Abstract
Biodegradable poly (lactic-co-glycolic acid) (PLGA) nanoparticles (NPs) encapsulating triplex-forming peptide nucleic acids (PNAs) and donor DNAs for recombination-mediated editing of the CCR5 gene were synthesized for delivery into human peripheral blood mononuclear cells (PBMCs). NPs containing the CCR5-targeting molecules efficiently entered PBMCs with low cytotoxicity. Deep sequencing revealed that a single treatment with the formulation resulted in a targeting frequency of 0.97% in the CCR5 gene and a low off-target frequency of 0.004% in the CCR2 gene, a 216-fold difference. NP-treated PBMCs efficiently engrafted immunodeficient NOD-scid IL-2rγ-/- mice, and the targeted CCR5 modification was detected in splenic lymphocytes 4 weeks posttransplantation. After infection with an R5-tropic strain of HIV-1, humanized mice with CCR5-NP–treated PBMCs displayed significantly higher levels of CD4+ T cells and significantly reduced plasma viral RNA loads compared with control mice engrafted with mock-treated PBMCs. This work demonstrates the feasibility of PLGA-NP–encapsulated PNA-based gene-editing molecules for the targeted modification of CCR5 in human PBMCs as a platform for conferring HIV-1 resistance.
Introduction
Individuals homozygous for a 32-bp deletion (CCR5-Δ32) in the CCR5 gene are almost completely resistant to HIV-1 infection, with no significant effects on health.1,2 In a ground-breaking report, an HIV-1–positive individual with acute myeloid leukemia was treated by transplant of hematopoietic stem and progenitor cells from a CCR5-Δ32 homozygous donor and was cured of HIV-AIDS, with no detectable HIV-1 despite discontinuation of antiretroviral therapy for more than 5 years.3,4 Notably, individuals heterozygous for this mutation also have a substantially reduced disease progression rate: hence ablating even a single allele of CCR5 can have a significant impact on disease susceptibility, making CCR5 an attractive target for gene therapy.5,6
We have developed triplex-forming peptide nucleic acids (PNAs) that specifically target the CCR5 gene by binding to the DNA and forming a PNA/DNA/PNA triple helix through a combination of Watson–Crick strand invasion and Hoogsteen bonding. This altered helical structure triggers recombination of short donor DNA fragments into the target gene in the vicinity of the triple helix to introduce an inactivating mutation.7 We hypothesize that the use of this technology to mimic the effect of the naturally occurring Δ32 mutation in primary human lymphocytes should make it possible to generate immune cells resistant to HIV-1 infection. In prior work, using electroporation to introduce the PNAs and donor DNAs into THP-1 cells (a human monocytic leukemia cell line), we showed that triplex-forming PNAs were able to bind in a sequence-specific manner to the CCR5 gene and induce recombination in the vicinity of the Δ32 mutation, resulting in reduced susceptibility to HIV-1 in culture.7
However, in view of the toxicity of electroporation on primary hematopoietic cells (the clinically relevant target), we tested the ability of biodegradable nanoparticles (NPs) to achieve delivery of encapsulated PNAs and donor DNAs into peripheral blood mononuclear cells (PBMCs), a modality that is also capable of increasing the bioavailability of the encapsulated mediators for in vivo applications.8,9 NPs composed of poly (lactic-co-glycolic acid) (PLGA) were used, as this polymer has been established to be safe in patients for over 30 years.10
We report here the characterization of these PLGA-NPs and their use in targeting the CCR5 gene in human PBMCs. We started with PBMCs heterozygous for the naturally occurring CCR5-Δ32 mutation, representing the genotypes of approximately 10% of the European-derived populations.11 Using PLGA-NPs, PNAs and donor DNAs were successfully delivered into the PBMCs, producing targeted modification of the CCR5 gene at a frequency in the range of 1% with minimal toxicity. Importantly, off-target effects in the highly homologous CCR2 gene were more than 200-fold lower. Engraftment of treated PMBCs was uncompromised in NOD-scid IL2rγ-/- mice, with the introduced CCR5 modification detected in splenic human leukocytes 28 days posttransplantation. Moreover, mice transplanted with the CCR5-modified PBMCs were resistant to HIV-1 infection, displaying preservation of CD4+ T-cell levels that was accompanied with reduced levels of plasma viral RNA at 10 days postchallenge with HIV-1. By contrast, mice transplanted with PBMCs treated with empty, blank NPs, showed a drastic depletion of CD4+ T cells and high levels of viremia, consistent with viral replication. This work demonstrates the utility of PLGA-NP–delivered PNAs and donor DNAs for the gene editing of CCR5 with a high specificity, providing the basis for a possible new therapeutic approach for HIV-1 infections.
Results
Formulation of NPs containing oligonucleotides targeting CCR5
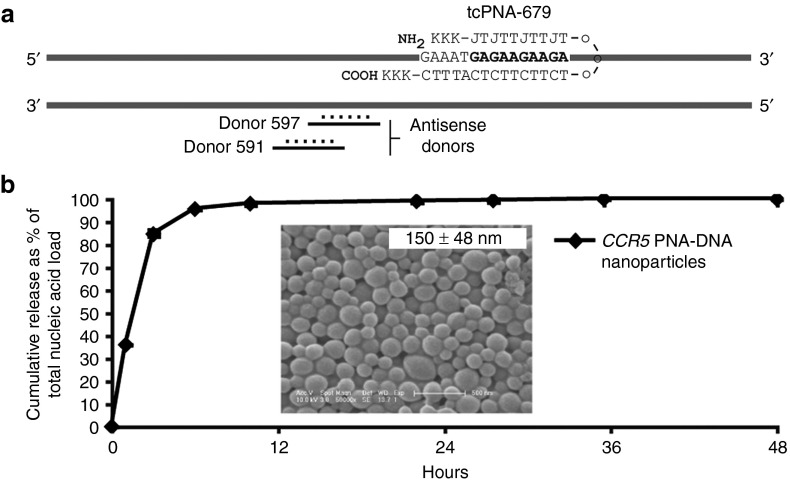
The sequences and characterization of the triplex-forming PNAs and donor DNAs used in this study were previously described in Schleifman et al. and are summarized here in Figure 1a.7 We previously reported an improved design of the triplex-forming PNA which resulted in a higher binding affinity in vitro and a 4.5-fold increase in targeted modification of the CCR5 gene in human cells. This improved PNA design, called a tail-clamp PNA (tcPNA), consists of two single strands of PNA connected by a flexible linker. As with triplex formation in general, it still requires a homopurine target site for the formation of a PNA/DNA/PNA triplex. The tcPNAs, however, also include additional bases (forming a “tail”) on the Watson–Crick-binding domain of the PNA, which not only serve to increase the targeting specificity by binding to a longer target site but also allow for binding to mixed sequences beyond the homopurine stretch (Figure 1a). We encapsulated this tcPNA (tcPNA-679) along with donor DNAs in PLGA-NPs for targeted modification and inactivation of the CCR5 gene in human PBMCs.
Figure 1.
Nucleic acid release from CCR5 nanoparticles. (a) Schematic of the CCR5 gene with the triplex-forming peptide nucleic acid, tcPNA-679, binding to the genomic DNA downstream of the two donor DNA oligonucleotides. K, lysine residue, J, pseudoisocytocine. (b) To calculate the kinetics of release of encapsulated nucleic acid, nanoparticles (NPs) were incubated in PBS at 37 °C and NP-free supernatants were collected for the analysis of total nucleic acid content by the absorbance at 260 nm at the indicated time points. At 48 hours, the residual nucleic acid in the NP pellet was extracted and the total nucleic acid load was calculated as a sum of absorbance obtained from the pellet and supernatant. Inset: SEM image of NPs. The average size of the NPs, calculated using the ImageJ software is depicted as mean ± SD. Scale bar: 500 nm.
PLGA-NPs containing PNAs and donor DNAs targeting the human CCR5 gene (CCR5-NPs) were formulated by a double-emulsion solvent evaporation technique, with a total of 1 nmol of nucleic acid per milligram of PLGA. Particles were generated with 0.25 nmol of each donor DNA per milligram of PLGA plus 0.5 nmol of the triplex-forming PNA per milligram of PLGA. NPs exhibited spherical morphology and size distributions in the 150-nm range as determined by scanning electron microscopy (Figure 1b, inset). Release of PNAs and donor DNAs from the NPs was quantified by measuring the absorbance of aliquots at 260 nm taken over time from particles incubated in PBS. The CCR5-NPs released greater than 90% of their contents within the first 12 hours, with almost complete release by 24 hours (Figure 1b).
Uptake and toxicity of NPs in PBMCs
Using the technique of triplex-induced homologous recombination, we sought to target and knockout CCR5 in PBMCs because this cell population contains the CD4+ lymphocytes that otherwise become depleted during progressive HIV-1 infection. This primary cell population, however, is very hard to transfect. We obtained single-donor human PBMCs that were either wild type at the CCR5 locus or heterozygous for the CCR5-Δ32 mutation. Heterozygous PBMCs were used to allow accurate quantification of the editing frequency at a single locus. Furthermore, ~10% of all northern Europeans carry one copy of the Δ32 allele and therefore represent a potential genotype in many HIV-1–affected individuals.11
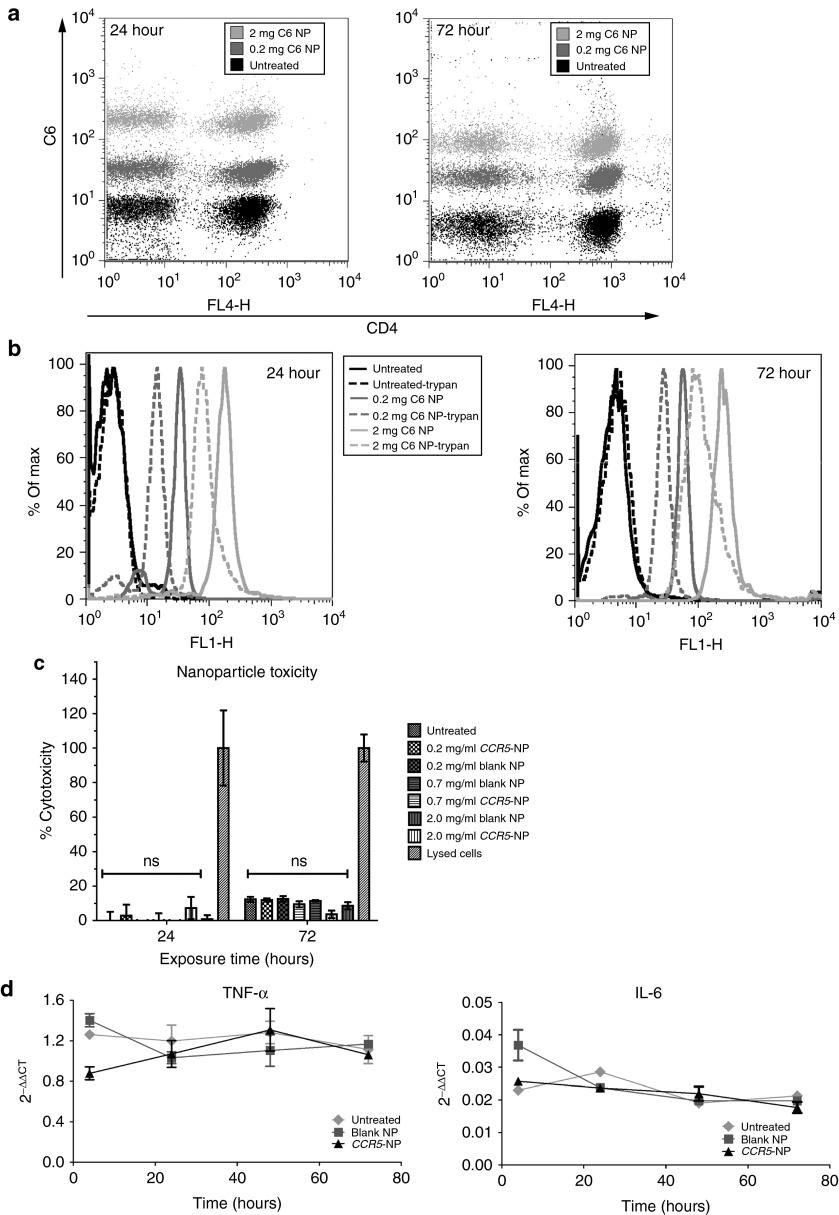
NPs were formulated to contain the fluorescent dye coumarin-6 (C6) to quantify NP uptake into human PBMCs, as C6 is not released substantially from the particles during the period of these experiments. C6-containing NPs were added to PBMCs at 0.2 or 2 mg/ml and 24 or 72 hours later; the samples were analyzed by flow cytometry. Almost 100% of PBMCs, with nearly all CD4+ T cells, showed C6 fluorescence, demonstrating association of the C6-NPs with the cells (Figure 2a). To distinguish adhesion from uptake and thus extracellular from intracellularly localized NPs, trypan blue was used before flow cytometry to quench the fluorescence in the externally accessible NPs. Treatment with trypan blue only marginally decreased the overall fluorescence, suggesting that most particles were internalized into the cells (Figure 2b).
Figure 2.
Characterization of CCR5 nanoparticles (NPs). (a) NPs containing the dye, coumarin 6 (C6) were added to wild-type peripheral blood mononuclear cells (PBMCs) (0.2 or 2 mg/ml), and fluorescence was measured by flow cytometric analysis 24 or 72 hours posttreatment. Cells were costained with anti-CD4-APC. (b) PBMCs treated as described above were quenched with trypan blue to assess internalized fluorescence versus external cell-associated fluorescence (uptake versus external association of NPs). Histograms of C6 fluorescence are shown. (c) Polyhydroxyalkanoate-activated PBMCs were treated with blank or CCR5-NPs at 0.2, 0.7, or 2.0 mg/ml, and culture supernatants were assayed for lactate dehydrogenase activity at 24 and 72 hours of culture. The positive control (lysed cells) for total lactate dehydrogenase release represents cells completely lysed with detergent. Repeated-measures one-way analysis of variance testing followed by a Dunnett's multiple comparisons test found no significant differences between the three groups treated with NPs and the untreated control cells (P > 0.05). ns, not significant. (d) Wild-type PBMCs were either untreated or treated with the indicated NPs and RNA was isolated at various time points. Quantitative reverse transcriptase polymerase chain reaction was performed to determine the mRNA levels of tumor necrosis factor-α or interleukin-6, and glyceraldehyde-3-phosphate dehydrogenase was used for normalization.
To evaluate the toxicity of the NP treatment, freshly isolated PBMCs were treated with C6-NPs at 0.2, 0.7, and 2 mg/ml and at 24 and 72 hours posttreatment; cell death was measured by assaying for lactate dehydrogenase release in culture supernatants. The C6-NPs did not significantly affect cell viability at any of the doses tested in comparison with untreated PBMCs (Figure 2c); the basal level of cytotoxicity observed is due to the culture of PBMCs in the absence of stimulatory cytokines. We also tested for NP-mediated induction of inflammatory responses. Quantitative reverse transcriptase polymerase chain reaction (PCR) was used to measure both TNF-α and IL-6 mRNA expression in PBMCs. Figure 2d shows that over the 3-day time course, no significant increases in either TNF-α or IL-6 mRNA levels were evident in PBMCs treated with either the blank NPs or CCR5-NPs compared with untreated cells, confirming that the NP preparations did not activate inflammatory pathways in primary human immune cells.
Targeted modification of CCR5 in human PBMCs
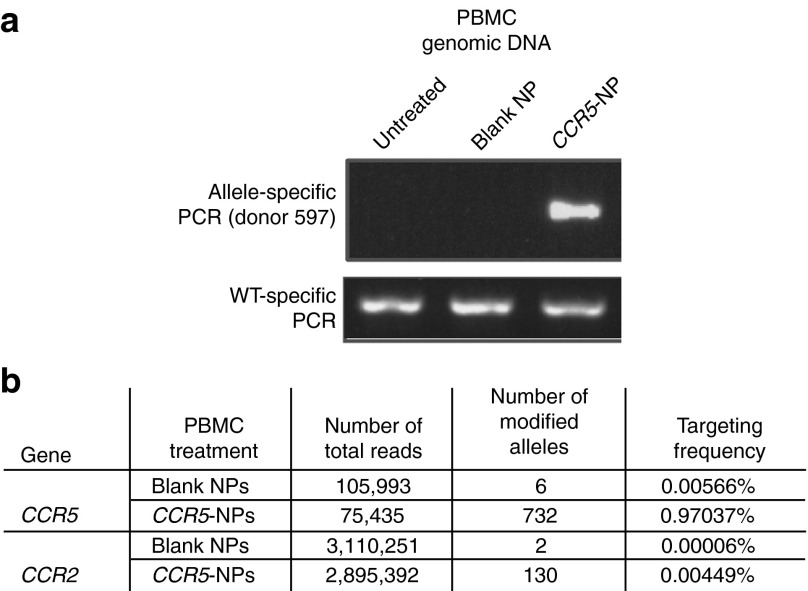
We assessed the ability of the CCR5-NPs to specifically modify the endogenous CCR5 gene in healthy human PBMCs. PBMCs, in the absence of treatment with stimulatory agents, were treated with blank particles or NPs containing the triplex-forming PNA and donor DNAs (donors 591 and 597), both designed to introduce an in-frame stop codon into the CCR5 gene leading to receptor knockout. Twenty-four hours posttreatment, genomic DNA was isolated from aliquots of the treated cell populations and analyzed by allele-specific PCR (AS-PCR).7 Targeted modifications of the CCR5 gene were detected only in the PBMCs treated with the PNA and donor DNA-containing NPs, indicating that efficient nuclear delivery of the effector nucleic acids was achieved producing site-specific modification at the endogenous CCR5 locus (Figure 3a).
Figure 3.
Triplex-mediated genomic modification in peripheral blood mononuclear cells (PBMCs) shows low off-target effects. (a) Blank nanopartcles (NPs) containing phosphate-buffered saline or CCR5-NPs containing donors and peptide nucleic acids were added to wild-type human PBMCs at a final concentration of 0.5 mg/ml. Twenty-four hours later, genomic DNA was isolated from the treated samples as well as untreated PBMCs, and targeted modification of the CCR5 gene was detected by AS-PCR. (b) Table depicting gene-targeted and off-targeted modification frequencies as determined by Illumina deep sequencing of the CCR5 and CCR2 gene in blank and CCR5-NP–treated PBMCS. The ratio of CCR5 to CCR2 targeted was determined by dividing the CCR5 modification frequency by the CCR2 modification frequency indicated in the table.
We next sought to determine the gene-targeting frequency and to evaluate for possible off-target effects in the genome after NP treatment. After confirming the presence of the targeted CCR5 modification in CCR5-NP–treated PBMCs by AS-PCR 48 hours posttreatment (data not shown), genomic DNA from these cell populations was subjected to deep-sequencing analysis to survey the CCR5, CCR2, CCR4, and CD4 alleles in the cell population by the Illumina pair-end deep-sequencing technique.12 CCR2 was chosen as an off-target control because it contains 86% sequence homology to CCR5 in the target region (donor and PNA-binding region) and therefore offers a stringent test for off-target effects.13 CCR4 was sequenced because it has up to 67% homology to CCR5 in various genomic regions and CD4 was chosen because although it has no homology to our target site, knockout of this receptor would also lead to resistance to HIV-1 infection. The raw sequence data were subjected to alignment and analysis, and the results revealed a CCR5 gene-targeting frequency of 0.97% (732 modified alleles of 75,435 sequenced) (Figure 3b) versus an off-target frequency in CCR2 of 0.004% (130 modified alleles of 2,895,392 sequenced), 0% in CCR4 (0 modified alleles of 5,035,475 sequenced), and 0% in CD4 (0 modified alleles of 4,353,167 sequenced). These quantitative results indicate that triplex-induced gene targeting is highly specific, with an on-target frequency that is 216-fold higher than the off-targeting frequency in a highly homologous target site, the CCR2 gene. In comparison, in a similar deep-sequencing analysis, zinc-finger nucleases (ZFNs) targeted to CCR5 produced off-target effects in the CCR2 gene in human cells at a frequency of 5.4%, more than 1,000-fold higher than what we have found for triplex-forming PNAs.13
CCR5-modified PBMCs resist HIV-1 challenge after engraftment in NOD-scid IL2rγ-/- mice
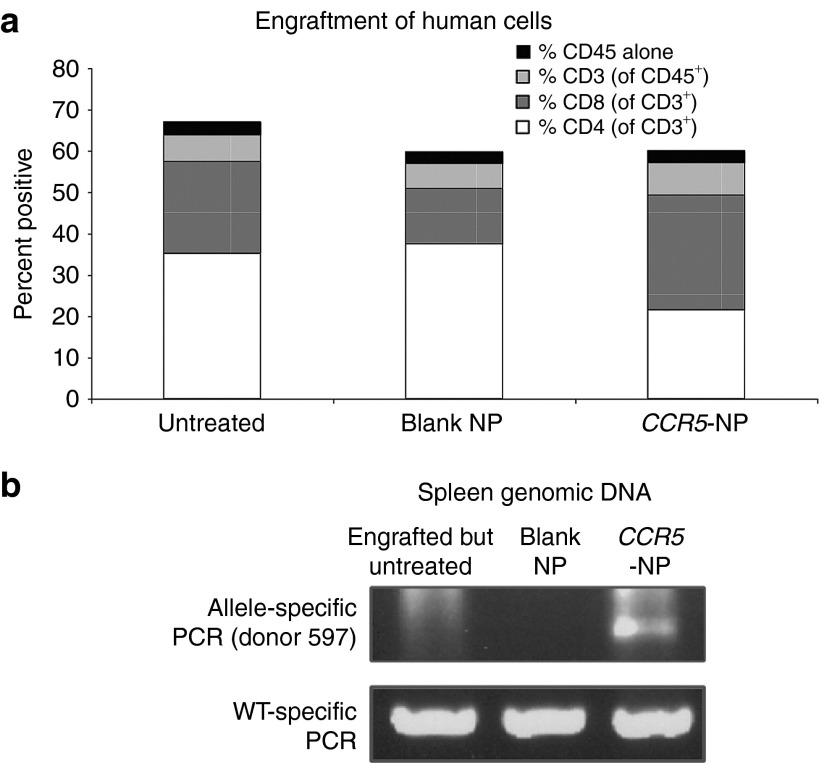
Human PBMCs are capable of engrafting and proliferating as T cells in NOD-scid IL2rγ-/- adult mice, and these engrafted mice can be challenged with live R5-tropic HIV-1.14 Engraftment and expansion of PBMCs treated ex vivo with NPs therefore allows for the in vivo functional evaluation of HIV-1 resistance conferred by triplex-mediated gene modification. To assess this, PBMC populations were treated with NPs and injected into NOD-scid IL2rγ-/- adult mice to evaluate their ability to engraft and expand in vivo. As shown in Figure 4a, engraftment of NP-treated PBMCs occurred at levels equal to those of untreated PBMCs with similar percentages of human leukocytes (CD45+) and human T-cell subsets detected in the mouse spleens 4 weeks posttransplant in all the treatment groups (as determined by flow cytometric staining with specific antibodies). Importantly, at 4 weeks posttransplantation, the targeted CCR5 modification was detected in splenic lymphocytes only from the mouse transplanted with PBMCs treated with CCR5-NPs but not in the cells from the engrafted mice in the control groups (Figure 4b).
Figure 4.
CCR5-nanoparticle (NP)–treated peripheral blood mononuclear cells (PBMCs) efficiently engraft NOD-scid IL2rγ-/- mice. (a) Bar graph depicting the percentages of individual human lymphocytic populations in spleens of adult NOD-scid IL2rγ-/- mice reconstituted with PBMCs that were untreated, treated with blank, or CCR5-targeted nanoparticles. %CD45 alone refers to the remainder of the CD45-positive cells that were not CD3+. A two-way analysis of variance with Tukey's multiple comparisons revealed no significant differences among the different groups. (b) Identification of targeted modification of the CCR5 gene in splenocytes of humanized mice reconstituted with human PBMCs (either untreated, treated with blank NPs, or with CCR5-NPs) at 4 weeks posttransplant. Allele-specific polymerase chain reaction was performed on the genomic DNA with the donor 1 primers.
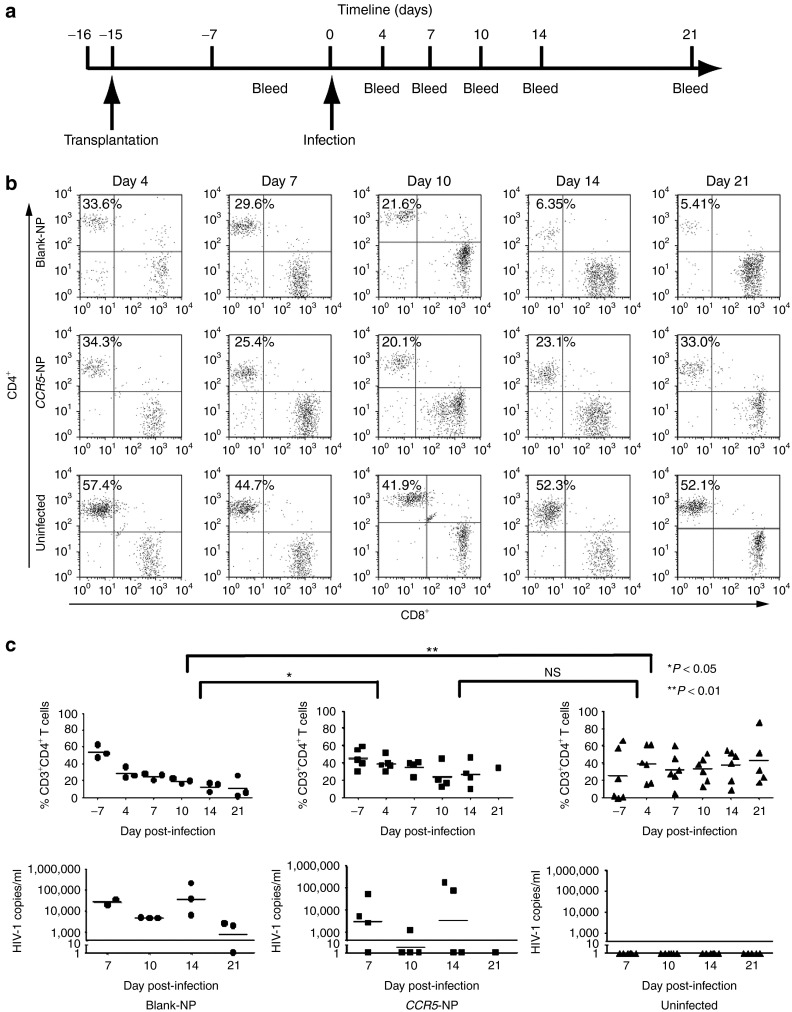
To ask whether targeted CCR5 disruption through PNA/DNA-containing NPs confers resistance of the modified PBMCs to HIV-1, PBMCs heterozygous for the CCR5-Δ32 mutation were treated with either blank or CCR5-NPs. Twenty-four hours posttreatment, an aliquot of both cell populations was harvested, and analysis of the genomic DNA by AS-PCR confirmed the presence of the CCR5-targeted modification in only the CCR5-NP–treated cells (consistent with the results above; data not shown). The experimental and control cell populations were then transplanted into adult NOD-scid IL2rγ-/- mice and allowed to engraft. Fourteen days posttransplantation, engraftment was confirmed by flow cytometric staining of mouse peripheral blood with antibodies specific for human T cells, and then the mice were infected with the CCR5-tropic HIV-1BaL by intraperitoneal injection (Figure 5a). Peripheral blood was analyzed periodically over the course of 3 weeks to assess the dynamics of human CD4+ T-cell levels in response to the viral infection by flow cytometry. HIV-1 RNA levels in the plasma were assessed by the Amplicor (Roche Molecular Diagnostics, Indianapolis, IN) assay to provide a quantitative measure of viral infection.15
Figure 5.
Mice reconstituted with CCR5-targeted nanoparticle (NP)-treated peripheral blood mononuclear cells (PBMCs) resist HIV-1 challenge. (a) Experimental timeline: PBMCs from a donor heterozygous for the Δ32 mutation were treated with either blank or CCR5-NPs and transplanted into NOD-scid IL2rγ-/- mice that were infected 2 weeks later with HIV-1BaL. (b) Representative fluorescence-activated cell sorting plots depicting the CD4+ and CD8+ T cells from one mouse of each treatment group over time. (c) Flow cytometric evaluation of peripheral T cells (upper panel) and plasma viral copy numbers measured by the Amplicor test (lower panel). CD4+ T-cell ratios were calculated as a ratio of the entire CD3 population (CD3+CD4+:CD3+). The solid black line in the lower panel represents the limit of detection of the Amplicor test. Statistical significance was analyzed by repeated-measures one-way Anova followed by Tukey's multiple comparisons test. NS, not significant.
Mice transplanted with the CCR5-NP–treated PBMCs maintained higher levels of human CD4+ T cells compared with the mice transplanted with PBMCs treated with blank NPs, at day 10 and day 14 postinfection (Figure 5b). Furthermore, the proportion of CD4+ T cells in the CCR5-NP-PBMC–engrafted mice continued to increase and reached levels similar to those seen in the uninfected mice by day 21 postinfection, in contrast to the blank NP-treated PBMC mice in which the CD4+ T cells declined and were almost completely lost by day 21 postinfection (P < 0.05 between CCR5-NP and blank-NP-PBMC mice) (Figure 5b,c, upper panel).
Concordant with the kinetics of CD4+ T-cell levels, the CCR5-NP-PBMC mice as a group consistently had lower copies of viral RNA in blood as compared with the blank-NP-PBMC mice at all time points tested, with some mice recording undetectable levels of viral RNA as early as day 7 postinfection (Figure 5c, lower panel). Collectively, the persistent maintenance of CD4+ cells and the low viral RNA levels demonstrate that the effective disruption of the CCR5 gene in the PBMCs treated with CCR5-NPs enables their maintenance and expansion in the face of HIV-1 viral infection in vivo. Importantly, this also validates that PLGA-NPs are a promising delivery system for the introduction of PNA-based gene-editing molecules into human T cells that are normally refractory to most nucleic acid transfection procedures.
Discussion
Gene-editing approaches to achieve permanent CCR5 gene disruption are gaining prominence as a means to eradicate HIV-1 infection. We report here the use of PLGA-NPs containing triplex-forming PNAs and donor DNAs for the targeted modification and permanent inactivation of the CCR5 gene in primary human PBMCs. This approach eliminates the risk of insertional mutagenesis associated with other common CCR5-targeting methods like the use of viral vectors for ZFN or shRNA expression.13,16 Furthermore, inherent toxicities are minimal as the approach does not necessitate the expression of exogenous nucleases and harnesses the natural host repair and recombination pathways. PBMCs efficiently internalized the formulated particles with minimal cytotoxicity, and the NP treatment did not elicit inflammatory responses or affect the ability of cells to engraft in a humanized mouse model. The frequency of site-specific modification of CCR5 in the PBMCs was 0.97% after a single treatment, with an off-target frequency of just 0.004% in CCR2, the most closely related gene to CCR5. HIV-1 infection of NOD-scid IL2rγ-/- mice engrafted with CCR5-NP–treated PBMCs demonstrated functional disruption of CCR5 as the mice showed recovery of CD4+ T-cell numbers with low to undetectable levels of viral RNA in the plasma, unlike mice engrafted with blank NP-treated cells. Stabilization of CD4+ T-cell levels was observed as early as 10 days postviral challenge and by day 21, xenogeneic expansion restored CD4+ T cells to levels similar to those in uninfected control mice. Importantly, preservation of CD4+ T-cell levels was achieved even with CCR5 modification at a frequency of ~1%, indicating that this level of CCR5 gene editing by triplex-forming PNAs and donor DNAs may be sufficient for a functional effect in vivo at least in cells already heterozygous for the CCR5-Δ32 mutation. We recognize that the rapidity of the CD4+ T-cell recovery might have been promoted by a quick expansion of the human T cells in a xenogeneic host environment. Nonetheless, this functional endpoint was achieved with an extremely low off-target frequency that may provide a substantial clinical advantage to this triplex-based approach as compared with nuclease-based strategies.
In the treated cell population of CCR5-Δ32 heterozygous PBMCs, a 1% overall modification frequency in the CCR5 target gene would, on average, render 0.5% of the cells homozygous, null for CCR5, assuming that either allele is equally susceptible to PNA-mediated targeting. The theoretical maximal yield of homozygous null cells would be 1% if all of the gene editing occurred on the wild-type allele, but this is not likely. Yet, even though only 0.5% (and at most 1.0%) of the NP-treated PBMCs were potentially rendered CCR5 null, these double knockout cells have a strong selective advantage in the face of HIV-1 infection in vivo, allowing the modified PBMCs to expand without being destroyed by the virus, leaving the unmodified cells to become infected and die off. Our results show that having 0.5% homozygous null cells in the engrafted population is sufficient to allow repopulation of CD4+ T cells in the face of HIV-1 infection because of the strong selective advantage in vivo in the mice. As shown in our data, this process occurs over several weeks in vivo and implies that if we were able to treat patient-specific CCR5-Δ32 T cells ex vivo and reinfuse them back into the patient, the resulting CCR5 null T cells could have a significant advantage that could lead to increased CD4 counts and reduced viral load. Furthermore, we have shown previously that we can directly modify human CD34+ stem cells in vivo in a similar mouse model by tail-vein injection of PNA-containing NPs.9 If such stem cells were modified, the T cells produced from these cells should have the same selective advantage in the face of viral challenge as the ex vivo modified T cells described here.
PBMCs are generally resistant to common transfection procedures. Due to their net neutral or positive charge, PNAs cannot be delivered by cationic lipids and instead have to be delivered by other means (such as electroporation, cell penetrating peptides, or microinjection), techniques which can be less effective or associated with considerable toxicity.17,18,19,20,21 Biodegradable PLGA-NPs enabled the delivery of PNA and donor DNA oligonucleotides into PBMCs in the absence of activating agents. PLGA is an FDA-approved material, and drug delivery systems based on PLGA are currently in clinical use.22 PLGA-NPs can also be modified on their surface to enable cell- and tissue-specific targeting to blood, lung, liver, and spleen; therefore, potentially allowing for cell- or tissue-specific delivery of PNAs and DNA donor oligonucleotides in vivo for specific gene-targeting applications.23,24
We previously demonstrated a targeting frequency of 2.46% in the CCR5 gene in human THP-1 cells with the same gene-targeting molecules used here but using electroporation as a means for delivery.7 However, we found that electroporation leads to very high toxicity in primary human PBMCs and abrogates their ability to successfully engraft using the same transplant protocol as described here (data not shown). In addition, in the THP-1 cells, direct sequencing of the CCR2 gene yielded an estimated off-target frequency of no more than 0.057%, with no mutations detected in the 1,740 cells analyzed, at least two orders of magnitude less than the reported 5.4% off-target frequency seen with ZFNs.7,13 In the work reported here, CCR5-NP treatment of PBMCs led to a precise genomic change in CCR5 at a frequency of 0.97%, as determined by Illumina deep sequencing. Furthermore, there were just 130 sequences changed in CCR2 in 2.8 million reads, an off-target frequency of 0.004%, which is more than three orders of magnitude less as compared with the off-target frequencies in CCR2 reported for ZFNs targeting CCR5.13
Although the targeted modification frequency achieved by the PNAs (0.97% in a single treatment) is about 15- to 30-fold lower than that seen with ZFNs, the PNAs still yielded functionally significant levels of gene editing in the target cell population, as evidenced by the HIV-1 resistance demonstrated in the engrafted mice. In addition, the CCR5 gene editing by the PNAs, in contrast to the ZFNs, was achieved without the use of viral vectors or activated T cells. However, the key point of comparison between PNA- and nuclease-based gene editing is the relatively high off-target genome effects of ZFNs, arising because ZFNs generate cleavage events throughout the genome at many other off-target sites.25 Comparatively, the much lower rate of off-target genome effects with triplex-forming PNAs at nonhomologous sites (more than 1,000-fold lower in comparison with ZFNs) has been determined by cumulative analyses of more than 1 × 107 alleles.26
It is also important to note that our methodology includes both the triplex-forming PNA to induce DNA repair and a donor DNA for the templated recombinational repair. We have reported previously7 that a single mismatch in the PNA-binding site (as is the case with CCR2) leads to a fourfold decrease in PNA binding to the target site. Although there may be other sites where the PNA can show partial or weak binding in the genome, even a single mismatch can reduce the binding efficiency that is needed for the induction of DNA repair. In prior work, we showed that weak-binding triplex-forming molecules do not effectively provoke DNA repair or recombination.27 For triplex-forming PNAs to be effective, they must bind very tightly and create a strong helical distortion that requires the nucleotide excision repair pathway for resolution.28 Several studies have shown that, otherwise, triplexes can simply be unwound and removed by helicases.29,30,31 Further to the point of specificity, the gene editing we report is the product of the combined activity of both the triplex-forming PNAs and the donor DNAs at their respective cognate sites in CCR5 and as such requires these sites to be nearby, at least within a few 100 base pairs.27 We have shown previously that donor DNAs, alone, can only mediate very low levels of targeted gene modification and that high-affinity triplex formation at a nearby site is needed to induce higher levels of recombination and gene editing. The combined action of both the donor DNA and the PNA molecules therefore requires the presence of nearby sites with homology to both molecules, providing a further level of stringency.
Importantly, therefore, although the triplex-forming PNAs are less efficient at inducing gene editing compared with engineered nucleases, they are also much less dangerous to other sites in the genome. The relative safety of triplex-forming PNAs is explained by the fact that they provoke recombination only by creating an altered helical structure that engages the cell's own repair mechanisms; they have no intrinsic nuclease or DNA-damaging activity. Triplex-forming PNAs can stimulate gene editing only by binding tightly to their matched target site. Because even one or two nucleotide mismatches strongly inhibit PNA binding, there is much less risk of mutations at nontargeted sites.27 This critical difference between triplex-forming PNAs and ZFNs is an important consideration for developing a clinical therapy, since off-target strand breaks could have deleterious consequences, with an increased risk of mutagenesis potentially leading to leukemias and other malignancies.
NP treatment of PBMCs had no deleterious effects on their ability to engraft and proliferate in mice. Multilineage repopulation was seen with levels equal to those produced by untreated cells. The presence of the targeted modification was confirmed in the splenic cells from mice transplanted with PBMCs treated with CCR5-NPs 4 weeks posttransplant, showing that the CCR5-modified cells are viable in vivo and that the targeted modification persists in the cell lineage during engraftment and expansion. Our recent publications also demonstrate that the triplex PNA approach can be extended to CD34+ hematopoietic stem cell precursors and does not affect their differentiation capacity.8,9
Overall, the work reported here demonstrates the feasibility of a NP and triplex-mediated strategy for permanently inactivating the CCR5 receptor in human T cells from HIV-1–infected individuals. Through autologous transplantation procedures, this could create a virus-resistant reservoir of cells, potentially leading to a ‘functional cure' for HIV-1.
Materials and methods
Design and synthesis of PNAs and single-stranded donor oligonucleotides. The PNA and donors used were previously characterized.7 Briefly, tcPNA-679 (N-Lys-Lys-Lys-JTJTTJTTJT-OOO-TCTTCTTCTCATTTC-Lys-Lys-Lys-C) was synthesized by Bio Synthesis (Lewisville, TX) or Panagene (Daejeon, Korea) and purified by RP-HPLC. Three lysine residues were conjugated to both the N- and C-terminal ends of the PNA for increased bioactivity, and 8-amino-2,6-dioxaoctanoic acid linkers were used as the flexible linker “O.” DNA oligonucleotides were synthesized by the Midland Certified Reagent Company (Midland, TX) and purified by RP-HPLC. All donor oligonucleotides were 5′ and 3′ end protected with three phosphorothioate internucleoside linkages.
NP formulation. PLGA-NPs were formulated by a double-emulsion solvent evaporation technique as previously described.32 Particles were stored at −20 °C following lyophilization.
NP characterization. Release of nucleic acids from particles was determined by incubating 4–6 mg of particles in 600 µl of PBS (Gibco, Grand Island, NY) in a 37 °C shaking incubator. Tubes were spun down and supernatant was removed at indicated time points and the absorbance at 260 nm was measured.
A sample of particles was analyzed using scanning electron microscopy (SEM). Samples were coated with 25-nm thick gold using a sputter coater and images were analyzed using ImageJ software (National Institutes of Health), with >500 particles analyzed per batch to determine size distribution. Brightness, contrast, and threshold were adjusted to enhance particle outlines, and ImageJ's “Analyze Particles” function was used to calculate the area of each particle.
Cell culture. Single-donor PBMCs were obtained from Cellular Technology (Shaker Heights, OH) and maintained in CTL-Test medium. Cells were thawed as per the Cellular Technology protocol and resuspended at 2 × 106 cells/ml in CTL media supplemented with L-glutamine (Gibco).
NP treatment of cells. NPs were resuspended in 500 µl of cold media. Resuspended particles were vortexed for 1 minute followed by sonication in an ice water bath for 30 seconds to ensure homogenous suspension of the particles. Resuspended particles were then added to the cells to the desired final concentration.
NP uptake in PBMCS. Uptake of C6-labeled NPs was determined by FACS, with trypan blue used to quench extracellular fluorescence as described previously.8,33
NP cytotoxicity. PBMCs were thawed and counted. Phytohemagglutinin of 5 µg/ml was added to the cells, and then PBMCs were seeded at 2 × 105 cells/well in a 96-well plate for overnight stimulation. The next morning, 20 U/ml of IL-2 was added to all the wells containing PBMCs. Later, in the afternoon, NPs were added to the cells in triplicate at the indicated final concentrations. Twenty-four hours later, 100 μl of the culture supernatant was removed from each well and added to a new plate to allow assay for lactate dehydrogenase activity (Cytotox-ONE; Promega, Madison, WI, according to the manufacturer's instructions). Cytotox-ONE substrate of 100 μl was added to each well and incubated for 10 minutes at room temperature. Cytotoxicity was calculated by the following formula: % cytotoxicity = (sample–culture medium background)/(lysed sample–culture medium background) where lysed sample corresponds to complete lysis of cells under identical conditions with a detergent. The experiment was done three times with three replicate wells per experiment for statistical considerations.
Genomic DNA isolation. Genomic DNA was isolated from cultured samples using the Wizard SV Genomic DNA Purification System (Promega). DNA was eluted with 100 μl of dH2O and diluted to 45 ng/μl for AS-PCR.
AS-PCR. AS-PCR was performed as previously detailed.7 The allele-specific forward primers were designed to contain the specific 6-bp mutations at the 3′ end while the wild-type forward primers contain the wild-type CCR5 sequence at the same position. Primer sequences and cycle parameters were available upon request. PCR products were separated on a 1% agarose gel and visualized using a gel imager. Wild-type forward primers paired with the universal reverse primer were used as a loading control.
Illumina sequencing preparation. Hundred base pair regions of CCR5 and CCR2 were amplified by PCR (including the site of the 6-bp mutation) from genomic DNA isolated from CCR5-NP and blank NP-treated PBMCs. PCR products were processed as per standard Illumina protocols to repair ends and add adapters. Equal amounts of each PCR product (CCR5 and CCR2) was pooled per treatment sample and given to the W.M. Keck Facility at Yale University for multiplexing and 75-bp, pair-end sequencing on the Illumina Genome Analyzer IIx platform.
Deep-sequencing data analysis. Paired-end Illumina sequencing reads were first demultiplexed and low-quality bases (Q2) were trimmed from the 3′ ends. Overlapping reads with at least 45 high-quality bases were combined into a single-consensus sequence and aligned against the amplicon sequences of CCR5 and CCR2 allowing one mismatch in the first 20 bases and up to 10 mismatches in subsequent sequence using Bowtie. Mutations were called using the default Bowtie output. Mutation frequencies were then scored for each gene by dividing the number of independent mutations by the total number of reads aligned to a gene, with adjacent mutations being counted as a single independent mutational event. Targeting frequencies were calculated as the number of occurrences of a target mutation divided by the total number of reads for each gene. Targets for CCR5 were a GCTGCT to CTAAGC substitution at positions 52–57 and a TGTCAT to CTGAGG substitution at positions 58–63. Cognate changes for CCR2 would be a GCTGCT to CTAAGC mutation at positions 27–32 and a CATCAT to CTGAGG substitution at positions 33–38.
Measurement of inflammatory cytokine mRNA production. PBMCs were collected through density-gradient centrifugation with Ficoll Histopaque (Sigma, St Louis, MO) and plated directly in CTL Test Media (Cellular Technology) supplemented with 1% L-glutamine. After 8 hours, nonadherent cells were replated at 2 million cells/ml and treated with 0.7 mg/ml of the indicated NPs. At various time points, samples were harvested and stored at −80 °C in RNAlater (Qiagen, Valencia, CA). RNA was extracted using the RNeasy Mini Kit (Qiagen) as per manufacturer's protocol, and cDNA was synthesized using the SuperScript II First-Strand Synthesis Kit (Invitrogen). Quantitative PCR was performed on cDNA with 20% Betaine (Sigma), 0.2 mmol/l dNTPs (American Bioanalytical, Natick, MA), Advantage 2 Polymerase mix (Clontech, Mountain View, CA), SYBR Green (Strategene, Santa Clara, CA), ROX (Strategene), and 2% Platinum Taq (Invitrogen). The following primers were used: TNF-α: 5′-gtggagatctcttcttgcac-3′ and 5′-cttgagaatgttaagggcact-3', IL6: 5′-actcacctcttcagaacgaa-3′ and 5′-tctggattcaatgaggagac-3′, and glyceraldehyde-3-phosphate dehydrogenase: 5′-gaaggtgaaggtcggagt-3′ and 3′-gaaatcccatcaccatcttc-5′. Primer sequences were obtained from the literature.34 The cycle conditions used were 94 °C for 2 minutes, followed by 40 cycles of 94 °C for 30 seconds, 50 °C for 30 seconds, and 72 °C for 1 minute. Relative gene expression was calculated using the 2−ΔCt method, with glyceraldehyde-3-phosphate dehydrogenase used as the reference gene.
Mouse transplantation with PBMCs. All the animals used were in accordance with the guidelines of the Institutional Animal Care and Use Committee of Yale University, The Jackson Laboratory, and conformed to the recommendations in the Guide for the Care and Use of Laboratory Animals (Institute of Laboratory Animal Resources, National Research Council, and National Academy of Sciences, 1996). NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ mice have been described previously and were obtained from the research colony maintained by L.D.S. at The Jackson Laboratory (Bar Harbor, ME).35
CCR5-Δ32 heterozygous or wild-type PBMCs were thawed as per CTL protocol, and 20 × 106 cells were treated with blank-NPs and 20 × 106 cells were treated with CCR5-NPs ~8 hours after thawing. Sixteen hours posttreatment, genomic DNA was isolated from an aliquot of each cell population and analyzed by AS-PCR for the presence of both donor-directed modifications. After confirmation of our desired modifications, cells were pelleted and resuspended at a concentration of 2.5 × 107 cells/ml in RPMI for injection into NOD-scid IL2rγ-/- mice. 5 × 106 PBMCs were transplanted into each NSG mouse through intraperitoneal injection.
Eight to 10 days after transplantation, mice were checked for reconstitution of human T cells by retoorbital venipuncture. Samples (100 µl) were layered onto ficoll-paque (GE Healthcare, Sunnyvale, CA) to separate mononuclear cells from erythrocytes. PBMCs were then assayed for lineage markers of human origin using antibodies purchased from BD Biosciences, San Jose, CA. Antibodies used were as follows: mouse anti-human CD45-APC, mouse anti-human CD3-FITC, mouse anti-human CD4-PerCP-Cy5.5, and mouse anti-human CD8-PE. Fluorescent data were acquired using a BD FACS Calibur machine, and data were analyzed using FlowJo 7.6 (Tree Star, Ashland, OR).
Four weeks after transplantation, a cohort of mice were killed, and various tissues were harvested and flash frozen. Genomic DNA was isolated from these tissues by phenol/chloroform extraction and analyzed by AS-PCR and quantitative AS-PCR.
Infection of humanized mice with HIV-1. Two weeks after transplantation with human PMBCs, mice were infected with 5,600 TCID50 HIV-1BaL by intraperitoneal injection. Mice were monitored for CD4 and CD8 counts and/or HIV-1 viremia by flow cytometry and Amplicor HIV-1 Monitor Test v1.5 (Roche Diagnostics, Indianapolis, IN), respectively. Peripheral blood samples were collected on days 4, 7, 10, 14, and 21 postinfection by retroorbital bleeding. PBMCs purified by ficoll-paque density centrifugation were stained as described above for the expression of human CD45, CD3, CD4, and CD8. Serum was stored at −80 °C until assayed for the presence of HIV-1 viral RNA. Peripheral T-cell ratios and plasma HIV-1 viremia were monitored by flow cytometry and the Amplicor assay for viral loads.
Statistical analysis. The data were analyzed using GraphPad Prism 5 (GraphPad, La Jolla, CA). Repeated-measures one-way analysis of variance with Tukey's multiple comparison testing were used to compare the treatment groups (for both in vitro and in vivo experiments) and to determine significance. All data with P < 0.05 were considered significant.
Acknowledgments
We thank Lisa Cabral (Yale University School of Medicine), Barbara Johnson (Yale University School of Medicine), Denise Hegan (Yale University School of Medicine), and Faye Rogers (Yale University School of Medicine) for their help. This work was supported by the Doris Duke Charitable Foundation Grant #2011102 (to P.M.G.), National Institutes of Health Grants R01HL082655 (to P.M.G.), R01HL085416 (to W.M.S.), AI46629 (to D.L.G., M.A.B., L.D.S.), DK32520 (to D.L.G., L.D.S.), by the George R. Pfeiffer Fellowship and NIH predoctoral Genetics training grant T32 GM007499 (to E.B.S), Ministry of Knowledge Economy under the KORUS Tech Program KT-2008-NT-APFS0-0001 (to P.K.), NIGMS Medical Scientist Training Program T32GM07205 (to N.A.M.) and F30HL110372 from the National Heart, Lung, and Blood institute (to N.A.M.), and the content is solely the responsibility of the authors and does not necessarily represent the official views of the funding organizations.
References
- Samson M, Libert F, Doranz BJ, Rucker J, Liesnard C, Farber CM, et al. Resistance to HIV-1 infection in caucasian individuals bearing mutant alleles of the CCR-5 chemokine receptor gene. Nature. 1996;382:722–725. doi: 10.1038/382722a0. [DOI] [PubMed] [Google Scholar]
- Liu R, Paxton WA, Choe S, Ceradini D, Martin SR, Horuk R, et al. Homozygous defect in HIV-1 coreceptor accounts for resistance of some multiply-exposed individuals to HIV-1 infection. Cell. 1996;86:367–377. doi: 10.1016/s0092-8674(00)80110-5. [DOI] [PubMed] [Google Scholar]
- Hütter G, Nowak D, Mossner M, Ganepola S, Müssig A, Allers K, et al. Long-term control of HIV by CCR5 Delta32/Delta32 stem-cell transplantation. N Engl J Med. 2009;360:692–698. doi: 10.1056/NEJMoa0802905. [DOI] [PubMed] [Google Scholar]
- Allers K, Hütter G, Hofmann J, Loddenkemper C, Rieger K, Thiel E, et al. Evidence for the cure of HIV infection by CCR5?32/?32 stem cell transplantation. Blood. 2011;117:2791–2799. doi: 10.1182/blood-2010-09-309591. [DOI] [PubMed] [Google Scholar]
- Ioannidis JP, Rosenberg PS, Goedert JJ, Ashton LJ, Benfield TL, Buchbinder SP, et al. International Meta-Analysis of HIV Host Genetics Effects of CCR5-Delta32, CCR2-64I, and SDF-1 3'A alleles on HIV-1 disease progression: An international meta-analysis of individual-patient data. Ann Intern Med. 2001;135:782–795. doi: 10.7326/0003-4819-135-9-200111060-00008. [DOI] [PubMed] [Google Scholar]
- O'Brien SJ, Nelson GW. Human genes that limit AIDS. Nat Genet. 2004;36:565–574. doi: 10.1038/ng1369. [DOI] [PubMed] [Google Scholar]
- Schleifman EB, Bindra R, Leif J, del Campo J, Rogers FA, Uchil P, et al. Targeted disruption of the CCR5 gene in human hematopoietic stem cells stimulated by peptide nucleic acids. Chem Biol. 2011;18:1189–1198. doi: 10.1016/j.chembiol.2011.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNeer NA, Chin JY, Schleifman EB, Fields RJ, Glazer PM, Saltzman WM. Nanoparticles deliver triplex-forming PNAs for site-specific genomic recombination in CD34+ human hematopoietic progenitors. Mol Ther. 2011;19:172–180. doi: 10.1038/mt.2010.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNeer NA, Schleifman EB, Cuthbert A, Brehm M, Jackson A, Cheng C, et al. Systemic delivery of triplex-forming PNA and donor DNA by nanoparticles mediates site-specific genome editing of human hematopoietic cells in vivo. Gene Ther. 2013;20:658–669. doi: 10.1038/gt.2012.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visscher GE, Robison RL, Maulding HV, Fong JW, Pearson JE, Argentieri GJ. Biodegradation of and tissue reaction to 50:50 poly(DL-lactide-co-glycolide) microcapsules. J Biomed Mater Res. 1985;19:349–365. doi: 10.1002/jbm.820190315. [DOI] [PubMed] [Google Scholar]
- Lucotte G. Frequencies of 32 base pair deletion of the (Delta 32) allele of the CCR5 HIV-1 co-receptor gene in Caucasians: a comparative analysis. Infect Genet Evol. 2002;1:201–205. doi: 10.1016/s1567-1348(02)00027-8. [DOI] [PubMed] [Google Scholar]
- Bashir A, Volik S, Collins C, Bafna V, Raphael BJ. Evaluation of paired-end sequencing strategies for detection of genome rearrangements in cancer. PLoS Comput Biol. 2008;4:e1000051. doi: 10.1371/journal.pcbi.1000051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez EE, Wang J, Miller JC, Jouvenot Y, Kim KA, Liu O, et al. Establishment of HIV-1 resistance in CD4+ T cells by genome editing using zinc-finger nucleases. Nat Biotechnol. 2008;26:808–816. doi: 10.1038/nbt1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson T, Greiner DL, Shultz LD. Creation of “humanized” mice to study human immunity. Curr Protoc Immunol. 2008;Chapter 15:Unit 15.21. doi: 10.1002/0471142735.im1521s81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allain JP, Laurian Y, Paul DA, Verroust F, Leuther M, Gazengel C, et al. Long-term evaluation of HIV antigen and antibodies to p24 and gp41 in patients with hemophilia. New Eng J Med. 1987. pp. 1114–1121. [DOI] [PubMed]
- Shimizu S, Hong P, Arumugam B, Pokomo L, Boyer J, Koizumi N, et al. A highly efficient short hairpin RNA potently down-regulates CCR5 expression in systemic lymphoid organs in the hu-BLT mouse model. Blood. 2010;115:1534–1544. doi: 10.1182/blood-2009-04-215855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braasch DA, Corey DR. Synthesis, analysis, purification, and intracellular delivery of peptide nucleic acids. Methods. 2001;23:97–107. doi: 10.1006/meth.2000.1111. [DOI] [PubMed] [Google Scholar]
- Koppelhus U, Nielsen PE. Cellular delivery of peptide nucleic acid (PNA). Adv Drug Deliv Rev. 2003;55:267–280. doi: 10.1016/s0169-409x(02)00182-5. [DOI] [PubMed] [Google Scholar]
- Maurisse R, De Semir D, Emamekhoo H, Bedayat B, Abdolmohammadi A, Parsi H, et al. Comparative transfection of DNA into primary and transformed mammalian cells from different lineages. BMC Biotechnol. 2010;10:9. doi: 10.1186/1472-6750-10-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papapetrou EP, Zoumbos NC, Athanassiadou A. Genetic modification of hematopoietic stem cells with nonviral systems: past progress and future prospects. Gene Ther. 2005;12 Suppl 1:S118–S130. doi: 10.1038/sj.gt.3302626. [DOI] [PubMed] [Google Scholar]
- Shiraishi T, Nielsen PE. Enhanced delivery of cell-penetrating peptide-peptide nucleic acid conjugates by endosomal disruption. Nat Protoc. 2006;1:633–636. doi: 10.1038/nprot.2006.92. [DOI] [PubMed] [Google Scholar]
- Lundström EA, Rencken RK, van Wyk JH, Coetzee LJ, Bahlmann JC, Reif S, et al. Triptorelin 6-month formulation in the management of patients with locally advanced and metastatic prostate cancer: an open-label, non-comparative, multicentre, phase III study. Clin Drug Investig. 2009;29:757–765. doi: 10.2165/11319690-000000000-00000. [DOI] [PubMed] [Google Scholar]
- Zhou J, Patel TR, Fu M, Bertram JP, Saltzman WM. Octa-functional PLGA nanoparticles for targeted and efficient siRNA delivery to tumors. Biomaterials. 2012;33:583–591. doi: 10.1016/j.biomaterials.2011.09.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng CJ, Saltzman WM. Enhanced siRNA delivery into cells by exploiting the synergy between targeting ligands and cell-penetrating peptides. Biomaterials. 2011;32:6194–6203. doi: 10.1016/j.biomaterials.2011.04.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattanayak V, Ramirez CL, Joung JK, Liu DR. Revealing off-target cleavage specificities of zinc-finger nucleases by in vitro selection. Nat Methods. 2011;8:765–770. doi: 10.1038/nmeth.1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers FA, Lin SS, Hegan DC, Krause DS, Glazer PM. Targeted gene modification of hematopoietic progenitor cells in mice following systemic administration of a PNA-peptide conjugate. Mol Ther. 2012;20:109–118. doi: 10.1038/mt.2011.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knauert MP, Lloyd JA, Rogers FA, Datta HJ, Bennett ML, Weeks DL, et al. Distance and affinity dependence of triplex-induced recombination. Biochemistry. 2005;44:3856–3864. doi: 10.1021/bi0481040. [DOI] [PubMed] [Google Scholar]
- Rogers FA, Vasquez KM, Egholm M, Glazer PM. Site-directed recombination via bifunctional PNA-DNA conjugates. Proc Natl Acad Sci USA. 2002;99:16695–16700. doi: 10.1073/pnas.262556899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopel V, Pozner A, Baran N, Manor H. Unwinding of the third strand of a DNA triple helix, a novel activity of the SV40 large T-antigen helicase. Nucleic Acids Res. 1996;24:330–335. doi: 10.1093/nar/24.2.330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain A, Bacolla A, Chakraborty P, Grosse F, Vasquez KM. Human DHX9 helicase unwinds triple-helical DNA structures. Biochemistry. 2010;49:6992–6999. doi: 10.1021/bi100795m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brosh RM, Majumdar A, Desai S, Hickson ID, Bohr VA, Seidman MM. Unwinding of a DNA triple helix by the Werner and Bloom syndrome helicases. J Biol Chem. 2001;276:3024–3030. doi: 10.1074/jbc.M006784200. [DOI] [PubMed] [Google Scholar]
- Fahmy TM, Samstein RM, Harness CC, Mark Saltzman W. Surface modification of biodegradable polyesters with fatty acid conjugates for improved drug targeting. Biomaterials. 2005;26:5727–5736. doi: 10.1016/j.biomaterials.2005.02.025. [DOI] [PubMed] [Google Scholar]
- Van Amersfoort ES, Van Strijp JA. Evaluation of a flow cytometric fluorescence quenching assay of phagocytosis of sensitized sheep erythrocytes by polymorphonuclear leukocytes. Cytometry. 1994;17:294–301. doi: 10.1002/cyto.990170404. [DOI] [PubMed] [Google Scholar]
- Park MC, Chung SJ, Park YB, Lee SK. Pro-inflammatory effect of leptin on peripheral blood mononuclear cells of patients with ankylosing spondylitis. Joint Bone Spine. 2009;76:170–175. doi: 10.1016/j.jbspin.2008.04.018. [DOI] [PubMed] [Google Scholar]
- Kumar P, Ban HS, Kim SS, Wu H, Pearson T, Greiner DL, et al. T cell-specific siRNA delivery suppresses HIV-1 infection in humanized mice. Cell. 2008;134:577–586. doi: 10.1016/j.cell.2008.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]