Abstract
Objective:
To explore potential causes of male infertility by determining the composition and structure of commensal bacterial communities in seminal fluids.
Design:
Microscopy of gram stained semen samples and classification of 16S rRNA gene sequences to determine the species composition of semen bacterial communities.
Setting(s):
Clinical andrology laboratory and academic research laboratories.
Patient(s):
19 sperm donors and 58 infertility patients.
Intervention(s):
None.
Main Outcome Measure(s):
Classification of 16S rRNA gene sequences, clustering of seminal microbial communities, and multiple statistical tests.
Result(s):
High numbers of diverse kinds of bacteria were present in most samples of both sperm donors and infertility patients. The bacterial communities varied widely between subjects, but they could be clustered into six groups based on similarities in composition and the rank abundances of taxa. Overall there were no significant differences between sperm donors and infertility patients. However, multiple statistical tests showed a significant negative association between sperm quality and the presence of Anaerococcus. The results also indicated that many of the bacterial taxa identified in semen also occur in the vaginal communities of some women, especially those with bacterial vaginosis, which suggests heterosexual sex partners may share bacteria.
Conclusion(s):
Diverse kinds of bacteria were present in the human semen, there were no significant differences between sperm donors and infertility patients, The presence of Anaerococcus might be a biomarker for low sperm quality.
Keywords: semen, microbiome, bacterial communities, fertility, reproduction
During the process of ejaculation in healthy men sperm pass through the ejaculatory ducts and mix with fluids from seminal vesicles, the prostate, and the bulbourethral glands to form semen that is transported through the entire male reproductive tract including the urethra (1, 2). Semen quality and quantity are both measures of fertility (3, 4) and can be classified as asthenozoospermia, oligoasthenozoospermia, severe oligoasthenozoospermia, and azoospermia (5).
Semen has been found to serve as a medium for the transmission of bacteria and viruses between men and women (6, 7) contributing to the development of sexually transmitted diseases (STDs) (8, 9). Moreover, certain bacteria, fungi, viruses and parasites are known to interfere with reproductive functions in both sexes and infections of the genitourinary tract account for about 15% of male infertility cases. This is generally accepted as one of the potentially correctable causes of male infertility (10). Infections and consequent inflammation in the male reproductive tract may compromise spermatogenesis and sperm cell function (11-13). The relationship between the presence of pathogenic microorganisms in the reproductive tract and infertility is widely documented (14-16). Several kinds of microorganisms found in the male urogenital tract are associated with sperm abnormalities, especially aberrant motility, deficient mitochondrial function, and loss of DNA integrity (17, 18). These microorganisms include Escherichia coli, Enterococcus faecalis, Ureaplasma urealyticum, Neisseria gonorrhoeae, Chlamydia trachomatis, Mycoplasma hominis, Candida albicans, and Trichomonas vaginalis. Most of these microorganisms are also associated with sexually transmitted infections (19, 20). Therefore, it is important to understand the bacterial species composition of seminal fluids to better understand the etiology and pathogenesis of urogenital tract infections and associations between urogenital infections and infertility.
Currently, little is known about the bacterial communities found in the male reproductive tract or the significance of bacteriospermia in asymptomatic men. Previous studies of seminal bacteria have largely focused on detecting and identifying overt pathogens, and heavily relied on cultivation-dependent methods that may have resulted in an incomplete census of the organisms present. The bacteria reportedly in seminal fluids including: Peptoniphilis, Anaerococcus, Finegoldia, Peptostreptococcus spp. Staphylococcus, Streptococcus, Corynebacteium, Enterococcus, Lactobacillus, Gardnerella, Prevotella and Escherichia coli (21-24). There are conflicting reports on the effect of indigenous bacteria on semen quality and their pathophysiologic role in male infertility has not been established (13, 25, 26). Previously studies have been done to test whether urinary tract pathogens such as Escherichia coli, Enterococcus faecalis and Ureaplasma urealyticum influence spermatogenesis and sperm function (27, 28). The results of these studies suggest that the simple presence of bacteria in semen samples may compromise sperm quality (2, 3, 29). However, the majority of the data on the interactions between spermatozoa and bacteria are derived from in vitro studies (30), under conditions that may not accurately mimic in vivo conditions, For example, the bacterial population densities used for in vitro experiments have been much higher than ever recovered from ejaculate specimens (31). In some other studies, these putative pathogenic bacteria were not only found in the reproductive tracts of infertile patients, but also in those of healthy men (25, 26). It remains unclear if the microorganisms found in semen necessarily signify infection and significantly contribute to male infertility.
Methods that require culturing of bacteria have traditionally been used to characterize bacteria of seminal fluid (21, 23, 24). While these have provided important insights to the microbiology of semen they are limited because many species of bacteria are recalcitrant to cultivation. To overcome this problem molecular methods that do not require the cultivation of organisms have been devised and used to investigate microbial diversity. The 16S rRNA gene is present in all bacteria and has regions of sequence conservation that can be targeted with broad range PCR primers. In addition, there are regions of sequence variation and these can be used to classify bacteria and infer phylogenetic relationships (32). The use of 16S rRNA gene sequence data is studies of bacterial diversity have been used to describe the species composition of various communities, including those in the human gastrointestinal tract, skin, oral and urogenital tracts (33-38). In this study, we used high-throughput DNA sequencing and newly developed bioinformatic tools to more fully characterize the bacterial species present in seminal fluids from both healthy sperm donors and infertility patients. As part of this we sought to determine if there were substantial differences in the composition and structure of bacterial communities in the seminal fluids of these two groups and to identify specific taxa that may be associated with low sperm quality.
MATERIALS AND METHODS
Clinical study design and subjects
In a cross-sectional clinical study conducted at Shanghai Jiao Tong University School of Medicine (Shanghai, China), seminal fluids were collected from 77 subjects who were between 18 and 40 years old. The study subjects were from four groups. Nineteen were from healthy sperm donors (Group 1). The other three groups included infertility patients with asthenozoospermia (Group 2), oligoasthenozoospermia (Group 3), and severe oligoasthenozoospermia and azoospermia (Group 4). These four groups were defined according to guidelines published by the World Health Organization (WHO) (39). In the normal control group the motile sperm demonstrated more than 50% progression, while the sperm counts were higher than 20×106 sperm per ml. In Group 2 the motility of sperm was abnormally low with less than 50% progression, while the sperm counts were no less than 20×106 sperm per ml. In Group 3 the sperm had less than 50% progression and the sperm counts were between 2×106 and 20×106 sperm per ml. In Group 4 the sperm counts were less than 2×106 or no any measurable level of sperm (Table 1). The demographics and other characteristics of the subjects are showed in Table S1. The distribution of subjects based on sperm concentration and sperm motility is shown at Figure S1.
Table 1.
Demographic data on study participants.
| Subjectsa | ||||
|---|---|---|---|---|
|
|
||||
| Group 1 | Group 2 | Group 3 | Group 4 | |
|
|
||||
| Number of subjects | 19 | 10 | 23 | 25 |
| Average age (y) | 25.4 ± 3.5 | 30.2 ± 6.1 | 31.1 ± 5.5 | 31.7 ± 4.0 |
| Semen volumes (ml) | 2.9 ± 0.9 | 2.6 ± 0.7 | 2.9 ± 1.6 | 2.5 ± 1.4 |
| Semen pH | 6.9 ± 1.7 | 7.1 ± 0.1 | 7.2 ± 0.1 | 7.2 ± 0.2 |
| Sperm counts | 1.63E8 ± 7.14E7 | 9.04E7 ± 8.17E7 | 8.10E6 ± 4.25E6 | NMb |
| Sperm motility (%) | 66.3 ± 7.5 | 37.1 ± 8.8 | 31.7 ± 12.8 | NAc |
Group 1 includes normal controls; Group 2 asthenospermia; Group 3 oligoasthenospermia; and Group 4 individuals with either azoospermia and severe oligozoospermia. The values shown are mean ± SD. .
NM indicates that individuals had sperm counts that were either very low or not measureable.
NA; not applicable.
Subjects were eligible for enrollment if they were willing to sign an informed consent and participate in this study; no genetic disease in subjects and their families; no systemic disease; no long-term exposure to radioactive ray and noxious substance; no drug taking in the past 2 months; had no history of STDs; had no systemic corticosteroids use. Subjects were not eligible if they were participating in another clinical study; had fever in the previous 2 months; had urinary tract infection or inflammation within the previous 2 months; used OTC or prescription antibiotics, immunosuppressive drugs, systemic corticosteroids, or cancer chemotherapy in the past 2 months; with seminal infections; had systemic diseases; were previous cryptorchidism or orchitis. The study protocol and informed consent document were reviewed and approved by the Institutional Review Board (IRB) of the Shanghai Jiao Tong University School of Medicine. Documented informed consent was obtained from all subjects prior to participation in this study.
Specimen collection
Semen specimens were collected by masturbation that was preceded by 3 ~ 7 days of abstinence. Prior to sample collection instructions were given to the subjects on procedures to be followed to prevent sample contamination. The subject’s hands were washed with soap 2 ~ 3 times. The penis, especially the glans and the coronal sulcus, were first cleaned with warm soapy water then swabbed with 75% alcohol 2 ~ 3 times. The semen was ejaculated directly into a sterile glass receptacle, avoiding contact with the interior of the sterile wall of the container. The freshly collected seminal fluid was used for routine semen clinical tests, Gram staining and microscopy. The remainder of semen samples were transferred to sterile microcentrifuge tubes and stored in −80 °C within 2 hours of collection.
Characterization of semen
Subsamples of each seminal fluid sample were used for a computer-assisted sperm motility analysis done using a semen auto-analyzer (Hamilton Thorne, Beverly, MA, USA). The analysis included measures of pH, semen liquefaction time, semen volume, sperm count, and percentage of motility sperm.
Thirty seminal fluid samples were randomly chosen to be examined by microscopy following Gram staining. A 100μl aliquot of each semen sample was concentrated by centrifugation at 14,000 rpm for 20 minutes, then washed with sterile PBS 2 ~ 3 times. After preparing a cell smear the samples were Gram stained with a Rapid Gram Stain kit according to the manufacturer’s protocol (Enkang, Shanghai, China). The slides were examined by microscope using 1000× magnification and oil immersion. Several fields of each slide were observed and some were photographed.
Genomic DNA extraction and purification
Genomic DNA was extracted from seminal fluids using a validated enzymatic lysis and bead-beating protocol, followed by purification using QIAamp DNA Mini Kit (Qiagen, Valencia, USA) that has been used in our previous human microbiome studies (36, 40). Briefly, 200 μl aliquots of semen samples were thawed on ice, and 750 μl of TE50 (10 mM Tris-HCl + 50 mM EDTA, pH 8.0) was added. 500 μl of each sample was transferred to a clean, sterile bead-beating tube (MP Biomedicals, Solon, USA) and kept on ice. A lytic enzyme cocktail was prepared at the time of extraction and added to each sample as follows: 50 μl of lysozyme (10 mg/ml), 6 μl of mutanolysin (25, 000 U/ml; Sigma-Aldrich) and 3 μl of lysostaphin (4,000 U/ml in sodium acetate; Sigma-Aldrich) and 41 μl of TE50 buffer (10 mM Tris-HCL, 50 mM EDTA, pH 8.0) for a final volume of 100 μl per sample. Samples were digested by incubation at 37°C for 60 min in a dry heat block. To each digested sample, 750 mg of sterile 0.1 mm diameter zirconia/silica beads (BioSpec Products, Bartlesville, OK) were added. Bead-beating was performed for 1 min at 36 oscillations/sec (2, 100 rpm) using a Mini-Beadbeater-96 (BioSpec Products, Bartlesville, OK). Following cell disruption, the tubes were centrifuged at 1200 rpm for 1 min. Aliquots of crude lysate from each sample were transferred to new, sterile microcentrifuge tubes and 50 μl of Proteinase K (20 mg/ml (> 600 mAU/ml)) and 500 μl of Qiagen buffer AL were added. Samples were mixed by pulse-vortexing for 15 sec and then incubated at 56°C for 30 min. After this step, 50 μl of 3 M sodium acetate (pH 5.5) was added, followed by 500 μl 100% ethanol at each sample. Vortexing was repeated for an additional 15 sec b Briefly, 200 μl aliquots of semen samples were thawed on ice, and 750 μl of TE50 (10 mM Tris-HCl + 50 mM EDTA, pH 8.0) was added. 500 μl of each sample was transferred to a clean, sterile bead-beating tube (MP Biomedicals, Solon, USA) and kept on ice. A lytic enzyme cocktail was prepared at the time of extraction and added to each sample as follows: 50 μl of lysozyme (10 mg/ml), 6 μl of mutanolysin (25, 000 U/ml; Sigma-Aldrich) and 3 μl of lysostaphin (4,000 U/ml in sodium acetate; Sigma-Aldrich) and 41 μl of TE50 buffer (10 mM Tris-HCL, 50 mM EDTA, pH 8.0) for a final volume of 100 μl per sample. Samples were digested by incubation at 37°C for 60 min in a dry heat block. To each digested sample, 750 mg of sterile 0.1 mm diameter zirconia/silica beads (BioSpec Products, Bartlesville, OK) were added. Bead-beating was performed for 1 min at 36 oscillations/sec (2, 100 rpm) using a Mini-Beadbeater-96 (BioSpec Products, Bartlesville, OK). Following cell disruption, the tubes were centrifuged at 1200 rpm for 1 min. Aliquots of crude lysate from each sample were transferred to new, sterile microcentrifuge tubes and 50 μl of Proteinase K (20 mg/ml (> 600 mAU/ml)) and 500 μl of Qiagen buffer AL were added. Samples were mixed by pulse-vortexing for 15 sec and then incubated at 56°C for 30 min. After this step, 50 μl of 3 M sodium acetate (pH 5.5) was added, followed by 500 μl 100% ethanol at each sample. Vortexing was repeated for an additional 15 sec before briefly centrifuging. From this point onward, purification of genomic DNA was done with QIAamp DNA Mini Kits (Qiagen) following manufacturer’s instructions. efore briefly centrifuging. From this point onward, purification of genomic DNA was done with QIAamp DNA Mini Kits (Qiagen) following manufacturer’s instructions.
Pyrosequencing V1-V2 regions of 16S rRNA gene
To characterize the composition and structure of bacterial communities indigenous to semen we sequenced the V1-V2 regions of 16S rRNA genes amplified by PCR from each sample. The amplicons were obtained by PCR using primers used flanked variable regions 1 and 2 of bacterial 16S rRNA genes (Escherichia coli positions 27F ~ 338R). The sequences of the primers used were
454_27F 5’-GCCTTGCCAGCCCGCTCAGTCAGAGTTTGATCCTGGCTCAG and
454_338R 5’-GCCTCCCTCGCGCCATCAGTGNNNNNNNNCATGCTGCCTCCCGTAGGAGT
where the underlined sequences are 454 Life Sciences® adapters B and A in 27F and 338R, respectively, and the bold font denotes the universal 16S rRNA primers 27F and 338R. The 338R primer included a unique sequence tag to barcode each of the samples denoted by the 8 N’s. Each PCR reaction (sample) contained a unique reverse primer, which allowed us to sequence the amplicons from all samples simultaneously, and afterwards assign each sequence to the sample from which they were obtained. Each PCR reaction contained 34.4 μl of PCR-grade water, 5.0 μl of 10× buffer (Applied Biosystems, Foster City, CA), 6.0 μl of 25 mM MgCl2 (Applied Biosystems), 0.4 μl of 25 mM dNTP (Amersham Bioscience, Piscataway, NJ), 0.5 μl of 20 μM forward primer 454_27f, and 0.5 μl of 20 μM reverse primer 454_338r, 0.2 μl of 5 U/μl Taq DNA polymerase (Applied Biosystems), and 1.0 μl of DNA template in a total volume of 50 μl. Amplification of fragments was done using an initial denaturation step of 94°C for 4 min, followed by 30 cycles of denaturation at 94°C for 1 min, annealing at 55°C for 1 min, and extension at 72°C for 2 min. A final extension step of 10 min at 72°C was done. The concentrations of amplicons were estimated using a GelDoc quantification system (BioRad, Hercules, CA) and roughly equal amounts (~100 ng) of all amplicons were mixed in a single tube. Amplification primers and reaction buffer were removed by processing the amplicon mixture with the AMPure Kit (Agencourt, Beverly, MA). Emulsion PCRs were done as described in Margulies, et al..(41) and sequencing was done with a Roche 454 GS-FLX (Roche-454 Life Sciences, Branford, CT).
DNA sequence data analysis and taxonomic classification
Raw unclipped DNA sequence reads were cleaned, assigned and filtered in the following manner. Raw SFF files were read directly into the R statistical programming language using the R package rSFFreader (Settles et al., https://github.com/msettles/rSFFreader, unpublished). Using full length sequence reads (unclipped) Cross Match (version 1.080806, parameters: min matches=15, min score=14) from the phred/phrap/consed application suite were used to identify Roche 454 adapter sequences, primer barcodes, and amplicon primer sequences. Cross Match alignment information was then read into R and processed to identify alignment quality, directionality, barcode assignment, and sequence quality clip points. Base quality clipping was then done using the application Lucy (version 1.20p, parameters: max average error=0.002, max error at ends=0.002), and the clipped reads were aligned to the SILVA bacterial sequence database using Mothur (version 1.12.1). Alignment end points were identified and used in subsequent filtering. Sequence reads were then filtered and only those that met the following criteria were analyzed further: (a) sequences were at least 100 bp in length; (b) max hamming distance of barcode = 1; (c) maximum number of matching error to forward primer sequences = 2; (d) had < 2 ambiguous bases; (e) had < 7 bp homopolymer run in sequence; (f) alignment to the SILVA bacterial database was within 75 bp of the expected alignment start position as identified by the trimmed mean of all read alignment (trim=10%); and (g) read alignment started within the first 5 bp and extended through read to within the final 5 bp. Each partial 16S rRNA gene sequence was classified using the RDP Naïve Bayesian Classifier (42). Reads were assigned to the first RDP level with a bootstrap score ?50. The proportion of a specific taxon in a community was calculated by dividing the number of DNA sequence reads assigned to that taxon by the total number of DNA sequence reads from that sample.
Statistical methods
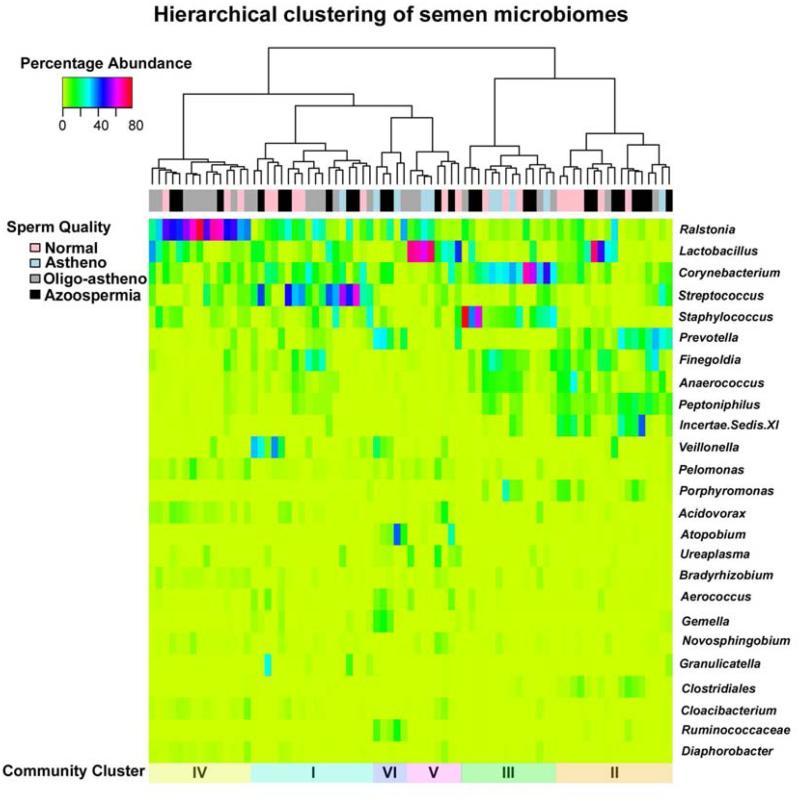
The clustering of communities based on community composition and abundance shown in Figure 2. was done using complete linkage hierarchical clustering using the R package (2). The figure was generated using a modified version of the heatmap routine in the R package. Hierarchical clustering arranges items (in this case semen bacterial communities) in a hierarchy with a tree-like structure that reflects the degree of dissimilarity between them in terms of relative bacterial species abundances. In this study the Bray-Curtis dissimilarities between communities were calculated and clustering was done using complete linkage in which the maximum distance between two clusters is computed (43). The tree-like structure that graphically depicts the resulting hierarchy is called a dendrogram.
Figure 2.
Hierarchical clustering of semen microbial communities in normal and infertile men and a heat map showing the proportions of various bacterial genera in each community. The sperm quality of each sample is indicated by the color-coded bar immediately below the dendrogram. Analysis of similarities in community composition and structure resulted in six clusters as indicated in the color bar below the heat map.
Similarities in the composition of semen bacterial communities were assessed by clustering using Ward’s linkage and Hellinger distance. Hellinger distance has been recommended for clustering of species abundance data, and it offers a compromise between linearity (reflected in abundant species) and resolution (reflected in rare species) that is better than chi-square metric or chi-square distance (44). We applied the pseudo F statistic developed by Calinski and Harabasz (45) to estimate the number of clusters. Milligan and Cooper (46) and Cooper and Milligan (47) compared thirty methods for estimating population clusters in simulation studies and the pseudo F statistic ranked the best in recovering true clusters from simulated data.
Given the optimum number of clusters identified in the previous step (six) and the four groups reflecting the fertility of the sampled individuals (see Clinical study design and subjects), we constructed a 6x4 contingency table (Table 2). This contingency table included groups as columns and clusters as rows. Counts within each cell represented the number of individuals that were sampled within a certain group and that had communities that belonged to a certain cluster. For example, the first cell included five sampled individuals from group 1 (healthy donors) with similar communities that belonged to cluster I. We used the approach described in Zhou et al. (34) to determine whether groups differed in their community composition. We compared a null model that assumed all groups had the same distribution of sampled individuals with similar clusters to one that assumed each group was different. The p-value of the resulting test was determined using the bootstrap to account for the small sample sizes observed within each group (34).
Table 2.
Numbers of subjects in various groups of semen microbial community types.
| Groups | ||||||
|---|---|---|---|---|---|---|
|
|
||||||
| Cluster | Total number of subjects |
Percent of subjects | Normal | Asthenozoospermia | Oligoasthenospermia | Oligospermia and azoospermia |
| I | 18 | 23.4 | 5 | 1 | 6 | 6 |
| II | 17 | 22.1 | 6 | 1 | 2 | 8 |
| III | 14 | 18.2 | 3 | 4 | 3 | 4 |
| IV | 15 | 19.5 | 3 | 0 | 9 | 3 |
| V | 8 | 10.4 | 2 | 2 | 2 | 2 |
| VI | 5 | 6.5 | 0 | 2 | 1 | 2 |
|
| ||||||
| Total number of subjects |
77 | 100.1 | 19 | 10 | 23 | 25 |
We used a proportional odds model, with a logit link function, to evaluate the association between sperm quality and specific taxa. The four groups identified above were ordered based on the presumed level and severity of decline in sperm quality. Accordingly, the categories normal, asthenozoospermia, oligoasthenozoospermia, and azoospermia were assigned numbers 0, 1, 2, 4, respectively, to differentiate their extent of worsening from the “normal” category. The resulting model is specified as:
| (1) |
Where Y is the response variable taking values 0, 1, 2, or 4; x is a continuous explanatory variable representing the relative abundance of a taxon. The R package “MASS” and its function “polr” were used to estimate the model parameters. A small p-value associated with the effect parameter “b” indicates potential association. This analysis was applied for the 50 most abundant taxa. P-values were adjusted to control for multiple testing. Two types of adjustments, controlling for family wise error rate and for false discovery rate (FDR) were applied.
We controlled family wise error rate using Wen and Lu’s approach (48). Wen and Lu use the number of principle components, resulting from principal components analysis (PCA), that explain most of the variation in the data to estimate the effective number of independent hypothesis tests to control for. This estimate iwas used in a Bonferroni correction to adjust the resulting p-values. In our case, PCA of the abundance data revealed 21 principal components that cumulatively explained 95% of the total variance. Twenty one was considered as the effective number of independent tests. Then the critical p-value was adjusted to 0.0024 (=0.05/21). Any individual p-values less than this threshold resulted in rejecting the null hypothesis of no association between the taxon and sperm quality.
RESULTS
Microscopy of Gram stained semen samples
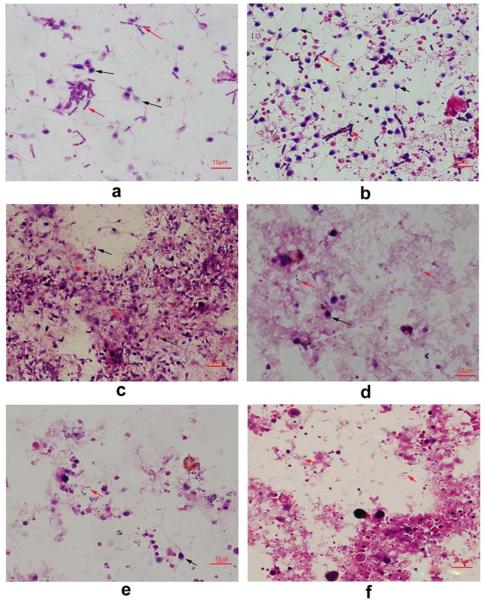
To obtain direct evidence for the presence of bacteria in semen samples we Gram stained randomly selected samples from each group of subjects and examined them by microscopy. The results showed that most of semen samples (28/30) had high numbers of bacteria present (Figure 1, Figure S2, Figure S3). The number of bacteria was higher than the number of sperm in most sperm donor samples. This indicates that the number of bacteria were greater than 106~107 bacterial cells per ml. Additionally, the quantity and diversity of bacteria (based on cellular morphology) varied between subjects. To our knowledge, this is the first demonstration that the number of the bacteria in semen exceeds the number of sperm and that semen bacterial communities contain diverse taxa that differ between men. This result was consistent with other studies showing that male reproductive tracts are not sterile and that the male reproductive tract harbors a large number of bacteria (25, 28).
Figure 1.
Gram stained semen samples from: semen donors NS8 and NS11 (panels a and b, respectively); (c) subject LM6 with asthenozoospermia; (d) subject OZ3 with oligoasthenozoospermia; (e) subject OZ13 with severe oligoasthenozoospermia; and (f) subject AS8 with azoospermia. The black arrows point to sperm and red arrows point to bacteria.
The bacterial community composition and structure in semen
The bacterial diversity in semen samples of healthy and infertile men were characterized by pyrosequencing the V1 ~ V2 region of 16S rRNA genes that had been amplified from total genomic DNA isolated from each sample. In total, 165,957 sequences were obtained with an average of 2155 reads from each of the 77 seminal fluid samples analyzed (Table S2). Taxonomic classification of bacteria in the samples was performed using the Ribosomal Database Project (RDP) Naïve Bayesian Classifier (42). The numerically important bacterial phylotypes that constituted more than 1% of semen communities are listed in Table S2. The results show that semen harbors diverse bacterial taxa, some of which were not previously known to occur in semen.
Communities that were similar in terms of species composition and structure were identified by cluster analysis (Figure 2). Using the pseudo F statistic (Calinski-Harabasz index) (4) we identified six groupings of semen community types present in the subjects sampled. Communities of cluster I were most common and present in 23.4% of the subjects sampled. Communities in this cluster were characterized by high proportions of Streptococcus, Corynebacterium, Finegoldia and Veillonella, in addition to various others including Lactobacillus, Prevotella, Staphylococcus, Anaerococcus, and Peptoniphilus. The communities of Cluster II were present in 22.1% of the subjects sampled and contained diverse species of anaerobes including Prevotella, Peptoniphilus, Lactobacillus, Incertae Sedis, Porphyromonas and Clostridiales. Cluster III communities were present in 18.2% of the subjects and these were typified by high proportions of Corynebacterium, Staphylococcus, Finegoldia and Anaerococcus while those of Cluster IV (19.5% of subjects), were predominated by Ralstonia but also had other appreciable proportions of other taxa including Lactobacillus, Corynebacterium, Streptococcus, Staphylococcus, Pelomonas and Acidovorax. Lactobacillus was the predominant species in Cluster V, which occurred in 10.4% of subjects. Cluster VI contained diverse bacterial species including Atopobium, Veillonella, Prevotella, Aerococcus and Gemella and was found in 6.49% of the semen samples analyzed (Table 2).
Overall, the most abundant bacteria in the semen communities were Ralstonia, Lactobacillus, Corynebacterium, Streptococcus, Staphylococcus, Prevotella, Finegoldia, Anaerococcus, Peptoniphilus, Incertae Sedis XI (family), Veillonella, Pelomonas, Porphyromonas, Acidovorax, Atopobium, Ureaplasma, Bradyrhizobium, Aerococcus, Gemella, Granulicatella, Clostridiales and Cloacibacterium. It should be noted that many of these bacterial species are opportunistic pathogens such as Ralstonia, Prevotella, Finegoldia, Anaerococcus, Peptoniphilus, Gemella, Granulicatella, Clostridiales, Pelomonas, Porphyromonas, Acidovorax and Cloacibacterium (25, 49-52). Moreover, many are also strictly anaerobic bacteria and closely related to those found in the human vagina, including members of the Clostridiales that have been associated with bacterial vaginosis (7). Other bacterial species in semen have been previously shown to be present in the urethra and the skin of the penis (20, 53).
Differences between the semen from sperm donors and infertile men
Table 2 shows the number of subjects from each infertility class present in each community cluster. Using the approach presented in Zhou et al. (34), we found no significant differences in the distribution of men of the four clinically defined groups among the various community clusters (p-value > 0.477), in other words, membership in a semen microbial community cluster was not predictive of a subject’s fertility. This suggests that infertile men do not have altered or unusual semen bacterial communities as compared to healthy semen donors.
To further identify the association between sperm quality and specific taxa, we built a proportional odds model and conducted multiple testing analyses. The results showed that among the 50 most abundant taxa present in these communities only Anaerococcus had a negative association (p = 0.0012) with sperm quality. This indicates that abundant Anaerococcus in semen microbial communities may be associated with male infertility.
DISCUSSION
In this study next generation DNA sequencing technology was exploited to characterize the diversity of bacteria in human semen communities to a greater depth and with less bias than had been done previously. The results showed that semen of both healthy sperm donors and infertile men harbor diverse bacterial communities that often include high proportions of fastidious or anaerobic bacterial species. Because most of the bacteria species in semen are not usually detected with traditional clinical cultivation methods, the medical and clinical importance of the bacterial species in semen needs to be reevaluated and better understood. We observed that the species composition of semen communities varied widely among men suggesting that each individual had unique and perhaps personalized bacterial communities in their semen. The origins of these bacteria are unknown, and likewise it is unknown whether the bacterial taxa present are transient organisms or persist for extended periods of time. However it is interesting to note that many of the bacteria present in semen are closely related to those found in urine (20), urethra (54, 55), coronal sulcus (53), and female vagina (36). This suggests that the bacteria in semen may have diverse origins.
The method used to characterize microbial diversity in this study is based on sequencing the V1-V2 regions of 16S rRNA genes amplified by PCR from each sample using so-called ‘universal primers’. It is known that although the primers theoretically anneal to the vast majority of known full-length bacterial 16S rRNA genes, they are not in fact universal and some taxa (phylotypes) may not be detected (56). Moreover, the efficiency of annealing to target sequences varies, and this will affect the apparent abundance of some populations in the community possibly causing some to be underestimated. These biases of PCR amplification of heterogeneous sequences are well-known (57) and perhaps unavoidable. Nonetheless they should be kept in mind when drawing conclusions about community composition and interpreting the apparent abundances of populations in a sample.
In this study, we also found that there were no overall patterns in bacterial community composition that could be used as a basis to discriminate between the semen communities of sperm donors and infertility patients. This suggests that infertile subjects did not have altered or unusual semen bacterial communities. However, analyses done to determine if specific community members were associated with sperm quality demonstrated a negative correlation between sperm quality and the presence of Anaerococcus (p-value = 0.0012). Members of the genus Anaerococcus are non-motile Gram-positive cocci that are strictly anaerobic and commonly found in the human vagina and various purulent secretions (53, 58). This species was also observed in the semen samples of infertility patients in another study (26) that showed the positive detection rate of Anaerococcus prevoti or Anaerococcus vaginalis was greater in infertility patients than those in a control group. In addition, Anaerococcus lactolyticus has been detected as a single pathogen in the urinary tract (50). The finding that Anaerococcus may affect sperm quality is worthy of further investigation to explore potential mechanisms by which this microorganism may affect sperm motility.
Heterosexual partners may share bacteria in their reproductive tracts. Recently, microbiota has been characterized in both female vagina and male semen using culture-independent methods. The studies on the microbial ecology of the human vagina have demonstrated that the indigenous bacterial communities differ in species composition among women, and exhibit complex temporal dynamics with continual and sometimes dramatic changes in species composition over short time scales (33, 34, 36). The basis for these differences among women has not been delineated, nor have the drivers of community dynamics been identified. On the other hand, this study along with several other studies confirmed that there were high numbers of bacteria present in semen (9, 10). As with vaginal communities, semen communities contained diverse taxa that varied between men. Interestingly, semen communities also included high proportions of strictly anaerobic bacteria, as do vaginal communities. The bacterial taxa common to these two habitats include Lactobacillus, Corynebacterium, Streptococcus, Staphylococcus, Prevotella, Finegoldia, Anaerococcus, Peptoniphilus, Atopobium, Veillonella, Porphyromonas, Gemella, Aerococcus, Ureaplasma, Acidovorax, Granulicatella, Clostridiales, Cloaclbacterium, and Gardnerella (36). In another investigation of sexual intercourse and the genital tract microbiota in infertile couples, it was reported that sexual intercourse caused significant shifts in vaginal microbiota and an increase in Nugent scores (6). This coincidence of events hints that some bacterial taxa may be shared between heterosexual sex partners. One can imagine that this ‘exchange’ might be either unidirectional (i.e., male to female or female to male) or bidirectional, thus altering the community composition of microbial communities in the reproductive tracts of one or both partners in either a transitory or long-term basis. These alterations may either cause or increase the risk of BV and infectious diseases including sexually transmitted diseases.
In sum, this study to explore potential causes of male infertility showed there were surprisingly high numbers of bacteria were present in most of the semen samples, including those from both infertile subjects as well as healthy sperm donors. Moreover, these semen bacterial communities contained diverse taxa that varied among men. No overall patterns of community composition distinguished semen of infertile men from that of semen donors. However, a detailed analysis of community composition suggested that Anaerococcus was more prevalent in the semen of infertile men and this warrants further investigation to determine if there is a causal link with male infertility. Several bacterial taxa found in semen are also found in the vaginal bacterial communities, which raise the intriguing, but perhaps not surprising possibility that bacteria are shared among heterosexual partners, and that partners might be influencing the species composition of their partner’s reproductive tract microbiome. The sharing of certain bacterial populations might influence risks to infertility, bacterial vaginosis and sexually transmitted infections. This observation could have important implications for reducing risks to these diseases if it is substantiated in future studies.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Wenjun Hu and Shaofeng Cao for their assistance of specimen collection. Yi Shi and Zheng Li who advised us on sample collection; G. Maria Schneider, Yanqin Hu, Meige Lu and Sanqing Yuan for their technical advice on sample analysis.
This work was supported by Shanghai Leading Academic Discipline Project (S30201), Shanghai Municipality Commission for Population and Family Planning (2009JG04), Science and Technology Commission of Shanghai Municipality (10DZ2270600) and Shanghai Basic Research Project (09DJ1400400). In addition, this work was also supported by the National Institute of Allergies and Infectious Diseases, National Institutes of Health (U19 AI084044) and a Center of Biomedical Research Excellence grant (P20 RR016448) from the National Center for Research Resources (NCRR) of the National Institutes of Health to LJF.
Footnotes
Conflicts of interest: The authors have no conflicts of interest to declare.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Capsule: High numbers of diverse kinds of bacteria were present in semen of both sperm donors and infertility patients. There were no significant differences between these two groups based on community composition.
REFERENCES
- 1.Willen M, Holst E, Myhre EB, Olsson AM. The bacterial flora of the genitourinary tract in healthy fertile men. Scand J Urol Nephrol. 1996;30:387–93. doi: 10.3109/00365599609181315. [DOI] [PubMed] [Google Scholar]
- 2.Hillier SL, Rabe LK, Muller CH, Zarutskie P, Kuzan FB, Stenchever MA. Relationship of bacteriologic characteristics to semen indices in men attending an infertility clinic. Obstet Gynecol. 1990;75:800–4. [PubMed] [Google Scholar]
- 3.Keck C, Gerber Schafer C, Clad A, Wilhelm C, Breckwoldt M. Seminal tract infections: impact on male fertility and treatment options. Human Reproduction Update. 1998;4:891–903. doi: 10.1093/humupd/4.6.891. [DOI] [PubMed] [Google Scholar]
- 4.Dohle GR, Colpi GM, Hargreave TB, Papp GK, Jungwirth A, Weidner W. EAU guidelines on male infertility. Eur Urol. 2005;48:703–11. doi: 10.1016/j.eururo.2005.06.002. [DOI] [PubMed] [Google Scholar]
- 5.Pant N, Upadhyay G, Pandey S, Mathur N, Saxena DK, Srivastava SP. Lead and cadmium concentration in the seminal plasma of men in the general population: correlation with sperm quality. Reprod Toxicol. 2003;17:447–50. doi: 10.1016/s0890-6238(03)00036-4. [DOI] [PubMed] [Google Scholar]
- 6.Gallo MF, Warner L, King CC, Sobel JD, Klein RS, Cu-Uvin S, et al. Association between Semen Exposure and Incident Bacterial Vaginosis. Infect Dis Obstet Gynecol. 2011;2011:842652. doi: 10.1155/2011/842652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Swidsinski A, Dorffel Y, Loening-Baucke V, Mendling W, Verstraelen H, Dieterle S, et al. Desquamated epithelial cells covered with a polymicrobial biofilm typical for bacterial vaginosis are present in randomly selected cryopreserved donor semen. FEMS Immunol Med Microbiol. 2010;59:399–404. doi: 10.1111/j.1574-695X.2010.00688.x. [DOI] [PubMed] [Google Scholar]
- 8.Elsner P, Hartmann AA. Gardnerella vaginalis in the male upper genital tract: a possible source of reinfection of the female partner. Sex Transm Dis. 1987;14:122–3. doi: 10.1097/00007435-198704000-00015. [DOI] [PubMed] [Google Scholar]
- 9.Ott MA, Katschke A, Tu W, Fortenberry JD. Longitudinal associations among relationship factors, partner change, and sexually transmitted infection acquisition in adolescent women. Sex Transm Dis. 2011;38:153–7. doi: 10.1097/OLQ.0b013e3181f2e292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pellati D, Mylonakis I, Bertoloni G, Fiore C, Andrisani A, Ambrosini G, et al. Genital tract infections and infertility. Eur J Obstet Gynecol Reprod Biol. 2008;140:3–11. doi: 10.1016/j.ejogrb.2008.03.009. [DOI] [PubMed] [Google Scholar]
- 11.Henkel R, Schill WB. Sperm separation in patients with urogenital infections. Andrologia. 1998;30(Suppl 1):91–7. doi: 10.1111/j.1439-0272.1998.tb02832.x. [DOI] [PubMed] [Google Scholar]
- 12.Urata K, Narahara H, Tanaka Y, Egashira T, Takayama F, Miyakawa I. Effect of endotoxin-induced reactive oxygen species on sperm motility. Fertil Steril. 2001;76:163–6. doi: 10.1016/s0015-0282(01)01850-7. [DOI] [PubMed] [Google Scholar]
- 13.Sanocka-Maciejewska D, Ciupinska M, Kurpisz M. Bacterial infection and semen quality. J Reprod Immunol. 2005;67:51–6. doi: 10.1016/j.jri.2005.06.003. [DOI] [PubMed] [Google Scholar]
- 14.Rehewy MS, Hafez ES, Thomas A, Brown WJ. Aerobic and anaerobic bacterial flora in semen from fertile and infertile groups of men. Arch Androl. 1979;2:263–8. doi: 10.3109/01485017908987323. [DOI] [PubMed] [Google Scholar]
- 15.Eggert-Kruse W, Rohr G, Strock W, Pohl S, Schwalbach B, Runnebaum B. Anaerobes in ejaculates of subfertile men. Hum Reprod Update. 1995;1:462–78. doi: 10.1093/humupd/1.5.462. [DOI] [PubMed] [Google Scholar]
- 16.Gdoura R, Kchaou W, Ammar-Keskes L, Chakroun N, Sellemi A, Znazen A, et al. Assessment of Chlamydia trachomatis, Ureaplasma urealyticum, Ureaplasma parvum, Mycoplasma hominis, and Mycoplasma genitalium in semen and first void urine specimens of asymptomatic male partners of infertile couples. J Androl. 2008;29:198–206. doi: 10.2164/jandrol.107.003566. [DOI] [PubMed] [Google Scholar]
- 17.La Vignera S, Vicari E, Condorelli RA, D'Agata R, Calogero AE. Male accessory gland infection and sperm parameters (review) Int J Androl. 2011;34:e330–47. doi: 10.1111/j.1365-2605.2011.01200.x. [DOI] [PubMed] [Google Scholar]
- 18.Merino G, Carranza-Lira S, Murrieta S, Rodriguez L, Cuevas E, Moran C. Bacterial infection and semen characteristics in infertile men. Arch Androl. 1995;35:43–7. doi: 10.3109/01485019508987852. [DOI] [PubMed] [Google Scholar]
- 19.Lacroix JM, Jarvi K, Batra SD, Heritz DM, Mittelman MW. PCR-based technique for the detection of bacteria in semen and urine. J Microbiol Meth. 1996;26:61–71. [Google Scholar]
- 20.Nelson DE, Van Der Pol B, Dong Q, Revanna KV, Fan B, Easwaran S, et al. Characteristic male urine microbiomes associate with asymptomatic sexually transmitted infection. Plos One. 2010;5:e14116. doi: 10.1371/journal.pone.0014116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Busolo F, Zanchetta R, Lanzone E, Cusinato R. Microbial flora in semen of asymptomatic infertile men. Andrologia. 1984;16:269–75. doi: 10.1111/j.1439-0272.1984.tb00282.x. [DOI] [PubMed] [Google Scholar]
- 22.Rodin DM, Larone D, Goldstein M. Relationship between semen cultures, leukospermia, and semen analysis in men undergoing fertility evaluation. Fertil Steril. 2003;79:1555–8. doi: 10.1016/s0015-0282(03)00340-6. [DOI] [PubMed] [Google Scholar]
- 23.Swenson CE, Toth A, Toth C, Wolfgruber L, O'Leary WM. Asymptomatic bacteriospermia in infertile men. Andrologia. 1980;12:7–11. doi: 10.1111/j.1439-0272.1980.tb00567.x. [DOI] [PubMed] [Google Scholar]
- 24.Virecoulon F, Wallet F, Fruchart-Flamenbaum A, Rigot JM, Peers MC, Mitchell V, et al. Bacterial flora of the low male genital tract in patients consulting for infertility. Andrologia. 2005;37:160–5. doi: 10.1111/j.1439-0272.2005.00673.x. [DOI] [PubMed] [Google Scholar]
- 25.Jarvi K, Lacroix JM, Jain A, Dumitru I, Heritz D, Mittelman MW. Polymerase chain reaction-based detection of bacteria in semen. Fertil Steril. 1996;66:463–7. [PubMed] [Google Scholar]
- 26.Kiessling AA, Desmarais BM, Yin HZ, Loverde J, Eyre RC. Detection and identification of bacterial DNA in semen. Fertil Steril. 2008;90:1744–56. doi: 10.1016/j.fertnstert.2007.08.083. [DOI] [PubMed] [Google Scholar]
- 27.De Francesco MA, Negrini R, Ravizzola G, Galli P, Manca N. Bacterial species present in the lower male genital tract: a five-year retrospective study. Eur J Contracept Reprod Health Care. 2011;16:47–53. doi: 10.3109/13625187.2010.533219. [DOI] [PubMed] [Google Scholar]
- 28.Imirzalioglu C, Hain T, Chakraborty T, Domann E. Hidden pathogens uncovered: metagenomic analysis of urinary tract infections. Andrologia. 2008;40:66–71. doi: 10.1111/j.1439-0272.2007.00830.x. [DOI] [PubMed] [Google Scholar]
- 29.Moretti E, Capitani S, Figura N, Pammolli A, Federico MG, Giannerini V, et al. The presence of bacteria species in semen and sperm quality. Journal of Assisted Reproduction and Genetics. 2009;26:47–56. doi: 10.1007/s10815-008-9283-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Berktas M, Aydin S, Yilmaz Y, Cecen K, Bozkurt H. Sperm motility changes after coincubation with various uropathogenic microorganisms: an in vitro experimental study. Int Urol Nephrol. 2008;40:383–9. doi: 10.1007/s11255-007-9289-4. [DOI] [PubMed] [Google Scholar]
- 31.Huwe P, Diemer T, Ludwig M, Liu J, Schiefer HG, Weidner W. Influence of different uropathogenic microorganisms on human sperm motility parameters in an in vitro experiment. Andrologia. 1998;30(Suppl 1):55–9. doi: 10.1111/j.1439-0272.1998.tb02827.x. [DOI] [PubMed] [Google Scholar]
- 32.Pace NR. A molecular view of microbial diversity and the biosphere. Science. 1997;276:734–40. doi: 10.1126/science.276.5313.734. [DOI] [PubMed] [Google Scholar]
- 33.Zhou X, Bent SJ, Schneider MG, Davis CC, Islam MR, Forney LJ. Characterization of vaginal microbial communities in adult healthy women using cultivation-independent methods. Microbiology. 2004;150:2565–73. doi: 10.1099/mic.0.26905-0. [DOI] [PubMed] [Google Scholar]
- 34.Zhou X, Brown CJ, Abdo Z, Davis CC, Hansmann MA, Joyce P, et al. Differences in the composition of vaginal microbial communities found in healthy Caucasian and black women. ISME J. 2007;1:121–33. doi: 10.1038/ismej.2007.12. [DOI] [PubMed] [Google Scholar]
- 35.Hou DS, Long WM, Shen J, Zhao LP, Pang XY, Xu C. Characterisation of the bacterial community in expressed prostatic secretions from patients with chronic prostatitis/chronic pelvic pain syndrome and infertile men: a preliminary investigation. Asian J Androl. 2012;14:566–73. doi: 10.1038/aja.2012.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ravel J, Gajer P, Abdo Z, Schneider GM, Koenig SS, McCulle SL, et al. Vaginal microbiome of reproductive-age women. Proc Natl Acad Sci U S A. 2010;108(Suppl 1):4680–7. doi: 10.1073/pnas.1002611107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gao Z, Tseng CH, Pei Z, Blaser MJ. Molecular analysis of human forearm superficial skin bacterial biota. Proc Natl Acad Sci U S A. 2007;104:2927–32. doi: 10.1073/pnas.0607077104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Palmer C, Bik EM, DiGiulio DB, Relman DA, Brown PO. Development of the human infant intestinal microbiota. PLoS Biol. 2007;5:e177. doi: 10.1371/journal.pbio.0050177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.WHO . WHO Laboratory manual for the examination of human semen and sperm-cervical mucus interaction. 4 Cambridge University Press; London: 1999. [Google Scholar]
- 40.Yuan S, Cohen DB, Ravel J, Abdo Z, Forney LJ. Evaluation of methods for the extraction and purification of DNA from the human microbiome. PLoS One. 2012;7:e33865. doi: 10.1371/journal.pone.0033865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA, et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–80. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang Q, Garrity GM, Tiedje JM, Cole JR. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol. 2007;73:5261–7. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.JR B, JT C. An Ordination of the Upland Forest Communities of Southern Wisconsin. Ecological Monographs. 1957;27:325–49. [Google Scholar]
- 44.Legendre P, Gallagher ED. Ecologically meaningful transformations for ordination of species data. Oecologia. 2001;129:271–80. doi: 10.1007/s004420100716. [DOI] [PubMed] [Google Scholar]
- 45.Calinski T, Harabasz J. A dendrite method for cluster analysis. Communications in Statistics. 1974;3:1–27. [Google Scholar]
- 46.Milligan GW, Cooper MC. An examination of procedures for determining the number of clusters in a data set. Psychometrika. 1985;50:159–79. [Google Scholar]
- 47.Milligan G, Cooper M. A study of variable standardization. J Classification. 1988;5:181–204. [Google Scholar]
- 48.Wen SH, Lu ZS. Factors affecting the effective number of tests in genetic association studies: a comparative study of three PCA-based methods. J Hum Genet. 2011;56:428–35. doi: 10.1038/jhg.2011.34. [DOI] [PubMed] [Google Scholar]
- 49.Carrell DT, Emery BR, Hamilton B. Seminal infection with Ralstonia picketti and Cytolysosomal spermophagy in a previously fertile man. Fertil Steril. 2003;79(Suppl 3):1665–7. doi: 10.1016/s0015-0282(03)00254-1. [DOI] [PubMed] [Google Scholar]
- 50.Domann E, Hong G, Imirzalioglu C, Turschner S, Kuhle J, Watzel C, et al. Culture-independent identification of pathogenic bacteria and polymicrobial infections in the genitourinary tract of renal transplant recipients. J Clin Microbiol. 2003;41:5500–10. doi: 10.1128/JCM.41.12.5500-5510.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Citron DM, Tyrrell KL, Goldstein EJ. Peptoniphilus coxii sp. nov. and Peptoniphilus tyrrelliae sp. nov. isolated from human clinical infections. Anaerobe. 2012;18:244–8. doi: 10.1016/j.anaerobe.2011.11.008. [DOI] [PubMed] [Google Scholar]
- 52.Hernandez-Rodriguez C, Romero-Gonzalez R, Albani-Campanario M, Figueroa-Damian R, Meraz-Cruz N, Hernandez-Guerrero C. Vaginal microbiota of healthy pregnant Mexican women is constituted by four Lactobacillus species and several vaginosis-associated bacteria. Infect Dis Obstet Gynecol. 2011;2011:851485. doi: 10.1155/2011/851485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Price LB, Liu CM, Johnson KE, Aziz M, Lau MK, Bowers J, et al. The Effects of Circumcision on the Penis Microbiome. Plos One. 2010;5:12. doi: 10.1371/journal.pone.0008422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Riemersma WA, van der Schee CJ, van der Meijden WI, Verbrugh HA, van Belkum A. Microbial population diversity in the urethras of healthy males and males suffering from nonchlamydial, nongonococcal urethritis. J Clin Microbiol. 2003;41:1977–86. doi: 10.1128/JCM.41.5.1977-1986.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Spaine DM, Mamizuka EM, Cedenho AP, Srougi M. Microbiologic aerobic studies on normal male urethra. Urology. 2005;56:207–10. doi: 10.1016/s0090-4295(00)00615-4. [DOI] [PubMed] [Google Scholar]
- 56.Forney LJ, Zhou X, Brown CJ. Molecular microbial ecology: land of the one-eyed king. Curr Opin Microbiol. 2004;7:210–20. doi: 10.1016/j.mib.2004.04.015. [DOI] [PubMed] [Google Scholar]
- 57.von Wintzingerode F, Gobel UB, Stackebrandt E. Determination of microbial diversity in environmental samples: pitfalls of PCR-based rRNA analysis. FEMS Microbiol Rev. 1997;21:213–29. doi: 10.1111/j.1574-6976.1997.tb00351.x. [DOI] [PubMed] [Google Scholar]
- 58.Ezaki T, Kawamura Y, Li N, Li ZY, Zhao L, Shu S. Proposal of the genera Anaerococcus gen. nov., Peptoniphilus gen. nov. and Gallicola gen. nov. for members of the genus Peptostreptococcus. Int J Syst Evol Microbiol. 2001;51:1521–8. doi: 10.1099/00207713-51-4-1521. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.