Abstract
Lactococci isolated from non-dairy sources have been found to possess enhanced metabolic activity when compared to dairy strains. These capabilities may be harnessed through the use of these strains as starter or adjunct cultures to produce more diverse flavor profiles in cheese and other dairy products. To understand the interactions between these organisms and the phages that infect them, a number of phages were isolated against lactococcal strains of non-dairy origin. One such phage, ΦL47, was isolated from a sewage sample using the grass isolate L. lactis ssp. cremoris DPC6860 as a host. Visualization of phage virions by transmission electron microscopy established that this phage belongs to the family Siphoviridae and possesses a long tail fiber, previously unseen in dairy lactococcal phages. Determination of the lytic spectrum revealed a broader than expected host range, with ΦL47 capable of infecting 4 industrial dairy strains, including ML8, HP and 310, and 3 additional non-dairy isolates. Whole genome sequencing of ΦL47 revealed a dsDNA genome of 128, 546 bp, making it the largest sequenced lactococcal phage to date. In total, 190 open reading frames (ORFs) were identified, and comparative analysis revealed that the predicted products of 117 of these ORFs shared greater than 50% amino acid identity with those of L. lactis phage Φ949, a phage isolated from cheese whey. Despite their different ecological niches, the genomic content and organization of ΦL47 and Φ949 are quite similar, with both containing 4 gene clusters oriented in different transcriptional directions. Other features that distinguish ΦL47 from Φ949 and other lactococcal phages, in addition to the presence of the tail fiber and the genome length, include a low GC content (32.5%) and a high number of predicted tRNA genes (8). Comparative genome analysis supports the conclusion that ΦL47 is a new member of the 949 lactococcal phage group which currently includes the dairy Φ949.
Keywords: Lactococcus lactis, non-dairy, phage, tail fiber, genome
Introduction
Cultures of lactic acid bacteria (LAB) used in cheese manufacture play a pivotal role in the formation of cheese flavor (Urbach, 1995). While the limited numbers of established dairy cultures in use have greatly reduced inconsistencies in cheese quality, it can be at the expense of cheese flavor. LAB isolated from non-dairy environments, such as plant material, often exhibit enhanced metabolic capabilities when compared to those of dairy origin and have been shown to contribute to a more diverse flavor profile in the cheese (Ayad et al., 1999, 2000; Morales et al., 2003). In addition to their role in flavor enhancement, cultures of non-dairy origin have also been shown to be naturally insensitive to bacteriophages which infect industrial strains (Ayad et al., 2000). Phage infection is the single largest cause of industrial milk fermentation problems, negatively impacting on the consistency of cheese quality and resulting in large economic losses (Coffey and Ross, 2002). Non-dairy lactococcal strains could potentially be exploited for use in cheese culture rotations as phage insensitive strains and may well reduce the negative consequences of phage infection within a production plant. However, given the high evolution rate of phages, it is highly likely that over time, phages would emerge to threaten these bacterial strains.
All lactococcal phages isolated to date are members of the Caudovirales order which comprises of three families: Siphoviridae (long, non-contractile tails), Myoviridae (long, contractile tails) and Podoviridae (short tails) [for review see Veesler and Cambillau (2011)]. Phage classification systems have been utilized in the design of anti-phage strategies to prevent phages amplifying to high titers in manufacturing plants (Jarvis et al., 1991). DNA hybridization assays and comparative genomic analysis coupled with visualization of virions has led to the division of lactococcal phages into one of ten groups (Deveau et al., 2006). Of these phage groups, the c2, 936 and P335 groups are predominantly associated with failed dairy fermentations and are frequently isolated from cheese manufacturing plants (Moineau et al., 1992; Murphy et al., 2013). Currently within the NCBI database, there are complete genome sequences for over 30 lactococcal bacteriophages. Phages classified within the same group can share a large degree of genome similarity as observed within the c2 and 936 groups (Lubbers et al., 1995; Chopin et al., 2001; Crutz-Le Coq et al., 2002). However, within the P335 group, there exists a high proportion of genetic diversity attributed to the presence of both temperate and virulent bacteriophages classified within this group (Labrie and Moineau, 2007; Garneau et al., 2008). Collectively, these groups account for the majority of sequenced lactococcal phage genomes, ranging in size from 22 to 40 kb. Larger lactococcal phages do also exist, with Φ949 at 114.8 kb being the largest sequenced lactococcal phage to date (Samson and Moineau, 2010). With the elucidation of the Φ949 genome, a genomic sequence is available for at least one member of each phage group.
The use of a wide range of different bacterial strains possessing an industrially important phenotype has presumably contributed to the diversity of lactococcal phages. Genome analysis of the rarer uncommon lactococcal phages has shed further light on this diversity. Comparative analysis of the structural proteins of Φ1358, show a high degree of similarity to 2 Listeria monocytogenes phages. Moreover this phage was also found to possess an uncommonly high %GC content of 51 (Dupuis and Moineau, 2010). Recent analysis of the Φ1706 genome, revealed that 22 ORFs shared similarities with proteins of Firmicutes, typically found in the human gut (Garneau et al., 2008). The ability of this phage to infect lactococci was attributed to the acquisition of a 4 gene module, allowing for host recognition of lactococcal cells (Garneau et al., 2008). Genomic analysis of the ΦKSY1 genome has suggested the exchange of genetic material between bacteria and phages from different environments (Chopin et al., 2007). ΦQ54 was found to possess a different modular configuration, thought to be derived from recombination events with 936 and c2 type phages (Fortier et al., 2006). It is probable that these rarer phages arose from recombination events with other lactococcal phages and phages infecting other Gram-positive bacteria (Dupuis and Moineau, 2010). Moreover, these phages appear to be less suited to thrive in milk fermentations in contrast to phages of the more common lactococcal phage groups which possess rapid reproduction rates believed to be driven by evolutionary pressure (Ferguson and Coombs, 2000; Dupuis and Moineau, 2010).
In this study we describe the isolation and characterization of ΦL47, a large lytic phage which infects the non-dairy isolate L. lactis ssp. cremoris DPC6860. We also report the complete genome sequence of ΦL47 which, to our knowledge, is the largest lactococcal phage reported to date. Due to the emergence of non-dairy lactococci as dairy cultures with enhanced flavor-forming activity, sufficient data needs to be generated with regards to these cultures and their phage resistance if they are to be successfully utilized in dairy processing. Therefore, the objective of this study was to provide a better understanding of phages of lactococci from non-dairy origins at a phenotypic and genomic level, thus offering further insight into phage-host interactions.
Materials and methods
Bacterial strains, bacteriophage and culture conditions
Dairy and non-dairy Lactococcus strains were supplied by the TFRC-Moorepark culture collection (Teagasc Food Research Centre, Moorepark, Ireland). L. lactis ssp. cremoris DPC6860 was previously isolated from grass (D. Cavanagh, unpublished) and characterized by 16S rRNA analysis as described by Alander et al. (1999). All lactococcal strains used in this study were cultured in M17 (Oxoid, Hampshire, England) media containing 0.5% lactose (wt./vol.) (VWR, Leuven, Belgium) (LM17) at 30°C for 18–24 h under aerobic conditions. Double-strength M17 broth, used for phage enrichment of sewage samples, was prepared by doubling the amount of dry M17 media and reconstituting it in the same amount of distilled water as the 1X media with the addition of 1% lactose (wt./vol.). Soft agar overlays and solid agar medium contained 0.75% and 1.5% agar, respectively. Lactococcal phages ΦKSY1, Φc2, ΦbIL170 and ΦP008 were originally obtained from the Felix d'Hérelle Reference Center for Bacterial Viruses (GREB, Pavillon de Médecine Dentaire Université Laval, QC, Canada) while phages Φ712, ΦHP, ΦebI and ΦML3 were obtained from the UCC culture collection (University College Cork, Cork, Ireland). Phages were propagated using their respective hosts at 30°C in M17 media containing 0.5% glucose (wt./vol.) (Sigma-Aldrich, Dublin, Ireland) and 10 mM CaCl2.
Spot assay
Bacterial infection by phages was assessed using spot plate assays with phage titers of 108 PFU/mL. Briefly, 10 μ L of phage lysate was spotted onto an LM17 soft agar overlay containing 10 mM CaCl2 and inoculated with 1 × 108 CFU/mL of the host organism. Spot plates were allowed to dry before incubation aerobically, at 30°C for 24 h.
Phage isolation and propagation
Raw sewage samples were collected from the sewage treatment facility in Mitchelstown, Co. Cork, Ireland. Phage isolation was conducted as described previously (Alemayehu et al., 2009). Individual plaques isolated following this method underwent 3 successive rounds of plaque purification, to ensure that a pure phage was isolated. Briefly, a single plaque was aseptically removed from an overlay plate using a sterile 1 mL pipette tip and added to 5 mL of mid-log phase host, containing 10 mM CaCl2. Following overnight incubation at 30°C, the mixture was centrifuged at 4,500 rpm for 15 min and filtered through a 0.45 μm pore filter (Sarstedt, Wexford, Ireland). The filtrate was diluted to 10−8 and plaqued on the appropriate media for 24 h at 30°C. These steps were repeated twice on the resulting plaques until a pure phage was obtained.
Lytic spectrum and adsorption to lactococcal cells
The lytic spectrum of ΦL47 was determined, as for dairy lactococcal phages, by spotting 10 μ L of phage lysate containing 107 PFU/mL onto soft agar seeded with a L. lactis strain. Adsorption of phage particles to lactococcal cells was determined as follows: 10 μ L of phage lysate (~107 PFU/mL) was mixed with 2 mL of late exponential cells. Samples were incubated at 30°C for 10 min with shaking, to allow the phage particles to attach. Each sample was centrifuged at 14,000 rpm for 5 min and filtered through a 0.2 μm filter (Sarstedt, Wexford, Ireland). The number of phages in the supernatant was determined by plaque assay and % adsorption was calculated using the formula [(initial titer—titer in supernatant)/ initial titer × 100%].
Electron microscopy
High titer phage suspensions for visualization of phage particles were prepared using CsCl gradient purification and ammonium acetate concentration. Pure phage samples were obtained using a CsCl gradient of polyethylene glycol (Sigma-Aldrich, Dublin, Ireland) precipitates as described by Sambrook and Russell (2001). For ammonium acetate precipitation, 1 L of fresh lysate was centrifuged at 10,000 rpm at 4°C for 10 min and filter sterilized using a 0.45 μm pore filter (Sarstedt). Phage particles were subsequently precipitated by centrifugation at 20,000 rpm for 1 h at 4°C. The supernatant was removed and the phage pellet re-suspended in 10 mL of ice cold 0.1 M ammonium acetate (Sigma-Aldrich). After pooling of phage samples, a further 10 mL of ammonium acetate was added and the sample centrifuged at 20,000 rpm for 1 h. The supernatant was again removed and the pellet suspended in a final volume of 1 mL ammonium acetate. Negative staining was performed on both phage samples using 2% uranyl acetate on carbon films. Each grid was examined at an 80 kV acceleration voltage using a Tecnai 10 transmission electron microscope (FEI Company, Eindhoven, The Netherlands). Micrograph images were captured using a MegaView 2 CCD-camera (Olympus SIS, Münster, Germany). Phage structure dimensions were determined based on the average of 10–15 measurements.
Structural analysis of phage proteins
Analysis of structural proteins was performed as described previously (Kelly et al., 2012) using high titer phage suspensions obtained from ammonium acetate concentration. Samples were mixed with 4X sample loading buffer and heated at 95°C prior to loading in a 12% SDS polyacrylamide gel. Protein bands were stained using Coomassie blue staining and excess dye removed using a de-staining solution (40% ethanol, 53% distilled water and 7% acetic acid). Protein size was estimated using a broad range protein ladder (New England Biolabs, Hertfordshire, UK) as a relative molecular weight marker (MWM).
DNA sequencing, annotation and comparative analysis
Bacteriophage DNA sequencing was performed using the Roche GS FLX+ system to >20× coverage (MWG, Ebersberg, Germany). The quality of the raw assembly reads were visualized and verified using the programme Hawkeye (Amos) (Schatz et al., 2013) and Consed (Gordon, 2003). To verify the genome structure, PCR amplicons were generated with the Platinum Hi-fidelity PCR Supermix (Invitrogen, Life Technologies, Dublin, Ireland) for various regions of the genome and at contig ends, followed by direct sequencing. The final phage genome was assembled using the Phred-Phrap-Consed package (Ewing and Green, 1998; Gordon, 2003). ORFs were predicted using the programs prodigal (Hyatt et al., 2010) and Glimmer (Delcher et al., 1999). Annotation was provided by the RAST annotation software (Aziz et al., 2008) and GAMOLA (Altermann and Klaenhammer, 2003). Genome annotation was verified manually using Artemis (http://www.sanger.ac.uk/resources/software/artemis/) (Rutherford et al., 2000) and detected open reading frames (ORFs) were functionally annotated using BLASTp (Altschul et al., 1990). Conserved domains were detected using InterProScan (http://www.ebi.ac.uk/InterProScan/) and DELTA-BLAST (Domain Enhanced Lookup Time Accelerated BLAST) (Marchler-Bauer et al., 2011). Identification of tRNAs was achieved using the software packages tRNA scan-SE (Lowe and Eddy, 1997) and ARAGORN v1.2.36 (Lowe and Eddy, 1997; Laslett and Canback, 2004). Comparative genome analysis of the Lactococcus ΦL47 with its most similar relative, Lactococcus Φ949, was performed using the Artemis Comparison Tool (ACT) programme (Carver et al., 2005). The genome sequence of ΦL47 is available from GenBank/EMBL under the accession number KF926093.
Results and discussion
Isolation of ΦL47 from sewage
Previous work in our laboratory led to the isolation of a bank of lactococcal strains from non-dairy origins, including grass, vegetable matter and bovine rumen samples. These strains, which included both L. lactis ssp. lactis and L. lactis ssp. cremoris, were evaluated for their flavor-forming ability in a mini-Gouda cheese model and were shown to have potential to diversify flavor in this system (D. Cavanagh, unpublished). Future use of these strains in a commercial setting will depend on a number of technological characteristics inherent in the strains, including their sensitivity to phages. The phage sensitivity of the non-dairy isolate bank was assessed using 8 common dairy lactococcal phages. Phage titers in excess of 107 PFU/mL were propagated and tested against the target strain using a spot plate assay performed in triplicate (Table 1). ΦPOO8, ΦbIL170, ΦHP, ΦC2 and ΦKSY1 were incapable of infecting all of the non-dairy isolates tested. However, ΦML3 was capable of infecting strains DPC6855 and DPC6856, isolated from grass and bovine rumen, respectively, while ΦebI infected the strains DPC6854 and DPC6855 but not DPC6856 (Table 1).
Table 1.
Phage resistance profile of dairy lactococcal phages to non-dairy Lactococcus strains.
| Strain | Phage | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Phage group | 936 | c2 | KSY 1 | ||||||
| Origin | Φ712 | ΦPOO8 | ΦbIL170 | ΦHP | ΦC2 | ΦebI | ΦML3 | ΦKSY1 | |
| L. lactis ssp. lactis DPC6853 | Corn | − | − | − | − | − | − | − | − |
| L. lactis ssp. cremoris DPC6854 | Grass | − | − | − | − | − | + | − | − |
| L. lactis ssp. cremoris DPC6855 | Grass | − | − | − | − | − | + | + | − |
| L. lactis ssp. cremoris DPC6856 | Rumen | + | − | − | − | − | − | + | − |
| L. lactis ssp. cremoris DPC6857 | Grass | − | − | − | − | − | − | − | − |
| L. lactis ssp. cremoris DPC6858 | Grass | − | − | − | − | − | − | − | − |
| L. lactis ssp. cremoris DPC6859 | Grass | − | − | − | − | − | − | − | − |
| L. lactis ssp. cremoris DPC6860 | Grass | − | − | − | − | − | − | − | − |
Overall, the non-dairy isolates displayed significant levels of insensitivity to the dairy phages. Given the high evolution rate of phages to overcome bacterial defence systems (Sturino and Klaenhammer, 2006), however, it is highly likely that over time, phages would emerge to threaten these bacterial strains. To understand the interaction of these isolates with the phages that infect them, a screening programme was initiated to identify phages specific for some of the non-dairy lactococci. Following 3 enrichment cycles of raw sewage samples with the grass isolate L. lactis DPC6860 as a host, phage ΦL47 was isolated. ΦL47 was found to form clear plaques of ~0.6 mm diameter when plaqued on L. lactis DPC6860. The formation of clear plaques would suggest that ΦL47 is virulent. Phage titers of 107–108 PFU/mL were recovered after a single propagation.
Limited host range of L. lactis DPC6860
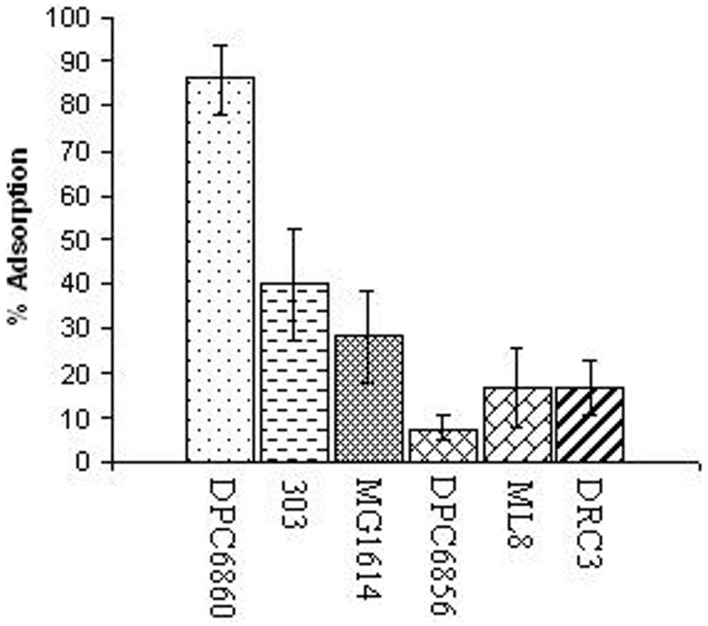
In order to determine the host range of ΦL47, a range of lactococcal strains were chosen for analysis including industrial dairy starters, strains of non-dairy origins and a strain from raw milk (Table 2). It was found that ΦL47 is capable of infecting 3 out of 7 non-dairy Lactococcus isolates but only 4 out of the 19 dairy strains, corresponding to hosts of 2 phage groups including the 949 group (Table 2). ΦL47 was unable to infect L. lactis IL1403, another host strain of Φ949. Contrastingly, although Φ949 was only able to infect 7 of 59 strains of Lactococcus, these corresponded to hosts belonging to 5 phage groups (Samson and Moineau, 2010). Levels of adsorption of ΦL47 to L. lactis DPC6860 were >85% compared to <40% for other strains tested (Figure 1). However, ΦL47 was capable of lysing strain ML8 but not 303 even though a higher level of phage adsorption was observed in the latter. This would suggest that L. lactis 303 possesses a form of anti-phage defence which prevents phage DNA entry or cleaves phage DNA.
Table 2.
Host range of Lactococcus phage ΦL47.
| Species | Origin/Use | Host celllysis |
|---|---|---|
| L. lactis ssp. cremoris DPC6855 | Grass | − |
| L. lactis ssp. cremoris DPC6859 | Grass | + |
| L. lactis ssp. cremoris DPC6854 | Grass | − |
| L. lactis ssp. cremoris DPC6858 | Grass | + |
| L. lactis ssp. cremoris DPC6857 | Grass | + |
| L. lactis ssp. cremoris DPC6856 | Rumen | − |
| L. lactis ssp. lactis DPC6853 | Corn | − |
| L. lactis ssp. cremoris HP | Dairy | + |
| L. lactis ssp. cremoris IE16 | Dairy | − |
| L. lactis ssp. cremoris MG1363 | Dairy | − |
| L. lactis ssp. lactis IL1403 | Dairy | − |
| L. lactis SMQ562 | Dairy | − |
| L. lactis ssp. cremoris UC509 | Dairy | − |
| L. lactis ssp. lactis biovar diacetylactis F7/2 | Dairy | − |
| L. lactis ssp. lactis ML8 | Cheese | + |
| L. lactis ssp. cremoris H88M1 | Raw milk | − |
| L. lactis ssp. lactis 303 | Cheese | − |
| L. lactis HT−2 | Dairy | − |
| L. lactis ssp. cremoris SK11 | Cheese | − |
| L. lactis U53 | Dairy | + |
| L. lactis ssp. lactis biovar diacetylactis DRC3 | Dairy | − |
| L. lactis 83 | Dairy | − |
| L. lactis BA1 | Dairy | − |
| L. lactis ssp. lactis biovar diacetylactis 938 | Dairy | − |
| L. lactis ssp. cremoris 310 | Cheese | + |
| L. lactis ssp. lactis 229 | Cheese | − |
| Lactobacillus plantarum | Grass | − |
| Lactobacillus brevis | Grass | − |
Figure 1.
Relative % adsorption of ΦL47 to Lactococcus strains of different environmental niches and subspecies designation.
Extended tail fiber of ΦL47
Visualization of phage particles was achieved by transmission electron microscopy performed on samples obtained from both CsCl gradient purification of PEG precipitations and ammonium acetate concentration. Images generated from samples obtained by CsCl gradient showed the majority of phage virions to have disintegrated, with broken tails and empty capsids. Images generated from ammonium acetate preparations displayed intact phage particles which allowed for the estimation of capsid diameter and tail length (Figure 2). It was determined that ΦL47 possesses an icosahedral capsid (diameter 75 nm) and a non-contractile tail (length 480 nm; width 15 nm) indicating that this phage is a member of the family Siphoviridae of the class Caudovirales. Morphologically, ΦL47 is very similar to the other lactococcal phage, Φ949 which possesses a non-contractile tail of 500 nm in length with an icosahedral capsid of 79 nm in diameter (Jarvis, 1984; Deveau et al., 2006; Samson and Moineau, 2010). However, ΦL47 appears to possess a distinctive tail fiber of 280 nm in length and tail fiber width of <10 nm (Figure 2B). Siphoviridae phages frequently possess a tail fiber involved in phage infection, however, a tail fiber of this length has not previously been reported in lactococcal phages (Vegge et al., 2005; Boulanger et al., 2008; Jakutytë et al., 2012). Enterococcus faecalis bacteriophage ΦEF24C-P2 was found to possess a long tail fiber which was associated with higher infectivity against bacterial strains than the un-mutated phage which does not have a tail fiber (Uchiyama et al., 2011). Similar to ΦEF24C-P2, an extended tail fiber was found in Bacteroides fragilis phage ATCC 51477-BI, which was isolated from wastewater (Hawkins et al., 2008). No bouquet-like arrangement was identified in ΦL47, as demonstrated by phage Φγ of Bacillus anthracis which possessed a 63 nm long tail fiber (Schuch and Fischetti, 2006).
Figure 2.
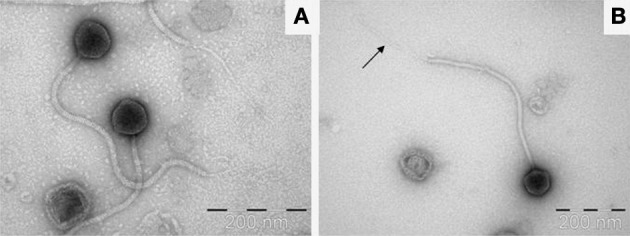
(A) Transmission electron micrograph image of ΦL47 negatively stained using 2% uranyl acetate. (B) Single view of ΦL47. Arrow shows the elongated tail fiber.
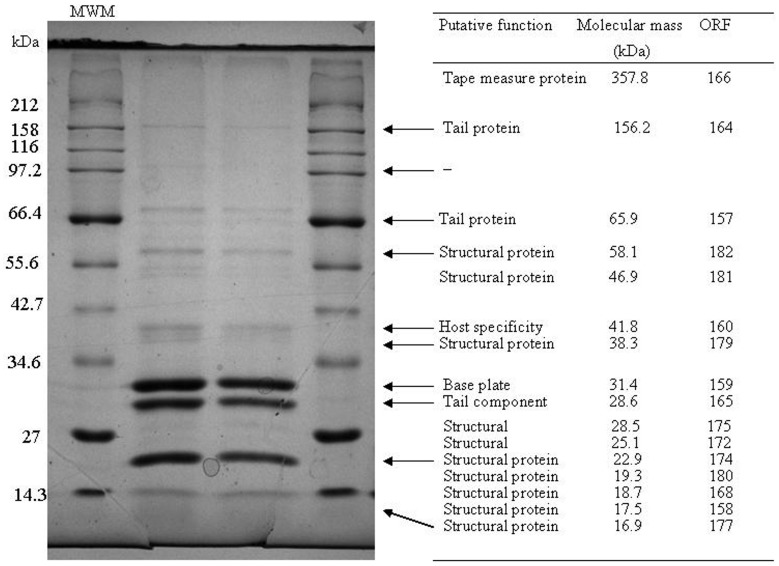
Structural protein analysis
SDS-PAGE was employed to isolate the structural proteins of ΦL47 and this led to the identification of 9 individual protein bands (Figure 3). Five proteins of low molecular weight (<30 kDa) identified from genome sequence analysis, were not identified on the gel. These proteins may not have been present at a sufficient concentration to allow for visualization. Molecular masses of proteins estimated by SDS-PAGE corresponded to the estimated molecular weight of structural proteins predicted from genome sequencing. Three major structural proteins were identified from SDS-PAGE, with the largest having a molecular weight of 31.4 kDa and may correspond to a base plate protein (ORF 176). One large minor protein of ~100 kDa was visualized on the gel but did not correspond to any identified ORF. Multiple bands were observed at ~50 kDa and ~30 kDa which could not be assigned to identified ORFs. It is possible that these proteins are proteolytic products of some other phage structural proteins, or they are of bacterial origin. Techniques such as N-terminal sequencing of protein bands would allow for a more definite relationship to be established between genome analyses and SDS-PAGE.
Figure 3.
Analysis of the structural proteins of ΦL47 obtained from ammonium acetate precipitations. A broad range ladder (New England Biolabs) was used as a relative molecular weight marker (MWM). Molecular masses of structural proteins was predicted from genome sequence data using the Compute pI/ Mw tool (Gasteiger et al., 2005).
Analysis of the L47 genome
General characteristics
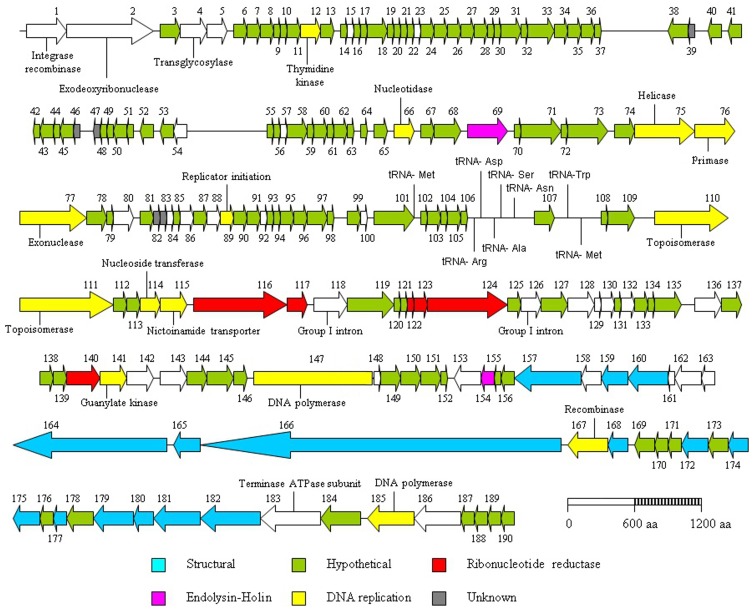
To date, all 59 sequenced L. lactis phages available in the public domain possess a dsDNA genome and are predominately 15 to 35 kb in size with the exception of the larger genomes of ΦKSY1 (79.2 kb), ΦP087 (60.1 kb), and Φ949 (114.8 kb). Analysis of the ΦL47 genome revealed a dsDNA molecule of 128, 932 bp, making it the largest sequenced lactococcal phage to date. ΦL47 was found to possess a molecular GC content of 32.5% which corresponds to that observed in the similarly sized Lactococcus phage, Φ949 (32.7%). The GC content of the host organism, L. lactis DPC6860, was calculated as 35.56%. A total of 190 ORFs were identified in the ΦL47 genome, 117 of which shared significant homology with L. lactis Φ949 (>50% amino acid identity). The majority of these genes were involved in capsid and tail morphogenesis as well as various hypothetical proteins. Φ949 was isolated from cheese whey in New Zealand over 35 years ago while ΦL47 was isolated from raw sewage indicating how this phage group thrives in their respective environments. Five ORFs were identified that shared no significant identity with proteins in the databases. Similar to the unusual genomic arrangement of Φ949, 4 gene clusters were identified in the ΦL47 genome, with 2 clusters transcribed in the opposite direction. Large inter-genic regions were identified between ORFs 37–38 and ORFs 54–55 which were preceded by a shift in the direction of gene transcription. The 190 ORFs identified in the ΦL47 genome (Table 3) account for 88% of the genome which is the same as that identified in Φ949. No putative function could be assigned to 132 ORFs (69.47%). The predominant start codon (89.71%) was ATG with only 8 ORFs starting with an uncommon start codon (TTG, GTG).
Table 3.
Predicted open reading frames (ORFs) of ΦL47a, b and predicted database matches.
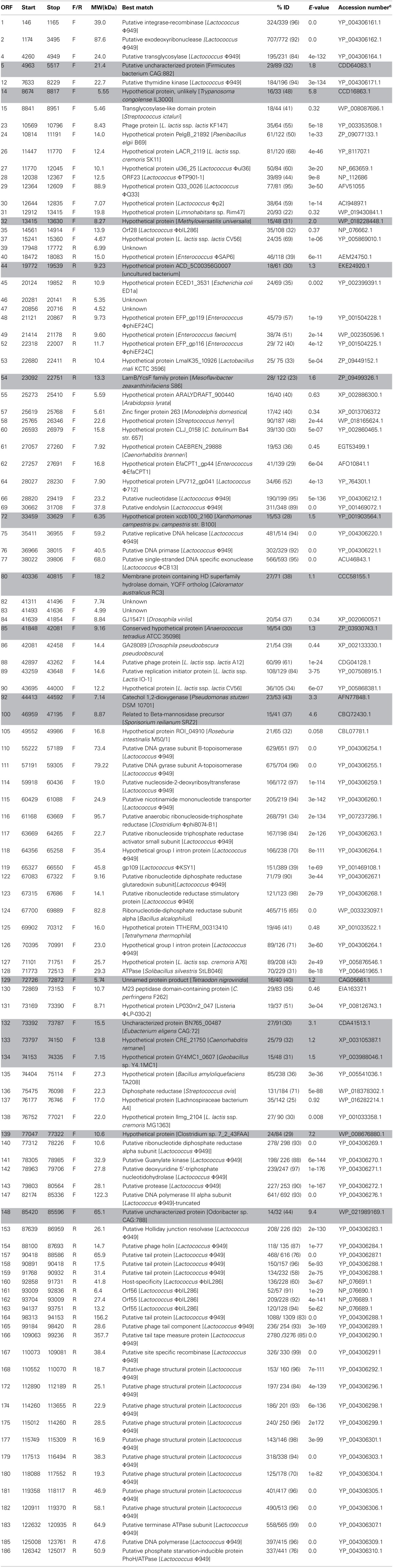
aHypothetical proteins with >60% identity to Φ949 have been omitted.
bORFs shaded gray possess E-values >1.
cAccession numbers corresponding to the NCBI database.
Abbreviations: F/ R, Forward/ Reverse; MW, Molecular weight; %ID, % Identity; E-value, Expect-value.
Tail morphology
As stated previously, ΦL47 possessed a long non-contractile tail of 480 nm in addition to a long tail fiber of 280 nm. Both capsid and tail structural genes were encoded from ORFs 157 to 182, and organized in a modular arrangement located downstream from the holin gene (Figure 4). The products of 14 of these ORFs encoded putative structural proteins sharing >70% amino acid identity with Φ949 (Table 3). This is not unusual as the major structural proteins are generally somewhat conserved in related phages (Suyama and Bork, 2001; Ceyssens et al., 2011). In contrast, proteins involved in adsorption to host cells, such as tail fibers, are expected to differ to a larger degree as they are modified to complement with surface receptors of bacterial hosts (Sandmeier et al., 1992; Hatfull, 2002; Desplats and Krisch, 2003; Silhavy et al., 2010). ORFs 160–163, located within the structural module, encode proteins sharing a high percentage identity (>60%) to proteins of ΦbIL286. ORF 159 encodes a protein with a high percentage identity to a putative tail protein of Lactococcus Φ949 but also the putative base plate protein of Lactococcus ΦP335, and the phage tail assembly protein of L. lactis ssp. cremoris A76. A conserved domain search of the putative tail protein of Φ949 reveals the presence of a prophage tail super family cl12123 domain which may possibly act as an endopeptidase.
Figure 4.
Genome organization of the lactococcal phage ΦL47. Each arrow represents an open reading frame (ORF), the orientation of which shows the direction of transcription. The predicted function of each ORF was determined by bioinformatic analyses (Carver et al., 2005).
ORF 160 encodes a putative host-specificity protein sharing a 60% identity at the amino acid level with that of the prophage ΦbIL286. This ORF also shares identity to the phage receptor binding protein (35%) and tail host specificity protein (36%) of the plant strain L. lactis KF147. ORFs 162 and 163 also indicate homology with ORF 55 of ΦbIL286, both with 89 and 94% identity, respectively. No conserved domains were identified within these two proteins; however, using BLASTp, both proteins were found to possess some identity to a putative tail fiber of Bacillus ΦphiNIt1. In order to assign a putative function to these ORFs of ΦL47, ORF 55 of ΦbIL286 was analyzed by InterProScan. The C-terminal region of this gene product was predicted to have an immunoglobulin-like domain IPR007110 as found in the major tail subunit of Enterobacteria ΦHK97 (Juhala et al., 2000) and other phages. Immunoglobulin-like domains participate in various functions, including cell-cell recognition and cell-surface receptors. Therefore, it is likely that these genes also form part of the ΦL47 tail.
Host cell lysis
In dsDNA phages, the combined action of the holin-lysin genes function in the release of new phage particles from an infected cell (Daniel et al., 2007). ORF 69 was found to encode a putative endolysin sharing 89% identity with that of Φ949, which possesses an amidase_2 domain pfam01510. This ORF also shows a high degree of similarity with gp073, sequence ID YP_001469072.1, of the temperate lactococcal phage ΦKSY1 (83% identity). No holin gene was identified in the same locus as the endolysin, however, a putative holin was located remotely from the endolysin (Figure 4). ORF 154 exhibits an 87% identity to the putative phage holin from Φ949, containing a holin LLH superfamily cl09890 domain, a conserved domain of ~100 amino acids found in prophage and phage of Gram-positive bacteria. The arrangement of the holin and endolysin genes in the ΦL47 genome is not typical of lactococcal phages but a similar organization has been reported in Φ949, ΦP087, and Φ1708 (Garneau et al., 2008; Villion et al., 2009).
DNA replication and nucleotide biosynthesis
The DNA replication module was located downstream from the endolysin gene and upstream from the holin gene. The predicted gene products for ORFs 75 (Helicase), 76 (Primase), 66 (Nucleotidase), 110 and 111 (DNA gyrase) and 114 (Deoxyribosyltransferase) displayed a large degree of homology with Φ949. ORF 89 was identified as a putative replication protein which shares 84% identity with the putative replication initiator protein of L. lactis ssp. lactis IO-1 (Kato et al., 2012). A DHH domain pfam01368 was identified in ORF 77 which includes the single-stranded DNA exonuclease RecJ. The protein encoded by this ORF exhibits a 95% identity with the single-stranded DNA exonuclease of Lactococcus ΦC13, Lactococcus ΦCaesusJM1 and Lactococcus Φ949. An exonuclease functions to catalyse the cleavage of a single nucleotide from the end of a polynucleotide chain and is involved in DNA repair, recombination and replication (Ceska and Sayers, 1998). No putative endonucleases were identified in the L47 genome, in contrast to two distinct HNH endonucleases in Φ949. Structure specific endonucleases cut at particular DNA structures and in some cases can give a competitive advantage to a given bacteriophage in a mixed infection (Goodrich-Blair and Shub, 1996). It is possible that ΦL47 possesses no HNH endonucleases as it has acquired other characteristics, such as possessing a reduced number of endonuclease recognition sites, that offer an advantage over other bacteriophages. Group I introns are frequently associated with endonucleases (Chevalier and Stoddard, 2001), however, ORFs 118 and 126 were both found to encode this type of intervening sequence (IVS) between ribonucleotide reductase proteins. Previously, two introns were found within the ribonucleotide reductase large subunit of Staphylococcus aureus ΦTwort (Landthaler et al., 2002). In Escherichia coli ΦT4, the aerobic Ribonucleotide reductase small subunit and the anaerobic ribonucleotide reductase were both found to possess group I introns (Sjöberg et al., 1986; Young et al., 1994). One explanation offered for the retention of introns in genes, such as those involved in ribonucleotide reduction, is that they encode functions replicated in the host organism that can be utilized for phage survival until such time as the insertion element and host environment adapt to one another (Derbyshire and Belfort, 1998).
The acquisition of ribonucleotide reductases (RNRs) is thought to arise from the host genome, to enable the adaptation to certain environmental conditions (Dwivedi et al., 2013). These enzymes function in DNA replication and repair via the conversion of ribonucleotides to deoxyribonucleotides (Dwivedi et al., 2013). The gene products of 7 ORFs were found to encode ribonucleotide di- and tri-phosphate reductases. ORF 116 encodes a putative anaerobic ribonucleoside-triphosphate reductase with a 25% identity to a putative anaerobic ribonucleoside-triphosphate reductase of Clostridium Φphi8074-B1. A class III ribonucleotide reductase domain cd01675 was identified within this gene product which uses a FeS cluster and S-adenosylmethionine to generate a glycyl radical. ORF 124 shares a 65% identity with the ribonucleotide-diphosphate reductase subunit alpha of Bacillus alcalophilus and contains a class I RNR domain cd01679. In contrast to class III, a class I RNR domain uses a di-iron-tyrosyl radical. The presence of class I and class III RNR genes in ΦL47 conforms with the RNR complement generally found in phages isolated from sewage (Dwivedi et al., 2013). Other ribonucleotide reductases shared a large degree of homology with Φ949 as well as other genes involved in nucleotide transport, modification and degradation.
tRNA encoding genes in L47 genome
The role of tRNAs in phage genomes is thought to be the encoding of codons that are less frequent in the host genome, thus allowing for the increased expression of phage proteins (Bailly-Bechet et al., 2007). In all, 8 tRNA genes (Table 4) were identified over a small region of the genome (48,524–53,674 bp) in ΦL47, using the tRNA-scan SE and Aragorn software (Lowe and Eddy, 1997; Laslett and Canback, 2004). tRNAs had a %GC content ranging from 39.4 to 50.7. Between ORFs 106 and 107, were positioned tRNAArg, tRNAAsp and tRNAAla while between ORFs 107 and 108 were tRNAAsn, tRNASer and tRNATrp. A 58 bp intron, from position 34 to 35, was identified in tRNASer. Two tRNAMet genes were identified, the first situated between ORFs 101 and 102, and the second between ORFs 107 and 108. The possession of more than one tRNA for a specific amino acid is a phenomenon that has been observed in Lactobacillus plantarum bacteriophage ΦLP65 (Chibani-Chennoufi et al., 2004b). The acquisition of tRNA genes from the host is proposed as being a random event and the genes are either retained, via a set of selection mechanisms, or they are lost (Bailly-Bechet et al., 2007). It is conceivable that ΦL47 has acquired two tRNAMet in order to gain a fitness advantage, due to the relatively harsh environment that it inhabits. In-situ burst sizes for organisms in a nutrient-poor environment, such as that from which the host was isolated, are generally smaller than when the host is infected in a nutrient rich, chemically defined media (Chibani-Chennoufi et al., 2004a). The retention of particular tRNA genes in phage genomes are thought to correspond to codons that are less abundant in the host genome (Bailly-Bechet et al., 2007) although this was not observed for Φ949 and its host L. lactis IL1403 (Samson and Moineau, 2010). Therefore, we may only hypothesize that these tRNA genes are associated with controlling phage protein production and are possibly involved in increasing reproduction rate and reducing latency time (Bailly-Bechet et al., 2007).
Table 4.
tRNA arrangement in ΦL47.
| tRNA | Amino acid | Anti-codon | Size (bp) | % GC | Start | End |
|---|---|---|---|---|---|---|
| 1 | Met | CAT | 73 | 47.9 | 48524 | 48596 |
| 2 | Arg | TCT | 73 | 39.7 | 50343 | 50415 |
| 3 | Asp | GTC | 75 | 50.7 | 51213 | 51287 |
| 4 | Ala | CGC | 71 | 50.7 | 51639 | 51709 |
| 5 | Ser | ACT | 92 | 43.5 | 51967 | 52116 |
| 6 | Asn | ATT | 76 | 39.5 | 52830 | 52905 |
| 7 | Trp | CCA | 71 | 39.4 | 53114 | 53184 |
| 8 | Met | CAT | 74 | 55.4 | 53400 | 53473 |
The arrangement of the tRNAs into distinct blocks suggests that they were obtained through separate recombination events with either host DNA, other phages or a combination of both (Weinbauer, 2004). The presence of tRNAs has been found to be particularly common in phages with a larger genome size (Bailly-Bechet et al., 2007). Six tRNA genes were identified in Φ949, the most identified in a lactococcal bacteriophage (Samson and Moineau, 2010). Similarly, in ΦLb338-1 and ΦK, both with a genome larger than 100 kb, 2 and 4 tRNAs were identified, respectively (O'Flaherty et al., 2004; Alemayehu et al., 2009). No tRNA encoding regions were found in 2 recently elucidated lactococcal phage genomes, belonging to a new P335 group, with sizes of ~31 kb (Mahony et al., 2013). However, 5 tRNA genes have been identified in ΦP087, a P087 species, of a smaller genome size (60,074 bp) than Φ949 and ΦL47 (Villion et al., 2009).
Comparative genome analysis
Numerous ORFs in the L47 genome were found to show identity to putative proteins of other lactococcal phages. ORFs 27–30 encode hypothetical proteins belonging to different lactococcal phages (Table 3). No conserved domains were detected within these ORFs. ORF 119, showing 39% identity with Lactococcus ΦKSY1, was found to encode a pfam01139 domain, a conserved domain of the uncharacterized protein family UPF0027. Putative proteins expressed by Enterococcus ΦphiEF24C and Enterococcus faecium are also found to be similar to those identified in the ΦL47 genome (3 ORFs; >40% identity). Previously, lactococcal ΦP087 was shown to possess genes with a high degree of similarity to structural genes of a prophage of Enterococcus faecalis V538 (Villion et al., 2009). Of note, ORF 23 was found to be highly similar to a phage protein of the plant Lactococcus strain, KF147. Upstream from the structural genes, ORF 130 encodes a M23 peptidase domain containing protein similar to Clostridium perfringens F262 (Nowell et al., 2012). The position of this gene with genes involved in DNA replication, suggests that it is not tail associated as in Lactococcus phages Tuc2009 and TP901-1 (Seegers et al., 2004; Stockdale et al., 2013). This peptidase would possibly hydrolyse peptidoglycan via D-Ala–D-Asp endopeptidase activity enabling the penetration of stationary phase lactococcal cells (Samson et al., 2013).
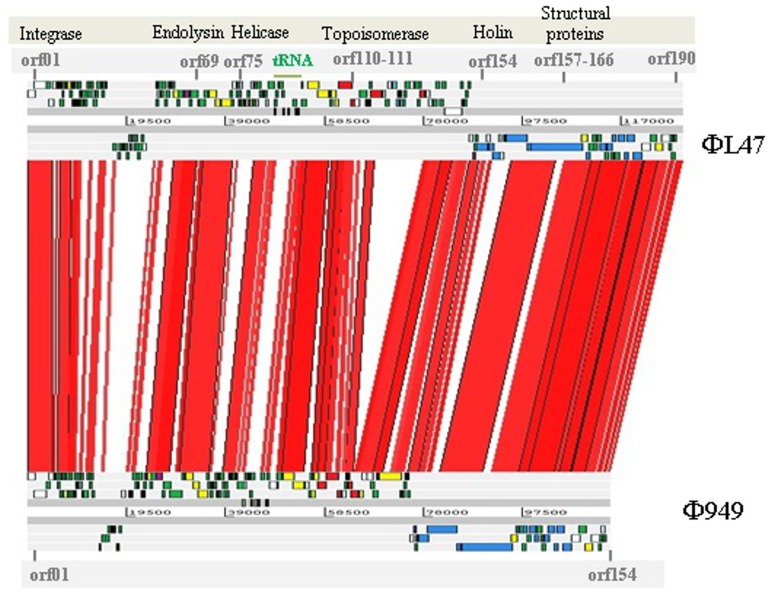
Comparative genomic analysis shows the high similarity at a nucleotide level between the ΦL47 and Φ949 phages (Figure 5). In addition, given the close amino acid identity at protein level across the genome (Table 3), suggesting a number of shared functions, it is likely that these phages have shared a common ancestor at some point. Studies of the more common lactococcal phage groups have indicated a large degree of homology between members of the c2 and 936 species while the P335 species displays a polythetic species theory (Deveau et al., 2006). Although much discussion surrounds phage evolution, in relation to virulent phages vertical lines of evolution are believed to be crucial in the development of certain phage families (Brussow and Kutter, 2005) and are particularly evident in dairy phages (Brüssow and Desiere, 2001). However, horizontal genetic exchange also plays a key role in the evolution of phages as exemplified by genetic mosaicism formed from a high occurrence of transfer events (Canchaya et al., 2003). Genetic exchange between bacteria and other phages from distinct niches is hypothesized to be pivotal in the evolution of rare lactococcal phages (Fortier et al., 2006; Chopin et al., 2007; Garneau et al., 2008; Villion et al., 2009). Due to the high degree of sequence homology between these two phages, it is expected that ΦL47 is another member of the 949 group. Analysis of Φ111 revealed this lactococcal phage to possess a genome size of ~134 kb and a long tail of 470 nm (Prevots et al., 1990). However, without comparative genome analysis it can only be speculated that Φ111 is a representative of the 949 group.
Figure 5.
BLASTn-based alignment (nucleotide identity >90% shown) of the genome sequences of L. lactis phage ΦL47 and Φ949 as displayed by the Artemis comparison tool (ACT). Red lines between genomes indicate orthologs in the same orientation. Coding sequences are color coded as described in Figure 4; structural (blue), hypothetical (green), DNA replication (yellow), ribonucleotide reductase (red), endolysin-holin (mauve), unknown (gray).
Concluding remarks
To date, numerous studies have investigated dairy lactococcal phages and how they interact with their respective hosts. Industrial Lactococcus strains, used in modern cheese production, are thought to have evolved from plant strains (Kelly et al., 2010). ΦL47, isolated from a non-dairy environment, possessed significant similarity to the rare, dairy phage Φ949, with both phages isolated from different environments almost 40 years apart. ΦL47 was found to possess a number of features which differentiate it from Φ949, most notably a long tail fiber, not previously reported in phages of Lactococcus. This tail fiber may play an important role in enabling ΦL47 to infect L. lactis DPC6860, which was largely resistant to dairy phages, and may account for the persistence of successful virulent phages in the wider environment as observed for ΦQ33 and ΦBM13 (Mahony et al., 2013). Further studies are required to establish the diversity of lactococcal bacteriophages from non-dairy origins and the similarity they may possess with dairy phages of other groups. This information could shed further light on the mechanisms they possess that allow them to thrive in harsher environments and may advance our understanding of host recognition and infection by lactococcal phages.
Author contributions
Daniel Cavanagh isolated phage, performed biological assays, genome analysis, data analysis and drafted the manuscript. Caitriona M. Guinane performed sequence assembly, analysis and assisted in manuscript drafting. Horst Neve performed transmission electron microscopy of phage particles. R. Paul Ross and Aidan Coffey advised on experimental design. Gerald F. Fitzgerald is a supervisor of this project. Olivia McAuliffe. is a supervisor of this project and assisted in the design of experiments, data analysis and manuscript drafting.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by the Irish Dairy Levy Research Trust. Daniel Cavanagh was supported by the Teagasc Walsh Fellowship Programme.
References
- Alander M., Satokari R., Korpela R., Saxelin M., Vilpponen-Salmela T., Mattila-Sandholm T., et al. (1999). Persistence of colonization of human colonic mucosa by a probiotic strain, Lactobacillus rhamnosus GG, after oral consumption. Appl. Environ. Microbiol. 65, 351–354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alemayehu D., Ross R. P., O'Sullivan O., Coffey A., Stanton C., Fitzgerald G. F., et al. (2009). Genome of a virulent bacteriophage Lb338-1 that lyses the probiotic Lactobacillus paracasei cheese strain. Gene 448, 29–39 10.1016/j.gene.2009.08.008 [DOI] [PubMed] [Google Scholar]
- Altermann E., Klaenhammer T. R. (2003). GAMOLA: a new local solution for sequence annotation and analyzing draft and finished prokaryotic genomes. OMICS 7, 161–169 10.1089/153623103322246557 [DOI] [PubMed] [Google Scholar]
- Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410 10.1016/S0022-2836(05)80360-2 [DOI] [PubMed] [Google Scholar]
- Ayad E. H. E., Verheul A., De Jong C., Wouters J. T. M., Smit G. (1999). Flavour forming abilities and amino acid requirements of Lactococcus lactis strains isolated from artisanal and non-dairy origin. Int. Dairy J. 9, 725–735 10.1016/s0958-6946(99)00140-5 [DOI] [Google Scholar]
- Ayad E. H. E., Verheul A., Wouters J. T. M., Smit G. (2000). Application of wild starter cultures for flavour development in pilot plant cheese making. Int. Dairy J. 10, 169–179 10.1016/s0958-6946(00)00041-8 [DOI] [Google Scholar]
- Aziz R. K., Bartels D., Best A. A., Dejongh M., Disz T., Edwards R. A., et al. (2008). The RAST server: rapid annotations using subsystems technology. BMC Genomics 9:75 10.1186/1471-2164-9-75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailly-Bechet M., Vergassola M., Rocha E. (2007). Causes for the intriguing presence of tRNAs in phages. Genome Res. 17, 1486–1495 10.1101/gr.6649807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulanger P., Jacquot P., Plançon L., Chami M., Engel A., Parquet C., et al. (2008). Phage T5 straight tail fiber is a multifunctional protein acting as a tape measure and carrying fusogenic and muralytic activities. J. Biol. Chem. 283, 13556–13564 10.1074/jbc.M800052200 [DOI] [PubMed] [Google Scholar]
- Brüssow H., Desiere F. (2001). Comparative phage genomics and the evolution of Siphoviridae: insights from dairy phages. Mol. Microbiol. 39, 213–223 10.1046/j.1365-2958.2001.02228.x [DOI] [PubMed] [Google Scholar]
- Brussow H., Kutter E. (2005). Genomics and evolution of tailed phages, in Bacteriophages: Biology and Applications, eds Kutter E., Sulakvelidze A. (Washington, DC: CRC Press; ), 91–128 [Google Scholar]
- Canchaya C., Fournous G., Chibani-Chennoufi S., Dillmann M.-L., Brüssow H. (2003). Phage as agents of lateral gene transfer. Curr. Opin. Microbiol. 6, 417–424 10.1016/S1369-5274(03)00086-9 [DOI] [PubMed] [Google Scholar]
- Carver T. J., Rutherford K. M., Berriman M., Rajandream M. A., Barrell B. G., Parkhill J. (2005). ACT: the Artemis Comparison Tool. Bioinformatics 21, 3422–3423 10.1093/bioinformatics/bti553 [DOI] [PubMed] [Google Scholar]
- Ceska T. A., Sayers J. R. (1998). Structure-specific DNA cleavage by 5′ nucleases. Trends Biochem. Sci. 23, 331–336 10.1016/S0968-0004(98)01259-6 [DOI] [PubMed] [Google Scholar]
- Ceyssens P.-J., Glonti T., Kropinski A. M., Lavigne R., Chanishvili N., Kulakov L., et al. (2011). Phenotypic and genotypic variations within a single bacteriophage species. Virol. J. 8, 1–5 10.1186/1743-422X-8-134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevalier B. S., Stoddard B. L. (2001). Homing endonucleases: structural and functional insight into the catalysts of intron/intein mobility. Nucleic Acids Res. 29, 3757–3774 10.1093/nar/29.18.3757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chibani-Chennoufi S., Bruttin A., Dillmann M.-L., Brüssow H. (2004a). Phage-host interaction: an ecological perspective. J. Bacteriol. 186, 3677–3686 10.1128/JB.186.12.3677-3686.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chibani-Chennoufi S., Dillmann M.-L., Marvin-Guy L., Rami-Shojaei S., Brüssow H. (2004b). Lactobacillus plantarum bacteriophage LP65: a new member of the SPO1-like genus of the family Myoviridae. J. Bacteriol. 186, 7069–7083 10.1128/JB.186.21.7069-7083.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chopin A., Bolotin A., Sorokin A., Ehrlich S. D., Chopin M. C. (2001). Analysis of six prophages in Lactococcus lactis IL1403: different genetic structure of temperate and virulent phage populations. Nucleic Acids Res. 29, 644–651 10.1093/nar/29.3.644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chopin A., Deveau H., Ehrlich S. D., Moineau S., Chopin M.-C. (2007). KSY1, a lactococcal phage with a T7-like transcription. Virology 365, 1–9 10.1016/j.virol.2007.03.044 [DOI] [PubMed] [Google Scholar]
- Coffey A., Ross R. P. (2002). Bacteriophage-resistance systems in dairy starter strains: molecular analysis to application. Antonie van Leeuwenhoek 82, 303–321 10.1023/A:1020639717181 [DOI] [PubMed] [Google Scholar]
- Crutz-Le Coq A.-M., Cesselin B., Commissaire J., Anba J. (2002). Sequence analysis of the lactococcal bacteriophage bIL170: insights into structural proteins and HNH endonucleases in dairy phages. Microbiology 148, 985–1001 [DOI] [PubMed] [Google Scholar]
- Daniel A., Bonnen P. E., Fischetti V. A. (2007). First complete genome sequence of two Staphylococcus epidermidis bacteriophages. J. Bacteriol. 189, 2086–2100 10.1128/JB.01637-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delcher A. L., Harmon D., Kasif S., White O., Salzberg S. L. (1999). Improved microbial gene identification with GLIMMER. Nucleic Acids Res. 27, 4636–4641 10.1093/nar/27.23.4636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derbyshire V., Belfort M. (1998). Lightning strikes twice: intron–intein coincidence. Proc. Natl. Acad. Sci. U.S.A. 95, 1356–1357 10.1073/pnas.95.4.1356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desplats C., Krisch H. M. (2003). The diversity and evolution of the T4-type bacteriophages. Res. Microbiol. 154, 259–267 10.1016/S0923-2508(03)00069-X [DOI] [PubMed] [Google Scholar]
- Deveau H., Labrie S. J., Chopin M.-C., Moineau S. (2006). Biodiversity and classification of lactococcal phages. Appl. Environ. Microbiol. 72, 4338–4346 10.1128/AEM.02517-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupuis M.-È., Moineau S. (2010). Genome organization and characterization of the virulent lactococcal phage 1358 and its similarities to listeria phages. Appl. Environ. Microbiol. 76, 1623–1632 10.1128/aem.02173-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dwivedi B., Xue B., Lundin D., Edwards R. A., Breitbart M. (2013). A bioinformatic analysis of ribonucleotide reductase genes in phage genomes and metagenomes. BMC Evol. Biol. 13:1 10.1186/1471-2148-13-33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewing B., Green P. (1998). Base-calling of automated sequencer traces usingPhred. II. error probabilities. Genome Res. 8, 186–194 10.1101/gr.8.3.175 [DOI] [PubMed] [Google Scholar]
- Ferguson P., Coombs D. (2000). Pulse-chase analysis of the in vivo assembly of the bacteriophage T4 tail. J. Mol. Biol. 297, 99–117 10.1006/jmbi.2000.3551 [DOI] [PubMed] [Google Scholar]
- Fortier L.-C., Bransi A., Moineau S. (2006). Genome sequence and global gene expression of Q54, a new phage species linking the 936 and c2 phage species of Lactococcus lactis. J. Bacteriol. 188, 6101–6114 10.1128/JB.00581-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garneau J. E., Tremblay D. M., Moineau S. (2008). Characterization of 1706, a virulent phage from Lactococcus lactis with similarities to prophages from other Firmicutes. Virology 373, 298–309 10.1016/j.virol.2007.12.002 [DOI] [PubMed] [Google Scholar]
- Gasteiger E., Hoogland C., Gattiker A., Wilkins M. R., Appel R. D., Bairoch A. (2005). Protein identification and analysis tools on the ExPASy server, in The Proteomics Protocols Handbook, ed Walker J. M. (New Jersey, NJ: Springer; ), 571–607 [Google Scholar]
- Goodrich-Blair H., Shub D. A. (1996). Beyond homing: competition between intron endonucleases confers a selective advantage on flanking genetic markers. Cell. 84, 211–221 10.1016/S0092-8674(00)80976-9 [DOI] [PubMed] [Google Scholar]
- Gordon D. (2003). Viewing and editing assembled sequences using Consed. Curr. Protoc. Bioinform. 2, 11.12.11–11.12.43 10.1002/0471250953.bi1102s02 [DOI] [PubMed] [Google Scholar]
- Hatfull G. F. (2002). A tail of two specifi-cities. Science. 295, 2031–2032 10.1126/science.1070586 [DOI] [PubMed] [Google Scholar]
- Hawkins S. A., Layton A. C., Ripp S., Williams D., Sayler G. S. (2008). Genome sequence of the Bacteroides fragilis phage ATCC 51477-B1. Virol. J. 5, 97 10.1186/1743-422X-5-97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyatt D., Chen G.-L., Locascio P., Land M., Larimer F., Hauser L. (2010). Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinform. 11:1 10.1186/1471-2105-11-119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakutytë L., Lurz R., Baptista C., Carballido-Lopez R., São-José C., Tavares P., et al. (2012). First steps of bacteriophage SPP1 entry into Bacillus subtilis. Virology 422, 425–434 10.1074/jbc.M301805200 [DOI] [PubMed] [Google Scholar]
- Jarvis A. W. (1984). Differentiation of lactic streptococcal phages into phage species by DNA-DNA homology. Appl. Environ. Microbiol. 47, 343–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvis A. W., Fitzgerald G. F., Mata M., Mercenier A., Neve H., Powell I. B., et al. (1991). Species and type phages of lactococcal bacteriophages. Intervirology 32, 2–9 [DOI] [PubMed] [Google Scholar]
- Juhala R. J., Ford M. E., Duda R. L., Youlton A., Hatfull G. F., Hendrix R. W. (2000). Genomic sequences of bacteriophages HK97 and HK022: pervasive genetic mosaicism in the lambdoid bacteriophages. J. Mol. Biol. 299, 27–51 10.1006/jmbi.2000.3729 [DOI] [PubMed] [Google Scholar]
- Kato H., Shiwa Y., Oshima K., Machii M., Araya-Kojima T., Zendo T., et al. (2012). Complete genome sequence of Lactococcus lactis IO-1, a lactic acid bacterium that utilizes xylose and produces high levels of L-Lactic acid. J. Bacteriol. 194, 2102–2103 10.1128/JB.00074-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly D., O'Sullivan O., Mills S., McAuliffe O., Ross P., Neve H., et al. (2012). Genome sequence of the phage clP1, which infects the beer spoilage bacterium Pediococcus damnosus. Gene 504, 53–63 10.1016/j.gene.2012.04.085 [DOI] [PubMed] [Google Scholar]
- Kelly W. J., Ward L. J. H., Leahy S. C. (2010). Chromosomal diversity in Lactococcus lactis and the origin of dairy starter cultures. Genome Biol. Evol. 2, 729–744 10.1093/gbe/evq056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labrie S. J., Moineau S. (2007). Abortive infection mechanisms and prophage sequences significantly influence the genetic makeup of emerging lytic lactococcal phages. J. Bacteriol. 189, 1482–1487 10.1128/JB.01111-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landthaler M., Begley U., Lau N. C., Shub D. A. (2002). Two self-splicing group I introns in the ribonucleotide reductase large subunit gene of Staphylococcus aureus phage Twort. Nucleic Acids Res. 30, 1935–1943 10.1093/nar/30.9.1935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laslett D., Canback B. (2004). ARAGORN, a program to detect tRNA genes and tmRNA genes in nucleotide sequences. Nucleic Acids Res. 32, 11–16 10.1093/nar/gkh152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe T. M., Eddy S. R. (1997). tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25, 0955–0964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubbers M. W., Waterfield N. R., Beresford T., Le Page R., Jarvis A. W. (1995). Sequencing and analysis of the prolate-headed lactococcal bacteriophage c2 genome and identification of the structural genes. Appl. Environ. Microbiol. 61, 4348–4356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahony J., Martel B., Tremblay D. M., Neve H., Heller K. J., Moineau S., et al. (2013). Identification of a new P335 subgroup through molecular analysis of lactococcal phages Q33 and BM13. Appl. Environ. Microbiol. 79, 4401–4409 10.1128/AEM.00832-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchler-Bauer A., Lu S., Anderson J. B., Chitsaz F., Derbyshire M. K., Deweese-Scott C., et al. (2011). CDD: a Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Res. 39, D225–D229 10.1093/nar/gkq1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moineau S., Fortier J., Ackermann H.-W., Pandian S. (1992). Characterization of lactococcal bacteriophages from Quebec cheese plants. Can. J. Microbiol. 38, 875–882 10.1139/m92-143 [DOI] [Google Scholar]
- Morales P., Fernández-GarcımathìA E., Gaya P., Nuñez M. (2003). Formation of volatile compounds by wild Lactococcus lactis strains isolated from raw ewes' milk cheese. Int. Dairy J. 13, 201–209 10.1016/s0958-6946(02)00151-6 [DOI] [Google Scholar]
- Murphy J., Royer B., Mahony J., Hoyles L., Heller K., Neve H., et al. (2013). Biodiversity of lactococcal bacteriophages isolated from 3 Gouda-type cheese producing plants. J. Dairy Sci. 96, 4945–4957 10.3168/jds.2013-6748 [DOI] [PubMed] [Google Scholar]
- Nowell V. J., Kropinski A. M., Songer J. G., Macinnes J. I., Parreira V. R., Prescott J. F. (2012). Genome sequencing and analysis of a Type A Clostridium perfringens isolate from a case of bovine clostridial abomasitis. PLoS ONE 7:e32271 10.1371/journal.pone.0032271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Flaherty S., Coffey A., Edwards R., Meaney W., Fitzgerald G. F., Ross R. P. (2004). Genome of staphylococcal phage K: a new lineage of Myoviridae infecting gram-positive bacteria with a low G+C content. J. Bacteriol. 186, 2862–2871 10.1128/JB.186.9.2862-2871.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prevots F., Mata M., Ritzenthaler P. (1990). Taxonomic differentiation of 101 lactococcal bacteriophages and characterization of bacteriophages with unusually large genomes. Appl. Environ. Microbiol. 56, 2180–2185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutherford K., Parkhill J., Crook J., Horsnell T., Rice P., Rajandream M. A., et al. (2000). Artemis: sequence visualization and annotation. Bioinformatics 16, 944–945 10.1093/bioinformatics/16.10.944 [DOI] [PubMed] [Google Scholar]
- Sambrook J., Russell D. W. (2001). Molecular Cloning: a Laboratory Manual. Vol. 1–3 Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press [Google Scholar]
- Samson J. E., Magadán A. H., Sabri M., Moineau S. (2013). Revenge of the phages: defeating bacterial defences. Nat. Rev. Microbiol. 11, 675–687 10.1038/nrmicro3096 [DOI] [PubMed] [Google Scholar]
- Samson J. E., Moineau S. (2010). Characterization of Lactococcus lactis phage 949 and comparison with other lactococcal phages. Appl. Environ. Microbiol. 76, 6843–6852 10.1128/aem.00796-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandmeier H., Iida S., Arber W. (1992). DNA inversion regions Min of plasmid p15B and Cin of bacteriophage P1: evolution of bacteriophage tail fiber genes. J. Bacteriol. 174, 3936–3944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schatz M. C., Phillippy A. M., Sommer D. D., Delcher A. L., Puiu D., Narzisi G., et al. (2013). Hawkeye and AMOS: visualizing and assessing the quality of genome assemblies. Brief. Bioinform. 14, 213–224 10.1093/bib/bbr074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuch R., Fischetti V. A. (2006). Detailed genomic analysis of the Wβ and γ phages infecting Bacillus anthracis: implications for evolution of environmental fitness and antibiotic resistance. J. Bacteriol. 188, 3037–3051 10.1128/JB.188.8.3037-3051.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seegers J. F., Mc Grath S., O'Connell-Motherway M., Arendt E. K., Van De Guchte M., Creaven M., et al. (2004). Molecular and transcriptional analysis of the temperate lactococcal bacteriophage Tuc2009. Virology. 329, 40–52 10.1016/j.virol.2004.07.003 [DOI] [PubMed] [Google Scholar]
- Silhavy T. J., Kahne D., Walker S. (2010). The bacterial cell envelope. Cold Spring Harb. Perspect. Biol. 2, 1–16 10.1101/cshperspect.a000414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjöberg B. M., Hahne S., Mathews C. Z., Mathews C. K., Rand K. N., Gait M. J. (1986). The bacteriophage T4 gene for the small subunit of ribonucleotide reductase contains an intron. EMBO J. 5, 2031–2036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stockdale S. R., Mahony J., Courtin P., Chapot-Chartier M.-P., Van Pijkeren J.-P., Britton R. A., et al. (2013). The lactococcal phages Tuc2009 and TP901-1 incorporate two alternate forms of their tail fiber into their virions for infection specialization. J. Biol. Chem. 288, 5581–5590 10.1074/jbc.M112.444901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturino J. M., Klaenhammer T. R. (2006). Engineered bacteriophage-defence systems in bioprocessing. Nat. Rev. Microbiol. 4, 395–404 10.1038/nrmicro1393 [DOI] [PubMed] [Google Scholar]
- Suyama M., Bork P. (2001). Evolution of prokaryotic gene order: genome rearrangements in closely related species. Trends Genet. 17, 10–13 10.1016/S0168-9525(00)02159-4 [DOI] [PubMed] [Google Scholar]
- Uchiyama J., Takemura I., Satoh M., Kato S.-I., Ujihara T., Akechi K., et al. Daibata M. (2011). Improved adsorption of an enterococcus faecalis bacteriophage Φ EF24C with a spontaneous point mutation. PLoS ONE 6: e26648 10.1371/journal.pone.0026648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbach G. (1995). Contribution of lactic acid bacteria to flavour compound formation in dairy products. Int. Dairy J. 5, 877–903 10.1016/0958-6946(95)00037-2 [DOI] [Google Scholar]
- Veesler D., Cambillau C. (2011). A common evolutionary origin for tailed-bacteriophage functional modules and bacterial machineries. Microbiol. Mol. Biol. Rev. 75, 423–433 10.1128/MMBR.00014-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vegge C. S., Brøndsted L., Neve H., Mc Grath S., Van Sinderen D., Vogensen F. K. (2005). Structural characterization and assembly of the distal tail structure of the temperate lactococcal bacteriophage TP901-1. J. Bacteriol. 187, 4187–4197 10.1128/JB.187.12.4187-4197.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villion M., Chopin M. C., Deveau H., Ehrlich S. D., Moineau S., Chopin A. (2009). P087, a lactococcal phage with a morphogenesis module similar to an Enterococcus faecalis prophage. Virology 388, 49–56 10.1016/j.virol.2009.03.011 [DOI] [PubMed] [Google Scholar]
- Weinbauer M. G. (2004). Ecology of prokaryotic viruses. FEMS Microbiol. Rev. 28, 127–181 10.1016/j.femsre.2003.08.001 [DOI] [PubMed] [Google Scholar]
- Young P., Öhman M., Xu M. Q., Shub D. A., Sjöberg B. M. (1994). Intron-containing T4 bacteriophage gene sunY encodes an anaerobic ribonucleotide reductase. J. Biol. Chem. 269, 20229–20232 [PubMed] [Google Scholar]