Abstract
Background
Colorectal adenomatous polyposis is associated with a high risk of colorectal cancer (CRC) and is frequently caused by germline mutations in APC or MUTYH. However, in about 20–30% of patients no underlying gene defect can be identified. In this study, we tested if recently identified CRC risk variants play a role in patients with >10 adenomas.
Methods
We analysed a total of 16 SNPs with a reported association with CRC in a cohort of 252 genetically unexplained index patients with >10 colorectal adenomas and 745 controls. In addition, we collected detailed clinical information from index patients and their first-degree relatives (FDRs).
Results
We found a statistically significant association with two of the variants tested: rs3802842 (at chromosome 11q23, OR=1.60, 95% CI 1.3 to 2.0) and rs4779584 (at chromosome 15q13, OR=1.50, 95% CI 1.2 to 1.9). The majority of index patients (84%) had between 10 and 100 adenomas and 15% had >100 adenomas. Only two index patients (1%), both with >100 adenomas, had FDRs with polyposis. Forty-one per cent of the index patients had one or more FDRs with CRC.
Conclusions
These SNPs are the first common, low-penetrant variants reported to be associated with adenomatous polyposis not caused by a defect in the APC, MUTYH, POLD1 and POLE genes. Even though familial occurrence of polyposis was very rare, CRC was over-represented in FDRs of polyposis patients and, if confirmed, these relatives will therefore benefit from surveillance.
INTRODUCTION
Adenomatous polyps are the primary precursor lesions of colorectal cancer (CRC).1 Patients with large numbers of adenomas (polyposis) have a high likelihood of carrying an inheritable high-penetrance germline defect, particularly those patients diagnosed at a young age. Germline mutations in APC and MUTYH are commonly found in patients with an adenomatous polyposis phenotype. More recently, mutations in the POLD1 and POLE genes were also shown to be a cause of polyposis.2 Nevertheless, a substantial number (ie, an estimated 20–30%, also depending on polyp count and age at diagnosis) of patients remain unexplained. Some susceptibility of these unexplained cases may be attributable to the SNPs that are known to increase CRC risk (table 1). Eight of these SNPs were recently shown to be over-represented in CRC-free patients with few (mean=2) adenomatous polyps.3 The main aim of our study was to investigate whether CRC-risk SNPs are associated with disease in patients with more than 10 colorectal adenomas but without a known causative mutation, that is, no germline mutations in the APC, MUTYH, POLD1 and POLE genes. In addition, the phenotype in index patients and their first-degree relatives (FDRs) was evaluated.
Table 1. Summary results for the tested SNPs in 252 patients with unexplained adenomatous polyposis and 745 healthy controls.
| Allele* |
MAF | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Chr | SNP | Base-pair position† | Minor | Major | Patients | Controls | p Value | OR (95% CI) | Gene | Reference |
| 1q41 | rs6691170 | 222045446 | T | G | 0.378 | 0.346 | 0.194 | 1.1 (0.9-1.4) | DUSP10 CICP13 | 32 |
| 1q41 | rs6687758 | 222164948 | G | A | 0.228 | 0.197 | 0.141 | 1.2 (0.9 to 1.5) | DUSP10 CICP14 | 32 |
| 3q26.2 | rs10936599 | 169492101 | T | C | 0.237 | 0.244 | 0.745 | 1.0 (0.8 to 1.2) | MYNN | 32 |
| 8q23.3 | rs16892766 | 117630683 | C | A | 0.086 | 0.086 | 0.998 | 1.0 (0.7 to 1.4) | TRPS1 EIF3H | 33 |
| 8q24.21 | rs6983267 | 128413305 | G | T | 0.575 | 0.530 | 0.078 | 1.2 (1.0 to 1.5) | POU5F1 MYC | 34 |
| 10p14 | rs10795668 | 8701219 | A | G | 0.290 | 0.324 | 0.154 | 0.9 (0.7 to 1.1) | KRT8P16 TCEB1P3 | 33 |
| 11q23 | rs3802842 | 111171709 | C | A | 0.355 | 0.256 | 0.00002 | 1.6 (1.3 to 2.0) | C11orf93 | 11 |
| 12q13.13 | rs7136702 | 50880216 | T | C | 0.357 | 0.327 | 0.222 | 1.1 (0.9 to 1.4) | TBX3 UBA52P7 | 32 |
| 12q13.13 | rs11169552 | 51155663 | T | C | 0.260 | 0.264 | 0.876 | 1.0 (0.8 to 1.2) | DIP2B ATF1 | 32 |
| 14q22.2 | rs4444235 | 54410919 | C | T | 0.444 | 0.437 | 0.763 | 1.0 (0.8 to 1.3) | RPS3AP46 BMP4 | 12 |
| 15q13 | rs4779584 | 32994756 | T | C | 0.256 | 0.187 | 0.001 | 1.5 (1.2 to 1.9) |
SCG5 GREM1
FMN1 CRAC1 |
35 |
| 16q22.1 | rs9929218 | 68820946 | A | G | 0.280 | 0.278 | 0.921 | 1.0 (0.8 to 1.3) | CDH1 | 12 |
| 18q21.1 | rs4939827 | 46453463 | C | T | 0.486 | 0.491 | 0.864 | 1.0 (0.8 to 1.2) | SMAD7 | 14 |
| 19q13.1 | rs10411210 | 33532300 | T | C | 0.090 | 0.092 | 0.918 | 1.0 (0.7 to 1.4) | RHPN2 | 12 |
| 20p12.3 | rs961253 | 6404281 | A | C | 0.386 | 0.375 | 0.661 | 1.0 (0.9 to 1.3) | TARDBPL BMP2 | 12 |
| 20q13.33 | rs4925386 | 60921044 | T | C | 0.297 | 0.316 | 0.423 | 0.9 (0.7 to 1.1) | LAMA5 | 32 |
For SNP rs6983267, the minor allele originally reported (G) is the most frequent allele, both in our cases and controls.
The SNP positions are according to Human Build 36.3.
Chr, chromosome; MAF, minor allele frequency.
METHODS
Patients
Patients were genotyped in a consecutive series of 1216 index patients tested for APC and/or MUTYH germline mutations at the Laboratory for Diagnostic Genome Analysis (LDGA) in Leiden, from 1986 to January 2009. Informed consent was given for further research at initial blood withdrawal. Exclusion criteria were less than 10 adenomas, low-quality DNA samples, a phenotype with predominantly hyperplastic polyposis or missing clinical information on the histopathology of the polyps.
Clinical data were collected from the Netherlands Foundation for the Detection of Hereditary Tumors (NFDHT) and from clinical genetics departments in the Netherlands. Collected data included date of birth, gender, date of diagnosis of polyposis, cumulative number of polyps counted at colonoscopy or in excised bowel, location and histology of polyps, presence of duodenal polyps, information on CRC, polyps/CRC in first-degree family members, date of last contact and status at last contact. The control cohort was described previously4 and comprised a total of 745 controls for which both gender and age at blood sampling was known were included.
Genotyping
The APC and MUTYH genes had been tested as published previously.5,6 Hotspot mutations for POLE (p.L424V) and POLD1 (p.S478N) were determined for all cases using an in-house developed KASPar assay (primers available upon request). Next, those cases without APC, MUTYH or the POLD1 or POLE hotspot mutations were selected for genotyping the 16 SNPs listed in table 1. Anonymised samples of leucocyte DNA from these cases and controls were tested using the Competitive Allele-Specific PCR (KASPar) assay as described previously.7 All KASPar genotyping assays were performed by KBiosciences, UK.
Statistical analysis
All statistical analyses were carried out using PLINK,8 including calculations of allele differences between cases and controls, genotype call rate per sample and per SNP, deviations from Hardy–Weinberg equilibrium (HWE), as well as multiple test correction. Genotypes of cases and controls were compared using a basic χ2 allelic test. SNP profiles of specific subcohorts are available upon request. Multiple test correction was done using false discovery rate estimation.9
RESULTS
Genotype
Of the 1216 index patients surveyed, 252 patients fulfilled our inclusion criteria for SNP genotyping. We analysed 16 SNPs and compared allele frequencies in our mutation-negative cohort and in the control population (table 1). The call rates were above 99%. No deviations from HWE were observed. The association was strongest for SNPs rs3802842 (p=2.0×10−5; OR=1.60, 95% CI 1.3 to 2.0) and rs4779584 (p=0.001; OR=1.50, 95% CI 1.2 to 1.9) that remained significant after adjustment for multiple testing (p=0.001 and 0.026, respectively).
In order to disentangle whether the identified association for these two SNPs was based on polyposis or CRC, we compared allele frequencies in index patients with (n=96) and without CRC (n=156) and in our control population. The association for SNPs rs3802842 (p=0.0037; OR=1.5 (95% CI 1.1 to 1.9)) and rs4779584 (p=0.023; OR=1.4 (95% CI 1.0 to 1.9)) remained statistically significant for patients without CRC, that is, with a polyposis-only phenotype. In addition, the 14 other SNPs were not significantly associated with CRC.
Phenotype
The clinical description of the 252 patients is listed in table 2. The majority of cases (63%) had a cumulative polyp count of between 10 and 50 (with a mean age at diagnosis of 53 years), 21% showed between 50 and 100 polyps, while 15% of the cases had more than 100 polyps (‘carpeted’, ‘numerous’, ‘classic polyposis’ definitions included). Histopathologic examination showed all or a vast majority of polyps as adenomas in 208 cases (83%), while 44 cases (17%) displayed a mixed phenotype of predominantly adenomas with some hyperplastic polyps. Our cohort included 64 patients that carried both SNPs. These patients did not appear to have a more exacerbated phenotype (ie, 41/160 (26%) in the 10–50 group, 10/53 (19%) in the 50–100 group and 13/39 (33%) in the >100 polyp count group).
Table 2. Clinical details of three categories of index patients: 10–50 adenomas, 50–100 adenomas and >100 adenomas.
| Panel A | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Index |
Personal history |
|||||||||
| Adenomas (n) | Number | Mean age | Men (%) | CRC | Men (%) | Age at CRC | Duodenal adenoma |
|||
| 10–50 | 160 | 63% | 53 (29–82) | 63 | 59 | 37% | 56 | 53 (26–75) | 7/20 | 35% |
| 50–100 | 53 | 21% | 51 (13–73) | 57 | 18 | 34% | 72 | 55 (37–73) | 1/17 | 6% |
| >100 | 39 | 15% | 44 (4–80) | 56 | 19 | 49% | 58 | 51 (29–80) | 7/44 | 16% |
| Total | 252 | 100% | 51 (4–82) | 60 | 96 | 38% | 59 | 53 (26–80) | 15/81 | 19% |
|
| ||||||||||
|
Panel B
Family history |
||||||||||
|
| ||||||||||
| Adenomas (n) | FH | FDRs with CRC (n) | Age at CRC | FDRs with polyps (n) | Age | |||||
|
| ||||||||||
| 10–50 | 95 | 59% | 72 | 61 (32–85) | 68 | 55 (30–85) | ||||
| 50–100 | 30 | 57% | 20 | 62 (44–91) | 21 | 55 (26–78) | ||||
| >100 | 18 | 46% | 12 | 56 (17–79) | 14 | 51 (15–70) | ||||
| Total | 143 | 57% | 104 | 60 (17–91) | 103 | 55 (15–85) | ||||
Panel A: Clinical details of the index patients. Number of patients, percentage and mean age at diagnosis; number and percentage of male patients; number, percentage, sex distribution and age at diagnosis of CRC in polyposis patients; number of patients with duodenal adenomas/patients who underwent upper gastrointestinal endoscopy.
Panel B: Clinical data of the first-degree relatives (FDRs) of the index patients. Number and percentage of index patients with a positive family history (FH), that is, FDRs with CRC and/or multiple adenomas (ie, <10 polyps).
CRC, colorectal cancer.
CRC was found in 96 patients (38%) at a mean age of 53 years. A substantial number of patients (17%) had two or more synchronous or metachronous CRCs (n=16), and three (n=1) or even four (n=1) synchronous CRCs were also seen. In terms of location, 48 CRCs were found distal to the splenic flexure (left-sided CRC), 23 CRCs were right sided and 9 patients had both left-sided and right-sided tumours. The exact tumour location was unknown for 16 CRCs. A total of 81 patients (32%) underwent upper gastrointestinal endoscopy. Fifteen patients (19%) had duodenal adenomas, and one patient had both duodenal and gastric polyps.
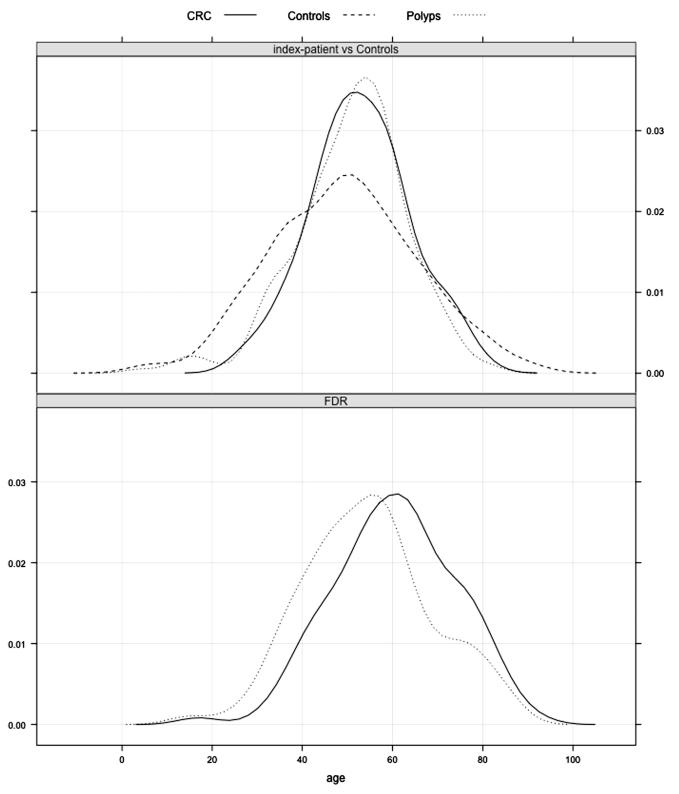
On investigation of family history, 143 (57%) index patients had close relatives with colorectal tumours; 104 patients (41%) had one or more FDRs with CRC, diagnosed at a mean age of 60 years (table 2). A steep increase in CRC incidence for FDRs was observed after the age of 30 (figure 1). Moreover, 103 patients (41%) had one or more FDRs with polyps. A family history with strong phenotypes was present in only two families, and in addition to index patients with >100 adenomas, one sister had >50 adenomas and the brother of the second case had >100 adenomas. The remaining index patients all had FDRs with low polyp counts (ie, less than 10 adenomas).
Figure 1. Age distribution as kernel density estimates.
Age of controls at blood withdrawal, age of index patients at polyposis diagnosis (Polyps) and age of index patients at colorectal cancer (CRC) diagnosis. The lower part shows the age of diagnosis of polyps and CRC in first-degree relatives (FDRs).
DISCUSSION
The present study provides new susceptibility loci for patients with colorectal adenomatous polyposis, defined here as a cumulative total of more than 10 colorectal adenomas, without an identified causative germline defect. Variants rs3802842 (on chromosome 11q23.1) and rs4779584 (on chromosome 15q13) are the first common, low-penetrant risk variants reported in this type of patients. The ORs we found (1.60 and 1.50) are much higher than values reported for patients with a few adenomas or for patients with CRC (typically ORs of 1.1 to 1.2).3,10–12 Our series is biased towards ascertaining index patients and their family members presenting with symptomatic CRC and is not a population-based series of polyposis patients. Therefore, prospective studies are needed to confirm our findings. Moreover, disentanglement of whether the identified association for these two SNPs was based on polyposis or CRC is difficult given how much these two phenotypes are interrelated. However, we found that the association still stood in the index patients with a polyposis-only phenotype, that is, after excluding the index patients with CRC. Therefore, even though the low-risk variants identified in the present study are unlikely to explain polyposis by themselves, they still might act as modifiers of so far unidentified high-risk factors or participate in a multifactorial cause of the phenotype.
The SNP rs3802842 appears to be involved in risk predisposition in hereditary forms of CRC and has previously been associated with early-onset CRC (diagnosed under the age of 50 years), low numbers of adenomas and familial aggregation of CRC.4,13,14 Moreover, rs3802842 was also shown to be associated with CRC risk in female patients with Lynch syndrome.15,16 Mechanistically, none of the transcripts (FLJ45803, LOC120376, C11orf53 and POU2AF1) mapping to the rs3802842 region at chromosome 11q23 showed possible causative exonic mutations, and rs3802842 is currently annotated to an intron of an uncharacterised protein, C11orf93. While the impact of this SNP might be via non-coding effects on gene expression, a clear mechanistic explanation for this association remains elusive despite extensive research.17
A more plausible mechanistic explanation may be available for the SNP rs4779584, as its chromosome 15q13.3 locus encompasses the genes SCG5, FMN1 and GREM1. A 40 kb duplication upstream of the GREM1 gene was previously identified in a large Ashkenazi family with a mixed polyposis phenotype.18 GREM1 is a component of the TGF-b superfamily signalling pathway acting as a bone morphogenetic protein (BMP) antagonist in the colon. Increased GREM1 expression is predicted to reduce BMP activity, a mechanism also found in juvenile polyposis.18 Given the reported association of 15q13 SNP with a mixed polyposis phenotype, we compared the allelic counts in patients with adenoma and patients with a phenotype of predominantly adenomas and some hyperplastic polyps (table 3). However, we did not find an over-representation of the 15q13 SNP in the subset with both adenomas and hyperplastic polyps (p=0.5).
Table 3. Analysis of the rs4779584 SNP on 15q13 in patients with adenoma and patients with a phenotype of predominantly adenomas and some hyperplastic polyps.
| Phenotype | CC | TC | TT | Total |
|---|---|---|---|---|
| Adenoma | 115 | 83 | 10 | 208 |
| Mixed | 24 | 15 | 5 | 44 |
| Total | 139 | 98 | 15 | 252 |
Phenotype
In our large cohort of patients with genetically unexplained polyposis, medical records were scrutinised in order to collect accurate phenotypic data. The majority of subjects exert a phenotype resembling that of attenuated adenomatous polyposis since they had a moderate number of polyps (ranging from 10 to 100) at an advanced age of diagnosis (mean age 52 years). The proportion of patients affected with duodenal adenomas was also in more agreement with MUTYH- (ie, 20%) than classic APC-associated polyposis (ie, 90%).19,20 Previous studies on APC and MUTYH mutation-negative polyposis patients show a clinically heterogeneous disease, with scarce information on family members.21-30
Regarding family history, strong phenotypes (>50 adenomas) were present in only two families (1%), and 41% of the index patients had one or more FDRs with single or multiple adenomas (ie, <10 polyps). Moreover, CRC in FDRs was observed in 41% of the index patients. This observation mimics studies on hyperplastic polyposis, where even in the absence of hyperplastic polyposis an increased CRC risk for FDRs of patients was found and for whom surveillance is advised.23,31
Clinical implications
Based on the clinical evaluation of our cohort, the following surveillance guidelines for FDRs of polyposis patients without known cause may be considered. First, to exclude the presence of polyposis—and despite the rare occurrence in the relatives of the patients in our series—we would recommend performing colonoscopy in parents, siblings and offspring at an early age. Based on the endoscopic findings, decisions can be made on the need for follow-up examinations. Second, based on the high CRC incidence in FDRs, we would also recommend colonoscopic surveillance, even in the absence of a polyposis phenotype on first colonoscopy at an early age. Given the age distribution of CRC onset in FDRs (figure 1), we suggest that surveillance commence at age 30, with intervals of 5–6 years dependent on endoscopic findings.
In conclusion, this is the first study to report an association between common, low-penetrance genetic variants and an unexplained adenomatous polyposis phenotype. Future studies should explore the mechanistic role of these variants in the development of polyposis. Whereas the familial occurrence of polyposis in relatives was rare, almost half of FDRs developed CRC. If over-representation of CRC in FDRs of polyposis patients is confirmed in other studies, these relatives should undergo colonoscopic surveillance, even in the absence of a polyposis phenotype upon first colonoscopy.
Acknowledgements
We thank the Mallorca Group for helpful discussion.
Funding: Association of International Cancer Research, grant 2010-0619 and Dutch Cancer Society, grant KWF-UL-2010-4656.
Footnotes
Competing interests: None.
Ethics approval: Medical ethics committee, LUMC.
Provenance and peer review: Not commissioned; externally peer reviewed.
REFERENCES
- 1.Kinzler KW, Vogelstein B. Lessons from hereditary colorectal cancer. Cell. 1996;87:159–70. doi: 10.1016/s0092-8674(00)81333-1. [DOI] [PubMed] [Google Scholar]
- 2.Palles C, Cazier JB, Howarth KM, Domingo E, Jones AM, Broderick P, Kemp Z, Spain SL, Guarino E, Almeida EG, Salguero I, Sherborne A, Chubb D, Carvajal-Carmona LG, Ma Y, Kaur K, Dobbins S, Barclay E, Gorman M, Martin L, Kovac MB, Humphray S, Lucassen A, Thomas HJ, Maher E, Evans G, Cummings C, Stevens M, Walker L, Halliday D, Armstrong R, Paterson J, Hodgson S, Homfray T, Side L, Izatt L, Donaldson A, Tomkins S, Morrison P, Goodman S, Brewer C, Henderson A, Davidson R, Murday V, Cook J, Haites N, Bishop T, Sheridan E, Green A, Marks C, Carpenter S, Broughton M, Greenhalge L, Suri M, Donnelly PC, Bell J, Bentley D, McVean G, Ratcliffe P, Taylor J, Wilkie A, Broxholme J, Buck D, Cornall R, Gregory L, Knight J, Lunter G, Tomlinson I, Wilkie A, Kingsbury Z, Grocock R, Hatton E, Holmes CC, Hughes L, Humburg P, Kanapin A, Murray L, Rimmer A, Petridis C, Roylance R, Sawyer EJ, Kerr DJ, Clark S, Grimes J, Kearsey SE, Houlston RS. Germline mutations affecting the proofreading domains of POLE and POLD1 predispose to colorectal adenomas and carcinomas. Nat Genet. 2013;45:136–44. doi: 10.1038/ng.2503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carvajal-Carmona LG, Zauber AG, Jones AM, Howarth K, Wang J, Cheng T, Riddell R, Lanas A, Morton D, Bertagnolli MM, Tomlinson I. Much of the genetic risk of colorectal cancer is likely to be mediated through susceptibility to adenomas. Gastroenterology. 2013;144:53–5. doi: 10.1053/j.gastro.2012.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Middeldorp A, Jagmohan-Changur S, van Eijk R, Tops C, Devilee P, Vasen HF, Hes FJ, Houlston R, Tomlinson I, Houwing-Duistermaat JJ, Wijnen JT, Morreau H, van Wezel T. Enrichment of low penetrance susceptibility loci in a Dutch familial colorectal cancer cohort. Cancer Epidemiol Biomarkers Prev. 2009;18:3062–7. doi: 10.1158/1055-9965.EPI-09-0601. [DOI] [PubMed] [Google Scholar]
- 5.Hes FJ, Nielsen M, Bik EC, Konvalinka D, Wijnen JT, Bakker E, Vasen HF, Breuning MH, Tops CM. Somatic APC mosaicism: an underestimated cause of polyposis coli. Gut. 2008;57:71–6. doi: 10.1136/gut.2006.117796. [DOI] [PubMed] [Google Scholar]
- 6.Nielsen M, Franken PF, Reinards TH, Weiss MM, Wagner A, van der Klift H, Kloosterman S, Houwing-Duistermaat JJ, Aalfs CM, Ausems MG, Brocker-Vriends AH, Gomez Garcia EB, Hoogerbrugge N, Menko FH, Sijmons RH, Verhoef S, Kuipers EJ, Morreau H, Breuning MH, Tops CM, Wijnen JT, Vasen HF, Fodde R, Hes FJ. Multiplicity in polyp count and extracolonic manifestations in 40 Dutch patients with MYH associated polyposis coli (MAP) J Med Genet. 2005;42:e54. doi: 10.1136/jmg.2005.033217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Middeldorp A, Jagmohan-Changur S, Helmer Q, van der Klift HM, Tops CM, Vasen HF, Devilee P, Morreau H, Houwing-Duistermaat JJ, Wijnen JT, van Wezel T. A procedure for the detection of linkage with high density SNP arrays in a large pedigree with colorectal cancer. BMC Cancer. 2007;7:6. doi: 10.1186/1471-2407-7-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–75. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Benjamini Y, Yekutieli D. The control of the false discovery rate in multiple testing under dependency. Ann Stat. 2001;29:1165–88. [Google Scholar]
- 10.Dunlop MG, Dobbins SE, Farrington SM, Jones AM, Palles C, Whiffin N, Tenesa A, Spain S, Broderick P, Ooi LY, Domingo E, Smillie C, Henrion M, Frampton M, Martin L, Grimes G, Gorman M, Semple C, Ma YP, Barclay E, Prendergast J, Cazier JB, Olver B, Penegar S, Lubbe S, Chander I, Carvajal-Carmona LG, Ballereau S, Lloyd A, Vijayakrishnan J, Zgaga L, Rudan I, Theodoratou E, Thomas H, Maher E, Evans G, Walker L, Halliday D, Lucassen A, Paterson J, Hodgson S, Homfray T, Side L, Izatt L, Donaldson A, Tomkins S, Morrison P, Brewer C, Henderson A, Davidson R, Murday V, Cook J, Haites N, Bishop T, Sheridan E, Green A, Marks C, Carpenter S, Broughton M, Greenhalge L, Suri M, Starr JM, Deary I, Kirac I, Kovacevic D, Aaltonen LA, Renkonen-Sinisalo L, Mecklin JP, Matsuda K, Nakamura Y, Okada Y, Gallinger S, Duggan DJ, Conti D, Newcomb P, Hopper J, Jenkins MA, Schumacher F, Casey G, Easton D, Shah M, Pharoah P, Lindblom A, Liu T, Edler D, Lenander C, Dalen J, Hjern F, Lundqvist N, Lindforss U, Pahlman L, Smedh K, Tornqvist A, Holm J, Janson M, Andersson M, Ekelund S, Olsson L, Smith CG, West H, Cheadle JP, Macdonald G, Samuel LM, Ahmad A, Corrie P, Jodrell D, Palmer C, Wilson C, O’Hagan J, Smith D, McDermott R, Walshe J, Cassidy J, McDonald A, Mohammed N, White J, Yosef H, Breathnach O, Grogan L, Thomas R, Eatock M, Henry P, Houston R, Johnston P, Wilson R, Geh I, Danwata F, Hindley A, Susnerwala S, Bradley C, Conn A, Raine A, Twelves C, Falk S, Hopkins K, Tahir S, Dhadda A, Maraveyas A, Sgouros J, Teo M, Ahmad R, Cleator S, Creak A, Lowdell C, Riddle P, Benstead K, Farrugia D, Reed N, Shepherd S, Levine E, Mullamitha S, Saunders M, Valle J, Wilson G, Jones A, Weaver A, Clark PI, Haylock B, Iqbal MI, Myint AS, Smith D, Beesley S, Sevitt T, Nicoll J, Daniel F, Ford V, Talbot T, Butt M, Hamid A, Mack P, Roy R, Osborne R, McKinna F, Alsab H, Basu D, Murray P, Sizer B, Azam FA, Neupane R, Waterston A, Glaholm J, Blesing C, Lowndes S, Medisetti A, Gaya A, Leslie M, Maisey N, Ross P, Dunn G, Al-Salihi O, Wasan HS, Palmer C, Tan LT, Dent J, Hofmann U, Joffe JK, Sherwin E, Soomal R, Chakrabarti A, Joseph S, Van d V, Wadd NJ, Wilson D, Anjarwalia S, Hall J, Hughes R, Polychronis A, Scarffe JH, Hill M, James RD, Shah R, Summers J, Hartley A, Carney D, McCaffrey J, Bystricky B, O’Reilly S, Gupta R, Al-Mishlab T, Gidden F, O’Hara R, Stewart J, Ashford R, Glynne-Jones R, Harrison M, Mawdsley S, Barlow H, Tighe M, Walther J, Neal J, Rees C, Bridgewater J, Karp S, McGovern U, Atherton PJ, El-Deeb H, Macmillan C, Patel K, Bessell EM, Dickinson PD, Potter V, Jephcott C, McAdam K, Wrigley J, Osborne R, Muthuramalingam S, O’Callaghan A, Bridgewater J, Melcher L, Braconi C, Geh JI, Palmer D, Narayana P, Steven N, Gaya A. Common variation near CDKN1A, POLD3 and SHROOM2 influences colorectal cancer risk. Nat Genet. 2012;44:770–6. doi: 10.1038/ng.2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tenesa A, Farrington SM, Prendergast JG, Porteous ME, Walker M, Haq N, Barnetson RA, Theodoratou E, Cetnarskyj R, Cartwright N, Semple C, Clark AJ, Reid FJ, Smith LA, Kavoussanakis K, Koessler T, Pharoah PD, Buch S, Schafmayer C, Tepel J, Schreiber S, Volzke H, Schmidt CO, Hampe J, Chang-Claude J, Hoffmeister M, Brenner H, Wilkening S, Canzian F, Capella G, Moreno V, Deary IJ, Starr JM, Tomlinson IP, Kemp Z, Howarth K, Carvajal-Carmona L, Webb E, Broderick P, Vijayakrishnan J, Houlston RS, Rennert G, Ballinger D, Rozek L, Gruber SB, Matsuda K, Kidokoro T, Nakamura Y, Zanke BW, Greenwood CM, Rangrej J, Kustra R, Montpetit A, Hudson TJ, Gallinger S, Campbell H, Dunlop MG. Genome-wide association scan identifies a colorectal cancer susceptibility locus on 11q23 and replicates risk loci at 8q24 and 18q21. Nat Genet. 2008;40:631–7. doi: 10.1038/ng.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Houlston RS, Webb E, Broderick P, Pittman AM, Di Bernardo MC, Lubbe S, Chandler I, Vijayakrishnan J, Sullivan K, Penegar S, Carvajal-Carmona L, Howarth K, Jaeger E, Spain SL, Walther A, Barclay E, Martin L, Gorman M, Domingo E, Teixeira AS, Kerr D, Cazier JB, Niittymaki I, Tuupanen S, Karhu A, Aaltonen LA, Tomlinson IP, Farrington SM, Tenesa A, Prendergast JG, Barnetson RA, Cetnarskyj R, Porteous ME, Pharoah PD, Koessler T, Hampe J, Buch S, Schafmayer C, Tepel J, Schreiber S, Volzke H, Chang-Claude J, Hoffmeister M, Brenner H, Zanke BW, Montpetit A, Hudson TJ, Gallinger S, Campbell H, Dunlop MG. Meta-analysis of genome-wide association data identifies four new susceptibility loci for colorectal cancer. Nat Genet. 2008;40:1426–35. doi: 10.1038/ng.262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Giraldez MD, Lopez-Doriga A, Bujanda L, Abuli A, Bessa X, Fernandez-Rozadilla C, Munoz J, Cuatrecasas M, Jover R, Xicola RM, Llor X, Pique JM, Carracedo A, Ruiz-Ponte C, Cosme A, Enriquez-Navascues JM, Moreno V, Andreu M, Castells A, Balaguer F, Castellvi-Bel S. Susceptibility genetic variants associated with early-onset colorectal cancer. Carcinogenesis. 2012;33:613–19. doi: 10.1093/carcin/bgs009. [DOI] [PubMed] [Google Scholar]
- 14.Broderick P, Carvajal-Carmona L, Pittman AM, Webb E, Howarth K, Rowan A, Lubbe S, Spain S, Sullivan K, Fielding S, Jaeger E, Vijayakrishnan J, Kemp Z, Gorman M, Chandler I, Papaemmanuil E, Penegar S, Wood W, Sellick G, Qureshi M, Teixeira A, Domingo E, Barclay E, Martin L, Sieber O, Kerr D, Gray R, Peto J, Cazier JB, Tomlinson I, Houlston RS. A genome-wide association study shows that common alleles of SMAD7 influence colorectal cancer risk. Nat Genet. 2007;39:1315–17. doi: 10.1038/ng.2007.18. [DOI] [PubMed] [Google Scholar]
- 15.Wijnen JT, Brohet RM, van Eijk R, Jagmohan-Changur S, Middeldorp A, Tops CM, van Puijenbroek M, Ausems MG, Gomez Garcia E, Hes FJ, Hoogerbrugge N, Menko FH, van Os TA, Sijmons RH, Verhoef S, Wagner A, Nagengast FM, Kleibeuker JH, Devilee P, Morreau H, Goldgar D, Tomlinson IP, Houlston RS, van Wezel T, Vasen HF. Chromosome 8q23.3 and 11q23.1 variants modify colorectal cancer risk in Lynch syndrome. Gastroenterology. 2009;136:131–7. doi: 10.1053/j.gastro.2008.09.033. [DOI] [PubMed] [Google Scholar]
- 16.Talseth-Palmer BA, Brenne IS, Ashton KA, Evans TJ, McPhillips M, Groombridge C, Suchy J, Kurzawski G, Spigelman A, Lubinski J, Scott RJ. Colorectal cancer susceptibility loci on chromosome 8q23.3 and 11q23.1 as modifiers for disease expression in Lynch syndrome. J Med Genet. 2011;48:279–84. doi: 10.1136/jmg.2010.079962. [DOI] [PubMed] [Google Scholar]
- 17.Pittman AM, Webb E, Carvajal-Carmona L, Howarth K, Di Bernardo MC, Broderick P, Spain S, Walther A, Price A, Sullivan K, Twiss P, Fielding S, Rowan A, Jaeger E, Vijayakrishnan J, Chandler I, Penegar S, Qureshi M, Lubbe S, Domingo E, Kemp Z, Barclay E, Wood W, Martin L, Gorman M, Thomas H, Peto J, Bishop T, Gray R, Maher ER, Lucassen A, Kerr D, Evans GR, van Wezel T, Morreau H, Wijnen JT, Hopper JL, Southey MC, Giles GG, Severi G, Castellvi-Bel S, Ruiz-Ponte C, Carracedo A, Castells A, Forsti A, Hemminki K, Vodicka P, Naccarati A, Lipton L, Ho JW, Cheng KK, Sham PC, Luk J, Agundez JA, Ladero JM, de la Hoya M, Caldes T, Niittymaki I, Tuupanen S, Karhu A, Aaltonen LA, Cazier JB, Tomlinson IP, Houlston RS. Refinement of the basis and impact of common 11q23.1 variation to the risk of developing colorectal cancer. Hum Mol Genet. 2008;17:3720–7. doi: 10.1093/hmg/ddn267. [DOI] [PubMed] [Google Scholar]
- 18.Jaeger E, Leedham S, Lewis A, Segditsas S, Becker M, Cuadrado PR, Davis H, Kaur K, Heinimann K, Howarth K, East J, Taylor J, Thomas H, Tomlinson I. Hereditary mixed polyposis syndrome is caused by a 40-kb upstream duplication that leads to increased and ectopic expression of the BMP antagonist GREM1. Nat Genet. 2012;44:699–703. doi: 10.1038/ng.2263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vogt S, Jones N, Christian D, Engel C, Nielsen M, Kaufmann A, Steinke V, Vasen HF, Propping P, Sampson JR, Hes FJ, Aretz S. Expanded extracolonic tumor spectrum in MUTYH-associated polyposis. Gastroenterology. 2009;137:1976–85. doi: 10.1053/j.gastro.2009.08.052. [DOI] [PubMed] [Google Scholar]
- 20.Bulow S, Bjork J, Christensen IJ, Fausa O, Jarvinen H, Moesgaard F, Vasen HF. Duodenal adenomatosis in familial adenomatous polyposis. Gut. 2004;53:381–6. doi: 10.1136/gut.2003.027771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jasperson KW, Tuohy TM, Neklason DW, Burt RW. Hereditary and familial colon cancer. Gastroenterology. 2010;138:2044–58. doi: 10.1053/j.gastro.2010.01.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mongin C, Coulet F, Lefevre JH, Colas C, Svrcek M, Eyries M, Lahely Y, Flejou JF, Soubrier F, Parc Y. Unexplained polyposis: a challenge for geneticists, pathologists and gastroenterologists. Clin Genet. 2012;81:38–46. doi: 10.1111/j.1399-0004.2011.01676.x. [DOI] [PubMed] [Google Scholar]
- 23.Cao X, Hong Y, Eu KW, Loi C, Cheah PY. Singapore familial adenomatous polyposis (FAP) patients with classical adenomatous polyposis but undetectable APC mutations have accelerated cancer progression. Am J Gastroenterol. 2006;101:2810–17. doi: 10.1111/j.1572-0241.2006.00842.x. [DOI] [PubMed] [Google Scholar]
- 24.Filipe B, Baltazar C, Albuquerque C, Fragoso S, Lage P, Vitoriano I, Mao de Ferro S, Claro I, Rodrigues P, Fidalgo P, Chaves P, Cravo M, Nobre Leitao C. APC or MUTYH mutations account for the majority of clinically well-characterized families with FAP and AFAP phenotype and patients with more than 30 adenomas. Clin Genet. 2009;76:242–55. doi: 10.1111/j.1399-0004.2009.01241.x. [DOI] [PubMed] [Google Scholar]
- 25.Rivera B, Gonzalez S, Sanchez-Tome E, Blanco I, Mercadillo F, Leton R, Benitez J, Robledo M, Capella G, Urioste M. Clinical and genetic characterization of classical forms of familial adenomatous polyposis: a Spanish population study. Ann Oncol. 2011;22:903–9. doi: 10.1093/annonc/mdq465. [DOI] [PubMed] [Google Scholar]
- 26.Bisgaard ML, Ripa R, Knudsen AL, Bulow S. Familial adenomatous polyposis patients without an identified APC germline mutation have a severe phenotype. Gut. 2004;53:266–70. doi: 10.1136/gut.2003.019042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Renkonen ET, Nieminen P, Abdel-Rahman WM, Moisio AL, Jarvela I, Arte S, Jarvinen HJ, Peltomaki P. Adenomatous polyposis families that screen APC mutation-negative by conventional methods are genetically heterogeneous. J Clin Oncol. 2005;23:5651–9. doi: 10.1200/JCO.2005.14.712. [DOI] [PubMed] [Google Scholar]
- 28.Cattaneo F, Molatore S, Mihalatos M, Apessos A, Venesio T, Bione S, Grignani P, Nasioulas G, Ranzani GN. Heterogeneous molecular mechanisms underlie attenuated familial adenomatous polyposis. Genet Med. 2007;9:836–41. doi: 10.1097/gim.0b013e31815bf940. [DOI] [PubMed] [Google Scholar]
- 29.Nielsen M, Hes FJ, Nagengast FM, Weiss MM, Mathus-Vliegen EM, Morreau H, Breuning MH, Wijnen JT, Tops CM, Vasen HF. Germline mutations in APC and MUTYH are responsible for the majority of families with attenuated familial adenomatous polyposis. Clin Genet. 2007;71:427–33. doi: 10.1111/j.1399-0004.2007.00766.x. [DOI] [PubMed] [Google Scholar]
- 30.Thirlwell C, Howarth KM, Segditsas S, Guerra G, Thomas HJ, Phillips RK, Talbot IC, Gorman M, Novelli MR, Sieber OM, Tomlinson IP. Investigation of pathogenic mechanisms in multiple colorectal adenoma patients without germline APC or MYH/ MUTYH mutations. Br J Cancer. 2007;96:1729–34. doi: 10.1038/sj.bjc.6603789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Boparai KS, Reitsma JB, Lemmens V, van Os TA, Mathus-Vliegen EM, Koornstra JJ, Nagengast FM, van Hest LP, Keller JJ, Dekker E. Increased colorectal cancer risk in first-degree relatives of patients with hyperplastic polyposis syndrome. Gut. 2010;59:1222–5. doi: 10.1136/gut.2009.200741. [DOI] [PubMed] [Google Scholar]
- 32.Houlston RS, Cheadle J, Dobbins SE, Tenesa A, Jones AM, Howarth K, Spain SL, Broderick P, Domingo E, Farrington S, Prendergast JG, Pittman AM, Theodoratou E, Smith CG, Olver B, Walther A, Barnetson RA, Churchman M, Jaeger EE, Penegar S, Barclay E, Martin L, Gorman M, Mager R, Johnstone E, Midgley R, Niittymaki I, Tuupanen S, Colley J, Idziaszczyk S, Thomas HJ, Lucassen AM, Evans DG, Maher ER, Maughan T, Dimas A, Dermitzakis E, Cazier JB, Aaltonen LA, Pharoah P, Kerr DJ, Carvajal-Carmona LG, Campbell H, Dunlop MG, Tomlinson IP. Meta-analysis of three genome-wide association studies identifies susceptibility loci for colorectal cancer at 1q41, 3q26.2, 12q13.13 and 20q13.33. Nat Genet. 2010;42:973–7. doi: 10.1038/ng.670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tomlinson IP, Webb E, Carvajal-Carmona L, Broderick P, Howarth K, Pittman AM, Spain S, Lubbe S, Walther A, Sullivan K, Jaeger E, Fielding S, Rowan A, Vijayakrishnan J, Domingo E, Chandler I, Kemp Z, Qureshi M, Farrington SM, Tenesa A, Prendergast JG, Barnetson RA, Penegar S, Barclay E, Wood W, Martin L, Gorman M, Thomas H, Peto J, Bishop DT, Gray R, Maher ER, Lucassen A, Kerr D, Evans DG, Schafmayer C, Buch S, Volzke H, Hampe J, Schreiber S, John U, Koessler T, Pharoah P, van Wezel T, Morreau H, Wijnen JT, Hopper JL, Southey MC, Giles GG, Severi G, Castellvi-Bel S, Ruiz-Ponte C, Carracedo A, Castells A, Forsti A, Hemminki K, Vodicka P, Naccarati A, Lipton L, Ho JW, Cheng KK, Sham PC, Luk J, Agundez JA, Ladero JM, de la Hoya M, Caldes T, Niittymaki I, Tuupanen S, Karhu A, Aaltonen L, Cazier JB, Campbell H, Dunlop MG, Houlston RS. A genome-wide association study identifies colorectal cancer susceptibility loci on chromosomes 10p14 and 8q23.3. Nat Genet. 2008;40:623–30. doi: 10.1038/ng.111. [DOI] [PubMed] [Google Scholar]
- 34.Tomlinson I, Webb E, Carvajal-Carmona L, Broderick P, Kemp Z, Spain S, Penegar S, Chandler I, Gorman M, Wood W, Barclay E, Lubbe S, Martin L, Sellick G, Jaeger E, Hubner R, Wild R, Rowan A, Fielding S, Howarth K, Silver A, Atkin W, Muir K, Logan R, Kerr D, Johnstone E, Sieber O, Gray R, Thomas H, Peto J, Cazier JB, Houlston R. A genome-wide association scan of tag SNPs identifies a susceptibility variant for colorectal cancer at 8q24.21. Nat Genet. 2007;39:984–8. doi: 10.1038/ng2085. [DOI] [PubMed] [Google Scholar]
- 35.Jaeger E, Webb E, Howarth K, Carvajal-Carmona L, Rowan A, Broderick P, Walther A, Spain S, Pittman A, Kemp Z, Sullivan K, Heinimann K, Lubbe S, Domingo E, Barclay E, Martin L, Gorman M, Chandler I, Vijayakrishnan J, Wood W, Papaemmanuil E, Penegar S, Qureshi M, Farrington S, Tenesa A, Cazier JB, Kerr D, Gray R, Peto J, Dunlop M, Campbell H, Thomas H, Houlston R, Tomlinson I. Common genetic variants at the CRAC1 (HMPS) locus on chromosome 15q13.3 influence colorectal cancer risk. Nat Genet. 2008;40:26–8. doi: 10.1038/ng.2007.41. [DOI] [PubMed] [Google Scholar]