during the first round of meiotic division suffer increased missegregation in meiosis II (Katis, 2004). During mitosis, kinetochore assembly is necessary and sufficient to direct accumulation of centromeric cohesin complexes (Weber, 2004). This cohesin enrichment depends on recruitment of the Scc2-Scc4 cohesin loading complex to centromeres by the kinetochore proteins Iml3 and Chl4 (Fernius, 2009; 2013). In addition, at anaphase of meiosis I, yeast rely on the pericentromeric protein Sgo1 for protection of cohesin against cleavage by separase (Kitajima, 2004), and Chl4 and Iml3 are in turn necessary for normal Sgo1 localization (Marston, 2004). Thus, Chl4 and Iml3 act upstream of both cohesin loading and Sgo1-mediated cohesin protection in establishing a pericentromeric region of sister chromatid cohesion during mitosis and meiosis.
Chl4 and Iml3 are part of a group of proteins called the Ctf19 complex in budding yeast, most members of which are orthologous to the human proteins that make up the so-called "constitutive centromere-associated network" (CCAN) (Schleiffer, 2012). The constituents of these kinetochore-associated complexes were identified in separate experiments by mass spectrometry of protein complexes isolated from cells (Cheeseman, 2002; De Wulf, 2003; Okada, 2006). Normal localization of a number of other Ctf19 complex members, including Ctf3 (in early anaphase) and Iml3, depends on Chl4 (Pot, 2003). Chl4 and Iml3 together are also necessary for efficient kinetochore localization of Cnn1, a histone-fold protein related in sequence and function to CENP-T (Schleiffer, 2012; Bock, 2012). Contact between a C-terminal region of the Chl4 orthologue, CENP-N, and a putative Iml3 orthologue, CENP-L, stabilizes CCAN proteins at the human kinetochore (Carroll, 2009). Chl4/CENP-N and Iml3/CENP-L thus appear to have a conserved function in recruiting inner kinetochore proteins.
We show here that Chl4 associates through a defined, C-terminal domain with Iml3 and that these proteins form a stable complex. Indeed, co-expression of Chl4 and Iml3 is necessary for reasonable recovery of recombinant Chl4 in any form – either the full-length polypeptide or the C-terminal interacting domain. Iml3 alone equilibrates rapidly between monomer and dimer, and in the presence of Chl4 it forms a stable Iml3-Chl4 heterodimer. We describe the crystal structures of both an Iml3 homodimer and an Iml3-Chl4361–458 heterodimer. The subunit interface in the Iml3 homodimer is essentially the same as the surface that contacts Chl4361–458 in the heterodimer. The purified Iml3-Chl4 protein complex interacts with both Mif2 and Sgo1. We suggest that the latter interaction is part of the mechanism that ensures protection of centromeric cohesins in meiosis and that the Mif2-Chl4-Sgo1 axis establishes a link between the kinetochore and sister chromatid cohesion.
Results
Reconstitution of an Iml3-Chl4 complex from Sacchoromyces cerevisiae
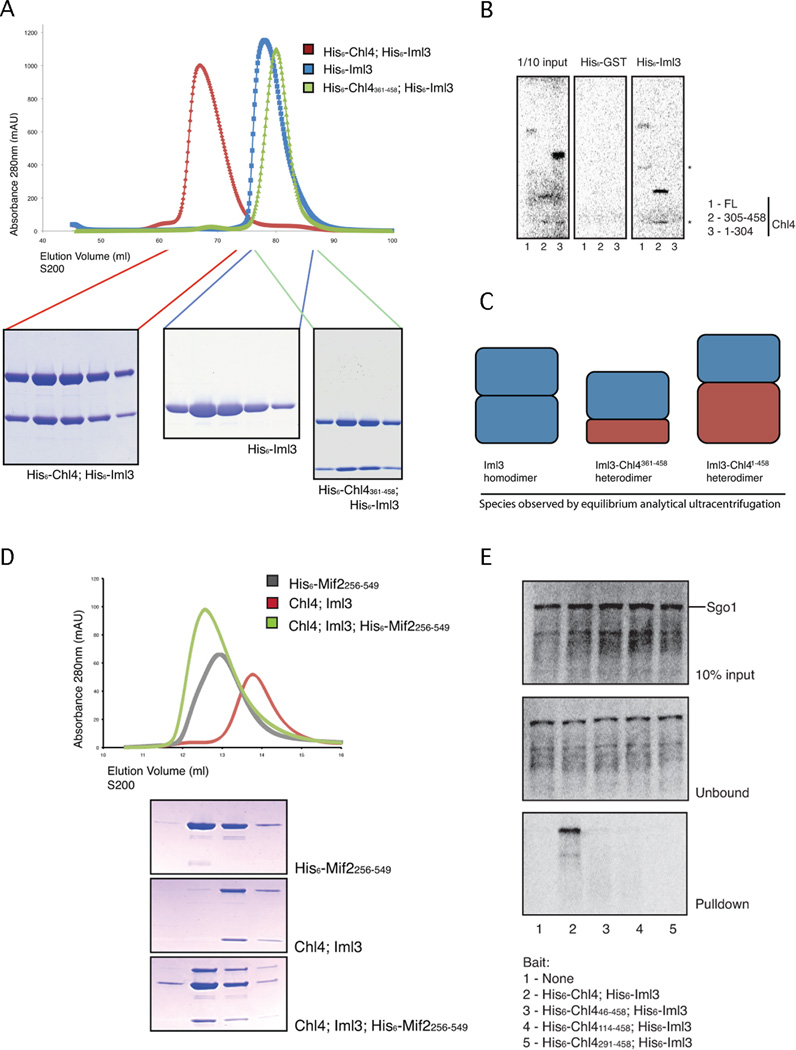
We could obtain soluble full-length Chl4 protein only by coexpressing Chl4 with Iml3, an observation consistent with a shared function for these proteins. The Iml3-Chl4 protein complex remained intact when subjected to size exclusion chromatography; Iml3 alone was also soluble (Fig. 1A). To determine the domain of Chl4 required for Iml3 binding, we produced untagged radiolabeled fragments of Chl4 in rabbit reticulocyte lysates for affinity pulldown experiments with purified His6-Iml3 as bait (Fig. 1B). Residues 305 to 458 of Chl4 were sufficient for binding to Iml3. We used partial tryptic digests and mass spectrometry to identify the N-and C-terminal domain boundaries of the associating fragment and purified a stable complex containing Iml3 and residues 361 to 458 of Chl4 (Chl4361–458, Fig. 1A).
Figure 1. Reconstitution of the Iml3-Chl4 complex and binding of Mif2 and Sgo1.
(A) Iml3 and Chl4 association. Recombinant protein complexes were analyzed by size exclusion chromatography. Chl4361–458 is a minimal fragment required for stable interaction with Iml3. (B) Iml3 binds the C-terminus of Chl4. His6-Iml3 was incubated with in vitro translated [35]S-labeled Chl4FL, Chl4305–458, or Chl41–304. Proteins that bound to Ni2+-NTA beads were visualized by gel radiography. Non-specific bands, presumably truncation products of Chl4, are marked with stars. (C) Schematic showing stoichiometries of various complexes determined by analytical ultracentrifugation (see also Table S1). (D) Iml3-Chl3 associates with a C-terminal fragment of Mif2. Iml3-Chl4 and Mif2256–549 were purified separately, mixed, and subjected to size exclusion chromatography. The trace shows absorbance at 280nm versus elution volume. Peak contents were analyzed by SDS-PAGE. All three proteins co-elute when mixed at an apparent 2:1:1 (Mif2256–549:Iml3:Chl4) molar ratio (see also Figure S1). (E) Iml3-Chl4 association with Sgo1. Recombinant Iml3-Chl4 proteins with His6 tags and the indicated Chl4 N-terminal truncations were purified and incubated with in vitro translated [35]Slabeled Sgo1. Proteins that bound to Ni2+-NTA beads were visualized by gel radiography.
Sedimentation equilibrium analytical ultracentrifugation of Iml3-Chl4, Iml3-Chl4361–458, and Iml3 showed that the Iml3-Chl4 and Iml3-Chl4361–458 complexes both had apparent molecular weights corresponding to heterodimers; Iml3 (without a Chl4 partner) showed evidence of exchange between monomer and dimer (Fig. 1C; Table S1).
Binding of Iml3-Chl4 to Mif2 and to Sgo1
The orthologues of Mif2 and Chl4, CENP-C and CENP-N, are both nucleosome-proximal (Carroll, 2009; 2010). We therefore looked for a potential association between these two components and found that a C-terminal fragment of Mif2 (residues 256–549) bound the Iml3-Chl4 complex (Fig. 1D). This interaction required the central region of Mif2 and the N-terminal domain of Chl4, which was sufficient to support Mif2 binding (Fig. S1). Multi-angle light scattering showed that this complex contains two copies of Mif2 and one copy each of Chl4 and Iml3. This stoichiometry is consistent both with the observation that Mif2 is a DNA-binding dimer (Cohen, 2008) and with native kinetochore protein copy numbers determined by light microscopy (Joglekar, 2006).
Because Chl4 and Iml3 are required for proper localization of Sgo1 to pericentromeres during meiosis, we tested for an interaction between the Iml3-Chl4 complex and Sgo1 protein (Fig. 1E). His-tagged full-length Iml3-Chl4 protein pulled down untagged radiolabeled, full-length Sgo1 presented as prey from an in vitro translation system. The interaction required a region near the N-terminus of Chl4 that is conserved in CENP-N of higher eukaryotes. We could not, however, express and purify a protein complex containing Iml3-Chl4 and Sgo1.
Structure of an Iml3 homodimer
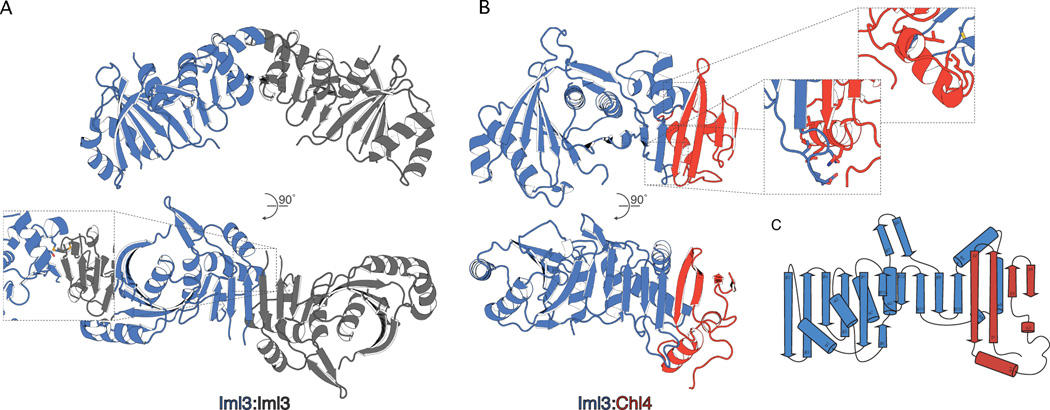
Crystals of full-length Iml3 grew in multiple conditions. We used selenomethionine derivatives of these crystals to record single-wavelength anomalous dispersion (SAD) diffraction data and to determine the structure of Iml3 at a resolution of 2.5Å (Fig. 2A). The structure shows a central β-sheet with ten strands that wrap around two α-helices. Four helices decorate the outside of this core. In our crystals, the β-sheets of Iml3 subunits related by a crystallographic dyad pair to form a continuous, 20-strand sheet.
Figure 2. Structures of the Iml3-Iml3 and Iml3-Chl4 complexes.
(A) Crystal structure of the Iml3-Iml3 homodimer (see Table S2 for crystallographic statistics). The contents of a single asymmetric unit of the crystal lattice are shown in blue. Inset shows a close-up and rotated view of the homodimer interface. Side chains for methionine 198 and serine 169 are shown as sticks (sulfur – yellow; oxygen – red). (B) Crystal structure, determined by molecular replacement, of the Iml3-Chl4361–458 heterodimer. Iml3 is blue and Chl4361–458 is red. Insets show close-up and rotated views of the heterodimer interface (see also Figure S2). (C) Secondary structure diagram of the Iml3-Chl4361–458 heterodimer. Beta augmentation occurs between Iml3 β10 and Chl4361–458 β2.
One entry in the structural database – the bacterial recombination-associated protein RdgC – has essentially the same fold as Iml3 (Ha, 2007; Holm, 2010). Several other proteins, including the "β-ear" of the clathrin adaptor complex AP-2 (Owen, 2000) spermine synthases (Wu, 2008) have domains with the same fold as the compact module at the N-terminus of Iml3 (residues 1–108); the TATA-binding protein, TBP, is a single-chain "pseudodimer" (Kim, 1993), of which each half has the structure of the Iml3 N-terminal module. No consistent sequence similarities link Iml3 with any of these proteins, however.
Structure of an Iml3-Chl4 heterodimer
We obtained crystals of the Iml3-Chl4361–458 complex, from which we recorded diffraction to a resolution of 2.3Å. We used the Iml3 coordinates to determine the structure of this complex by molecular replacement (Fig. 2B). The Iml3-Chl4 heterodimer relies on β-augmentation at the same surface that mediates Iml3 homodimerization (Fig. 2C). Homodimer and heterodimer formation are therefore mutually exclusive. Alignments of Chl4 and CENP-N sequences from various organisms show patches of strong similarity; the segment that contacts Iml3 (residues 395–431) is the only one of these in the C-terminal part of the protein.
A Chl4 homodimer could in principle form without steric clash, but a cluster of four lysines at the dyad would probably destabilize it. Therefore, while Iml3 may undergo exchange between monomeric and dimeric species in solution, Chl4 appears to have Iml3 as an obligate heterodimeric partner. The Iml3-Chl4 heterodimer buries roughly twice the solvent-accessible surface area as does the Iml3-Iml3 homodimer (3010Å 2 versus 1739Å2), consistent with our observation that in the presence of Chl4, Iml3 heterodimerizes preferentially.
Both CENP-L and CENP-M have been proposed as candidate orthologues of Iml3 (McClelland, 2006. Schleiffer, 2012). Multiple sequence alignments that include metazoan CENP-L orthologues show that the Iml3 dimerization interface is more conserved than the rest of the protein (Fig. S2). Structural evidence therefore favors orthology between Iml3 and CENP-L.
Effects of point mutations at the dimer interface
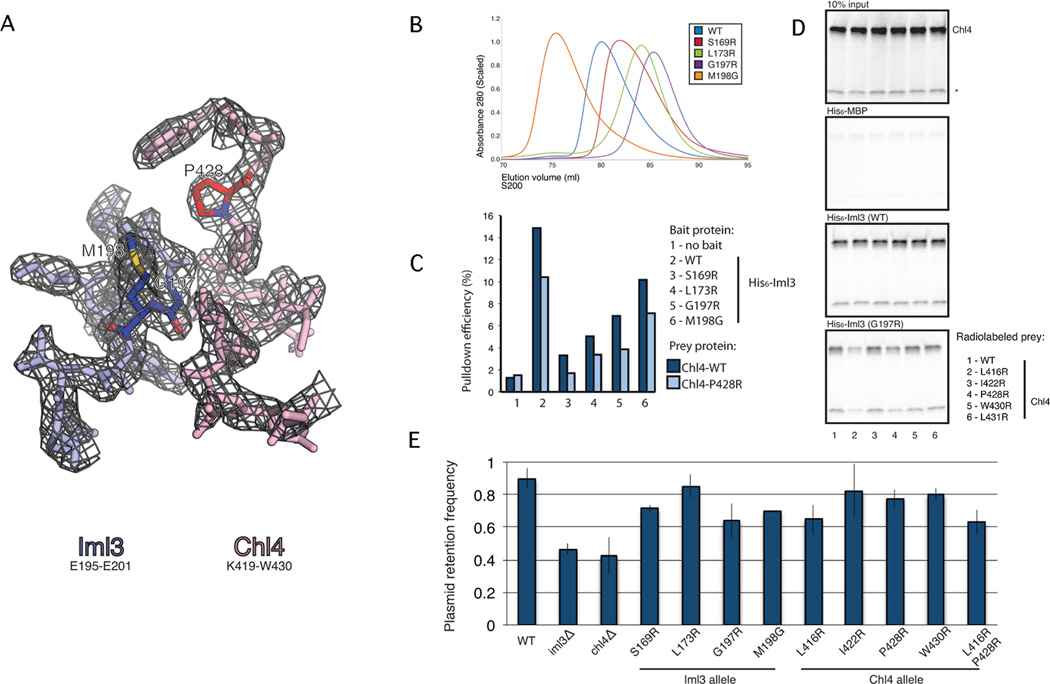
We tested the structural determinants of dimerization with amino acid substitutions in a part of the Chl4-Iml3 dimerization interface distinct from the augmented β-sheet but for which we had unambiguous electron density (Fig. 3A). Three mutations (S169R, L173R, and G197R) in Iml3 reduced homodimerization as assessed by size exclusion chromatography (Fig. 3B) and multi-angle light scattering (Fig. S3A). These mutations also reduced the affinity of Iml3 for Chl4 (Fig. 3C). One substitution (M198G) in Iml3 stabilized the homodimer, with only a small effect on heterodimerization. The Chl4:Iml3 heterodimer is more stable than the homodimer, and single point mutations on the Chl4 side of the interface have effects too weak to detect reliably. When coupled with a G197R substitution in Iml3, however, both L416R and P428R in Chl4 measurably weaken the heterodimer (Fig. 3D). The effects of these various mutations confirm the correspondence of the crystallographically determined contacts and the associations we have examined in solution.
Figure 3. Point mutations that disrupt Iml3-Chl4 and Iml3-Iml3 interactions.
(A) Refined electron density at the Iml3-Chl4 dimer interface. 2Fo-Fc map contoured to 1.5σ for Iml3195–201 (blue) and Chl4419–430(red). Iml3G197, Iml3M198, and Chl4P428 are colored darker and by atom type. (B) Point mutations in the dimer interface impede Iml3 homodimerization (see also Figure S3). Iml3 proteins carrying His6 tags and the indicated point mutations were purified and subjected to size exclusion chromatography. The point mutations Iml3S169R, Iml3L173R, and Iml3G197R disrupt homodimer formation to varying degrees. Iml3M198G forms an Iml3 homodimer that is more stable than wild-type. (C) Iml3 point mutations in the dimer interface impede Iml3-Chl4 complex formation. Iml3 proteins carrying His6 tags and the indicated point mutations were purified and incubated with in vitro translated [35]S-labeled Chl4 or Chl4P428R. Ni2+-affinity pulldowns were performed, and bound protein was visualized by gel radiography. Integrated band intensities are plotted for the indicated pulldowns, and pulldown efficiency is given as the percent of input material recovered. All Iml3 point mutants disrupt Chl4 binding to varying degrees. Chl4P428R shows a mild defect in complex formation for all Iml3 proteins tested. (D) Chl4 point mutations that disrupt Iml3-Chl4 binding. [35]S-labeled Chl4 proteins (WT and five mutants) were translated as for (C), and Ni2+-affinity pulldowns were performed using His6-tagged Iml3 or His6-tagged Iml3G197R as bait. Chl4L416R and Chl4P428R disrupt binding to Iml3G197R. A non-specific band, presumably a truncation product of Chl4, is marked with a star. (E) Point mutations in the dimer interface partially recapitulate the plasmid missegregation phenotype seen in iml3- or chl4- strains. Haploid yeast strains carrying the indicated Iml3 or Chl4 alleles were tested for the ability to segregate a minichromosome absent auxotrophic selection. All point mutants tested except Iml3L173R showed plasmid segregation defects (three biological replicates, p-value < .05, Student’s t-test, error bars show +/− SD).
Heterodimer formation is necessary for accurate plasmid segregation
To determine whether Iml3-Chl4 dimerization is essential for their function, we tested in vivo the effects of the mutations described above. S. cerevisiae strains that lack Chl4 or Iml3 have elevated levels of minichromosome missegregation (Fernius, 2009). We therefore determined whether point mutations that alter Iml3-Chl4 dimer formation could mimic this phenotype. We constructed haploid strains with amino acid substitutions in Iml3 or Chl4, encoded at their native loci, and monitored the segregation of a centromere-bearing plasmid in the absence of selection for an auxotrophic marker. With one exception, haploid strains carrying the point mutations Iml3S169R, Iml3L173R, Iml3G197R, Iml3M198G, Chl4L416R, Chl4P428R, and Chl4W430R were measurably poorer than wild-type strains at maintaining a plasmid that contained the CENVI sequence (Fig. 3E). We conclude that the Iml3-Chl4 interaction is important for accurate segregation of genetic material. In meiosis II, chromosome segregation is acutely sensitive to Iml3-Chl4 status, and Iml3 deletion lowers sproulation efficieincy (Gosh, 2004). We examined the meiotic products of diploid cells and found that cells lacking Iml3 and cells expressing only Iml3G197R had decreased sporulation efficiencies (Fig. S3C).
Discussion
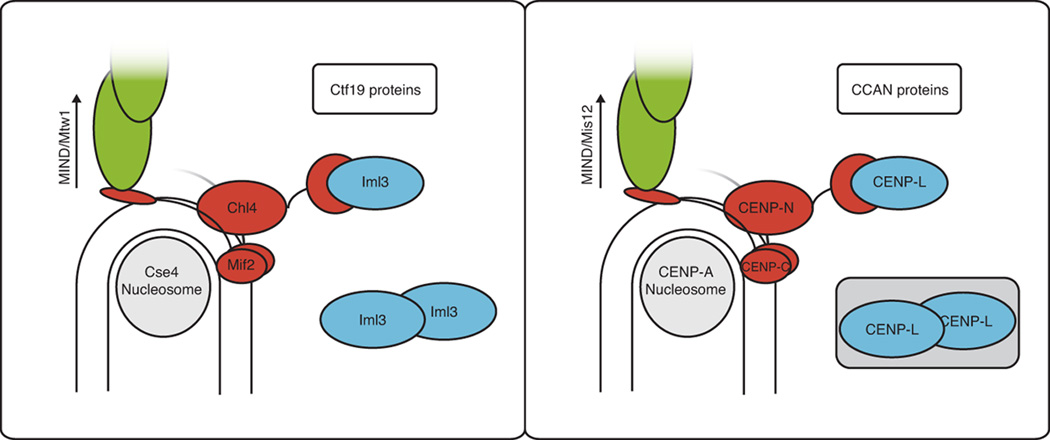
Conservation of kinetochore proteins from yeast to humans is now a well-established inference from genome sequencing and genetic studies, but conservation of their spatial organization and physiological functions has only recently become a feasible target of structural and biochemical analyses. In human cells, CENP-N (the Chl4 orthologue) and CENP-C (Mif2) both contact CENP-A (Cse4) nucleosomes (Carroll, 2009; 2010). Moreover, when associated with centromeric nucleosomes, CENP-N, together with CENP-L (Iml3), recruits further CCAN components. We describe here structures, interactions, and phenotypic consequences of interfering with those interactions. Our results are consistent with the notion that, Chl4, like CENP-N, directs assembly of functionally related kinetochore proteins when it is localized to the core centromere. Stability of Chl4 depends on Iml3, which appears to function principally as a regulatory chaperone. We further show that the Iml3-Chl4 heterodimer interacts in vitro with the centromere-binding protein Mif2 and with the regulator of cohesin protection, Sgo1. Our results are evidence for a conserved organizational hierarchy that extends from the specialized centromeric nucleosome through CENP-C/Mif2 and Chl4/CENP-N to include other CCAN/Ctf19 proteins (Fig. 4).
Figure 4. Conservation at the inner kinetochore.
Schematic representation of the inner kinetochore in budding yeast (left) and in humans (right). A human CENP-L dimer is shown in a shaded box to reflect uncertainty.
For Chl4, strong conservation appears at odds with its dispensability in dividing yeast cells. Iml3 and Chl4 null strains are viable, as are strains missing almost any other member of the Ctf19 group of proteins. Nearly all of these deletion strains suffer, however, from a diminished ability to accurately segregate whole chromosomes. For Iml3 and Chl4, our data and data from others suggest three possible molecular explanations for this phenotype. First, strains with defective Iml3-Chl4 dimers may experience weakened kinetochore-MT attachment as a result of inadequate recruitment of Cnn1 (the yeast orthologue of CENP-T). Cnn1 does not localize to kinetochores in Iml3-Chl4-deficient strains (Bock, 2012), and Cnn1-deficient strains have plasmid segregation defects (Schleiffer, 2012). Cnn1 binds the Ndc80 complex, which in turn connects to microtubules (Schleiffer, 2012). This conserved connection improves chromosome transmission fidelity, but it is not required for kinetochore-MT attachment. Second, because of inadequate recruitment of Sgo1, strains with defective Iml3-Chl4 dimers may be less likely to achieve kinetochore biorientation. Sgo1 promotes mitotic kinetochore biorientation (Indjeian, 2007), by a mechanism still not known. Third, strains with defective Iml3-Chl4 dimers may fail to accumulate cohesins at the centromere and pericentromere, as has been demonstrated for Iml3- and Chl4-deficient strains (Fernius, 2009). All three of these possibilities may contribute to crippling normal chromosome segregation.
Centromeric localization of Sgo1 is subject to complex regulation. During mitosis and meiosis, Sgo1 associates with both pericentromeric regions and the core centromere. Iml3 and Chl4 associate only with the latter (Kiburz, 2005; Fernius, 2009), but their presence at the centromere is necessary for the pericentromeric (and centromeric) localization of Sgo1 (Kiburz, 2005; Marston, 2005). The checkpoint kinase Bub1 is also required for normal Sgo1 localization during both mitosis and meiosis (Kiburz, 2005). During mitosis, Sgo1 localization requires phosphorylation of histone H2A by Bub1 at serine 121 (Fernius, 2007; Kawashima, 2010). Our data show that Iml3 binds and stabilizes the C-terminal domain of Chl4 and that the Chl4 N-terminal region contacts both Sgo1 and Mif2. We propose that binding to Chl4 contributes to Sgo1 localization.
How does Chl4/CENP-N recruit other Ctf19/CCAN proteins to the kinetochore? In a model for contacts among inner-kinetochore components shown in Fig. 4, Chl4 binds to Mif2 associated with the core centromere (and, by inference from properties of CENP-N, to the CENP-A histone, Cse4) and directs the assembly of other Ctf19 components. In human cells, this recruitment requires the C-terminal domain of CENP-N (Carroll, 2009). It is therefore likely that the Chl4 C-terminal domain or Iml3 interacts directly with one or more budding yeast orthologues of these human CCAN proteins. Our failure to observe binding between Iml3-Chl4 and other Ctf19 group proteins, however, leads us to propose that post-translational modification of either Iml3-Chl4 or its binding partner(s) is required for these interactions. This proposal is consistent with cell-cycle dependent kinetochore recruitment of the CENP-I orthologue Ctf3 (Pot, 2003). Modification or association with other factors may also influence the equilibrium between Iml3 homodimerization and Iml3-Chl4 heterodimerization, creating a potential point of regulation for Chl4-mediated recruitment of binding partners to the kinetochore.
Dipterans and nematodes do not have obvious Chl4/CENP-N orthologues (Schleiffer, 2012). Because insects other than dipterans do have identifiable versions of Chl4/CENP-N, these absences likely arose from separate gene-loss events. We see two possible explanations for this observation. First, the proteins that Chl4/CENP-N recruits to kinetochores may be absent from dipterans and nematodes, so Chl4 is not needed to recruit them. Second, proteins that Chl4/CENP-N recruits to kinetochores may have developed other ways to achieve correct localization in these organisms. The former possibility applies to several Ctf19/CCAN components, including CENP-H/I/K proteins (Ctf3/Mcm16/Mcm22 in budding yeast), which have not been found in worms or flies. How do these species manage accurate chromosome segregation without this functional subdomain of the kinetochore (assuming comparable chromosome transmission fidelity across species)? For many nematodes, the answer may be that their holocentric chromosomes provide extra MT connections that ensure accurate biorientation and segregation. For dipterans, the answer is less clear, especially given the observation that the fly orthologue of Sgo1, Mei-S332, seems to have a conserved function in meiosis (Katis, 2004). As is the case for Scm3/HJURP, which has apparently been replaced with Cal1 (Mellone BG, 2011), dipterans may have arrived at a separate but equal functional substitute for the Iml3/CENP-L-Chl4/CENP-N kinetochore substructure.
Experimental Procedures
Protein expression and purification
Coding regions for all genes were cloned from S. cerevisia genomic DNA into E. coli expression vectors with T7 polymerase promoters, N-terminal His6 tags, and TEV protease cleavage sites. For coexpression, coding regions were cloned into a single polycistron using a PCR-based method. All proteins were expressed in the SoluBL21 E. coli strain (Genlantis) and purified by Ni2+ affinity, ion exchange (HiTrap SP HP for Iml3-Chl4 full-length, HiTrap Q for all others, GE Bioscience), and gel filtration chromatography (Superdex-200, GE Bioscience). Gel filtration buffer contained 20mM Tris-HCl, pH 8.5, 1mM TCEP, and 100mM NaCl (Iml3, Iml3-Chl4361–458) or 200mM NaCl (Iml3-Chl4, Mif2256–549). His6 tags were cleaved following ion exchange with His6-tagged TEV protease for two hours at room temperature. Protease, cleaved tags, and uncleaved proteins were removed by Ni2+ affinity before gel filtration.
Crystallization and structure determination
All crystals were obtained by the hanging drop vapor diffusion method at 18°C. Well solution was mixed 1:1 (v/v) with protein. Iml3 formed hexagonal plates within two days, and rhombohedral Iml3-Chl4361–458 crystals were seen within 24 hours. Crystallization condition (mother liquor) for Iml3 was as follows: 3.75M sodium formate, 4% 2,5-diaminopentane. Larger Iml3 crystals were obtained by microseeding with smaller crystals of the same morphology. Crystallization condition for Iml3-Chl4361–458 was as follows: 0.35M Li3-citrate pH9, 25% (w/v) PEG-3350. In both cases, crystals were washed in concentrated drop solution supplemented with 30% (v/v) glycerol before freezing in liquid nitrogen. All data sets were collected on NE-CAT beamline 24ID–E. Data were indexed and scaled with HKL2000 (Otwinowski, 1997).
The structure of Iml3 was determined by single-wavelength anomalous dispersion (SAD) at 2.5Å resolution. An initial model was then used to determine by molecular replacement the phases for the Iml3-Chl4361–458 structure at 2.3Å resolution. SAD phases for the Iml3 crystals were determined with Phaser as implemented by Phenix Autobuild (Adams, 2010). For nine expected selenium atoms per asymetric unit, Phaser found ten with a mean figure of merit of .394. Inspection revealed the extra selenium to be on a twofold axis of symmetry, and it was removed from further phasing calculations. Autobuild density-modified maps for Iml3 and Iml3-Chl4 were used for model building. Models were improved by iterative rounds of refinement with Phenix Refine. Riding hydrogens were included as implemented by Phenix. During late stages of refinement, 10 TLS groups were invoked for Iml3 in both the Iml3-alone and the Iml3-Chl4361–458 structures (Painter, 2006). The final models include residues 1–43, 51–60, 71–151, and 159–242 of Iml3 (for the Iml3 crystals: Rwork/Rfree = 22.4/25.6) and residues 1–242 of Iml3 and residues 374–450 of Chl4 (for the Iml3-Chl4361–458 crystals: Rwork/Rfree = 18.2/21.1). The coordinates have been deposited in the protein database, accession numbers 4IT3 and 4JE3, for Iml3 and Iml3-Chl4361–458, respectively.
Analytical ultracentrifugation and multi-angle light scattering
For analytical ultracentrifugation, purified proteins were prepared at three concentrations in gel filtration buffer and spun to equilibrium at five different speeds (15,19, 25, 30, and 43 × 1000rpm) in a Beckman Optima XL-A centrifuge. Results were fit to a single-species model (Iml3-Chl4, Iml3-Chl4361–458) or to a monomer-dimer exchange model (Iml3) using Sedphat (Vistica, 2004).
For multi-angle light scattering (MALS), samples were injected onto a 24ml size exclusion column (S200, GE) mounted on a high-pressure liquid chromatography system (1260 Infinity, Agilent). Light scattering and refractive index were measured in real time (Helios-II, T-rEX, Wyatt). The Astra software package was used for data analysis (Wyatt). dn/dc values were computed as the mass-weighted average of those for complex constituents, assuming the indicated stoichiometries.
In vitro translation and pulldown assay
Coding sequences for prey proteins were cloned into a vector containing a T7 promoter and a eukaryotic Kozak sequence. [35]S-labeled proteins were produced using an in vitro tranlation system (Promega). Purified His6 tagged proteins were mixed with untagged [35]S-labeled proteins in binding buffer (20mM Tris pH 8, 150mM NaCl, 20mM imidazole, 5% gycerol, 2mM beta-mercaptoethanol, and .05% (v/v) NP-40) on ice for one hour. Binding reactions were then incubated with agitation at 4°C for one hour in the presence of 15μl of NiNTA resin (Qiagen), washed three times with binding buffer, and eluted with SDS-PAGE loading dye supplemented with 400mM imidazole. Loaded and bound proteins were analyzed by SDS-PAGE electrophoresis followed by visualization with a phosphorimaging screen (BioRad).
Yeast strains and plasmid segregation
Yeast strains containing chromosomal point mutations and deletions indicated in the main text were constructed by PCR methods as previously described (Longtine, 1998). Primer sequences and constructs are available upon request. Strains were verified by colony PCR and sequencing. For plasmid segregation assays, a plasmid containing a CEN-ARS sequence from chromosome VI and the leu2 gene was transformed, and transformants were selected for by growth in SC(-leu) for 24 hours. Transformants were then grown in YPD for two hours and plated for single colonies on YPD agar. Colonies were replica plated onto SC and SC(-leu) agar and counted. Sporulation was carried out in liquid culture as described (Padmore, 1991).
Supplementary Material
Highlights.
Yeast kinetochore protein, Chl4 (CENP-N ortholog), heterodimerizes with Iml3 (CENP-L).
X-ray crystal structure of Iml3-Chl4361–458 heterodimer shows key interface.
Mutations at Iml3-Chl4 interface disturb plasmid segregation.
Iml3-Chl4 heterodimer binds Mif2 and Sgo1 in vitro.
Acknowledgments
We thank Peter De Wulf, Carol Baisden, Natalie Nannas, and Erin Loeliger for their generosity with yeast strains and plasmids. We benefitted from stimulating discussions with Kevin Corbett, Andrew Murray, Adele Marston, and Kostadin Petrov. We also thank members of the Harrison group for help with crystallography, especially Uhn-Soo Cho and Yoana Dimitrova for their help with data collection. David King carried out mass-spectrometry to identify tryptic peptides, and the staff at the Nikon Imaging Facility at Harvard Medical School supported our microscopy efforts. This work was supported by funding from the National Science Foundation (SMH), and HHMI (SCH).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, et al. PHENIX: A comprehensive python-based system for macromolecular structure solution. Acta Cryst. D. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bock LJ, Pagliuca C, Kobayashi N, Grove RA, Oku Y, Shrestha K, et al. Cnn1 inhibits the interactions between the KMN complexes of the yeast kinetochore. Nat. Cell Biol. 2012;14:614–624. doi: 10.1038/ncb2495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll CW, Milks KJ, Straight AF. Dual recognition of CENP-A nucleosomes is required for centromere assembly. J.Cell Biol. 2010;189:1143–1155. doi: 10.1083/jcb.201001013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll CW, Silva MC, Godek KM, Jansen LE, Straight AF. Centromere assembly requires the direct recognition of CENP-A nucleosomes by CENP-N. Nat. Cell Biol. 2009;11:896–902. doi: 10.1038/ncb1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman IM, Anderson S, Jwa M, Green EM, Kang JS, Yates JR, 3rd, Chan CS, Drubin DG, Barnes G. Hierarchical assembly of the budding yeast kinetochore from multiple subcomplexes. Cell. 2002;111:163–172. doi: 10.1016/s0092-8674(02)00973-x. [DOI] [PubMed] [Google Scholar]
- Cohen RL, Espelin CW, De Wulf P, Sorger PK, Harrison SC, Simons KT. Structural and functional dissection of Mif2p, a conserved DNA-binding kinetochore protein. Mol. Biol. Cell. 2008;19:4480–4491. doi: 10.1091/mbc.E08-03-0297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Wulf P, McAinsh AD, Sorger PK. Hierarchical assembly of the budding yeast kinetochore from multiple subcomplexes. Genes Dev. 2003;17:2902–2921. doi: 10.1101/gad.1144403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernius J, Hardwick KG. Bub1 kinase targets Sgo1 to ensure efficient chromosome biorientation in budding yeast mitosis. PLoS Genet. 2007;3:e213. doi: 10.1371/journal.pgen.0030213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernius J, Marston AL. Establishment of cohesion at the pericentromere by the Ctf19 kinetochore subcomplex and the replication fork-associated factor, Csm3. PLoS Genet. 2009;5(9):E1000629. doi: 10.1371/journal.pgen.1000629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernius J, Nerusheva OO, Galander S, Alves FD, Rappsilber J, Marston AL. Cohesin-Dependent association of Scc2/4 with the centromere initiates pericentromeric cohesion establishment. Curr. Biol. 2013;23:599–606. doi: 10.1016/j.cub.2013.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh SK, Sau S, Lahiri S, Lohia A, Sinha P. The Iml3 protein of the budding yeast is required for the prevention of precocious sister chromatid separation in meiosis I and for sister chromatid disjunction in meiosis II. Curr. Genet. 2004;46:82–91. doi: 10.1007/s00294-004-0516-6. [DOI] [PubMed] [Google Scholar]
- Ha JY, Kim HK, Kim, do J, Kim KH, Oh SJ, Lee HH, Yoon HJ, Song HK, Suh SW. The recombination-associated protein RdgC adopts a novel toroidal architecture for DNA binding. Nucleic Acids Res. 2007;35:2671–2681. doi: 10.1093/nar/gkm144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm L, Rosenström P. Dali server: conservation mapping in 3D. Nucleic Acids Res. 2010:W545–W549. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh WK, Falvo JV, Gerke LC, Carroll AS, Howson RW, Weissman JS, et al. Global analysis of protein localization in budding yeast. Nature. 2003;425:686–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- Indjeian VB, Murray AW. Budding yeast mitotic chromosomes have an intrinsic bias to biorient on the spindle. Curr. Biol. 2007;17:1837–1846. doi: 10.1016/j.cub.2007.09.056. [DOI] [PubMed] [Google Scholar]
- Joglekar AP, Bouck DC, Molk JN, Bloom KS, Salmon ED. Molecular architecture of a kinetochore-microtubule attachment site. Nat. Cell Biol. 2006;8:581–585. doi: 10.1038/ncb1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katis VL, Galova M, Rabitsch KP, Gregan J, Nasmyth K. Maintenance of cohesin at centromeres after meiosis I in budding yeast requires a kinetochore-associated protein related to MEI-S332. Curr. Biol. 2004;14:560–572. doi: 10.1016/j.cub.2004.03.001. [DOI] [PubMed] [Google Scholar]
- Kawashima SA, Yamagishi Y, Honda T, Ishiguro K, Watanabe Y. Phosphorylation of H2A by Bub1 prevents chromosomal instability through localizing shugoshin. Science. 2010;327:172–177. doi: 10.1126/science.1180189. [DOI] [PubMed] [Google Scholar]
- Kiburz BM, Amon A, Marston AL. Shugoshin promotes sister kinetochore biorientation in saccharomyces cerevisiae. Mol. Biol.Cell. 2007;19:1199–1209. doi: 10.1091/mbc.E07-06-0584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiburz BM, Reynolds DB, Megee PC, Marston AL, Lee BH, Lee TI, Levine SS, Young RA, Amon A. The core centromere and Sgo1 establish a 50-kb cohesin-protected domain around centromeres during meiosis I. Genes Dev. 2005;19:3017–3030. doi: 10.1101/gad.1373005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JL, Nikolov DB, Burley SK. Co-crystal structure of TBP recognizing the minor groove of a TATA element. Nature. 1993;365:520–527. doi: 10.1038/365520a0. [DOI] [PubMed] [Google Scholar]
- Kitajima TS, Kawashima SA, Watanabe Y. The conserved kinetochore protein shugoshin protects centromeric cohesion during meiosis. Nature. 2004;427:510–517. doi: 10.1038/nature02312. [DOI] [PubMed] [Google Scholar]
- Longtine MS, McKenzie A, 3rd, Demarini DJ, Shah NG, Wach A, Brachat A, et al. Additional modules for versatile and economical PCR-based gene deletion and modification in saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Marston AL, Tham WH, Shah H, Amon A. A genome-wide screen identifies genes required for centromeric cohesion. Science. 2004;303:1367–1370. doi: 10.1126/science.1094220. [DOI] [PubMed] [Google Scholar]
- McClelland SE, Borusu S, Amaro AC, Winter JR, Belwal M, McAinsh AD, et al. The CENP-A NAC/CAD kinetochore complex controls chromosome congression and spindle bipolarity. EMBO J. 2007;26:5033–5047. doi: 10.1038/sj.emboj.7601927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellone BG, Grive KJ, Shteyn V, Bowers SR, Oderberg I, Karpen GH. Assembly of drosophila centromeric chromatin proteins during mitosis. PLoS Genet. 2011;7(5):e1002068. doi: 10.1371/journal.pgen.1002068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng TM, Waples WG, Lavoie BD, Biggins S. Pericentromeric sister chromatid cohesion promotes kinetochore biorientation. Mol. Biol. Cell. 2009;20:3818–3827. doi: 10.1091/mbc.E09-04-0330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada M, Cheeseman IM, Hori T, Okawa K, McLeod IX, Yates JR, 3rd, Desai A, Fukagawa T. The CENP-H-I complex is required for the efficient incorporation of newly synthesized CENP-A into centromeres. Nat. Cell Biol. 2006;8:446–457. doi: 10.1038/ncb1396. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z, Minor V. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Owen DJ, Vallis Y, Pearse BM, McMahon HT, Evans PR. The structure and function of the beta 2-adaptin appendage domain. EMBO J. 2000;19:4216–4227. doi: 10.1093/emboj/19.16.4216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padmore R, Cao L, Kleckner N. Temporal comparison of recombination and synaptonemal complex formation during meiosis in S. cerevisiae. Cell. 1991;66:1239–1256. doi: 10.1016/0092-8674(91)90046-2. [DOI] [PubMed] [Google Scholar]
- Painter J, Merritt EA. Optimal description of a protein structure in terms of multiple groups undergoing TLS motion. Acta Cryst. D. 2006;62:439–450. doi: 10.1107/S0907444906005270. [DOI] [PubMed] [Google Scholar]
- Pot I, Measday V, Snydsman B, Cagney G, Fields S, Davis TN, Muller EG, Hieter P. Chl4p and iml3p are two new members of the budding yeast outer kinetochore. Mol. Biol. Cell. 2003;14:460–476. doi: 10.1091/mbc.E02-08-0517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Przewloka MR, Venkei Z, Bolanos-Garcia VM, Debski J, Dadlez M, Glover DM. CENP-C is a structural platform for kinetochore assembly. Curr. Biol. 2011;21:399–405. doi: 10.1016/j.cub.2011.02.005. [DOI] [PubMed] [Google Scholar]
- Schleiffer A, Maier M, Litos G, Lampert F, Hornung P, Mechtler K, Westermann S. CENP-T proteins are conserved centromere receptors of the Ndc80 complex. Nat. Cell Biol. 2012;14:604–613. doi: 10.1038/ncb2493. [DOI] [PubMed] [Google Scholar]
- Screpanti E, De Antoni A, Alushin GM, Petrovic A, Melis T, Nogales E, et al. Direct binding of cenp-C to the Mis12 complex joins the inner and outer kinetochore. Curr. Biol. 2011;21:391–398. doi: 10.1016/j.cub.2010.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T, Fuchs J, Loidl J, Nasmyth K. Cohesin ensures bipolar attachment of microtubules to sister centromeres and resists their precocious separation. Nat. Cell Biol. 2000;2:492–499. doi: 10.1038/35019529. [DOI] [PubMed] [Google Scholar]
- Vistica J, Dam J, Balbo A, Yikilmaz E, Mariuzza RA, Rouault TA, et al. Sedimentation equilibrium analysis of protein interactions with global implicit mass conservation constraints and systematic noise decomposition. Anal. Biochem. 2004;326:234–256. doi: 10.1016/j.ab.2003.12.014. [DOI] [PubMed] [Google Scholar]
- Weber SA, Gerton JL, Polancic JE, DeRisi JL, Koshland D, Megee PC. The kinetochore is an enhancer of pericentric cohesin binding. PLoS Biol. 2004;2:E260. doi: 10.1371/journal.pbio.0020260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Min J, Zeng H, McCloskey DE, Ikeguchi Y, Loppnau P, Michael AJ, Pegg AE, Plotnikov AN. Crystal structure of human spermine synthase: implications of substrate binding and catalytic mechanism. J. Biol. Chem. 2008;283:16135–16146. doi: 10.1074/jbc.M710323200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.