Abstract
BACKGROUND
Immunosuppression is associated with a variety of idiopathic clinical syndromes that may have infectious causes. It has been hypothesized that the cord colitis syndrome, a complication of umbilical-cord hematopoietic stem-cell transplantation, is infectious in origin.
METHODS
We performed shotgun DNA sequencing on four archived, paraffin-embedded endoscopic colon-biopsy specimens obtained from two patients with cord colitis. Computational subtraction of human and known microbial sequences and assembly of residual sequences into a bacterial draft genome were performed. We used polymerase-chain-reaction (PCR) assays and fluorescence in situ hybridization to determine whether the corresponding bacterium was present in additional patients and controls.
RESULTS
DNA sequencing of the biopsy specimens revealed more than 2.5 million sequencing reads that did not match known organisms. These sequences were computationally assembled into a 7.65-Mb draft genome showing a high degree of homology with genomes of bacteria in the bradyrhizobium genus. The corresponding newly discovered bacterium was provisionally named Bradyrhizobium enterica. PCR identified B. enterica nucleotide sequences in biopsy specimens from all three additional patients with cord colitis whose samples were tested, whereas B. enterica sequences were absent in samples obtained from healthy controls and patients with colon cancer or graft-versus-host disease.
CONCLUSIONS
We assembled a novel bacterial draft genome from the direct sequencing of tissue specimens from patients with cord colitis. Association of these sequences with cord colitis suggests that B. enterica may be an opportunistic human pathogen. (Funded by the National Cancer Institute and others.)
Allogeneic hematopoietic stem-cell transplantation (HSCT) is a cornerstone of therapy for patients with certain hematologic diseases and is associated with a risk of serious complications.1,2 Conditioning and antimicrobial therapy can have direct toxic effects and alter the gut microbiome.3 Immunosuppression and the limited efficacy of immunologically naive stem cells in umbilical-cord HSCT can result in life-threatening infections, especially in the first year after transplantation.4 Gastrointestinal toxicity is common after HSCT and can be manifested clinically as colitis.5–8 Several types of colitis affect patients undergoing transplantation; these include bacterial, viral, and parasitic types as well as colitis associated with graft-versus-host disease (GVHD).7,8
Recently, a syndrome of colitis that appears to be unique to patients undergoing umbilical-cord HSCT has been described.9 The syndrome termed “cord colitis” is clinically and histopathologically distinct from other known causes of colitis in patients undergoing HSCT. This syndrome of non-bloody, frequent stools developed 3 to 11 months after umbilical-cord HSCT in 11 of 104 patients (11%) at a single center. Histopathological evaluation of colon-biopsy samples revealed chronic active colitis and epithelioid granulomas, without evidence of known microbial pathogens, viral cytopathic changes, or signs of GVHD. A traditional infectious-disease evaluation did not reveal a cause for this syndrome. The patients were eventually treated with metronidazole (in 11 patients) and a fluoroquinolone (in 9 patients); all 11 patients had a response to antibacterial therapy. A subset of patients had a relapse after the cessation of antibiotics, and all of these patients had a subsequent response to the reinitiation of antibacterial therapy.
It has been hypothesized that cord colitis is a manifestation of GVHD (rather than a distinct clinical syndrome),10 a transfusion-mediated colitis,11 colonic infection with Tropheryma whipplei,12 or an inflammatory disorder due to interactions between cord-blood stem cells in patients with double-umbilical-cord transplants.13 We pursued the hypothesis that this unusual form of colitis may be caused by an infectious organism. This hypothesis was based on several factors, including the anticipated alterations in the gut microbiome, the immunologic deficits associated with umbilical-cord stem cells, and the unique epidemiologic, clinical, and histologic characteristics of the syndrome.2–4
Investigators have previously used nucleic acid–based approaches for the identification of pathogens, resulting, for example, in the discovery of T. whipplei (the causative agent in Whipple's disease) and the Merkel-cell polyomavirus (the proposed causative agent in Merkel-cell carcinoma of the skin).14–16 Such approaches have also shown other disease–microbe associations, such as that between Fusobacterium nucleatum and colorectal cancer.17,18
We performed shotgun DNA sequencing of biopsy specimens obtained from patients with cord colitis, followed by computational subtraction of human sequences and known microbial sequences. The resultant microbial and viral analysis of the biopsy-tissue components allowed for clear characterization of the metagenome in this idiopathic, antibiotic-responsive syndrome.
METHODS
SAMPLE SELECTION
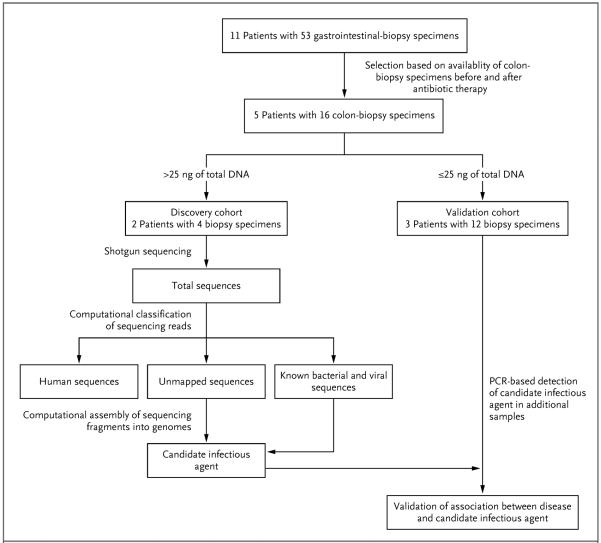
We chose all 11 affected patients in the original cohort at Brigham and Women's Hospital9 for nucleic acid–based investigation. We initially limited the study to samples from a single institution, since there is substantial variation among centers in clinical approaches that are likely to affect the gut microbiome (e.g., the use of antithymocyte globulin, prophylactic and empirical antibiotic use, and GVHD prophylaxis). During review of the gastrointestinal-biopsy specimens from the original cohort, we noted that 5 patients had undergone lower gastrointestinal endoscopy with biopsy both before and after the initiation of antibiotic treatment for cord colitis; 16 of these 23 colon-biopsy specimens were selected for further investigation (Table 1). As controls, formalin-fixed, paraffin-embedded colon samples from 5 healthy persons who had undergone screening colonoscopy and 3 patients who had undergone umbilical-cord HSCT and had pathologically confirmed intestinal GVHD were used, as well as DNA from colon specimens obtained from 5 patients undergoing resection for colon cancer, as described previously.18 In addition, we obtained upper gastrointestinal–biopsy specimens from the duodenum and stomach from 3 patients from the initial cohort with cord colitis.
Table 1.
Clinicopathological Data Regarding Patients with Cord Colitis Included in the Metagenomic Discovery and Validation Cohorts.*
| Patient No. | Sex | Diagnosis | Transplantation Details | Onset of Cord Colitis Syndrome after HSCT | Timing and Type of Antibiotic Therapy after HSCT | Sample No. | Timing of Biopsy after HSCT | Biopsy Site | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Initiation | Termination | Initiation after Relapse | Termination after Relapse | Stomach | Duodenum | Ileum | Colon | Sigmoid | Rectum | |||||||
| no. of days | day (type of therapy) | no. of days | ||||||||||||||
| 4 | F | Acute myeloid leukemia | Myeloablative UC-HSCT | 103 | 111 (M, C) | 121 | 125 (M, C) | 855 | 4a | 30 | x | |||||
| 4b | 120 | x | ||||||||||||||
| 4c | 180 | x | ||||||||||||||
| 4d | 236 | x | x | x | x | |||||||||||
| 4e | 358 | x | x | |||||||||||||
| 5 | F | Chronic myeloid leukemia | Myeloablative UC-HSCT | 158 | 181 (M, C) | 271 | 278 (M, C) | ongoing | 5a | 64 | x | |||||
| 5b† | 105 | x | ||||||||||||||
| 5c† | 209 | x | x | x | ||||||||||||
| 5d | 526 | x | x | x | x | |||||||||||
| 6 | M | Myelodysplastic syndrome | Myeloablative UC-HSCT | 167 | 177 (M, C) | 191 | NA | NA | 6a | 55 | x | |||||
| 6b | 205 | x | x | |||||||||||||
| 9 | M | Chronic lymphocytic leukemia | RIC UC-HSCT | 314 | 375 (M) | 385 | NA | NA | 9a | −5553‡ | ||||||
| 9b | 257 | x | ||||||||||||||
| 9c | 312 | x | x | x | ||||||||||||
| 9d | 371 | x | ||||||||||||||
| 9e | 481 | x | ||||||||||||||
| 9f | 560 | x | ||||||||||||||
| 9g | 642 | x | ||||||||||||||
| 9h | 668 | x | ||||||||||||||
| 11 | M | Hodgkin's disease | RIC UC-HSCT | 298 | 298 (M, L) | 358 | 376 (M, L) | 436 | 11a | 205 | x | |||||
| 11b† | 266 | x | x | x | ||||||||||||
| 11c | 285 | x | ||||||||||||||
| 11d† | 342 | x | x | |||||||||||||
Nucleic acid–based analysis was performed on 16 of the 23 biopsy samples listed here: Patient 4, samples a, b, c, and e; Patient 5, samples b, c, and d; Patient 9, samples b, c, d, e, and f; Patient 6, samples a and b; and Patient 11, samples b and d. Diagnosis refers to the diagnosis for which stem-cell transplantation was performed. C denotes ciprofloxacin, HSCT hematopoietic stem-cell transplantation, L levofloxacin, M metronidazole, NA not applicable, RIC reduced-intensity conditioning, and UC umbilical cord.
This sample was one of four samples included in the discovery cohort. Details regarding the DNA analysis are provided in Tables S1 and S2 in the Supplementary Appendix.
Patient 9 underwent an appendectomy several years before transplantation.
We also obtained gastrointestinal-biopsy specimens from a patient who was treated at Massachusetts General Hospital for colitis after undergoing HSCT and who had some features that were consistent with cord colitis, in order to investigate the microbiome of a patient who had undergone HSCT at another institution. (Details are provided in the Supplementary Appendix, available with the full text of this article at NEJM.org.) The institutional review board at each institution approved the study. A waiver of the requirement for informed consent was granted by the human research committee at Partners HealthCare, which also approved the study.
DNA EXTRACTION AND SEQUENCING
After removal of the first 20 μm of each formalin-fixed, paraffin-embedded block, we extracted DNA from two 20-μm sections with the use of a RecoverAll Total Nucleic Acid Isolation kit (Ambion). In cases in which extraction yielded more than 25 ng of DNA (in Patients 5 and 11),9 we pursued DNA sequencing. In cases in which extraction yielded 25 ng or less of DNA, we reserved the samples for validation studies (Fig. 1). Bar-coded libraries were prepared from pairs of samples obtained before and after the initiation of antibiotic therapy in Patients 5 and 11, as described previously.18 Paired-end 76-bp or 101-bp massively parallel sequencing was performed at separate sequencing centers for each patient in order to control for possible contamination (see the Supplementary Appendix for a detailed description of the contamination analysis).
Figure 1. Sample Selection and Analyses.
Formalin-fixed, paraffin-embedded samples obtained from patients with cord colitis were selected for molecular analysis on the basis of clinical criteria. Patients for whom colon-biopsy samples were available for the period ranging from 120 days before antibiotic therapy to 200 days after such therapy were selected for inclusion in the initial cohort. DNA extraction and sequencing were followed by PathSeq analysis,19 in which computational subtraction was applied for the removal of human and known microbial sequences. The remaining unmapped sequencing reads and the reads with a high degree of homology with known microbial sequences were then computationally assembled into longer contiguous overlapping sequences (contigs) representing genomic fragments of a novel organism. Candidate pathogens, which were predicted by PathSeq analysis of the discovery cohort, were detected by targeted methods, such as polymerase-chain-reaction (PCR) assay, in the validation cohort.
COMPUTATIONAL SUBTRACTION AND ASSEMBLY OF UNMAPPABLE READS
We used PathSeq software, version 1.2 (www.broadinstitute.org/software/pathseq), to perform iterative computational subtraction of human reads, known microbial reads, and viral reads, as described previously.19 We used the Velvet software package for de novo short-read assembly (assembly without the use of a reference genome) to generate contiguous overlapping sequences (contigs) from reads that were not mapped in the PathSeq analysis.20 We used the Basic Local Alignment Search Tool (BLAST) for nucleotide sequences (BLASTN) and proteins (BLASTX) to compare all contigs with the National Center for Biotechnology Information (NCBI) databases of nucleotide and protein sequences, which include all human, nonhuman eukaryotic, prokaryotic, and viral nucleic acid and amino acid sequences from nondraft genomes.
Nonhuman reads from samples 11b and 11d (from Patient 11) were pooled and subjected to de novo assembly with the use of two different software packages: Velvet and ALLPATHS.20,21 Contigs that formed the novel genome were aligned to the NCBI nucleotide-sequence database with the use of BLASTN.22 Contigs with a high degree of homology with bradyrhizobium species were represented to a similar depth of coverage, suggesting a common origin. These contigs also had percentages of GC (guanine–cytosine) content similar to one another (Fig. S2 in the Supplementary Appendix). We linked contigs to one another using paired reads to generate supercontigs (see the Supplementary Appendix for a detailed description of assembly methods).
COMPARATIVE GENOMIC ANALYSIS
The supercontigs that were generated by the de novo assembly formed the draft genome of a novel organism, called Bradyrhizobium enterica (deposited as NCBI Bioproject PRJNA174084; accession number, AMFB00000000; strain name, B. enterica DFCI-1; www.broadinstitute.org/annotation/genome/Bradyrhizobium_enterica.1/MultiHome.html). We used the Prodigal annotation tool23 to annotate the B. enterica genome. We used the PhyloPhlAn tool (http://huttenhower.sph.harvard.edu/phylophlan) to perform rooted phylogenetic analysis on a subset of 400 core genes, followed by bootstrap analysis to determine the strength of the predicted phylogenetic associations.
Comparative genomic analysis was performed (for details, see the Supplementary Appendix). Global alignment of amino acid sequences was performed with the use of the Needleman–Wunsch algorithm,24 and percentage identity between each B. enterica gene and its closest homologue in B. japonicum was determined.
PCR AMPLIFICATION OF A B. ENTERICA TARGET AND HUMAN ACTIN CONTROL
Primers for polymerase-chain-reaction (PCR) assays were designed against a poorly conserved region of the provisional B. enterica genome with the use of PrimerQuest (Integrated DNA Technologies) and were generated. These primers (forward primer, 5′-TCGAGGGCTACGGCTTGAAGATTT-3′; reverse primer, 5′-ACAACGTGTTGCCGCCAATATGAG-3′) amplify a 367-bp target, which spans an intergenic region (supercontig 17 at base-pair position 152,156–152,522). Primers that target the human actin gene (forward primer, 5′-GCGAGAAGATGACCCAGATC-3′; reverse primer, 5′-CCAGTGGTACGGCCAGAGG-3′) amplify a 102-bp target. A detailed description of PCR conditions is provided in the Supplementary Appendix.
FISH ANALYSIS
We used fluorescence in situ hybridization (FISH) to perform experiments on formalin-fixed, paraffin-embedded colon-biopsy specimens obtained from patients with cord colitis and from controls. The experiments were carried out according to the method of Swidsinski, with both a bradyrhizobium-specific probe and a eubacterial (universal bacterial) probe.25,26 (See the Supplementary Appendix for detailed descriptions of FISH methods and competition experiments performed to show the specificity of the bradyrhizobium probe.)
RESULTS
SHOTGUN SEQUENCING AND PathSeq ANALYSIS
DNA was extracted from the temporally distinct colon-biopsy specimens from Patients 5 and 11 (samples 5b and 5c and samples 11b and 11d) (Table 1) and was used for massively parallel sequencing (Fig. 1). Bar-coded libraries were prepared and subjected to sequencing on the Illumina V3 Platform.19 Sequential computational subtraction of human reads and known microbial reads (including bacteria, archaea, viruses, and fungi) was performed with the use of PathSeq (Table S1 in the Supplementary Appendix).19 All data regarding nonhuman reads are deposited in the NCBI Sequence Read Archive (submission number, SRS386798). More than 2.5 million reads remained unmapped, suggesting the presence of abundant sequences that were absent from the reference databases used.
GENOME ASSEMBLY AND COMPARATIVE GENOMIC ANALYSIS
A pooled set of nonhuman reads from samples 11b and 11d was subjected to de novo assembly.20,21 The ALLPATHS software generated the largest number of total contigs that were over 2.5 kb in length. Ninety-nine contigs that were generated by this method were assembled into 89 supercontigs and manually reviewed; 1 supercontig (3621 bp) was removed, since it showed high sequence similarity to a SEN virus.27 A 126-kb circular supercontig (contig 32, supercontig 25) had a high degree of homology with a plasmid element from bradyrhizobium species BTAi1 (pBBta01; accession number, CP000495.1); this plasmid is absent in B. japonicum. The 88 remaining supercontigs all contained regions with a high degree of homology with B. japonicum, which consists of a single circular chromosome of 9,105,828 bp; 86 of the 88 supercontigs had a GC content of 60 to 66%. The resulting draft genome size (including the plasmid) was 7,645,871 bp, with a 64.4% GC content. (The genome sizes of most bradyrhizobium species range from approximately 7.5 to 10 Mb.) Given the known limitations of massively parallel sequencing in genome assembly,28 small areas of the genome probably remain unassembled. The high coverage of the genome suggests that most of it has been discovered. With the use of the Prodigal genome annotation tool, 7112 protein-encoding genes were predicted within the provisional genome23 (see the Supplementary Appendix for details regarding genome assembly and annotation).
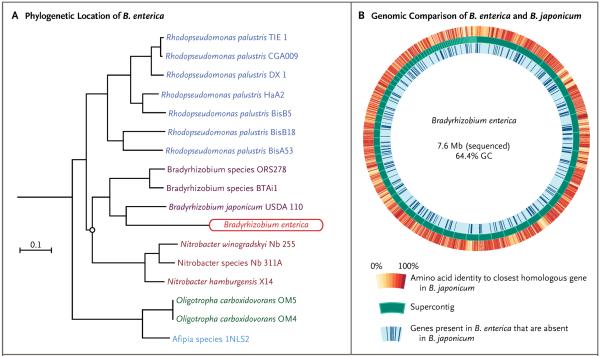
Phylogenetic analysis with the use of PhyloPhlAn generated a rooted phylogenetic tree (Fig. 2A). Bootstrap analysis revealed more than 99% consensus at all branch points except for one (circled in Fig. 2A), where the bootstrap value was 0.181. The organism was provisionally named B. enterica, given the close phylogenetic relationship with B. japonicum and the human anatomical location where the organism was discovered. The amino acid sequence identity between homologous proteins in B. enterica and B. japonicum is shown in Figure 2B.29 A list of genes that are present in B. enterica and absent in B. japonicum is provided in the Supplementary Appendix.
Figure 2. Phylogenetic Analysis and Genomic Assembly of the Organism Associated with Cord Colitis and Comparison of the Provisional Genome for Bradyrhizobium enterica with the Genome of B. japonicum USDA 110.
A rooted phylogenetic tree showing the predicted evolutionary relationship between B. enterica and related species was constructed by means of multisequence alignment of 400 core, protein-coding genes (Panel A). Bootstrap analysis was performed showing more than 99% consensus at all branch points except the circled one, where the bootstrap value was 18.1%. Shown is a Circos plot29 of the draft B. enterica genome, which was assembled with the use of unmappable reads from shotgun whole-genome sequencing of samples of cord colitis (Panel B). The draft genome, which is composed of linear contigs, is represented circularly in the middle track in order of descending contig size. A circular contig that probably represents a plasmid similar to that found in bradyrhizobium species BTAi1 was excluded from this representation. On the inner track, dark blue lines that are perpendicular to the circular genome plot indicate genes present in B. enterica that are not present in B. japonicum USDA 110. On the outer track, the amino acid sequence identity of each B. enterica protein to its closest B. japonicum homologue is represented. In the middle track, each contig is represented by a dark green block, with its borders outlined in light green.
CHARACTERIZATION OF MICROBIAL SEQUENCES IN CORD COLITIS SAMPLES
Assembly of long contigs corresponding to the draft genome of a novel organism was technically possible because of the inferred high abundance of the novel organism and the oligoclonality of the microbiome in the samples obtained from patients with cord colitis. To determine the ratio of B. enterica to total bacterial reads in the four index samples, we once more performed PathSeq analysis, with the addition of the draft B. enterica genome to the reference database. The most abundant bacterial reads are presented in Table 2.
Table 2.
Sequencing Reads for the 27 Most Abundant Bacteria in Four Samples from Patients with Cord Colitis in the Discovery Cohort.*
| Organism | Sample No. | |||
|---|---|---|---|---|
| 5b | 5c | 11b | 11d | |
| number of reads | ||||
| Bradyrhizobium enterica | 631,276 | 119,122 | 1,669,087 | 1,360,422 |
| Delftia acidovorans | 5,006 | 7,517 | 137 | 39 |
| Stenotrophomonas maltophilia | 2,740 | 3,615 | 130 | 50 |
| Other delftia species† | 2,103 | 2,975 | 371 | 113 |
| Propionibacterium acnes | 1,070 | 355 | 5,985 | 1,087 |
| Bradyrhizobium japonicum | 695 | 180 | 1,520 | 1,177 |
| Ralstonia pickettii | 678 | 148 | 1,295 | 419 |
| Other bradyrhizobium species | 542 | 110 | 1,146 | 887 |
| Pseudomonas mendocina | 460 | 76 | 1,276 | 417 |
| Pseudomonas fluorescens | 375 | 76 | 126 | 56 |
| Rhodopseudomonas palustris | 311 | 77 | 627 | 451 |
| Acidovorax ebreus | 208 | 101 | 682 | 290 |
| Xanthomonas campestris | 208 | 97 | 127 | 252 |
| Alicycliphilus denitrificans | 174 | 91 | 300 | 139 |
| Ralstonia solanacearum | 137 | 26 | 333 | 103 |
| Other acidovorax species | 123 | 78 | 349 | 174 |
| Mesorhizobium opportunistum | 99 | 94 | 111 | 50 |
| Burkholderia gladioli | 95 | 39 | 409 | 248 |
| Pseudomonas aeruginosa | 89 | 80 | 149 | 39 |
| Methylobacterium radiotolerans | 79 | 99 | 114 | 15 |
| Escherichia coli | 74 | 81 | 472 | 295 |
| Rubrivivax gelatinosus | 67 | 28 | 240 | 177 |
| Other agrobacterium species† | 46 | 11 | 769 | 269 |
| Cupriavidus taiwanensis | 46 | 60 | 29 | 19 |
| Asticcacaulis excentricus | 45 | 34 | 289 | 72 |
| Agrobacterium tumefaciens | 22 | 8 | 489 | 135 |
| Fusobacterium nucleatum | 8 | 2 | 142 | 217 |
| Rothia mucilaginosa | 5 | 5 | 313 | 14 |
| Clostridium difficile | 3 | 1 | 4 | 114 |
| Streptococcus sanguinis | 3 | 6 | 582 | 3 |
| Lactobacillus reuteri | 1 | 0 | 433 | 0 |
PathSeq taxonomic classification was performed on all four sequenced samples obtained from Patients 5 and 11 with the inclusion of the Bradyrhizobium enterica draft genome in the reference database. Organisms are listed according to their abundance in sample 5b. Additional details regarding the viral reads are provided in Table S4 in the Supplementary Appendix.
Only one species was identified.
In Patient 5, the relative abundance of B. enterica reads in the posttreatment sample, as compared with the pretreatment sample, obtained from Patient 5 decreased by 84.4%. The posttreatment sample was obtained 28 days after the initiation of antibiotic therapy for the cord colitis syndrome (see the Supplementary Appendix for details). Similarly, in Patient 11, the relative abundance of B. enterica reads in the posttreatment sample, as compared with the pretreatment sample, decreased by 60.5%. The posttreatment sample was obtained 44 days after the initiation of antibiotic therapy for relapsed cord colitis syndrome. B. enterica was the predominant bacterium in all four samples (Table 2). The most abundant human viruses that were identified on whole-genome sequencing are presented in Table S2 in the Supplementary Appendix.
In contrast to the microbiome of colon samples from healthy controls30 and samples of normal colonic tissue obtained adjacent to colorectal tumors,18 known intestinal commensals (e.g., Propionibacterium acnes) were present at a much lower level than B. enterica, with the number of P. acnes reads ranging from 0.08 to 0.3% of the total number of B. enterica reads. Cytomegalovirus colitis had previously been diagnosed and treated in Patient 11. There was no histopathological evidence of cytopathic changes associated with cytomegalovirus at the time that cord colitis was diagnosed.9 Of note, the number of cytomegalovirus reads was low and decreased further in the analysis of the second biopsy sample (Table S4 in the Supplementary Appendix).
DETECTION OF B. ENTERICA IN CONTROLS AND ADDITIONAL PATIENTS WITH CORD COLITIS
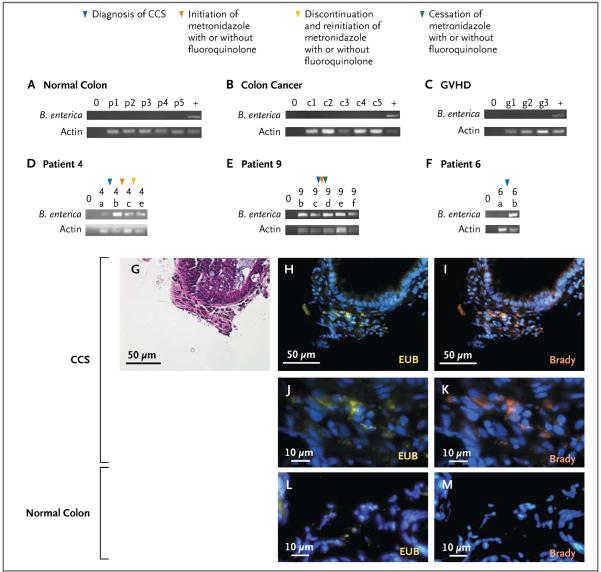
PCR analysis was performed to investigate the differential abundance of B. enterica, as compared with total bacteria and total human cells, in patients with cord colitis as compared with healthy controls, patients with colon cancer, and patients who had undergone umbilical-cord HSCT and had pathologically confirmed GVHD. In addition to samples from these controls, colon-biopsy samples were obtained within 120 days before treatment and 200 days after treatment in three other patients with cord colitis. Given the very limited amount of DNA available in these samples, quantitative PCR studies were not possible. B. enterica was undetectable in all specimens from the three types of control tissue (Fig. 3A, 3B, and 3C). The abundance of B. enterica in colon-biopsy samples from three additional patients with the cord colitis syndrome was inferred by the intensity of the band corresponding to the B. enterica PCR product, as compared with the actin PCR product (Fig. 3D, 3E, and 3F). According to this qualitative measurement, B. enterica was less abundant in samples obtained before the onset of cord colitis, was present in all samples obtained close to the time of diagnosis of cord colitis, and in some cases, decreased in abundance after antimicrobial treatment.
Figure 3. Abundance of Bradyrhizobium enterica in Samples Obtained from Patients with Cord Colitis, as Compared with Control Samples.
B. enterica was more abundant in samples obtained from patients with the cord colitis syndrome (CCS) than in samples obtained from healthy controls, patients with colon cancer, or patients with graft-versus-host disease (GVHD). In addition, B. enterica was present in colon-biopsy samples from three additional patients with cord colitis. In Panels A through F, the top lanes show amplification of a B. enterica target after 35 cycles of a polymerase-chain-reaction (PCR) assay, and the bottom lanes show amplification of a human actin target after 35 PCR cycles. A lane containing reagents but no DNA (no-template control) is indicated by 0. Shown are results for five healthy control specimens (p1 through p5) (Panel A), five colon-cancer specimens (c1 through c5) (Panel B), three colon-biopsy specimens from patients with pathologically confirmed GVHD (g1 through g3) (Panel C), and DNA from temporally distinct colon-biopsy samples from Patient 4 (4a, 4b, 4c, and 4e) (Panel D), from Patient 9 (9b through 9f) (Panel E), and from Patient 6 (6a and 6b) (Panel F). Samples are displayed chronologically. Milestones in therapy for the three patients with cord colitis are indicated by colored arrowheads. Microscopical images of colon tissue obtained from a patient with cord colitis are shown, including a section stained with hematoxylin and eosin (Panel G) and a corresponding section (Panels H through K), along with colon tissue from healthy controls (Panels L and M) stained with either a universal eubacterial probe (EUB, yellow) or a bradyrhizobium-specific probe (Brady) and counterstained with 4',6-diamidino-2-phenylindole (DAPI, orange).
B. enterica was also detected in samples obtained on stomach and duodenal biopsy from three patients with cord colitis (Patients 5, 6, and 11) who had upper gastrointestinal tract involvement at the time of diagnosis (see the Supplementary Appendix). Sequencing reads from B. enterica were also identified in samples obtained on duodenal and gastric biopsy from a patient with colitis after HSCT at Massachusetts General Hospital (see the Supplementary Appendix). This patient had some clinical and histopathological features that were consistent with a diagnosis of cord colitis.
FISH was used to visualize B. enterica in formalin-fixed, paraffin-embedded colon samples from patients with cord colitis. Hybridization with both universal bacterial and bradyrhizobium probes showed the presence of a specific fluorescence signal representing B. enterica within affected tissue (Fig. 3G through 3K). This signal was absent in normal colon samples (Fig. 3L and 3M). All samples that were examined with the use of FISH were counterstained with 4′,6-diamidino-2-phenylindole (DAPI) to visualize host-cell nuclei.
DISCUSSION
Conventional microbiologic tools can be used successfully to detect many clinically significant infectious organisms. However, many potentially infectious syndromes remain idiopathic. Determining a candidate causal agent in these diseases can be challenging and is often unsuccessful. Several observers have predicted that new genomic methods that are sensitive and unbiased may illuminate candidate agents in a subset of diseases, as they have in selected circumstances previously.14–16,31
We used genomic tools to identify a new bacterial species, provisionally named B. enterica, in samples obtained from a cohort of patients with an idiopathic, antibiotic-responsive colitis syndrome with distinct histopathological features. Without a priori knowledge of the organism, we assembled a novel bacterial genome and candidate human pathogen from a specimen of disease tissue. The unusual lack of diversity in the colonic microbiome after HSCT that we noted in these samples has been described previously.3 The abundance of B. enterica in the samples suggests that the syndrome is distinct from other known transplantation-associated colitis syndromes. The organism appeared to be specific to patients with cord colitis; it was not present in various controls, including patients with intestinal GVHD.
The phylogenetic analysis showed that B. enterica was taxonomically related to plant endosymbionts such as B. japonicum, a nitrogen-fixing bacterium that has been used extensively, along with related organisms, in commercial agriculture.32 To date, bradyrhizobium species have not been associated with human disease. The draft genome of B. enterica appears to lack several genes that are critical for nitrogen fixation but does code for at least four filamentous hemagglutinin genes; these genes encode proteins that have been implicated in the binding of pathogenic bacteria, such as Bordetella pertussis, to human airway epithelium.33 Although B. enterica has yet to be cultured and its natural habitat has not yet been defined, it may not be an institution-specific organism. Our findings, though compelling, are insufficient to confirm an association between B. enterica and cord colitis. To assess the generalizability of such an association, future studies will need to focus on the identification of patients with cord colitis at other institutions and investigate the microbiome in these patients and in controls. We hypothesize that variations in institutional HSCT practices probably affect the composition of the gut microbiome and thus the incidence of cord colitis. We anticipate that the identification of additional cases of this rare disease will require a multicenter effort.
Organisms related to B. enterica have shown direct or inferred sensitivity to fluoroquinolones34 and metronidazole,35,36 the therapy that was effective in the treatment of patients with cord colitis in the original cohort.9 Genes encoding pyruvate–ferredoxin oxidoreductase, which are predicted to have a critical role in the reduction of metronidazole and thus its activity,37,38 are present in the genome of B. enterica, a finding that supports the hypothesis that B. enterica is the target of metronidazole therapy in patients with cord colitis. However, on the basis of the available data, it remains possible that bacterial clearance over time and resolution of symptoms may be the result of maturation of the transplanted immune system in affected patients and not a direct effect of antibiotic therapy.
Although we have not shown that B. enterica is the cause of cord colitis,39 we have demonstrated the usefulness of sequencing-based technologies for the unbiased identification of previously undiscovered candidate human pathogens. We anticipate that this reverse microbiology approach, in which the discovery of a novel microorganism is possible without a priori knowledge of the organism or its genome, will allow for the identification of additional potential pathogens.
Our initial work supports the presence of this novel bacterium at the genomic level, but additional biologic characterization is clearly needed. Isolation and culture of B. enterica and the generation and purification of native or recombinant target antigens for antibody development, for example, will greatly aid in the determination of the medical significance and epidemiologic features of this organism. Investigation of the association between this organism and known colitis syndromes (e.g., idiopathic human immunodeficiency virus–associated diarrheal illnesses, inflammatory bowel disease, and the irritable bowel syndrome) could be clinically informative. A description of the epidemiologic features of B. enterica in normal and diseased tissues and the identification of its natural habitat may lead to a better biologic and biomedical understanding of this organism. Finally, we must caution that although we have suggested a possible association between B. enterica and cord colitis, we have not shown that the association is the cause or the consequence of the clinical syndrome, nor have we shown the generalizability of the association in patients other than those in the original, single-institution cohort.
Supplementary Material
Acknowledgments
Supported by grants from the American Association for Cancer Research, the American Society for Blood and Marrow Transplantation, and the Bladder Cancer Advocacy Network (all to Dr. Bhatt); and by grants from the Starr Cancer Consortium and National Cancer Institute (RC2CA148317) (both to Dr. Meyerson).
We thank Daniel Auclair, James Gomez, Rhonda O'Keefe, Deborah Hung, Roby Bhattacharyya, Ann Hirsch, Nikhil Wagle, Frederick Wilson, Doyle Ward, Joshua Francis, Nancy Berliner, and Robert Mayer for helpful discussions.
Footnotes
Disclosure forms provided by the authors are available with the full text of this article at NEJM.org.
REFERENCES
- 1.Copelan EA. Hematopoietic stem-cell transplantation. N Engl J Med. 2006;354:1813–26. doi: 10.1056/NEJMra052638. [DOI] [PubMed] [Google Scholar]
- 2.Rocha V, Labopin M, Sanz G, et al. Transplants of umbilical-cord blood or bone marrow from unrelated donors in adults with acute leukemia. N Engl J Med. 2004;351:2276–85. doi: 10.1056/NEJMoa041469. [DOI] [PubMed] [Google Scholar]
- 3.Taur Y, Xavier JB, Lipuma L, et al. Intestinal domination and the risk of bacteremia in patients undergoing allogeneic hematopoietic stem cell transplantation. Clin Infect Dis. 2012;55:905–14. doi: 10.1093/cid/cis580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Klein AK, Patel DD, Gooding ME, et al. T-cell recovery in adults and children following umbilical cord blood transplantation. Biol Blood Marrow Transplant. 2001;7:454–66. doi: 10.1016/s1083-8791(01)80013-6. [DOI] [PubMed] [Google Scholar]
- 5.Schulenburg A, Turetschek K, Wrba F, et al. Early and late gastrointestinal complications after myeloablative and non-myeloablative allogeneic stem cell transplantation. Ann Hematol. 2004;83:101–6. doi: 10.1007/s00277-003-0756-4. [DOI] [PubMed] [Google Scholar]
- 6.van Kraaij MG, Dekker AW, Verdonck LF, et al. Infectious gastro-enteritis: an uncommon cause of diarrhoea in adult allogeneic and autologous stem cell transplant recipients. Bone Marrow Transplant. 2000;26:299–303. doi: 10.1038/sj.bmt.1702484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McDonald GB, Shulman HM, Sullivan KM, Spencer GD. Intestinal and hepatic complications of human bone marrow transplantation. Gastroenterology. 1986;90:770–84. doi: 10.1016/0016-5085(86)91137-6. [DOI] [PubMed] [Google Scholar]
- 8.Idem Intestinal and hepatic complications of human bone marrow transplantation. Gastroenterology. 1986;90:460–77. doi: 10.1016/0016-5085(86)90949-2. [DOI] [PubMed] [Google Scholar]
- 9.Herrera AF, Soriano G, Bellizzi AM, et al. Cord colitis syndrome in cord-blood stem-cell transplantation. N Engl J Med. 2011;365:815–24. doi: 10.1056/NEJMoa1104959. [DOI] [PubMed] [Google Scholar]
- 10.van Bekkum D, Vossen J, Zurcher C. Cord colitis syndrome in cord-blood stem-cell transplantation. N Engl J Med. 2011;365:2336. doi: 10.1056/NEJMc1111264. [DOI] [PubMed] [Google Scholar]
- 11.Cid J, Lozano M. Cord colitis syndrome in cord-blood stem-cell transplantation. N Engl J Med. 2011;365:2336. doi: 10.1056/NEJMc1111264. [DOI] [PubMed] [Google Scholar]
- 12.Matuchansky C. Cord colitis syndrome in cord-blood stem-cell transplantation. N Engl J Med. 2011;365:2336–7. doi: 10.1056/NEJMc1111264. [DOI] [PubMed] [Google Scholar]
- 13.Masuda S. Cord colitis syndrome in cord-blood stem-cell transplantation. N Engl J Med. 2011;365:2337. doi: 10.1056/NEJMc1111264. [DOI] [PubMed] [Google Scholar]
- 14.Feng H, Shuda M, Chang Y, Moore PS. Clonal integration of a polyomavirus in human Merkel cell carcinoma. Science. 2008;319:1096–100. doi: 10.1126/science.1152586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Relman DA, Schmidt TM, MacDermott RP, Falkow S. Identification of the uncultured bacillus of Whipple's disease. N Engl J Med. 1992;327:293–301. doi: 10.1056/NEJM199207303270501. [DOI] [PubMed] [Google Scholar]
- 16.Relman DA. Microbial genomics and infectious diseases. N Engl J Med. 2011;365:347–57. doi: 10.1056/NEJMra1003071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Castellarin M, Warren RL, Freeman JD, et al. Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma. Genome Res. 2012;22:299–306. doi: 10.1101/gr.126516.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kostic AD, Gevers D, Pedamallu CS, et al. Genomic analysis identifies association of Fusobacterium with colorectal carcinoma. Genome Res. 2012;22:292–8. doi: 10.1101/gr.126573.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kostic AD, Ojesina AI, Pedamallu CS, et al. PathSeq: software to identify or discover microbes by deep sequencing of human tissue. Nat Biotechnol. 2011;29:393–6. doi: 10.1038/nbt.1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zerbino DR, Birney E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008;18:821–9. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Butler J, MacCallum I, Kleber M, et al. ALLPATHS: de novo assembly of whole-genome shotgun microreads. Genome Res. 2008;18:810–20. doi: 10.1101/gr.7337908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–10. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 23.Hyatt D, Chen GL, Locascio PF, Land ML, Larimer FW, Hauser LJ. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics. 2010;11:119. doi: 10.1186/1471-2105-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Needleman SB, Wunsch CD. A general method applicable to the search for similarities in the amino acid sequence of two proteins. J Mol Biol. 1970;48:443–53. doi: 10.1016/0022-2836(70)90057-4. [DOI] [PubMed] [Google Scholar]
- 25.Swidsinski A. Standards for bacterial identification by fluorescence in situ hybridization within eukaryotic tissue using ribosomal rRNA-based probes. Inflamm Bowel Dis. 2006;12:824–7. doi: 10.1097/00054725-200608000-00018. [DOI] [PubMed] [Google Scholar]
- 26.Swidsinski A, Loening-Baucke V, Lochs H, Hale LP. Spatial organization of bacterial flora in normal and inflamed intestine: a fluorescence in situ hybridization study in mice. World J Gastroenterol. 2005;11:1131–40. doi: 10.3748/wjg.v11.i8.1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tanaka Y, Primi D, Wang RY, et al. Genomic and molecular evolutionary analysis of a newly identified infectious agent (SEN virus) and its relationship to the TT virus family. J Infect Dis. 2001;183:359–67. doi: 10.1086/318091. [DOI] [PubMed] [Google Scholar]
- 28.Pop M. Genome assembly reborn: recent computational challenges. Brief Bioinform. 2009;10:354–66. doi: 10.1093/bib/bbp026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Krzywinski M, Schein J, Birol I, et al. Circos: an information aesthetic for comparative genomics. Genome Res. 2009;19:1639–45. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Human Microbiome Project Consortium Structure, function and diversity of the healthy human microbiome. Nature. 2012;486:207–14. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Virgin HW, Todd JA. Metagenomics and personalized medicine. Cell. 2011;147:44–56. doi: 10.1016/j.cell.2011.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kaneko T, Nakamura Y, Sato S, et al. Complete genomic sequence of nitrogen-fixing symbiotic bacterium Bradyrhizobium japonicum USDA110 (supplement) DNA Res. 2002;9:225–56. doi: 10.1093/dnares/9.6.225. [DOI] [PubMed] [Google Scholar]
- 33.Relman DA, Domenighini M, Tuomanen E, Rappuoli R, Falkow S. Filamentous hemagglutinin of Bordetella pertussis: nucleotide sequence and crucial role in adherence. Proc Natl Acad Sci U S A. 1989;86:2637–41. doi: 10.1073/pnas.86.8.2637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gober JW, Kashket ER. Role of DNA superhelicity in regulation of bacteroid-associated functions of Bradyrhizobium sp. strain 32H1. Appl Environ Microbiol. 1989;55:1420–5. doi: 10.1128/aem.55.6.1420-1425.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kelley BC, Nicholas DJ. Inhibition of nitrogenase activity by metronidazole in rhodopseudomonas capsulata. Arch Microbiol. 1981;129:344–8. doi: 10.1007/BF00406459. [DOI] [PubMed] [Google Scholar]
- 36.Jjemba PK. The effect of chloroquine, quinacrine, and metronidazole on both soybean plants and soil microbiota. Chemosphere. 2002;46:1019–25. doi: 10.1016/s0045-6535(01)00139-4. [DOI] [PubMed] [Google Scholar]
- 37.Edwards DI. Reduction of nitroimidazoles in vitro and DNA damage. Biochem Pharmacol. 1986;35:53–8. doi: 10.1016/0006-2952(86)90554-x. [DOI] [PubMed] [Google Scholar]
- 38.Müller M. Reductive activation of nitroimidazoles in anaerobic microorganisms. Biochem Pharmacol. 1986;35:37–41. doi: 10.1016/0006-2952(86)90552-6. [DOI] [PubMed] [Google Scholar]
- 39.Fredericks DN, Relman DA. Sequence-based identification of microbial pathogens: a reconsideration of Koch's postulates. Clin Microbiol Rev. 1996;9:18–33. doi: 10.1128/cmr.9.1.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.