Abstract
Diatoms often dominate planktonic communities in the ocean and phototrophic biofilms in streams and rivers, greatly contributing to global biogeochemical fluxes. In pelagic ecosystems, these microscopic algae can form chain-like microcolonies, which seem advantageous for nutrient uptake and protect against grazing, and at the same time reduce sinking. Despite the capability of many diatoms to form chains, their contribution to the architecture of phototrophic biofilms remains elusive. Here we propose a computational model to simulate the growth and behaviour of Diatoma chains in contrasting flow environments. This mass-spring mechanical model captures the natural behaviour of Diatoma chains well, emphasising the relevance of chain growth and entanglement for biofilm morphogenesis. The model qualitatively describes formation of intricate dome-shaped structures and of dreadlock-type streamers as observed in nature in multidirectional and unidirectional flow, respectively. The proposed model is a useful tool to study the effect of fluid dynamics on biofilm morphogenesis.
Diatoms are the most successful microscopic algae and greatly contribute to the global biogeochemical machinery1. In the ocean, small-scale turbulence that affects the replenishment of nutrients2 and constrains microcolony formation3,4 is key to the ecology and evolution of planktonic diatoms. In streams and rivers, diatoms are essential building-blocks of benthic phototrophic biofilms — surface-attached and matrix-enclosed microbial communities — and are relevant, for instance, for primary production and sediment stabilization5,6. Overall, benthic biofilms dominate microbial life in streams and rivers, where they control vital ecosystem functions and even influence large-scale carbon fluxes7,8.
As a seemingly universal feature, biofilms can form, depending on the fine-scale hydrodynamics, conspicuous physical structures such as filamentous streamers that oscillate in the flow9,10 or star-like features11. In monospecies bacterial biofilms, the morphogenesis of such structures depends only on the constituent bacterial cells and their extracellular polymeric substances (EPS)12. In streams and rivers, however, biofilms are not monospecies communities; they include prokaryotes, microalgae, small metazoans and various organic and inorganic particles, and their organisation is therefore very complex. Yet unresolved questions remain: how can apparently universal structures emerge in the phototrophic biofilms governing benthic life, and what is the contribution of diatoms to the architecture of these biofilms?
In this study, we used empirical observations on the formation of phototrophic biofilms dominated by chain-forming diatoms and created a model of diatom chain growth and biofilm morphogenesis in different hydrodynamic regimes. Biofilms were grown in streamside flumes under contrasting flow conditions reflecting the full complexity occurring in low-emergence streams. The growth and behaviour of the Diatoma chains were modelled assuming a system of particles connected by springs as is often done to model flexible structures. This approach has proven successful to model, for example, the behaviour of hair strips13, protein folding14 and realistic rendering of cloth15. Mass-spring models have also been used to explain patterns which develop during the social movement of myxobacteria16,17. Here, populations consisting of thousands of flexible rod-shaped cells gliding on a substratum propelled by a motility engine were shown to form specific patterns, function of the diverse forces acting on each bacterial body and multiple cell-cell interactions. A similar approach was used to represent trichomes of gliding cyanobacteria and to quantify the effectiveness of photophobic responses in large populations of long flexible filaments18. For biofilm modelling, several mass-spring models have also been reported. A mass-spring model was used to study cell attachment in flow channels19 and a coupled particle-spring mechanical model with immersed boundary method was built to study biofilm deformation under various flow conditions20.
In this study, a mass-spring model of flexible Diatoma chains was exposed to processes of movement due to flow, cell growth, chain collisions, sticking and cell attachment. The model is a useful theoretical tool to study the interaction between fluid flow and biofilm morphogenesis. Our study explains how under slow and multidirectional flow, intricate dome-shaped Diatoma structures are formed, whereas elongated, dreadlock-type filamentous structures develop in fast and mostly unidirectional flow.
Results
Empirical observations
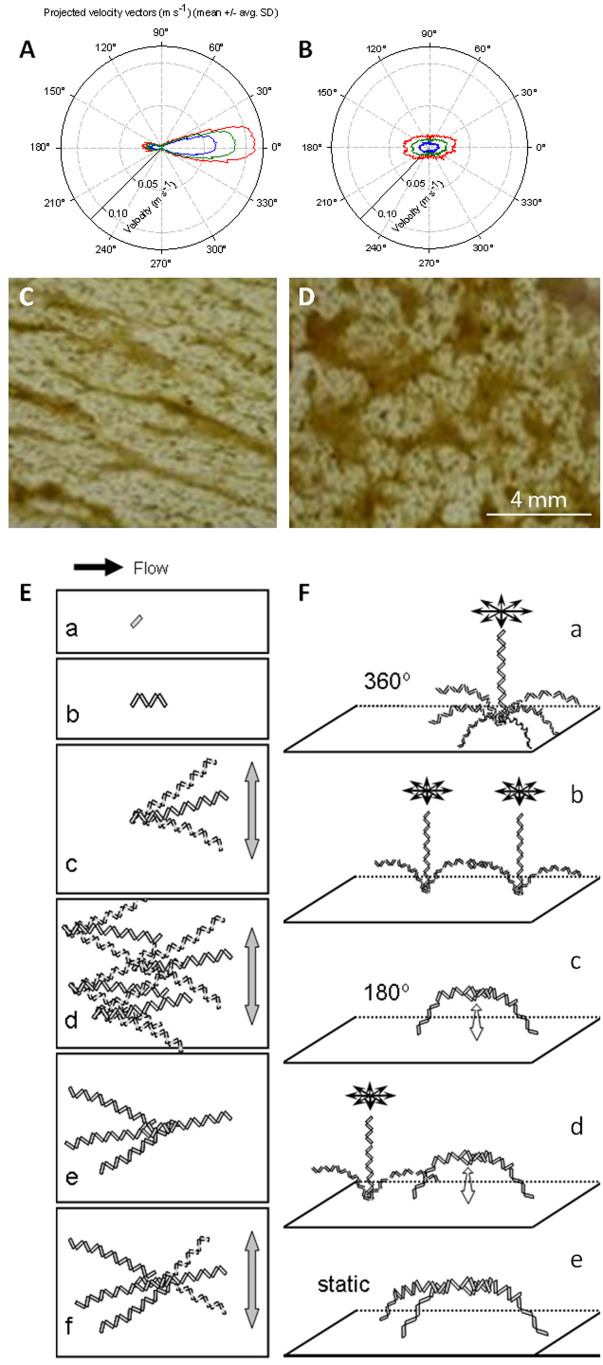
Triangular bedforms (8 cm high) were used to induce varying hydrodynamic conditions in experimental flumes11,21. At the crest of these bedforms, the flow was largely unidirectional with an average speed of 0.13±0.01 m s−1 (Figure 1A). In the trough between consecutive bedforms, eddies imposed multidirectionality on the flow, including variation in the vertical dimension. The flow velocity averaged 0.04±0.01 m s−1 (Figure 1B). In both microenvironments, Diatoma cells (length: ca. 50 μm; width: ca. 5 μm) dominated nascent phototrophic biofilms and developed chains up to several millimetres in length. However, these chains ultimately developed differing architectures in the contrasting hydrodynamic microenvironments. Ripple-like structures and streamers characterised biofilms at the crest (Figure 1C,E) whereas Y-shaped structures were abundant in the biofilms growing in the trough (Figure 1D,F).
Figure 1.
(A,B) Polar charts of flow velocity magnitude and orientation on the ridge (A) and valley (B) at different heights from the river bottom. The green line indicates the mean velocity measured at 5 mm above the sediment; the blue and red lines indicate the standard deviation, SD (blue = mean - SD; red = mean + SD). (C,D) Ortho-photographic images of biofilms from the crest (C, ridge) and trough (D, valley) between two consecutive bedforms are also shown. Flow is from the left; view is from the top. Adapted from Singer et al.24. The conceptual model for colony architecture formation at the respective hydrodynamic flow conditions is shown in (E, ridge) and (F, valley). (E) At the ridge, there is fast quasi-unidirectional flow. (a) A single Diatoma cell has attached to the substratum. (b) Cell growth has occurred. (c) As the chain length increases, it begins to move with the water flow. (d) Multiple chains move back and forth, affected by the water. (e) Eventually, motion results in chains sticking together in a so-called dreadlock. (f) Although constrained, the filament dreadlock is still able to move with the water flow. (F) At the valley, there is slow multidirectional flow. (a) Multidirectional flow results in 360° filament movement. (b) Two freely moving filaments entangle. (c) A bow-shaped structure results. Movement is constrained to a 180° flapping motion. (d) A free filament near the bow-shaped structure moves in 360°, still unaffected by the bow. (e) Once the filament and the bow entangle, an almost static Y-shaped structure results.
We observed that in multidirectional flow, individual Diatoma chains can rotate 360° around their base (Figure 1Fa) thus increasing the probability to encounter neighbouring chains (Figure 1Fb). The entanglement of two neighbouring Diatoma chains produces an arch which is restricted to a 180° flapping movement in the water (Figure 1Fc). This arch can further grow in height due to cell division but can also expand in width due to particle sequestration from streaming water. Next, if an arch becomes further entangled with a neighbouring Diatoma chain that rotates in the flow, a Y-shaped structure emerges forming the backbone of the observed irregular quasi-polygonal structures (Figure 1Fd). In contrast, unidirectional flow at the crest constrains the movement of Diatoma chains forcing them to entangle into elongated streamers that still float above the substratum (Figure 1E).
Model processes
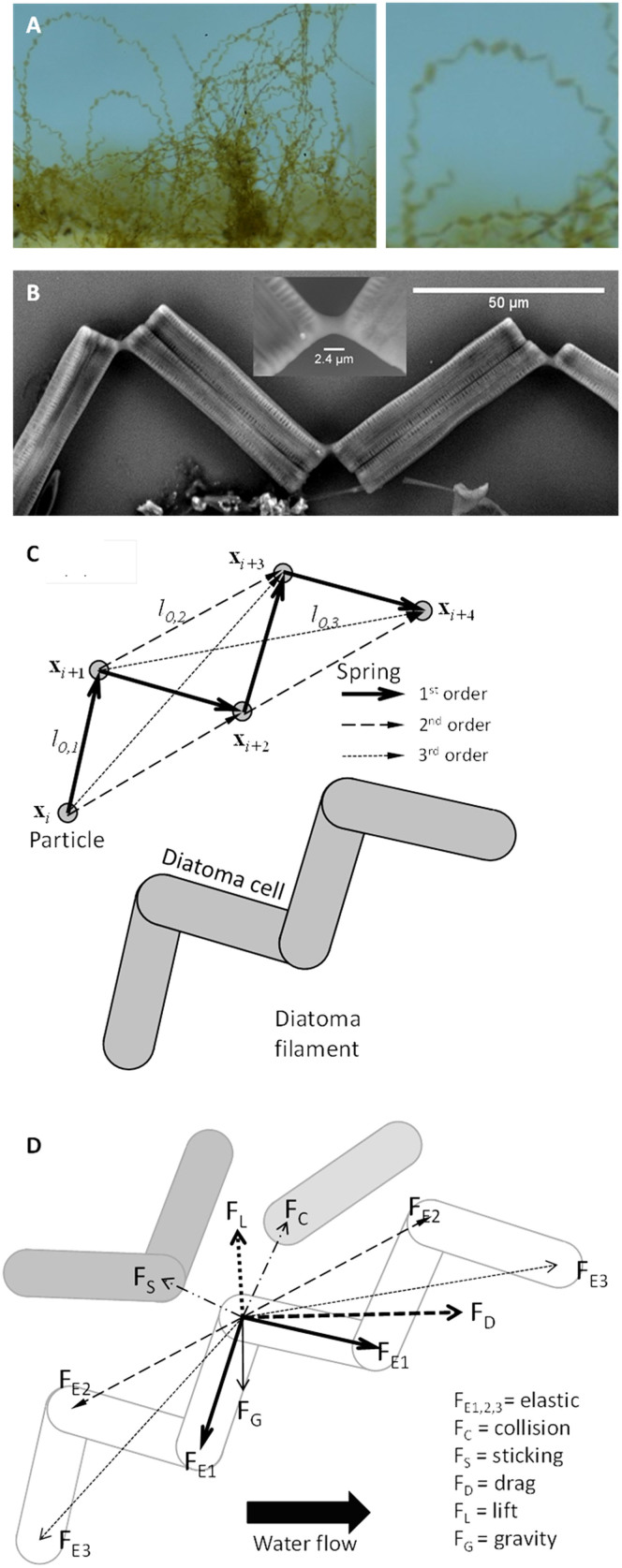
To explain chain formation and behaviour of Diatoma cells in contrasting flow environments, a mass-spring numerical model, summarized in the Methods section and detailed in the Supplementary Methods online, has been developed. In the model, individual Diatoma cells are connected by an EPS pad, which results in a zigzagging chain (Figure 2A,B). Such a chain, made up of n cells, is modelled as an array of n+1 particles with mass, which are connected by three springs of different type (Figure 2C). First-order springs connect adjacent particles to construct the body of each Diatoma cell. These springs are stiff because the silicate cell wall of Diatoma cells does not allow for elongation or deformation. Second-order springs connect every second particle to form the zigzagging shape of the chain. These springs are more flexible than first-order springs because the intercellular EPS pad allows for angles within a measured range of 115.7° to 128.2° between two cells. Third-order springs connect every third particle in the chain to prevent rotation and deformation of the generated zigzags. The movement of individual Diatoma cells was modelled taking into account not only drag and lift forces from the fluid flow, elastic forces of the Diatoma cell, and collision forces between Diatoma chains, but also chain growth and chain-substratum attachment phenomena in a three-dimensional Cartesian coordinate system reflecting flow environment conditions measured in situ as described in the Methods section.
Figure 2.
(A) At the trough, Diatoma form bows in the flow. A magnified version of a bow is shown. (B) Scanning electron micrograph of a chain zigzag. The stiff mucilage pads between individual cells are evident. (C) Mass-spring model of zigzagged chains of Diatoma cells. First-order springs oppose the tensile deformation of a cell, second-order springs keep the angles between cells by opposing bending and third-order springs impede torsion in the zigzag chain. Each Diatoma cell makes up one portion of the chain. (D) Representative schematic of the overall force balance acting on a particle. Elastic forces are exerted by primary, secondary and tertiary springs (FE). The drag (FD) and lift (FL) forces are exerted by the water flow. Gravity (FG) is applied as well on apparent masses, thus accounting also for buoyancy. Repulsive collision forces (FC) and sticking spring (FS) may also act between different cells.
Studies exist to determine the mechanical properties of zigzagged diatom chains4. Measurement of the flexural stiffness of the Diatoma cells and of the intercellular EPS bonds is not likely to provide accurate constants for all three springs required for the simulations. Spring constants were thus chosen empirically by performing sensitivity analyses (Supplementary Table S1 and Figure S1). The constants for the wall-, collision- and sticking-springs were set ten-fold weaker than the constitutive springs to allow for flexibility in the contacts. Setting the springs too stiff resulted in chains of cells that did not respond to hydrodynamics, while setting the springs too weak resulted in “escape” of the chains, unrealistic stretching and loss of the characteristic zigzagging pattern. A moderate value was therefore retained, such that filaments resembled realistic microcolonies.
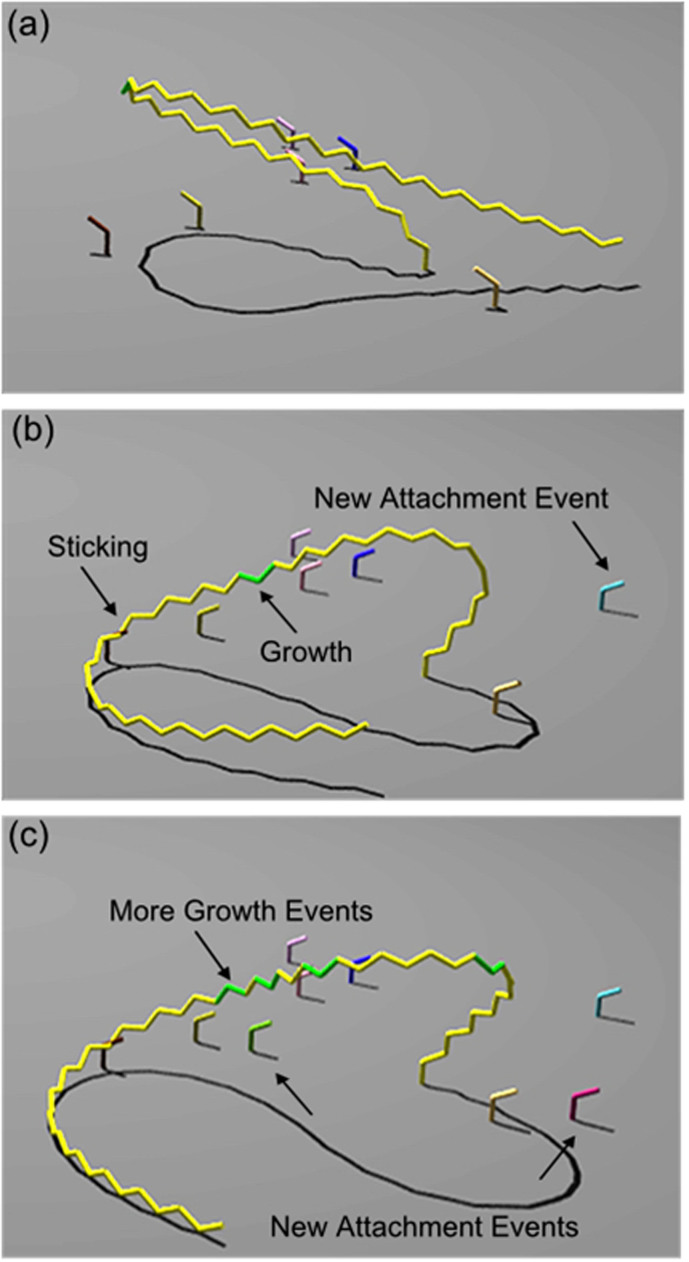
Simulations showed that once Diatoma chains oscillating in the flow collide, they irreversibly connect and thereby increase the chance of further collisions with neighbouring chains. This leads to the formation of complicated structures. The probability to encounter and to stick together was set low enough in the model to ensure that this process did not visibly overwhelm the system with massive knotting of the Diatoma chains, but it was high enough to allow “dreadlock” formation, for instance. Sticking leads not only to intertwining of chains, but indeed also to the formation of arches as observed in natural biofilms. Figure 3 (a–c) shows the formation of such an arch upon sticking of a long chain to a short attached chain.
Figure 3. Creation of a Diatoma bow at valley conditions through sticking to a newly attached filament.
Attachment and growth events can also be seen. Movement time is: (a) 3.142 s, the long (yellow) chain of cells moves unrestricted; (b) 3.353 s, the long chain sticks to another shorter chain, while other cells attach to substratum; (c) 4.322 s, the bow has formed and new cells (green) have grown within the chain.
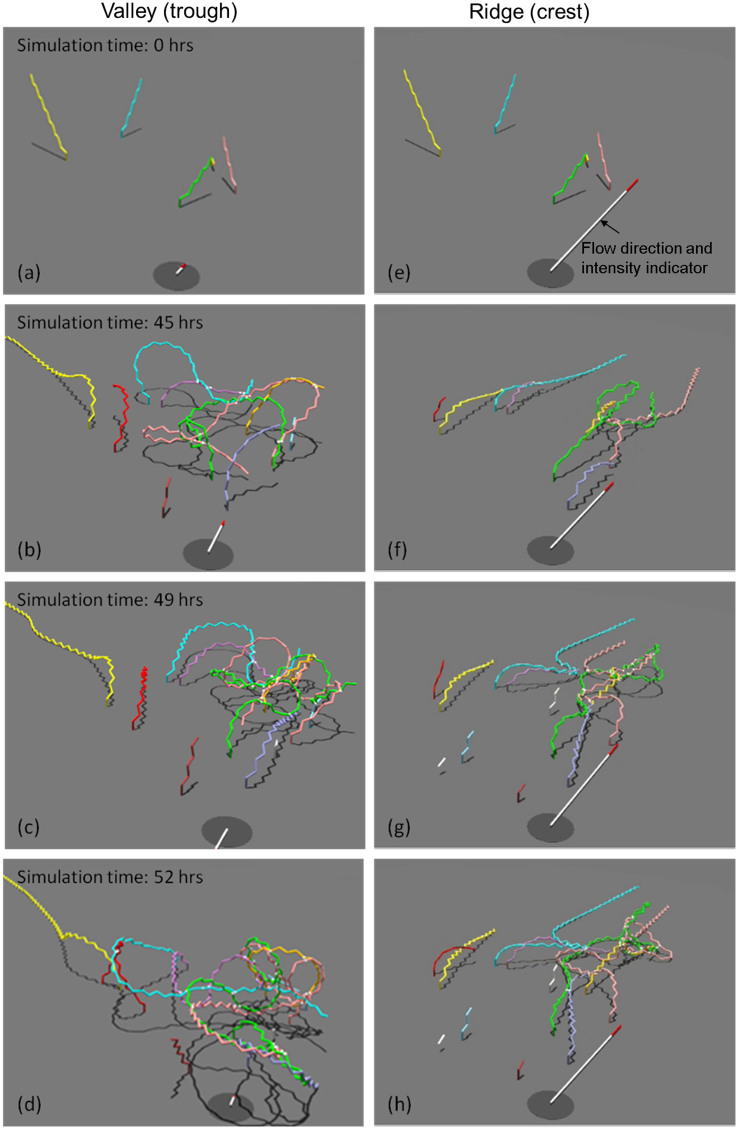
When embedded in the hydrodynamic microenvironment characteristic for the crest and the trough, respectively, the proposed model provided a reasonably realistic picture of the Diatoma chain architecture and behaviour (Figure 4, and Supplementary Videos SV1 and SV2). In the trough, zigzagged Diatoma chains developed rapidly over two days and moved following the flow patterns of water. After a lag phase of growth and interactions, chains of cells that moved in the water flow started to become entangled with neighbouring chains. Shorter Diatoma cell chains were persistently entangled even when bent over by water flow. Model simulations suggest that long chains with a potentially larger interaction radius had higher chance to become entangled and to form prominent arch-like structures as those observed in natural biofilms. After this threshold, arch-formation became more frequent eventually resulting in complex dome-shaped configurations. Some of the arches pressed down to the substratum by other extending filaments were no longer able to move in the flow and a rather static structure emerged that reasonably well mimicked observed patterns (Figure 2A).
Figure 4.
(a–d) River bottom valley (trough) colony formation. (e–h) River bottom ridge (crest) results. (a,e) Initial configuration (five Diatoma chains of different lengths). (b,f) During the first hours of the simulation, the chains of cells extended until collision and attachment events could result in the specific architecture formation. Once the chains were long enough for river hydrodynamics to bring them into close proximity, progressive sticking and entanglement could be seen. The multidirectional flow led to dome-shaped colony formation (b–d), while sticking and entanglement of the filaments was decreased in the mainly unidirectional flow (f–h). Rather, the flow led to ‘dreadlock' formation. Filaments aligned with the much stronger flow (note the greater length of the white with red tip flow indicator). Animations of these simulations are presented in the Supplementary Videos SV1 and SV2.
At the crest, simulations were run using the same input parameters (Supplementary Methods and Supplementary Table S1) as in the trough scenario, except the water flow velocity and its spatial distribution were altered. Simulations reflected natural conditions where unidirectional and faster flow bent growing Diatoma chains over and pulled them in one preferential direction. The flow environment prevented the development of an arch formation from the entanglement of neighbouring chains and ultimately the dome-like structures. Rather, the long filamentous chains aligned themselves with the flow. Generally, interactions between chains were reduced compared to the trough, though inevitably, as chains grew longer, entanglement and sticking did occur to form elongated dreadlock-like streamers that oscillated in the flow.
Discussion
Diatoms greatly contribute to the biomass and structure of phototrophic biofilms in streams and rivers where they are involved in key processes including whole-ecosystem primary production, autotrophic respiration and organic matter degradation22,23. Depending on the local flow environment, these benthic biofilms develop complex architectures, which in turn can affect local hydrodynamics but also carbon and nutrient dynamics8,11,24. Understanding mesoscale (millimetre range) biofilm morphogenesis and resulting architectures is necessary to better evaluate mass transfer and related phenomena in biofilms25,26. The model constructed in this study, in conjunction with empirical observations, unravels the morphogenesis of phototrophic biofilms dominated by colony-forming diatoms as often encountered in pristine streams. Embedded in the local hydrodynamics, a simple mechanical mass-spring model qualitatively simulated the growth and behaviour of Diatoma chains and their assembly to higher complexity structures.
Numerical simulations illustrated how colonies and chains develop from single Diatoma cells that randomly adhere to the substratum. Short evolving chains were characterized by a relatively high flexural stiffness and were only marginally affected by hydrodynamics. However, as Diatoma chains elongated, their flexibility and the flow momentum allowed their tips to explore a larger territory, thereby increasing the probability to encounter and to stick to adjacent chains. It seems that this phase is a tipping point in the morphogenesis as, dependent on the hydrodynamic environment, Diatoma chains can now assemble into different morphologies depending on the hydrodynamic microenvironments at the crest and in the trough.
At the crest, adjacent Diatoma chains entangle and, guided by predominantly unidirectional flow, form dreadlock-like streamers. Streamers are universal biofilm architectures that have been reported from monospecies bacterial biofilm growing under turbulent27 and laminar10,28 flow, but also from complex stream biofilms containing bacteria, diatoms and EPS29. Elevated flow velocities at the crest may improve nutrient flux and mass transfer, besides the mass transfer enhancement due to the oscillatory movement of streamers30,31. Only those filaments that are securely anchored can flourish in this environment. However, this elevated shear stress is also likely to damage weaker streamers and may impede the attachment of unicellular organisms.
In the trough, morphogenesis results in networks with Y-shaped structures as basic elements. Similar structures were reported from cyanobacterial mats where motile Pseudoanabena cells collide, align and clump to produce intersecting ridges that surround areas with low cell density32. Furthermore, using a size-structured population model, Hödl et al. (2013) showed how short-range dispersal of cells and coalescence of adjacent microbial clusters may result, depending on the prevailing direction of water flow, in networked or longitudinal structures in complex stream biofilms. Our results now identify diatoms as key architects of these apparently universal biofilm structures. Their important role in guiding physical structure may explain in part why diatoms have been so successful in colonizing most aquatic ecosystems1,33.
Of major evolutionary advantage to Diatoma is their encasement in a silica shell and capability to form chains, which may improve nutrient supply, protect from grazing and counteract sinking in pelagic ecosystems2,4,33. Numerical simulations suggest that stiff diatom chains experience higher nutrient flux in a turbulent flow environment than flexible chains2. Our simulations show that nascent and short Diatoma chains bend less compared to longer chains. This would thus provide an advantageous nutrient environment to these cells during initial and fast growth, as stiff chains may maintain a larger effective size, cover a larger territory and are likely to encounter more random nutrient sources2. As further revealed by the simulations, elongated chains exhibit less resistance to the flow and bend more, which, according to Musielak et al.2, would induce a less favourable nutrient environment. The entanglement into arch-like and Y-like structures counteracts the high flexibility of individual chains thereby potentially enhancing mass flux per cell at this later stage of biofilm growth. Furthermore, our simulations suggest that Diatoma structures, whether entangled in filamentous streamers or forming the more complex three-dimensional scaffolds, reversibly respond to the hydrodynamic forces and thus exhibit rheological properties similar to the viscoelasticity of biofilm EPS34. This would be a further advantage to the Diatoma chains now forming the scaffold for further biofilm growth.
Modelling revealed the importance of growth rate and sticking frequency on Diatoma chain formation. Spatial proximity of chains to one another at initialization affected whether and when the flow would bring these filaments together; careful adjustment of sticking probability was necessary to ensure that chains did not become over-tangled. In fact, in nature, flow is likely to sometimes detach chains that are attached to one another or the substratum. In numerical models, biofilm development is usually described by a balance between microbial growth and attachment (contributing to increase in biofilm volume) and biofilm detachment (leading to biomass loss)35. This balance is essential when explaining the formation of different biofilm morphologies36. Implementation of detachment in future models should provide further insight into the effect of physical dispersal on chain formation. Additional detail (e.g., processes such as sticking, microbial motility, selective attachment places, or specific cell-cell interactions) can also be added using an individual-based modelling approach as the one developed in this study.
Flow shapes not only architecture but also community composition37,38. It would thus also be of great value to extend the model to include the effect of biodiversity on streamer and colony formation. Though diatom chains seem to form the structural scaffolding of phototrophic biofilms, additional microorganisms have a large impact on growth and extension of colonies. At low flow, there may be higher diversity due to increased aggregation, with structure oscillations increasing particle trapping39. At high flow, however, an increased flux of microorganisms may occur from the bulk liquid to the biofilm29. Extending the mass-spring framework to account for differential attachment of microbes at the crest and trough should provide additional insight into community succession at different hydrodynamic conditions.
Methods
Hydrodynamic environment
A streamside flume (length: 40 m) was constructed to capture the diversity of biofilms from various hydrodynamic environments and growth stages, mimicking a flow landscape typical for streams with low submergence21. The experimental set-up has been described in detail previously11. Briefly, graded and periodically installed bedforms (bottom length: 1 m; width: 0.40 m; ascending slope: 0.75 m; descending slope: 0.25 m; maximum elevation: 0.08 m) were installed in the flume, which was continuously fed in a once-through mode with stream water (Oberer Seebach); flow rate was adjusted to 2.25 L s−1 with an average flume-scale flow velocity of 0.08 m s−1; average residence time of water was 8 minutes in the flume. The crest and the trough were selected as hydrodynamic extremes. High-resolution Acoustic Doppler Velocimetry (ADV, Nortek Vectrino, side-looking probe) was used to capture the three-dimensional flow velocity over the bedforms.
Biofilm growth
Biofilms were grown on initially sterile glass slides that were ignited (450°C, 4 h) to remove organics40. Replicate glass slides were exposed at the crest and in the trough of repeated bedforms. Growth of complex biofilms including bacteria, algae and non-living particles typically occurred within two weeks and reflects the communities in the streamwater24,37,40.
Mass-spring computational model
Diatoma chains were modelled as a collection of masses connected by springs and exposed to model processes of: (1) movement and deformation within the flow; (2) growth by division of Diatoma cells; (3) collision between different chains; (4) attachment of new cells to the support material; and (5) sticking of the chains to one another. General details are provided below; for more information, the reader is referred to the Supplementary Methods.
Diatoma chain movementA Diatoma chain is modelled as an array of particles with mass, connected by three spring types, which build rigid cells, keep angles between cells, and ensure the zigzag conformation of the chain (Figure 2b). Chain movement is determined by the motion of each individual particle of that particular chain. The particle movement is governed by the resultant of all the forces acting on it, according to Newton's laws. Each particle i is characterised by position xi = [xi,yi,zi] and velocity vi = [vxi,vyi,vzi] vectors, which concatenated form a phase space, and the mass mi. The first particle of a Diatoma chain is irreversibly attached to the substratum and the other n particles can move. For the whole set of f chains, equations of motion can be written in a compact vector form as the system
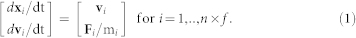 of 6×n×f ordinary differential equations (ODE). To construct the phase space, the forces acting on each particle were computed and resolved into components in the three directions. Each particle has several different forces acting on it: drag FD,i and lift FL,i resulting from the applied force of flowing water, gravity and buoyancy combined in FG,i, as well as elastic forces from the first- (FE1,i), second- (FE2,i) and third-order (FE3,i) springs, from collisions (FC,i), and from sticking-springs (FS,i). The contributions of each of these forces were added to form the combined force Fi used in the equation of motion (1). A representation of the forces acting on a particle is given in Figure 3B and more information on force computations is in the Supplementary Methods.
of 6×n×f ordinary differential equations (ODE). To construct the phase space, the forces acting on each particle were computed and resolved into components in the three directions. Each particle has several different forces acting on it: drag FD,i and lift FL,i resulting from the applied force of flowing water, gravity and buoyancy combined in FG,i, as well as elastic forces from the first- (FE1,i), second- (FE2,i) and third-order (FE3,i) springs, from collisions (FC,i), and from sticking-springs (FS,i). The contributions of each of these forces were added to form the combined force Fi used in the equation of motion (1). A representation of the forces acting on a particle is given in Figure 3B and more information on force computations is in the Supplementary Methods.Chain growthChain growth in length is due to reproduction of Diatoma cells. These cells are inserted in the Diatoma zigzag chain over time. This process was split into two phases in the model: cell ageing and cell division. No cell growth limitation by light or nutrients was taken into account. Therefore, the filament chain growth follows an exponential relationship.
Chain collisionMovement of the chain necessitated implementation of a collision detection and response algorithm to ensure that, during movement, each chain does not pass through itself, the other chains, or the substratum. A collision response is triggered when the shortest distance between each two segments lies below a threshold value (i.e., twice the Diatoma cell radius). The response takes the form of a repulsive force FC applied weighted to the two ends of each colliding cell.
Chain attachmentAttachment is the process in which cells stick to the substratum. Attachment involved adding a new chain consisting of three particles (i.e., two Diatoma cells connected under the characteristic angle) to a randomly chosen position (standard uniform distribution in both Lx and Ly) on the substratum base. Such an event was implemented to occur at the end of each growth step, provided that the maximum number of daily attachments was not exceeded (one at the ridge, two in the valley). These values were based on measured attachment frequencies at the crest and in the trough (see model parameters in the Supplementary Methods).
Chain stickingTo realistically simulate sticking between chains, the assumption was made that within a given growth time step, sticking occurs in 1 in 10,000 collisions (standard uniform distribution), with a maximum number of three sticking events per movement time step. These values result in visually realistic colony architecture formation for the given system size and flow parameters. Sticking results in the creation of an elastic spring (FS) between the two particles that have collided, which keeps the two sticking chains together.
Model solution
Modelling movement involved the solution of the ordinary differential equations for each particle in the system by an explicit eighth-order Dormand-Prince routine41. Integration results were saved at intervals Δtm,s within the movement time step Δtm. In order to visualize the results, the freely available ray-tracing software Persistance of Vision (POV-Ray, www.povray.org) was used.
Author Contributions
K.C., A.S. and C.P. developed the mathematical model. I.H. designed and performed experiments and the conceptual model. T.B. and C.P. arranged funding, designed experiments, and supervised all work. K.C., T.B. and C.P. wrote the manuscript.
Supplementary Material
Supplementary Methods, Tables and Figures
Supplementary Material Video SV1
Supplementary Material Video SV2
Acknowledgments
CP acknowledges the financial support by the Netherlands Organization for Scientific Research (NWO, VIDI grant 864.06.003). TJB was supported by the Austrian Science Fund (START Y420-B17) and IH by the PhD School “Symbiosis and Biotic Interactions”, University of Vienna.
References
- Bowler C., Vardi A. & Allen A. E. Oceanographic and biogeochemical insights from diatom genomes. Ann. Rev. Mar. Sci. 2, 333–365 (2010). [DOI] [PubMed] [Google Scholar]
- Musielak M. M., Karp-Boss L., Jumars P. A. & Fauci L. J. Nutrient transport and acquisition by diatom chains in a moving fluid. J. Fluid. Mech. 638, 401–421 (2009). [Google Scholar]
- Jumars P. A., Trowbridge J. H., Boss E. & Karp-Boss L. Turbulence-plankton interactions: a new cartoon. Mar. Ecol-Evol. Persp. 30, 133–150 (2009). [Google Scholar]
- Young A. M., Karp-Boss L., Jumars P. A. & Landis E. N. Quantifying diatom aspirations: mechanical properties of chain-forming species. Limnol. Oceanogr. 57, 1789–1801 (2012). [Google Scholar]
- Gerbersdorf S. U., Jancke T., Westrich B. & Paterson D. M. Microbial stabilization of riverine sediments by extracellular polymeric substances. Geobiology 6, 57–69 (2008). [DOI] [PubMed] [Google Scholar]
- Decho A. W. Microbial biofilms in intertidal systems: an overview. Cont. Shelf Res. 20, 1257–1273 (2000). [Google Scholar]
- Battin T. J. et al. Biophysical controls on organic carbon fluxes in fluvial networks. Nat. Geosci. 1, 95–100 (2008). [Google Scholar]
- Battin T. J., Kaplan L. A., Denis Newbold J. & Hansen C. M. Contributions of microbial biofilms to ecosystem processes in stream mesocosms. Nature 426, 439–442 (2003). [DOI] [PubMed] [Google Scholar]
- Stoodley P., Lewandowski Z., Boyle J. D. & Lappin-Scott H. M. Oscillation characteristics of biofilm streamers in turbulent flowing water as related to drag and pressure drop. Biotechnol. Bioeng. 57, 536–544 (1998). [DOI] [PubMed] [Google Scholar]
- Rusconi R., Lecuyer S., Autrusson N., Guglielmini L. & Stone H. A. Secondary flow as a mechanism for the formation of biofilm streamers. Biophys. J. 100, 1392–1399 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hödl I. et al. Biophysical controls on cluster dynamics and architectural differentiation of microbial biofilms in contrasting flow environments. Environ. Microbiol. (2013). 10.1111/1462-2920.12205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsek M. R. & Tolker-Nielsen T. Pattern formation in Pseudomonas aeruginosa biofilms. Curr. Opin. Microbiol. 11, 560–566 (2008). [DOI] [PubMed] [Google Scholar]
- Taşkiran H. D. & Gūdūkbay U. Physically based simulation of hair strips in real time. Proc. 13th Central Europe Conf. Computer Graphics, Visualization and Computer Vision, 153–156 (2005). [Google Scholar]
- Windisch B., Bray D. & Duke T. Balls and chains - a mesoscopic approach to tethered protein domains. Biophys. J. 91 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Provot X. Deformation constraints in a mass-spring model to describe rigid cloth behavior. Graphics Interface, 147–155 (1995). [Google Scholar]
- Janulevicius A., van Loosdrecht M. C. M., Simone A. & Picioreanu C. Cell Flexibility Affects the Alignment of Model Myxobacteria. Biophys. J. 99, 3129–3138 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey C. W. et al. Study of elastic collisions of Myxococcus xanthus in swarms. Phys. Biol. 8 (2011). [DOI] [PubMed] [Google Scholar]
- Tamulonis C., Postma M. & Kaandorp J. Modeling filamentous cyanobacteria reveals the advantages of long and fast trichomes for optimizing light exposure. PLoS One 6 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillon R., Fauci L., Fogelson A. & Gaver D. Modeling biofilm processes using the immersed boundary method. J. Comput. Phys. 129, 57–73 (1996). [Google Scholar]
- Alpkvist E. & Klapper I. Description of mechanical response including detachment using a novel particle model of biofilm/flow interaction. Water Sci. Technol. 55, 265–273 (2007). [DOI] [PubMed] [Google Scholar]
- Singer G. et al. Microcosm design and evaluation to study microbial biofilms. Limnol. Oceanogr. Meth. 4, 436–447 (2006). [Google Scholar]
- Cummins K. W. Structure and function of stream ecosystems. BioScience 24, 632–641 (1974). [Google Scholar]
- Danger M. et al. Benthic algae stimulate leaf litter decomposition in detritus-based headwater streams: a case of aquatic priming effect? Ecology 94, 1604–1613 (2013). [DOI] [PubMed] [Google Scholar]
- Singer G., Besemer K., Schmitt-Kopplin P., Hödl I. & Battin T. J. Physical heterogeneity increases biofilm resource use and its molecular diversity in stream mesocosms. PLoS One 5 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milferstedt K., Pons M. N. & Morgenroth E. Analyzing characteristic length scales in biofilm structures. Biotechnol. Bioeng. 102, 368–379 (2009). [DOI] [PubMed] [Google Scholar]
- Wagner M., Taherzadeh D., Haisch C. & Horn H. Investigation of the mesoscale structure and volumetric features of biofilms using optical coherence tomography. Biotechnol. Bioeng. 107, 844–853 (2010). [DOI] [PubMed] [Google Scholar]
- Stoodley P., Debeer D. & Lewandowski Z. Liquid flow in biofilm systems. Appl. Environ. Microbiol. 60, 2711–2716 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drescher K., Shen Y., Bassler B. L. & Stone H. A. Biofilm streamers cause catastrophic disruption of flow with consequences for environmental and medical systems. Proc. Natl. Acad. Sci. USA. 110, 4345–4350 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besemer K. et al. Biophysical controls on community succession in stream biofilms. Appl. Environ. Microbiol. 73, 4966–4974 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taherzadeh D., Picioreanu C. & Horn H. Mass transfer enhancement in moving biofilm structures. Biophys. J. 102, 1483–1492 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taherzadeh D. et al. Computational study of the drag and oscillatory movement of biofilm streamers in fast flows. Biotechnol. Bioeng. 105, 600–610 (2010). [DOI] [PubMed] [Google Scholar]
- Shepard R. N. & Sumner D. Y. Undirected motility of filamentous cyanobacteria produces reticulate mats. Geobiology 8, 179–190 (2010). [DOI] [PubMed] [Google Scholar]
- Kooistra W. H. et al. Global diversity and biogeography of Skeletonema species (bacillariophyta). Protist 159, 177–193 (2008). [DOI] [PubMed] [Google Scholar]
- Flemming H. C. & Wingender J. The biofilm matrix. Nat. Rev. Microbiol. 8, 623–633 (2010). [DOI] [PubMed] [Google Scholar]
- Wanner O. & Reichert P. Mathematical modeling of mixed-culture biofilms. Biotechnol. Bioeng. 49, 172–184 (1996). [DOI] [PubMed] [Google Scholar]
- Van Loosdrecht M. C. M., Picioreanu C. & Heijnen J. J. A more unifying hypothesis for biofilm structures. FEMS Microbiol. Ecol. 24, 181–183 (1997). [Google Scholar]
- Besemer K., Singer G., Hödl I. & Battin T. J. Bacterial community composition of stream biofilms in spatially variable-flow environments. Appl. Environ. Microbiol. 75, 7189–7195 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoodley P., Sauer K., Davies D. G. & Costerton J. W. Biofilms as complex differentiated communities. Annu. Rev. Microbiol. 56, 187–209 (2002). [DOI] [PubMed] [Google Scholar]
- Rickard A. H., McBain A. J., Stead A. T. & Gilbert P. Shear rate moderates community diversity in freshwater biofilms. Appl. Environ. Microbiol. 70, 7426–7435 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hödl I. et al. Voronoi tessellation captures very early clustering of single primary cells as induced by interactions in nascent biofilms. PLoS One 6 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Press W. H., Teukolsky S. A., Vetterling W. T. & Flannery B. P. Numerical Recipes: the Art of Scientific Computing. 3rd edn, (Cambridge University Press, 2007).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Methods, Tables and Figures
Supplementary Material Video SV1
Supplementary Material Video SV2