Abstract
Effective monitoring of glucose levels is necessary for patients to achieve greater control over their diabetes. However, only about a quarter of subjects with diabetes who requires close serum glucose monitoring, regularly check their serum glucose daily. One of the potential barriers to patient compliance is the blood sampling requirement. Saliva and its protein contents can be altered in subjects with diabetes, possibly due to changes in glycemic control. We propose here that salivary proteomes of subjects with diabetes may be different based on their glycemic control as reflected in A1C levels. A total of 153 subjects with type 1 or 2 diabetes were recruited. Subjects in each type of diabetes were divided into 5 groups based on their A1C levels; <7, 7–8, 8–9, 9–10, >10. To examine the global proteomic changes associated with A1C, the proteomic profiling of pooled saliva samples from each group was created using label-free quantitative proteomics. Similar proteomic analysis for individual subjects (N=4, for each group) were then applied to examine proteins that may be less abundant in pooled samples. Principle component analysis (PCA) and cluster analysis (p<0.01 and p<0.001) were used to define the proteomic differences. We, therefore, defined the salivary proteomic changes associated with A1C changes. This study demonstrates that differences exist between salivary proteomic profiles in subjects with diabetes based on the A1C levels.
Keywords: A1C, Diabetes, Mass Spectrometry, Saliva, Proteome
Introduction
Diabetes affects approximately 346 million people worldwide and over 25.8 million people in the US [http://www.cdc.gov/diabetes/pubs/estimates11.htm] [http://www.who.int/mediacentre/factsheets/fs312/en/]. Type 2 diabetes (T2DM) is a major public health problem that accounts for nearly 90% of all those diagnosed with diabetes and is often associated with obesity and insulin resistance condition. Type 1 diabetes (T1DM) affects a relatively smaller number of patients and is often associated with autoimmune destruction of insulin producing beta cells in the pancreas. The potential cures for T1DM, including beta-cell transplantation/replacement therapy, are still in the experimental stages. Sustainable and reliable methods of monitoring glycemic conditions are necessary in managing both T2DM and T1DM1–7. Clinically, Fasting Plasma Glucose (FPG), Oral Glucose Tolerance Test (OGTT), and hemoglobin A1C (A1C) are used to determine the patient’s overall glycemic control. FPG and OGTT reflect the glycemic control at a single point in time. FPG usage is constrained by difficulties arising from the need for fasting. OGTT requires a large consumption of glucose. While FPG is still used widely, the American Diabetes Association recommended against using the OGTT, because of its cumbersome technique and low reproducibility8. A1C is a non-enzymatically glycosylated or glycated form of hemoglobin through Amadori reaction of a lysine residue and a glucose molecule. The A1C value corresponds to an average value of plasma glucose over a 120-day time period of circulation survival time of erythrocytes9–14. A1C is, therefore, clinically, a more reliable method in giving an overall long-term picture of glycemic control than FPG and OGTT—which only allow one point measurement of plasma glucose14. In addition to clinical measurement of plasma glucose, patients with diabetes, especially T1DM, require daily monitoring of plasma glucose using either Home Self-Monitoring of Blood Glucose (SMBG) or Continuous Monitoring of Blood Glucose (CMBG)15. Patients with T1DM monitor their own plasma glucose using SMBG on a regular basis 3–5 times a day16. SMBG if properly used can be effective in the management of T1DM-related hyperglycemia; however, SMBG can be expensive without health insurance and is often not used due to poor patient adherence. It was estimated that only about a quarter of patients that need to monitor their plasma glucose, in particular young children and teenagers with T1DM, monitor their serum glucose every day17–19. Similar problems in monitoring serum glucose are also found in elderly patients with T2DM19. CMBG is a subcutaneous sensor constantly measuring plasma glucose. Cost, inconvenience, and surgical requirement are among the main drawbacks of CMBG9–14. Blood sampling is perhaps one of the major factors hindering the use of SMBG and CMBG. There is a need to find more simple and non-invasive methods to supplement the current methods and, therefore, enhance patient compliance.
Several groups, including ours, have shown that there are changes in salivary proteomes as a result of type 1 or 2 diabetes (T1DM or T2DM). Rao et al, 2009 showed that there are changes in salivary proteomes between controls and subjects with pre-diabetes and T2DM20. The results from our group suggested that the differences in salivary proteomes can be found in subjects without teeth (edentulous)21. Similar to T2DM proteomics, Cabras et al, 2010 and Hirtz et al, 2006 demonstrated that there are differences in salivary proteomes of subjects with T1DM and controls22, 23.
It is clear that diabetic conditions, both T1DM and T2DM, affect the salivary proteome and perhaps different degrees of the condition may also alter the salivary proteome. However, none of these previous studies looked at the relationship between salivary proteomes and known clinical measures, in particular A1C. In this study, we attempted to define the salivary proteomic changes in subjects with T1DM and T2DM based on their glycemic control reflected in the A1C value. Our goals were to determine if there are salivary proteomic changes based on A1C and if there are similarities or differences between the changes in A1C associated salivary proteomes between each type of diabetes.
Results
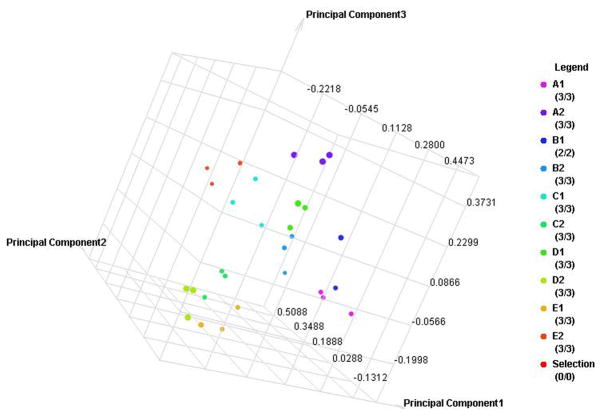
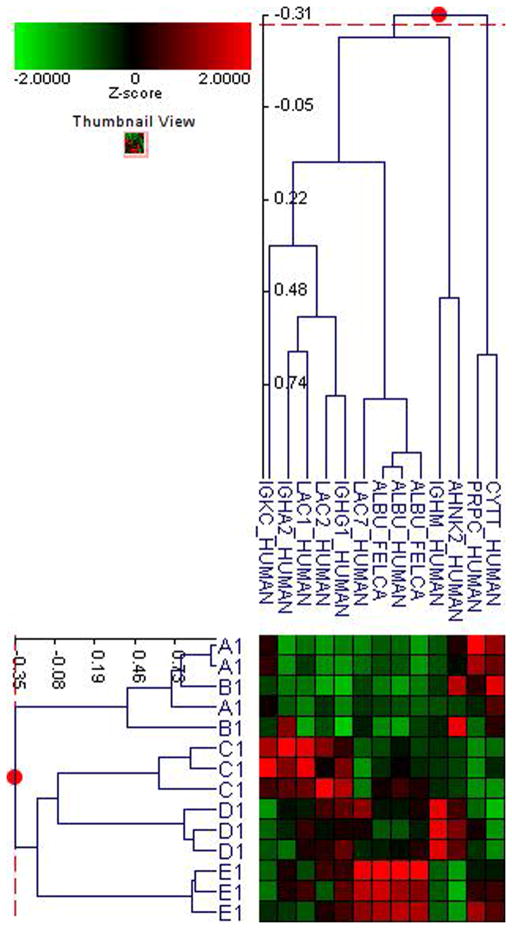
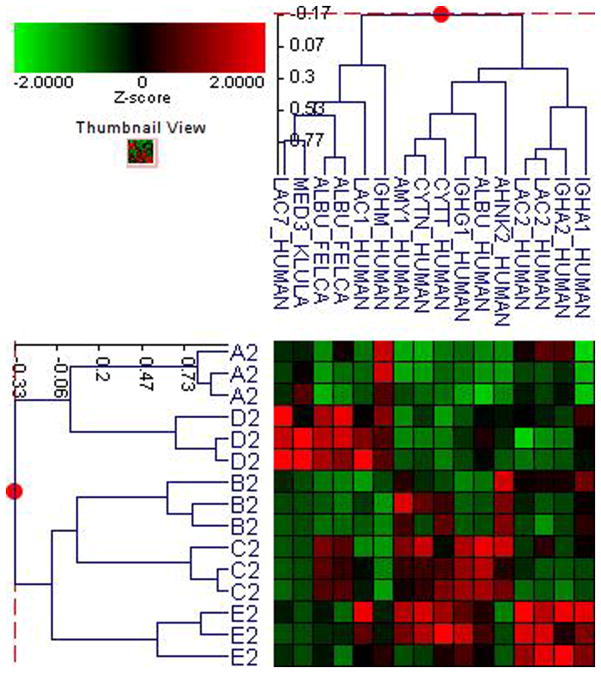
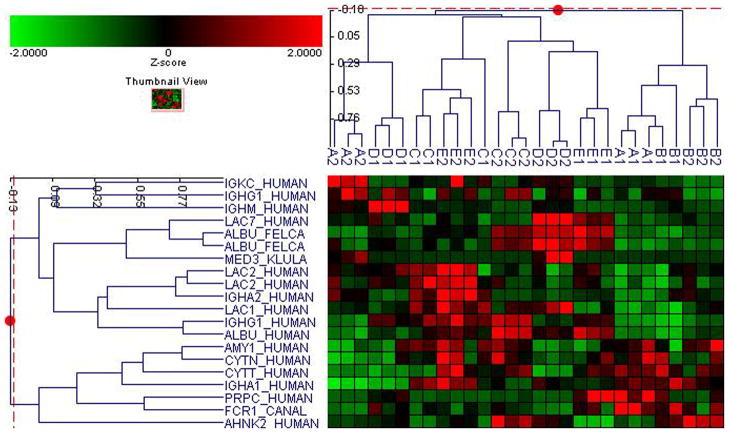
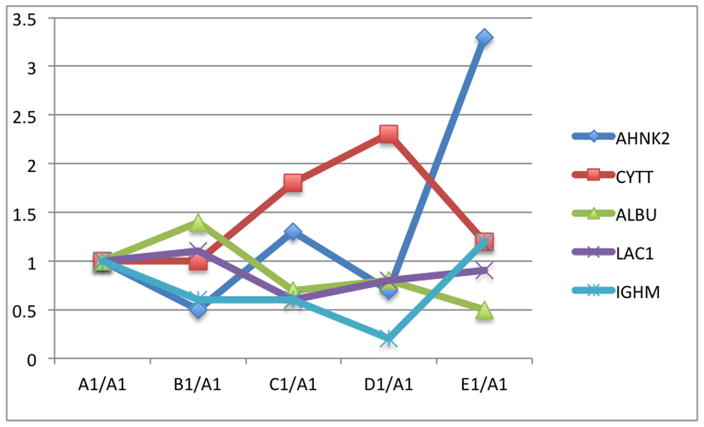
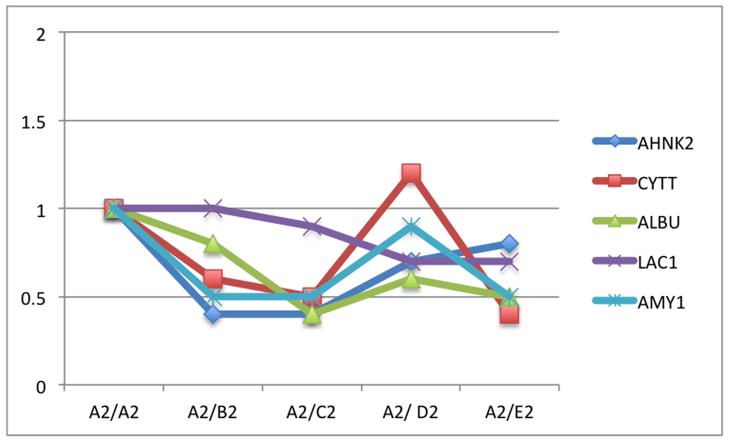
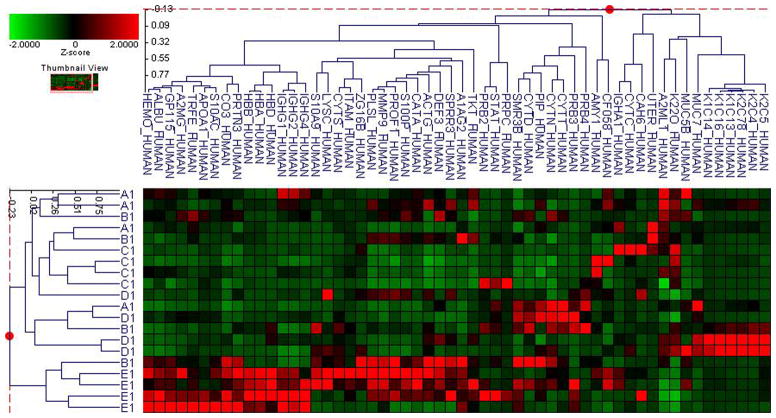
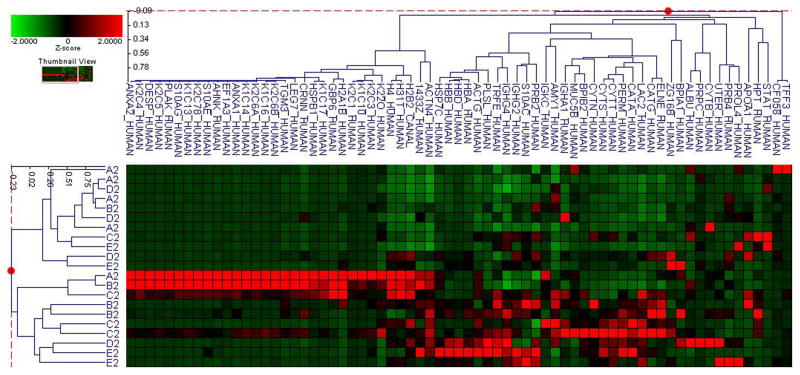
Fifty-six subjects with T1DM and ninety-seven subjects with T2DM (total of 153 subjects) were recruited. Based on the A1C level, we divided subjects into five groups based on A1C levels, <7, 7–8, 8–9, 9–10, and >10; these groups were referred to as A1, B1, C1, D1, E1 for T1DM and A2, B2, C2, D2, E2 for T2DM. First, to explore the global proteomic changes associated with A1C, pooled saliva from each group were analyzed using a label-free quantitative proteomic approach. A label-free differential expression LC/MS/MS method similar to our previous study was used to qualitatively compare protein expression levels between the five sample groups from each type of diabetes analyzed in triplicate (3 pooled samples per each group and 15 pooled samples per type of diabetes). Mass signals were aligned and the results were processed using the Rosetta Elucidator software package, ANOVA (p<0.05). Mass signals were annotated with the corresponding peptide and protein information based on the database search results using a 1% false discovery rate cut off. Data processing of the Thermo Orbitrap data resulted in detection of 148 proteins. Principle component analysis (PCA) was performed on the collected data set to evaluate the data for outliers and sample trends (Figure 1a). An outlier in one of the triplicates in the B1 group was observed and removed from further statistical analysis. An error-weighted ANOVA and a standard ANOVA were used to determine mass patterns that correlated to the A1C levels; low (A), medium (C) and high (E). The B and D groups were left out from the analysis to simplify the analysis in the biomarker exploration and to enhance the possible differences in biomarker expression. Signals with a p<0.05 for the error-weighted ANOVA were selected as tentative biomarkers and summarized by protein. After selection of the biomarkers, we utilized cluster analysis for all five groups to compare the pattern of expression for each biomarker (Figure 1b–d). The corresponding tentative differentially expressed proteins for each type of diabetes are summarized in Table 1 and 2. For T1DM, based on the error-weighted ANOVA, thirteen peptides were found differentially expressed including four serum albumin peptides, seven immunoglobulin fragments, one cystatin-SA peptide and one salivary acidic phosphoprotein peptide. For T2DM, based on the error-weighted ANOVA, sixteen peptides were found differentially expressed, including three peptides of serum albumin, eight immunoglobulin fragments, one cystatin-SA, one cystatin-SN, one protein AHNK2, and one Mediator of RNA polymerase II transcription subunit 3. The fold expression trends of some biomarkers compared with the lowest A1C group (A1 or A2) were shown in Figure 2a and 2b. The expression pattern of certain proteins including serum albumin (ALB), alpha-amylase 1 (AMY1), cystatin-SA (CYTT), and cystatin-SN (CYTN) were shown using western blot analysis (Figure 2c).
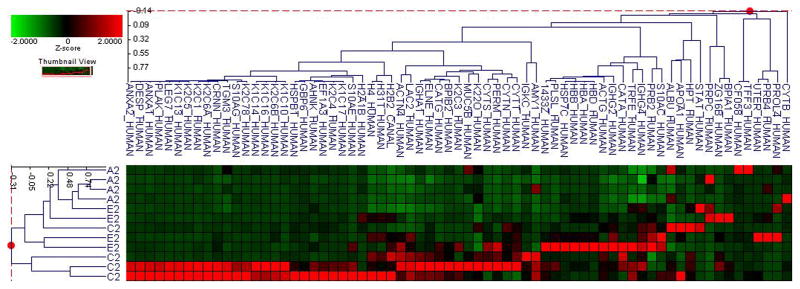
Figure 1. Pooled sample proteomic analysis.
A. Principle component analysis (PCA) of the proteomes from pooled saliva samples demonstrates differentially expressed peptides for each type of diabetes in each A1C group (A–E refer to levels of A1C <7, 7–8, 8–9, 9–10 and 1 for type 1 and 2 for type 2 diabetes).
B. Cluster analysis of type 1 diabetes demostrates differential expression of proteins based on ANOVA (p<0.05).
C. Cluster analysis of type 2 diabetes demostrates differential expression of proteins based on ANOVA (p<0.05).
D. Cluster analysis of both type 1 and 2 diabetes demostrates differential expression of proteins based on ANOVA (p<0.05).
Table 1.
Differentially expressed peptides from T1DM pooled samples
| Protein Description | Peptide Name | P-value (ANOVA) | B1/A1 | C1/A1 | D1/A1 | E1/A1 | Highest | Lowest |
|---|---|---|---|---|---|---|---|---|
| 385369 IGKC_HUMAN P01834 Ig kappa chain C region |
CLLNNFYPR | 8.49E-04 | 1.3 | 0.7 | 1.6 | 1.4 | C1 | D1 |
| 385401 LAC7_HUMAN A0M8Q6 Ig lambda-7 chain C region |
KATLVCLV SDFYPGAVTVAWK | 6.35E-04 | 0.7 | 1.2 | 0.5 | 0.3 | E1 | C1 |
| 385434 IGHM_HUMAN P01871 Ig mu chain C region |
NVPLPVIAELPPK | 3.42E-10 | 0.6 | 0.6 | 0.2 | 1.2 | D1 | E1 |
| 385456 IGHA2_HUMAN P01877 Ig alpha-2 chain C region |
LLPPPSEELALNELVTLTCLAR | 0.0181 | 0.7 | 0.5 | 0.8 | 0.7 | C1 | A1 |
| 385471 PRPC_HUMAN P02810 Salivary acidic proline-rich phosphoprotein |
GRPQGPPQ QGGHQQG | 9.41E-05 | 2.5 | 84.7 | 2.8 | 1.4 | A1 | C1 |
| 385496 AHNK2_HUMAN Q8IVF2-3 Isoform 3 of Protein AHNAK2 |
LPEGQVPEGAGLK | 1.61E-06 | 0.5 | 1.3 | 0.7 | 3.3 | B1 | E1 |
| 385523 CYTT_HUMAN P09228 Cystatin-SA |
SQPNLDTCAFHEQPELQK | 6.41E-04 | 1.0 | 1.8 | 2.3 | 1.2 | A1/B1 | D1 |
| 385543 LAC2_HUMAN, LAC3_HUMAN P0CG05 Ig lambda-2 chain C regions |
AAPSVTLFPPSSEELQANK | 0.0196 | 1.0 | 0.5 | 0.6 | 0.7 | C1 | A1/B1 |
| 385652 LAC1_HUMAN P0CG04 Ig lambda-1 chain C regions |
ANPTVTLFPPSSEELQANK | 0.001 | 1.1 | 0.6 | 0.8 | 0.9 | C1 | B1 |
| 385686 IGHG1_HUMAN P01857 Ig gamma-1 chain C region |
EPQVYTLPPSR | 1.16E-06 | 1.3 | 0.5 | 0.6 | 0.6 | C1 | B1 |
| 385688 ALBU_HUMAN P02768 Serum albumin precursor. |
QTALVELVK | 5.50E-06 | 1.4 | 0.7 | 0.8 | 0.5 | E1 | B1 |
Table 2.
Differentially expressed peptides from T2DM pooled samples
| Protein Description | Peptide Name | P-value (ANOVA) | A2/B2 | A2/C2 | A2/D2 | A2/E2 | Highest | Lowest |
|---|---|---|---|---|---|---|---|---|
| 385401 LAC7_HUMAN A0M8Q6 Ig lambda-7 chain C region |
KATLVCLVSDFYPGAVTVAWK | 3.12E-06 | 1.1 | 1.4 | 0.3 | 1.0 | D2 | C2 |
| 385434 IGHM_HUMAN P01871 Ig mu chain C region |
NVPLPVIAELPPK | 9.82E-06 | 1.9 | 2.1 | 1.1 | 1.4 | A2 | C2 |
| 385447 AMY1_HUMAN P04745 Alpha-amylase 1 |
ALVFVDNHDNQR | 0.0047 | 0.5 | 0.5 | 0.9 | 0.5 | B2/C2/E2 | A2 |
| 385450 LAC2_HUMAN P0CG05 Ig lambda-2 chain C regions |
YAASSYLSLTPEQWK | 0.0041 | 1.0 | 1.1 | 1.3 | 0.6 | E2 | D2 |
| 385456 IGHA2_HUMAN P01877 Ig alpha-2 chain C region |
LLPPPSEELALNELVTLTCLAR | 0.0038 | 1.2 | 1.4 | 1.4 | 0.6 | E2 | C2/D2 |
| 385469 CYTN_HUMAN P01037 Cystatin-SN |
ALHFAISEYNK | 0.0081 | 0.5 | 0.4 | 0.7 | 0.4 | C2/E2 | A2 |
| 385487 IGHA1_HUMAN P01876 Ig alpha-1 chain C region |
TPLTATLSK | 3.27E-06 | 0.4 | 0.6 | 0.5 | 0.4 | B2/E2 | A2 |
| 385496 AHNK2_HUMAN Q8IVF2-3 Isoform 3 of Protein AHNAK2 |
LPEGQVPEGAGLK | 2.74E-05 | 0.4 | 0.4 | 0.7 | 0.8 | B2/C2 | A2 |
| 385523 CYTT_HUMAN P09228 Cystatin-SA |
SQPNLDTCAFHEQPELQK | 6.04E-04 | 0.6 | 0.5 | 1.2 | 0.4 | E2 | D2 |
| 385543 LAC2_HUMAN P0CG05 Ig lambda-2 chain C regions |
AAPSVTLFPPSSEELQANK | 0.0098 | 1.2 | 1.1 | 1.2 | 0.6 | E2 | B2/D2 |
| 385652 LAC1_HUMAN P0CG04 Ig lambda-1 chain C regions |
ANPTVTLFPPSSEELQANK | 0.0148 | 1.0 | 0.9 | 0.7 | 0.7 | D2/E2 | A2/B2 |
| 385686 IGHG1_HUMAN P01857 Ig gamma-1 chain C region |
EPQVYTLPPSR | 4.05E-06 | 0.9 | 0.5 | 0.8 | 0.5 | C2/E2 | A2 |
| 385688 ALBU_HUMAN P02768 Serum albumin precursor. |
QTALVELVK | 2.78E-06 | 0.8 | 0.4 | 0.6 | 0.5 | C2 | A2 |
| 386406 MED3_KLULA Mediator of RNA polymerase II transcription subunit 3 |
FFVQETLGYTQLASNGSAAGITKTSSGNDGNTTGSTANTMAMAK | 2.33E-06 | 131.9 | 213.1 | 0.3 | 1000.0 | D2 | E2 |
Figure 2.
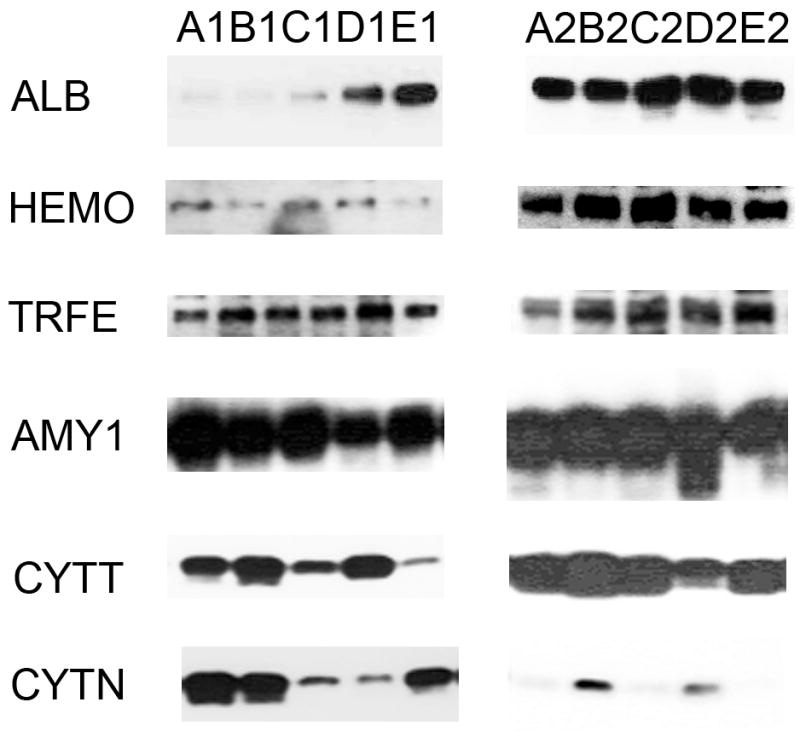
The relative levels of expression of certain proteins in pooled saliva samples showing a comparison of the relative levels of expression of serum albumin precursor, cystatin-SA, immunoglobulin fragments, and protein AHNK2 with the increased A1C levels (A–E). The levels of expression were calibrated using the level of expression of the group with lowest A1C (A1 or A2). The graphs were plotted using the log expression level of each group divided by A1 for type 1 diabetes and A2 for type 2 diabetes.
A. Relative expression levels of selected peptide masses in type 1 diabetes
B. Relative expression levels of selected peptide masses in type 2 diabetes
C. Western blot analyses
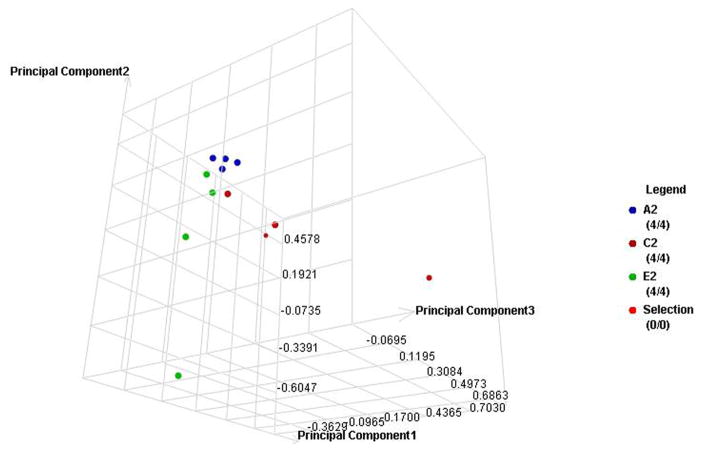
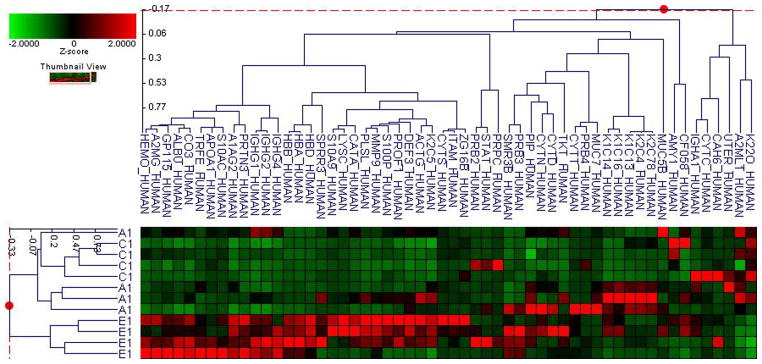
The label-free quantitative analysis of the pooled samples allows a global view of the prominent proteins associated with changes in A1C in each type of diabetes. However, it is possible that the results might be different in an individual level. Without an analysis of individual subjects, we might have missed some biomarkers with a lower level of expression. More importantly, the results may be influenced by particular samples that may have higher overall protein content or higher level of expression for certain proteins. We therefore selected four samples from each group using all of the subjects in the smallest group (E1) and four each from other groups using demographically matched subjects for each type of diabetes. Then we utilized a label-free quantitative LC/MS/MS method and statistical analysis similar to the one used with the pooled samples. Similarly, the data were processed using the Rosetta Elucidator software package, error-weight ANOVA (p<0.05). Mass signals were screened using a 1% false discovery rate cut off. In the similar fashion as the pool sample analysis, principle component analysis (PCA) and cluster analysis were then performed on three groups with low (A), medium (C), and high (E) A1C levels (Figure 3a–d), and then all five groups (Figure 4a–d). Out of total 93 proteins from both types, 24 and 37 proteins are unique to T1DM and T2DM, respectively. 32 proteins are common in both types. Out of these 32 common proteins, eight are found in the pooled sample analysis (Table 3). In addition to the error-weight ANOVA, we also used standard ANOVA to examine the signals at p<0.01 and p<0.001 (Table 4). Using western analysis (Figure 2c), we also demonstrate the expression of hemopexin (HEMO) and transferrin (TRFE) in pooled saliva samples.
Figure 3. Individual sample proteomic analysis of the low (A), medium (C) and high (E) A1C groups in each type of diabetes.
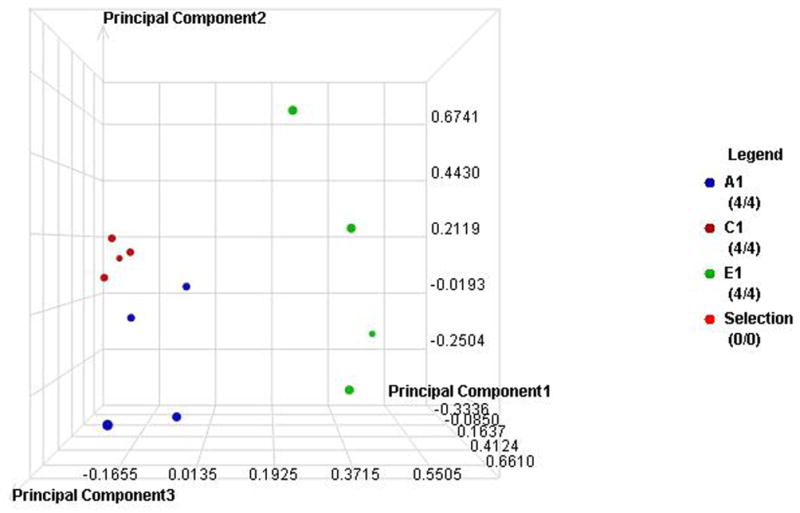
A. PCA of the proteomes from individual samples with type 1 diabetes
B. Cluster anlysis of type 1 diabetes
C. PCA of the proteomes from individual samples with type 2 diabetes
D. Cluster anlysis of type 2 diabetes
Figure 4. Individual sample proteomic analysis of five groups in each type of diabetes.
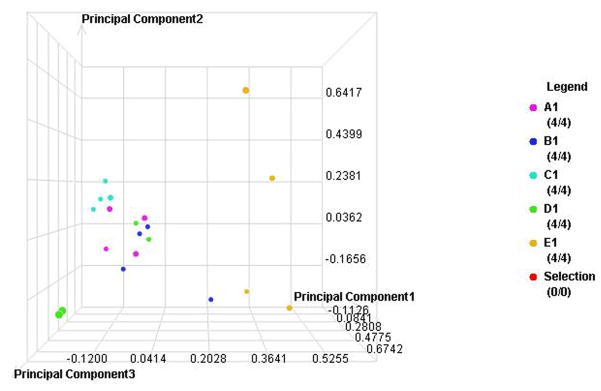
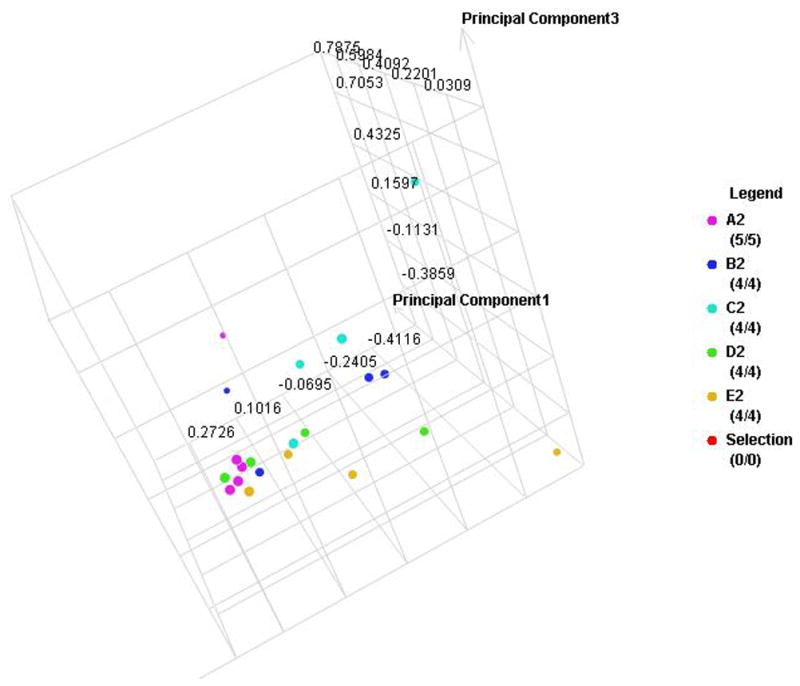
A. PCA of the proteomes from individual samples with type 1 diabetes
B. Cluster anlysis of type 1 diabetes
C. PCA of the proteomes from individual samples with type 2 diabetes
D. Cluster anlysis of type 2 diabetes
Table 3.
Selected Salivary Proteins Associated with A1C Changes in Individual Sample Analysis
| Serum originating proteins | Functions | Relation to diabetes |
|---|---|---|
| Serum albumin (ALB) | Regulation of the colloidal osmotic pressure of blood.60 Antioxidant agent.61 |
Increased glycation resulting from prolonged hyperglycemia of serum albumin destabilizes albumin structure27 Increased glycosylation of serum albumin in Diabetes Mellitus.62 Structural changes of albumin may reduce antioxidant properties and contribute to vascular and metabolic morbidities.61 |
| Hemoglobin (Hb) | Role of oxygen transport from the lungs to the rest of the body. | Glycated hemoglobin, or hemoglobin A1c, is used to identify average plasma glucose levels over time. In diabetes, higher HbA1c levels indicate poor control of glucose levels.63 Increased hemoglobin degradation can directly alter iron metabolism. However, iron can increase through several pathways related to insulin, oxidative stress, and inflammatory responses.28 |
| Alpha-2-macroglobulin (A2MG) | Proteinase inhibitor. Is able to “trap” enzymes, with the trapped enzyme remaining active against low M.W. substrates, and a greatly reduced activity against larger substrates.21 | May be found in higher levels in poorly controlled diabetes.20, 21 Higher level of A2MG is higher in T2DM49 and was thought that it reflects a degradation of basement membrane of blood vessels indicating the diabetes-associated vascular complications50. |
| Serum amyloid A (SAA) | A major acute phase reactant.36 A polipoprotein of the HDL complex.21 | While SAA is known to be a predictor for coronary artery disease and cardiovascular complications resulting from diabetes, the predictive value of SAA was controversial in diabetes.37, 38 Elevated blood concentration in T2DM. Has been implicated in the development of T2DM and atherosclerosis.64 |
| Serotransferrin (TRFE) | Serotransferrin is responsible for the transport of iron from sites of absorption and heme degradation to those of storage and utilization. Serum transferrin may also have a further role in stimulating cell proliferation.65 | Due to increased glycation of hemoglobin and increased iron metabolism in poorly controlled diabetes, transferrin—acting in transportation of iron and heme degradation—can be found in higher levels in saliva and urine.21, 23, 31, 66 Serotransferrin, along with haptoglobin and hemopexin have shown to be increased in the livers of obese mice.32 Excretion of Serotransferrin has been cited as an indicator of diabetic nephropathy.20,29 |
| Hemopexin (HEMO) | Heme recycle and degradation.67 | Increased iron metabolites result in increased proteins related to iron metabolism, in particular serotranferrin and hemopexin.30 Hemopexin’s involvement in heme degradation means that it will be expressed in higher levels with uncontrolled diabetes.31 Chen et al proposed that the increased hemopexin is a part of the glucose-induced oxidative stress.33–35 They, therefore, suggest that increased hemopexin in the serum may indicate the progression of T1DM that may predict future complications— most importantly retinopathy. |
| Haptoglobin (HPT) | Haptoglobin functions in hepatic recycling by combining with free plasma hemoglobin, which prevents kidney damage. | Haptoglobin 2-2 genotype was linked to obesity, hypertension, and T2DM in some populations39 and was determined to be an important risk factor for T2DM related cardiovascular complications.40–44 Serum haptoglobin in both T1DM and T2DM increases with other serum acute phase proteins.45–48 |
| Complement C3 (CPAMD1) | Following proteolytic degradation of Complement C3, C3a is a mediator of local inflammatory process. C3b can bind to cell surface carbohydrates or immune aggregates. | C3 is linked to T2DM development and seems to correlate with the serum glucose and insulin level.56 Engström et al, 2005 suggested that the increased level of complement C3 is an important risk factor for T2DM. Complement C3 deficient mice seem to be immune to induce T1DM.57 Hertle et al, 2012 pointed out that increased serum complement C3 is a sign of diabetes-associated cardiovascular complications.58 Gao et al 2008 found that complement C3 is increased in patients with diabetic retinopathy.59 |
| Salivary gland originating proteins | Functions | Relation to diabetes |
| Alpha amylase 1 (AMY1) and Amylase 2B (AMY2B) | Binds 1 Calcium ion per subunit.21 Plays a role in the initial digestion of starch.68 | Found to be downregulated in patients with diabetes.21, 23 But alpha amylase was found to be upregulated in type II diabetics.69 Alpha Amylase 2B was found upregulated in diabetics.20 |
| Proteins PLUNC, LPLUNC2 | May be involved in the airway inflammatory response to irritants.70 | Lower levels of concentration detected in saliva of diabetic patients.21 However, in Rao et al, PLUNC was shown to be upregulated in diabetic patients.20 |
| Carbonic Anhydrase 6 (CAH6) | Reversible hydration of carbon dioxide. Undetermined role in saliva.21 | Lower level of expression in saliva of diabetic patients in Border et al.21 Higher level of expression in saliva of diabetic patients in Rao et al.20 |
| Cystatin S (CYTS) | Expressed in saliva, tears, urine, and seminal fluid. Cystatin S has been shown to be an inhibitor of several cysteine proteinases.71 | Found to be upregulated in diabetic subjects.21 |
| Cystatin SN (CYTN) | Expressed in saliva, tears, urine, and seminal fluid. Like Cystatin S, Cystatin SN acts as an inhibitor of several cysteine proteinases, but it has been shown to be a better inhibitor of papain and dipeptidyl peptidase I than Cystatin S.72 | Found to be downregulated in diabetic subjects.21, 23 |
| Prolactin inducible protein (PIP) | Prolactin-inducible protein functions in the regulation of water transport in the apocrine glands.73 PIP has the ability to bind potentially with CD4-T cell receptor, IgG, actin, ZAG, fibronectin and enamel pellicle. 74 | PIP’s binding ability suggests a range of immunological functions.74 |
| Uteroglobulin (UTER) | Secreted from the Clara cells of the pulmonary airways. Binds phosphatidylcholine, phosphatidylinositol, polychlorinated biphenyls (PCB) and weakly progesterone, potent inhibitor of phospholipase A2.75 | Found to be downregulated in Diabetic subjects in Border et al.21 |
| Tissue/mucosal originating proteins | Functions | Relation to Diabetes |
| GAPDH | Participates in nuclear events including transcription, RNA transport, DNA replication and apoptosis. Nuclear functions are probably due to the nitrosylase activity that mediates cysteine S-nitrosylation of nuclear target proteins such as SIRT1, HDAC2 and PRKDC. Glyceraldehyde-3-phosphate dehydrogenase is a key enzyme in glycolysis that catalyzes the first step of the pathway by converting D-glyceraldehyde 3-phosphate (G3P) into 3-phospho-D-glyceroyl phosphate.76, 77 | Upregulated in diabetic subjects.21, 78 |
| Immunoglobulins | Function | Relation to Diabetes |
| IgG | Most prevalent antibody isotype found in the body. IgG is important in fighting infections.79 | Glycosylation of IgG has been shown to affect immune function in Type II diabetes.51 Glycations of immunoglobulins especially serum IgG have been shown to effect the antibody activities in T2DM.51 These glycated IgGs can induce cell neuoronalapoptosis in vitro.52 Glycation of IgG, similar to hemoglobin and albumin, may have caused an increase in the salivary IgG level in poorly controlled diabetes21, 53–55 |
Table 4.
Differential expressed proteins analyzed using individual saliva samples
| Protein ID | Protein function | T1 or T2 | ## peptides w/p-value <0.001<0.001 | # peptides w/p-value <0.01 |
|---|---|---|---|---|
| HBB | Hemoglobin subunit beta OS=Homo sapiens | T1 | 12 | 16 |
| HBA | Hemoblobin subunit alpha OS=Homo sapiens | T1 | 5 | 7 |
| HBD | Hemoglobin subunit delta OS=Homo sapiens | T1 | 1 | 1 |
| ALBU | Serum albumin OS=Homo sapiens | T1 | 5 | 41 |
| CO3 | Complement C3 OS=Homo sapiens | T1 | 5 | 16 |
| A1AG2 | Alpha-1-acid glycoprotein 2 OS=Homo sapiens | T1 | 1 | 1 |
| CF058 | Uncharacterized protein C6orf58 OS=Homo sapiens | T1 | 1 | 1 |
| FETUA | Alpha-2-HS-glycoprotein OS=Homo sapiens | T1 | 2 | |
| A2MG | Alpha-2-macroglobulin OS=Homo sapiens | T1 | 4 | |
| AMY1 | Alpha-amylase 1 OS=Homo sapiens | T1 | 1 | |
| ANXA1 | Annexin A1 OS=Homo sapiens | T1 | 1 | |
| CAH6 | Carbonic Anyhydras 6 OS=Homo sapiens | T1 | 1 | |
| COF1 | Cofilin-1 OS=Homo sapiens | T1 | 1 | |
| CFAB | Complement factor B OS=Homo sapiens | T1 | 1 | |
| FIBG | Firbinogen gamma chain OS=Homo sapiens | T1 | 1 | |
| HSPB1 | Heat shock protein beta-1 OS=Homo sapiens | T1 | 3 | |
| HEMO | Hemopexin OS=Homo sapiens | T1 | 4 | |
| IGHG1 | Ig gamma-1 chain C region OS=Homo sapiens | T1 | 3 | |
| IGHG2 | Ig gamma-2 chain C region OS=Homo sapiens | T1 | 3 | |
| IGHG4 | Ig gamma-4 chain C region OS=Homo sapiens | T1 | 2 | |
| PLAK | Junction plakoglobin OS=Homo sapiens | T1 | 1 | |
| K22O | Keratin, type II cytoskeletal 2 oral OS=Homo sapiens | T1 | 1 | |
| K2C78 | Keratin, type II cystoskeletal 78 OS=Homo sapiens | T1 | 1 | |
| PERL | Lactoperoxidase OS=Homo sapiens | T1 | 1 | |
| MMP9 | Matrix meallopreoteinase-9 OS=Homo sapiens | T1 | 2 | |
| PRTN3 | Myeloblastin OS=Homo sapiens | T1 | 1 | |
| PLSL | Plastin-2 OS=Homo sapiens | T1 | 1 | |
| GP115 | Probable G-protein couples receptor 115 OS=Homo sapiens | T1 | 1 | |
| KPYM | Pyruvate kinase isozymes M1/M2 OS=Homo sapiens | T1 | 1 | |
| TRFE | Serotransferrin OS=Homo sapiens | T1 | 3 | |
| SPRL1 | SPARC-like protein 1 OS=Homo sapiens | T1 | 2 | |
| TBA4A | Tubulin alpha-4A chain OS=Homo sapiens | T1 | 2 | |
| IGHG4 | Ig gamma-4 chain C region OS=Homo sapiens | T2 | 2 | 4 |
| IGHG2 | Ig gamma-2 chain C region OS=Homo sapiens | T2 | 2 | |
| HPT | Haptoglobin OS=Homo sapiens | T2 | 1 | |
| TRFE | Serotransferrin OS=Homo sapiens | T2 | 1 |
Discussion
This study is perhaps the first study to look at the salivary proteomic changes of both T1DM and T2DM based on the individual A1C level. We attempt to address the issue of patient’s compliance in monitoring their serum glucose. We see that saliva, with its low-level of invasiveness and easy accessibility, can be an attractive fluid to use in monitoring glycemic control. This may, therefore, complement current blood glucose monitoring methods and improve patient compliance. Our proteomic analyses showed that the biomarkers can originate from serum, the host immune system, salivary glands, and oral mucosal tissues (Table 3). It is important to note here that several proteins have at least one biological pathway related to diabetes. Measuring the changes of these biomarkers may allow insight into clinical situations of individual patients and may, in the future, provide greater personalized health care for patients. In this study, we separated the subjects into T1DM and T2DM groups and divided them based on HbA1C levels. Note that while the NGSP (National Glycohemoglobin Standardization Program) guidelines recommend HbA1C (and serum glucose) as followed; 6.2–6.8 (110–130) for well controlled group, 6.9–7.3 (130–160) for sufficiently controlled group, 7.4–8.3 for insufficiently controlled, and >8.3 (>160) for poorly controlled group; we chose to analyze the samples here based on the availability of the samples to explore the trend of HbA1C and salivary proteins. Our pooled and individual saliva sample proteomic analyses provide a proof-of-principle that there is a global proteomic change in saliva based on different A1C levels. The salivary proteomes appear to be distinct when compared with low, medium and high A1C levels. Proteomic changes based on A1C, as seen in PCA and cluster analysis, are stronger in T1DM than T2DM. This is not surprising since most patients with T2DM are older and often have several other health conditions, e.g. obesity, high blood pressure, cardiovascular disease, etc. We, therefore, will focus our discussion on T1DM A1C associated protein biomarkers and later give comments on the T2DM. We performed label-free quantitative proteomics of pooled and individual samples to examine the higher abundant proteins as well as the lower abundant proteins. The pooled sample analysis allows a global examination and naturally averages all the confounding effects in each group. However, pooled sample analyses can lead to missing important biomarkers with a relatively low level of expression. While there is little trend in the expression of salivary peptides to A1C level in the analysis of pooled samples (Figure 2a–b), it is clear that some peptides such as AHNK2 are highly expressed in group E (compared to other groups). Note that while the proteomic analysis suggested changes in HEMO, TRFE, CYTT or CYTN, only changes in ALB and AMY1 expression can be definitively confirmed by Western analysis of the pooled samples (Figure 1C). The inconsistency of proteomic analysis and antibody-based assays is common in this type of study.
Individual sample analysis reveals several protein biomarkers, including serum albumin (ALB), cystatin-SA (CYTT), cystatin-SN (CYTN), and alpha-amylase 1 (AMY1), similar to the pooled sample analysis. We also saw other important biomarkers from other studies, including hemoglobin (Hb), serotransferrin (TRFE), alpha-2-macroglobulin (A2MG), hemopexin (HEMO), PLUNC protein, carbonic anhydrase 6 (CAH6), uteroglobulin, PIP, cystatin-S (CYTS), and GAPDH (Table 3). Our analysis demonstrate changes in the salivary proteomic level of important serum originating proteins including hemoglobin (Hb), albumin (ALB), hemopexin (HEMO), haptoglobin (HPT), serotransferrin (TRFE), alpha-2-macroglobulin (A2MG), complement C3 (CO3) and serum amyloid proteins (SAA), associated with increased A1C in subjects with diabetes, especially T1DM (Table 3). Prolonged elevation of serum glucose can lead to glycation of serum proteins. The glycation modification increases protein’s local hydrophilicity and, therefore, destabilizes the protein by making the globular protein unfold and, therefore, prone to enzymatic degradation25, 26. These serum proteins play a critical role in oxidative pathway and iron/heme metabolism (see details in Table 3). Interestingly, there are also alterations in the expression of several immunoglobulin fragments (Figure 2a–b).
Besides the serum originating proteins and immunoglobulin fragments, our proteomic results are similar to our previous finding in edentulous T2DM21 that hyperglycemia resulting from diabetes can alter the expression of salivary proteins including salivary amylases (AMY-1), BPI or PLUNC family proteins, CAH6, cystatins, PIP, UTER, and GAPDH. We previously proposed that prolonged hyperglycemia could inhibit protein production of salivary glands through oxidative stress or disruption of enzymes/proteins essential to salivary protein production due to glycation21. Main candidates from salivary gland originating proteins associated with A1C changes include AMY-1, PLUNC protein, and Cystatin-S family proteins. This suggests that salivary gland originating proteins related to digestion, innate immunity, and protease inhibition show signs of lower expression in relation to higher A1C level. We believe that individual subjects may have different baselines for each salivary protein and therefore intra-subject evaluation in a longitudinal manner is needed for future verification of identified potential HbA1C-related salivary proteins.
Conclusion
Diabetes requires long-term or lifelong serum glucose monitoring. Use of blood samples hinders the monitoring of glycemic control. Saliva proteins may be used to supplement current methods to improve patient compliance. There is a potential for developing an antibody-based tool to monitor changes in certain salivary proteins. A prospective longitudinal study with a larger population comparing the salivary protein biomarkers with known serum markers will be needed.
Materials & Methods
Subject recruitment and sample collection
The subject recruitment process and study protocol was approved by the Office of Human Research Ethics, the University of North Carolina (UNC) Institutional Review Board (IRB), No. 10-0492. All subjects were recruited from the UNC Diabetes Care Center and were given a written informed consent for saliva sample collection, storage, and analysis for this study. Subjects were screened to minimize oral and systemic confounding factors (Supplementary Table 1). There were 56 T1DM and 97 T2DM. Patient population includes 77 males and 76 females, ages ranging from 19 to 79 (Supplementary Table 2 and 3). The sample collection protocol was similar to our previous study21. Briefly, subjects were asked to refrain from drinking, eating, and practicing any oral hygiene habits 2 hours before sample collection. They were also asked to complete the sample collection within 30 min between 9 am to 12 pm (before lunch time). The subject was asked to spit into 15 ml falcon tube within 30–60 minutes to provide about 4 ml of saliva samples. The falcon tube was then placed in a centrifuge at 4,000 rpm at 4C for 15 minutes to remove food debris. About 750 ul of the supernatant was then aliquoted into a 1.5 ml cryotube. The tube was then fast frozen in liquid nitrogen and placed in −80C for storage prior to analysis. The subjects were grouped based on their levels of A1C; <7 (A), 7–8 (B), 8–9 (C), 9–10 (D) and >10 (E); and their type of diabetes (1 or 2). For example Group A1 refers to type 1 diabetes with A1C<7. This grouping was done arbitrarily to achieve even distribution of subjects. Samples were analyzed two times as pooled samples and as individual samples. Pooled samples were aliquoted from all of the samples in each group. The E1 group was the smallest group of 4 subjects. For the individual analysis, we, therefore, selected four subjects from each group. Demographic data were used to match subjects within the same type of diabetes.
Sample preparation and Mass Spectrometry Analysis
Similar protocol described by Border et al, 201221 and Al-Tarawneh et, 201124, was used for both pooled and individual sample analyses. The total concentration of protein in each sample was determined using the Thermo Scientific Micro BCA Protein Assay kit. The concentration of the sample was adjusted to the working range of 5–200 mg/ml based on absorbance values compared to a BSA standard curve. A volume of each sample corresponding to 35μg of protein (based on the protein quantitation results) was used. The sample volumes were made equal by adding 50mM ammonium bicarbonate to a volume of 29.8 μL. A 1% solution of Rapigest was added to each sample to denature the proteins, and the mixture was placed in a shaking heated mixer at 40°C for 10 minutes. Disulfide bonds were reduced by adding 200mM dithiothreitol (DTT) to each sample and heating the tubes to 80°C for 15 minutes. Free sulfur atoms were alkylated with 400mM iodoacetamide (IA) by placing the tubes in the dark for 30 minutes at room temperature. A tryptic digest was performed by adding 0.7μg Gold-Mass Spectrometry grade Trypsin to each tube and incubating at 37°C overnight. Alcohol dehydrogenase (ADH) digest from yeast was added to a final concentration of 50 fmol/μg protein. The trypsin reaction was stopped and the Rapigest was degraded with the addition of 10% TFA/20% acetonitrile/70% water that was then heated to 60°C for 2 hours. The samples were centrifuged and the supernatant pipetted into autosampler vials. The processed samples were analyzed on a Thermo Scientific LTQ Orbitrap XL mass spectrometry system coupled to a Waters nanoACQUITY UPLC system. Peptides were separated on a Waters nanoACQUITY UPLC Column (1.7 μm BEH 130 C18, 75 μm × 250 mm) using a linear gradient from 5 to 60% B over 60 min and then from 60 to 95% B over 5 min where A is 99.9/0.1 water/formic acid and B is 99.9/0.1 acetonitrile/formic acid. Mass spectra were acquired using Data Dependent scans (Nth order double play) on the LTQ Orbitrap XL system over 90 min.
The study sequence consisted of the study sample injections bracketed by a pair of QC injections. Data from all study samples were acquired using Data Dependent™ scans (Nth order double play) on the LTQ Orbitrap XL. Database searches were performed in Elucidator (Rosetta Biosoftware) using MASCOT (Matrix Sciences, London, UK). Analytical results were also viewed in Scaffold (Proteome Software, Portland, OR). QC and study samples were evaluated to confirm data quality. Liquid chromatography total ion current (TIC) outputs were assessed for signal quality and changes in signal intensity. Results were also monitored for signal trends, such as a consistent increase or decrease in TIC maximum values, and MASCOT search results were used to monitor the quality of the mass spectrometry (MS) data.
Raw MS data files for the study samples, collected on the Thermo Orbitrap XL system, were processed in Elucidator. MS data was grouped in Elucidator based on sample group (Type and A1C level) and aligned. Sample groups are used to assist in data alignment, feature identification, and can be utilized for QC assessment and group comparisons. Thermo Orbitrap data files were searched using the Mascot search engine against the SwissProt human/candida database (appended with yeast ADH, March 23, 2012 for pooled samples and August 3, 2012 for individual samples). The aligned mass features were annotated with these database search results using the results from the system Peptide Tellers and a predicted error rate of 1%. MS data were summarized to the feature level, normalized, and an error-weighted ANOVA test was performed to compare the expression results between sample groups. Candidate differentially expressed markers were determined based on a p<0.05. Features were summarized by peptide based on the results of the database search. Principle component analyses and cluster analyses were performed, first for the low (A), medium (C), and high (E) A1C groups in each type of diabetes, then for all groups with each type of diabetes.
Western blot analysis
Pooled saliva samples from each group were used. These individual samples for western analysis are not included in the proteomic study. Each sample was centrifuged for 15 min at 13,000 rpm at 4°C to remove debris and precipitated proteins. The total protein content in the clear supernatant was measured using a Bradford assay with bovine serum albumin as a standard. Supernatants in total of ~2–4 mg of protein per gel lane were subjected to SDS-PAGE on a 4–15% gradient gel. We chose to use the same amount of volume in each lane (rather than the same amount of total protein) to evaluate the absolute expression difference. We used the same volume instead of the same amount of protein because of varying availability of each protein in saliva sample. Normally, our sample has about 1–5 mg/ml of proteins. 2 mg concentration worked well with majority of high abundant proteins, e.g. AMY, ALB, etc. For most serum proteins except for ALB, we have to increase the total load of proteins to 4 mg to see any bands in western analysis. The separated proteins were transferred to nitrocellulose membrane, and the membranes were probed with serum albumin, Cystatin SA, SN, anti-human PLUNC monoclonal mouse IgG2B antibody (R&D Systems), anti-human salivary amylase sheep polyclonal antibody (Abcam), respectively. Horseradish peroxidase-conjugated goat polyclonal secondary antibody to mouse IgG (Abcam) and horseradish peroxidase-conjugated rabbit polyclonal secondary antibody to sheep IgG (Abcam) were, respectively, used at a 1:5000 dilution to detect PLUNC and AMY-1. Enhanced chemiluminescence (Amersham Biosciences, Inc.) was used to visualize detected bands.
Supplementary Material
Acknowledgments
This work was partly supported by the National Institute of Health (NIH) grant HL092338 (to S.B.), the North Carolina Translational and Clinical Sciences Institute Pilot grants (supported by Award Number UL1RR025747 from the National Center for Research Resources); 2KR80905 (to H.K) and 10KR81002 (to S.B.), and the University of North Carolina Junior Faculty Development Award (to S.B.). The authors thank David Barrow, Ivandario Saldarriaga, Aiden Vu, members of the University of North Carolina Diabetes Care Center, and General-Oral Health Clinic for subject recruitment, sample collection, and experimental assistance.
Footnotes
Author Contributions
Authors SB, SSB, JC, SO, ELHW, MCW and JBB played a major role in experimental designs. Authors JC, WCB, MVM, MBB, HK and SB wrote human subject approval, recruited subjects, collected samples and prepared samples for mass spectrometry analysis. Authors SSB and JC performed mass spectrometry experiments. Authors SSB, JC, SB, WCB, and MBB analyzed the mass spectrometry analysis. Authors EE and MCW did statistical analysis. Authors MVM and SB performed western blot analysis. Authors SB and WCB wrote, organized and prepared the manuscript for publication. All authors contributed to and approved the present manuscript.
Conflict of Interest
The authors declare no financial conflict of interest.
References
- 1.Gallagher EJ, Le Roith D, Bloomgarden Z. J Diabetes. 2009;1:9–17. doi: 10.1111/j.1753-0407.2009.00009.x. [DOI] [PubMed] [Google Scholar]
- 2.Goldstein DE, Little RR, Lorenz RA, Malone JI, Nathan D, Peterson CM, Sacks DB. Diabetes Care. 2004;27:1761–1773. doi: 10.2337/diacare.27.7.1761. [DOI] [PubMed] [Google Scholar]
- 3.Harris MI. Diabetes Care. 2001;24:979–982. doi: 10.2337/diacare.24.6.979. [DOI] [PubMed] [Google Scholar]
- 4.Hempe JM, Gomez R, McCarter RJ, Jr, Chalew SA. J Diabetes Complications. 2002;16:313–320. doi: 10.1016/s1056-8727(01)00227-6. [DOI] [PubMed] [Google Scholar]
- 5.Nathan DM, Buse JB, Davidson MB, Ferrannini E, Holman RR, Sherwin R, Zinman B. Diabetologia. 2009;52:17–30. doi: 10.1007/s00125-008-1157-y. [DOI] [PubMed] [Google Scholar]
- 6.Pugliese A, Reijonen HK, Nepom J, Burke GW., 3rd Diabetes Manag (Lond) 2011;1:229–238. doi: 10.2217/dmt.10.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weir GC, Cavelti-Weder C, Bonner-Weir S. Genome Med. 2011;3:61. doi: 10.1186/gm277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Riccardi G, Vaccaro O, Rivellese A, Pignalosa S, Tutino L, Mancini M. Am J Epidemiol. 1985;121:422–429. doi: 10.1093/oxfordjournals.aje.a114014. [DOI] [PubMed] [Google Scholar]
- 9.Bennett CM, Guo M, Dharmage SC. Diabet Med. 2007;24:333–343. doi: 10.1111/j.1464-5491.2007.02106.x. [DOI] [PubMed] [Google Scholar]
- 10.Buell C, Kermah D, Davidson MB. Diabetes Care. 2007;30:2233–2235. doi: 10.2337/dc07-0585. [DOI] [PubMed] [Google Scholar]
- 11.Cheng YJ, Gregg EW, Geiss LS, Imperatore G, Williams DE, Zhang X, Albright AL, Cowie CC, Klein R, Saaddine JB. Diabetes Care. 2009;32:2027–2032. doi: 10.2337/dc09-0440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Peters AL, Davidson MB, Schriger DL, Hasselblad V. JAMA. 1996;276:1246–1252. [PubMed] [Google Scholar]
- 13.Rohlfing CL, Little RR, Wiedmeyer HM, England JD, Madsen R, Harris MI, Flegal KM, Eberhardt MS, Goldstein DE. Diabetes Care. 2000;23:187–191. doi: 10.2337/diacare.23.2.187. [DOI] [PubMed] [Google Scholar]
- 14.Selvin E, Steffes MW, Gregg E, Brancati FL, Coresh J. Diabetes Care. 2011;34:84–89. doi: 10.2337/dc10-1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pickup JC, Freeman SC, Sutton AJ. BMJ. 2011;343:d3805. doi: 10.1136/bmj.d3805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ziegler R, Heidtmann B, Hilgard D, Hofer S, Rosenbauer J, Holl R. Pediatr Diabetes. 2011;12:11–17. doi: 10.1111/j.1399-5448.2010.00650.x. [DOI] [PubMed] [Google Scholar]
- 17.Ellis DA, Frey MA, Naar-King S, Templin T, Cunningham P, Cakan N. Diabetes Care. 2005;28:1604–1610. doi: 10.2337/diacare.28.7.1604. [DOI] [PubMed] [Google Scholar]
- 18.Mazze RS, Shamoon H, Pasmantier R, Lucido D, Murphy J, Hartmann K, Kuykendall V, Lopatin W. Am J Med. 1984;77:211–217. doi: 10.1016/0002-9343(84)90693-4. [DOI] [PubMed] [Google Scholar]
- 19.Kitis Y, Emiroglu ON. Appl Nurs Res. 2006;19:134–143. doi: 10.1016/j.apnr.2005.07.007. [DOI] [PubMed] [Google Scholar]
- 20.Rao PV, Reddy AP, Lu X, Dasari S, Krishnaprasad A, Biggs E, Roberts CT, Nagalla SR. J Proteome Res. 2009;8:239–245. doi: 10.1021/pr8003776. [DOI] [PubMed] [Google Scholar]
- 21.Border MB, Schwartz S, Carlson J, Dibble CF, Kohltfarber H, Offenbacher S, Buse JB, Bencharit S. Mol Biosyst. 2012;8:1304–1310. doi: 10.1039/c2mb05079j. [DOI] [PubMed] [Google Scholar]
- 22.Cabras T, Pisano E, Mastinu A, Denotti G, Pusceddu PP, Inzitari R, Fanali C, Nemolato S, Castagnola M, Messana I. Mol Cell Proteomics. 2010;9:2099–2108. doi: 10.1074/mcp.M110.001057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hirtz C, Chevalier F, Sommerer N, Raingeard I, Bringer J. M Rossignol and D de Périere Clin Proteomics. 2006;2:117–127. [Google Scholar]
- 24.Al-Tarawneh SK, Border MB, Dibble CF, Bencharit S. OMICS. 2011;15:353–361. doi: 10.1089/omi.2010.0134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cussimanio BL, Booth AA, Todd P, Hudson BG, Khalifah RG. Biophys Chem. 2003;105:743–755. doi: 10.1016/s0301-4622(03)00100-5. [DOI] [PubMed] [Google Scholar]
- 26.Sen S, Kar M, Roy A, Chakraborti AS. Biophys Chem. 2005;113:289–298. doi: 10.1016/j.bpc.2004.05.005. [DOI] [PubMed] [Google Scholar]
- 27.Vetter SW, Indurthi VS. Clin Chim Acta. 2011;412:2105–2116. doi: 10.1016/j.cca.2011.07.022. [DOI] [PubMed] [Google Scholar]
- 28.Fernandez-Real JM, Penarroja G, Castro A, Garcia-Bragado F, Hernandez-Aguado I, Ricart W. Diabetes. 2002;51:1000–1004. doi: 10.2337/diabetes.51.4.1000. [DOI] [PubMed] [Google Scholar]
- 29.Bernard A, Amor AO, Goemare-Vanneste J, Antoine JL, Lauwerys R, Colin I, Vandeleene B, Lambert A. Clin Chim Acta. 1990;190:249–262. doi: 10.1016/0009-8981(90)90178-u. [DOI] [PubMed] [Google Scholar]
- 30.Van Campenhout A, Van Campenhout C, Lagrou AR, Abrams P, Moorkens G, Van Gaal L, Manuel-y-Keenoy B. Diabetes Metab Res Rev. 2006;22:444–454. doi: 10.1002/dmrr.635. [DOI] [PubMed] [Google Scholar]
- 31.Varghese SA, Powell TB, Budisavljevic MN, Oates JC, Raymond JR, Almeida JS, Arthur JM. J Am Soc Nephrol. 2007;18:913–922. doi: 10.1681/ASN.2006070767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang X, Choi JW, Oh TS, Choi DK, Mukherjee R, Liu H, Yun JW. Proteomics. 2012;12:284–299. doi: 10.1002/pmic.201100271. [DOI] [PubMed] [Google Scholar]
- 33.Chen CC, Lu YC, Chen YW, Lee WL, Lu CH, Chen YH, Lee YC, Lin ST, Timms JF, Lee YR, Chou HC, Chan HL. J Proteomics. 2012;75:3760–3777. doi: 10.1016/j.jprot.2012.04.047. [DOI] [PubMed] [Google Scholar]
- 34.Trougakos IP, Gonos ES. Free Radic Res. 2006;40:1324–1334. doi: 10.1080/10715760600902310. [DOI] [PubMed] [Google Scholar]
- 35.Kim JH, Jun HO, Yu YS, Min BH, Park KH, Kim KW. Invest Ophthalmol Vis Sci. 2010;51:561–566. doi: 10.1167/iovs.09-3774. [DOI] [PubMed] [Google Scholar]
- 36.Urieli-Shoval S, Linke RP, Matzner Y. Curr Opin Hematol. 2000;7:64–69. doi: 10.1097/00062752-200001000-00012. [DOI] [PubMed] [Google Scholar]
- 37.Johnson BD, Kip KE, Marroquin OC, Ridker PM, Kelsey SF, Shaw LJ, Pepine CJ, Sharaf B, Bairey Merz CN, Sopko G, Olson MB, Reis SE. Circulation. 2004;109:726–732. doi: 10.1161/01.CIR.0000115516.54550.B1. [DOI] [PubMed] [Google Scholar]
- 38.Helmersson J, Vessby B, Larsson A, Basu S. Circulation. 2004;109:1729–1734. doi: 10.1161/01.CIR.0000124718.99562.91. [DOI] [PubMed] [Google Scholar]
- 39.Quaye IK, Ababio G, Amoah AG. J Atheroscler Thromb. 2006;13:90–94. doi: 10.5551/jat.13.90. [DOI] [PubMed] [Google Scholar]
- 40.Levy AP, Roguin A, Hochberg I, Herer P, Marsh S, Nakhoul FM, Skorecki K. N Engl J Med. 2000;343:969–970. doi: 10.1056/NEJM200009283431313. [DOI] [PubMed] [Google Scholar]
- 41.Levy AP, Hochberg I, Jablonski K, Resnick HE, Lee ET, Best L, Howard BV. J Am Coll Cardiol. 2002;40:1984–1990. doi: 10.1016/s0735-1097(02)02534-2. [DOI] [PubMed] [Google Scholar]
- 42.Roguin A, Koch W, Kastrati A, Aronson D, Schomig A, Levy AP. Diabetes Care. 2003;26:2628–2631. doi: 10.2337/diacare.26.9.2628. [DOI] [PubMed] [Google Scholar]
- 43.Suleiman M, Aronson D, Asleh R, Kapeliovich MR, Roguin A, Meisel SR, Shochat M, Sulieman A, Reisner SA, Markiewicz W, Hammerman H, Lotan R, Levy NS, Levy AP. Diabetes. 2005;54:2802–2806. doi: 10.2337/diabetes.54.9.2802. [DOI] [PubMed] [Google Scholar]
- 44.Milman U, Blum S, Shapira C, Aronson D, Miller-Lotan R, Anbinder Y, Alshiek J, Bennett L, Kostenko M, Landau M, Keidar S, Levy Y, Khemlin A, Radan A, Levy AP. Arterioscler Thromb Vasc Biol. 2008;28:341–347. doi: 10.1161/ATVBAHA.107.153965. [DOI] [PubMed] [Google Scholar]
- 45.Ganrot PO, Gydell K, Ekelund H. Acta Endocrinol (Copenh) 1967;55:537–544. doi: 10.1530/acta.0.0550537. [DOI] [PubMed] [Google Scholar]
- 46.Jonsson A, Wales JK. Diabetologia. 1976;12:245–250. doi: 10.1007/BF00422091. [DOI] [PubMed] [Google Scholar]
- 47.McMillan DE. Metabolism. 1989;38:1042–1046. doi: 10.1016/0026-0495(89)90038-3. [DOI] [PubMed] [Google Scholar]
- 48.Asleh R, Guetta J, Kalet-Litman S, Miller-Lotan R, Levy AP. Circ Res. 2005;96:435–441. doi: 10.1161/01.RES.0000156653.05853.b9. [DOI] [PubMed] [Google Scholar]
- 49.James K, Merriman J, Gray RS, Duncan LJ, Herd R. J Clin Pathol. 1980;33:163–166. doi: 10.1136/jcp.33.2.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Brownlee M. Lancet. 1976;1:779–780. doi: 10.1016/s0140-6736(76)91614-7. [DOI] [PubMed] [Google Scholar]
- 51.Kaneshige H. Diabetes. 1987;36:822–828. doi: 10.2337/diab.36.7.822. [DOI] [PubMed] [Google Scholar]
- 52.Srinivasan S, Stevens MJ, Sheng H, Hall KE, Wiley JW. J Clin Invest. 1998;102:1454–1462. doi: 10.1172/JCI2793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tenovuo J, Lehtonen OP, Viikari J, Larjava H, Vilja P, Tuohimaa P. J Dent Res. 1986;65:62–66. doi: 10.1177/00220345860650011101. [DOI] [PubMed] [Google Scholar]
- 54.Ardawi MS, Nasrat HA, Bahnassy AA. Diabet Med. 1994;11:384–387. doi: 10.1111/j.1464-5491.1994.tb00290.x. [DOI] [PubMed] [Google Scholar]
- 55.Rodriguez-Segade S, Camina MF, Paz JM, Del Rio R. Clin Chim Acta. 1991;203:135–142. doi: 10.1016/0009-8981(91)90285-k. [DOI] [PubMed] [Google Scholar]
- 56.Engstrom G, Hedblad B, Eriksson KF, Janzon L, Lindgarde F. Diabetes. 2005;54:570–575. doi: 10.2337/diabetes.54.2.570. [DOI] [PubMed] [Google Scholar]
- 57.Gao X, Liu H, Ding G, Wang Z, Fu H, Ni Z, Ma J, Liu F, Fu Z. Clin Immunol. 2011;140:236–243. doi: 10.1016/j.clim.2011.02.004. [DOI] [PubMed] [Google Scholar]
- 58.Hertle E, van Greevenbroek MM, Stehouwer CD. Diabetologia. 2012;55:881–884. doi: 10.1007/s00125-012-2462-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gao BB, Chen X, Timothy N, Aiello LP, Feener EP. J Proteome Res. 2008;7:2516–2525. doi: 10.1021/pr800112g. [DOI] [PubMed] [Google Scholar]
- 60.Lu J, Stewart AJ, Sadler PJ, Pinheiro TJ, Blindauer CA. Biochem Soc Trans. 2008;36:1317–1321. doi: 10.1042/BST0361317. [DOI] [PubMed] [Google Scholar]
- 61.Faure P, Wiernsperger N, Polge C, Favier A, Halimi S. Clin Sci (Lond) 2008;114:251–256. doi: 10.1042/CS20070276. [DOI] [PubMed] [Google Scholar]
- 62.Dolhofer R, Wieland OH. FEBS Lett. 1979;103:282–286. doi: 10.1016/0014-5793(79)81345-9. [DOI] [PubMed] [Google Scholar]
- 63.Selvin E, Marinopoulos S, Berkenblit G, Rami T, Brancati FL, Powe NR, Golden SH. Ann Intern Med. 2004;141:421–431. doi: 10.7326/0003-4819-141-6-200409210-00007. [DOI] [PubMed] [Google Scholar]
- 64.Walder K, Kantham L, McMillan JS, Trevaskis J, Kerr L, De Silva A, Sunderland T, Godde N, Gao Y, Bishara N, Windmill K, Tenne-Brown J, Augert G, Zimmet PZ, Collier GR. Diabetes. 2002;51:1859–1866. doi: 10.2337/diabetes.51.6.1859. [DOI] [PubMed] [Google Scholar]
- 65.Konen J, Shihabi Z, Newman J. Am J Kidney Dis. 1993;22:791–797. doi: 10.1016/s0272-6386(12)70336-0. [DOI] [PubMed] [Google Scholar]
- 66.Li RX, Chen HB, Tu K, Zhao SL, Zhou H, Li SJ, Dai J, Li QR, Nie S, Li YX, Jia WP, Zeng R, Wu JR. PLoS One. 2008;3:e3224. doi: 10.1371/journal.pone.0003224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ratra R, Kar-Roy A, Lal SK. Biochemistry. 2008;47:1957–1969. doi: 10.1021/bi7016552. [DOI] [PubMed] [Google Scholar]
- 68.Ramasubbu N, Paloth V, Luo Y, Brayer GD, Levine MJ. Acta Crystallogr D Biol Crystallogr. 1996;52:435–446. doi: 10.1107/S0907444995014119. [DOI] [PubMed] [Google Scholar]
- 69.Aydin S. J Biochem Mol Biol. 2007;40:29–35. doi: 10.5483/bmbrep.2007.40.1.029. [DOI] [PubMed] [Google Scholar]
- 70.Lindahl M, Stahlbom B, Tagesson C. Electrophoresis. 2001;22:1795–1800. doi: 10.1002/1522-2683(200105)22:9<1795::AID-ELPS1795>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 71.Isemura S, Saitoh E, Ito S, Isemura M, Sanada K. J Biochem. 1984;96:1311–1314. doi: 10.1093/oxfordjournals.jbchem.a134952. [DOI] [PubMed] [Google Scholar]
- 72.Isemura S, Saitoh E, Sanada K. FEBS Lett. 1986;198:145–149. doi: 10.1016/0014-5793(86)81201-7. [DOI] [PubMed] [Google Scholar]
- 73.Mazoujian G, Pinkus GS, Davis S, Haagensen DE., Jr Am J Pathol. 1983;110:105–112. [PMC free article] [PubMed] [Google Scholar]
- 74.Hassan MI, Waheed A, Yadav S, Singh TP, Ahmad F. Cell Mol Life Sci. 2009;66:447–459. doi: 10.1007/s00018-008-8463-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Matsunaga A, Numakura C, Kawakami T, Itoh Y, Kawabata I, Masakane I, Suzuki T, Suzuki M, Goto T, Itoh K, Hayasaka K. Am J Kidney Dis. 2002;39:36–41. doi: 10.1053/ajkd.2002.29875. [DOI] [PubMed] [Google Scholar]
- 76.Tisdale EJ. J Biol Chem. 2002;277:3334–3341. doi: 10.1074/jbc.M109744200. [DOI] [PubMed] [Google Scholar]
- 77.Ercolani L, Florence B, Denaro M, Alexander M. J Biol Chem. 1988;263:15335–15341. [PubMed] [Google Scholar]
- 78.Hittel DS, Hathout Y, Hoffman EP, Houmard JA. Diabetes. 2005;54:1283–1288. doi: 10.2337/diabetes.54.5.1283. [DOI] [PubMed] [Google Scholar]
- 79.Janeway CA, Jr, Travers P, Walport M. Garland Science. 5. New York: 2001. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.