Abstract
Hypericum perforatum L. is a medicinal plant considered as an important natural source of secondary metabolites with a wide range of pharmacological attributes. Hairy roots (HR) were induced from root segments of in vitro grown seedlings from H. perforatum after cocultivation with Agrobacterium rhizogenes A4. Investigations have been made to study the production of phenolic compounds in dark-grown (HR1) and photoperiod-exposed (HR2) cultures. The chromatographic analysis of phenolic acids, flavonols, flavan-3-ols, and xanthones revealed marked differences between HR1 and HR2 cultures. The production of quinic acid, kaempferol, and seven identified xanthones was increased in HR2. Moreover, HR2 showed a capability for de novo biosynthesis of two phenolic acids (3-p-coumaroylquinic acid and 3-feruloylquinic acid), three flavonol glycosides (kaempferol hexoside, hyperoside, and quercetin acetylglycoside), and five xanthones (tetrahydroxy-one-methoxyxanthone, 1,3,5-trihydroxy-6-methoxyxanthone, 1,3,5,6-tetrahydroxy-2-prenylxanthone, paxanthone, and banaxanthone E). On the other side, HR1 cultures were better producers of flavan-3-ols (catechin, epicatechin, and proanthocyanidin dimers) than HR2. This is the first comparative study on phenolic profile of H. perforatum HR cultures grown under dark and photoperiod conditions.
1. Introduction
Hypericum perforatum L. (St. John's wort) is a traditional medicinal plant with a complex mixture of secondary metabolites. Phenolic compounds as naphthodianthrones, acylphloroglucinols, flavonoids, and xanthones are the main bioactive metabolites commonly described for this plant [1]. In phytomedicine, Hypericum extracts are responsible for a plethora of pharmacological activities including antidepressant, antiviral, antioxidant, anti-inflammatory, and antimicrobial properties [2]. To meet the increasing demand for plants utilized in the pharmaceutical industry, much of the recent research has focussed on the development of new in vitro culture techniques as a useful alternative to improve the yield of bioactive metabolites in plant material.
Agrobacterium rhizogenes-mediated plant transformation represents a convenient experimental system for establishment of hairy roots (HR). Transformed root cultures represent an attractive model system for the production of high-value secondary metabolites, including pharmaceuticals and other biologically active substances of commercial importance [3]. Namely, HR cultures may synthesize higher levels of secondary metabolites or amounts comparable to those of the intact plant and offer a promising approach to the industrial exploitation of HR for production of novel metabolites [4, 5]. Until now, only A. rhizogenes- [6, 7] and biolistic-mediated [8] transformation methods have been applied. In recent years, it has been shown that HR are responsive to physical stimuli such as exposure to light which is known to regulate a number of plant developmental processes [9], as well as primary and secondary metabolite production [10]. These findings indicate that the exposure of HR to light leads to alternations in their biosynthetic potentials. Although several studies investigated secondary metabolite production in root cultures [11, 12], the capacity of H. perforatum HR to produce phenolic compounds has never been explored.
This study describes the phenolic profile of transformed roots (HR) from H. perforatum transformed with A. rhizogenes strain A4, grown in constant dark (HR1) or in light/dark photoperiod (HR2) conditions. Phenolic compounds in HR were analyzed using high-performance liquid chromatography (HPLC) coupled with diode array detection (DAD) and tandem mass spectrometry (MSn) with electrospray ionization (ESI). All present derivatives of phenolic acids, flavonol glycosides, flavonoid aglycones, flavan-3-ols, and xanthones were identified from corresponding UV and MS spectra and quantified by HPLC-DAD.
2. Material and Methods
2.1. Plant Material
Seeds from H. perforatum were collected from wild plants growing in a natural population in the Pelister National Park at about 1394 m. Voucher specimen (number 060231) of H. perforatum is deposited in the Herbarium at the Faculty of Natural Sciences and Mathematics, University “Ss. Cyril and Methodius,” Skopje, Macedonia. As for a previous study [13], seeds were surface sterilized and in vitro germinated seedlings were maintained in a growth chamber at 25 ± 1°C under a photoperiod of 16 h light, irradiance at 50 μmol · m2 · s−1, and 50 to 60% relative humidity.
2.2. Establishment of Hairy Roots
The wild type Agrobacterium rhizogenes agropine strain A4 (obtained from Institut National de la Recherche Agronomique-INRA, Versailles, France) was used for H. perforatum transformation experiments [14]. Transformation protocol was performed according to Di Guardo et al. [6] with the modifications described in our previous study [15]. Briefly, the HR cultures were induced by A. rhizogenes A4 from root segments of one-month-old in vitro germinated seedlings from H. perforatum. Transgenic status of the HR was confirmed by PCR analysis using rolB specific primers [15]. Transformed root cultures were maintained by subculturing at one-month intervals on MS/B5 hormone-free medium. The subculture was carried out at 25 ± 1°C in the dark (HR1) and under photoperiod (HR2) of 16 h light (50 μmol · m2 · s−1). One-month-old HR1 and HR2 cultures were harvested (1 g) and then frozen in liquid nitrogen or lyophilized and stored at −80°C, until analysis.
2.3. HPLC/DAD/ESI-MSn Analysis
Phenolic compounds extraction from freeze-dried lyophilized and powdered root cultures was performed as previously reported by Tusevski et al. [15]. The HPLC system was equipped with an Agilent 1100 series diode array and mass detector in series (Agilent Technologies, Waldbronn, Germany). It consisted of a G1312A binary pump, a G1313A autosampler, a G1322A degasser, and a G1315B photodiode array detector, controlled by ChemStation software (Agilent, v.08.03). Chromatographic separations were carried out on 150 mm × 4.6 mm, 5 μm XDB-C18 Eclipse column (Agilent, USA). The mobile phase consisted of two solvents: water-formic acid (A; 99 : 1, v/v) and methanol (B) in the following gradient program: 90% A and 10% B (0–20 min), 80% A and 20% B (20–30 min), 65% A and 35% B (30–50 min), 50% A and 50% B (50–70 min), and 20% A and 80% B (70–80 min) and continued with 100% B for a further 10 min. Each run was followed by an equilibration period of 10 min. The flow rate was 0.4 mL/min and the injection volume was 10 μL. All separations were performed at 38°C. Formic acid (HCOOH) and methanol (CH3OH) were HPLC grade solvents (Sigma-Aldrich, Germany). The HPLC-water was purified by a PURELAB Option-Q system (Elga LabWater, UK). The commercial standards chlorogenic acid, rutin, quercetin, kaempferol, catechin, epicatechin, and xanthone (Sigma-Aldrich, Germany) were used as reference compounds. The reference compounds were dissolved in 80% methanol in water. The concentration of the stock standard solutions was 1 mg · mL−1 and they were stored at −20°C. Spectral data from all peaks were accumulated in the range of 190–600 nm, and chromatograms were recorded at 260 nm for xanthones, at 280 nm for flavan-3-ols, at 330 nm for phenolic acids, and at 350 nm for flavonols. Peak areas were used for quantification at wavelengths where each group of phenolic compounds exhibited an absorption maximum. The HPLC system was connected to the Agilent G2445A ion-trap mass spectrometer equipped with electrospray ionization (ESI) system and controlled by LCMSD software (Agilent, v.6.1.). Nitrogen was used as nebulizing gas at a pressure-level of 65 psi and the flow was adjusted to 12 L · min−1. Both the heated capillary and the voltage were maintained at 350°C and 4 kV, respectively. MS data were acquired in the negative ionization mode. The full scan mass covered the mass range from m/z 100 to 1200. Collision-induced fragmentation experiments were performed in the ion trap using helium as a collision gas, with voltage ramping cycle from 0.3 up to 2 V. Maximum accumulation time of the ion trap and the number of MS repetitions to obtain the MS average spectra were set at 300 ms and 3, respectively. Identification of the component peaks was performed by the UV/Vis, MS, and MS2 spectra and retention times of the abovementioned available standards.
2.4. Statistical Analysis
The experiments were independently repeated twice under the same conditions and all analyses were performed in triplicate. Secondary metabolite contents were expressed as mg · 100 g−1 dry weight (DW). Standard deviation of mean value was shown as ± S.D. The statistical analyses including calculations of means and standard deviations were performed applying Excel (Microsoft Office, 2007).
3. Results and Discussion
3.1. Establishment of Hairy Roots
As previously reported [15], H. perforatum HR were initiated by inoculation of root explants with A. rhizogenes A4. On the basis of culture conditions, selected dark-grown (HR1) and photoperiod-exposed (HR2) cultures showed differences in the morphology. Dark-grown hairy root cultures were thinner and whitish in colour showing rapid plagiotropic growth with active branching and vigorous production of elongated lateral roots. Present results confirmed that transformed roots of H. perforatum had characteristic traits of HR previously described by Tepfer [16]. In contrast, HR2 cultures began to turn pale green after 7 days of culture and continued to acquire green coloration during the course of subsequent growth period. Moreover, HR2 appeared intense greenish-brown after one month of culture. It was seen that the growth of HR was generally most vigorous between the 3rd and 4th weeks of the cultivation period (1 month), but their growth declined after the 5th week due to the nutrient depletion. For HPLC analysis, one-month-old HR cultures were further evaluated.
3.2. HPLC/DAD/ESI-MSn Analysis
The HPLC/DAD/ESI-MSn technique was used to analyse the phenolic profile of H. perforatum HR1 and HR2 cultures. Four groups of phenolic compounds such as phenolic acids, flavonols, flavan-3-ols, and xanthones were recorded in HR cultures (Tables 1 and 2). The identification of phenolic compounds (Tables 1 and 2, Figure 1) was based on the typical UV/Vis spectral data and LC/MS in the negative ionization mode [M–H]− with the subsequent MS2, MS3, and MS4 analysis for further identification with reference to similar data previously reported [15, 17–26]. The HPLC analysis of phenolic compounds revealed marked differences between HR1 and HR2 cultures (Tables 1 and 2, Figure 1).
Table 1.
Retention times, UV, and mass spectral data of phenolic acids, flavonols, and flavan-3-ols in Hypericum perforatum dark-grown (HR1) and photoperiod-exposed (HR2) hairy root culture extractsa.
| Peak no. | Compounds | t R (min) | UV (nm) | [M–H]–
(m/z) |
–MS2 [M–H]–
(m/z) |
HR1 (mg·100 g−1 DW ± S.D.) | HR2 (mg·100 g−1 DW ± S.D.) |
|---|---|---|---|---|---|---|---|
| Phenolic acids | |||||||
| F1 | Quinic acid | 3.9 | 262, 310 | 191 | 173, 127 | 166.77 ± 1.20 | 233.14 ± 19.31 |
| F3 | 3-p-Coumaroylquinic acid | 19.9 | 314 | 337 | 191, 163 | n.d. | 12.18 ± 0.92 |
| F5 | 3-Feruloylquinic acid | 25.3 | 314 | 367 | 193 | n.d. | 5.87 ± 0.26 |
|
| |||||||
| Flavonols | |||||||
| F8 | Quercetin 6-C-glucoside | 33.9 | 256, 356 | 421 | 331, 301 | 2.99 ± 0.79 | 1.74 ± 0.11 |
| F10 | Isorhamnetin O-hexoside | 38.1 | 254, 356 | 477 | 316, 315, 271 | 11.80 ± 0.94 | 1.74 ± 0.09 |
| F11 | Kaempferol hexoside | 41.2 | 256, 266, 350 | 447 | 285 | n.d. | 1.99 ± 0.26 |
| F12 | Hyperoside (quercetin 3-O-galactoside) | 43.8 | 264, 296 sh, 354 | 463 | 301 | n.d. | 2.77 ± 0.18 |
| F13 | Rutin (quercetin 3-O-rutinoside) | 44.9 | 263, 298 sh, 356 | 609 | 301 | 14.72 ± 2.16 | 5.46 ± 0.43 |
| F14 | Quercetin acetylglycoside | 48.1 | 254, 298, 358 | 505 | 463, 445, 301 | n.d. | 3.94 ± 0.10 |
| F15 | Kaempferol | 59.5 | 256, 266, 350 | 285 | / | 3.92 ± 0.38 | 6.26 ± 0.37 |
|
| |||||||
| Flavan-3-ols | |||||||
| F2 | Catechin | 19.5 | 280 | 289 | 245, 205 | 27.28 ± 3.20 | 2.62 ± 0.08 |
| F6 | (Epi)catechin | 29.9 | 280 | 289 | 245, 205 | 184.85 ± 12.92 | 133.36 ± 15.19 |
| F4 | Proanthocyanidin dimer | 24.5 | 280 | 577 | 559, 451, 425, 407, 289 | 146.95 ± 9.13 | 56.61 ± 2.65 |
| F7 | Proanthocyanidin dimer | 33.4 | 280 | 577 | 559, 451, 425, 407, 289 | 41.43 ± 1.03 | 0.76 ± 0.08 |
| F9 | Proanthocyanidin dimer | 36.8 | 280 | 577 | 559, 451, 425, 407, 289 | 29.24 ± 2.47 | 24.93 ± 0.15 |
an.d.: not detected; DW: dry weight; sh: shoulder; t R: retention time. MS2 ions in bold indicate the base peak. For information on peak numbers, see Figure 1.
Table 2.
Retention times, UV, and mass spectral data of xanthones in Hypericum perforatum dark-grown (HR1) and photoperiod-exposed (HR2) hairy root culture extractsa.
| Peak no. |
Compounds | t R (min) | UV (nm) | [M–H]–
(m/z) |
–MS2 [M–H]–
(m/z) |
HR1 (mg·100 g−1 DW ± S.D.) | HR2 (mg·100 g−1 DW ± S.D.) |
|---|---|---|---|---|---|---|---|
| X1 | Mangiferin | 37.3 | 238, 256, 312, 362 | 421 | 331, 301, 258 | 1383.25 ± 88.91 | 669.67 ± 24.12 |
| X2 | Xanthone derivative 1 | 45.8 | 208, 257, 322, 374 | 441 | 423, 397, 373, 305, 257, 229 | 109.47 ± 9.81 | n.d. |
| X3 | Xanthone derivative 2 | 46.2 | 242, 306 | 367 | 287 | 635.06 ± 18.52 | 600.59 ± 39.62 |
| X4 | 1,3,5,6-Tetrahydroxyxanthone dimer | 50.2 | 252, 284, 328 | 517 | 499, 468, 446, 391, 365 | 821.61 ± 28.39 | 692.94 ± 19.28 |
| X5 | 1,3,6,7-Tetrahydroxyxanthone dimer | 53.9 | 238, 254, 312, 364 | 517 | 517, 469, 447, 379, 257 | 522.56 ± 25.44 | 88.31 ± 2.88 |
| X6 | 1,3,5,6-Tetrahydroxyxanthone | 55.4 | 250, 282, 328 | 259 | 229, 213, 187 | 190.17 ± 20.73 | 949.35 ± 51.71 |
| X7 | 1,3,6,7-Tetrahydroxyxanthone | 55.8 | 236, 254, 314, 364 | 259 | 231, 215, 187, 147 | 167.14 ± 9.52 | 874.85 ± 31.24 |
| X8 | Tetrahydroxy-one-methoxyxanthone | 57.7 | 254, 286, 328 | 289 | 274, 175 | n.d. | 448.65 ± 9.44 |
| X9 | Xanthone derivative 3 | 59.2 | 244, 280, 316 | 353 | 273 | n.d. | 276.57 ± 9.29 |
| X10 | 1,3,5-Trihydroxy-6-methoxyxanthone | 63.3 | 250, 284, 326 | 273 | 258,225 | n.d. | 150.86 ± 12.62 |
| X11 | Mangiferin C-prenyl isomer | 73.5 | 238, 260, 312, 372 | 489 | 399, 369, 327 | 433.68 ± 82.56 | n.d. |
| X12 | 1,3,6,7-Tetrahydroxyxanthone 8-prenylxanthone | 73.9 | 248, 312, 366 | 327 | 325, 297, 258, 201 | 547.65 ± 15.21 | 737.48 ± 65.39 |
| X13 | 1,3,5,6-Tetrahydroxyxanthone 8-prenylxanthone | 74.9 | 242, 260, 320, 368 | 327 | 325, 297, 258, 201 | 368.17 ± 21.70 | n.d. |
| X14 | 1,3,7-Trihydroxy-2-(2-hydroxy-3-methyl-3-butenyl)xanthone | 75.3 | 238, 260, 314, 388 | 327 | 309, 257 | 588.66 ± 49.31 | 854.53 ± 31.88 |
| X15 | Toxyloxanthone | 76.2 | 242, 262, 330, 384 | 325 | 307, 283, 272 | 577.03 ± 5.09 | 1542.09 ± 129.21 |
| X16 | 1,3,7-Trihydroxy-6-methoxy-8-prenylxanthone | 76.5 | 240, 260, 318, 370 | 341 | 326, 311, 297, 285 | 650.13 ± 34.77 | n.d. |
| X17 | 1,3,6,7-Tetrahydroxyxanthone 2-prenylxanthone | 76.7 | 248, 312, 368 | 327 | 325, 283, 271 | 1402.03 ± 85.98 | 656.33 ± 37.25 |
| X18 | γ-Mangostin isomer | 77.1 | 254, 286, 324 | 395 | 326, 283, 271 | 1226.31 ± 185.52 | 1480.32 ± 130.06 |
| X19 | 1,3,6-Trihydroxy-7-methoxy-8-prenylxanthone | 77.2 | 240, 256, 312, 370 | 341 | 293, 256 | 3240.28 ± 140.14 | n.d. |
| X20 | 1,3,5,6-Tetrahydroxyxanthone 2-prenylxanthone | 77.4 | 238, 260, 318, 372 | 327 | 297, 258 | n.d. | 699.36 ± 49.61 |
| X21 | Paxanthone | 78.0 | 244, 264, 324, 386 | 339 | 324, 307 | n.d. | 4040.70 ± 209.82 |
| X22 | γ-Mangostin isomer | 78.9 | 260, 316, 370 | 395 | 351, 339, 326, 283 | 3629.15 ± 338.08 | n.d. |
| X23 | Trihydroxy-1-methoxy-C-prenylxanthone | 79.4 | 260, 286, 314 | 341 | 326 | 11314.34 ± 469.01 | 10067.14 ± 561.72 |
| X24 | Xanthone derivative 4 | 79.9 | 260, 308, 374 | 295 | 277, 251, 195, 171 | n.d. | 2778.02 ± 81.11 |
| X25 | γ-Mangostin | 80.0 | 246, 262, 320 | 395 | 351, 339, 326, 283 | 7861.71 ± 415.11 | n.d. |
| X26 | Banaxanthone D | 80.2 | 244, 268, 332 | 461 | 393, 341, 297 | 1784.69 ± 88.90 | n.d. |
| X27 | Xanthone derivative 5 | 80.5 | 254, 310 | 355 | 340, 325, 297, 285, 271 | 2266.19 ± 191.89 | 1765.42 ± 36.19 |
| X28 | Garcinone E | 81.2 | 256, 286, 332 | 463 | 394, 351, 339, 297, 285 | 8229.95 ± 537.14 | 10844.13 ± 288.29 |
| X29 | Xanthone derivative 6 | 82.2 | 262, 288, 322 | 393 | / | 421.44 ± 36.66 | 370.43 ± 45.16 |
| X30 | Banaxanthone E | 82.6 | 252, 302, 330 | 477 | 419, 393, 339, 297 | n.d. | 499.91 ± 38.44 |
| X31 | Xanthone derivative 7 | 83.6 | 270, 330, 400 | 467 | 398, 383, 327, 271, 234 | n.d. | 429.57 ± 7.82 |
| X32 | Garcinone C | 83.9 | 286, 340 | 413 | 369, 344, 301, 233 | 1185.94 ± 149.05 | 943.63 ± 55.98 |
| X33 | Xanthone derivative 8 | 84.4 | 254, 284, 326 | 481 | 412, 397, 327, 271, 234 | 562 ± 38.99 | 126.80 ± 1.69 |
an.d.: not detected; DW: dry weight; sh: shoulder; t R: retention time. MS2 ions in bold indicate the base peak. For information on peak numbers, see Figure 1.
Figure 1.
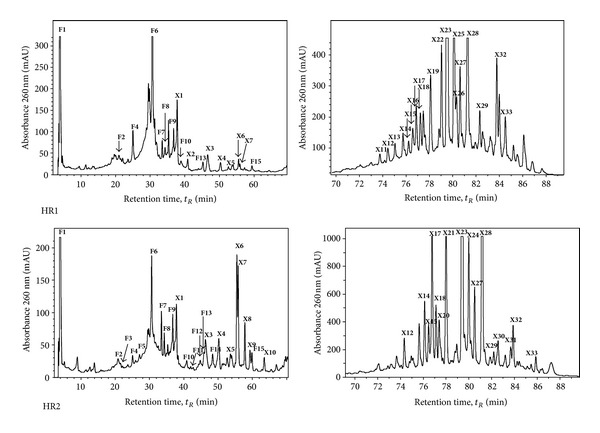
Chromatograms of Hypericum perforatum dark-grown (HR1) and photoperiod-exposed (HR2) hairy root culture extracts monitored at 260 nm for detection of phenolic compounds. Compound symbols correspond to those indicated in Tables 1 and 2.
3.2.1. Phenolic Acids
Compound F1 occurred at retention time of 3.9 min and exhibited a [M–H]− ion at m/z 191 (Table 1, Figure 1). Its MS2 fragmentation produced a [M–H–CO–2H2O]− ion at m/z 127 as a base peak. A [M–H–H2O]− ion at m/z 173 was also observed. Compound F1 was identified as quinic acid, taking into account its MSn fragmentation pattern and the literature data [17]. Quinic acid (F1) was the only detectable phenolic acid in both HR cultures. A 1.4-fold increase of quinic acid was observed in HR2 compared to HR1 cultures.
Two peaks, 3-p-coumaroylquinic acid (F3) and 3-feruloylquinic acid (F5), were detected only in HR2 cultures with identical UV spectra characterized by absorption band at 314 nm. Compounds F3 and F5 were readily distinguished by their cinnamic acid-derived MS2 base peaks at m/z 163 and at m/z 193, respectively. Quinic acid is the most important component as a key intermediate in the biosynthesis of aromatic compounds. The condensation between quinic acid and caffeic acid leads to the formation of chlorogenic acid in the shikimic acid pathway [27]. Chlorogenic acid is an important antioxidative compound recently produced by H. perforatum adventitious roots cultivated in bioreactor [11], shoot cultures [28], and transgenic plantlets [29].
3.2.2. Flavonols
The flavonols were observed to be qualitatively and quantitatively different in both H. perforatum HR cultures (Table 1, Figure 1). A major identified group of compounds belonged to flavonols according to their characteristic UV spectra of flavonols glycosylated at C3 (257, 265 sh, 355 nm). The detected compound F8 can be identified as C-glycoside of quercetin. The deprotonated molecular ion [M–H]− of compound F8 was detected at m/z 421. It showed MS2 fragmentation characteristic of mono-C-hexosyl flavones, with losses of 90 and 120 amu [19], giving m/z 301 ion characteristic for quercetin. The compound F10 had a molecular ion [M–H]− at m/z 477. The MS2 spectra of this compound showed fragmentation ions at m/z 315 (loss of 162 amu), suggesting a presence of hexose residue. So, compound F10 was tentatively identified as isorhamnetin O-hexoside. Compound F13 had a molecular ion [M–H]− at m/z 609, and its MS2 gave a single ion at m/z 301, indicating quercetin derivative with rutinose at C3 [20]. The absence of intermediate fragmentation between the deprotonated molecular ion and the aglycone ion is indicative of an interglycosidic linkage 1 → 6 [21]; therefore, this compound was identified as quercetin 3-O-rutinoside (rutin). Flavonol glycosides as F8, F10, and F13 detected in HR2 cultures were in lower amounts compared with those in HR1.
The compound F11 was identified as kaempferol derivative with glycosylation in position 3 according to its UV-spectra (256, 266, 350 nm). The MS and MS2 spectra were consistent with the presence of a hexose residue and confirm the kaempferol aglycone. Therefore, this compound was identified as kaempferol hexoside. Compound F12 had a deprotonated molecular ion [M–H]− at m/z 463 and its MS2 gave a single ion at m/z 301, indicating quercetin hexose derivative, most probably hyperoside (quercetin 3-O-galactoside) [30]. Compound F14 gave deprotonated molecular ion at m/z 505. Its MS2 fragmentation produced a [M–H–42]− ion at m/z 463 and [M–H–42–162]− ion at m/z 301 as a base peak, indicating quercetin acetylglycoside. It is worth noting, that flavonol glycosides as F11, F12, and F14 were synthesized only in HR2 cultures.
With regard to the class of flavonol glycosides, our results showed that both HR cultures had capability to produce quercetin and kaempferol derivatives. However, there is no available study for the potential of H. perforatum root cultures to produce flavonol derivatives. Several differences can be pointed out when comparing the composition of flavonol glycosides in HR cultures with those of H. perforatum in vitro cultures. In our previous work [31, 32], we indicated that H. perforatum cells, calli, and shoots demonstrate a considerable potential for producing quercetin, isoquercitrin, and quercitrin upon elicitation with jasmonic acid and salicylic acid. The LC-MS screening of twelve H. perforatum HR transgenic plants showed a large variability in the content of rutin, hyperoside, quercetrin, and quercetin [29]. Moreover, the abovementioned flavonol glycosides had been identified in H. perforatum regenerated plantlets [33] and H. undulatum shoot cultures [34].
The HPLC-MS analysis of flavonoid aglycones in HR cultures resulted in the identification of kaempferol (F15). Its molecular ion at m/z 285 corresponded to that of kaempferol. The identification was made by comparing its UV and MS spectra to analytical standards and literature data [28]. A 1.6-fold increase of kaempferol was observed in HR2 compared to HR1 cultures.
3.2.3. Flavan-3-ols
Flavan-3-ols (catechins) were identified as the main flavonoid fraction in HR cultures. The HPLC analysis confirmed the presence of 5 flavan-3-ols: F2, F4, F6, F7, and F9 in both HR cultures (Table 1, Figure 1). The mass spectrum in full scan mode showed the deprotonated molecules [M–H]− of catechin (F2) and epicatechin (F6) at m/z 289, with characteristic MS2 ions at m/z 245 and 205 and UV maximum at 280 nm. Compounds F4, F7, and F9 had [M–H]− at m/z 577 and main fragmentation with loss of 152 amu, characteristic fragmentation pathway by retro Diels-Alder reaction [22], and were recognized as proanthocyanidin dimers. Dark-grown HR were better producers of catechins and proanthocyanidin dimers than HR2. Literature data about the production of catechin derivatives in in vitro cultures of H. perforatum are scarce. In our previous work [35], we indicated that H. perforatum root cultures may be considered as a promising source of proanthocyanidin dimers. Nevertheless, catechin, epicatechin, and proanthocyanidin dimers had been previously identified in shoots and calli of H. erectum [36] and H. undulatum shoot cultures [34].
3.2.4. Xanthones
Xanthones comprise the majority of the phenolic compounds detected by HPLC/DAD/ESI-MSn (Table 2, Figure 1). They include simple oxygenated xanthones or derivatives with prenyl, pyran, or methoxy groups. Compound X1 was putatively identified as mangiferin. The HPLC-MS/MS analysis of this compound gave a molecular ion m/z [M–H]− of 421 and major −MS2 fragments at m/z 331 [M–H–90]− and 301 [M–H–120]−, losses characteristics of C-hexosyl compounds [19]. Compounds X4, X6, X8, X13, and X20 showed UV spectral characteristics of the 1,3,5,6 oxygenated xanthones, with band IV reduced to shoulder [37], while most of the other identified xanthones had UV spectra similar to mangiferin typical of the 1,3,6,7 oxygenation pattern with a very well-defined band IV [23, 38]. Compounds X6 and X7 were identified as 1,3,5,6-tetrahydroxyxanthone and 1,3,6,7-tetrahydroxyxanthone aglycones, respectively (single intense molecular ion [M–H]− at m/z 259) [20, 25]. Compounds X4 and X5 gave molecular ions [M–H]− at m/z 517. Major −MS2 fragments at m/z 365 and 257, respectively, characterized them as dimers of 1,3,5,6-tetrahydroxyxanthone and 1,3,6,7-tetrahydroxyxanthone. Compound X10 gave a molecular ion [M–H]− at m/z 273 and major MS2 fragment at m/z 258 [M–H–15]−. The MS analysis indicates the presence of one methoxy and three hydroxyl groups. These together with comparison to literature data gave its identification as 1,3,5-trihydroxy-6-methoxyxanthone [26]. Compound X8 had the same UV spectra as compound X11 but contained one hydroxy group more and molecular ion [M–H]− at 289, base peak at m/z 274. We tentatively assigned this compound as tetrahydroxy-one-methoxyxanthone (X8). Compound X11 was putatively identified as mangiferin-C-prenyl isomer. The HPLC-MS/MS analysis of this compound gave molecular ions [M–H]− at m/z 489 and major MS2 fragments at m/z 399 [M–H–90]−, 369 [M–H–120]− with losses characteristics of C-hexosyl compounds [21] and 327 as a base peak (1,3,6,7-tetrahydroxyxanthone-C-prenyl residue). Compounds X12 and X17 had UV spectra characteristic of 1,3,6,7-oxygenated xanthones and molecular ions [M–H]− at 327. So, these compounds were identified as 1,3,6,7-tetrahydroxyxanthone-C-prenyl isomers. In the literature, it is found that in some Hypericum species, the C-prenyl moiety can be in position 2 or 8 [24]. They can be tentatively assigned as 1,3,6,7-tetrahydroxy-8-prenyl xanthone (X12) and 1,3,6,7-tetrahydroxy-2-prenyl xanthone (X17). Compounds X13 and X20 had same fragmentation pattern as X12 and X17 but different UV spectra, characteristic of 1,3,5,6-tetrahydroxyxanthone, leading to their assignment as 1,3,5,6-tetrahydroxy-C-prenylxanthone. These compounds were identified as 1,3,5,6-tetrahydroxy-8-prenylxanthone (X13) and 1,3,5,6-tetrahydroxy-2-prenylxanthone (X20). Compound X14 gave molecular ion [M–H]− at m/z 327 but showed a different fragmentation pattern in comparison with the other compounds with the same mass. In the MS2, a loss of a hydroxyl group [M–H2O]− to give the base peak at m/z 309 is exhibited, indicating that the OH group is not linked to the xanthone aglycone but to the prenyl group. In the next MS3 step, after the loss of the prenyl moiety, the base peak at m/z 257 was detected. According to this behavior and literature data [26], it is evident that this compound is 1,3,7-trihydroxy-2-(2-hydroxy-3-methyl-3-butenyl)-xanthone. Xanthones X16 and X19 were identified as 1,3,7-trihydroxy-6-methoxy-8-prenyl xanthone and 1,3,6-trihydroxy-7-methoxy-8-prenyl xanthone (molecular ions [M–H]− at m/z 341), respectively, using the obtained spectral data and comparison to previously published data [20, 24, 25]. Compound X23 had similar fragmentation pattern as compound X16, indicating that compound X23 has similar nature as compound X16. We can tentatively term compound X23 as trihydroxy-1-metoxy-C-prenyl xanthone. The comparison to previously published data [39] for UV and MS spectra indicates that compound X25 is γ-mangostin (molecular ion [M–H]− at m/z 395). Compounds X18 and X22 were putatively identified as isomers of γ-mangostin (1,3,6,7-tetrahydroxyxanthone-C-bis-prenyl), since they have a similar molecular ion [M–H]− of 395 but different UV spectra and retention times. Compound X15 gave a [M–H]− peak at m/z 325. The UV spectrum was characteristic of 1,3,5,6-tetraoxygenated xanthone. A distinct shoulder at 365 nm revealed conjugation with a pyran ring. The MSn and UV spectra were in complete agreement with those of toxyloxanthone [25]. Compound X21 gave a [M–H]− peak at m/z 339, which results from methylation of toxyloxanthone giving paxanthone [23, 25, 40]. Compounds X26, X28, X30, and X32 gave deprotonated molecular ions [M–H]− at m/z 461, 463, 477, and 413, respectively. Their MS2 spectra were generated by the loss of a prenyl residue C4H8 (56 amu) and two prenyl residues (112 amu). So, compounds X26, X28, X30, and X32 were identified as banaxanthone D, garcinone E, banaxanthone E, and garcinone C, respectively. Several other peaks (X2, X3, X9, X24, X27, X29, X31, and X33) were categorized as xanthone derivatives by HPLC-DAD-MS/MS analysis but were not fully identified.
Among the twenty-five identified xanthones, seven (X6, X7, X12, X14, X15, X18, and X28) were upregulated in HR2 compared to HR1 cultures. Moreover, five xanthones (X8, X10, X20, X21, and X30) were synthesized only in HR2 cultures. Recent studies showed that Hypericum in vitro cultures have the potential to accumulate xanthones and their production can be manipulated by the hormonal supplementation [28], or/and by the culture type [33]. It is probable that phytohormones either facilitate or hamper the expression and activity of specific xanthone enzymes that influence xanthone accumulation in H. perforatum callus [33], cells [28], and root cultures [12]. Namely, Tocci et al. [12] suggested that root cultures grow continuously on nutrient media supplemented with auxins, but sometimes repetitive subcultures may induce loss of morphogenetic potential, resulting in poor or negligible secondary metabolite production. On the other hand, our results showed that H. perforatum HR cultures successfully grow on hormone-free media and represent a continuous source for high-level xanthone production.
Taken together, results in our study showed distinct phenolic profile between dark-grown (HR1) and photoperiod-exposed (HR2) cultures. Namely, phenolic compounds identified in HR2 cultures compared to HR1 could be distinguished in four groups: (i) compounds whose quantity increased (F1, F15, X6, X7, X12, X14, X15, X18, and X28), (ii) compounds whose quantity decreased (F2, F4, F6–F10, F13, X1, X4, X5, X17, X23, and X32), (iii) compounds that were not detectable (X11, X13, X16, X19, X22, X25, and X26), and (iv) compounds that were de novo synthesized (F3, F5, F11, F12, F14, X8, X10, X20, X21, and X30). Consequently, results from our experiments demonstrated that the exposure of HR2 cultures to photoperiod leads to alternations in their biosynthetic potentials.
Recent study showed that the phenolic biosynthesis and flavonoids formation are light-dependent processes [41]. Moreover, changes in light intensity are capable of inducing the production of flavonoids and total phenolics in plants [42]. Therefore, de novo biosynthesis and accumulation of phenolic acids, flavonols, and xanthones in HR2 cultures are not surprising since considerable evidence now shows that many of the enzymes in the phenylpropanoid/flavonoid pathway could be upregulated by light. In addition, Abbasi et al. [43] demonstrated light-stimulated accumulation of phenolic acids and phenylalanine ammonia lyase (PAL) activity in Echinacea purpurea HR cultures. Considering results from our study, we could hypothesize that shifting the dark-grown HR to photoperiod might induce a short-term “light-stress” response. In this view, the presence of light could induce a variety of responses along with metabolic changes that directly or indirectly trigger a “later” increase in xanthone accumulation. On the other hand, our results showed that photoperiod has an inhibitory effect on the accumulation of flavan-3-ols in HR2 cultures. Possible reasons for downregulation of flavan-3-ols could be due to the activation of their catabolism and/or reaction to unidentified products that exist in photoperiod-exposed cultures. Therefore, photoregulation of phenolic compounds biosynthesis in H. perforatum HR may offer additional advantages of quantitative and qualitative improvements of these medicinally important metabolites.
4. Conclusions
In conclusion, H. perforatum HR cultures provided a promising system for the production of various groups of phenolic compounds. Distinct phenolic profile between dark-grown and photoperiod-exposed HR cultures was shown as detailed for the first time. HR cultures grown under photoperiod can be proposed as a useful source for accumulation of phenolic acids and flavonols, while dark-adapted HR represent an alternative tool for flavan-3-ol production. More importantly, both HR cultures synthesized and stored significant quantities of xanthones. The use of the results reported here might contribute to further study on photoregulation and optimal control of secondary metabolite production in H. perforatum HR cultures.
Abbreviations
- DAD:
Diode array detection
- ESI-MS:
Electrospray ionisation-mass spectrometry
- HPLC:
High-performance liquid chromatography
- HR:
Hairy roots
- LC-MS:
Liquid chromatography-mass spectrometry
- [M–H]−:
Negative molecular ion
- MSn:
Collision fragment ions
- PCR:
Polymerase chain reaction.
References
- 1.Nahrstedt A, Butterweck V. Lessons learned from herbal medicinal products: the example of St. John’s wort. Journal of Natural Products. 2010;73(5):1015–1021. doi: 10.1021/np1000329. [DOI] [PubMed] [Google Scholar]
- 2.di Carlo G, Borrelli F, Ernst E, Izzo AA. St John’s wort: prozac from the plant kingdom. Trends in Pharmacological Sciences. 2001;22(6):292–297. doi: 10.1016/s0165-6147(00)01716-8. [DOI] [PubMed] [Google Scholar]
- 3.Giri A, Narasu ML. Transgenic hairy roots: recent trends and applications. Biotechnology Advances. 2000;18(1):1–22. doi: 10.1016/s0734-9750(99)00016-6. [DOI] [PubMed] [Google Scholar]
- 4.Kim YJ, Weathers PJ, Wyslouzil BE. Growth of Artemisia annua hairy roots in liquid- and gas-phase reactors. Biotechnology and Bioengineering. 2002;80(4):454–464. doi: 10.1002/bit.10389. [DOI] [PubMed] [Google Scholar]
- 5.Georgiev MI, Agostini E, Ludwig-Müller J, Xu J. Genetically transformed roots: from plant disease to biotechnological resource. Trends in Biotechnology. 2012;30(10):528–537. doi: 10.1016/j.tibtech.2012.07.001. [DOI] [PubMed] [Google Scholar]
- 6.Di Guardo A, Cellarova E, Koperdáková J, et al. Hairy root induction and plant regeneration in Hypericum perforatum L. Journal of Genetics and Breeding. 2003;57(3):269–278. [Google Scholar]
- 7.Vinterhalter B, Ninković S, Cingel A, Vinterhalter D. Shoot and root culture of Hypericum perforatum L. transformed with Agrobacterium rhizogenes A4M70GUS. Biologia Plantarum. 2006;50(4):767–770. [Google Scholar]
- 8.Franklin G, Oliveira M, Dias ACP. Production of transgenic Hypericum perforatum plants via particle bombardment-mediated transformation of novel organogenic cell suspension cultures. Plant Science. 2007;172(6):1193–1203. [Google Scholar]
- 9.Halliday KJ, Fankhauser C. Phytochrome-hormonal signalling networks. New Phytologist. 2003;157(3):449–463. doi: 10.1046/j.1469-8137.2003.00689.x. [DOI] [PubMed] [Google Scholar]
- 10.Hemm MR, Rider SD, Ogas J, Murry DJ, Chapple C. Light induces phenylpropanoid metabolism in Arabidopsis roots. The Plant Journal. 2004;38(5):765–778. doi: 10.1111/j.1365-313X.2004.02089.x. [DOI] [PubMed] [Google Scholar]
- 11.Cui X-H, Chakrabarty D, Lee E-J, Paek K-Y. Production of adventitious roots and secondary metabolites by Hypericum perforatum L. in a bioreactor. Bioresource Technology. 2010;101(12):4708–4716. doi: 10.1016/j.biortech.2010.01.115. [DOI] [PubMed] [Google Scholar]
- 12.Tocci N, ’Auria FDD, Simonetti G, Panella S, Palamara AT, Pasqua G. A three-step culture system to increase the xanthone production and antifungal activity of Hypericum perforatum subsp. angustifoliumin vitro roots. Plant Physiology and Biochemistry. 2012;57:54–58. doi: 10.1016/j.plaphy.2012.04.014. [DOI] [PubMed] [Google Scholar]
- 13.Gadzovska S, Maury S, Ounnar S, et al. Identification and quantification of hypericin and pseudohypericin in different Hypericum perforatum L. in vitro cultures. Plant Physiology and Biochemistry. 2005;43(6):591–601. doi: 10.1016/j.plaphy.2005.05.005. [DOI] [PubMed] [Google Scholar]
- 14.Slightom JL, Durand-Tardif M, Jouanin L, Tepfer D. Nucleotide sequence analysis of TL-DNA of Agrobacterium rhizogenes agropine type plasmid. Identification of open reading frames. The Journal of Biological Chemistry. 1986;261(1):108–121. [PubMed] [Google Scholar]
- 15.Tusevski O, Stanoeva JP, Stefova M, et al. Hairy roots of Hypericum perforatum L.: a promising system for xanthone production. Central European Journal of Biology. 2013;8(10):1010–1022. [Google Scholar]
- 16.Tepfer D. Transformation of several species of higher plants by Agrobacterium rhizogenes: sexual transmission of the transformed genotype and phenotype. Cell. 1984;37(3):959–967. doi: 10.1016/0092-8674(84)90430-6. [DOI] [PubMed] [Google Scholar]
- 17.Zhang Y, Shi P, Qu H, Cheng Y. Characterization of phenolic compounds in Erigeron breviscapus by liquid chromatography coupled to electrospray ionization mass spectrometry. Rapid Communications in Mass Spectrometry. 2007;21(18):2971–2984. doi: 10.1002/rcm.3166. [DOI] [PubMed] [Google Scholar]
- 18.Papetti A, Daglia M, Aceti C, et al. Hydroxycinnamic acid derivatives occurring in Cichorium endivia vegetables. Journal of Pharmaceutical and Biomedical Analysis. 2008;48(2):472–476. doi: 10.1016/j.jpba.2008.04.014. [DOI] [PubMed] [Google Scholar]
- 19.Ferreres F, Andrade PB, Valentão P, Gil-Izquierdo A. Further knowledge on barley (Hordeum vulgare L.) leaves O-glycosyl-C-glycosyl flavones by liquid chromatography-UV diode-array detection-electrospray ionisation mass spectrometry. Journal of Chromatography A. 2008;1182(1):56–64. doi: 10.1016/j.chroma.2007.12.070. [DOI] [PubMed] [Google Scholar]
- 20.Conceição LFR, Ferreres F, Tavares RM, Dias ACP. Induction of phenolic compounds in Hypericum perforatum L. cells by Colletotrichum gloeosporioides elicitation. Phytochemistry. 2006;67(2):149–155. doi: 10.1016/j.phytochem.2005.10.017. [DOI] [PubMed] [Google Scholar]
- 21.Cuyckens F, Rozenberg R, De Hoffmann E, Claeys M. Structure characterization of flavonoid O-diglycosides by positive and negative nano-electrospray ionization ion trap mass spectrometry. Journal of Mass Spectrometry. 2001;36(11):1203–1210. doi: 10.1002/jms.224. [DOI] [PubMed] [Google Scholar]
- 22.Miketova P, Schram KH, Whitney JL, et al. Mass spectrometry of selected components of biological interest in green tea extracts. Journal of Natural Products. 1998;61(4):461–467. doi: 10.1021/np9703959. [DOI] [PubMed] [Google Scholar]
- 23.Ishiguro K, Nakajima M, Fukumoto H, Isoi K. Co-occurrence of prenylated xanthones and their cyclization products in cell suspension cultures of Hypericum patulum . Phytochemistry. 1995;38(4):867–869. [Google Scholar]
- 24.Franklin G, Conceição LFR, Kombrink E, Dias ACP. Xanthone biosynthesis in Hypericum perforatum cells provides antioxidant and antimicrobial protection upon biotic stress. Phytochemistry. 2009;70(1):60–68. doi: 10.1016/j.phytochem.2008.10.016. [DOI] [PubMed] [Google Scholar]
- 25.Dias ACP, Seabra RM, Andrade PB, Ferreres F, Ferreira MF. Xanthone production in calli and suspended cells of Hypericum perforatum . Journal of Plant Physiology. 2001;158(7):821–827. [Google Scholar]
- 26.Tanaka N, Takaishi Y. Xanthones from Hypericum chinense . Phytochemistry. 2006;67(19):2146–2151. doi: 10.1016/j.phytochem.2006.05.043. [DOI] [PubMed] [Google Scholar]
- 27.Niggeweg R, Michael AJ, Martin C. Engineering plants with increased levels of the antioxidant chlorogenic acid. Nature Biotechnology. 2004;22(6):746–754. doi: 10.1038/nbt966. [DOI] [PubMed] [Google Scholar]
- 28.Dias ACP, Seabra RM, Andrade PB, Fernandes-Ferreira M. The development and evaluation of an HPLC-DAD method for the analysis of the phenolic fractions from in vivo and in vitro biomass of Hypericum species. Journal of Liquid Chromatography and Related Technologies. 1999;22(2):215–227. [Google Scholar]
- 29.Bertoli A, Giovannini A, Ruffoni B, et al. Bioactive constituent production in St. John’s Wort in vitro hairy roots. Regenerated plant lines. Journal of Agricultural and Food Chemistry. 2008;56(13):5078–5082. doi: 10.1021/jf0729107. [DOI] [PubMed] [Google Scholar]
- 30.Silva BA, Ferreres F, Malva JO, Dias ACP. Phytochemical and antioxidant characterization of Hypericum perforatum alcoholic extracts. Food Chemistry. 2005;90(1-2):157–167. [Google Scholar]
- 31.Gadzovska S, Maury S, Delaunay A, Spasenoski M, Joseph C, Hagège D. Jasmonic acid elicitation of Hypericum perforatum L. cell suspensions and effects on the production of phenylpropanoids and naphtodianthrones. Plant Cell, Tissue and Organ Culture. 2007;89(1):1–13. [Google Scholar]
- 32.Gadzovska S, Maury S, Delaunay A, et al. The influence of salicylic acid elicitation of shoots, callus, and cell suspension cultures on production of naphtodianthrones and phenylpropanoids in Hypericum perforatum L. Plant Cell, Tissue AndOrgan Culture. 2013;113(1):25–39. [Google Scholar]
- 33.Pasqua G, Avato P, Monacelli B, Santamaria AR, Argentieri MP. Metabolites in cell suspension cultures, calli, and in vitro regenerated organs of Hypericum perforatum cv. Topas. Plant Science. 2003;165(5):977–982. [Google Scholar]
- 34.Rainha N, Koci K, Coelho AV, Lima E, Baptista J, Fernandes-Ferreira M. HPLC-UV-ESI-MS analysis of phenolic compounds and antioxidant properties of Hypericum undulatum shoot cultures and wild-growing plants. Phytochemistry. 2013;86:83–91. doi: 10.1016/j.phytochem.2012.10.006. [DOI] [PubMed] [Google Scholar]
- 35.Tusevski O, Gadzovska Simic S. Phenolic acids and flavonoids in Hypericum perforatum L. hairy roots. International Journal of Pharma and Bio Sciences. 2013;4(3):737–748. [Google Scholar]
- 36.Yazaki K, Okuda T. Procyanidins in callus and multiple shoot cultures of Hypericum erectum . Planta Medica. 1990;56(5):490–491. doi: 10.1055/s-2006-961020. [DOI] [PubMed] [Google Scholar]
- 37.Chaudhuri RK, Ghosal S. Xanthones of Canscora decussata schult. Phytochemistry. 1971;10(10):2425–2432. [Google Scholar]
- 38.Nielsen H, Arends P. Xanthone constituents of Hypericum androsaemum . Journal Of Natural Products. 1979;42(3):301–304. [Google Scholar]
- 39.Ishiguro K, Fukumoto H, Nakajima M, Isoi K. Xanthones in cell suspension cultures of Hypericum paturum . Phytochemistry. 1993;33(4):839–840. [Google Scholar]
- 40.Dias ACP, Seabra RM, Andrade PB, Ferreres F, Fernandes-Ferreira M. Xanthone biosynthesis and accumulation in calli and suspended cells of Hypericum androsaemum . Plant Science. 2000;150(1):93–101. [Google Scholar]
- 41.Xie BD, Wang HT. Effects of light spectrum and photoperiod on contents of flavonoid and terpene in leaves of Ginkgo biloba L. Journal of Nanjing Forestry University. 2006;30:51–54. [Google Scholar]
- 42.Graham TL. Flavonoid and flavonol glycoside metabolism in Arabidopsis. Plant Physiology and Biochemistry. 1998;36(1-2):135–144. [Google Scholar]
- 43.Abbasi BH, Tian C-L, Murch SJ, Saxena PK, Liu C-Z. Light-enhanced caffeic acid derivatives biosynthesis in hairy root cultures of Echinacea purpurea . Plant Cell Reports. 2007;26(8):1367–1372. doi: 10.1007/s00299-007-0344-5. [DOI] [PubMed] [Google Scholar]


