Abstract
Anaplastic lymphoma kinase (ALK) physiologically expressed only by nervous system cells displays remarkable capacity to transform CD4+ T lymphocytes and other types of non-neural cells. Here we report that activity of nucleophosphmin (NPM)/ALK chimeric protein, the dominant form of ALK expressed in T-cell lymphomas (ALK+TCL), closely resembles cell activation induced by interleukin 2 (IL-2), the key cytokine supporting growth and survival of normal CD4+ T lymphocytes. Direct comparison of gene expression by ALK+TCL cells treated with an ALK inhibitor and IL-2-dependent ALK-TCL cells stimulated with the cytokine revealed a very similar, albeit inverse, gene regulation pattern. Depending on the analysis method, up to 67% of the modulated genes could be defined as modulated in common by NPM/ALK and IL-2. Based on the gene expression patterns, Jak/STAT and IL-2 signaling pathways topped the list of pathways identified as affected by both IL-2 and NPM/ALK. The expression dependence on NPM/ALK and IL-2 of the five selected genes: CD25 (IL-2Rα), Egr-1, Fosl-1, SOCS3, and Irf-4 was confirmed at the protein level. In both ALK+TCL and IL-2-stimulated ALK-TCL cells, CD25, SOCS3, and Irf-4 genes were activated predominantly by the STAT5 and STAT3 transcription factors, while transcription of Egr-1 and Fosl-1 was induced by the MEK-ERK pathway. Finally, we found that Egr-1, a protein not associated previously with either IL-2 or ALK, contributes to the cell proliferation. These findings indicate that NPM/ALK transforms the target CD4+ T lymphocytes, at least in part, by utilizing the pre-existing, IL-2-dependent signaling pathways.
Introduction
Anaplastic lymphoma kinase (ALK) is physiologically expressed only in certain immature neuronal cells (1). However, aberrant expression of ALK has been identified in a number of histologically diverse malignancies including T- and B-cell lymphomas, inflammatory myofibroblastic tumors, neuroblastomas and carcinomas of lung and other organs (1–3). T-cell lymphomas (TCL) that express ALK are recognized as a distinct category of lymphoma. Ectopic expression of ALK in the affected CD4+ T lymphocytes is the result of chromosomal translocations involving the ALK gene and several different partners, most frequently by far the nucleophosmin (NPM) gene (3). The NPM/ALK chimeric protein is not only constitutively expressed but also is persistently activated through autophosphorylation (4, 5). NPM/ALK displays potent cell-transforming properties as demonstrated both in vitro (4, 6) and in vivo (7, 8). NPM/ALK mediates its oncogenicity by activating a number of cell-signaling pathways including STAT3, STAT5, and MEK/ERK (9–11). Chronic activation of these signal transmitters leads to persistent expression of genes, the protein products of which are involved in such key cell functions as promotion of cell proliferation and protection from apoptotic cell death. However, the fundamental question of how ALK, the tyrosine kinase physiologically expressed exclusively by the neural cells, is able to transform non-neural cells such as CD4+ T lymphocytes remains unanswered.
IL-2 and functionally related cytokines signal through receptors that share the common γ (IL-2Rγ) chain (12, 13) and are critical for maturation, proliferation and survival of the normal CD4+ T lymphocytes and other immune cells (14–17). Analysis of the intracellular signaling pathways indicate that many of these IL-2-regulated cell functions are primarily mediated by the MEK-ERK, phosphatidylinositol 3-kinase (PI3K)-Akt, and STAT5 pathways (18–27).
Here we report that NPM/ALK-induced cell signaling results in a distinct gene expression pattern that closely resembles the gene expression changes induced by IL-2. These findings indicate that NPM/ALK succeeds in transforming the target CD4+ T lymphocytes at least in part by mimicking the effects of IL-2, the natural key regulator of these cells.
Materials and Methods
ALK- and ALK+ALCL Cell Lines
NPM/ALK-expressing Sudhl-1, JB6, Sup-M2, Karpas 299 cell lines were derived from ALK+TCL patients (9–11, 28, 29). IL-2-dependent T cell lines Sez-4 and SeAx were derived from ALK-TCL patients (30). The cell lines were cultured at 37 °C and 5% CO2 in RPMI medium 1640, supplemented with 2-mM L-glutamine, 10% heat-inactivated FBS (FBS), 1% penicillin/streptomycin mixture and, where applicable, 200 units of IL-2 (Bender MedSystems).
Microarray Analysis
The ALK+TCL Sudhl-1 cell line was treated in triplicates with the ALK inhibitor, CEP-14083, or the compound’s solvent for 6 h. The Sez-4 cell line was starved of IL-2 for 16 h and placed into six-well plates in 10 mL RPMI/10% FBS for 2 h followed by addition of IL-2 (200 U) or medium alone for 4 h. The isolated RNA was reverse-transcribed, biotin-labeled, and hybridized to the U133 Plus 2.0 array chips (Affymetrix) as described (30). Microarray data were normalized using the MAS5 algorithm and analyzed using Partek GS (Partek). Differentially expressed genes were identified using ANOVA. A gene list with a 5% false discovery rate (FDR = 0.05) was used to identify the target genes common for both NPM/ALK and IL-2. Hierarchical clustering was performed by Cluster 3.0 and presented using Java Tree View 1.1.0. The Ingenuity and Gene Ontology data bases were used for functional analysis of gene lists.
Western blot and antibodies
The cells were washed, centrifuged, and lysed in lysis buffer supplemented with 0.5 mmol/L phenylmethylsulfonyl fluoride, phosphatase inhibitor cocktails I and II from Sigma and protease inhibitor cocktail from Roche, according to the manufacturer’s specifications. For normalization of gel loading, the protein extracts were assayed by the Lowry method (Bio-Rad Dc protein assay). Typically, 10 to 50 μg of the protein per lane was loaded. To detect total proteins, we used anti-Egr-1 (Cell Signaling); anti-Fosl-1, -Socs3, -Irf-4 and anti-actin (all Santa Cruz) antibodies. To examine protein phosphorylation, the membranes were incubated with the antibodies specific for STAT3 Y705, STAT5 Y694 and ERK1/2 T202/Y204 (all from Cell Signaling). The membranes were incubated with the appropriate secondary peroxidase-conjugated antibodies. The blots were developed using the enhanced chemiluminescence (ECL) plus Western blotting detection system from Amersham.
Flow Cytometry
Cells (0.5 × 106) were washed in 1xPBS and stained for 20 min with mouse antibody against CD25 (dilution 1:10, FITC, BD PharMingen) or FITC-labeled mouse IgG1 isotype controls. The stained cells after double washing in 1xPBS were applied to the flow cytometer (FACSCalibur; Becton Dickinson), and 20,000 events were analyzed. Results of the cell staining are presented as the value of mean channel signal from three experiments.
Enzyme immunoassay (EIA)
To detect the secreted, soluble form of CD25 (sCD25), we used commercially available sCD25 (sIL-2Rα) human ELISA kit (Quantikine, DR2A00, R&D Systems) according to the manufacturer’s instructions. Briefly, samples were diluted 10-fold, applied in duplicates in 50 μl per well of the microplate pre-coated with first CD25 antibody, and incubated for 3 h with 100 ul of second CD25 antibody-peroxidase conjugate. After washing, substrate solution (200 ul) was added for 20 mins, followed by addition of stop solution (50 μl). Optical Density (O.D.) was measured at 450nm, using a microplate reader (Titertek Multiskan). Standards of known sCD25 concentrations were analysed and used as reference.
Kinase inhibitors
A potent ALK inhibitor CEP-14083 and its structurally-related ALK noninhibitory counterpart CEP-11988, both used at the dose of 175 nM, have been described previously (28). Pan-Jak (Jak I; Calbiochem) is a quinolin derivative with the structure 2-(1,1-dimethylethyl)-9-fluoro-3,6-dihydro-7H-benz(h)-imidaz(4,5-f) isoquinolin-7-one (30). It inhibits enzymatic function of all four members of the Jak family with an IC50 of 15 nmol/L for Jak1, 1 nmol/L for Jak2, 5 nmol/L for Jak3, and 1 nmol/L for Tyk2 in in vitro kinase activity inhibition assay. The inhibitor preferentially targeting Jak3 (30) displays an in vitro IC50 kinase activity inhibition of Jak3 at 2 nmol/L for Jak3, 20 nmol/L for Jak2, and 100 nmol/L for Jak1. U0126 (Promega, Madison, WI, USA) and PD98059 (Calbiochem, La Jolla, CA, USA) are MEK1/2 inhibitors and were used at concentrations of 15 and 20 μM, respectively (29).
Small interfering RNA assay
A mixture of four ERK1, ERK2, STAT3, STAT5a or STAT5b-specific small interfering RNA (siRNA) or nonsense siRNA (all purchased from Dharmacon) was introduced into cells at 0.1–10 nmol/L by lipofection with lipofectamine (DMRIE-C; Invitrogen). Cells were harvested after 48–96h culture in the medium containing IL-2, in the case of Sez-4 cell line.
Cell proliferation assay
Forty eight h after the knock-down of Egr-1 cell proliferation was evaluated by detection of bromodeoxyuridine (BrdU) incorporation using the commercially available kit cell proliferation ELISA (Roche) according to the manufacturer’s protocol. In brief, cells were seeded into 96-well plates (Corning) at a concentration of 5 × 103 cells per well in RPMI medium supplemented with 10% FBS and labeled with BrdU (Roche) for 4 h. After centrifugation (10′ at 300 x g), supernatant removal, and plate drying, the cells were fixed and the DNA was denaturated by the addition of 200 mL FixDenat reagent. The amount of incorporated BrdU was determined by incubation with a specific antibody conjugated with peroxidase followed by colorimetric conversion of the substrateand absorbance evaluation in the ELISA plate reader.
Results
Gene expression profile is similar in IL-2-stimulated ALK-TCL and ALK+TCL cells
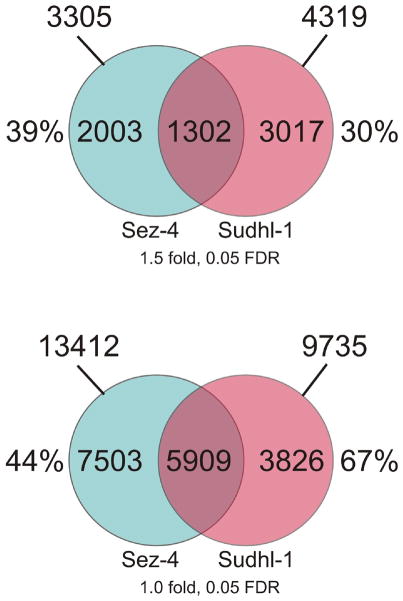
Previous studies indicate that several key signal transduction pathways including Jak-STAT, MEK-ERK, and mTORC1 are aberrantly activated in both IL-2-dependent ALK-TCL and ALK+TCL cells (9–11, 28–30). To further explore the apparent similarity, we analyzed these and additional pathways by comparing genome-wide gene expression patterns in ALK-TCL cells in response to IL-2 and in ALK+TCL cells in response to ALK inhibition. By applying a filter threshold of at least 1.5 fold change in gene expression and a false discovery rate (FDR) of 0.05, we identified 3305 genes modulated by IL-2 in the ALK-TCL-derived Sez-4 cells and 4319 genes affected by ALK inhibition in ALK+TCL Sudhl-1 cells (Fig. 1A). Among these two sets of gene transcripts, 1302 were shared and accounted for 39% and 30% of all modulated genes in the Sez-4 and Sudhl-1 cells, respectively. When no fold treshhold and a FDR coefficient lower than 0.05 were applied, we found expression of 13412 and 9735 genes to be changed in Sez-4 and Sudhl-1, respectively. Strikingly, 5909 of these genes were regulated simultaneously in both cell types, representing 44% and 67% of all modulated genes in Sez-4 and Sudhl-1 cells, respectively (Fig. 1B). Not surprisingly given the emerging similarity between IL-2-induced and NPM/ALK signaling, almost all of the jointly regulated genes were modulated in the opposite direction in response to IL-2 stimulation and ALK inhibition. Fig. 2 depicts the set of 155 common genes, expression of which was the most affected (> 2 fold change in expression; FDR 0.05) by the cell modulation. For some of the genes up to four different probes were present in the array and, as can be seen, they tend to cluster together indicating the high technical quality of the data.
FIGURE 1.
Genes regulated in common by IL-2 and NPM/ALK. The ALK-TCL derived, IL-2 dependent Sez-4 cell line was cultured in medium deprived of IL-2 for 18h and subsequently re-stimulated with IL-2 (200 units) for 4h. The ALK+TCL derived Sudhl-1 cell line was treated with an ALK-specific inhibitor (175nM) for 6h. Cells were harvested and total RNA reverse transcribed to cDNA and hybrydized to Affymetrix HG-U133 Plus 2 arrays containing DNA probes for >47,000 genes. The results were analyzed using GeneSpring and Partek Genomics Suite software. Blue circles: genes modulated in the Sez-4 cell line in response to IL-2 (cut off: Fold change >1.5(A), no fold change used as selection criteria (B) and FDR>0.05 (A and B). Pink circles: genes modulated in the Sudhl-1 cell line in response to the ALK inhibitor (cut off: Fold change >1.5(A), no fold change used as selection criteria (B) and FDR>0.05 (A and B)). Areas where two circles overlap represents the commonly regulated 1302 and 5909 genes between both cell lines. The experiments were performed in triplicates (N=3). The entire microarray data sets are available in the NIH database GEO at www.ncbi.nlm.nih.gov/geo under the accession numbers GSE8685 and GSE50803.
FIGURE 2.
Hierarchical clustering of 155 commonly regulated transcripts. The graph represents an intensity plot of 155 overlapping transcripts with opposite direction of regulation of expression in Sez-4 and Sudhl-1 cell lines after tratement. Genes were selected using a 1.5 fold cut off and p values < 0.05. Analysis was performed with Cluster 3.0 and Java TreeView 1.1.0. The results are derived from the two triplicate studies summarized in Figure 1.
IL-2 and NPM/ALK activate overlapping spectrum of signaling pathways
By analyzing the genes regulated in common using Gene Ontology and KEGG databases, we assigned these genes into cell signaling pathways (Table 1). When sorted according to the highest probability (the lowest p-value) two signaling pathways: Jak/STAT and IL-2 topped the list. Although reassuring, activation of IL-2 signaling in Sez-4 was totally predictable given that IL-2 was indeed used as the stimulus in these cells. However, the similar designation of the gene expression re-programming induced by NPM/ALK further indicated that this oncogenic kinase activates the same signaling pathways as IL-2. Interestingly, another top jointly regulated pathways was the Circardian Rhythm signaling reported to be involved in the pathogenesis of leukemias and lymphomas (31). Finally, identification of p53 signaling as another highly regulated pathway is in agreement with the reported ability of NPM/ALK to inhibit p53 via MDM2 and JNK (32).
Table 1.
Signaling pathways activated in common by IL-2 and NPM/ALK
| Pathway | p-value |
|---|---|
| JAK/Stat Signaling | 1.33E-02 |
| IL-2 Signaling | 1.49E-02 |
| Circadian Rhythm Signaling | 1.61E-02 |
| p53 Signaling | 1.92E-02 |
IL-2- and NPM/ALK- modulated genes are translated into proteins
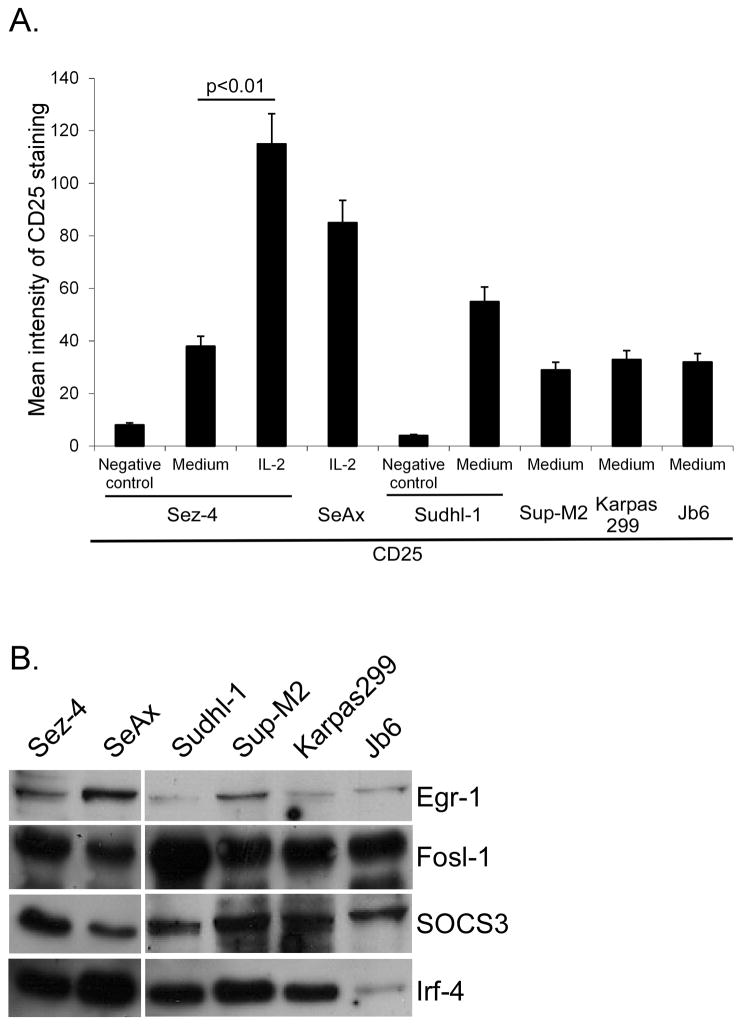
To affirm the biological relevance of the genomic analysis we examined the expression of five selected genes regulated by both IL-2 and ALK on the protein level. These genes: CD25 (IL-2Rα), Egr-1, Fosl-1, SOCS3, and Irf-4 were selected based on either their known functional importance or novelty, at least in the context of IL-2 and ALK signaling. By using flow cytometry to detect CD25 and Western blotting to detect the other remaing genes, we identified expression of all five proteins not only in Sez-4 and Sudhl-1 cells but also in another IL-2 dependent ALK-TCL cell line SeAx as well as three additional ALK+TCL cell lines listed in Fig. 3A and 3B.
FIGURE 3.
Expression of proteins encoded by genes regulated in common by both IL-2 and NPM/ALK. IL-2 -dependent ALK-TCL derived cell lines Sez-4 and SeAx were starved of IL-2 for 16 h (A) and subsequently stimulated by medium alone or IL-2 (200U) for 12 h. ALK+TCL derived cell lines Sudhl-1, Sup-M2, Karpas299 and Jb6 were cultured in normal growth media (A and B). Expression levels of CD25 were assesed by surface staining with FITC-labeled mouse antibody against CD25 or mouse IgG1 isotype controls and analysed using flow cytometry. The cell lysates were directly analyzed by Western blot with specific antibodies against Egr-1, Fosl-1, SOC3 and Irf-4 (B). The depicted experiments are representative of three independent experiments (N=3).
IL-2 and NPM/ALK induce CD25 expression through STAT5 and STAT3
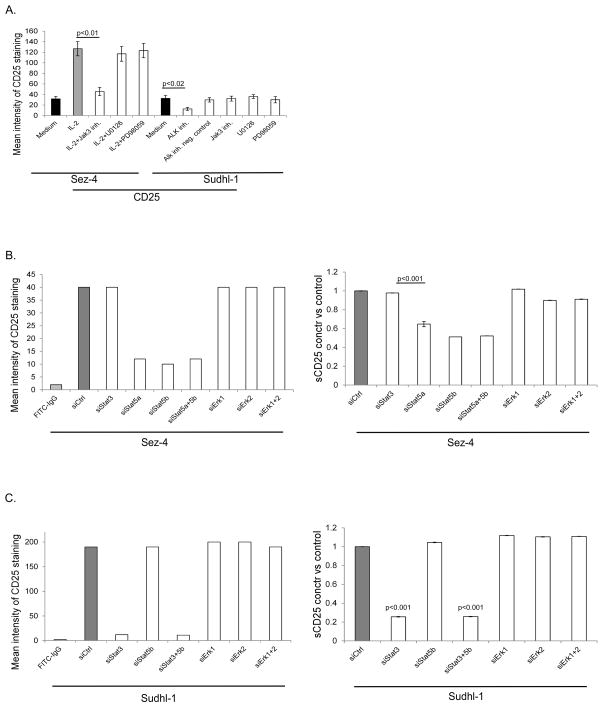
To confirm the role of IL-2 and ALK in inducing expression of CD25, we exposed Sez-4 cells to IL-2 and Sudhl-1 cells to ALK inhibitor. Indeed, IL-2 stimulation markedly enhanced and ALK inhibition greatly diminished expression of CD25 (Fig. 4A). To identify cell signaling pathways involved in induction of CD25 expression down-stream of IL-2R and NPM/ALK, we applied an inhibitor against the Jak family with the highest activity against Jak3 as well as two different inhibitors of MEK, all used at the pre-tested effective doses (Fig. 4A). While the Jak inhibitor profoundly diminished CD25 expression in the IL-2-treated Sez-4 cells in accordance with the key role of Jak3 in IL-2 signaling (28), it had no effect on CD25 expression in Sudhl-1 cells, confirming our previous finding that NPM-ALK signaling is Jak3-independent (29). Inhibition of MEK by two different inhibitors did not affect CD25 expression in either Sez-4 or Sudhl-1 cells, indicating that the MEK-ERK pathway is not involved in the regulation of CD25 expression.
FIGURE 4.
Involvement of STAT and MEK-ERK pathways in regulation of CD25 expression. The IL-2-dependent Sez-4 cell line was starved of IL-2 for 16 h and stimulated by medium alone or IL-2 for 12 h. Inhibitors of Jak and MEK (U0126 and PD98059) were added to the culture media 1 h before the IL-2 stimulation (A). The Sudhl-1 cell line was cultured for 8 h in the presence of an ALK inhibitor, a non-active formulation of the ALK inhibitor (negative control) or Jak/STAT and MAPK inhibitors (A). Alternatively, siRNA-induced specific depletion of the listed targets in both cell types with nonsense siRNA serving as control (B and C). Surface expression of CD25 was visualized by specific staining with FITC-labeled antibody against CD25 and analysed by flow cytometry (left panels). The concentration of soluble CD25 (sCD25) was determined by EIA (right panels). The depicted flow cytometry experiments are representative of three independent experiments (N=3); EIA to detect sCD25 was performed twice (N=2), each time the samples were analyzed in duplicates (N=2).
Because STATs, in particular STAT5, have been implicated in the induction of CD25 (33, 34), we next examined if their siRNA-induced depletion affects CD25 expression in the Sez-4 and Sudhl-1 cells. Indeed, depletion of STAT5a and STAT5b (supplementary Fig. S1, upper panel), markedly diminished CD25 expression in Sez-4 cells, examined at both cell-surface (Fig. 4B, left panel and Fig. S2, left panel column) and soluble (sCD25) level (Fig. 4B, right panel). In agreement with the MEK inhibition data (Fig. 4A), depletion of ERK1 and ERK2 had no effect on CD25 expression. Whereas ALK+TCL including Sudhl-1 typically fail to express STAT5A due to epigenetic silencing of its gene (35), they weakly express STAT5B (35, 36) and very strongly STAT3 (9, 28). While depletion of STAT5B in Sudhl-1 cells (supplementary Fig. S1, lower panel), did not significantly affect CD25, depletion of STAT3 had a profound inhibitory effect on CD25 expression (Fig. 4C and Fig. S2, right panel column). As in Sez-4 cells, depletion of ERK1 and ERK2 exerted no significant effect on CD25. These data indicate that the STAT family members, but not the MEK-ERK pathway, induce CD25 expression in both ALK-TCL and ALK+TCL. While STAT5 plays a key role in the former cell type, STAT3 is critical for CD25 expression in the latter cell type.
STAT and MEK-ERK pathways play similar role in modulation of specific genes in both ALK-and ALK+TCL
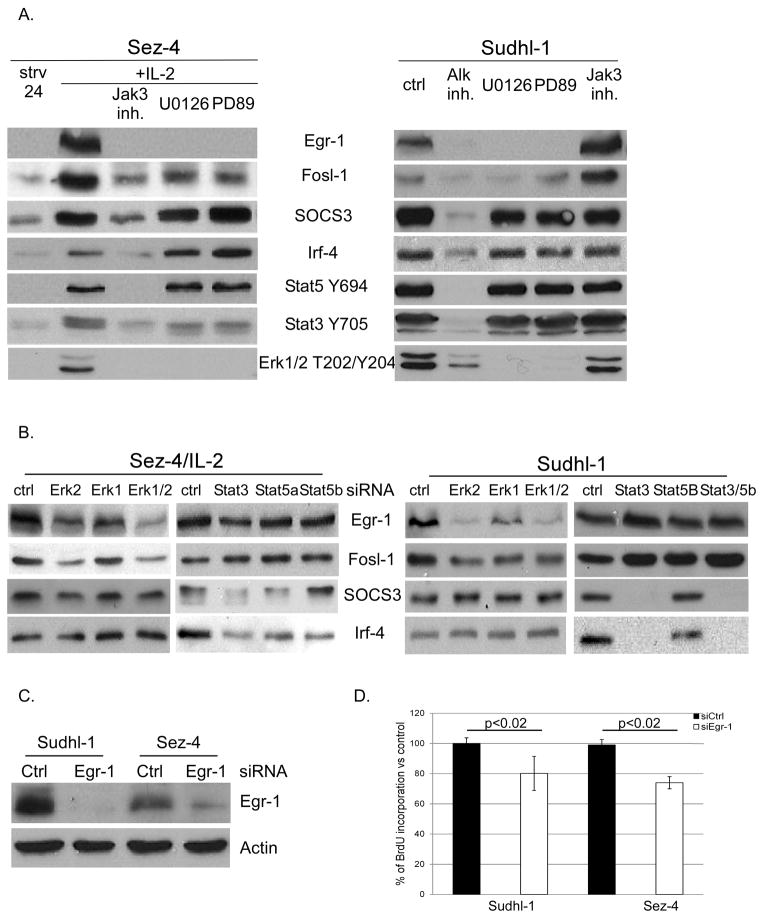
To identify the pathways triggering expression of the other selected genes: Egr-1, Fosl-1, SOCS3 and Irf-4, we also employed small molecule inhibitors and siRNA technology. Exposure of the IL-2-stimulated Sez-4 cells to the Jak inhibitor preferentially active against Jak3, profoundly suppressed expression of all four of the gene protein products with inhibition of STAT5, STAT3, and ERK phosphorylation serving as positive control (Fig. 5A, left panel). In addition, expression of Egr-1 and Fosl-1, but not SOCS3 and Irf-4, was inhibited by both MEK inhibitors. Exposure of the NPM/ALK-expressing Sudhl-1 cells to the ALK inhibitor, but not the Jak inhibitor, profoundly suppressed expression of all four proteins as well as phosphorylation of STAT5, STAT3, and ERK (Fig. 5A, right panel). Similar to Sez-4 cells, expression of both Egr-1 and Fosl-1, but not SOCS3 and Irf-4, was suppressed by the MEK inhibitors. siRNA-mediated depletion of ERK1 and ERK2 in Sez-4 and Sudhl-1 cells also adversly affected expression of Egr-1 and Fosl-1, but not of SOCS3 and Irf-4 (Fig. 5B). In contrast, depletion of STAT3 suppressed expression of SOCS3 and Irf-4 but not Egr-1 and Fosl-1 in Sez-4 and Sudhl-1 cells. These data indicate that expression of Egr-1 and Fosl-1 is regulated by the MEK-ERK pathway and of SOCS3 and Irf-4 by STAT pathway. Furthermore, expression of these selected and representative genes is indeed IL-2 and NPM/ALK dependent.
FIGURE 5.
Involvement of STAT and MEK-ERK pathways in regulation of selected IL-2- and NPM/ALK-dependent genes. Sez-4 and Sudhl-1 cell lines were treated as described in Fig. 4. Cells were lysed and analyzed by Western blot with specific antibodies against Egr-1, Fosl-1, SOC3 and Irf-4. The extent of inhibition of Jak/STAT and MAPK pathways was visualized using phospho-specific antibodies against STAT3, STAT5 and ERK1/2 (A, B). Depletion of Egr-1 was achieved using the specific siRNA with nonsense siRNA serving as control (C). The effect of Egr-1 depletion on cell proliferation was assesed in BrdU incorporation assay (D). The Western blot experiments are representative of at least three (N=3; panel A) or two (N=2; panel B) independent experiments. BrdU uptake was measured twice (N=2), each time performed in triplicates (N=3).
Egr-1 plays a role in growth of ALK- and ALK+TCL cells
Egr-1 is one of the most strongly regulated genes in both ALK- and ALK+TCL cells with its upregulation of up to 25 times in response to IL-2 and downregulation of up to 28 times in response to the ALK inhibition (Fig. 2 and data not presented). Egr-1 has been implicated in the carcinogenesis (37) as well as biology of normal T lymphocytes (38). However, its role in malignant T lymphocytes remains undefined. To explore Egr-1 function in the lymphoma cells, we depleted Egr-1 in Sez-4 and Sudhl-1 cells (Fig. 5C) and evaluated its impact on their proliferative rate. As can be seen in Fig. 5D, Egr-1 loss decreased to a similar degree proliferation of both cell types indicating that this transcription factor contributes to the cell growth-promoting properties of both IL-2 and NPM/ALK.
Discussion
While the oncogenic properties of aberrantly expressed and activated ALK, in particular of its NPM/ALK variant are well recognized (1–3), the question of how this kinase normally expressed solely in neural cells suceeds in transforming CD4+ T lymphocytes and other non-neural target cells remains only partially answered.
Previous studies indicate that NPM/ALK mediates its oncogenicity by activating a number of cell-signaling pathways including STAT3, STAT5, MEK/ERK, PI3K/Akt and mTORC1 (9–11). In retrospect, these signaling pathways are also activated by IL-2 (18–27). Through a series of focused experiments using ALK+TCL and IL-2-dependent ALK-TCL cells, we now provide direct evidence that NPM/ALK activity closely resembles the effects of IL-2 stimulation. As we show, NPM/ALK and IL-2 activate similar signal transduction pathways and regulate expression of many of the same genes. It can, therefore, be argued that NPM/ALK “hijacks” in target CD4+ T lymphocytes the pre-existing IL-2-triggered cell signaling pathways that are critical for growth and survival of these cells under physiological conditions. Given that NPM/ALK is constitutively activated through autophosphorylation, its expression leads to chronic activation of the pathways, rather than the transient one as is the case with IL-2 and other cytokines. In consequence, the NPM/ALK-transformed cells display perpetual growth and survival; the key features of malignant cells.
It is interesting in this context that ALK+TCL cells display loss of the common γ (IL-2Rγ) chain (39), the key component of the cell-surface receptor complex activated by IL-2 and functionally similar cytokines. This IL-2Rγ expression loss is due to epigenetic silencing of the IL-2Rγ gene induced by the NPM/ALK itself (39). Together with our current observation of NPM/ALK-mediated cell signaling recapitulating IL-2 function this finding indicates that NPM/-ALK transforms the target CD4+ T cells by rendering them independent of external stimuli provided by IL-2 type cytokines, on one hand, and by constitutively activating intracellular signaling pathways normally regulated by these cytokines, on the other hand. These combined features may explain to a large degree the remarkable potency of NPM/ALK as an oncogene (1–4, 6–8).
Whereas strikingly similar, the activities of NPM/ALK and IL-2 in ALK+TCL and ALK-TCL, respectively, do not seem to be identical. While STAT3 plays the critical role in NPM/ALK-induced oncogenesis, STAT5 signaling seems more important in the IL-2 signaling including the IL-2-dependent ALK-TCL cells. Of note, the STAT5A gene is epigenetically silenced in ALK+TCL (35). Our data demonstrating that in IL-2-stimulated Sez-4 cells STAT5 (Fig. 4B) and in the NPM-ALK-transformed Sudhl-1 cells STAT3 (Fig. 4C) play the key role in inducing CD25 expression, indicates that STATs may, at least in some instances, substitute for each other, possibly depending on their relative concentraion and activation status part compensate for loss of STAT5A expression. Furthermore, a previous study has found similarities between signaling induced by NPM/ALK and the T-cell receptor (40), most notaby activation by both of the NFAT transcription factor. This finding indicates that, in addition to IL-2-type signaling, NPM/ALK mimics some aspects of signaling from another receptor complex that is critical for function of normal CD4+ T lymphocytes. Similar to IL-2Rγ, ALK+TCL cells display NPM/ALK-dependent epigenetic gene silencing and, hence, loss of expression of several members of the T-cell receptor complex including the T-cell receptor itself, its partner CD3, and the key down-stream kinase ZAP-70 (41). This observation suggests that NPM/ALK inhibits in the transformed CD4+ T lymphocytes also the stimuli generated by the T-cell receptor complex, in addition to inhibiting cell signals normally provided by IL-2-type cytokines.
Among the genes regulated in common by NPM/ALK and IL-2 that attracted our attention was Egr-1. This transcription factor can be induced in diverse tissues by a spectrum of growth factors and stress stimuli (37, 38). Interesingly, Egr-1 has been implicated in both promotion and inhibition of carcinogenesis indicting that its activity is complex and dependent on the cellular and signaling context. Accordingly, Egr-1 has been shown to induce proliferation of prostatic carcinoma cells (37). However, in colorectal carcinoma cells exposed to Cox inhibitors Egr-1 expression promotes cell apoptosis (42). In regard to normal T lymphocytes, Egr-1 is induced in response TCR signaling and promotes synthesis of cytokines including IL-2 and TNFa (38). Our findings that Egr-1 expression can be induced by IL-2 as well as NPM/ALK are novel and have potential basic and translational implications. While the role of such induction in normal T lymphocytes needs to be established, our observation that Egr-1 contributed to growth of malignant T cells from both ALK- and ALK+TCL (Fig. 5D) suggests that Egr-1 may become a therapeutic target in these and, possibly, other types of T-cell lymphoma.
There is accumulating evidence that IL-2-type signaling may play a key role in the pathogenesis of not only ALK+TCL but also other types of T-cell lymphoma. These lymphomas include cutaneous T-cell lymphoma (43, 44) and adult type T-cell leukemia/lymphoma (ATLL; 45) in which aberrant activation of the IL-2 dependent pathways, in particular of the Jak/STAT pathway is well documented. Of interest, activating mutations of Jak3 in its FERM domain have been identified in tumors from four out of thirty six ATLL patients examined (46) providing additional evidence that this kinase, and thus IL-2-dependent signaling, promotes lymphomagenesis. More recently, another set of Jak3 activating mutations within the pseudokinase domain has been found in tumor tissues from twenty three out of sixty five patients with NK/T-cell lymphoma (47) further expanding the list of lymphomas with dysregulation of IL-2-type signaling. In retrospect, this emerging reliance on IL-2 signaling by the various types of T-cell malignancies may not be that suprising, given the critical role of cytokines from the IL-2 family in maturation and functional differentiation of normal T lymphocytes. This dependence on IL-2 signaling has potentially important translational implications with the key kinases in the pathway representing attractive therapeutic targets. While in ALK+TCL NPM/ALK itself can be effectively targeted by small molecule inhibitors with the preliminary clinical findings being very encouraging (48), in the other types of lymphoma Jak1 and Jak3, the kinases transmitting the stimuli generated by IL-2 and similar cytokines, should be equally attractive targets. This may be particularly true for the lymphomas which carry activating mutation of Jak3 (46, 47).
Supplementary Material
Acknowledgments
Supported by the NCI grants R01-CA89194 and R01-CA96856, Leukemia and Lymphoma Society grant 6100-09, and Fran and Jim Maguire Fund.
Footnotes
Disclosures
The authors have no conflicts of interest.
References
- 1.Li R, Morris SW. Development of anaplastic lymphoma kinase (ALK) small-molecule inhibitors for cancer therapy. Med Res Rev. 2008;28:372–412. doi: 10.1002/med.20109. [DOI] [PubMed] [Google Scholar]
- 2.Chiarle R, Voena C, Ambrogio C, Piva R, Inghirami G. The anaplastic lymphoma kinase in the pathogenesis of cancer. Nat Rev Cancer. 2008;8:11–23. doi: 10.1038/nrc2291. [DOI] [PubMed] [Google Scholar]
- 3.Wasik MA, Zhang Q, Marzec M, Kasprzycka M, Wang HY, Liu X. Anaplastic lymphoma kinase (ALK)-induced malignancies: novel mechanisms of cell transformation and potential therapeutic approaches. Semin Oncol. 2009;36:S27–S35. doi: 10.1053/j.seminoncol.2009.02.007. [DOI] [PubMed] [Google Scholar]
- 4.Fujimoto J, Shiota M, Iwahara T, Seki N, Satoh H, Mori S, Yamamoto T. Characterization of the transforming activity of p80, a hyperphosphorylated protein in a Ki-1 lymphoma cell line with chromosomal translocation t(2;5) Proc Natl Acad Sci U S A. 1996;93:4181–4186. doi: 10.1073/pnas.93.9.4181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Morris SW, Kirstein MN, Valentine MB, Dittmer KG, Shapiro DN, Saltman DL, AT Fusion of a kinase gene, ALK, to a nucleolar protein gene, NPM, in non-Hodgkin’s lymphoma. Science. 1994;263:1281–1284. doi: 10.1126/science.8122112. [DOI] [PubMed] [Google Scholar]
- 6.Bischof D, Pulford K, Mason DY, Morris SW. Role of the nucleophosmin (NPM) portion of the non-Hodgkin’s lymphoma-associated NPM-anaplastic lymphoma kinase fusion protein in oncogenesis. Mol Cell Biol. 1997;17:2312–2325. doi: 10.1128/mcb.17.4.2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kuefer MU, Look AT, Pulford K, Behm FG, Pattengale PK, Mason DY, Morris SW. Retrovirus-mediated gene transfer of NPM-ALK causes lymphoid malignancy in mice. Blood. 1997;90:2901–2910. [PubMed] [Google Scholar]
- 8.Chiarle R, Gong JZ, Guasparri I, Pesci A, Cai J, Liu J, Simmons WJ, Dhall G, Howes J, Piva R, Inghirami G. NPM-ALK transgenic mice spontaneously develop T-cell lymphomas and plasma cell tumors. Blood. 2003;101:1919–1927. doi: 10.1182/blood-2002-05-1343. [DOI] [PubMed] [Google Scholar]
- 9.Zhang Q, Raghunath PN, Xue L, Majewski M, Carpentieri DF, Odum N, Morris S, Skorski T, Wasik MA. Multilevel dysregulation of STAT3 activation in anaplastic lymphoma kinase-positive T/null-cell lymphoma. J Immunol. 2002;168:466–474. doi: 10.4049/jimmunol.168.1.466. [DOI] [PubMed] [Google Scholar]
- 10.Kasprzycka M, Marzec M, Liu X, Zhang Q, Wasik MA. Nucleophosmin/anaplastic lymphoma kinase (NPM/ALK) oncoprotein induces the T regulatory cell phenotype by activating STAT3. Proc Natl Acad Sci U S A. 2006;103:9964–9999. doi: 10.1073/pnas.0603507103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marzec M, Kasprzycka M, Liu X, El-Salem M, Halasa K, Raghunath PN, Bucki R, Wlodarski P, Wasik MA. Oncogenic tyrosine kinase NPM/ALK induces activation of the rapamycin-sensitive mTOR signaling pathway. Oncogene. 2007;26:5606–5614. doi: 10.1038/sj.onc.1210346. [DOI] [PubMed] [Google Scholar]
- 12.Rochman Y, Spolski R, Leonard WJ. New insights into the regulation of T cells by gamma(c) family cytokines. Nat Rev Immunol. 2009;9:480–490. doi: 10.1038/nri2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Overwijk WW, Schluns KS. Functions of gammaC cytokines in immune homeostasis: current and potential clinical applications. Clin Immunol. 2009;132:153–165. doi: 10.1016/j.clim.2009.03.512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Smith KA. T-cell growth factor. Immunol Rev. 1980;51:337–357. doi: 10.1111/j.1600-065x.1980.tb00327.x. [DOI] [PubMed] [Google Scholar]
- 15.Smith KA. Interleukin-2: inception, impact, and implications. Science. 1988;240:1169–1176. doi: 10.1126/science.3131876. [DOI] [PubMed] [Google Scholar]
- 16.Hatakeyama M, Tsudo M, Minamoto S, Kono T, Doi T, Miyata T, Miyasaka M, Taniguchi T. Interleukin-2 receptor beta chain gene: generation of three receptor forms by cloned human alpha and beta chain cDNA’s. Science. 1989;244:551–556. doi: 10.1126/science.2785715. [DOI] [PubMed] [Google Scholar]
- 17.Smith KA. Interleukin-2. Curr Opin Immunol. 1992;4:271–276. doi: 10.1016/0952-7915(92)90076-q. [DOI] [PubMed] [Google Scholar]
- 18.Bessoles S, Fouret F, Dudal S, Besra GS, Sanchez F, Lafont V. IL-2 triggers specific signaling pathways in human NKT cells leading to the production of pro- and anti-inflammatory cytokines. J Leukoc Biol. 2008;84:224–233. doi: 10.1189/jlb.1007669. [DOI] [PubMed] [Google Scholar]
- 19.Mitchell TJ, John S. Signal transducer and activator of transcription (STAT) signalling and T-cell lymphomas. Immunology. 2005;114:301–312. doi: 10.1111/j.1365-2567.2005.02091.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Evans GA, Howard OM, Erwin R, Farrar WL. Interleukin-2 induces tyrosine phosphorylation of the vav protooncogene product in human T cells: lack of requirement for the tyrosine kinase lck. Biochem J. 1993;294:339–342. doi: 10.1042/bj2940339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Friedmann MC, Migone TS, Russell SM, Leonard WJ. Different interleukin 2 receptor beta-chain tyrosines couple to at least two signaling pathways and synergistically mediate interleukin 2-induced proliferation. Proc Natl Acad Sci U S A. 1996;93:2077–2082. doi: 10.1073/pnas.93.5.2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang Q, Nowak I, Vonderheid EC, Rook AH, Kadin ME, Nowell PC, Shaw LM, Wasik MA. Activation of Jak/STAT proteins involved in signal transduction pathway mediated by receptor for interleukin 2 in malignant T lymphocytes derived from cutaneous anaplastic large T-cell lymphoma and Sezary syndrome. Proc Natl Acad Sci U S A. 1996;93:9148–9153. doi: 10.1073/pnas.93.17.9148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ravichandran KS, Igras V, Shoelson SE, Fesik SW, Burakoff SJ. Evidence for a role for the phosphotyrosine-binding domain of Shc in interleukin 2 signaling. Proc Natl Acad Sci U S A. 1996;93:5275–5280. doi: 10.1073/pnas.93.11.5275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gaffen SL, Lai SY, Ha M, Liu X, Hennighausen L, Greene WC, Goldsmith MA. Distinct tyrosine residues within the interleukin-2 receptor beta chain drive signal transduction specificity, redundancy, and diversity. J Biol Chem. 1996;271:21381–21390. doi: 10.1074/jbc.271.35.21381. [DOI] [PubMed] [Google Scholar]
- 25.Fujii H, Ogasawara K, Otsuka H, Suzuki M, Yamamura K, Yokochi T, Miyazaki T, Suzuki H, Mak TW, Taki S, Taniguchi T. Functional dissection of the cytoplasmic subregions of the IL-2 receptor betac chain in primary lymphocyte populations. EMBO J. 1998;17:6551–6557. doi: 10.1093/emboj/17.22.6551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gadina M, Sudarshan C, Visconti R, Zhou YJ, Gu H, Neel BG, O’Shea JJ. The docking molecule gab2 is induced by lymphocyte activation and is involved in signaling by interleukin-2 and interleukin-15 but not other common gamma chain-using cytokines. J Biol Chem. 2000;275:26959–26966. doi: 10.1074/jbc.M004021200. [DOI] [PubMed] [Google Scholar]
- 27.Bensinger SJ, Walsh PT, Zhang J, Carroll M, Parsons R, Rathmell JC, Thompson CB, Burchill MA, Farrar MA, Turka LA. Distinct IL-2 receptor signaling pattern in CD4+CD25+ regulatory T cells. J Immunol. 2004;172:5287–5296. doi: 10.4049/jimmunol.172.9.5287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marzec M, Kasprzycka M, Ptasznik A, Wlodarski P, Zhang Q, Odum N, Wasik MA. Inhibition of ALK enzymatic activity in T-cell lymphoma cells induces apoptosis and suppresses proliferation and STAT3 phosphorylation independently of Jak3. Lab Invest. 2005;85:1544–1554. doi: 10.1038/labinvest.3700348. [DOI] [PubMed] [Google Scholar]
- 29.Marzec M, Kasprzycka M, Liu X, Raghunath PN, Wlodarski P, Wasik MA. Oncogenic tyrosine kinase NPM/ALK induces activation of the MEK/ERK signaling pathway independently of c-Raf. Oncogene. 2007;26:813–821. doi: 10.1038/sj.onc.1209843. [DOI] [PubMed] [Google Scholar]
- 30.Marzec M, Halasa K, Kasprzycka M, Wysocka M, Liu X, Tobias JW, Baldwin D, Zhang Q, Odum N, Rook AH, Wasik MA. Differential effects of interleukin-2 and interleukin-15 versus interleukin-21 on CD4+ cutaneous T-cell lymphoma cells. Cancer Res. 2008;68:1083–1091. doi: 10.1158/0008-5472.CAN-07-2403. [DOI] [PubMed] [Google Scholar]
- 31.Gery S, Koeffler HP. Circadian rhythms and cancer. Cell Cycle. 2010;9:1097–1103. doi: 10.4161/cc.9.6.11046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cui YX, Kerby A, McDuff FK, Ye H, Turner SD. NPM-ALK inhibits the p53 tumor suppressor pathway in an MDM2 and JNK-dependent manner. Blood. 2009;113:5217–5227. doi: 10.1182/blood-2008-06-160168. [DOI] [PubMed] [Google Scholar]
- 33.Nakajima H, Liu XW, Wynshaw-Boris A, Rosenthal LA, Imada K, Finbloom DS, Hennighausen L, Leonard WJ. An indirect effect of Stat5a in IL-2-induced proliferation: a critical role for Stat5a in IL-2-mediated IL-2 receptor alpha chain induction. Immunity. 1997;7:691–701. doi: 10.1016/s1074-7613(00)80389-1. [DOI] [PubMed] [Google Scholar]
- 34.Kim HP, Kelly J, Leonard WJ. The basis for IL-2-induced IL-2 receptor alpha chain gene regulation: importance of two widely separated IL-2 response elements. Immunity. 2001;15:159–172. doi: 10.1016/s1074-7613(01)00167-4. [DOI] [PubMed] [Google Scholar]
- 35.Zhang Q, Wang HY, Liu X, Wasik MA. Stat5a is epigenetically silenced in the NPM/ALK-transformed T lymphocytes and acts in such cells as tumor suppressor by inhibiting expression of the NPM/ALK oncogene. Nature Med. 2007;13:1341–1348. doi: 10.1038/nm1659. [DOI] [PubMed] [Google Scholar]
- 36.Nieborowska-Skorska M, Slupianek A, Xue L, Zhang Q, Raghunath PN, Hoser G, Wasik MA, Morris SW, Skorski T. Role of signal transducer and activator of transcription 5 in nucleophosmin/anaplastic lymphoma kinase-mediated malignant transformation of lymphoid cells. Cancer Res. 2001;61:6517–6523. [PubMed] [Google Scholar]
- 37.Gitenay D, V, Baron T. Is EGR1 a potential target for prostate cancer therapy? Future Oncol. 2009;5:993–1003. doi: 10.2217/fon.09.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Decker EL, Nehmann N, Kampen E, Eibel H, Zipfel PF, Skerka C. Early growth response proteins (EGR) and nuclear factors of activated T cells (NFAT) form heterodimers and regulate proinflammatory cytokine gene expression. Nucleic Acids Res. 2003;31:911–921. doi: 10.1093/nar/gkg186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang Q, Wang HY, Liu X, Bhutani G, Kantekure K, Wasik M. IL-2R common gamma-chain is epigenetically silenced by nucleophosphin-anaplastic lymphoma kinase (NPM-ALK) and acts as a tumor suppressor by targeting NPM-ALK. Proc Natl Acad Sci U S A. 2011;108:11977–11982. doi: 10.1073/pnas.1100319108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Turner SD, Yeung D, Hadfield K, Cook SJ, Alexander DR. The NPM-ALK tyrosine kinase mimics TCR signalling pathways, inducing NFAT and AP-1 by RAS-dependent mechanisms. Cell Signal. 2007;19:740–747. doi: 10.1016/j.cellsig.2006.09.007. [DOI] [PubMed] [Google Scholar]
- 41.Ambrogio C, Martinengo C, Voena C, Tondat F, Riera L, di Celle PF, Inghirami G, Chiarle R. NPM-ALK oncogenic tyrosine kinase controls T-cell identity by transcriptional regulation and epigenetic silencing in lymphoma cells. Cancer Res. 2009;69:8611–8619. doi: 10.1158/0008-5472.CAN-09-2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mahalingam D, Natoni A, Keane M, Samali A, Szegezdi E. Early growth response-1 is a regulator of DR5-induced apoptosis in colon cancer cells. Br J Cancer. 2010;102:754–764. doi: 10.1038/sj.bjc.6605545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang Q, Nowak I, Vonderheid EC, Rook AH, Kadin ME, Nowell PC, Shaw LM, Wasik MA. Activation of Jak/STAT proteins involved in signal transduction pathway mediated by receptor for interleukin 2 in malignant T lymphocytes derived from cutaneous anaplastic large T-cell lymphoma and Sezary syndrome. Proc Natl Acad Sci U S A. 1996;93:9148–9153. doi: 10.1073/pnas.93.17.9148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Abraham RM, Zhang Q, Odum N, Wasik MA. The role of cytokine signaling in the pathogenesis of cutaneous T-cell lymphoma. Cancer Biol Ther. 2011;12:1019–1022. doi: 10.4161/cbt.12.12.18144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Takemoto S, Mulloy JC, Cereseto A, Migone TS, Patel BK, Matsuoka M, Yamaguchi K, Takatsuki K, Kamihira S, White JD, Leonard WJ, Waldmann T, Franchini G. Proliferation of adult T cell leukemia/lymphoma cells is associated with the constitutive activation of JAK/STAT proteins. Proc Natl Acad Sci U S A. 1997;94:13897–13902. doi: 10.1073/pnas.94.25.13897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Elliott NE, Cleveland SM, Grann V, Janik J, Waldmann TA, Davé UP. FERM domain mutations induce gain of function in JAK3 in adult T-cell leukemia/lymphoma. Blood. 2011;118:3911–3921. doi: 10.1182/blood-2010-12-319467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Koo GC, Tan SY, Tang T, Poon SL, Allen GE, Tan L, Chong SC, Ong WS, Tay K, Tao M, Quek R, Loong S, Yeoh KW, Yap SP, Lee KA, Lim LC, Tan D, Goh C, Cutcutache I, Yu W, Ng CC, Rajasegaran V, Heng HL, Gan A, Ong CK, Rozen S, Tan P, Teh BT, Lim ST. Janus kinase 3-activating mutations identified in natural killer/T-cell lymphoma. Cancer Discov. 2012;2:591–597. doi: 10.1158/2159-8290.CD-12-0028. [DOI] [PubMed] [Google Scholar]
- 48.Gambacorti-Passerini C, Messa C, Pogliani EM. Crizotinib in anaplastic large-cell lymphoma. N Engl J Med. 2011;364:775–776. doi: 10.1056/NEJMc1013224. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.