Abstract
Clarin-1 (CLRN1) is the causative gene in Usher Syndrome type 3A, an autosomal recessive disorder characterized by progressive vision and hearing loss. CLRN1 encodes Clarin-1, a glycoprotein with homology to the tetraspanin family of proteins. Previous cell culture studies suggest that Clarin-1 localizes to the plasma membrane and interacts with the cytoskeleton. Mouse models demonstrate a role for the protein in mechanosensory hair bundle integrity, but the function of Clarin-1 in hearing remains unclear. Even less is known of its role in vision, because the Clrn1 knockout mouse does not exhibit a retinal phenotype and expression studies in murine retinas have provided conflicting results. Here, we describe cloning and expression analysis of the zebrafish clrn1 gene, and report protein localization of Clarin-1 in auditory and visual cells from embryonic through adult stages. We detect clrn1 transcripts as early as 24 hours post-fertilization, and expression is maintained through adulthood. In situ hybridization experiments show clrn1 transcripts enriched in mechanosensory hair cells and supporting cells of the inner ear and lateral line organ, photoreceptors, and cells of the inner retina. In mechanosensory hair cells, Clarin-1 is polarized to the apical cell body and the synapses. In the retina, Clarin-1 localizes to lateral cell contacts between photoreceptors and is associated with the outer limiting membrane and subapical processes emanating from Müller glial cells. We also find Clarin-1 protein in the outer plexiform, inner nuclear and ganglion cell layers of the retina. Given the importance of Clarin-1 function in the human retina, it is imperative to find an animal model with a comparable requirement. Our data provide a foundation for exploring the role of Clarin-1 in retinal cell function and survival in a diurnal, cone-dominant species.
1. Introduction
Usher syndrome (USH) is the most common hereditary form of combined deafness and blindness. To date, 11 different genes have been implicated as causative of USH (Bonnet and El Amraoui, 2012, Riazuddin et al, 2012, Puffenberger et al, 2012). Although the acquisition rate for molecular data has accelerated over the past decade, the mechanisms of USH pathology remain mysterious. The complex genetic profile of USH may explain its clinical heterogeneity: Usher patients are typically categorized into one of three clinical subtypes, USH1, USH2, or USH3, based on variations in the age of onset and severity of auditory and visual symptoms. Mutations in the CLRN1 gene cause Usher type 3A, unique among the USH subtypes for its high degree of symptomatic variability. Unlike USH1 and USH2, where hearing impairment is congenital and usually stable, USH3 patients experience hearing loss that worsens over time. The age of onset of this progressive hearing loss is variable, but occurs postlingually in all documented cases. Vision loss due to retinal degeneration is also progressive in USH3 patients, but neither the onset nor rate of degeneration is temporally correlated with the hearing loss.
Clarin-1 is a four-pass membrane domain protein with homology to the tetraspanin family of proteins (Adato et al, 2002), which include Connexins and Claudins, known to cause deafness in humans when mutated (Duman and Tekin, 2012). Cell culture experiments with HA-tagged Clarin-1 have shown trafficking to the plasma membrane (Isosomppi et al., 2009) and interactions with the actin cytoskeleton (Tian et al., 2009, Geng et al., 2012), a relationship consistent with the disrupted morphology of actin-based hair bundles in the cochleae of Clrn1 mutant mice (Geng et al., 2009, 2012). In mouse tissues (Zallocchi et al., 2009, 2012), Clarin-1 localization is reported in cochleae at hair cell synapses, in afferent neurons, and transiently in stereocilia. Retinal localization is restricted to photoreceptor cell synapses and the base of the connecting cilium. In disagreement with the previous cell culture study showing association between Clarin-1 and actin (Isosomppi et al. 2009), Zallocchi et al. instead reported Clarin-1 colocalized with microtubules, possibly indicating a role in trans-Golgi vesicle trafficking.
Based on the clinical symptoms of USH3A, it is apparent that Clarin-1 plays an important role in human vision, yet mouse knockouts of Clrn1 do not show a retinal phenotype at any point in their lives. Mice have rod-dominated vision conducive to low light conditions, and although night blindness caused by rod degeneration is often the first symptom of vision loss in human Usher syndrome, this is not always the case with USH3 patients (Pakarinen et al., 1995). Clarin-1 may be required for proper cone function or cone connections and interactions with other retinal cell types in retinas specialized for daylight conditions. Investigating the molecular functions of Clarin-1 in an alternative model system with cone-dominant vision is required for furthering our understanding of USH3A pathology. Here, we report expression and localization of the zebrafish ortholog of CLRN1 in both the inner ear and cone-dominant retina.
Results
2.1 Cloning and expression analysis of zebrafish clrn1
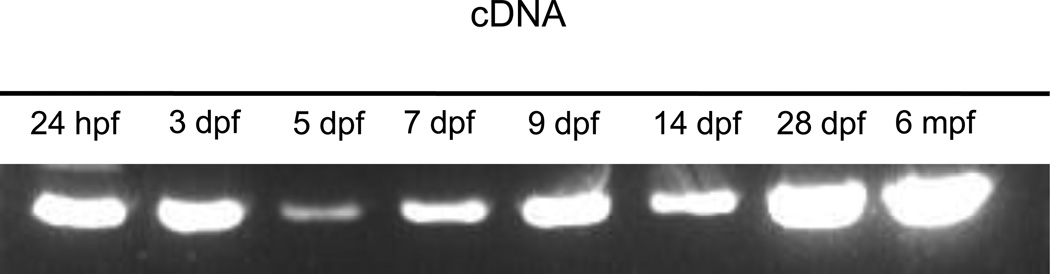
We used the human Clarin-1 sequence in a protein BLAST search to identify the zebrafish ortholog of CLRN1. We discovered a single gene on zebrafish chromosome 15, composed of three exons and encoding a 232 amino acid protein with 61% identity and 77% similarity to human Clarin-1. Residue homology is highest in the extracellular loops and at the intracellular C-terminus, and lowest in the transmembrane domains, suggesting conservation of exposed regions that may function in protein-protein interactions. Although multiple CLRN1 splice variants have been reported in human (Västinsalo et al, 2010), we isolated only a single transcript of zebrafish clrn1 corresponding to the main human splice variant, despite extensive 5’ and 3’ RACE experiments to identify additional isoforms. We designed primers in the 5’ UTR and at the 3’ end of the predicted coding sequence for RT-PCR amplification. We amplified a single band of approximately 700 base pairs in cDNA templates from animals ranging in age from 24 hours to 28 days post-fertilization (dpf), as well as from adult eye (Fig. 1). Sequencing confirmed that all fragments corresponded to the full-length clrn1 transcript.
Fig. 1.
clrn1 transcript is detectable from embryonic to adult stages in zebrafish by RT-PCR. cDNA sequences were reverse transcribed from total RNA isolate from whole fish between 24hpf and 28dpf and from adult retinas. cDNA was amplified with primers specific to the full-length clrn1 transcript with 35 cycles of PCR. Equal volumes of the PCR reactions were loaded into each lane on a 1% agarose gel containing Ethidium Bromide. Bands were visualized with ultraviolet light. Each lane revealed a single band of 702 base pairs. Bands were subsequently extracted and purified using a Qiagen Gel extraction kit. The resulting product was sequenced and confirmed to be the full-length clrn1 transcript.
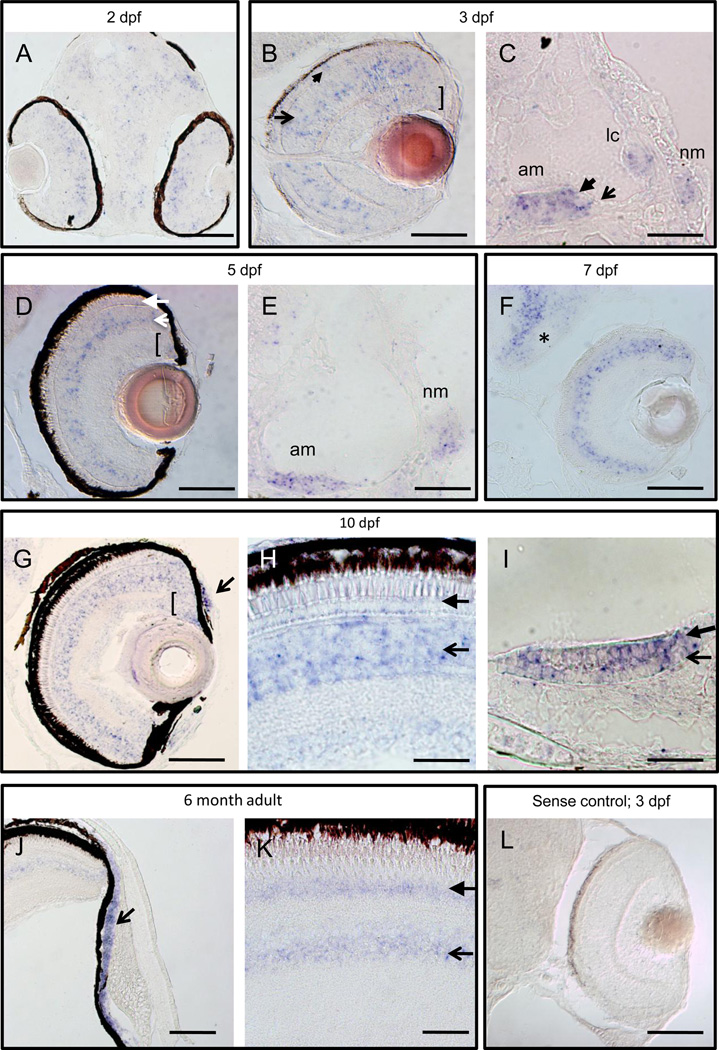
We cloned full-length clrn1 from 5 dpf cDNA into the pCR®-blunt II-Topo Vector and used it to generate an antisense RNA probe that we applied to sectioned tissue for in situ hybridization. clrn1 transcripts were expressed in sensorineural cells, consistent with previously published data on other zebrafish Usher genes (Ernest et al., 2000; Söllner et al., 2004; Seiler et al., 2005; Phillips et al., 2011). We observed weak expression in the eye, ear, and brain tissue of 2 dpf embryos (Fig. 2a and data not shown). By 3 dpf, cells in the outer and inner nuclear layers (ONL and INL, respectively) of the retina were positive for clrn1 transcript, with additional expression noted in the ciliary marginal zone (CMZ) and ganglion cell layer (Fig. 2b). Expression was maintained in mechanosensory hair cells of the inner ear and lateral line at this stage (Fig. 2c). The expression pattern in the ONL persisted through all stages examined. In contrast, the INL expression of clrn1 appeared throughout this cell layer at larval stages, but became more restricted in adult retinas, confined primarily to a region consistent with the location of amacrine cell bodies. GCL and CMZ signal was detectable at all larval stages examined (Fig 2d–h). Additional signal was detected in the supporting cells of the acoustic organ (Fig. 2i) and the anterior chamber of the eye Fig. 2g, j) from 10dpf onward. Outside these sensory organs, clrn1 transcript was present in the brain (Fig. 2f) and the gonad (data not shown).
Fig. 2.
clrn1 is expressed in the retina, inner ear, and lateral line at embryonic, larval, and adult stages. A digoxigenin-labeled cRNA probe was used to detect clrn1 transcript on sectioned tissue from 2 dpf to 6 months of age. (A–C) Developing eyes and ears are enriched for clrn1 in the outer (closed arrow) and inner (open arrow) nuclear layers and ganglion cell layer (bracket) of the retina, (A, B), in mechanosensory hair cells (closed arrowhead in C) and supporting cells (open arrow in C) of the ear and in the neuromast cells (nm). (D–F) Retinal expression is retained through larval stages in all nuclear layers of the retina and in the proliferating cells of the ciliary marginal zone (open and closed arrows in D indicate inner nuclear layer and outer nuclear layer cells, respectively; bracket indicates the CMZ), all sensory patches of ear, and neuromasts (E). Additional expression is noted in brain (asterisk in F). 7 dpf larva in F was raised in PTU to suppress melanocyte formation. (G–I) 10 dpf zebrafish larvae express clrn1 in the anterior chamber of the retina (arrow in G) and continued expression is present in the photoreceptor layer (closed arrowhead in H) inner retina (open arrowhead in H) and CMZ (bracket in G). clrn1 expression is detected both in mechanosensory hair cells (closed arrowhead in I) and supporting cells (open arrow in I) of the sensory patches (anterior macula shown in I). (J–K) Continued expression of clrn1 transcript in the anterior chamber (open arrowhead in J), photoreceptors (closed arrow in K), and inner retinal cells (open arrow in K) of adult retinas. No signal is detected in tissues incubated with a sense probe (shown in L: 3 dpf larval retina treated with PTU). C,E,I: lateral views with anterior to the left; all others horizontal views, with anterior to the bottom. Abbreviations: am: anterior macula; lc: lateral crista; nm: neuromast. Scale bars: A–C, E, H, I and L: 20 µm. D, G, J and K: 50 µm. F: 100 µm.
2.2 Detection of Clarin-1 protein
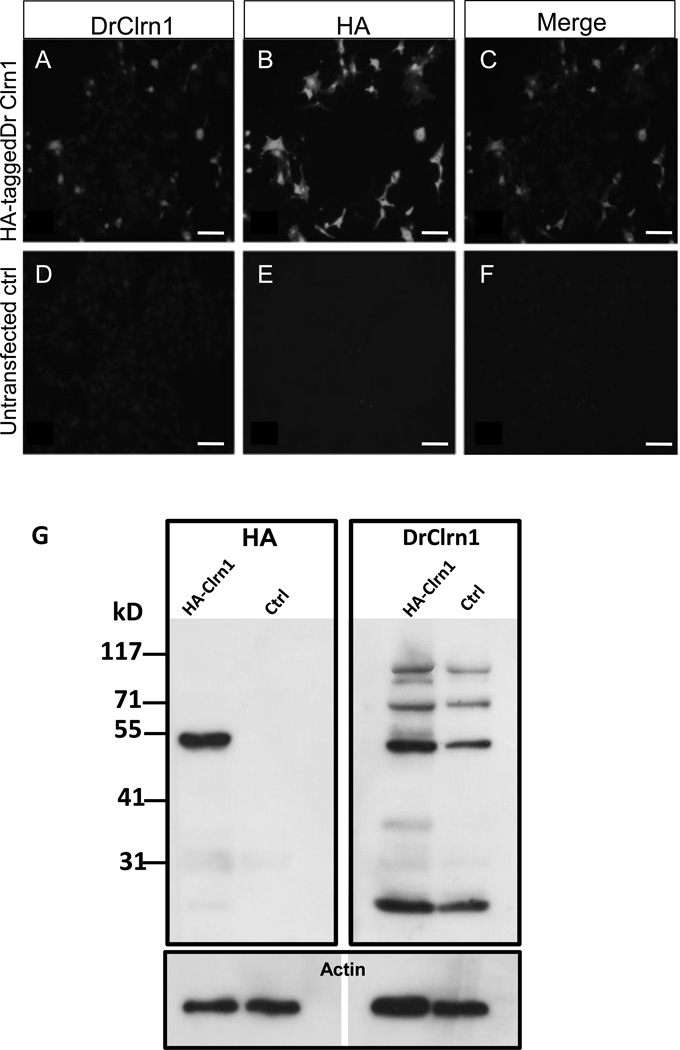
We obtained a custom peptide antibody generated via Genomic Antibody Technology (GAT) by Strategic Diagnostics Inc. (method described in Brown et al., 2011). This antibody was designed to recognize the first extracellular loop of zebrafish Clarin-1 (DrClrn1). We demonstrated specific reactivity of this reagent in two ways. First, we expressed clrn1 in vitro by transiently transfecting BHK cells with an expression vector containing the sequence encoding DrClrn1 fused to an HA-tag at its C-terminus. We observed complete colocalization between DrClrn1 and the HA-tag (Fig. 3a–f). We then took advantage of the Gal4/upstream activation sequence (UAS) system to express zebrafish clrn1 in vivo. We injected one-cell stage zebrafish embryos from a heat-shock inducible Gal4 transgenic line with an expression vector containing the sequence of zebrafish clrn1-HA under the control of a UAS promoter. Protein extracts from 30 hpf Tg(hsp70l:Gal4) embryos subjected to heat shock at 24 hpf were probed with an antibody against HA or DrClrn1 (Fig. 3g). With the DrClrn1 antibody, multiple products were observed, with prominent bands at approximately 52 kD and 25 kD. With the anti-HA antibody, we also observed a strong band at approximately 52 kD and a faint signal at approximately 25 kD in extracts from heat-shocked embryos injected with the construct, but not in heat-shocked uninjected control embryos. These bands were consistent with the most prominent bands observed when blotting these samples with DrClrn1 antibody. A similar banding pattern was obtained from HA-tagged human Clarin-1 using an antibody to HA (Isosomppi et al, 2009), where 27 kD and 48 kD bands were noted as Clarin-1 monomers and homodimers, respectively. Other tetraspanins are known to form homodimers, trimers, and tetramers that persist in stringent detergent conditions (Hemler 2005). The predicted size of the zebrafish Clarin-1 monomer is 26 kD. Although 52 kD is slightly larger than what would be predicted for a DrClrn1 homodimer, the corresponding band size obtained with the HA antibody in the presence of DrClrn1-HA indicates that the DrClrn1 antibody specifically recognizes zebrafish Clarin-1.
Fig. 3.
DrClrn1 antibody specifically recognizes Clarin-1 protein. Full-length zebrafish clrn1 was cloned into an HA-tag containing vector and transfected into BHK cells. Transfected cells were positively identified when incubated with DrClrn1 (A) or HA (B) antibodies. Clarin-1 and HA merged image shown in C. No signal was detected with either antibody in untransfected cell cultures (D–F). (G) Western blot of protein extracts from whole 30 hpf Tg(hsp70l:Gal4) larvae using an anti HA-antibody (left blot) or DrClrn1 antibody (right). Protein extracts from embryos injected with a Clrn1-HA construct under the control of a UAS promoter and from uninjected Tg(hsp70l:Gal4) embryos were obtained 6 hours after heat shock treatment. The anti-HA recognizes an approximately 52 kD band in the injected sample, which is absent from the control lane, and faintly labels a band at 25 kD. An additional faint band is noted at approximately 31 kD on the membrane incubated with anti-HA, but as this is also detectable in the uninjected control lane, it appears to be nonspecific. Multiple bands are present in extracts from injected and uninjected embryos incubated with DrClrn1. The band migrating at approximately 25 kD is consistent with the predicted size of the Clarin-1 protein. The approximately 52 kD band is consistent with the presence of Clarin-1 homodimers, and bands of higher weights are consistent with multimers. Membranes were subsequently probed with anti-pan actin as a loading control (lower panel). Scale bars: 50 µm.
2.3 Clarin-1 localization in zebrafish mechanosensory cells
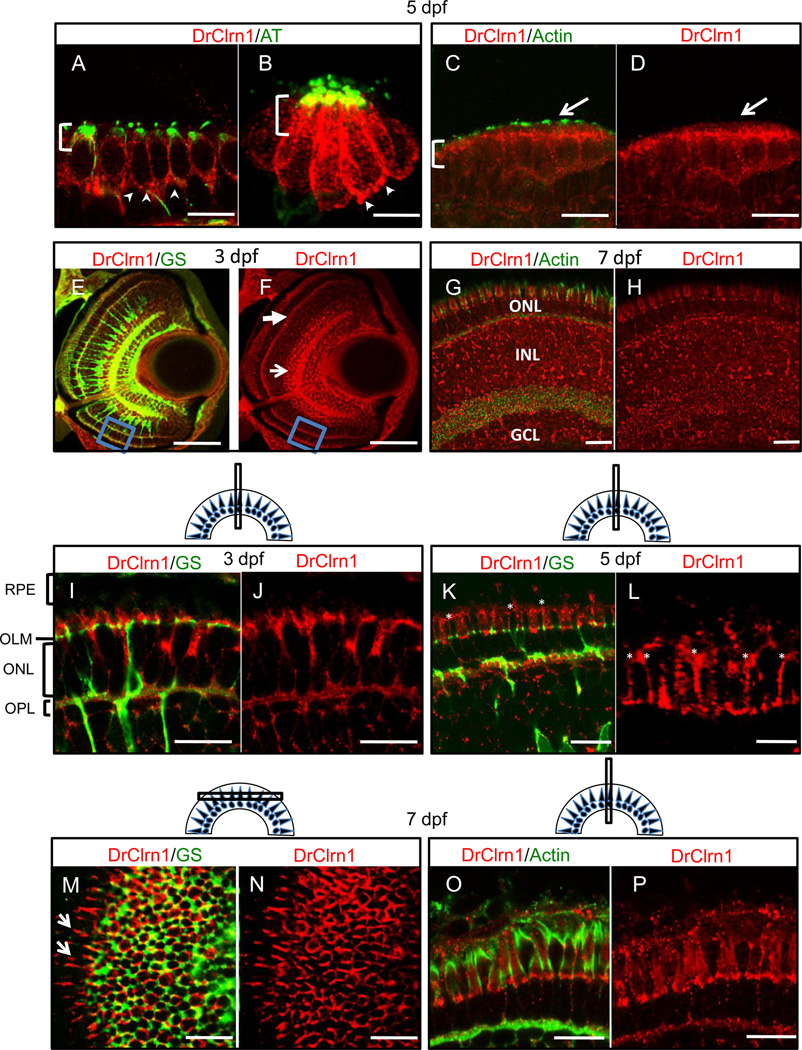
To examine the distribution of Clarin-1 in developing and mature sensory organs, we used the zebrafish specific antibody to label sectioned tissues from zebrafish between 3 dpf and 6 months of age. Sensory patches of the larval inner ear were strongly labeled with Clarin-1 antibodies, with enriched signal in the apical cell body and at synapses of hair cells (Fig. 4a). The hair cell localization pattern observed in sensory patches of the ear was recapitulated in the mechanosensory cells of the neuromasts (Fig. 4b). All hair cell localization detected in larval ear and lateral line was contained within the cell bodies and at cell margins. We did not observe Clarin-1 signal in the stereocilia (Fig. 4c,d, and data not shown), in contrast to data reported from mouse studies (Zallocchi et al, 2009, Geng et al., 2012).
Fig. 4.
Clarin-1 localizes in mechanosensory hair cell bodies and at the outer limiting membrane and lateral contacts between photoreceptors in the larval retina. (A–D) Cross sections showing Clarin-1 enrichment in apical regions of cell bodies (brackets) and at synapses (arrowheads) of hair cells of the ear (anterior macula shown in A,C) and neuromasts (B). Clarin-1 does not colocalize with actin filaments in the hair cell stereocilia (white arrows in C,D). Kinocilia are labeled with Acetylated Tubulin antibody (AT) and stereocilia are labeled with Actin antibody. (E–H) Cross sections through the retina of 3 dpf (E,F) and 7 dpf (G,H) larvae labeled with DrClrn1 and Glutamine synthetase (GS, panel E) or Actin (panel G) antibodies. Clarin-1 is enriched at synapses (OPL indicated with white closed arrow in F), and in the inner nuclear layer (white open arrow in F) at 3dpf. Blue box in ventral region of E and F indicates region magnified in panels I and J. A full retina view at 7dpf shows Clarin-1 label in the photoreceptors (ONL), Inner nuclear layer (INL) and ganglion cell layer (GCL). (I–L) High magnification views of 3dpf (I, J), and 5dpf (K, L) retinas. Clarin-1 is enriched at the outer limiting membrane (OLM) and in the outer plexiform layer (OPL). Glial cell processes are seen passing through the layer of the photoreceptor nuclei (ONL). At 3 dpf, the apical structures of photoreceptors, the inner and outer segments, are rudimentary and the retinal pigmented epithelium (RPE) lies close to the OLM. By 5 dpf, cone apices have extended such that space between the OLM and the RPE has increased, and fine, filamentous enrichments of Clarin-1 can be observed between cone inner segments (asterisks in K,L; high magnification image shown in L). (M,N) Transverse section through the 7dpf retina to visualize the photoreceptor layer from the top down. The meshwork of cell junctions that make up the OLM derive from Müller cell processes labeled by Glutamine synthetase (GS). Clarin-1 localization partially overlaps, and is also enriched at the lateral interfaces extending from the periphery of the transverse cut (white arrows in I). (O,P) Clarin-1 is seen in close proximity to actin at the lateral contacts between photoreceptors in the 7 dpf retina. Scale bars: E, F: 50 µm; A, C, D, G–P: 10 µm. B and L: 5 µm. Schematics show the plane of sectioning for retinal tissue.
2.4 Clarin-1 localization in the zebrafish retina
Consistent with the expression data, we found that Clarin-1 protein was present, in the ONL, the basal portion of the INL, and the GCL in larval retinas (Fig. 4e–h). Because Usher syndrome specifically affects photoreceptor survival in humans, we concentrated our analyses on the subcellular localization of Clarin-1 in and around these cells. At 3 dpf, photoreceptor outer segments are short and still developing. As they extend apically toward the retinal pigmented epithelium (RPE), projections from the Müller cells grow upward from the outer limiting membrane (OLM), a meshwork of Müller glial cell processes that provides structural and biochemical support through the formation of adherens junctions with photoreceptors (Gosens et al., 2008, Omri et al, 2010). These processes interdigitate between photoreceptor cell bodies in a domain known as the subapical region (SAR). Like the OLM, the SAR accumulates proteins known to form cell junctions and is thought to be important for photoreceptor stability. We detected significant levels of Clarin-1 protein in the outer retina at the level of the outer limiting membrane (OLM) (Fig. 4i–p). Clarin-1 partially colocalized with the Müller cell marker Glutamine synthetase (GS) in this region, with the Clarin-1 signal extending more apically than GS. Some colocalization was also observed in the outer plexiform layer and along Müller cell processes in the inner nuclear layer.
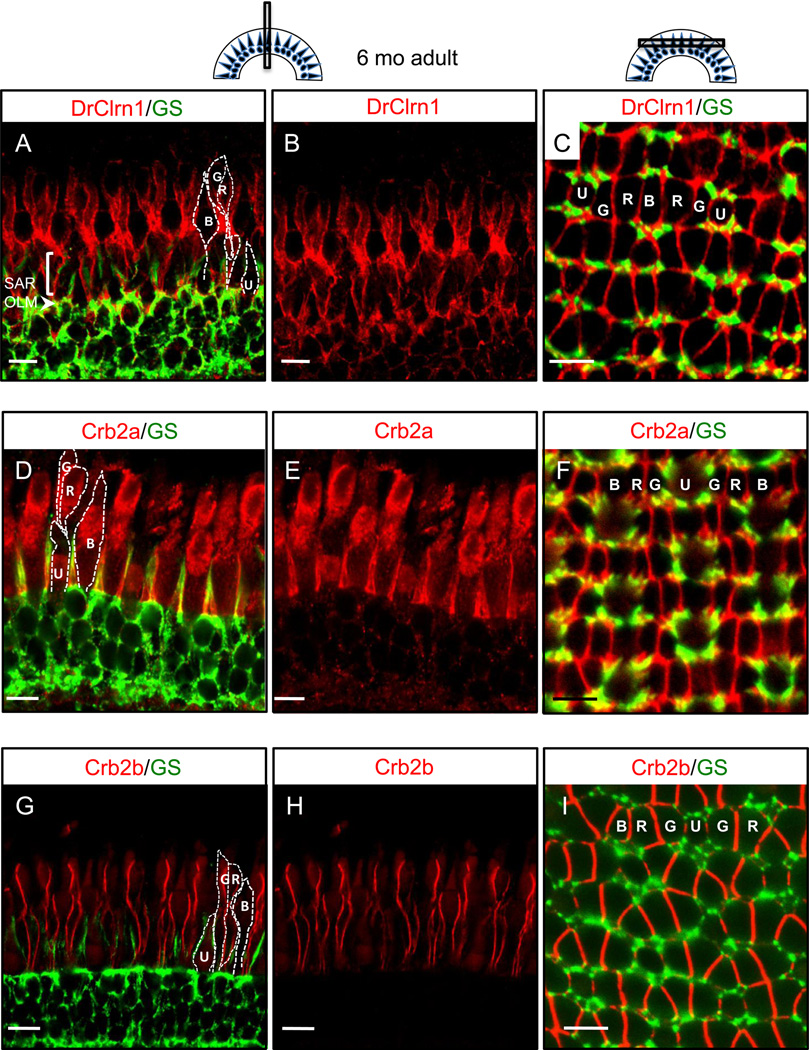
In the adult retina, Clarin-1 localization remains strong at the OLM and SAR, and additional labeling is noted at the lateral cell contacts between cone photoreceptor inner segments (Fig. 5a,c). In adult zebrafish retinas, photoreceptors are arranged in a consistent mosaic pattern across the retina (Fadool, 2003; Raymond and Barthel, 2004; illustrated in figure 5a,c). Recent work has shown filamentous contacts between the inner segments of neighboring photoreceptors (Zou et al, 2012). Proteins of the Crumbs pathway have been shown to localize in these regions of lateral cell contact, and mutations in genes encoding Crumbs and associated proteins are known to cause disorganized photoreceptor cell polarity in zebrafish (Wei et al., 2006, Zou et al., 2010, 2012). Because of the similarity between Clarin-1 localization at cone inner segments and the localization reported for zebrafish Crumbs proteins, we labeled retinal sections with Crb2a or Crb2b antibodies so that direct comparisons could be made.
Fig. 5.
Clarin-1 localizes at interfaces between photoreceptor inner segments in the adult retina. (A–C) Cross (A,B) and transverse (C) sections through adult retinas show Clarin-1 localization partially overlapping with glial cells labeled with Glutamine Synthetase (GS) at the OLM (arrowhead). Enrichment of Clarin-1 is also noted in the subapical region (SAR) and between all cone inner segments in the outer retina. The silhouettes of the four cone subtypes are outlined in white in A,D and G and the mosaic cone arrangement is labeled in C, F and I. (D–I) Cross (D–E, G–H) and transverse (F, I) sections of adult retinas labeled with Crb2a (D–F) and Crb2b (G–I) antibodies recapitulate results published by Zou et al, 2012, and provide points of reference for the Clarin-1 (A–C) localization pattern at lateral cone interfaces. Abbreviations: OLM: outer limiting membrane; G: green opsin cone, R: red opsin cone; B: blue opsin cone; SAR (subapical region) U: UV opsin cone. Scale bars: 10 µm.
We obtained results similar to the published data for these antibodies (Zou et al, 2012). In our hands, Crb2a largely colocalized with GS in Müller cells, with some additional localization at lateral cell contacts between photoreceptors (Fig. 5d–f), whereas Crb2b was restricted to photoreceptor inner segment interfaces and appeared largely exclusive of Müller cells (Fig. 5g–i). Compared to these largely non-overlapping Crumbs protein domains, Clarin-1 appeared intermediate between the two, showing some partial overlap with Müller cell processes in addition to pronounced localization at juxtaposing cell membranes of the photoreceptor inner segments (Fig. 5a–c). Because the three antibodies, Clarin-1, Crb2a, and Crb2b, were raised in rabbit, it was not possible to use them in co-labeling experiments. However, comparisons of these identically staged tissues with the same counter-stain indicate a close association between Clarin-1 and Crumbs proteins, which has not been previously reported. In contrast to the findings of Zallocchi et al (2009), we did not observe Clarin-1 in proximity to the connecting cilia of photoreceptors at any time point examined.
3. Discussion
Our data show distinct and sustained expression of clrn1 in zebrafish from early larval through post-developmental time points. Specific expression of the transcript in sensory patches is corroborated by protein localization in hair cells and supporting cells of the inner ear. The presence of zebrafish clrn1 in the inner retina is consistent with in situ results reported in young mice (Geller et al, 2009). However, Geller and colleagues found that the transcript in mouse retinas reduces dramatically after postnatal day 7, suggesting that Clarin-1 is not essential for murine visual function. By contrast, clrn1 is expressed in the zebrafish retina at all larval and adult stages examined, predicting function in both photoreceptors and in inner retinal cells throughout the life of the animal.
Although we cannot completely rule out the possibility that the DrClrn1 antibody detects nonspecific protein products, our cell culture and Western blot experiments provide strong support that this antibody specifically recognizes zebrafish Clarin-1. Western blot analysis from published results (Isosomppi et al., 2009) demonstrated the persistence of several Clarin-1 products in extracts from transfected cell cultures, despite the denaturing conditions used sample preparation and protein separation. Similarly, although we see multiple bands with the DrClrn1 antibody, the prominent 52 kD band on Western Blots produced with this reagent is consistent with the presence of a Clarin-1 homodimer, and the more prominent smaller and larger bands are consistent with the presence of monomers and multimers, respectively. Isosomppi and colleagues showed that HA-tagged human Clrn1 recognized a larger running band representative of the glycosylated Clarin-1 homodimer. It is possible that the larger than expected size of the predominant product seen with both the DrClrn1 and the HA antibody in our analyses is the glycosylated form. However, in contrast to the previous study, we do not see a product consistent with the size of the unglycosylated state. It may be that the glycosylated Clarin-1 homodimer is the predominant form in vivo and the unglycosylated form is not detectable with our methods, but further studies will be necessary to address this issue.
We note abundant localization of Clarin-1 associated with the outer limiting membrane in the retina, from late embryonic stages of development through adulthood. Clarin-1 localization is proximal to the actin cytoskeleton in the larval outer retina at lateral cell contacts, consistent with previous findings in cell culture (Isosomppi et al, 2009). However, no apparent association between Clarin-1 and actin in stereocilia was observed in vivo, as previously reported in vitro. Further, we did not observe Clarin-1 associated with cilia in either the retina or the hair cells, in contrast to the previous report showing murine Clarin-1 at the connecting cilia of photoreceptors (Zallocchi et al, 2009). This discrepancy may be due to species-specific differences in the spatiotemporal localization of Clarin-1.
The presence of Clarin-1 at the lateral contacts between photoreceptor inner segments in the adult retina is intriguing due to its similarity to the distribution of Crumbs proteins at these locations (van Rossum et al., 2006, Gosens et al., 2008, Zou et al., 2012). Previous studies have suggested an association between the scaffold protein Whirlin, the affected protein in Usher type 2d, and the Crumbs associated protein MPP5/Pals 1 (Gosens et al., 2007). Zebrafish Whirlin localizes in the SAR and is colocalized with Clarin-1 in that region (not shown). Combined with our previous observation that Harmonin (Ush1c) localizes to Müller cells (Phillips et al., 2011), these novel subcellular distributions may indicate roles for Usher proteins in establishing or maintaining lateral cell contacts between photoreceptor subtypes and between photoreceptors and Müller glial cells.
In humans and model organisms, defects in the Crumbs protein network lead to severe disruptions of apicobasal cell polarity and congenital vision problems (Richard et al, 2006, Gosens et al, 2008, Wei et al., 2006, Jensen and Westerfield, 2004; Zou et al, 2010). By comparison, the slow, progressive vision loss experienced by Usher patients is mild, but suggestive of an instability in cell function or structure that causes degeneration throughout the lifetime of the patient. The potential function of the Usher proteins in ciliary trafficking has been extensively studied to date (Märker et al., 2008, Yang et al., 2010, Sahly et al., 2012), but a comprehensive approach to studying molecular localizations and interactions in other regions of the retina will be important for illuminating the full complexity of this disease and for developing therapies to preserve vision in Usher patients. The Clrn1 knockout mouse has provided a tractable model to test gene replacement therapies for the hearing and balance defects of USH3 (Aarnisalo et al., 2007), but lack of a retinal phenotype (Geller, et al., 2009), despite the reported localization of Clarin-1 in adult mouse retinas (Zallocchi et al., 2009), limits its usefulness in testing similar rescue tools for vision loss. Development of a zebrafish model in which Clarin-1 is present and functional in a cone-rich retinal milieu throughout the life of the organism will expand our potential to understand and defeat vision loss associated with USH3.
4. Experimental Procedures
4.1 Zebrafish strains and maintenance
Adult zebrafish were maintained as described (Westerfield, 2007), according to University of Oregon IACUC guidelines. For all histological experiments, embryos were collected from natural crosses of F1 hybrids from AB x TU matings and raised at 28.5°C prior to fixation. In some experiments, a 0.15 mM solution of PTU (1-phenyl-2-thio-urea) was added to the embryo medium to suppress retinal pigment. For Western blot experiments, embryos were obtained from Tg(hsp70l:Gal4)1.5kca4 (Scheer and Campos-Ortega, 1999) incrosses and maintained as described before heat shock treatment and subsequent euthanization.
4.2 Cloning of zebrafish clrn1
Total RNA was extracted from embryonic, larval, and juvenile fish, and from enucleated adult retinas, using Trizol (Invitrogen) and the RNeasy mini kit (Qiagen). The resulting RNA was reverse transcribed using Oligo d(T) primers and Superscript II reverse transcriptase (Invitrogen). Full-length clrn1 transcript was amplified from cDNAs with primers designed for the Ensembl transcript ENSDART00000062909 (Forward primer: 5’ccgtgtccaaacacaatgc 3’; Reverse primer: gtacatgagatctgcagctccggt 3’) using Phusion Taq polymerase (NEB). The resulting 702 base pair fragments were subcloned into the pCR®-blunt II-Topo Vector (Invitrogen). Rapid amplification of cDNA ends (RACE) was performed to identify the 5’ and 3’ ends of clrn1 splice variants using RACE system Version 2.0 (Invitrogen). Purified fragments were submitted to Genewiz (Plainfield, NJ) for sequencing.
4.3 In situ hybridization
A full-length digoxigenin-labeled cRNA probe was generated from the plasmid insert using an SP6 RNA polymerase (Roche). In situ hybridization on sectioned tissue was performed as described in Ebermann et al. (2010). Images were collected on a Zeiss Axioplan 2 microscope using a Zeiss Axiocam HRc and Axiovision software.
4.4 Cell culture and transfections
The full-length zebrafish clrn1 sequence was inserted into hemagglutinin (HA)-tag containing phCMV3 Xi expression vector (Gene Therapy Systems Inc.). Baby hamster kidney (BHK-21, CCL-10) cells were transfected with the expression vector using Fugene®6 (Roche). Transfected cells were fixed with 4 % paraformaldehyde (PFA) and permeabilized by treatment with 0.2 % saponin (Sigma-Aldrich) after 24 hours of transfection. The cells were treated with 5% bovine serum albumin (BSA) (Jackson ImmunoResearch). The cells were incubated with antibodies against HA (HA.11 [MMS-101R]; Covance) diluted 1:700 and Clrn1 (Novus) diluted 1:2000. Secondary antibodies were conjugated either with Cy2 or Cy3 (Jackson ImmunoResearch). Cells were mounted with Gel/Mount (Biomeda) and analyzed with a Zeiss Axioplan 2 microscope.
4.5 Microinjection and Heat Shock
Embryos were injected as described (Westerfield, 2007) with 1 nl of solution containing 50 ng/µl of a construct consisting of the full-length sequence of zebrafish clrn1 fused to an HA-tag in its 3' end inserted under the control of an Upstream Activation Sequence (UAS) promoter using the MultiSite Gateway® technology (Invitrogen) combined with 200 ng/µl Transposase RNA synthesized from a pCS2 vector. Injected embryos and uninjected control siblings were maintained at 28.5°C for 24 hours, then heat shocked in a water bath at 39°C for 30 minutes, and returned to maintenance temperature for 6 hours until collection for protein extraction.
4.5 Western Blot
Tissues were homogenized in 10 mM Tris pH 7.4, 150 mM NaCl, 1 mM EDTA, 0.1% SDS, 1% Triton, 1% Sodium Deoxycholate, and 5 mg/ml protease inhibitor cocktail (Pierce). Samples were run on an 8% acrylamide gel and transferred to immobilon PVDF membranes (Millipore) with Nupage reagents (Invitrogen). Membranes were washed in Phosphate Buffered Saline with 0.1% Tween-20 (PBS-T) 3 × 10 minutes, incubated in blocking solution (10% NFDM in PBS-T) for 1 hour at room temperature, then in primary antibodies [Clrn1 (Novus) 1:800; mouse monoclonal HA (Covance) 1:500; mouse monoclonal pan-actin (Millipore) 1:5000] diluted in blocking solution overnight at 4°C. Membranes were rinsed three times for 5 minutes each in PBS-T, incubated in donkey anti-rabbit or donkey-anti mouse HRP (Jackson) for 1 hour at room temperature, then rinsed three times for 5 minutes each in PBS-T. Signal was detected through HRP catalysis with a chemiluminescent substrate (ECL Plus detection system; Pierce). To strip blots for reprobing with actin as a loading control, membranes were rehydrated and incubated in 0.1 M glycine pH 2.2 twice for 10 minutes followed by 2 × 10 minute washes in PBS-T before blocking as described.
4.5 Immunohistochemistry
Euthanized embryos and larvae were fixed in 4% PFA in 1× PBS-T overnight at 4°C, washed 2 × 10 minutes in PBS-T, dehydrated in an ascending series of methanol, and stored at −20°C. In preparation for sectioning, tissues were rehydrated in descending methanol series and cryoprotected in 10% sucrose followed by 30% sucrose in PBS-T prior to embedding in a 1% agarose, 0.5% agar, 5% sucrose solution. 16 µm sections were cut onto APTES-coated slides and stored at −20°C if not used immediately. Slides were hydrated in PBS-T and heated in a pressure cooker in 10 mM Sodium Citrate solution (pH 8.5) for antigen retrieval. Antibody labeling proceeded as described (Phillips et al, 2011), using the following antibody concentrations: Clrn1 (Novus), 1:800; Crb2a and Crb2b (a gift from Xiangyun Wei), 1:300; Acetylated Tubulin (Santa Cruz), 1:1000; Glutamine Synthetase (Millipore), 1:1000; Pan-actin (Millipore), 1:5000. Goat anti-mouse 488 and Goat anti-rabbit 568 secondary antibodies from Molecular Probes were used at a concentration of 1:1000. Fluorescent Images were collected on a Zeiss LSM5 confocal microscope.
-
*
We examine gene expression and protein localization of Clrn1(USH3A) in zebrafish
-
*
We report novel subcellular localization at photoreceptor cell boundaries
-
*
We correlate Clarin-1 protein localization with Crumbs proteins in the retina
-
*
We establish zebrafish as a potential model for Usher syndrome type 3A
Acknowledgements
We thank Xiangyun Wei for providing aliquots of Crb2a and Crb2b antibodies, Sabrina Toro for comments on this manuscript, Steve Seredick for technical assistance, and Reetta Jalkanen for technical assistance.
This work was supported by NIH Grants DC004186, DC010447, and HD22486, as well as De Blindas Vänner Foundation, European Molecular Biology Organization (EMBO), Finnish Eye and Tissue Bank Foundation, Foundation Fighting Blindness, Hope for Vision, Maud Kuistila Memorial Foundation, Oskar Öflunds Foundation, Otto A Malm Foundation, Paulo Foundation, Research Foundation of the University of Helsinki, Retina Finland, the Usher III Initiative and Understödsförening Liv och Hälsa.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aarnisalo AA, Pietola L, Joensuu J, Isosomppi J, Aarnisalo P, Dinculescu A, Lewin AS, Flannery J, Hauswirth WW, Sankila E-M, Jero J. Anti-clarin-1 AAV-delivered ribozyme induced apoptosis in the mouse cochlea. Hear. Res. 2007;230:9–16. doi: 10.1016/j.heares.2007.03.004. [DOI] [PubMed] [Google Scholar]
- Adato A, Vreugde S, Joensuu T, Avidan N, Hamalainen R, Belenkiy O, Olender T, Bonne-Tamir B, Ben-Asher E, Espinos C, Millán JM, Lehesjoki A-E, Flannery JG, Avraham KB, Pietrokovski S, Sankila E-M, Beckmann JS, Lancet D. USH3A transcripts encode clarin-1, a four pass transmembrane-domain protein with a possible role in sensory synapses. Eur. J. Hum. Genet. 2002;10:339–350. doi: 10.1038/sj.ejhg.5200831. [DOI] [PubMed] [Google Scholar]
- Bonnet Crystel, El-Amraoui A. Usher syndrome (sensorineural deafness and retinitis pigmentosa): pathogenesis, molecular diagnosis and therapeutic approaches. Curr. Opin. Neurol. 2012;25(1):42–49. doi: 10.1097/WCO.0b013e32834ef8b2. [DOI] [PubMed] [Google Scholar]
- Brown MC, Joaquim TR, Chambers R, Onisk DV, Yin F, et al. Impact of Immunization Technology and Assay Application on Antibody Performance – A Systematic Comparative Evaluation. PLoS ONE. 2011;6(12):e28718. doi: 10.1371/journal.pone.0028718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duman D, Tekin M. Autosomal recessive nonsyndromic deafness genes: a review. Front Biosci. 2012. 2012 Jun 1;17:2213–2236. doi: 10.2741/4046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebermann I, Phillips JB, Liebau MC, Koenekoop RK, Schermer B, Lopez I, Schäfer E, Roux A-F, Dafinger C, Bernd A, Zrenner E, Claustres M, Blanco B, Nürnberg G, Nürnberg P, Ruland R, Westerfield M, Benzing T, Bolz HJ. PDZD7 is a modifier of retinal disease and a contributor to digenic Usher syndrome. J. Clin. Inv. 2010;120:1812–1823. doi: 10.1172/JCI39715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernest S, Rauch GJ, Haffter P, Geisler R, Petit C, Nicolson T. Mariner is defective in myosin VIIA: a zebrafish model for human hereditary deafness. Hum. Mol. Genet. 2000;9:2189–2196. doi: 10.1093/hmg/9.14.2189. [DOI] [PubMed] [Google Scholar]
- Fadool JM. Development of a rod photoreceptor mosaic revealed in transgenic zebrafish. Dev. Biol. 2003;258:277–290. doi: 10.1016/s0012-1606(03)00125-8. [DOI] [PubMed] [Google Scholar]
- Geller SF, Guerin KI, Visel M, Pham A, Lee ES, Dror AA, Avraham KB, Hayashi T, Ray CA, Reh TA, Bermingham-McDonogh O, Triffo WJ, Bao S, Isosomppi J, Västinsalo H, Sankila E-M, Flannery JG. CLRN1 is nonessential in the mouse retina but is required for cochlear hair cell development. PLoS Genet. 2009;5(8):e1000607. doi: 10.1371/journal.pgen.1000607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng R, Geller SF, Hayashi T, Ray CA, Reh TA, Bermingham-McDonogh O, Jones SM, Wright CG, Melki S, Imanishi Y, Palczewski K, Alagramam KN, Flannery JG. Usher syndrome IIIA gene clarin-1 is essential for hair cell function and associated neural activiation. Hum. Mol. Genet. 2009;18(15):2748–2760. doi: 10.1093/hmg/ddp210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng R, Melki S, Chen DH-C, Tian G, Furness DN, Oshima-Takago T, Neef J, Moser T, Askew C, Horwitz G, Holt JR, Imanishi Y, Alagramam KN. The mechanosensory structure of the hair cell requires Clarin-1, a protein encoded by Usher syndrome III causative gene. J. Neurosci. 2012;32(28):9485–9498. doi: 10.1523/JNEUROSCI.0311-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gosens I, van Wijk E, Kersten FF, Krieger E, van der Zwaag B, Märker T, Letterboer SJ, Klooster J, Cremers FP, Roepman R, Wijnholds J. MPP1 links the Usher protein network and the Crumbs protein complex in the retina. Invest. Ophthalmol. Vis. Sci. 2007;46(6):2192–2201. [Google Scholar]
- Gosens I, den Hollander AI, Cremers FPM, Roepman R. Composition and function of the Crumbs protein complex in the mammalian retina. Exp. Eye Res. 2008;86:713–726. doi: 10.1016/j.exer.2008.02.005. [DOI] [PubMed] [Google Scholar]
- Hemler ME. Tetraspanin functions and associated microdomains. Nat. Rev. Mol. Cell Biol. 2005;6(10):801–811. doi: 10.1038/nrm1736. [DOI] [PubMed] [Google Scholar]
- Isosomppi J, Västinsalo H, Geller SF, Heon E, Flannery JG, Sankila EM. Disease-causing mutations in the CLRN1 gene alter normal CLRN1 protein. Mol. Vis. 2009;15:1806–1818. [PMC free article] [PubMed] [Google Scholar]
- Jensen AM, Westerfield M. Zebrafish mosaic eyes is a novel FERM protein required for retinal lamination and retinal pigmented epithelial tight junction formation. Curr. Biol. 2004;14:711–717. doi: 10.1016/j.cub.2004.04.006. [DOI] [PubMed] [Google Scholar]
- Maerker T, van Wijk E, Overlack N, Kersten FF, McGee J, Goldmann T, Sehn E, Roepman R, Walsh EJ, Kremer H, et al. A novel Usher protein network at the periciliary reloading point between molecular transport machineries in vertebrate photoreceptor cells. Hum. Mol. Genet. 2008;17:71–86. doi: 10.1093/hmg/ddm285. [DOI] [PubMed] [Google Scholar]
- Omri S, Omri B, Savoldelli M, Jonet L, Thillaye-Goldenberg B, Thuret G, Gain P, Jeanny JC, Cristanti P, Behar-Cohen F. The outer limiting membrane (OLM) revisited: clinical implications. Clin. Ophthalmol. 2010;4:183–195. doi: 10.2147/opth.s5901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pakarinen L, Tuppurainen K, Laippala P, Mäntyjärvi M, Puhakka H. The ophthalmological course of Usher syndrome type III. Int. Ophthalmol. 1995–1996;19(5):307–311. doi: 10.1007/BF00130927. [DOI] [PubMed] [Google Scholar]
- Phillips JB, Blanco-Sanchez B, Lentz JJ, Tallafuss A, Khanobdee K, Sampath S, Jacobs ZG, Han PF, Mishra M, Titus TA, Williams DS, Keats BJ, Washbourne P, Westerfield M. Harmonin (Ush1c) is required in zebrafish Müller glial cells for photoreceptor synaptic development and function. Dis. Model. Mech. 2011;4(6):786–800. doi: 10.1242/dmm.006429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puffenberger EG, Jinks RN, Sougnez C, Cibulskis K, Willert RA, Achilly NP, Cassidy RP, Fiorentini CJ, Heiken KF, Lawrence JJ, Mahoney MH, Miller CJ, Nair DT, Politi KA, Worcester KN, Setton RA, Dipiazza R, Sherman EA, Eastman JT, Francklyn C, Robey-Bond S, Rider NL, Gabriel S, Morton DH, Strauss KA. Genetic mapping and exome sequencing identify variants associated with five novel diseases. PLoS One. 2012;7(1):e28936. doi: 10.1371/journal.pone.0028936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiners J, Nagel-Wolfrum K, Jurgens K, Märker T, Wolfrum U. Molecular basis of human Usher syndrome: deciphering the meshes of the Usher protein network provides insights into the pathomechanisms of the Usher disease. Exp. Eye Res. 2006;83:97–119. doi: 10.1016/j.exer.2005.11.010. [DOI] [PubMed] [Google Scholar]
- Riazuddin S, Belyantseva IA, Giese AP, Lee K, Indzhykulian AA, Nandamuri SP, Yousaf R, Sinha GP, Lee S, Terrell D, Hegde RS, Ali RA, Anwar S, Andrade-Elizondo PB, Sirmaci A, Parise LV, Basit S, Wali A, Ayub M, Ansar M, Ahmad W, Khan SN, Akram J, Tekin M, Riazuddin S, Cook T, Buschbeck EK, Frolenkov GI, Leal SM, Friedman TB, Ahmed ZM. Alterations of the CIB2 calcium- and integrin-binding protein cause Usher syndrome type 1J and nonsyndromic deafness DFNB48. Nat. Genet. 2012;44(11):1265–1271. doi: 10.1038/ng.2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard M, Roepman R, Aartsen WM, van Rossum AG, den Hollander AI, Knust E, Wijnholds J, Cremers FP. Towards understanding CRUMBS function in retinal dystrophies. Hum. Mol. Genet. 2006;15(Spec No 2):R235–R243. doi: 10.1093/hmg/ddl195. [DOI] [PubMed] [Google Scholar]
- Scheer N, Campos-Ortega JA. Use of the Gal4-UAS technique for targeted gene expression in the zebrafish. Mech. Dev. 1999;80:153–158. doi: 10.1016/s0925-4773(98)00209-3. [DOI] [PubMed] [Google Scholar]
- Seiler C, Finger-Baier KC, Rinner O, Makhankov YV, Schwarz H, Neuhauss SC, Nicolson T. Duplicated genes with split functions: independent roles of protocadherin15 orthologs in zebrafish hearing and vision. Development. 2005;132:615–623. doi: 10.1242/dev.01591. [DOI] [PubMed] [Google Scholar]
- Söllner C, Rauch GJ, Siemens J, Geisler R, Schuster SC, Muller U, Nicolson T. Mutations in cadherin 23 affect tip links in zebrafish sensory hair cells. Nature. 2004;428:955–959. doi: 10.1038/nature02484. [DOI] [PubMed] [Google Scholar]
- Tian G, Zhou Y, Hajkova D, Miyagi M, Dinculescu A, Hauswirth WW, Palczewski K, Geng R, Alagramam KN, Isosomppi J, Sankila E-M. Clarin-1, encoded by the Usher syndrome III causative gene, forms a membranous microdomain. J. Biochem. 2009;284:18980–18993. doi: 10.1074/jbc.M109.003160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Rossum AG, Aartsen WM, Meuleman J, Klooster J, Malysheva A, Versteeg I, Arsanto JP, Le Bivic A, Wijnholds J. Pals1/Mpp5 is required for correct localization of Crb1 at the subapical region in polarized Muller glia cells. Hum Mol Genet. 2006;15(18):2659–2672. doi: 10.1093/hmg/ddl194. [DOI] [PubMed] [Google Scholar]
- Västinsalo H, Jalkanen R, Dinculescu A, Isosomppi J, Geller S, Flannery JG, Hauswirth WW, Sankila E-M. Alternative splice variants of the USH3A gene Clarin 1 (CLRN1) Eur. J. Hum. Genet. 2011;19(1):30–35. doi: 10.1038/ejhg.2010.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei X, Zou J, Takechi M, Kawamura S, Li L. Nok plays an essential role in maintaining the integrity of the outer nuclear layer in the zebrafish retina. Exp Eye Res. 2006;83(1):31–44. doi: 10.1016/j.exer.2005.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westerfield M. The Zebrafish Book: A Guide for the Laboratory Use of Zebrafish (Danio rerio) Eugene: University of Oregon Press; 2007. [Google Scholar]
- Yang J, Liu X, Zhao Y, Adamian M, Pawlyk B, Sun X, McMillan DR, Liberman MC, Li T. Ablation of Whirlin long isoform disrupts the USH2 protien complex and causes vision and hearing loss. PLoS Genet. 2010;6:e1000955. doi: 10.1371/journal.pgen.1000955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zallocchi M, Meehan DT, Delimont D, Askew C, Garige S, Gratton MA, Rothermund-Franklin CA, Cosgrove D. Localization and expression of clarin-1, the Clrn1 gene product, in auditory hair cells and photoreceptors. Hear. Res. 2009;255:109–120. doi: 10.1016/j.heares.2009.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zallocchi M, Meehan DT, Delimont D, Rutledge J, Gratton MA, Flannery J, Cosgrove D. Role for a novel Usher protein complex in hair cell synaptic maturation. PLoS One. 2012;7(2):e30573. doi: 10.1371/journal.pone.0030573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou J, Yang X, Wei X. Restricted localization of ponli, a novel zebrafish MAGUK-family protein, to the inner segment interface areas between green, red, blue cones. Invest. Ophthalmol. Vis. Sci. 2010;51(3):1738–1746. doi: 10.1167/iovs.09-4520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou J, Wang X, Wei X. Crb apical polarity proteins maintain zebrafish retinal cone mosaics via intercellular binding of their extracellular domains. Dev. Cell. 2012;22(6):1261–1274. doi: 10.1016/j.devcel.2012.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]