Abstract
While Streptococcus pyogenes is consistently susceptible toward penicillin, therapeutic failure of penicillin treatment has been reported repeatedly and a considerable number of patients exhibit allergic reactions to this substance. At the same time, streptococcal resistance to alternative antibiotics, e.g., macrolides, has increased. Taken together, these facts demand the development of novel therapeutic strategies. In this study, S. pyogenes growth was inhibited by application of peptide-conjugated antisense-peptide nucleic acids (PNAs) specific for the essential gyrase A gene (gyrA). Thereby, HIV-1 Tat peptide-coupled PNAs were more efficient inhibitors of streptococcal growth as compared with (KFF)3K-coupled PNAs. Peptide-anti-gyrA PNAs decreased the abundance of gyrA transcripts in S. pyogenes. Growth inhibition by antisense interference was enhanced by combination of peptide-coupled PNAs with protein-level inhibitors. Antimicrobial synergy could be detected with levofloxacin and novobiocin, targeting the gyrase enzyme, and with spectinomycin, impeding ribosomal function. The prospective application of carrier peptide-coupled antisense PNAs in S. pyogenes covers the use as an antimicrobial agent and the employment as a knock-down strategy for the investigation of virulence factor function.
Keywords: antimicrobial, antisense, cell-penetrating peptides, HIV-1 peptide, PNA, Streptococcus pyogenes
Introduction
Streptococcus pyogenes (group A streptococci (GAS)) is an exclusively human pathogen, which causes a wide spectrum of infectious diseases ranging from mild superficial infections of the skin and the mucosal membranes to invasive diseases like necrotizing fasciitis (flesh-eating disease) or streptococcal toxic shock syndrome. Typically, superficial infections are associated with spontaneous recovery. However, if mild infections remain untreated, severe invasive infections or autoimmune sequelae can develop as a consequence.1,2 Therefore, antibiotic therapy is strongly indicated upon streptococcal infections. Currently, penicillin is the standard treatment of streptococcal pharyngitis. Reasons are the continuing susceptibility of GAS toward penicillin, its efficiency, safety, and the comparably low costs of penicillin treatment.3,4,5 However, penicillin-related treatment failure has been reported repeatedly in cases of streptococcal pharyngitis.6,7 Factors that have been discussed to be responsible for this phenomenon include the coexistence of β-lactamase–producing bacteria,8 biofilm formation by GAS,9 and internalization of GAS into epithelial host cells.10,11,12 Another problem poses the rising occurrence of macrolide resistance in GAS,13,14 which limits the use of macrolides to patients with significant penicillin allergies.15 Consequently, the development of novel therapeutic strategies remains an imperative.
Among the innovative therapeutic approaches, antisense molecules gain increasing importance. One advantage of antisense interference is the specific effect on target molecules. Another is the lack of already established bacterial resistance mechanisms toward antisense agents. Peptide nucleic acids (PNAs) have been tested as antimicrobial agents in the past decade in a variety of bacterial species. Their chemical properties place PNAs between peptides and nucleic acids. Nucleobases, which are capable of sequence-specific base pairing, are present in PNAs. However, peptide bonds replace the nucleic acid-specific sugar-phosphate backbone.16 PNAs show a high stability in organic solutions as well as in water and their hybrid characteristics add to their stability in biological environments. So far, no nuclease or protease is known to be capable of hydrolyzing PNAs. Consequently, PNAs proved to be very stable in human serum and cellular extracts.17
PNA uptake is limited by the outer cell membrane in Gram-negative bacteria.18 Cell-penetrating peptides (CPPs) are naturally occurring or synthetic peptides containing positively charged residues that are able to enter eukaryotic cells and bacteria. They can be employed for the transduction of cargos into target cells.19,20 Transport of PNAs into Gram-negative bacteria could be facilitated by (KFF)3K CPPs coupled to the PNA molecules.21,22,23,24 The mRNA of several essential genes has been targeted by PNA antisense interference to achieve inhibition of bacterial growth, including the gene for an RNA polymerase primary sigma factor (rpoD), the gene for the gyrase A subunit (gyrA), the gene coding for the antiacyl carrier protein (acpP), and the ompA gene, coding for an outer membrane protein.24,25,26 In a different approach, bacterial protein biosynthesis has been inhibited by targeting with PNAs specific for the 16S or the 23S RNA.23,27
In a limited number of studies, CPP-conjugated PNAs have been tested in Gram-positive bacteria. In general, the antisense effect was less pronounced in Gram-positive species than in E. coli and a higher PNA concentration was required to cause growth reduction.23,28
Here, we aimed at inhibiting growth of GAS M49, which is a generalist known to be responsible for skin and throat infections, by antisense targeting of the essential gene gyrA. Its gene product represents the subunit A of the DNA topoisomerase gyrase, which is involved in replication and is thus required for bacterial growth. Growth reduction was achieved employing PNAs specific for gyrA, which were either coupled to (KFF)3K- or to Tat-peptides, respectively. Tat-conjugated anti-gyrA PNAs inhibited the growth of GAS M49 more efficiently than (KFF)3K-coupled anti-gyrA PNAs, while showing a lower unspecific CPP-related toxicity. Combination testing revealed antimicrobial synergy between antisense-PNAs and conventional antibiotics.
Results
GAS M49 growth reduction by carrier peptide-coupled anti-gyrA PNAs
Anti-gyrA PNAs were designed complementary to nucleotides covering the start codon of gyrA and three regions throughout the coding sequence of the gene (Table 1). We tested anti-gyrA PNAs with and without coupling to the synthetic (KFF)3K peptide, which had been used successfully before to support PNA uptake in a variety of bacterial species. In Figure 1a, a schematic of (KFF)3K-coupled anti-gyrA PNAs complementary to the start region is shown as a representative example. First, the four different target sequences within gyrA were compared. PNAs lacking the (KFF)3K-carrier peptide did not influence bacterial growth at all (data not shown). Also, (KFF)3K-coupled PNAs complementary to gyrA nucleotides 91-105, 867-881, and 1925-1939, respectively, did not interfere with GAS M49 growth (Table 1). Exclusively, (KFF)3K-coupled PNAs complementary to the start codon region of the gyrA transcript (nucleotides −5 to 5) led to a concentration-dependent reduction of GAS M49 growth (Table 1; Figure 2a).
Table 1. Peptide nucleic acids (PNAs) and inhibitory concentrations.
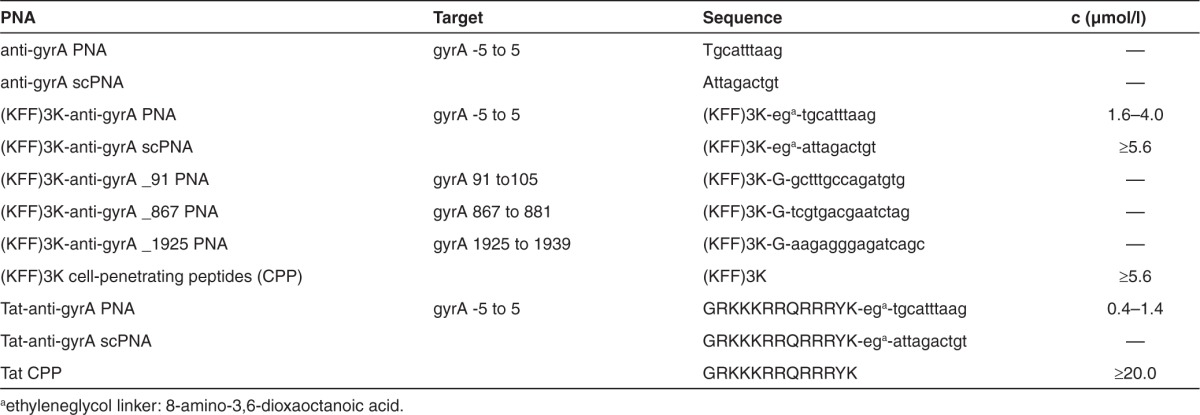
Figure 1.

Design of antisense peptide nucleic acids (PNAs) specific for gyrA in GAS M49. Upper case: partial sequence of the gyrase A gene, start-codon marked in bold; lower case, respective PNA sequences, e.g., 8-amino-3,6-dioxaoctanoic acid. (a) (KFF)3K-coupled PNAs complementary to the start-codon region of gyrA. (b) (KFF)3K-coupled PNAs composed of the same bases as in A but in a randomized (scrambled) sequence. Scrambled PNAs show no complementarity to the target region.
Figure 2.
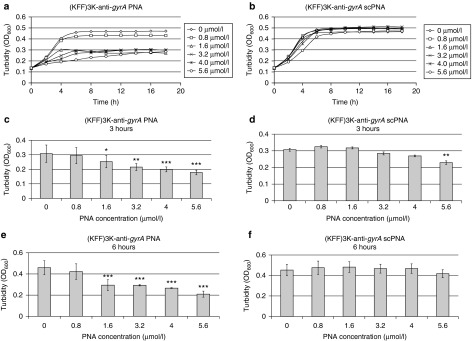
Effect of (KFF)3K-coupled peptide nucleic acids (PNAs) on GAS M49 growth. (a) Concentration-dependent growth inhibition of GAS M49 by (KFF)3K-anti-gyrA PNA. (b) No growth inhibition of GAS M49 by (KFF)3K-anti-gyrA scPNA (0.8–4.0 µmol/l). At PNA concentrations ≥5.6 µmol/l, unspecific toxic effects of the scPNAs were observed. (c/d) Statistic evaluation of the growth inhibition effect of (KFF)3K-anti-gyrA PNA (c) and (KFF)3K-anti-gyrA scPNA (d) at 3 hours. Error bars represent the mean ± SD, n = 5. (e/f): statistic evaluation of the growth inhibition effect of (KFF)3K-anti-gyrA PNA (e) and (KFF)3K-anti-gyrA scPNA (f) at 6 hours. Error bars represent the mean ± SD, n = 5.
To control for specificity of the interaction, scrambled PNAs (scPNAs) were designed, which shared the same base composition with sequence-specific anti-gyrA PNAs but exhibited a randomized sequence (Figure 1b). Comparison with (KFF)3K-anti-gyrA scPNA revealed that growth reduction caused by treatment with (KFF)3K-anti-gyrA PNA was sequence specific (Figure 2b). No growth inhibition was achieved by addition of scPNA within a concentration range of 0.8–4.0 µmol/l. At PNA concentrations ≥5.6 µmol/l, unspecific toxic effects of the scPNAs were observed (Figure 2b). Application of the (KFF)3K peptide alone also resulted in the reduction of bacterial growth at concentrations ≥5.6 µmol/l (data not shown). It is likely that growth inhibition upon application of high concentrations of (KFF)3K peptide-coupled PNAs is mediated at least in part by toxic effects of the leader peptide. The minimal inhibitory concentration (MIC) of (KFF)3K-anti-gyrA PNAs was 10 µmol/l. At this concentration, inhibition is caused by a combination of a sequence-specific action of the antisense molecule and an unspecific toxic effect of the leader peptide. Consequently, the following experiments were performed in the sub-MIC sequence-specific effective concentration range of (KFF)3K-anti-gyrA PNA. The observed effect upon treatment with (KFF)3K-anti-gyrA PNA was statistically relevant in the exponential (3 hours) as well as in the early stationary growth phase (6 hours) (Figure 2c–f). Dose dependency of GAS M49 growth inhibition became evident upon comparison of growth rates between samples (Supplementary Figure S1).
(KFF)3K-anti-gyrA PNA effects the abundance of gyrA transcripts in GAS M49
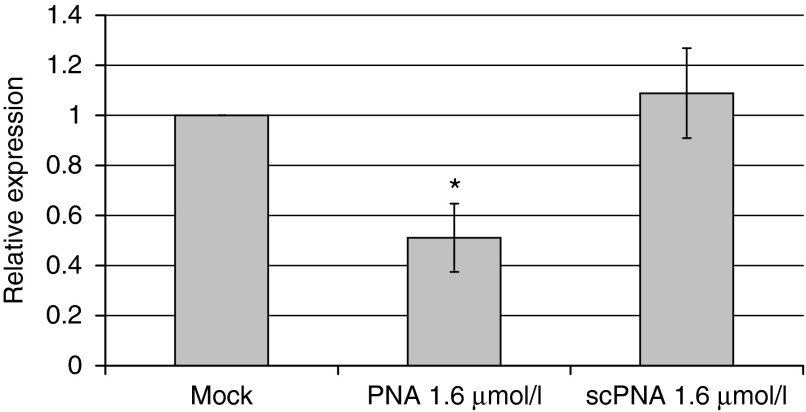
We asked whether antisense binding of (KFF)3K-anti-gyrA PNA to the gyrA mRNA was reflected by changes of the gyrA transcript level. The influence of the presence of sequence-specific PNAs on the amount of gyrA mRNA was tested by reverse transcription followed by quantitative reverse transcription polymerase chain reaction (Figure 3). Transcript abundance of the 5S RNA gene was used for normalization. The gyrA mRNA level in mock-treated GAS samples served as control. Upon treatment with 1.6 µmol/l (KFF)3K-anti-gyrA PNA, the amount of gyrA transcript was reduced to 50% of the amount detected in the untreated control sample. Addition of scPNA did not influence the relative gyrA mRNA level detected by quantitative reverse transcription polymerase chain reaction.
Figure 3.
Effect of (KFF)3K-anti-gyrA peptide nucleic acid (PNA) treatment on the abundance of gyrA transcripts. RNA was extracted from PNA-treated and scPNA-treated samples. The graph shows the relative gyrA expression compared to mock treated samples. Error bars represent the mean ± SD, n = 3.
The HIV-1 Tat peptide supports the antimicrobial activity of anti-gyrA PNA more efficiently than the (KFF)3K-carrier
In GAS M49, PNA uptake and PNA-mediated growth inhibition were not achieved when PNAs lacking a leader peptide were employed. Fusion of the CPP (KFF)3K to gyrA-specific PNA enabled sequence-specific growth inhibition of GAS M49. The uptake efficiency of every given CPP varies between bacterial species. Similar to the cationic and hydrophobic (KFF)3K peptide, the HIV-1 Tat protein-derived arginine-rich recombinant peptide YGRKKRRQRRR (Tat) translocates by destabilizing phospholipid bilayers and thereby induces transient pores in the respective membrane.29 In contrast to (KFF)3K, Tat has not been used before for transduction into bacteria. To test whether the Tat peptide could be employed as a leader peptide for the uptake of peptide oligonucleotides into GAS M49, it was coupled to gyrA antisense PNA. The effect of Tat-anti-gyrA PNA on growth of GAS M49 was compared with the growth inhibition mediated by Tat-anti-gyrA scPNA. Significant growth reduction in the presence of Tat-anti-gyr PNA was observed in the concentration range of 0.4–1.4 mol/l (Figure 4a,c,d). Addition of higher concentrations of Tat-PNA anti-gyrA did not lead to a further increase of the observed effects (data not shown). Application of Tat-anti-gyrA scPNA did not lead to growth inhibition of GAS M49 (Figure 4b), indicating a sequence-specific inhibitory effect. Addition of Tat peptide alone did not lead to GAS M49 growth inhibition up to a concentration of 10 µmol/l Tat peptide (data not shown). In comparison with anti-gyrA PNAs coupled to the (KFF)3K leader peptide, Tat-PNA anti-gyrA showed a much more efficient growth inhibition of GAS M49 (Table 1).
Figure 4.
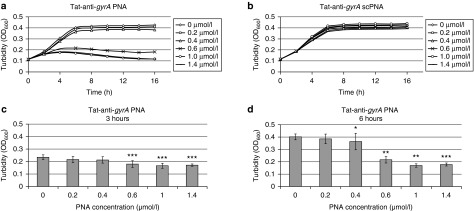
Effect of Tat-coupled peptide nucleic acids (PNAs) on GAS M49 growth. (a) Concentration-dependent growth inhibition of GAS M49 by Tat-anti-gyrA PNA. (b) No growth inhibition of GAS M49 by Tat-anti-gyrA scPNA. (c,d) statistic evaluation of the growth inhibition effect of Tat-anti-gyrA PNA at 3 (c) and at 6 hours (d). Error bars represent the mean ± SD, n = 6.
Antimicrobial synergy between peptide-coupled anti-gyrA PNAs and peptide-level antibiotics
The bacterial gyrase protein is a well-known target of antibiotics including aminocoumarins and quinolones. Given the fact that antisense-PNA treatment is reducing bacterial growth effectively but not completely, we wanted to test whether the combined application of peptide-coupled anti-gyrA PNAs with conventional antibiotics leads to synergistic effects. Gyrase peptide-targeting antibiotics were compared with the antibiotics targeting proteins unrelated to DNA replication. A widely used test for the determination of antimicrobial synergy between different inhibitors is the chequerboard assay.30 Following this protocol, the MIC for each compound is determined independently. Following serial dilution of the individual inhibitors, each concentration of one agent is then combined with each concentration of the second agent. Bacterial growth is determined for the combinations after appropriate incubation of the samples. For analyses of the inhibitor interactions, the fractional inhibitory concentration indices (FICI) are calculated.31 Checkerboard assay data have been interpreted variably, leading to ambiguous results.32 Furthermore, a linear dose dependency, which cannot be assumed for all antimicrobial substances, is a prerequisite for correct synergy determination using this method. Moreover, as pointed out before, at the MIC determined for (KFF)3K-anti-gyrA PNA, the sequence-specific effect of PNA was superimposed by toxic effects of the carrier peptide. For these reasons, the checkerboard assay was not feasible for combination testing of PNAs with antibiotics in GAS M49. To be able to work within the sequence-specific effective concentration range, spectrophotometric assessment of dose–response curves for antimicrobial combinations was employed.33,34,35
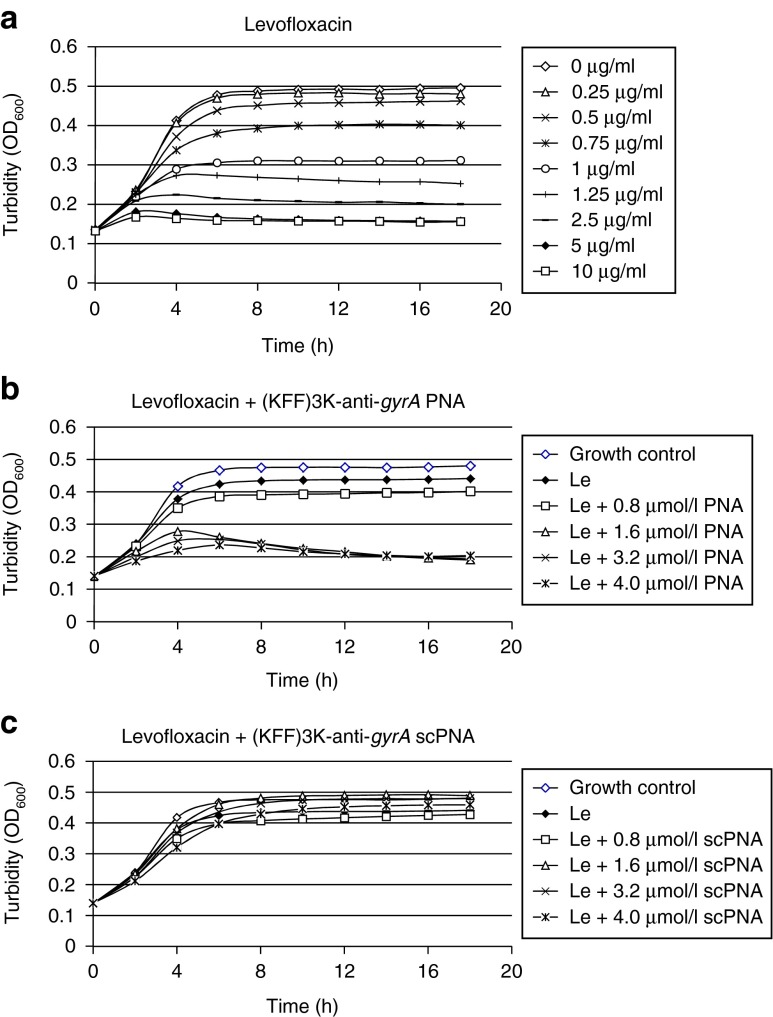
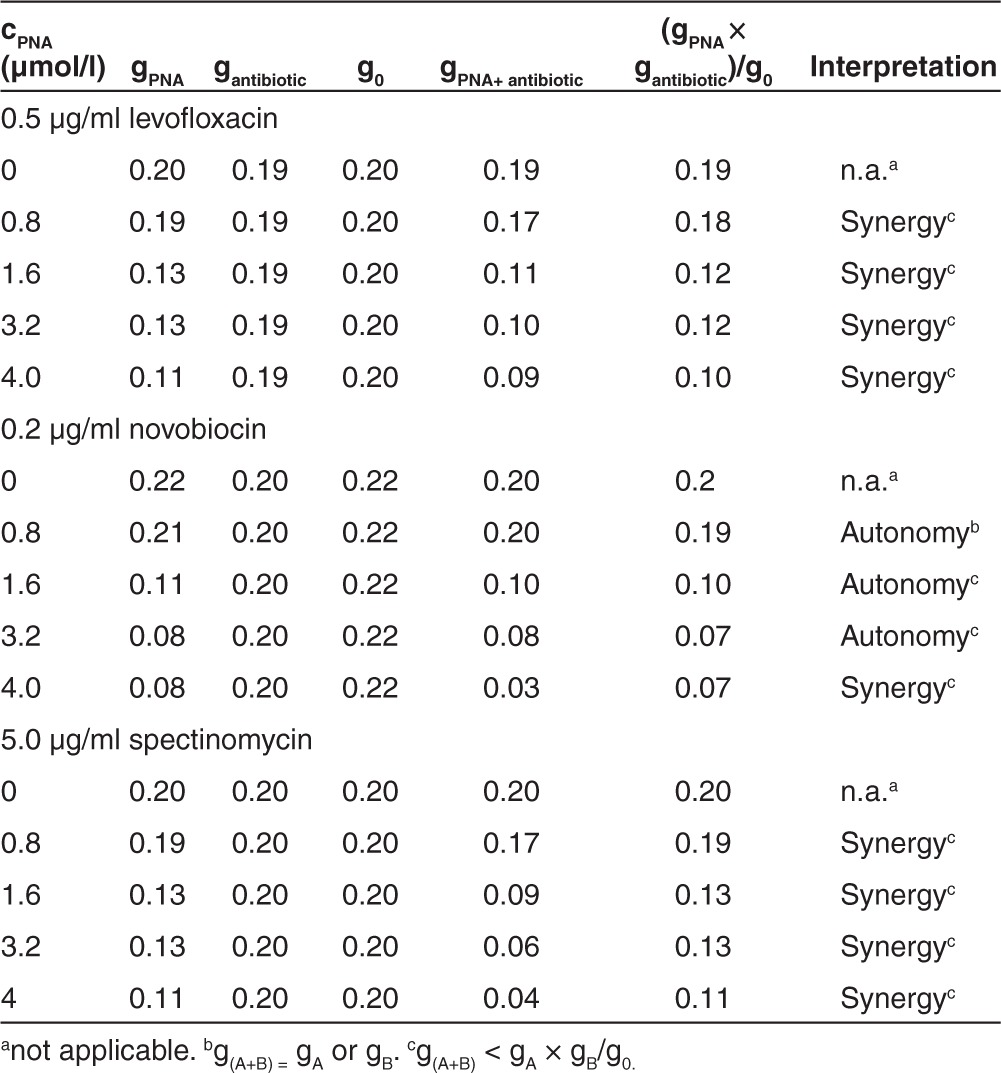
As examples for antibiotics interfering with the same pathway as anti-gyrA PNAs, inhibitors of gyrase A (levofloxacin) and gyrase B (novobiocin) were chosen. These were compared with spectinomycin, which binds to the ribosomal 30S subunit and thereby inhibits a pathway distinct from DNA replication. In Figure 5a–c, the spectrophotometric recordings for the interaction of (KFF)3K-anti-gyrA PNAs with levofloxacin are shown as one representative example. Reduced growth of GAS M49 in the presence of levofloxacin (0.5–10 µg/ml) could be detected (Figure 5a). At a concentration of 0.5 µg/ml levofloxacin, a slight growth repression was observed repeatedly. This concentration was chosen for combination testing. GAS M49 was incubated in the presence of 0.5 µg/ml levofloxacin and 0.8–4.0 µmol/l (KFF)3K-anti-gyrA PNAs, respectively. Upon addition of (KFF)3K-anti-gyrA PNAs to the culture, an increased inhibition of growth could be observed in comparison with samples containing levofloxacin alone (Figure 5b). Reduction of the growth constant g = In (ODtx/ODt0)/tx in the presence of one inhibitor or following application of a combination of the two agents served as a measure of interaction of the two effectors. For the combination of levofloxacin with (KFF)3K-anti-gyrA PNA, synergy was observed over the entire PNA concentration range tested in this experiment (Table 2). (KFF)3K-anti-gyrA scPNA did not enhance the effect of levofloxacin under these conditions (Figure 5b). The corresponding data for novobiocin and spectinomycin are presented in Supplementary Figures S2 and S3. Following treatment with novobiocin (0.2 µg/ml), synergy with (KFF)3K-anti-gyrA PNA was detected only at 4.0 µmol/l PNA (Table 2). Incubation of 5 µg/ml spectinomycin in combination with (KFF)3K-anti-gyrA PNA (0.8–4.0 µmol/l) led to synergistic effects at all PNA concentrations tested (Table 2). Again, no synergy was observed upon addition of (KFF)3K-anti-gyrA scPNA (Supplementary Figures S2c and S3c).
Figure 5.
Combination testing of levofloxacin with (KFF)3K-anti-gyrA peptide nucleic acid. (a) Dose-dependent growth inhibition of GAS M49 by levofloxacin. (b) Growth behavior of GAS M49 in the presence of 0.5 µg/ml levofloxacin and increasing concentrations of (KFF)3K-anti-gyrA peptide nucleic acid (PNA) (0.8–4.0 µmol/l). (c) Growth behavior of GAS M49 in the presence of 0.5 µg/ml levofloxacin and increasing concentrations of (KFF)3K-anti-gyrA scPNA (0.8–4.0 µmol/l).
Table 2. Interpretation of the decreasing growth rate constant (6 hours) (KFF)3K-anti-gyrA peptide nucleic acid (PNA).
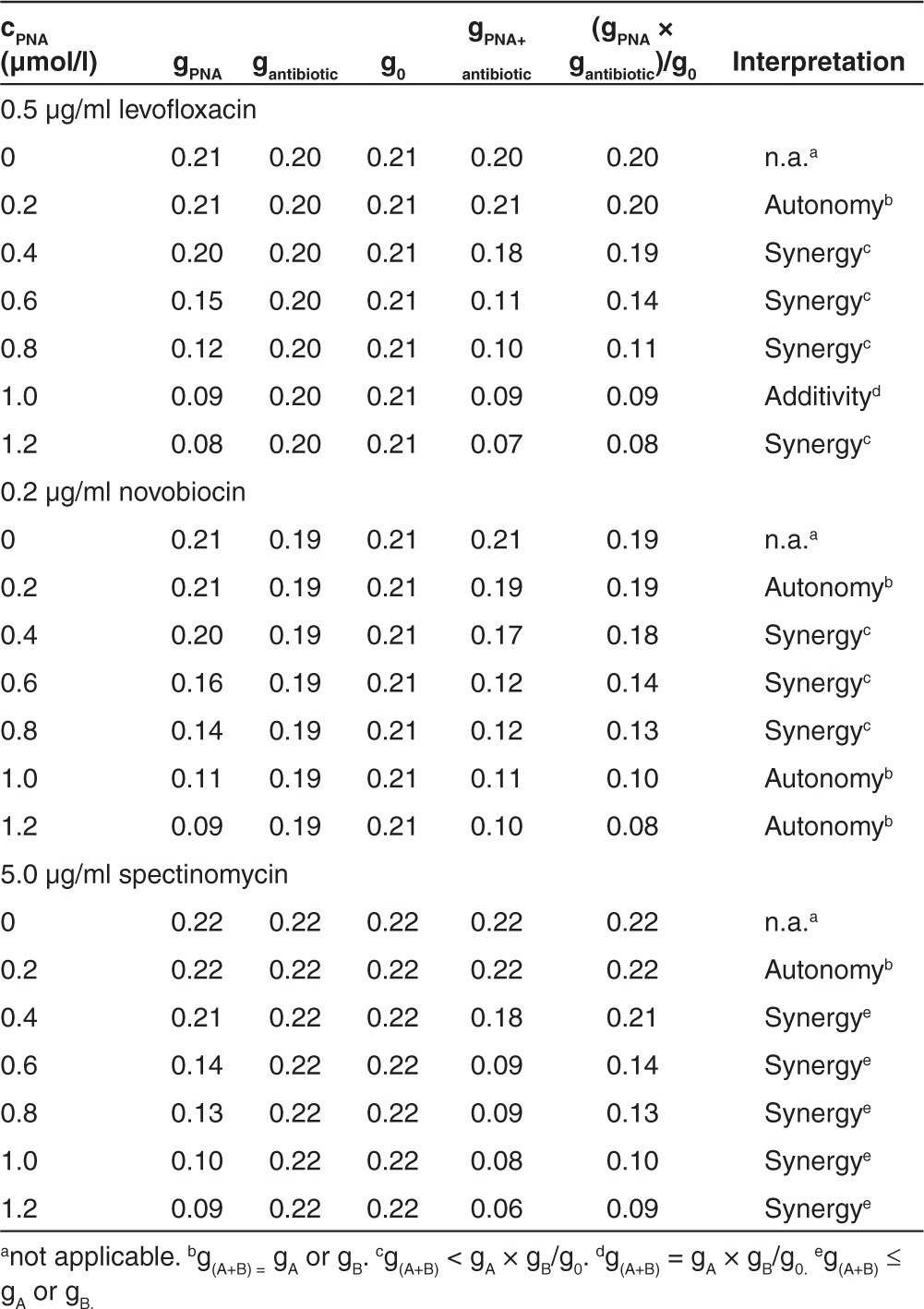
Combination of Tat-anti-gyrA PNA with peptide antibiotics led to comparable results (Supplementary Figures S4–S6; Table 3). Upon combination of Tat-anti-gyrA PNA (0.2–1.2 µmol/l) with levofloxacin (0.5 µg/ml), a synergistic effect was observed at 0.4–0.8 and 1.2 µmol/l PNA (Table 3). Treatment with novobiocin (0.2 µg/ml) led to synergy with 0.4–0.8 µmol/l PNA (Table 3). Application of 5 µg/ml spectinomycin in combination with Tat-anti-gyrA PNA led to synergistic effects from 0.4 to 1.2 µmol/l PNA (Table 3). Tat-conjugated scPNA did not show interaction with the antibiotics tested (Supplementary Figures S4b, S5b, and S6b).
Table 3. Interpretation of the decreasing growth rate constant (6 hours) Tat-anti-gyrA peptide nucleic acid (PNA).
Both (KFF)3K-anti-gyrA PNA and Tat-anti-gyrA PNA showed synergy with all three antibiotics tested in this study. The effect was independent of the pathway targeted by the antimicrobial substance. Combination of peptide-coupled anti-gyrA PNAs with antibiotics achieved a stronger overall growth inhibition than application of PNA alone.
Discussion
In the era of increasing drug resistance, it is more important than ever to assure timely development of innovative antimicrobial agents. Among antisense molecules, PNAs are particularly promising candidates due to their specific structural features. They are known for their strong pairing to DNA as well as to RNA and their pseudopeptide backbone confers stability while allowing molecular modifications, e.g., the conjugation of CPPs. The potency of antisense PNAs has been studied most thoroughly in Escherichia coli. Few reports are available about the effectivity of PNAs in Gram-positive pathogens. We set out to test whether PNA antisense targeting of a gene coding for an essential enzyme in GAS M49 will pose an impediment to its growth. We choose gyrA, because the gyrase enzyme represents a well-characterized target of antibiotics, including aminocoumarins and quinolones, and because gyrA has been successfully used before for PNA antisense-studies in other species.24,28
We observed growth reduction in GAS M49 following the application of anti-gyrA PNAs. However, conjugation of anti-gyrA PNAs with CPPs was required for antimicrobial activity of the antisense-PNAs. This is in accordance with the previous studies. PNA uptake by the bacterial cell has repeatedly been described as a limiting factor in antimicrobial PNA action. To overcome this obstacle, a variety of different strategies has been pursued in the past. The LPS layer-defective E. coli-mutant AS1936 has been used frequently for PNA studies, because of its increased permeability. Another widely employed option is the conjugation of synthetic cationic peptides, which are able to penetrate the outer membrane of Gram-negative bacteria.20 One well-studied example is the (KFF)3K peptide, which has been used as a leader peptide to facilitate PNA uptake into E. coli.21,37 It has also been shown to support PNA function in other Gram-negative bacteria and in some Gram-positive species, e.g., S. aureus.22,23,24,28 We tested (KFF)3K-PNA conjugates, which proved to be effective in GAS M49. While no activity of mere anti-gyrA PNAs could be observed, GAS M49 growth was inhibited in the presence of micromolar concentrations of (KFF)3K-coupled anti-gyrA PNA. The (KFF)3K-anti-gyrA PNA concentration range needed for a dose-dependent growth reduction in GAS M49 (1.6–4.0 µmol/l) was comparable with those reported from other species. In S. aureus, (KFF)3K-anti-gyrA PNA showed a growth inhibitory effect at 2–10 µmol/l,28 in Klebsiella pneumoniae, 10–40 µmol/l (KFF)3K-anti-gyrA PNA were needed for a dose-dependent inhibition.24 Growth inhibition by (KFF)3K-coupled antisense PNAs specific for rpoD, coding for the RNA polymerase primary sigma factor s70, required a PNA concentration of 12.5 µmol/l in S. aureus and 40 µmol/l in K. pneumoniae, respectively.25,38
Even though involvement of antisense binding has been shown in the inhibition of gene expression by PNAs,39 the exact mechanism is not known. The start codon region was found to be the most efficient target region to achieve inhibition of gene expression in E. coli, and impairment of translation initiation was considered to be a likely mechanism.40 On the other hand, reduction of the corresponding mRNA has been observed, indicating mRNA degradation upon binding of complementary PNAs.24,25 In GAS M49, a decrease of gyrA mRNA abundance to 50% could be detected upon treatment with (KFF)3K-anti-gyrA PNAs (Figure 3). Similar results have been obtained in E. coli, where application of (KFF)3K-anti-acpP-PNAs caused a decrease of acpP mRNA abundance to about 60% of the untreated control.26 Since ribosomal binding during translation acts as protective barrier against cleavage and thereby stabilizes mRNA,41 a hampered gyrA mRNA translation initiation due to PNA binding might be responsible for a moderate destabilization of the gyrA transcript in GAS M49.
The (KFF)3K-anti-gyrA PNA concentration required for the implementation of growth inhibition in GAS M49 was much higher than reported from E. coli inhibition studies. The MIC in E. coli K12 varied between 2 and 6 µmol/l, depending on the target,42 whereas the apparent MIC observed for GAS M49 was about 10 µmol/l, which is an approximate value, because it is influenced by toxic effects of the (KFF)3K leader. We speculated that import of PNAs into M49 could be improved by PNA coupling to a different CPP, preferentially to a peptide exhibiting lower toxicity. Besides the synthetic (KFF)3K peptide, there are many naturally occurring CPPs known, which represent short sequences of amino acids that are capable of entering most mammalian cells.19 CPPs are often highly cationic and hydrophilic. Translocation of cargos across the cell membrane seems to involve destabilization of the membrane and formation of a pore by the cationic peptides.29 The HIV-1 Tat protein contains a small region corresponding to residues 47YGRKKRRQRR57R, which is capable of membrane translocation by an apparently energy-independent mechanism.43 This fragment of the basic protein domain was shown to exhibit no cytotoxicity in HeLa cells at concentrations up to 100 µmol/l.44 However, translocation efficiency and toxicity have not been tested in bacteria, yet.
We used a synthetic HIV-1 Tat peptide derivative (Table 1) for conjugation to anti-gyrA PNA. Growth reduction of GAS M49 in a dose-dependent manner was achieved with 0.4–1.4 µmol/l Tat-anti-gyrA PNA (Figure 4a–d). Thereby, Tat-conjugated anti-gyrA PNA showed an enhanced antimicrobial activity compared with PNA coupled to (KFF)3K. Growth inhibition of GAS M49 was already detectable at Tat-PNA concentrations below 1 µmol/l. Upon application of the Tat-peptide alone, no toxicity was observed in GAS M49 cultures up to a concentration of 10 µmol/l Tat-peptide. Improved antisense effects in combination with low general toxicity are very desirable properties. It has been speculated that treatment failure and recurrent infections are caused by the internalization of S. pyogenes into host cells.10 Antisense agents coupled to Tat, which allows import into host cells in order to target intracellular bacteria and at the same time exhibits low cytotoxicity, might help to circumvent these therapeutic obstacles. Recently, growth of Brucella suis in infected macrophages was shown to be inhibited by 30 µmol/l (KFF)3K peptide-coupled PNAs.45 Application of Tat-coupled PNAs might reduce the required PNA concentration for the targeting of intracellular bacteria.
In the near future, we plan to use Tat-conjugated antisense-PNAs in GAS for the directed knock down of putative virulence genes.46 Experimental regulation of gene expression by antisense technology will help to study the function of virulence factors, including regulatory small RNAs, and will allow straight-forward screening for virulence specific phenotypes. Antisense-mediated downregulation of virulence-related streptococcal genes may provide also a therapeutic advantage. GAS with attenuated virulence could be targeted by the host immune system leading to a subsequent clearance of infection without the requirement of further antimicrobial treatment.
Combination of antimicrobial drugs is an interesting option for the improvement of therapy. The development of resistance to the respective drugs can be delayed or prevented by combinatorial administration. Another advantage is that the killing rate of bacteriostatic agents can be potentially increased in combination with a second drug. Moreover, in some cases, antimicrobial synergy can be observed upon combined treatment.30,47 Combination therapy may also be advantageous when resistance to a single agent develops rapidly.48 Combined in vitro application of antisense PNAs and peptide-targeting antibiotics have been tested in E. coli and in S. aureus.49 The authors described synergistic antimicrobial effects for combinations of drugs sharing the same genetic targets. By contrast, we found synergistic effects between peptide-coupled anti-gyrA PNA and three peptide level antibiotics, independent of the pathway targeted by the antibiotics (Tables 2 and 3). We speculate that during growth, inhibition of translation by ribosomal deficiency will interfere with DNA replication, because the required proteins are not replenished. Enhanced growth inhibition by (KFF)3K-anti-gyrA PNAs as well as Tat-anti-gyrA PNAs upon coapplication with conventional antibiotics suggests that antisense PNAs are promising candidates for combination therapy.
Materials and Methods
PNA synthesis. PNAs and PNAs with carrier peptide conjugates were synthesized and purified by HPLC at the DKFZ (Heidelberg, Germany). All PNAs used in this work are listed in Table 1.
Bacterial strains and growth conditions. GAS serotype M49, strain 591, a clinical isolate from a skin infection, was kindly provided from R. Lütticken (Aachen, Germany). GAS M49 was cultured in Todd–Hewitt broth (Invitrogen, Life Technologies GmbH, Darmstadt, Germany) supplemented with 0.5% yeast extract (THY; Invitrogen) at 37 °C under a 5% CO2 to 20% O2 atmosphere.
Testing of bacterial growth. Overnight cultures of GAS M49 were diluted 1:20 in THY and allowed to grow to mid-exponential growth phase. Bacteria were diluted to ~1 × 107 CFU/ml in THY. All inhibition experiments were performed in 96-well microtiter plates (Greiner Bio-One, Kremsmünster, Austria). PNAs of 20 µl diluted in H2O to a final PNA concentration of 0.2–20 µmol/l, as indicated, were added per well. In experiments testing the effect of levofloxacin, novobiocin, or spectinomycin on the growth of GAS M49, 20 µl of the respective component diluted in the appropriate solvent to concentrations as indicated were added to each well. Bacterial suspension of 180 µl was added to a total volume of 200 µl/well. As a contamination control, 20 µl of H2O was added to at least three wells on the same plate and 180 µl of THY was added. The microtiter plates were incubated at 37 °C, ambient air, in a temperature-controlled plate reader (SpectraMax M2; Molecular Devices, Sunnyvale, CA), which was set to shake the plate every 5 minutes before measuring absorption at 600 nm. Viable cell counts were determined by plating appropriate dilutions on THY agar plates. The plates were incubated overnight at 37 °C under a 5% CO2-20% O2 atmosphere. CFUs were determined by visual inspection. At time point 0, the viable cell count corresponded to 1–3 × 106 CFU/well. Each sample was prepared in triplicate; each experiment has been performed in at least four independent biological replicates. Growth rates were determined in the exponential growth phase: µ = logx2 – logx1/log(e) × (t2 – t1). t1 = 3 hours; t2 = 3.5 hours; x1 = OD600 at t1, x2 = OD600 at t2. The interaction of antimicrobial substances was determined in combination testing by calculation of the growth constant g = In (ODtx/ODt0)/tx in the presence of one inhibitor or following application of a combination of the two agents. Thereby, autonomy was defined as g(A+B) = gA or gB and synergy as g(A + B) <gA × gB/g0.
RNA isolation. For RNA isolation, five wells were prepared for each experimental condition, treated as indicated, and pooled after 6 hours of incubation. Total bacterial RNA was then isolated using the FastRNAProBlue Kit from MP Biomedicals, Illkirch, France as outlined in the protocol provided by the manufacturer. The purified total RNA was extracted with acidic phenol and digested with DNaseI (Ambion, Life Technologies GmbH, Darmstadt, Germany) to remove remaining traces of chromosomal DNA. The RNA preparation was treated with 10 U of DNase1 for 30 minutes at 37 °C. The enzyme was subsequently heat inactivated at 72 °C for 5 minutes.
Quantitative reverse transcription polymerase chain reaction. Fifty nanogram of acidic phenol-extracted and DNAseI-treated total RNA was reverse transcribed to generate cDNA using the First-Strand cDNA Synthesis Kit from Invitrogen following the protocol provided by the manufacturer. For reverse transcription, two reactions were performed using random hexamer primers provided by the kit. One reaction contained reverse transcriptase, a second control reaction was performed without enzyme to exclude product formation from residual genomic DNA templates in the following gene-specific polymerase chain reaction. All reactions were performed in triplicates. The real-time polymerase chain reaction amplification was performed with SYBR Green (Fermentas, Fisher Scientific-Germany GmbH, Schwerte, Germany) using an ABI PRISM 7000 Sequence Detection system (Applied Biosystems, Life Technologies GmbH, Darmstadt, Germany). The level of 5S RNA gene transcription was used for normalization.50 Primers were designed based on the full genome sequence of S. pyogenes M49 strain NZ131 (NCBI accession number: NC011375). gyrA-specific primers: 5′-TGAGTGTCATTGTGGCAAGAGC-3′ and 5′-AGAGAATACGACGATGCACAGG-3′ 5S-specific primers: 5′-AGCGACTACCTTATCTCACAG-3′ and 5′-GAGATACACCTGTACCCATG-3′. Primer efficiency was tested on genomic GAS M49 DNA before use in reverse transcription reactions. Relative gene expression was determined by the ΔΔCT method.51
Statistics. Statistical significance was determined using the two-tailed Mann–Whitney U test. Differences between samples were expressed as “not significant” (P ≥ 0.05), marginally significant (P < 0.05)*, significant (P < 0.01)**, highly significant (P < 0.001)***.
SUPPLEMENTARY MATERIAL Figure S1. Dose-dependent inhibition of the GAS M49 growth rate (µ) by (KFF)3K-anti-gyrA PNA. Figure S2. Combination testing of novobiocin with (KFF)3K-anti-gyrA PNA. Figure S3. Combination testing of spectinomycin with (KFF)3K-anti-gyrA PNA. Figure S4. Combination testing of levofloxacin with Tat-anti-gyrA PNA. Figure S5. Combination testing of novobiocin with Tat-anti-gyrA PNA. Figure S6. Combination testing of spectinomycin with Tat-anti-gyrA PNA.
Acknowledgments
The work of N.P. and B.K. was supported by a BMBF grant in the framework of the ERA-Net PathoGenoMics 2 program (FKZ 0315437B). The work of M.M. and T.H. was supported by a BMBF grant ERA-NET PathoGenoMics program sncRNAomics project (62080061). An application has been filed for a patent relating to the small RNAs mentioned in this manuscript and their prospective inhibition by antisense PNAs. German Patent and Trade Mark Office (DPMA), official file number: 10 2012 104 814.2.
Supplementary Material
References
- Carapetis JR, Steer AC, Mulholland EK, Weber M. The global burden of group A streptococcal diseases. Lancet Infect Dis. 2005;5:685–694. doi: 10.1016/S1473-3099(05)70267-X. [DOI] [PubMed] [Google Scholar]
- Cunningham MW. Pathogenesis of group A streptococcal infections and their sequelae. Adv Exp Med Biol. 2008;609:29–42. doi: 10.1007/978-0-387-73960-1_3. [DOI] [PubMed] [Google Scholar]
- Bisno AL, Gerber MA, Gwaltney JM, Jr, Kaplan EL, Schwartz RH. Infectious Diseases Society of America Practice guidelines for the diagnosis and management of group A streptococcal pharyngitis. Infectious Diseases Society of America. Clin Infect Dis. 2002;35:113–125. doi: 10.1086/340949. [DOI] [PubMed] [Google Scholar]
- Dajani A, Taubert K, Ferrieri P, Peter G, Shulman S. Treatment of acute streptococcal pharyngitis and prevention of rheumatic fever: a statement for health professionals. Committee on Rheumatic Fever, Endocarditis, and Kawasaki Disease of the Council on Cardiovascular Disease in the Young, the American Heart Association. Pediatrics. 1995;96 4 Pt 1:758–764. [PubMed] [Google Scholar]
- van Driel ML, De Sutter AI, Keber N, Habraken H, Christiaens T. Different antibiotic treatments for group A streptococcal pharyngitis. Cochrane Database Syst Rev. 2010. p. CD004406. [DOI] [PubMed]
- Kuhn SM, Preiksaitis J, Tyrrel GJ, Jadavji T, Church D, Davies HD. Evaluation of potential factors contributing to microbiological treatment failure in Streptococcus pyogenes pharyngitis. Can J Infect Dis. 2001;12:33–39. doi: 10.1155/2001/297304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macris MH, Hartman N, Murray B, Klein RF, Roberts RB, Kaplan EL, et al. Studies of the continuing susceptibility of group A streptococcal strains to penicillin during eight decades. Pediatr Infect Dis J. 1998;17:377–381. doi: 10.1097/00006454-199805000-00006. [DOI] [PubMed] [Google Scholar]
- Brook I, Gober AE. Failure to eradicate streptococci and beta-lactamase producing bacteria. Acta Paediatr. 2008;97:193–195. doi: 10.1111/j.1651-2227.2007.00610.x. [DOI] [PubMed] [Google Scholar]
- Baldassarri L, Creti R, Recchia S, Imperi M, Facinelli B, Giovanetti E, et al. Therapeutic failures of antibiotics used to treat macrolide-susceptible Streptococcus pyogenes infections may be due to biofilm formation. J Clin Microbiol. 2006;44:2721–2727. doi: 10.1128/JCM.00512-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa T, Terao Y, Okuni H, Ninomiya K, Sakata H, Ikebe K, et al. Biofilm formation or internalization into epithelial cells enable Streptococcus pyogenes to evade antibiotic eradication in patients with pharyngitis. Microb Pathog. 2011;51:58–68. doi: 10.1016/j.micpath.2011.03.009. [DOI] [PubMed] [Google Scholar]
- Sela S, Neeman R, Keller N, Barzilai A. Relationship between asymptomatic carriage of Streptococcus pyogenes and the ability of the strains to adhere to and be internalised by cultured epithelial cells. J Med Microbiol. 2000;49:499–502. doi: 10.1099/0022-1317-49-6-499. [DOI] [PubMed] [Google Scholar]
- Podbielski A, Kreikemeyer B. Persistence of group A streptococci in eukaryotic cells–a safe place. Lancet. 2001;358:3–4. doi: 10.1016/S0140-6736(00)05296-X. [DOI] [PubMed] [Google Scholar]
- Michos AG, Bakoula CG, Braoudaki M, Koutouzi FI, Roma ES, Pangalis A, et al. Macrolide resistance in Streptococcus pyogenes: prevalence, resistance determinants, and emm types. Diagn Microbiol Infect Dis. 2009;64:295–299. doi: 10.1016/j.diagmicrobio.2009.03.004. [DOI] [PubMed] [Google Scholar]
- Richter SS, Heilmann KP, Dohrn CL, Beekmann SE, Riahi F, Garcia-de-Lomas J, et al. Increasing telithromycin resistance among Streptococcus pyogenes in Europe. J Antimicrob Chemother. 2008;61:603–611. doi: 10.1093/jac/dkm525. [DOI] [PubMed] [Google Scholar]
- Logan LK, McAuley JB, Shulman ST. Macrolide treatment failure in streptococcal pharyngitis resulting in acute rheumatic fever. Pediatrics. 2012;129:e798–e802. doi: 10.1542/peds.2011-1198. [DOI] [PubMed] [Google Scholar]
- Nielsen PE, Egholm M. An introduction to peptide nucleic acid. Curr Issues Mol Biol. 1999;1:89–104. [PubMed] [Google Scholar]
- Demidov VV, Potaman VN, Frank-Kamenetskii MD, Egholm M, Buchard O, Sönnichsen SH, et al. Stability of peptide nucleic acids in human serum and cellular extracts. Biochem Pharmacol. 1994;48:1310–1313. doi: 10.1016/0006-2952(94)90171-6. [DOI] [PubMed] [Google Scholar]
- Good L, Sandberg R, Larsson O, Nielsen PE, Wahlestedt C. Antisense PNA effects in Escherichia coli are limited by the outer-membrane LPS layer. Microbiology (Reading, Engl) 2000;146 (Pt 10):2665–2670. doi: 10.1099/00221287-146-10-2665. [DOI] [PubMed] [Google Scholar]
- Gupta B, Levchenko TS, Torchilin VP. Intracellular delivery of large molecules and small particles by cell-penetrating proteins and peptides. Adv Drug Deliv Rev. 2005;57:637–651. doi: 10.1016/j.addr.2004.10.007. [DOI] [PubMed] [Google Scholar]
- Vaara M, Porro M. Group of peptides that act synergistically with hydrophobic antibiotics against gram-negative enteric bacteria. Antimicrob Agents Chemother. 1996;40:1801–1805. doi: 10.1128/aac.40.8.1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriksson M, Nielsen PE, Good L. Cell permeabilization and uptake of antisense peptide-peptide nucleic acid (PNA) into Escherichia coli. J Biol Chem. 2002;277:7144–7147. doi: 10.1074/jbc.M106624200. [DOI] [PubMed] [Google Scholar]
- Ghosal A, Nielsen PE. Potent antibacterial antisense peptide-peptide nucleic acid conjugates against Pseudomonas aeruginosa. Nucleic Acid Ther. 2012;22:323–334. doi: 10.1089/nat.2012.0370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatamoto M, Nakai K, Ohashi A, Imachi H. Sequence-specific bacterial growth inhibition by peptide nucleic acid targeted to the mRNA binding site of 16S rRNA. Appl Microbiol Biotechnol. 2009;84:1161–1168. doi: 10.1007/s00253-009-2099-0. [DOI] [PubMed] [Google Scholar]
- Kurupati P, Tan KS, Kumarasinghe G, Poh CL. Inhibition of gene expression and growth by antisense peptide nucleic acids in a multiresistant beta-lactamase-producing Klebsiella pneumoniae strain. Antimicrob Agents Chemother. 2007;51:805–811. doi: 10.1128/AAC.00709-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai H, You Y, Yan H, Meng J, Xue X, Hou Z, et al. Antisense inhibition of gene expression and growth in gram-negative bacteria by cell-penetrating peptide conjugates of peptide nucleic acids targeted to rpoD gene. Biomaterials. 2012;33:659–667. doi: 10.1016/j.biomaterials.2011.09.075. [DOI] [PubMed] [Google Scholar]
- Nikravesh A, Dryselius R, Faridani OR, Goh S, Sadeghizadeh M, Behmanesh M, et al. Antisense PNA accumulates in Escherichia coli and mediates a long post-antibiotic effect. Mol Ther. 2007;15:1537–1542. doi: 10.1038/sj.mt.6300209. [DOI] [PubMed] [Google Scholar]
- Xue-Wen H, Jie P, Xian-Yuan A, Hong-Xiang Z. Inhibition of bacterial translation and growth by peptide nucleic acids targeted to domain II of 23S rRNA. J Pept Sci. 2007;13:220–226. doi: 10.1002/psc.835. [DOI] [PubMed] [Google Scholar]
- Nekhotiaeva N, Awasthi SK, Nielsen PE, Good L. Inhibition of Staphylococcus aureus gene expression and growth using antisense peptide nucleic acids. Mol Ther. 2004;10:652–659. doi: 10.1016/j.ymthe.2004.07.006. [DOI] [PubMed] [Google Scholar]
- Herce HD, Garcia AE, Litt J, Kane RS, Martin P, Enrique N, et al. Arginine-rich peptides destabilize the plasma membrane, consistent with a pore formation translocation mechanism of cell-penetrating peptides. Biophys J. 2009;97:1917–1925. doi: 10.1016/j.bpj.2009.05.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eliopoulos G, Moellering R.1996Antimicrobial combinations. 4330–396.Williams & Wilkins; Baltimore, MD; Antibiotics in Laboratory Medicine, 4th edn, Lorian, V. Ref Type: Edited Book. [Google Scholar]
- den Hollander JG, Mouton JW, Verbrugh HA. Use of pharmacodynamic parameters to predict efficacy of combination therapy by using fractional inhibitory concentration kinetics. Antimicrob Agents Chemother. 1998;42:744–748. doi: 10.1128/aac.42.4.744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonapace CR, Bosso JA, Friedrich LV, White RL. Comparison of methods of interpretation of checkerboard synergy testing. Diagn Microbiol Infect Dis. 2002;44:363–366. doi: 10.1016/s0732-8893(02)00473-x. [DOI] [PubMed] [Google Scholar]
- Geisler N, Gattringer R, Graninger W, Georgopoulos A. Wirksamkeit von teicoplanin oder vancomycin in kombination mit linezolid gegenüber methicillin-resistentem Staphylococcus aureus mittels spektrophotometrischer Trübungsmessung. Antibiotika Monitor. 2003;6 [Google Scholar]
- King TC, Schlessinger D, Krogstad DJ. The assessment of antimicrobial combinations. Rev Infect Dis. 1981;3:627–633. doi: 10.1093/clinids/3.3.627. [DOI] [PubMed] [Google Scholar]
- King TC, Krogstad DJ. Spectrophotometric assessment of dose-response curves for single antimicrobial agents and antimicrobial combinations. J Infect Dis. 1983;147:758–764. doi: 10.1093/infdis/147.4.758. [DOI] [PubMed] [Google Scholar]
- Sekiguchi M, Iida S. Mutants of Escherichia coli permeable to actinomycin. Proc Natl Acad Sci USA. 1967;58:2315–2320. doi: 10.1073/pnas.58.6.2315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Good L, Awasthi SK, Dryselius R, Larsson O, Nielsen PE. Bactericidal antisense effects of peptide-PNA conjugates. Nat Biotechnol. 2001;19:360–364. doi: 10.1038/86753. [DOI] [PubMed] [Google Scholar]
- Bai H, Sang G, You Y, Xue X, Zhou Y, Hou Z, et al. Targeting RNA polymerase primary s70 as a therapeutic strategy against methicillin-resistant Staphylococcus aureus by antisense peptide nucleic acid. PLoS ONE. 2012;7:e29886. doi: 10.1371/journal.pone.0029886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Good L, Nielsen PE. Antisense inhibition of gene expression in bacteria by PNA targeted to mRNA. Nat Biotechnol. 1998;16:355–358. doi: 10.1038/nbt0498-355. [DOI] [PubMed] [Google Scholar]
- Dryselius R, Aswasti SK, Rajarao GK, Nielsen PE, Good L. The translation start codon region is sensitive to antisense PNA inhibition in Escherichia coli. Oligonucleotides. 2003;13:427–433. doi: 10.1089/154545703322860753. [DOI] [PubMed] [Google Scholar]
- Deana A, Belasco JG. Lost in translation: the influence of ribosomes on bacterial mRNA decay. Genes Dev. 2005;19:2526–2533. doi: 10.1101/gad.1348805. [DOI] [PubMed] [Google Scholar]
- Hatamoto M, Ohashi A, Imachi H. Peptide nucleic acids (PNAs) antisense effect to bacterial growth and their application potentiality in biotechnology. Appl Microbiol Biotechnol. 2010;86:397–402. doi: 10.1007/s00253-009-2387-8. [DOI] [PubMed] [Google Scholar]
- Herce HD, Garcia AE. Molecular dynamics simulations suggest a mechanism for translocation of the HIV-1 TAT peptide across lipid membranes. Proc Natl Acad Sci USA. 2007;104:20805–20810. doi: 10.1073/pnas.0706574105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vivès E, Brodin P, Lebleu B. A truncated HIV-1 Tat protein basic domain rapidly translocates through the plasma membrane and accumulates in the cell nucleus. J Biol Chem. 1997;272:16010–16017. doi: 10.1074/jbc.272.25.16010. [DOI] [PubMed] [Google Scholar]
- Rajasekaran P, Alexander JC, Seleem MN, Jain N, Sriranganathan N, Wattam AR, et al. Peptide nucleic acids inhibit growth of Brucella suis in pure culture and in infected murine macrophages. Int J Antimicrob Agents. 2013;41:358–362. doi: 10.1016/j.ijantimicag.2012.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Järver P, Langel K, El-Andaloussi S, Langel U. Applications of cell-penetrating peptides in regulation of gene expression. Biochem Soc Trans. 2007;35 Pt 4:770–774. doi: 10.1042/BST0350770. [DOI] [PubMed] [Google Scholar]
- Amaral L, Viveiros M. Why thioridazine in combination with antibiotics cures extensively drug-resistant Mycobacterium tuberculosis infections. Int J Antimicrob Agents. 2012;39:376–380. doi: 10.1016/j.ijantimicag.2012.01.012. [DOI] [PubMed] [Google Scholar]
- Eliopoulos GM, Moellering RC., Jr Antibiotic synergism and antimicrobial combinations in clinical infections. Rev Infect Dis. 1982;4:282–293. doi: 10.1093/clinids/4.2.282. [DOI] [PubMed] [Google Scholar]
- Dryselius R, Nekhotiaeva N, Good L. Antimicrobial synergy between mRNA- and protein-level inhibitors. J Antimicrob Chemother. 2005;56:97–103. doi: 10.1093/jac/dki173. [DOI] [PubMed] [Google Scholar]
- Patenge N, Billion A, Raasch P, Normann J, Wisniewska-Kucper A, Retey J, et al. Identification of novel growth phase- and media-dependent small non-coding RNAs in Streptococcus pyogenes M49 using intergenic tiling arrays. BMC Genomics. 2012;13:550. doi: 10.1186/1471-2164-13-550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.