Abstract
The present study confirms the occurrence of Chilli veinal mottle virus (ChiVMV) under the genus Potyvirus in Naga chilli (Capsicum chinense) in Meghalaya based on mechanical transmission assay, transmission electron microscopy, RT-PCR and sequence analysis. This is the first record of Chivmv in Naga chilli in North-East India.
Keywords: Naga chilli, Chilli veinal mottle virus, Molecular detection, Cylindrical inclusion protein
Naga chilli (Capsicum chinense Jacq.) is widely grown in North-east India. During 2011–2012, disease symptoms like mottling, vein banding, narrowing and distortion of leaves followed by stunted growth were observed in C. chinense plants growing in experimental fields at Umiam, Meghalaya (altitude 1000 m). The disease incidence was about 85–90 % in all experimental fields surveyed. Therefore, an attempt was made to identify the virus associated with the characteristic symptoms.
Leaf samples of Naga chilli plants were collected randomly from all the fields during August–September, 2012. Leaf sap of symptomatic young leaves [diluted (1:2 v/v) in phosphate buffer (0.2 M, pH 7.4)] was inoculated onto five healthy Naga chilli plants at 2–3 leaf stage by rubbing carborundum-dusted leaves. Inoculated plants were then maintained in insect proof cages at 18–22 °C and observed every 2 days post inoculation for symptom expression. The mechanical inoculation produced typical systemic mottling symptom (Fig. 1a) on all artificially inoculated plants during 18–21 days post-inoculation. The inoculated symptomatic leaf samples were examined under electron microscope (EM) following the leaf dip method using 2 % aqueous uranyl acetate (UA). Under EM flexuous filamentous virus particle was observed (Fig. 1b). Based on symptoms, transmission assay and electron microscopy, Potyvirus [2] was suspected to be associated with the disease. Therefore, attempt was made to detect the virus applying reverse transcription polymerase chain reaction (RT-PCR)-based methods.
Fig. 1.
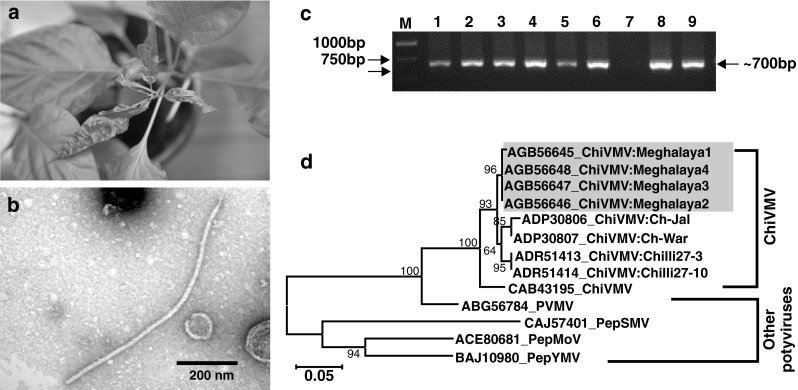
a Veinal mottle symptom in mechanically inoculated chilli plant, b Transmission electron micrograph of flexous filamentous Potyvirus particles in infected leaf tissues c PCR-detection of Potyvirus using degenerate primers; M = 1 kb DNA ladder, lane 7 = healthy plant, d Phylogenetic tree based on amino acid sequences of CI domain of four Meghalaya isolates of ChiVMV (in box) along with reported ChiVMV, PVMV and other potyviruses. The tree was generated using NJ algorithm in MEGA5.0. The bootstrap (1000 replicates) values are shown next to the branches (only when >50 %). The scale bar represents a genetic distance of 0.05
Total RNA was extracted from both symptomatic and symptomless (collected from net-house grown healthy plants) leaf samples (RNeasy Plant Mini Kit, Qiagen, Valencia, CA) and complementary DNA (cDNA) was synthesized (RevertAid™, Fermentas, India). PCR assay was carried out using Potyvirus specific degenerate primer CIFor/CIRev, designed to amplify a ~700 bp region of cylindrical inclusion protein (CI) domain of Potyvirus open reading frame (ORF) [3]. All the symptomatic leaf samples showed virus-specific amplification of ~700 bp in RT-PCR assay. No amplification was found in asymptomatic leaf samples collected from net house grown plants (Fig. 1c). The RT-PCR also showed specific amplicon in RNA extracts from the mechanically inoculated chilli plants.
The RT-PCR amplicons from four samples: three from different experimental fields and one from artificially inoculated plant were gel purified (GeneJET, Fermentas, India) and each fragment was sequenced bi-directionally (Chromous Biotech, Bangalore, India) and deposited in NCBI GenBank (Accession numbers: KC119086–KC119089 for isolate 1–4, respectively). The partial sequences of CI (583 bp) domain of four Meghalaya isolates shared 98.0–100.0 and 99.0–100.0 % identity at nucleotide and amino acid level, respectively, which confirmed that the four isolates belonged to the same species, as the threshold of 85.0 % nucleotide sequence identity proposing to differentiate species within Potyvirus [3]. The sequences showed maximum nucleotide identity of 90.0 % and amino acid identities of 98.0 % with the earlier reported Chilli veinal mottle virus (ChiVMV) isolate (GU294791) from India. Further four Meghalaya isolates were compared with published sequences of Chilli infecting potyviruses such as ChiVMV, Pepper veinal mottle virus (PVMV), Pepper mottle virus (PepMoV), Pepper yellow mosaic virus (PepYMV) and Pepper severe mosaic virus (PepSMV). The pair-wise multiple alignment (CLUSTAL W, DNASTAR Inc.7.1, USA) showed nucleotide sequence identity of 82.2–89.4, 73.8–73.9 and 59.7–63.1 % with ChiVMV, PVMV and other potyviruses, respectively. Whereas, the corresponding values at amino acid level was 94.3–97.9, 87.0–87.6 and 60.3–65.8 %, respectively. In amino acid based neighbour-joining (NJ) phylogeny, four Meghalaya isolates grouped with reported isolates of ChiVMV (Fig. 1d). Similar pattern of clustering was also observed at nucleotide level (data not shown). In NJ tree, the PVMV isolate showed close association with ChiVMV, whereas, other potyviruses such as PepMoV, PepSMV and PepYMV formed a separate cluster (Fig. 1d). Similar clustering pattern was also reported by Moury et al. [5], suggesting molecular distinction between ChiVMV and PVMV.
The virus incidence in Naga chilli is an emerging problem in North-east India. The current study confirmed the incidence of ChiVMV in Naga chilli. The incidence of ChiVMV in C. annum was reported from South India [1, 4, 6]. To the best of our knowledge, this is the first report and molecular evidence of ChiVMV infection in Naga chilli from North-east India.
Acknowledgements
The work was funded by Indian Council of Agricultural Research. The authors are grateful to Director, NBPGR, New Delhi and Officer-In-Charge, NBPGR, Regional Station, Umiam, Meghalaya for providing chilli seeds used in this study.
References
- 1.Anindya R, Joseph J, Gowri TDS, Savithri HS. Complete genomic sequence of Pepper vein banding virus (PVBV): a distinct member of the genus Potyvirus. Arch Virol. 2004;149:625–632. doi: 10.1007/s00705-003-0236-0. [DOI] [PubMed] [Google Scholar]
- 2.Berger PH, Adams MJ, Barnett OW, Brunt AA, Hammond J, Hill JH, Jordan RL, Kashiwazaki S, Rybicki E, Spence N, Stenger DC, Ohki ST, Uyeda I, van Zaayen A, Valkonen J, Vetten HJ. Family potyviridae. In: Fauquet CM, Mayo MA, Maniloff J, Desselberger U, Ball L, editors. Virus taxonomy eighth report of the international committee on taxonomy of virus. London: Elsevier Academic Press; 2005. pp. 385–396. [Google Scholar]
- 3.Ha C, Coombs S, Revill PA, Harding RM, Vu M, Dale JL. Design and application of two novel degenerate primer pairs for the detection and complete genomic characterization of potyviruses. Arch Virol. 2008;153:25–36. doi: 10.1007/s00705-007-1053-7. [DOI] [PubMed] [Google Scholar]
- 4.Joseph J, Savithri HS. Determination of 3′-terminal nucleotide sequence of pepper vein banding virus RNA and expression of its coat protein in Escherichia coli. Arch Virol. 1999;144:1679–1687. doi: 10.1007/s007050050696. [DOI] [PubMed] [Google Scholar]
- 5.Moury B, Palloix A, Caranta C, Gognalons P, Souche S, Gebre Selassie K, Marchoux G. Serological, molecular, and pathotype diversity of Pepper veinal mottle virus and Chili veinal mottle virus. Phytopathology. 2005;95:227–232. doi: 10.1094/PHYTO-95-0227. [DOI] [PubMed] [Google Scholar]
- 6.Ravi K, Joseph J, Nagaraju N, Krishnaprasad S, Reddy H, Savithri HS. Characterization of a pepper vein banding virus from chilli pepper in India. Plant Dis. 1997;81:673–677. doi: 10.1094/PDIS.1997.81.6.673. [DOI] [PubMed] [Google Scholar]


