Abstract
Background/Aims
Obesity is a major contributor to the global burden of chronic disease and disability, though current knowledge of causal biologic underpinnings is lacking. Through the regulation of energy homeostasis and interactions with adiposity and gut signals, the brain is thought to play a significant role in the development of this disorder. While neuroanatomic variation has been associated with obesity, it is unclear if this relationship is influenced by common genetic mechanisms. In this study, we sought genetic components that influence both brain anatomy and body mass index (BMI) to provide further insight into the role of the brain in energy homeostasis and obesity.
Methods
MRI images of brain anatomy were acquired in 839 Mexican American individuals from large extended pedigrees. Bivariate linkage and quantitative analyses were performed in SOLAR.
Results
Genetic factors associated with increased BMI were also associated with reduced cortical surface area and subcortical volume. We identified two genome-wide quantitative trait loci that influenced BMI and ventral diencephalon volume, and BMI and supramarginal gyrus surface area, respectively.
Conclusions
This study represents the first genetic analyses seeking evidence of pleiotropic effects acting on both brain anatomy and BMI. Results suggest that a region on chromosome 17 contributes to the development of obesity, potentially through leptin-induced signaling in the hypothalamus, and that a region on chromosome 3 appears to jointly influences food-related reward circuitry and the supramarginal gyrus.
Keywords: BMI, obesity, imaging, brain, pleiotropy
Obesity is a major contributor to chronic disease and disability worldwide. In the US alone, one third of the adult population is obese, and more than two thirds are overweight or obese. Youth prevalence has tripled in the past 10 years, with 17% of children and adolescents now considered obese [1,2]. In 2009 only two US states had an obesity prevalence rate of less than 20% and a staggering 33 states had prevalence rates greater than 25% [1]. Of major importance for providing insight into this epidemic is the characterization of a poorly understood genetic component to disease susceptibility. Obesity-related phenotypes are 40% to 70% heritable, [2,3,4], yet risk genes remain elusive. The latest update of the Obesity Gene Map reports 127 candidate genes identified for common human obesity. However only 22 of these genes (or more accurately, gene regions) are supported by multiple studies. Furthermore, the established body mass index (BMI) loci detected from large-scale genome-wide association analyses jointly account for <2% of inter-individual variation in BMI [5,6].
Several of the most strongly associated and replicated gene regions for obesity including FTO, BDNF, SH2B1 and NEGR1 have also been shown to influence aspects of neuronal function, particularly in the hypothalamus, reinforcing the view that obesity is in part a brain-related disorder [6]. Further supporting a role for a significant link between the brain and obesity is the increased risk of brain atrophy, cognitive dysfunction and dementia later in the life of obese individuals. In an imaging study of 94 elderly subjects, Raji and colleagues showed that higher BMI was significantly correlated with reduced grey matter and white matter volumes throughout the brain, and that atrophy (indicating an increased risk for dementia) was evident in people with greater percentages of body fat tissue. Similar results were observed for individuals with high fasting insulin levels and type 2 diabetes [7]. A third imaging study of 50 adults demonstrated that increased BMI is associated with axonal and/or myelin abnormalities in frontal white matter and neuronal injury in frontal grey matter [8]. In a fourth study by Ho and colleagues, subjects with higher BMI had significantly lower regional brain volumes and structural brain atrophy was observed in carriers of at least one copy of a risk allele correlated with a marker in the FTO gene, although it still remains to be established that FTO is the underlying causal gene [9]. Additionally, individuals carrying this risk allele showed brain tissue deficits in the frontal and occipital lobe regions; the same areas associated with brain volume reductions in subjects with high BMI. These results suggest an underlying common susceptibility variant for obesity that was independently associated with brain atrophy [9]. Together, these neuroanatomical studies demonstrate a consistent link between increasing obesity and brain atrophy and suggest that this relationship may be mediated by genetic factors. However, formal genetic experiments to implicate pleiotropy between obesity and neuroanatomic phenotypes have not been reported.
The current study represents an attempt to examine the potential pleiotropic basis of obesity and brain structural variation in a human population. Using an extended pedigree design, we examine the evidence for such shared genetic influence and attempt to exploit it to identify likely genomic locations harboring causal loci.
Research Design and Methods
Human Subject Selection
All subjects included in this analysis were Mexican American individuals from the Genetics of Brain Structure and Function Study (GOBS) that were originally recruited to participate in the San Antonio Family Heart Study (SAFHS) [10] or the San Antonio Family Gallbladder Study [11]. At baseline, the SAFHS included 1,431 individuals in 42 extended families. Ascertainment occurred by way of a single adult Mexican American proband selected at random, without regard to presence or absence of disease and almost exclusively from Mexican American census tracts in San Antonio. To ensure large, multigenerational pedigrees, probands had to have at least 6 age-eligible offspring and/or siblings living in San Antonio. All first, second, and third degree relatives of the proband and of the proband's spouse, aged 16 years or above, were eligible to participate in the study. The SAFGS cohort currently includes 907 individuals from 39 large Mexican American families. Recruitment was similar to that of the SAFHS, with probands recruited from a random sample of Mexican American individuals, but in the SAFGS probands were selected for type 2 diabetes [12]. All 1st, 2nd, and 3rd degree relatives of the proband, ≥18 years of age were invited to participate. The near identity of the ascertainment designs for SAFHS and SAFGS has been confirmed via comparisons of all diabetes- and obesity-related phenotypes.
For the analyses herein, we utilized 839 GOBS subjects [13] from whom we have acquired MRI-based anatomical brain measures and BMI as a measure of obesity. Our sample is composed of 514 females with an average age of 43.0 ± 14.2 years and 325 males with an average age of 43.7 ± 15.1 years from 57 extended families. Of these families, 19 represent large extended pedigrees, each with 18 or more participants (max pedigree size = 125).
All participants gave written informed consent. The Institutional Review Boards of the University of Texas Health Science Center at San Antonio and Yale University approved all protocols.
MRI Acquisition and Processing
Magnetic resonance images were acquired on a Siemens 3T TIM Trio scanner with an 8-channel head coil in the Research Imaging Institute, University of Texas Health Science Center at San Antonio. The anatomic imaging protocol included seven T1-weighted 3D Turbo-flash scans with an adiabatic inversion contrast pulse and the following parameters: TE/TR/TI = 3.04/2100/785 ms, flip angle=13°, and 800 micron isotropic voxel resolution. These images underwent a retrospective motion correction [14] and were averaged to achieve optimal gray/white matter contrast for each subject.
Anatomic (gray matter) image processing was based on surface representations of the cortex using FreeSurfer [15,16] as implemented in our group [17]. First, images underwent inhomogeneity corrections and intensity normalization, linear alignment to a common atlas space, and were skull-stripped. Next, white matter voxels were identified and the borders between gray and white matter were defined. Hemispheres were separated and a tessellated mesh was built around white matter voxels. This mesh was smoothed and topological defects were corrected to accurately model the white matter surface. The gray matter (pial) surface was generated by expanding the white matter surface to the gray matter/CSF boundary while constraining the smoothness of the surface, resulting in a continuous polygonal tessellations spanning the cortex. Surfaces were visually inspected and manually edited if necessary. The pial surface was inflated into a sphere, registered to an atlas, and parcellated into regions of interest defined by the Desikan atlas [18]. Cortical surface area was calculated as the sum of the areas of each tessellation falling within a given region, in the subject’s native space. Subcortical regions were parceled using analogous procedures and volumetric measures were calculated accordingly, also in the subject’s native space. FreeSurfer measurements have been validated against histological [19] and manual measurements [20,21].
High Density SNP Typing
More than one million SNPs, using the Illumina HumanHap550 BeadChip (in tandem with the supplemental HumanHap450S BeadChip) or the 1M BeadChip were analysed. These SNPs capture ~90% of the common variation in humans. Raw genotype data were processed using standard quality control procedures, and variants were checked for consistency of Mendelian transmission, an advantage of family-based designs. Missing genotypes were imputed using the pedigree-exploiting procedure developed by Burdick and colleagues [22] as implemented in MERLIN [23].
Quantitative Genetic Analysis
Heritability of each trait was estimated using SOLAR [24]. SOLAR employs variance component methods to analyse family-based quantitative data by partitioning the observed covariance into genetic and environmental components. The demographic covariates sex, age, age2 and their interactions were included in all genetic analyses. Prior to analysis, each phenotype was transformed to approximate normality using a direct inverse Gaussian transformation.
Genetic correlation represents the common genetic covariance between two traits, referred to as pleiotropy [25]. Bivariate quantitative genetic analysis was used to estimate the genetic (ρg) and environmental (ρe) correlations between each potential obesity-related and brain-related phenotype. The phenotypic correlation (ρp), which quantifies the overall relationship between the two traits, can be derived from the genetic and environmental correlations as ρp = ρg√(h2eh2i) + ρe√[(1-h2e)(1-h2i)]. These parameters were estimated by jointly utilizing all available familial relationships with a multivariate normal threshold model for combined dichotomous and continuous traits [25,26].
Results from bivariate quantitative analyses of obesity and brain measures were used to assess the overall potential for identifying overlapping influence of causal genetic variants across these two major phenotypic domains. Specifically, we assessed the endophenotypic value of quantitative brain measures vis-à-vis obesity using the Endophenotype Ranking Value (ERV), which represents the standardized genetic covariance between trait and illness [27]. Mathematically, the ERV is defined as the absolute value of the product of the square-root of illness heritability (h2i), the square-root of the endophenotype heritability (h2e), and their genetic correlation (ρg): ERVie = |√h2i√h2eρg|, where heritability is the portion of phenotypic variance accounted for by additive genetic variance (h2=σ2g/σ2p). In this context, a brain-based endophenotype is both heritable and genetically correlated with obesity. Therefore, ranking endophenotypes served as an a priori theoretical mechanism for placing focus on the action of gene sets that affect both brain and susceptibility to obesity.
The ERV varies between 0 and 1, where higher values indicate that the endophenotype and illness are more strongly influenced by common genetic factors. All brain imaging traits were ranked for relevance to obesity using this approach to identify those pairs of traits that showed the most significant evidence for being jointly influenced by common gene sets. Only these pairs of traits underwent QTL localization and identification analyses.
Bivariate QTL Localization Analysis of Brain-Related Endophenotypes and Obesity
Quantitative trait linkage analysis was used to identify specific chromosomal regions harboring quantitative trait loci (QTLs) that influence both brain and obesity phenotypic domains simultaneously. Bivariate linkage analysis exploits the genetic covariance between two traits to improve the power to localize QTLs and to detect QTL-specific pleiotropic effects [26]. In the current experiment, we perfomed bivariate linkage analyses (again using the SOLAR package) between cortical surface area or subcotial volume measures selected via the ERV analysis and BMI to search for chomorsomal regions that influnce both neuroanatomic variability and body composition. More specifically, after addressing (by blanking, recalling, or retyping) mistyping errors identified using Simwalk II [28], genotype data were used to compute maximum likelihood estimates of allele frequencies. Matrices of empirical estimates of identity-by-descent (IBD) allele sharing at points throughout the genome for every relative pair were computed using the Loki package [29] based on a selection of approximately 15,000 common SNPs that were in linkage equilibrium (define as r2 < 0.2) with one another. SNPs are only used for their combined information on IBD status; we do not test individual SNPs. We used high-resolution chromosomal maps provided by deCODE genetics [30] to project physical genomic distances onto recombination-relevant genetic distances. For the localization of QTLs, we performed both univariate and bivariate variance components linkage analyses. All QTL analyses included covariates for age, age2, sex and their interactions. Bivariate LOD scores were adjusted for additional degrees of freedom incurred, making them directly comparable to traditional univariate LOD scores. Once a genome-wide significant localization was made, formal single degree of freedom likelihood ratio tests for pleiotropy were performed to test the specific hypothesis that a QTL at that location influenced a given brain measure and BMI risk jointly.
We employed the theoretical method of Feingold et al [31] to calculate genome-wide significance levels for our linkage analyses. For our pedigree structure (which determines the average non-independence between IBD vectors involving approximately 388 effective tests), genome-wide significance requires a LOD score of 2.9 (nominal p-value = 1.3×10−4) while suggestive evidence (i.e., expected to occur only once by chance in a genome scan) requires a LOD of 1.68 (nominal p = 0.0027). We do not additionally correct for the testing of multiple phenotypes since each bivariate analysis also includes BMI and represents a separate hypothesis with prior support from our ERV analyses.
Results
Heritability of BMI and Brain Phenotypes
Using the complete extended pedigree information, the heritability of BMI in the Mexican American sample was estimated to be 0.565 ± 0.057 (p = 1.8 × 10−30), indicating a substantial genetic component and suggesting it is reasonable to search for genes responsible for the observed variance. As we have previously shown, all of our cortical surface area and subcortical volumes are significantly heritable (Tables 1 and 2) [32].
Table 1.
Bivariate Analyses between BMI and Cortical Surface Area
| Lobe | Region of Interest | Heritability | Environmental Correlation (ρe) |
Genetic Correlation (ρg) |
Phenotypic Correlation (ρp) |
|---|---|---|---|---|---|
| Cingulate | Caudal anterior | 0.529 | −0.022 | −0.034 | −0.028 |
| Cingulate | Isthmus | 0.707 | 0.250 | −0.013 | 0.088 |
| Cingulate | Posterior | 0.684 | 0.241 | −0.133 | 0.017 |
| Cingulate | Rostral anterior | 0.512 | 0.086 | −0.071 | 0.006 |
| Frontal | Middle frontal gyrus, caudal | 0.673 | 0.081 | 0.005 | 0.035 |
| Frontal | Frontal pole | 0.152 | 0.009 | −0.022 | 0.000 |
| Frontal | Orbitofrontal cortex, lateral | 0.670 | 0.484 | −0.293 | 0.020 |
| Frontal | Orbitofrontal cortex, medial | 0.568 | 0.250 | −0.187 | 0.015 |
| Frontal | Paracentral gyrus | 0.644 | 0.203 | −0.036 | 0.065 |
| Frontal | Inferior frontal gyrus, pars opercularis | 0.565 | 0.321 | −0.155 | 0.066 |
| Frontal | Inferior frontal gyrus, pars orbitalis | 0.491 | 0.219 | −0.300 | −0.042 |
| Frontal | Inferior frontal gyrus, pars triangularis | 0.648 | 0.336 | −0.183 | 0.037 |
| Frontal | Precentral gyrus | 0.717 | 0.131 | 0.047 | 0.078 |
| Frontal | Middle frontal gyrus, rostral | 0.719 | 0.296 | −0.247 | −0.039 |
| Frontal | Superior frontal gyrus | 0.755 | 0.265 | −0.058 | 0.056 |
| Insula | Insular cortex | 0.712 | 0.449 | −0.242 | 0.021 |
| Occipital | Cuneus cortex | 0.612 | 0.268 | −0.296 | −0.047 |
| Occipital | Lateral occipital cortex | 0.731 | 0.361 | −0.231 | −0.010 |
| Occipital | Lingual gyrus | 0.728 | 0.227 | −0.191 | −0.032 |
| Occipital | Pericalcarine cortex | 0.684 | 0.052 | −0.135 | −0.058 |
| Parietal | Inferior parietal cortex | 0.590 | 0.367 | −0.321 | −0.015 |
| Parietal | Postcentral gyrus | 0.648 | 0.252 | −0.207 | −0.014 |
| Parietal | Precuneus cortex | 0.749 | 0.430 | −0.116 | 0.080 |
| Parietal | Superior parietal cortex | 0.677 | 0.268 | −0.065 | 0.070 |
| Parietal | Supramarginal gyrus | 0.645 | 0.359 | −0.274 | −0.006 |
| Temporal, Lateral | Banks of the superior temporal sulcus | 0.497 | 0.099 | −0.157 | −0.029 |
| Temporal, Lateral | Inferior temporal gyrus | 0.621 | 0.104 | −0.208 | −0.072 |
| Temporal, Lateral | Middle temporal gyrus | 0.640 | 0.319 | −0.315 | −0.048 |
| Temporal, Lateral | Superior temporal gyrus | 0.738 | 0.566 | −0.228 | 0.060 |
| Temporal, Lateral | Transverse temporal cortex | 0.670 | 0.375 | −0.200 | 0.034 |
| Temporal, Medial | Entorhinal cortex | 0.612 | 0.030 | −0.140 | −0.065 |
| Temporal, Medial | Fusiform gyrus | 0.623 | 0.411 | −0.385 | −0.040 |
| Temporal, Medial | Parahippocampal gyrus | 0.637 | 0.267 | −0.166 | 0.019 |
| Temporal, Medial | Temporal pole | 0.477 | 0.003 | −0.145 | −0.070 |
Table 2.
Bivariate Analyses between BMI and Subcortical Volumes
| Region of Interest | Heritability | Environmental Correlation (ρe) |
Genetic Correlation (ρg) |
Phenotypic Correlation (ρp) |
|---|---|---|---|---|
| Accumbens Area | 0.495 | 0.109 | −0.255 | −0.075 |
| Amygdala | 0.793 | 0.365 | −0.116 | 0.041 |
| Caudate | 0.761 | 0.139 | −0.178 | −0.064 |
| Hippocampus | 0.740 | 0.401 | −0.197 | 0.020 |
| Pallidum | 0.727 | 0.137 | −0.136 | −0.032 |
| Putamen | 0.758 | 0.208 | −0.085 | 0.019 |
| Thalamus | 0.777 | 0.144 | −0.185 | −0.069 |
| Ventral Diencephalon | 0.726 | 0.191 | −0.274 | −0.098 |
Bivariate Correlations between BMI and Cortical Surface Area
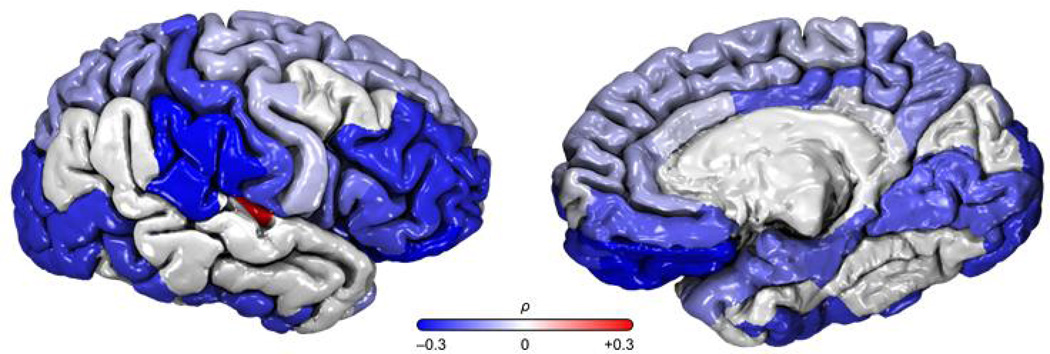
We performed bivariate quantitative genetic analyses examining the phenotypic, genetic and environmental correlations between BMI and brain traits for the 839 GOBS participants with available neuroanatomic images (Table 1). Genetic correlations were typically negative (Figure 1) suggesting the same genetic factors that increase BMI act to reduce cortical surface area. The strongest genetic correlations were observed in mesial and lateral temporal, frontal, occipital and parietal lobes and the insular cortex, including the following gyri: fusiform (ρg= −0.385), middle temporal (ρg= −0.321), inferior parietal (ρg= −0.376), lateral orbitofrontal (ρg= −0.293), and supramarginal (ρg= −0.293). In contrast to the genetic correlations, environmental correlations were typically positive, suggesting a potential dynamic feedback mechanism between the genetic component and an environmental stressor.
Figure 1.
A very similar pattern of results was observed for subcortical volumes (Table 2). The strongest genetic correlations between BMI and subcortical regions included the ventral diencephalon (ρg=−0.274) and accumbens area (ρg=−0.255), with each region showing positive environmental correlations (ρe=0.191 and ρe=0.109, respectively). The ventral diencephalon is primarily composed of the hypothalamus, which regulates food intake and is strongly associated with obesity [6].
Endophenotype Ranking
To select a subset of brain traits for linkage analyses, we utilized the Endophenotype Ranking Value (ERV) approach to select neuroanatomic traits that likely share causal mechanisms with BMI [27]. The top 10 ranked ERV statistics for brain traits and BMI are presented in Table 3. After this objective prioritization, bivariate linkage analysis was run on each trait combination to search for specific genome locations that harbor pleiotropic QTLs influencing brain and obesity.
Table 3.
ERV and Bivariate Linkage Results
| Brain Trait | ERV | ERV p-value |
Max LOD Score |
QTL nominal p−value |
Chromosomal Region |
|---|---|---|---|---|---|
| Fusiform | 0.217 | 0.002 | 2.248 | 6.5×10−4 | 6q13 |
| Inferior Parietal | 0.184 | 0.009 | 2.200 | 7.3×10−4 | 2q34 |
| Middle Temporal | 0.178 | 0.009 | 1.817 | 0.0019 | 17p11 |
| Lateral Orbitofrontal | 0.173 | 0.014 | 1.890 | 0.0016 | 6q16 |
| Cuneus | 0.166 | 0.019 | 2.292 | 5.8×10−4 | 5p15 |
| Supramarginal | 0.164 | 0.024 | 3.257 | 5.4×10−5 | 3q22 |
| Parsorbitalis | 0.152 | 0.031 | 2.505 | 3.4×10−4 | 15q14 |
| Rostral Middle Frontal | 0.152 | 0.033 | 1.966 | 0.0013 | 6q14 |
| Insula | 0.152 | 0.035 | 2.037 | 0.0011 | 10p13 |
| Ventral Diencephalon | 0.168 | 0.051 | 3.271 | 5.2×10−5 | 17p13 |
Bivariate Linkage Analysis
Two significant QTLs and a number of suggestive regions were observed after performing bivariate linkage on the top 10 ERV-ranked brain traits and BMI. We identified a QTL with a LOD of 3.27 for BMI and ventral diencephalon on chromosome 17p13.1. This chromosomal region was previously implicated in obesity and related disorders [33,34], and the ventral diencephalon directly supports the regulation of food intake. Another QTL, with a genome-wide significant LOD of 3.26 on chromosome 3q22.1, was identified for BMI and the surface area of the supramarginal gyrus. Both the genomic region [35] and brain region [36] were independently associated with obesity in previous reports. Thus, our joint analyses of brain structure and obesity build on these findings and provide unequivocal evidence for two location-specific pleiotropic QTLs. In addition, we observed a number of genome-wide suggestive scores (LOD>1.68; Table 3), including a locus at chromosome 6q13-16 that included the surface area of the fusiform gyrus (LOD=2.2, 6q13), the lateral orbitofrontal gyrus (LOD = 1.9, 6q16) and the rostral middle frontal gyrus (LOD = 2.0, 6q14).
Discussion
This is the first report to quantify shared genetic factors of brain anatomy and obesity and localize significant pleiotropic influence. Specifically, we performed bivariate linkage analyses with a subset of brain traits and BMI to localize two genome-wide significant QTLs, on chromosomal regions were 3q22.1 and 17p13.1. Region 17p13.1 harbors genes that pleiotropically influenced ventral diencephalon volume and BMI. Meyre and colleagues reported suggestive evidence for linkage in this region for childhood obesity [37]. Additionally, strong evidence for a QTL influencing plasma leptin levels (LOD = 5) [34] and marginal evidence for linkage with both hip and waist circumference (LOD=1.1) was also reported in this region [33]. The second QTL we identified was 3q22.1, which significantly influenced the surface area of the supramarginal gyrus and BMI. This genomic region was previously associated with obesity by Wu and colleagues, who found significant evidence for linkage with BMI (LOD 3.45) in a sample of African Americans [35]. Our results identify regions of the genome that are directly related to both brain structure and obesity. Therefore, the same causal biologic pathway likely influences the underlying function of these brain regions and the outcome measure of obesity.
The ventral diencephalon houses the hypothalamus, which is involved in the regulation of eating tendencies. Although feeding behavior is complex, the lateral hypothalamus is commonly referred to as the “feeding center” of the brain because animal models show that stimulation increases food intake yet a lesion inhibits motivation to feed [38]. This suggests functional plausibility for our finding on chromosome 17. Furthermore, leptin, which is secreted by fat cells, triggers signalling from neurons of the arcuate nucleus to the paraventricular nucleus, lateral hypothalamus and lateral horn of the spinal cord. Depending on the particular type of peptide carried by these neurons, metabolism is increased (or decreased), along with sympathetic tone, while feeding is decreased (or increased, respectively). Additionally, neurons containing these peptides project to widespread areas of the cortex and may be involved in other components of feeding strategies [38]. Therefore, our findings and those of Kissebah and colleagues [34] provide strong circumstantial evidence that the region identified on chromosome 17 contributes to the development of obesity through leptin-induced signaling in the hypothalamus.
The supramarginal gyrus has been linked to brain function differences in overweight and healthy weight individuals. Frankort and colleagues reported that both left (p = 0.00025) and right (p = 0.00083) gyri, amongst other regions, showed significant activation differences in response to passively viewing versus imagining the taste of high-calorie food pictures [36]. This paradigm was designed to depict reward related brain activity. Similarly, Rothemund and colleagues reported that, in obese compared to normal-weight women, viewing high-calorie food stimuli differentially activated the dorsal striatum, insula, hippocampus and an area just deep to the left supramarginal gyrus (T = 3.0) [38]. This was interpreted as a habit-forming system associated with lowered dopamine receptor availability in obese subjects. While the role of the supramarginal gyrus is poorly understood in this context, our findings in conjunction with these reports implicate its involvement in the pathogenesis of obesity due to shared influence of a region on chromosome 3.
The initial results from this study suggested that genetic factors associated with increasing BMI were also associated with reduced cortical surface area, with the strongest correlations observed in mesial and lateral temporal, frontal, occipital and parietal lobes and the insular cortex. We also observed the same pattern of negative genetic correlations between BMI and subcortical volumes, indicating that genes associated with increased BMI are also associated with reduced subcortical volumes. This observation is conceptually similar to that of Ho and colleagues, who reported significantly lower regional brain volumes with increasing BMI (p=0.02) [9]. Specifically, with every 1-unit increase in BMI there was an associated 1–1.5% average brain tissue reduction in several brain regions including the frontal, occipital and parietal lobe regions. However, this work was based exclusively on phenotypic correlations and did not contain a genetic component. Together, these results provide substantial evidence for an underlying biologic mechanism and our pedigree-based design enabled us to go beyond the inferential limits of traditional epidemiologic studies that enlist only unrelated individuals.
It should be noted that the neuroanatomic data used here does not directly index neuronal activity specifically or neurophysiological function more generally. Rather, measures of cortical surface area or subcortical volume are highly heritable traits that have been previously related to obesity (e.g. [9]). Unlike previous investigations of neuroanatomic variation and BMI, our strategy involved prioritizing brain traits by only selecting those with common genetic effects related to obesity (via the ERV analysis [27]). Indeed, the ERV analysis significantly reduced the number of brain traits tested and ensured that linkage was only performed on traits with high levels of genetic covariance with BMI. However, this prioritization strategy, combined with the fact that BMI was always one of the two traits used in the bivariate analyses, makes it very difficult to correct for the number of linkage analyses performed. Yet, given that both of the identified genome-wide significant QTLs have been previously linked to BMI, it is unlikely that these results are spurious.
The final results from this study localized two chromosomal regions whose genes pleiotropically influenced BMI and brain structure. Future use of whole genome sequence data in these regions provides a powerful approach to find causal variants and inform potential obesity treatments. Indeed, by discovering genes that predispose obesity risk, it is our eventual goal to speed the development of drug targets to slow the epidemic advancement of obesity and its sequalae.
Acknowledgements
We are grateful to the participants in the San Antonio Family Study. Financial support for this study was provided by the National Institute of Mental Health grants MH078143, MH078111 and MH083824; and the National Institute for Heart, Lungs and Blood grant HL045222. The development of the analytical methods and SOLAR software used in this study was supported by National Institute of Mental Health grant R37 MH059490. The high density SNP genotyping data used in this study was generated with generous support from the Azar and Shepperd Families of San Antonio and in part by ChemGenex Pharmaceuticals. Parts of this investigation were conducted in facilities constructed with support from the Research Facilities Improvement Program grants C06 RR013556 and C06 RR017515 from the National Center for Research Resources, National Institutes of Health. The AT&T Genomics Computing Center supercomputing facilities used for this work were supported in part by a gift from the AT&T Foundation and with support from the National Center for Research Resources Grant Number S10 RR029392.
References
- 1.Centers for Disease Control and Prevention. Vital Signs: State-Specific Obesity Prevalence Among Adults – United States, 2009. Morbidity and Mortality Weekly Report. 2010 Aug 3;Vol 50 [PubMed] [Google Scholar]
- 2.Walley AJ, Blakemore AIF, Froguel P. Genetics of obesity and the prediction of risk of health. Hum Mol Genet. 2006;15:R124–R130. doi: 10.1093/hmg/ddl215. [DOI] [PubMed] [Google Scholar]
- 3.Comuzzie AG, Higgins PB, Voruganti S, Cole S. Cutting the fat: The genetic dissection of body weight. In: Bouchard C, editor. Progress in Molecular Biology and Translational Science. vol 94. Elsevier Inc. Academic Press; 2010. [DOI] [PubMed] [Google Scholar]
- 4.Loos RF. Recent progress in the genetics of common obesity. Br J Clin Pharmacol. 2009;68:811–829. doi: 10.1111/j.1365-2125.2009.03523.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rankinen T, Zuberi A, Chagnon YC, Weisnagel SJ, Argyropoulos G, Walts B, Perusse L, Bouchard L. The human obesity gene map: the 2005 update. Obesity. 2006;14:529–644. doi: 10.1038/oby.2006.71. [DOI] [PubMed] [Google Scholar]
- 6.Cheung WW, Mao P. Recent advances in obesity: Genetics and beyond. ISRN Endocrinol. 2012;2012:536905. doi: 10.5402/2012/536905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Raji CA, Ho AJ, Parikshak N, Becker JT, Lopez OL, Kuller LH, Hua X, Leow AD, Toga AW, Thompson PM. Brain structure and obesity. Hum Brain Mapp. 2010;31(3):353–364. doi: 10.1002/hbm.20870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gazdzinski S, Kornak J, Weiner MW, Meyerhoff DJ. Body mass index and magnetic resonance markers of brain integrity in adults. Ann Neurol. 2008;63(5):652–657. doi: 10.1002/ana.21377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ho AJ, Stein JL, Hua X, Lee S, Hibar DP, Leow AD, Dinov ID, Toga AW, Saykin AJ, Shen L, Foroud T, Pankratz N, Huentelman MJ, Craig DW, Gerber JD, Allen AN, Corneveaux JJ, Stephan DA, DeCarli CS, DeChairo BM, Potkin SG, Jack CR, Jr, Weiner MW, Raji CA, Lopez OL, Becker JT, Carmichael OT, Thompson PM. Alzheimer's Disease Neuroimaging Initiative. A commonly carried allele of the obesity-related FTO gene is associated with reduced brain volume in the healthy elderly. Proc Natl Acad Sci USA. 2010;107(18):8404–8409. doi: 10.1073/pnas.0910878107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mitchell BD, Kammerer CM, Blangero J, Mahaney MC, Rainwater DL, Dyke B, Hixson JE, Henkel RD, Sharp RM, Comuzzie AG, VandeBerg JL, Stern MP, MacCluer JW. Genetic and environmental contributions to cardiovascular risk factors in Mexican Americans. The San Antonio Family Heart Study. Circulation. 1996;94:2159–2170. doi: 10.1161/01.cir.94.9.2159. [DOI] [PubMed] [Google Scholar]
- 11.Puppala S, Dodd GD, Fowler S, Arya R, Schneider J, Farook VS, Granato R, Dyer TD, Almasy L, Jenkinson CP, Diehl AK, Stern MP, Blangero J, Duggirala R. A genomewide search finds major susceptibility loci for gallbladder disease on chromosome 1 in Mexican Americans. Am J Hum Genet. 2006;78:377–392. doi: 10.1086/500274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Duggirala R, Blangero J, Almasy L, Dyer TD, Williams KL, Leach RJ, O'Connell P, Stern MP. Linkage of type 2 diabetes mellitus and of age at onset to a genetic location on chromosome 10q in Mexican Americans. Am J Hum Genet. 1999;64:1127–1140. doi: 10.1086/302316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Olvera RL, Bearden CE, Velligan DI, Almasy L, Carless MA, Curran JE, Williamson DE, Duggirala R, Blangero J, Glahn DC. Common genetic influences on depression, alcohol, and substance use disorders in Mexican American families. Am J Genet B Neuropsychiatr Genet. 2011;156:561–568. doi: 10.1002/ajmg.b.31196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kochunov P, Lancaster JL, Glahn DC, Purdy D, Laird AR, Gao F, Fox P. Retrospective motion correction protocol for high-resolution anatomical MRI. Hum Brain Mapp. 2006;27:957–962. doi: 10.1002/hbm.20235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dale AM, Fischl B, Sereno MI. Cortical surface-based analysis. I. Segmentation and surface reconstruction. Neuroimage. 1999;9:179–194. doi: 10.1006/nimg.1998.0395. [DOI] [PubMed] [Google Scholar]
- 16.Fischl B, Sereno MI, Dale AM. Cortical surface-based analysis. II: Inflation, flattening, and a surface-based coordinate system. Neuroimage. 1999;9:195–207. doi: 10.1006/nimg.1998.0396. [DOI] [PubMed] [Google Scholar]
- 17.Winkler A, Kochunov P, Blangero J, Almasy L, Zilles K, Fox P, Duggirala R, Glahn D. Cortical thickness or grey matter volume? The importance of selecting the phenotype for imaging genetics studies. Neuroimage. 2010;53:1135–1146. doi: 10.1016/j.neuroimage.2009.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Desikan RS, Segonne F, Fischl B, Quinn BT, Dickerson BC, Blacker D, Buckner RL, Dale AM, Maguire RP, Hyman BT, Albert MS, Killiany RJ. An automated labeling system for subdividing the human cerebral cortex on MRI scans into gyral based regions of interest. Neuroimage. 2006;31:968–980. doi: 10.1016/j.neuroimage.2006.01.021. [DOI] [PubMed] [Google Scholar]
- 19.Rosas HD, Liu AK, Hersch S, Glessner M, Ferrante RJ, Salat DH, van der Kouwe A, Jenkins BG, Dale AM, Fischl B. Regional and progressive thinning of the cortical ribbon in Huntington's disease. Neurology. 2002;58:695–701. doi: 10.1212/wnl.58.5.695. [DOI] [PubMed] [Google Scholar]
- 20.Kuperberg GR, Broome MR, McGuire PK, David AS, Eddy M, Ozawa F, Goff D, West WC, Williams SC, van der Kouwe AJ, Salat DH, Dale AM, Fischl B. Regionally localized thinning of the cerebral cortex in schizophrenia. Arch Gen Psychiatry. 2003;60:878–888. doi: 10.1001/archpsyc.60.9.878. [DOI] [PubMed] [Google Scholar]
- 21.Salat DH, Buckner RL, Snyder AZ, Greve DN, Desikan RS, Busa E, Morris JC, Dale AM, Fischl B. Thinning of the cerebral cortex in aging. Cereb Cortex. 2004;14:721–730. doi: 10.1093/cercor/bhh032. [DOI] [PubMed] [Google Scholar]
- 22.Burdick JT, Chen WM, Abecasis GR, Cheung VG. In silico method for inferring genotypes in pedigrees. Nat Genet. 2006;38:1002–1004. doi: 10.1038/ng1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Abecasis GR, Cherny SS, Cookson WO, Cardon LR. Merlin--rapid analysis of dense genetic maps using sparse gene flow trees. Nat Genet. 2002;30:97–101. doi: 10.1038/ng786. [DOI] [PubMed] [Google Scholar]
- 24.Almasy L, Blangero J. Multipoint quantitative-trait linkage analysis in general pedigrees. Am J Hum Genet. 1998;62:1198–1211. doi: 10.1086/301844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Almasy L, Dyer TD, Blangero J. Bivariate quantitative trait linkage analysis: pleiotropy versus co-incident linkages. Genet Epidemiol. 1997;14:953–958. doi: 10.1002/(SICI)1098-2272(1997)14:6<953::AID-GEPI65>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 26.Williams JT, Begleiter H, Porjesz B, Edenberg HJ, Foroud T, Reich T, Goate A, Van Eerdewegh P, Almasy L, Blangero J. Joint multipoint linkage analysis of multivariate qualitative and quantitative traits. II. Alcoholism and event-related potentials. Am J Hum Genet. 1999;65:1148–1160. doi: 10.1086/302571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Glahn DC, Curran JE, Winkler AM, Carless MA, Kent JW, Jr, Charlesworth JC, Johnson MP, Goring HHH, Cole SA, Dyer TD, Moses EK, Olvera RL, Kochunov P, Duggirala R, Fox PT, Almasy L, Blangero J. High dimensional endophenotype ranking in search for major depression risk genes. Biol Psychiatry. 2012;71:6–14. doi: 10.1016/j.biopsych.2011.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sobel E, Papp JC, Lange K. Detection and integration of genotyping errors in statistical genetics. Am J Hum Genet. 2002;70:496–508. doi: 10.1086/338920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Heath S. Markov chain Monte Carlo methods for radiation hybrid mapping. J Comput Biol. 1997;4:505–515. doi: 10.1089/cmb.1997.4.505. [DOI] [PubMed] [Google Scholar]
- 30.Kong A, Gudbjartsson DF, Sainz J, Jonsdottir GM, Gudjonsson SA, Richardsson B, Sigurdardottir S, Barnard J, Hallbeck B, Masson G, Shlien A, Palsson ST, Frigge ML, Thorgeirsson TE, Gulcher JR, Stefansson K. A high-resolution recombination map of the human genome. Nat Genet. 2002;31:241–247. doi: 10.1038/ng917. [DOI] [PubMed] [Google Scholar]
- 31.Feingold E, Brown PO, Siegmund D. Gaussian models for genetic linkage analysis using complete high-resolution maps of identity by descent. Am J Hum Genet. 1993;53(1):234–251. [PMC free article] [PubMed] [Google Scholar]
- 32.Winkler A, Kochunov P, Blangero J, Almasy L, Zilles K, Fox P, Duggirala R, Glahn D. Cortical thickness or grey matter volume? The importance of selecting the phenotype for imaging genetics studies. Neuroimage. 2010;53:1135–1146. doi: 10.1016/j.neuroimage.2009.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Belligni EF, Di Gregorio E, Biamino E, Calcia A, Molinatto C, Talarico F, Ferrero GB, Brusco A, Silengo MC. 790 Kb microduplication in chromosome band 17p13.1 associated with intellectual disability, afebrile seizures, dysmorphic features, diabetes, and hypothyroidism. Eur J Med Genet. 2012;55(3):222–224. doi: 10.1016/j.ejmg.2012.01.016. [DOI] [PubMed] [Google Scholar]
- 34.Kissebah AH, Sonnenberg GE, Myklebust J, Goldstein M, Broman K, James RG, Marks JA, Krakower GR, Jacob HJ, Weber J, Martin L, Blangero J, Comuzzie AG. Quantitative trait loci on chromosomes 3 and 17 influence phenotypes of the metabolic syndrome. Proc Natl Acad Sci USA. 2000;97:14478–14483. doi: 10.1073/pnas.97.26.14478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu X, Cooper RS, Borecki I, Hanis C, Bray M, Lewis CE, Zhu X, Kan D, Luke A, Curb D. A combined analysis of genomewide linkage scans for body mass index from the National Heart, Lung, and Blood Institute Family Blood Pressure Program. Am J Hum Genet. 2002;70(5):1247–1256. doi: 10.1086/340362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Frankort A, Roefs A, Siep N, Roebroeck A, Havermans R, Jansen A. Reward activity in satiated overweight women is decreased during unbiased viewing but increased when imagining taste: an event-related fMRI study. Int J Obes (Lond) 2012;36(5):627–637. doi: 10.1038/ijo.2011.213. [DOI] [PubMed] [Google Scholar]
- 37.Meyre D, Lecoeur C, Delplanque J, Francke S, Vatin V, Durand E, Weill J, Dina C, Froguel P. A genome-wide scan for childhood obesity-associated traits in French families shows significant linkage on chromosome 6q22.31-q23.2. Diabetes. 2004;53(3):803–811. doi: 10.2337/diabetes.53.3.803. [DOI] [PubMed] [Google Scholar]; Hardy SGP, Chronister RB, Parent AD. The Hypothalamus. In: Haines D, editor. Fundamental Neuroscience for Basic and Clinical Applications – 3rd edition. Philadelphia: Churchill-Livingstone; 2006. [Google Scholar]
- 38.Rothemund Y, Preuschhof C, Bohner G, Bauknecht HC, Klingebiel R, Flor H, Klapp BF. Differential activation of the dorsal striatum by high-calorie visual food stimuli in obese individuals. Neuroimage. 2007;37(2):410–421. doi: 10.1016/j.neuroimage.2007.05.008. [DOI] [PubMed] [Google Scholar]