Abstract
In budding yeast, a single double-strand break (DSB) triggers extensive ATM (Tel1) and ATR (Mec1)-dependent phosphorylation of histone H2A (γ-H2AX) around the DSB. We describe Mec1- and Tel1-dependent phosphorylation of γ-H2B at H2B-T129. γ-H2B formation is impaired by γ-H2AX and its binding partner, Rad9. High-density microarray analyses show similar γ-H2AX and γ-H2B distributions, but γ-H2B is absent near telomeres. Both γ-H2AX and γ-H2B are strongly diminished over highly transcribed regions. When transcription of GAL genes are turned off, γ-H2AX is restored within 5 min, in a Mec1 dependent manner; when these genes are again induced, γ-H2AX is rapidly lost. Moreover, when a DSB is induced near CEN2, γ-H2AX spreads to all other centromeric regions, again depending on Mec1. Taken together, our data provide new insights on the function and establishment of the phosphorylation events occurring onto chromatin after DSB induction.
Introduction
In budding yeast, as in mammals, a double-strand break (DSB) provokes a DNA damage response that rapidly leads to phosphorylation of a histone H2A isoform over a large domain around the DSB 1,2. In mammals, about 10% of H2A isoforms are histone H2AX, whose C-terminal S139 can be phosphorylated (termed γ-H2AX), while 90% of H2A lacks this serine. In Saccharomyces cerevisiae, histone H2A comprises the great majority of H2A isoforms and is phosphorylated on S129; we will refer to this modification also as γ-H2AX. Both in yeast and in mammals, γ-H2AX rapidly spreads on large chromatin domain (on more than a megabase in mammals and about 50 kb in yeast).
Although γ-H2AX was originally analyzed after inflicting insults to the DNA by γ-irradiation or laser irradiation, a more detailed analysis employed site-specific endonucleases to create defined DSBs in nearly all cells in the population (reviewed in 3). In budding yeast, HO endonuclease, which normally only cleaves a single site within the MAT locus, has been used to create one or more DSBs at other sites by inserting recognition sites at different locations. These studies have shown that γ-H2AX spreads roughly symmetrically and intensely around a DSB, in each instance about the same 50-kb distance on each side 1,4. Spreading of γ-H2AX is not blocked by the presence of yeast’s very small centromere or by the presence of an ~3-kb heterochromatic HMR locus, although heterochromatic sequences themselves are refractory to γ-H2AX modification 4. γ-H2AX is also found at telomeres, in spite of their partially heterochromatic nature 4,5,6; this modification may be provoked as telomeres are replicated. Recently, we carried out a detailed analysis of the γ-H2AX landscape in mammalian cells upon sequence-specific DSBs induction 7, which revealed that within the large chromatin domains covered by this modification, strongly transcribed regions were markedly reduced in γ-H2AX. Whether these transcribed loci were physically excluded – for example, looped out - from the domain where γ-H2AX spreads or whether the act of transcription itself prevented γ-H2AX formation or maintenance has not been established, although the contribution of cohesins in this process 8 would suggest a role of chromosome conformation in γ-H2AX exclusion at active genes.
In both yeast and mammals, γ-H2AX can be created by either the Tel1 (ATM) or Mec1 (ATR) checkpoint protein kinase. In yeast, Tel1 associates with the nearly-blunt ends of an HO endonuclease-induced DSB by virtue of its association with the MRX complex, whereas Mec1, through its association with the ATRIP homolog, Ddc2, will bind to single-stranded DNA (ssDNA) created by 5′ to 3′ resection of the DSB ends 9–11. In G1 arrested yeast cells, where resection is inhibited, only Tel1 promotes γ-H2AX formation 1. Conversely, as resection proceeds at a rate of about 4 kb/h and more ssDNA is revealed, γ-H2AX can slowly spread further down the damaged chromosome, beyond the initial ~50 kb. This late extension of γ-H2AX can only be carried out by Mec1 4.
In the present study, we examined in greater detail how γ-H2AX is propagated from the DSB site in yeast, and the functions of each kinase in these process. As described below, we have characterized a second DNA-damage-dependent histone phosphorylation, the phosphorylation of the terminal threonine in histone H2B (T129), which can also be created by either Mec1 or Tel1 kinase, and whose formation is impaired by the presence of γ-H2AX, mediated by the BRCT repeat-containing binding partner, Rad9.
Using chromatin immunoprecipitation combined with high-density microarray analyses (ChIP-chip), we investigated the distributions of both γ-H2AX and γ-H2B after induction of multiple DSBs in budding yeast. We find that, as in mammalian cells, strongly transcribed genes show little γ-H2AX or γ-H2B modification but that the histones in these regions can be rapidly phosphorylated - primarily by the Mec1 kinase - if transcription is subsequently inhibited. Moreover, γ-H2AX is able to spread in trans, on physically adjacent chromosomes, as exemplified by the detection of γ-H2AX at all centromeric regions when a DSB is induced close to CEN2. Importantly, this trans phosphorylation is performed by Mec1, underlying major differences in the activity of both kinases on chromatin.
Results
Phosphorylation of H2A inhibits phosphorylating histone H2B
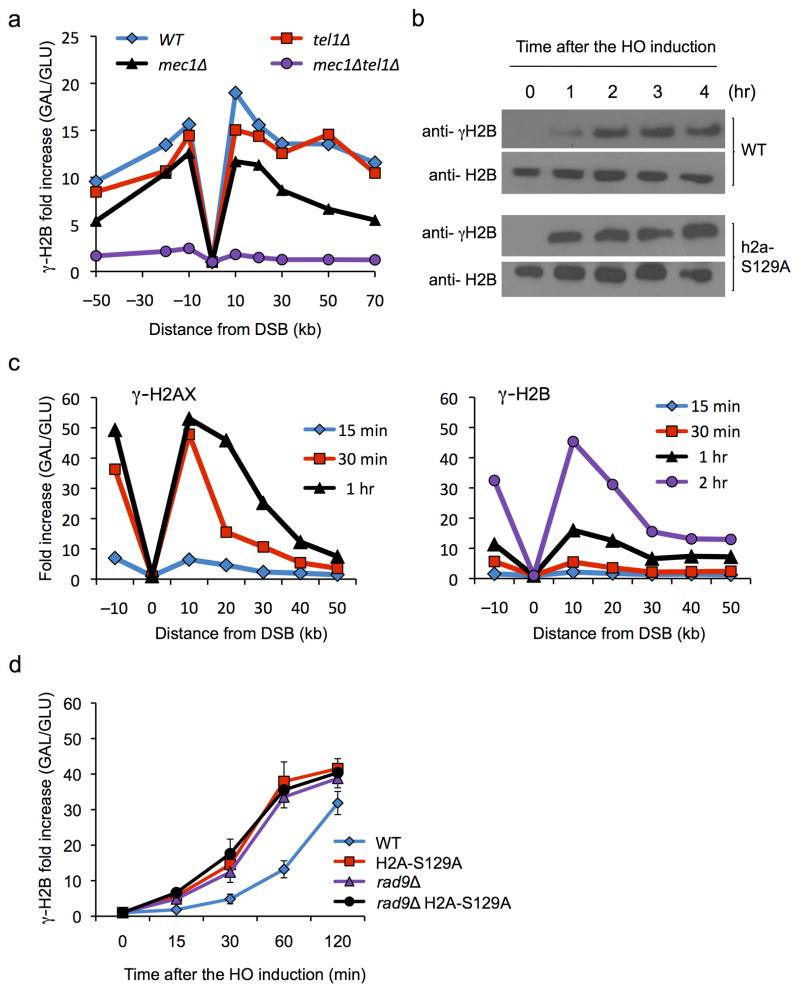
We have identified a second DNA-damage-dependent histone phosphorylation, at the terminal threonine (T129) in histone H2B (γ-H2B). Using an antibody specific for the phosphorylated histone H2B (Suppl. Fig. 1a), we showed that the phosphorylation of histone H2B-T129 is independent of phosphorylation of histone H2A, as the signal is evident in a strain lacking H2A phosphorylation (Suppl. Fig. 1a). This modification can also be created by either Mec1 (ATR-like) or Tel1 (ATM-like) checkpoint protein kinase (Fig. 1a). Similar to γ-H2AX, γ-H2B is also able to spread beyond 50 kb from the DSB (Fig. 1a,c). Whereas γ-H2AX rapidly increases after the induction of a DSB (which takes about 20 min) the appearance of γ-H2B is markedly slower, although it eventually reaches a similar level (Fig. 1b,c). The slow accumulation of γ-H2B is apparently caused by the formation of γ-H2AX, because when we examine γ-H2B induction in a strain lacking γ-H2AX (i.e. in a strain 4 carrying only H2A-S129A), it appears with more rapid kinetics, although still slower than seen for γ-H2AX, suggesting that γ-H2AX somehow impairs γ-H2B establishment (Fig. 1b,d).
Figure 1. Spreading of γ-H2AX and γ-H2B around an HO-induced DSB.
a. γ-H2B spreads extensively around an unrepaired DSB at MATα in strain JKM179, as measured by ChIP 2 hr after HO endonuclease expression was induced. Data for cells lacking Mec1 and/or Tel1 are shown b. Western blot analysis shows that the kinetics of γ-H2B formation is more rapid in a h2a-S129A strain lacking γ-H2AX phosphorylation. Levels of phosphorylation are normalized to the amount of total H2B protein. c. Slow kinetics of γ-H2B formation compared to γ-H2AX are seen in ChIP around the MAT DSB. d. The slow formation of γ-H2B 10 kb distal to the DSB site is accelerated both by eliminating γ-H2AX and by deleting RAD9. For ChIP assays in a, c and d, the fold increase was calculated by normalization of IP values with non-IP (input) values and then further normalized with reference to the values prior to induction of the DSB (0 hr). Error bars represent S.D. from three independent experiments.
One possibility to explain how γ-H2AX would impair the kinetics of γ-H2B modification would be if there were a protein or proteins that bind to γ-H2AX and occlude access to the C-terminal tail of H2B. We postulated that one such protein could be the Rad9 protein that binds to γ-H2AX through its BRCT domains 12. Indeed, deleting RAD9 in an otherwise wild type strain resulted in more rapid modification of γ-H2B (Fig. 1d) without changing the kinetics of modification of γ-H2AX (unpublished). It is of course possible that Rad9’s effect is more indirect, reflecting its role in activating downstream checkpoint kinases; however the behavior of a rad9Δ h2a-S129A double mutant strain was identical to either single mutant (Fig. 1d).
Spreading of γ-H2AX and γ-H2B in strains with three DSBs
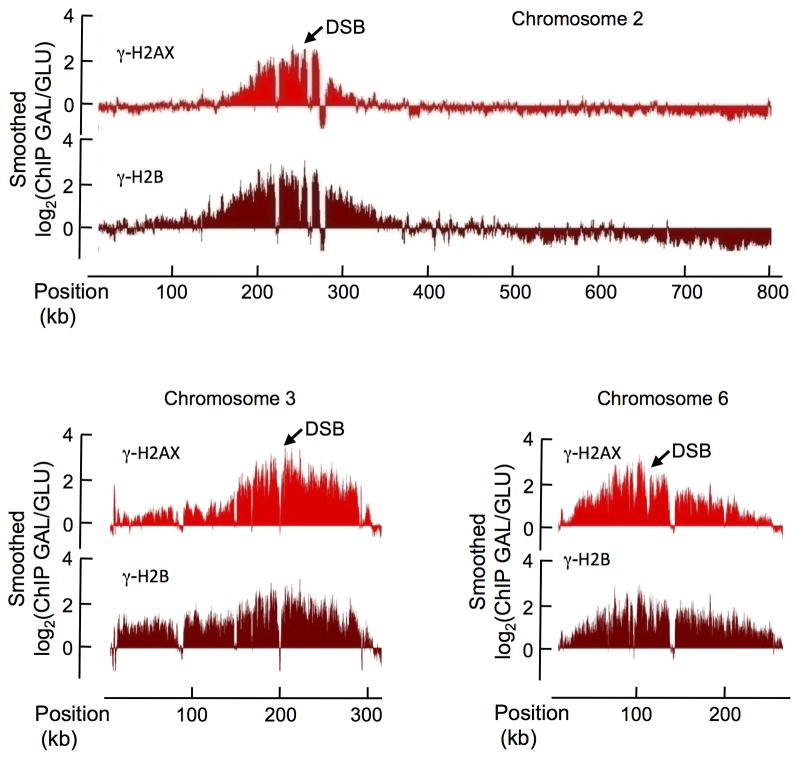
To further investigate the spreading mechanism of DSB-induced histone phosphorylation, we created a budding yeast strain carrying three HO cleavage sites, on chromosomes 2, 3, and 6 in a strain lacking HML or HMR templates to repair such breaks. Thus when GAL::HO is induced, cells suffer three unrepairable DSBs. All three sites are cleaved efficiently (Suppl. Fig. 1b). We performed γ-H2B and γ-H2AX ChIP experiments, before and after DSB induction that we hybridized on Affymetrix yeast tiling arrays, covering the entire yeast genome with 5-bp resolution. Upon HO induction, we observed a highly significant increase of γ-H2AX and γ-H2B around the three induced DSBs, rising to 15 times over the background level. Both γ-H2AX and γ-H2B domains covered about 200 kb around the DSB (Fig. 2), consistent with our previous findings 4.
Figure 2. Profiles of γ-H2AX and γ-H2B in the three HO-cuts strain after DSB induction.
ChIP-chip measurements were performed with antibodies against γ-H2AX (light red) and γ-H2B (dark red), using yeast grown on galactose containing media for 1 hour (DSB induction). The log2 ratio of ChIP-chip signals obtained on Galactose versus Glucose growth is shown (GAL/GLU), across the chromosomes 2, 3 and 6, carrying HO cuts sites (indicated by arrows).
The γ-H2AX profile before DSB induction was highly similar to the γ-H2AX profile already reported 5 validating our ChIP-chip dataset (Suppl Fig. 2a). We compared the ChIP data for the phosphorylated histones to the published RNA polymerase (PolII) distribution, and calculated for each gene the average PolII enrichment, reflecting their transcriptional state. As expected 5, γ-H2AX was found to be enriched on repressed genes (Suppl. Fig. 2b).
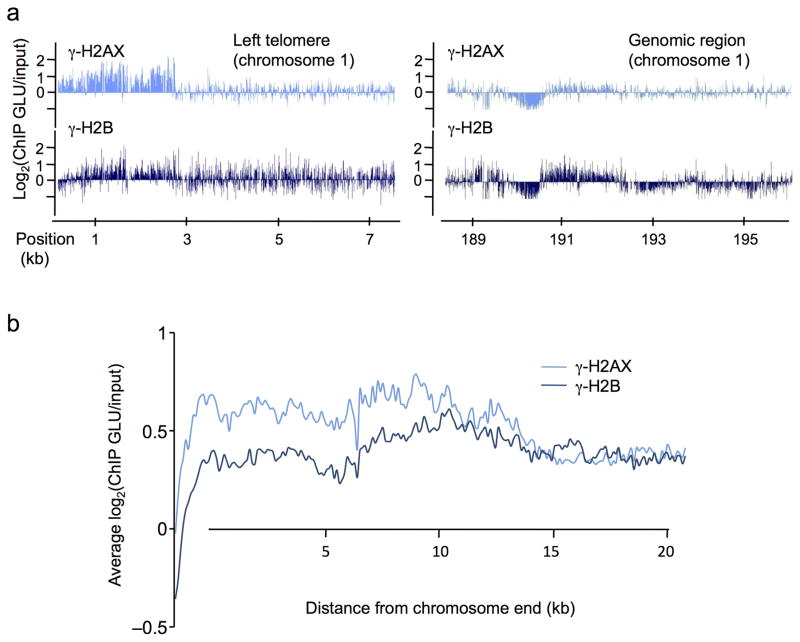
We also examined the distribution of γ-H2B. Overall, γ-H2B distribution was very similar to γ-H2AX (Fig. 2, Suppl. Fig. 2a), and also accumulated on silent genes (Suppl. Fig. 2b); however, at telomeres, γ-H2AX showed a clear enrichment whereas there was much lower γ-H2B accumulation (Fig. 3a and Suppl. Fig. 2c left panels), compared to equivalent amounts of these modifications on most genomic regions (Fig. 3a and Suppl. Fig. 2c right panel). This was confirmed by averaging γ-H2AX and γ-H2B signals over 20 kb, at all yeast telomeres (right and left ends combined) (Fig. 3b). These data suggest that these two phosphorylated forms of histones may possess different properties and functions and may also be a reflection of the difference in the kinetics of formation of the two modifications as shown in Fig. 1b,c.
Figure 3. Profiles of γ-H2AX and γ-H2B in undamaged cells.
a. The profiles of γ-H2AX (light blue) and γ-H2B (dark blue) obtained at the left telomere of chromosome 1 (left panel) and on a genomic region farther on the same chromosome (right panel) are shown. Note that γ-H2B is less enriched than γ-H2AX at the telomere, while both signals are equivalent further away. b. γ-H2AX and γ-H2B ChIP-chip signals were averaged on 20 kb at all chromosomes ends (left and right arms combined). Note the higher level of γ-H2AX at telomeres compared to γ-H2B.
Interestingly, strains lacking γ-H2AX have slightly longer telomeres (approximately an additional 100 bp) than a wild type strain (Suppl. Fig. 3a). These longer telomeres are present despite the fact that 5′ to 3′ resection of an HO–induced DSB ends is accelerated almost two-fold in a strain with H2A-S129A compared to wild-type strain 13,14. The rules for processing telomere ends may be different and γ-H2AX may in fact recruit telomere end-processing factors or alter the binding of some telomere-associated proteins. In contrast, the absence of γ-H2B appeared to have no effect on telomere length, but a strain carrying mutations of both phosphorylation sites appears to have shorter telomeres than H2A-S129A alone.
Phosphorylation of H2A alters γ-H2B distribution
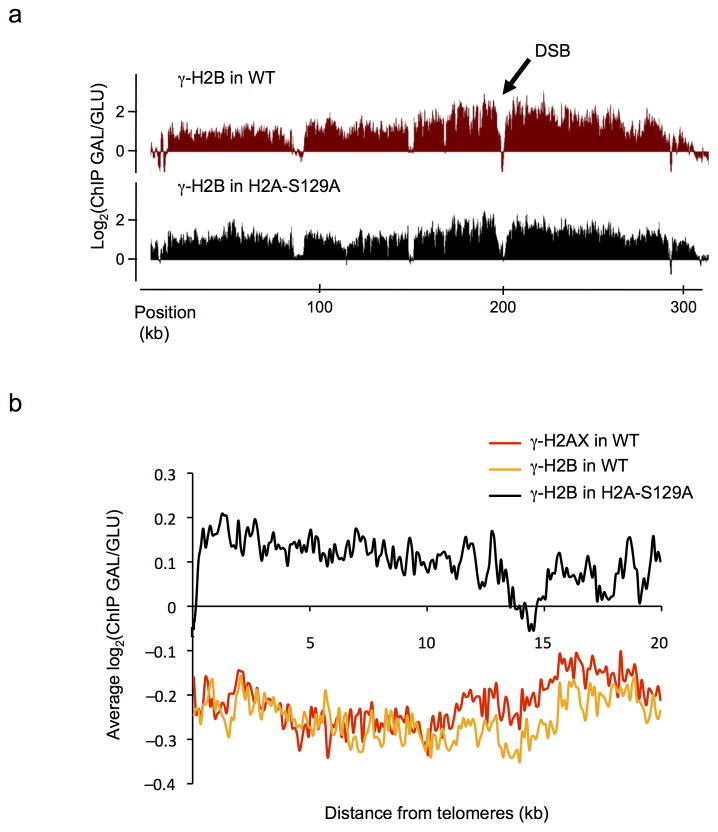
To further investigate the relationship between γ-H2B and γ-H2AX, we analyzed γ-H2B distribution by ChIP-chip in an H2A-S129A mutant strain. Upon growth on glucose (no DSB induction), the distribution of γ-H2B in the mutant strain was highly similar to that observed in WT (Suppl. Fig. 3b–c), except at telomeres, where γ-H2B was markedly reduced (Suppl. Fig. 3d top panel). After DSB induction at MAT on chromosome 3, γ-H2B spreads over a roughly identical domain both in WT and in H2A-S129A mutant strains (Fig. 4a). However, the level of γ-H2B increased dramatically at all subtelomeric regions in the mutant strain upon growth on galactose compared to growth on glucose (Fig. 4b). Such an increase of γ-H2B at subtelomeric regions is not seen when γ-H2AX can form, or for γ-H2AX itself (Fig. 4b and Suppl. Fig. 3d). We suggest two explanations for these observations. First, γ-H2B will more rapidly appear in cells lacking γ-H2AX (Fig. 1b,d). We know that telomere regions are constitutively modified so that there must be transient activations of Mec1 or Tel1 near telomeres 4. Induction of a full-blown checkpoint response by inflicting a DSB could therefore present telomeres with more active Mec1 or Tel1, so that γ-H2B can now appear. Second, induction of a single unrepaired DSB at MAT has previously been shown to cause an alteration in telomere structure, with the dissociation of Sir3 and other silencing proteins 15,16. Thus the telomeric regions may become more accessible to γ-H2B modification.
Figure 4. Differential spreading of γ-H2B in a H2A-S129A mutant strain.
a. γ-H2B ChIP-chip experiments were performed in a WT (in red) or H2A-S129A mutant (in black) background, carrying a DSB at the MAT locus on chromosome 3 before and after DSB induction. The profile of the ratio γ–H2AX GAL/GLU is shown. b. The GAL/GLU ratio obtained for γ-H2AX (red), γ-H2B in WT (orange) and γ-H2B in H2A mutant strain (black) were averaged on 20 kb at telomeres.
Dynamics of γ-H2AX modification in a transcribed region
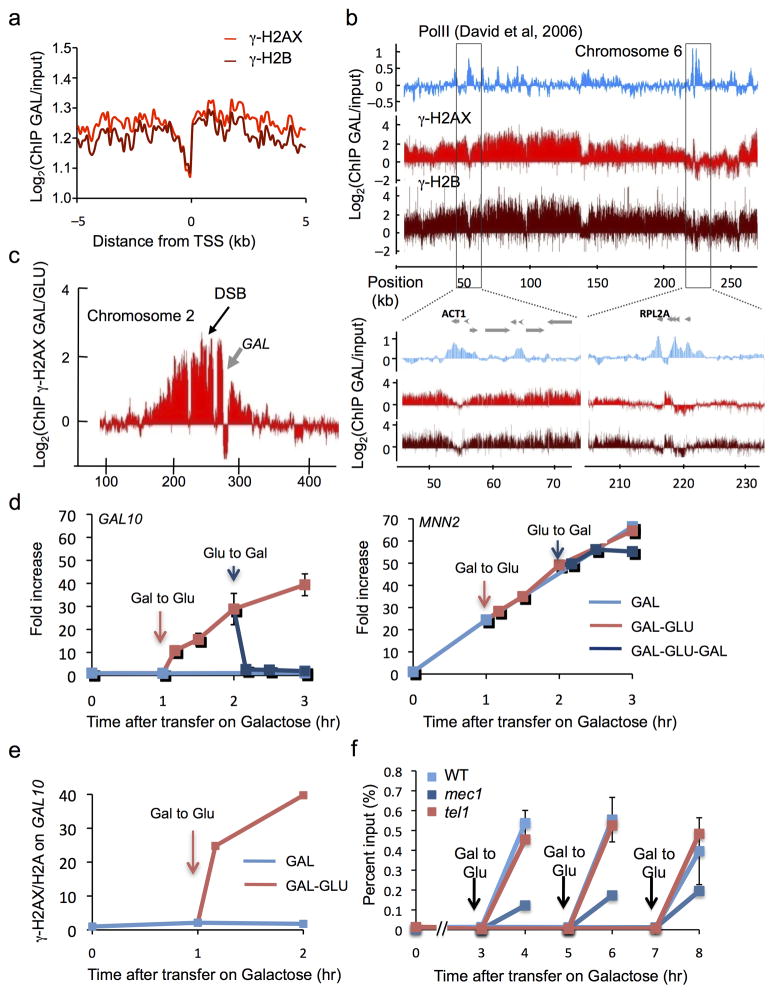
In mammalian cells, γ-H2AX is excluded from promoters and does not accumulate on transcribed genes encompassed in these large chromatin domains covered by γ-H2AX 7. These constraints are also seen in yeast. We retrieved the gene positions encompassed in yeast DSB-induced γ-H2AX and γ-H2B domains, and averaged the signal obtained for these two modifications around the transcription start site (TSS). Similar to our finding in mammalian cells, γ-H2AX and γ-H2B signals decrease at TSS (Fig. 5a) in budding yeast. To assess the relationship between transcription and the accumulation of these marks, we next compared our data with previously published Pol II distribution along the yeast chromosomes 17. Within DSB-induced γ-H2AX domains, loci enriched in PolII presented a clear lack of γ-H2AX and γ-H2B (Fig. 5b). As an example, the highly transcribed actin and ribosomal subunit genes (ACT1 and RPL2) were strongly devoid of both γ-H2AX and γ-H2B (Fig. 5b, bottom panels).
Figure 5. Transcriptionally active units are refractory to γ-H2AX and γ-H2B.
a. Averaged γ-H2AX (light red) and γ-H2B (dark red) ChIP-chip signals upon galactose growth around the TSS of genes encompassed in γ-H2AX domains (supplemental methods). b. γ-H2AX, γ-H2B (this study) and RNA-Pol II (previously reported 17) profiles are shown across chromosome 6. Bottom panel shows a magnification of two regions carrying highly transcribed genes (ACT1 and RPL2A). c. γ-H2AX profile on a part of chromosome 2, one hour after HO induction. The GAL gene cluster (gray arrow), highly transcribed in this growth condition, is strongly depleted in γ-H2AX. d. Left panel: γ-H2AX levels (measured by ChIP-qPCR) within GAL10 are very low for 3 h DSB induction, but rise rapidly if cells are transferred to glucose that shuts off GAL10 transcription (red arrow). γ-H2AX levels rapidly drop when cells are again placed on galactose (blue arrow). Right panel: at the MNN2 gene (not regulated by galactose), γ-H2AX levels rise continuously after transfer to galactose. e. The ratio between γ-H2AX and H2A is shown at GAL10 under different growth conditions (measured by ChIP-qPCR). f. ChIP-qPCR were performed in wildtype, Tel1 and/or Mec1-deleted strains, and transfer on glucose was performed at different time points as indicated. The rapid increase in γ-H2AX in GAL10 is seen if cells are transferred on glucose at 3, 5 or 7 h after DSB induction. This increase can be carried out by either Tel1 or Mec1. Error bars represent the range from two independent experiments.
To examine the lack of γ-H2AX modification in transcribed regions in greater detail, we focused our attention on the cluster of GAL7, GAL10, and GAL1 genes located about 26 kb from the DSB on chromosome 2. The galactose-dependent induction of the DSB is accompanied by the induction of transcription of these genes. One hour after induction of the DSB, γ-H2AX has spread > 50 kb on either side of the break, with a notable “hole” over the GAL gene cluster. These genes also seems to act as a partial boundary for γ-H2AX spreading, since the signal is much lower on the distal side of the active GAL cluster (Fig. 5c). The lack of γ-H2AX over the GAL genes was confirmed by qPCR for four sites in GAL7, GAL10, and GAL1 (Suppl. Fig. 4a). For example, one hour after galactose induction of HO endonuclease cleavage, the level of γ-H2AX in the middle of the GAL10 gene was only 2-fold over the 0 h point (Suppl. Fig. 4a and Fig. 5d left panel, see time point 1h). This is in contrast to the very high level of γ-H2AX detected one hour after HO induction on the MNN2 gene that lies between the DSB and the GAL cluster (Fig. 5d right panel, see time point 1h). γ-H2AX remained at a very low level at GAL10 even 3 h after HO induction, when the modification rose to 70-fold at MNN2 (Fig. 5d left and right panel respectively). Importantly, this absence of γ-H2AX on GAL genes cannot be explained primarily by the decrease of H2A over the transcribed genes, since transcription activation by galactose induction only led to only a two-fold reduction of nucleosome occupancy over the transcribed GAL10 gene, as observed by ChIP using an anti-H2A antibody (Suppl. Fig. 4c). Previous studies have shown that there is a high level of histone H2B (and presumably H2A) turnover over the transcribed GAL genes 18. Our data suggest that either the turnover of nucleosomes is more rapid than the ability of the checkpoint kinases to phosphorylate H2A and/or there is intrinsically lower activity of the kinases in transcribed regions.
Once the DSB has been induced for several hours, the break will persist even when GAL::HO expression is repressed and the HO protein is degraded 19. This enabled us to ask what happens to γ-H2AX over the GAL genes when their transcription is repressed. We monitored γ-H2AX occupancy over GAL10 after cells were rapidly removed from YEP-GAL medium and transferred to YEPD (dextrose), which turns off transcription. Within 10 min after turning off GAL gene transcription, γ-H2AX occupancy rose significantly and by 30–60 min had almost reached the level of the non-transcribed MNN2 adjacent gene (Fig. 5d left and right panel respectively). Again, changes in nucleosome occupancy on GAL10 gene upon glucose treatment (about two fold, Suppl. Fig. 4c) cannot account for such an increase of γ-H2AX. Normalization of γ-H2AX against H2A enrichment showed a similar response of γ-H2AX at GAL10 (Fig. 5e). Thus, the checkpoint kinases are able to restore γ-H2AX to a region devoid of modification even several hours after the region has been initially modified.
By re-inducing transcription of the GAL genes we could also assess the dynamics of γ-H2AX loss. Indeed, within 10 min after transferring cells from dextrose to galactose-containing medium, γ-H2AX declined rapidly over GAL10 (Fig. 5d left panel). The rapid loss of the modification is interestingly different from the “transcriptional memory” exhibited by some other post-translational histone modifications of these genes, which persist for several hr20,21.
The rapid loss of γ-H2AX from transcribed genes is not caused by dephosphorylation of histones by any of the three phosphatases – Pph3, Ptc2 and Ptc3 - that have been implicated in turning off the DNA damage response and, in the case of Pph3, dephosphorylating γ-H2AX22–24 (Suppl. Fig. 4b top panel). It is likely that γ-H2AX is dephosphorylated after its displacement, as is the case in the removal of γ-H2AX after DSB repair 22.
There also has been the suggestion that γ-H2AX can be replaced by the minor histone H2A variant, H2A.Z, encoded by the HTZ1 gene, and that there are alterations of transcription regulation without H2A.Z 25–27. However, there was no change in removing γ-H2AX in a strain deleted for HTZ1 (Suppl. Fig. 4b bottom panel). These data support the idea that – although overall nucleosome occupancy does not dramatically decrease when transcription is re-initiated - the drastic loss of γ-H2AX upon transcription activation is likely due to a transcription-dependent increase in H2A-H2B dimer exchange.
We then asked whether both Mec1 and Tel1 can restore γ-H2AX to the nontranscribed GAL genes. Removing Tel1 had no discernable effect on the accumulation of γ-H2AX at GAL10 gene after transfer to glucose, but this restoration was severely affected in a strain lacking Mec1 (Fig. 5f). Interestingly, there was still partial restoration in mec1 mutant strain, even 7 h after induction of the DSB, suggesting that – even though Tel1 is no longer detected by live cell imaging or by ChIP 11,28 - it is still associated with the DSB sufficiently to add some γ-H2AX. To be certain that the reduced modification in mec1Δ was not a consequence of the failure of these cells to arrest prior to anaphase, in contrast to the extended arrest of wild type and tel1Δ strains suffering a DSB, we examined the accumulation of γ-H2AX in nocodazole arrested cells by inducing a DSB and then shifting cells to dextrose. The results (Suppl. Fig. 4d) are consistent with those shown in Figure 5f, but we made the unexpected discovery that nocodazole-arrested mec1Δ cells have a surprisingly low level of γ-H2AX modification. We do not know why Tel1 is unable to compensate at this stage in the cell cycle. Previous studies have ruled out the simplest explanation, that 5′ to 3′ resection of DSB ends is faster at this stage of the cell cycle, so that Tel1 could not associate with the ends29.
Altogether these data indicate that γ-H2AX phosphorylation is a highly dynamic process, involving rapid removal (by nucleosome exchange rather than by dephosphorylation) as well as rapid phosphorylation, primarily by Mec1 and to a lesser extent by Tel1. The process of transcription counteracts γ-H2AX accumulation probably by influencing this dynamic balance, leading to a lack of γ-H2AX on transcribed units.
Spreading of γ-H2AX and γ-H2B to all CEN regions
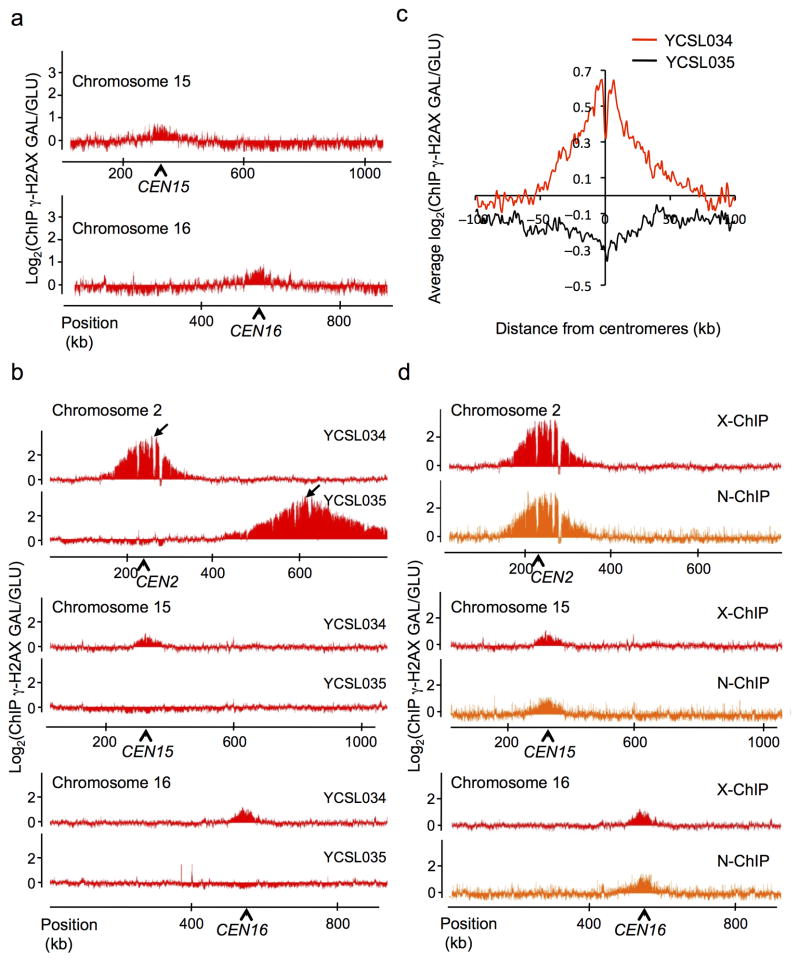
During the course of our investigation of γ-H2AX and γ-H2B profiles, we noticed that the regions surrounding each centromere were reproducibly enriched for γ-H2AX and γ-H2B, and that this occurred only after DSB induction (Fig. 6a, Suppl. Fig. 5). One of the HO cleavage sites is located 14 kb from CEN2. In view of previous reports that centromeres cluster together at the spindle pole body 30, the increase of γ-H2AX and γ-H2B around the centromeres of undamaged chromosomes could reflect the ability of the checkpoint kinases to phosphorylate targets on other chromosomes that are in close proximity but themselves have no damage. To explore this hypothesis, we created two new yeast strains that carry a single HO cleavage site on chromosome 2, one with the DSB site 14 kb from CEN2 (YCSL034) and one where the cut site is located 387 kb away (YCSL035). Induction of an unrepairable DSB in these two strains resulted in γ-H2AX spreading ~200 kb surrounding the DSB (Fig. 6b top panel). In the strain carrying the single HO site close to the centromere, there was a marked enrichment of γ-H2AX at pericentromeric regions of all others chromosomes; however this trans-spreading was not seen in the strain carrying the DSB far from the centromere (Fig. 6b middle and bottom panels, Fig. 6c). These results suggest that Mec1 and/or Tel1 checkpoint kinases can phosphorylate histone targets in trans, on undamaged chromosomes, if the targets are in close physical proximity. Similar results have recently been reported by Renkawitz et al. 31.
Figure 6. γ-H2AX can spread in trans on physically close loci.
a. Profile of γ-H2AX around centromeres of chromosomes 15 and 16 (indicated by arrows), after one-hour DSB induction in the 3-HO cuts strain, shown as the log2 ratio of the signal obtained after galactose versus glucose growth (GAL/GLU). b. γ-H2AX ChIP-chip was performed before and after galactose growth, either in strain YCSL034 carrying a single HO cut site at 13.5 kb from CEN2 or in strain YCSL035 carrying a single HO cut site 387 kb from CEN2. Profiles (ratio of GAL/GLU) obtained on chromosome 2 (carrying the DSB) and on chromosomes 15 and 16 are shown. c. Averaged γ-H2AX signal obtained after DSB induction, around centromere of undamaged chromosomes (i.e. chromosome 2 is excluded), using the data obtained with YCSL034 (red line) or YCSL035 (black line) strain. d. γ-H2AX ChIP-chip was performed before and after DSB induction, either using a protocol involving a crosslinking agent (X-ChIP in red) or in native conditions (N-ChIP in orange). Profiles obtained on chromosome 2 (carrying the DSB) and on chromosomes 15 and 16 are shown.
Since all yeast centromeres are clustered together by their association with the spindle pole body 30, it is possible that they might be crosslinked to each other during the process of ChIP, in which case the γ-H2AX signals on other centromeric regions we detected might simply be artifacts of crosslinking. To rule out the possibility, we developed a native ChIP assay (N-ChIP), in which no cross-linker is used, to demonstrate that γ-H2AX is being spread to other centromeric regions if a DSB is close to a centromere. As shown in Fig. 6d (top panel), γ-H2AX spreads around the HO-induced DSB, similar to the standard X-ChIP procedure (using a crosslinking agent). Importantly, we also observed a clear enrichment of γ-H2AX around the other centromeres in these native conditions (Fig. 6d middle and bottom panels), indicating that the observed γ-H2AX spreading in trans is not an artifact of crosslinking, but really reflects the ability of the kinases to phosphorylate nucleosomes located on physically close regions, such as clustered centromeres.
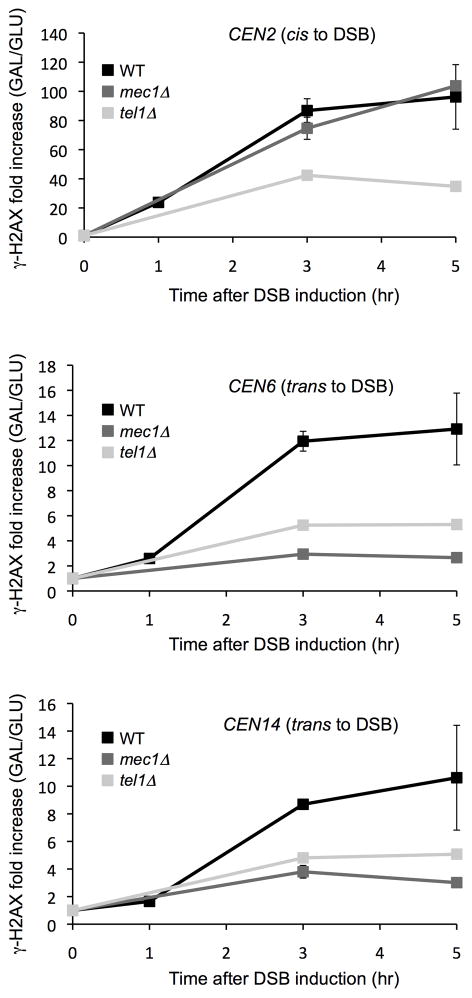
The spreading to different centromeres is catalyzed predominantly by Mec1. Accumulation of γ-H2AX at CEN6 and CEN14 was nearly absent in a mec1Δ strain, indicating that trans spreading is severely affected in this condition (Fig. 7 middle and bottom panels). In a tel1Δ strain, there is a general reduction of γ-H2AX levels, but the reduction is not greater in trans than in cis. The generally lower level of γ-H2AX modification in tel1Δ strains could reflect a role for the ATM homolog (Tel1) in fully activating the ATR homolog (Mec1). Mec1 has 9 SQ or TQ sites that might be modified by Tel1 or autophosphorylated by Mec1; alternatively Tel1 may facilitate Mec1 dimerization and interaction with broken DNA ends as has been documented in mammalian cells32. We note that we measured spreading in both cis and trans 2 hr after HO induction, before cells had fully activated the DNA damage checkpoint or arrested in the G2/M phase of the cell cycle 33,34. As noted above, when we repeated this experiment in nocodazole-arrested mec1Δ cells the level of γ-H2AX spreading is very low even in cis, but again there was little trans-spreading (unpublished).
Figure 7. Effect of Mec1 and Tel1 kinases on spreading in cis and trans from a DSB induced near CEN2.
An HO-induced DSB was created by galactose-mediated induction at a site 13.5 kb from CEN2. Increases in γ-H2AX were measured at the times indicated for a site 17.7 kb distal to the DSB on the same chromosome (a), 9.9 kb from CEN6 (b) and overlapping CEN14 (c). Fold increase is calculated by normalizing IP value to input and then to 0hr. Note that in tel1Δ strains there is a lower level of spreading in both cis and trans, whereas the absence of MEC1 specifically affects spreading in trans. Error bars represent the range from two independent experiments.
Discussion
By investigating the landscape and dynamics of γ-H2AX and γ-H2B phosphorylation in budding yeast we have discovered several important facets of the DNA damage response. First, the “holes” in the landscape of γ-H2AX, which correlate with transcribed regions, are rapidly reversible as transcription is shut off. The appearance of γ-H2AX modification is as fast as observed changes in chromatin structure 35,36. Depletion of γ-H2AX in transcribed regions is not primarily due to low histone occupancy but to a marked deficiency in modification. This may occur because the rate of histone turnover is faster than the rate of modification, but studies on the retention of “old” histones in transcribed regions argues against this point of view 37. Rather it seems that the checkpoint kinases appear to be unable to gain access to histones in strongly transcribed regions.
Second, the reestablishment of γ-H2AX in transcribed regions occurs very rapidly, within 5 min after shutting off transcription. Phosphorylation of these regions is done primarily by Mec1. However, Tel1 can partially compensate the loss of Mec1, even 7 h after the DSB was induced, despite the fact that it has generally been reported to disappear from DSB ends within a few hours11,28.
Third, γ-H2B behaves differently from γ-H2AX, despite being phosphorylated by the same two checkpoint kinases. The kinetics of γ-H2B modification are regulated by the earlier appearance of γ-H2AX, enforced by Rad9. This difference is reflected in the paucity of γ-H2B in subtelomeric regions and the notable increase in occupancy in these regions when checkpoint kinases are activated after a DSB.
Finally, we demonstrate that checkpoint kinases can modify sequences that are physically adjacent but not engaged in a direct interaction with a DSB. Previously we showed that a region around the Recombination Enhancer, which binds to the vicinity of a DSB, could be modified by either Mec1 or Tel138. Here, we show that a DSB near one centromere results in γ-H2AX and γ-H2B on all the other centromeres, which cluster around the spindle pole body 30, but is largely Mec1-dependent.
What the role of γ-H2B might be remains unclear. It clearly does not substitute for γ-H2AX in several critical functions. For example, recruitment of cohesins to regions around DSBs is lost in a H2A-S129A strain where γ-H2B still forms 39,40. Cells with H2B-T129A have very little sensitivity to several DNA damaging agents, except for a mild sensitivity to phleomycin (Suppl. Fig. 6b); but the h2b-T129A mutation appears to suppress several phenotypes displayed by h2a-S129A mutants, including increased telomere length (Suppl. Fig. 3a), faster 5′ to 3′ resection of DSB ends (Suppl. Fig. 6a) and sensitivity to MMS (Suppl. Fig. 6b).
It is of particular interest that only Mec1 is able to ensure the phosphorylation on other physically proximal chromosomes. Tel1 (ATM) may be more confined to act in cis because of its tethering to DSB ends via its association with Mre11-Rad50-Xrs2 (NBS1 in mammals), whereas Mec1 may diffuse in a “cloud” round the DSB, as proposed by Bakkenist and Katsten41.
Online Methods
Yeast Strains
All strains are isogenic and derived from JKM179 1 and have the genotype hoΔ hmlΔ::ADE1 MATα hmrΔ::ADE1 ade1-100 leu2-3,112 lys5 trp1::hisG ura3-52 ade3::GAL10::HO. A galactose-inducible HO endonuclease gene is integrated at the ADE3locus 3. Some derivatives of strain JKM179 carry additional HO endonuclease cleavage sites (HOcs), as previously described 2. All strains are described in Suppl. Table 1. Cells were pre-cultured in YP-lactate medium until cell density reaches 5–10×106 cells/ml and then HO expression is induced by adding 2% galactose (final concentration) to the medium.
Crosslinked Chromatin Immunoprecipitation (X-ChIP)
Procedures for X-ChIP analysis were described previously 4. Briefly, 45ml of cell culture was harvested and incubated with formaldehyde (final concentration of 1%) at RT for 10min. Crosslinking was stopped by adding 2.5M glycine to a final concentration of 125mM. Cells were suspended in lysis buffer (HEPES pH7.5, 1mM EDTA, 140mM NaCl, 1% TritonX-100, 0.1% NaDOC, 1mg/ml Bacitracin, 1mM Benzamidine, 1mM PMSF) and lyzed by bead beating at 4 °C for 1hr. Sonication was utilized to shear chromatin into fragments of the size of ~500bp. The chromatin fragments were incubated with 1 μl of anti-γ-H2AX antibody (ab15083) or 1 μl of anti-γ-H2B antibody (ab92973) and then pulled down with protein G-agarose (Roche). Validation of ab15083 is provided on the manufacturer’s website; validation for ab92973 is provided in this paper. The anti-γH2B antibody was created by Abcam in collaboration with the Haber lab to test the hypothesis that H2B-T129 might be phosphorylated after DNA damage, as hypothesized by Jessica Downs 5. After removing crosslinks and extracting DNA, quantitative PCR was carried out to examine the enrichment of the modified histones around DSBs with a various set of primers, listed in Suppl. Table 2.
Native Chromatin Immunoprecipitation (N-ChIP)
250 ml of cell culture was harvested without crosslinking at each time point of galactose induction. Cells were incubated in 1 ml of spheroplasting solution (1,000U/ml lyticase (Sigma), 1M sorbitol, 1mM PMSF, and 1mM fresh β-mercaptoethanol) at 37°C for 8min. Spheroplasts were spun down, resuspended in Micrococcal nuclease (MNase, Sigma) digestion buffer (1M sorbitol, 50mM NaCl, 10mM Tris pH7.4, 1mM CaCl2, 0.5mM spermidine, 0.075% Igepal CA-630, 1mM PMSF), and then aliquoted into two tubes for titration of MNase digestion. Chromatin was fragmented by MNase digestion at 37°C for 5min and stopped by adding EDTA. The degree of digestion was examined by running 1ul of chromatin on an agaose gel. The procedures for immunoprecipitation were described in45.
Western blots
10ml of cultures were grown in YPD medium were harvested at each time point and washed with 20% TCA. Cell pellets were resuspended in 250ml of 20% TCA and followed by 0.5ml glass bead lysis. The glass beads were removed, 0.3ml of 5% TCA was added, and precipitated proteins collected by centrifugation. The samples were transferred to 1.5ml tubes and 0.7ml of 5% TCA was added. Cells were centrifuged at 13,000 rpm at 4 °C for 10 min and supernatant was discarded. The cell pellet was washed with 100% of cold EtOH and resuspended in 100μl SDS loading buffer with 50μl of 1M Tris pH8.0. After 10 minutes at 95°C, insoluble material was removed by centrifugation and the supernatant were fractionated by SDS-PAGE and transferred to a protein membrane for overnight with 20V. The membranes were cropped around the histone sized region (15 KDa). After incubation with 5% nonfat milk in TBST (10 mM Tris, pH 8.0, 150 mM NaCl, 0.5% Tween 20) for 1 hr, the membrane was washed once with TBST and incubated with 1:3000 dilution of anti-γ-H2B antibody (ab92973) or a 1:3000 dilution of anti-γ-H2A antibody (ab15083) at 25 °C for 2 h. Membranes were then washed three times for 10 min and incubated with a 1:10000 dilution of anti-rabbit antibody for 1 h at 25 °C. Blots were washed with TBST three times and developed with the ECL reagent (GE Healthcare Life Sciences)
ChIP-chip
For ChIP-chip, DNA was amplified, labelled, and hybridized to high density oligonucleotide tiling arrays covering yeast chromosomes (Affymetrix), using the standard Affymetrix procedure, by the GeneCore facility at EMBL Heidelberg. All microarray data are available through Array Express (E-MTAB-1409).
Scanned array data (.CEL files) were normalized using TAS (quantile normalization, scale set to 500). The γ-H2AX or γ-H2B ChIPs upon Glucose growth, were used as “Control” samples, while the ChIP performed upon Galactose growth were used as “Treatment” samples. To analyse the distribution of both modifications without break induction (Fig. 3) input DNA samples were used as “Control” and ChIP samples as “treatment”. Enrichment values were calculated with a bandwidth of 0, and the signal scale set as Log2. For the Figures showing distribution on whole chromsomes, data were further smoothed using a 50 probes slidding window for easier vizualisation on the Integrated Genome Browser (IGB, http://bioviz.org/igb/). All calculations (Fig. 3c, 4b, 5a, 6c and Suppl. Fig. 2b, and 3b) were performed on unsmoothed, raw data. Boundaries of γ-H2AX domains were manually determined.
In order to plot data with respect to transcription start sites (TSS), telomeres, and centromeres, annotations were obtained from the SGD database http://www.yeastgenome.org/. All genomic coordinates were from the genome assembly of 2003 (SacSer1).
Supplementary Material
Acknowledgments
We thanks V. Benes and T. Ivacevic from the EMBL Genomic Core facility for hybridization with Affymetrix arrays. Funding in the Legube laboratory was provided by grants from the Association Contre le Cancer (ARC), Agence Nationale pour la Recherche (ANR-09-JCJC-0138), Canceropole GSO, and Research Innovation Therapeutic Cancerologie (RITC). Research in the Haber lab was supported by National Institutes of Health grants GM61766, GM20056 and GM76020.
Footnotes
Accession Codes
All microarray data are available through Array Express (E-MTAB-1409).
Author contributions
C-S Lee designed, executed and analyzed experiments shown in Figures 5, 6 and 7 and related Supplemental Data and prepared ChIP samples for experiments shown in Figures 2–6. K Lee designed, executed and analyzed experiments shown in Figure 1 and related Supplemental Data and prepared ChIP samples for experiments shown in Figures 2–6. G Legube performed ChIP-chip analysis and analyzed the data shown in Figures 2–6 and related Supplemental Data and analyzed the data. JE Haber designed experiments and analyzed data and was the principal author of the manuscript, with contributions from all three co-authors.
References
- 1.Shroff R, et al. Distribution and dynamics of chromatin modification induced by a defined DNA double-strand break. Curr Biol. 2004;14:1703–1711. doi: 10.1016/j.cub.2004.09.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rogakou EP, Boon C, Redon C, Bonner WM. Megabase chromatin domains involved in DNA double-strand breaks in vivo. J Cell Biol. 1999;146:905–916. doi: 10.1083/jcb.146.5.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Harrison JC, Haber JE. Surviving the breakup: the DNA damage checkpoint. Annu Rev Genet. 2006;40:209–235. doi: 10.1146/annurev.genet.40.051206.105231. [DOI] [PubMed] [Google Scholar]
- 4.Kim JA, Kruhlak M, Dotiwala F, Nussenzweig A, Haber JE. Heterochromatin is refractory to gamma-H2AX modification in yeast and mammals. J Cell Biol. 2007;178:209–218. doi: 10.1083/jcb.200612031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Szilard RK, et al. Systematic identification of fragile sites via genome-wide location analysis of gamma-H2AX. Nat Struct Mol Biol. 2010;17:299–305. doi: 10.1038/nsmb.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kitada T, et al. gammaH2A is a component of yeast heterochromatin required for telomere elongation. Cell Cycle. 2011;10:293–300. doi: 10.4161/cc.10.2.14536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Iacovoni JS, et al. High-resolution profiling of gammaH2AX around DNA double strand breaks in the mammalian genome. EMBO J. 2010;29:1446–1457. doi: 10.1038/emboj.2010.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Caron P, et al. Cohesin protects genes against gammaH2AX Induced by DNA double-strand breaks. PLoS Genet. 2012;8:e1002460. doi: 10.1371/journal.pgen.1002460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Melo JA, Cohen J, Toczyski DP. Two checkpoint complexes are independently recruited to sites of DNA damage in vivo. Genes Dev. 2001;15:2809–2821. doi: 10.1101/gad.903501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zou L, Elledge SJ. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science. 2003;300:1542–1548. doi: 10.1126/science.1083430. [DOI] [PubMed] [Google Scholar]
- 11.Nakada D, Matsumoto K, Sugimoto K. ATM-related Tel1 associates with double-strand breaks through an Xrs2-dependent mechanism. Genes Dev. 2003;17:1957–1962. doi: 10.1101/gad.1099003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hammet A, Magill C, Heierhorst J, Jackson SP. Rad9 BRCT domain interaction with phosphorylated H2AX regulates the G1 checkpoint in budding yeast. EMBO Rep. 2007;8:851–857. doi: 10.1038/sj.embor.7401036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen X, et al. The Fun30 nucleosome remodeler promotes resection of DNA double-strand break ends. Nature. 2012;489:576–580. doi: 10.1038/nature11355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eapen VV, Sugawara N, Tsabar M, Wu WH, Haber JE. The Saccharomyces cerevisiae chromatin remodeler Fun30 regulates DNA end resection and checkpoint deactivation. Mol Cell Biol. 2012;32:4727–4740. doi: 10.1128/MCB.00566-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martin S, Laroche T, Suka N, Grunstein M, Gasser SM. Relocalization of telomeric Ku and SIR proteins in response to DNA strand breaks in yeast. Cell. 1999;97:621–633. doi: 10.1016/s0092-8674(00)80773-4. [DOI] [PubMed] [Google Scholar]
- 16.Mills K, Sinclair D, Guarente L. MEC1-dependent redistribution of the Sir3 silencing protein from telomeres to DNA double-strand breaks. Cell. 1999;97:609–620. doi: 10.1016/s0092-8674(00)80772-2. [DOI] [PubMed] [Google Scholar]
- 17.David L, et al. A high-resolution map of transcription in the yeast genome. Proc Natl Acad Sci U S A. 2006;103:5320–5325. doi: 10.1073/pnas.0601091103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jamai A, Imoberdorf RM, Strubin M. Continuous histone H2B and transcription-dependent histone H3 exchange in yeast cells outside of replication. Mol Cell. 2007;25:345–355. doi: 10.1016/j.molcel.2007.01.019. [DOI] [PubMed] [Google Scholar]
- 19.Moore JK, Haber JE. Cell cycle and genetic requirements of two pathways of nonhomologous end-joining repair of double-strand breaks in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:2164–2173. doi: 10.1128/mcb.16.5.2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ng HH, Robert F, Young RA, Struhl K. Targeted recruitment of Set1 histone methylase by elongating Pol II provides a localized mark and memory of recent transcriptional activity. Mol Cell. 2003;11:709–719. doi: 10.1016/s1097-2765(03)00092-3. [DOI] [PubMed] [Google Scholar]
- 21.Zhou BO, Zhou JQ. Recent transcription-induced histone H3 lysine 4 (H3K4) methylation inhibits gene reactivation. J Biol Chem. 2011;286:34770–34776. doi: 10.1074/jbc.M111.273128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Keogh MC, et al. A phosphatase complex that dephosphorylates gammaH2AX regulates DNA damage checkpoint recovery. Nature. 2006;439:497–501. doi: 10.1038/nature04384. [DOI] [PubMed] [Google Scholar]
- 23.Kim JA, Hicks WM, Li J, Tay SY, Haber JE. Protein phosphatases pph3, ptc2, and ptc3 play redundant roles in DNA double-strand break repair by homologous recombination. Mol Cell Biol. 2011;31:507–516. doi: 10.1128/MCB.01168-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leroy C, et al. PP2C phosphatases Ptc2 and Ptc3 are required for DNA ceckpoint inactivation after a double-strand break. Mol Cell. 2003;11:827–835. doi: 10.1016/s1097-2765(03)00058-3. [DOI] [PubMed] [Google Scholar]
- 25.Morillo-Huesca M, Clemente-Ruiz M, Andujar E, Prado F. The SWR1 histone replacement complex causes genetic instability and genome-wide transcription misregulation in the absence of H2A.Z. PLoS One. 2010;5:e12143. doi: 10.1371/journal.pone.0012143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Papamichos-Chronakis M, Krebs JE, Peterson CL. Interplay between Ino80 and Swr1 chromatin remodeling enzymes regulates cell cycle checkpoint adaptation in response to DNA damage. Genes Dev. 2006;20:2437–2449. doi: 10.1101/gad.1440206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Halley JE, Kaplan T, Wang AY, Kobor MS, Rine J. Roles for H2A.Z and its acetylation in GAL1 transcription and gene induction, but not GAL1-transcriptional memory. PLoS Biol. 2010;8:e1000401. doi: 10.1371/journal.pbio.1000401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lisby M, Barlow JH, Burgess RC, Rothstein R. Choreography of the DNA damage response: spatiotemporal relationships among checkpoint and repair proteins. Cell. 2004;118:699–713. doi: 10.1016/j.cell.2004.08.015. [DOI] [PubMed] [Google Scholar]
- 29.Ira G, et al. DNA end resection, homologous recombination and DNA damage checkpoint activation require CDK1. Nature. 2004;431:1011–1017. doi: 10.1038/nature02964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Duan Z, et al. A three-dimensional model of the yeast genome. Nature. 2010;465:363–367. doi: 10.1038/nature08973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Renkawitz J, Lademann CA, Kalocsay M, Jentsch S. Monitoring homology search during DNA double-strand break repair in vivo. Mol Cell. 2013;50:261–272. doi: 10.1016/j.molcel.2013.02.020. [DOI] [PubMed] [Google Scholar]
- 32.Myers JS, Cortez D. Rapid activation of ATR by ionizing radiation requires ATM and Mre11. J Biol Chem. 2006;281:9346–9350. doi: 10.1074/jbc.M513265200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pellicioli A, Lee SE, Lucca C, Foiani M, Haber JE. Regulation of Saccharomyces Rad53 checkpoint kinase during adaptation from DNA damage-induced G2/M arrest. Mol Cell. 2001;7:293–300. doi: 10.1016/s1097-2765(01)00177-0. [DOI] [PubMed] [Google Scholar]
- 34.Lee SE, et al. Saccharomyces Ku70, mre11/rad50 and RPA proteins regulate adaptation to G2/M arrest after DNA damage. Cell. 1998;94:399–409. doi: 10.1016/s0092-8674(00)81482-8. [DOI] [PubMed] [Google Scholar]
- 35.Bash R, Lohr D. Yeast chromatin structure and regulation of GAL gene expression. Prog Nucleic Acid Res Mol Biol. 2001;65:197–259. doi: 10.1016/s0079-6603(00)65006-7. [DOI] [PubMed] [Google Scholar]
- 36.Cavalli G, Thoma F. Chromatin transitions during activation and repression of galactose-regulated genes in yeast. EMBO J. 1993;12:4603–4613. doi: 10.1002/j.1460-2075.1993.tb06149.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Radman-Livaja M, et al. Patterns and mechanisms of ancestral histone protein inheritance in budding yeast. PLoS Biol. 2011;9:e1001075. doi: 10.1371/journal.pbio.1001075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li J, et al. Regulation of budding yeast mating-type switching donor preference by the FHA domain of Fkh1. PLoS Genet. 2012;8:e1002630. doi: 10.1371/journal.pgen.1002630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Unal E, et al. DNA damage response pathway uses histone modification to assemble a double-strand break-specific cohesin domain. Mol Cell. 2004;16:991–1002. doi: 10.1016/j.molcel.2004.11.027. [DOI] [PubMed] [Google Scholar]
- 40.Strom L, Lindroos HB, Shirahige K, Sjogren C. Postreplicative recruitment of cohesin to double-strand breaks is required for DNA repair. Mol Cell. 2004;16:1003–1015. doi: 10.1016/j.molcel.2004.11.026. [DOI] [PubMed] [Google Scholar]
- 41.Bakkenist CJ, Kastan MB. DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature. 2003;421:499–506. doi: 10.1038/nature01368. [DOI] [PubMed] [Google Scholar]
References for Online Methods
- 1.Lee SE, et al. Saccharomyces Ku70, mre11/rad50 and RPA proteins regulate adaptation to G2/M arrest after DNA damage. Cell. 1998;94:399–409. doi: 10.1016/s0092-8674(00)81482-8. [DOI] [PubMed] [Google Scholar]
- 2.Kim JA, Kruhlak M, Dotiwala F, Nussenzweig A, Haber JE. Heterochromatin is refractory to gamma-H2AX modification in yeast and mammals. J Cell Biol. 2007;178:209–218. doi: 10.1083/jcb.200612031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sandell LL, Zakian VA. Loss of a yeast telomere: arrest, recovery, and chromosome loss. Cell. 1993;75:729–739. doi: 10.1016/0092-8674(93)90493-a. [DOI] [PubMed] [Google Scholar]
- 4.Sun K, Coic E, Zhou Z, Durrens P, Haber JE. Saccharomyces forkhead protein Fkh1 regulates donor preference during mating-type switching through the recombination enhancer. Genes Dev. 2002;16:2085–2096. doi: 10.1101/gad.994902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Foster ER, Downs JA. Methylation of H3 K4 and K79 is not strictly dependent on H2B K123 ubiquitylation. J Cell Biol. 2009;184:631–638. doi: 10.1083/jcb.200812088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wagschal A, Delaval K, Pannetier M, Arnaud P, Feil R. Chromatin Immunoprecipitation (ChIP) on Unfixed Chromatin from Cells and Tissues to Analyze Histone Modifications. CSH Protoc 2007. 2007 doi: 10.1101/pdb.prot4767. pdb prot4767. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.