Abstract
Newcastle disease virus (NDV) hemagglutinin–neuraminidase (HN) is a multifunctional protein, which possesses both the receptor recognition and neuraminidase activities. The fusion (F) protein is a type I membrane glycoprotein that mediates the merger of the viral envelope to the host cell membrane. Although the functions of the HN and F proteins have been well studied, however, the factors shaping synonymous codon usage bias and nucleotide composition in HN and F genes have been few reported. In our study, we analyzed synonymous codon usage using the 69 NDV HN and F genes, respectively. The general correlation between base composition and codon usage bias suggests that mutational pressure rather than natural selection is the main factor that determines the codon usage bias in HN and F genes. In addition, other factors, such as the aromaticity and hydrophobicity, also influence the codon usage variation among HN and F genes. This study represents the most comprehensive analysis to date of NDV HN and F genes codon usage patterns and provides a basic understanding of the mechanisms for codon usage bias.
Electronic supplementary material
The online version of this article (doi:10.1007/s13337-013-0175-7) contains supplementary material, which is available to authorized users.
Keywords: Newcastle disease virus, Hemagglutinin–neuraminidase, Fusion protein, Synonymous codon usage, Mutational pressure, Natural selection
It is well known that synonymous codons are not used randomly. Some codons are used more frequently than others [5, 14, 17]. Compositional constraints and natural selection are thought to be the two main factors accounting for codon usage variation among genes in different organisms [11, 13, 20]. The different patterns of codon usage in mammals may arise from compositional constraints of the genomes [2]. In contrast to some unicellular organisms, low expressed genes displayed a more uniform pattern of codon usage, whereas high expressed genes have a strong selective preference for codons with a high concentration of the corresponding acceptor tRNA molecule [3, 4, 7, 8, 13, 20]. Recently, codon usage was also found to be related to gene function [15], protein secondary structure [19], replicational and translational selection [21], dinucleotide bias [25], gene length [10], tRNA abundance [1], codon–anticodon [5] interaction [18], and tissue or organ specificity. In addition, mutational pressure and translational selection were thought to be the main factors that account for codon usage variation among genes in some RNA virus. Studies the extent and causes of biases in codon usage is essential to the understanding of viral evolution, particularly the interplay between viruses and the immune response [19]. In contrast to many virus such influenza virus, where codon usage bias and nucleotide composition have been studied in great detail, the factors shaping synonymous codon usage bias and nucleotide composition in NDV have been studied only to a limited extent.
Newcastle disease virus, also known as avian paramyxovirus serotype-1 (APMV-1), a member of the genus Avulavirus within the Paramyxoviridae family [12], is a negative-sense, single stranded, non-segmented, enveloped RNA virus [9]. The NDV genome is composed of six genes and encodes their corresponding six structural proteins: nucleoprotein (NP), phosphoprotein (P), matrix (M), fusion (F), hemagglutinin–neuraminidase (HN), and the RNA polymerase (L). The HN protein of NDV is a multifunctional protein. It possesses both the receptor recognition and neuraminidase (NA) activities associated with the virus [16]. Fusion protein has been reported to media the fusion of viral envelope with cell membrane and considered as the major determinant of virulence [22]. Although the functions of the HN and F protein in NDV infection have been well studied, their roles in NDV evolution is not known at present. Recently, recombination was found to play a more important role than positive selection in the formation of genetic diversity in NDV genome [23]. Previous studies of NDV HN and F genes have mainly been limited to phylogenic analysis. However, few synonymous codon usage analyses have been applied. In order to better understand the characteristics of the NDV HN and F genes and to reveal more information about the NDV virus, we have analyzed the bias of codon usage. In this report, we sought to address the codon usage in NDV HN and F genes. Spearman’s rank correlation analysis and multiple regression analysis were performed to determine the role of different factors in shaping the codon usage biases in the various NDV viruses. All statistical analyses, as well as cluster analysis, were carried out using the statistical analysis software SPSS Version 15.0.
Relative synonymous codon usage (RSCU) values are largely independent of amino acid composition and are particularly useful in comparing codon usage between genes, or sets of genes that differ in their size and amino acid composition. RSCU values of different codon in each HN and F ORFs were calculated to investigate the extent of codon bias in NDV HN and F genes. The details of each ORF and the overall RSCU values of 59 codons in 69 NDV HN and F genes are, respectively, represented in Tables 1, 2, 3, and 4 of Electronic supplementary material. In HN gene, the preferentially used codons were A-ended (6 ones), U-ended (7 ones) codons, C-ended (4 ones) codons, and G-ended (4 ones) codons. In F gene, the preferentially used codons were A-ended (7 ones), U-ended (10 ones) codons, C-ended (2 ones) codons, and G-ended (2 ones) codons. The GC index was used to calculate the overall GC content in the ORF, while the index GC3s was used to calculate the fraction of GC nucleotides at the synonymous third codon position (excluding Met, Trp, and the termination codons). The average GC content of all NDV HN gene was 46.3 % (from 44.4 to 47.3 %, with a S.D. of 0.49 %), and the average GC3s content in codons was 43.5 % (from 37.3 to 45.1 %, with a S.D. of 1.3 %). Similarly, the average GC content of all NDV F gene was 44.7 % (from 43.6 to 46.2 %, with a S.D. of 0.45 %), and the average GC3s content in codons was 42.2 % (From 39.5 to 46.7 %, with a S.D. of 1.2 %). This is consistent with previous observations that NDV viruses are GC-moderate genomes [23], and so it is expected that the third-ended codons are not preferentially used. In order to investigate whether these 69 coding sequences of NDV HN and F genes display similar compositional features, ENC (effective number of codons) values were calculated (Tables 1, 2 of Electronic supplementary material). The effective number of codons of a gene (ENC) is generally used to quantify the codon usage bias of a gene, which is essentially independent of gene length. The ENC values range from 20 to 61. The larger the extent of codon preference in a gene, the smaller the ENC value is. In an extremely biased gene where only one codon is used for each amino acid, this value would be 20; in an unbiased gene, it would be 61. The ENC values of NDV HN genes vary from 54.5 to 58.7, with a mean of 57.7 and S.D. of 0.803, and the ENC values of F genes vary from 53.7 to 59.4, with a mean of 55.7 and S.D. of 1.3. We found that all the ENC values for HN and F ORFs are much higher (ENC >50). Based on this finding, together with published data on codon usage bias among some RNA viruses [6, 21, 25], we conclude that the codon usage bias in both NDV HN and F genes are less.
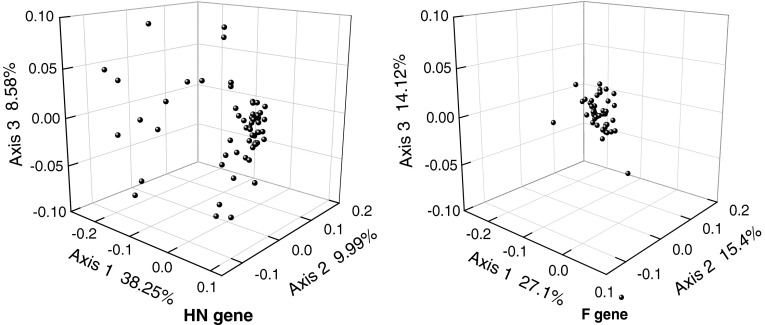
To investigate synonymous codon usage variation among NDV HN and F genes, correspondence analysis (COA) was implemented for 69 NDV HN and F ORFs selected for this study. Multivariate statistical analysis can be used to explore the relationships between variables and samples. The major trend in codon usage variation among ORFs can be investigated using COA. In order to minimize the effects of amino acid composition on codon usage, each ORF is represented as a 59-dimensional vector; each dimension corresponds to the RSCU value of one sense codon (excluding AUG, UGG, and stop codons). Major trends within this dataset can be determined using measures of relative inertia and genes ordered according to their positions along the axis of major inertia. Figure 1 depicts the position of each ORF on the plane defined by the first, second and third principal axes generated by COA on RSCU values of ORFs. As for HN gene, the first principal axis accounts for 38.25 % of the total variation. The next three axes account for 9.99, 8.58, and 6.36 % of the variation, respectively. In F gene, the first principal axis accounts for 27.1 % of the total variation, and the next three axes account for 15.4, 14.12, and 8.63 % of the variation. This observation indicates that although the first major axis explains a substantial amount of variation in trends in codon usage of HN and F genes, the second major axis also has an appreciable impact on total variation in synonymous codon usage. If not specifically mentioned, the values of the first two axes of this COA were used for correlation and regression analysis hereafter.
Fig. 1.
A plot of value of the first, second and third axis of each ORF in COA. As for HN gene, the first axis accounts for 38.25 % of all variation among ORFs, the second axis accounts for 9.99 % and third axis accounts for 8.58 % of total variation. In F gene, the first principal axis accounts for 27.1 % of the total variation, and the next two axes account for 15.4, 14.12 % of the variation
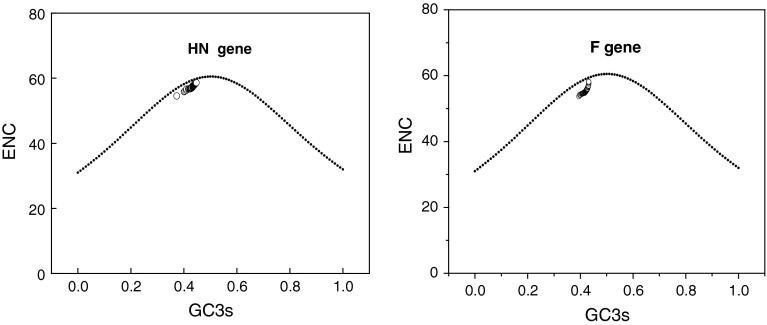
In addition, mutational pressure and translational selection were thought to be the main factors that account for codon usage variation among genes in some RNA virus [11]. The plot of ENC and GC3s is an effective way to explore codon usage variation among genes [24]. In order to further find whether codon usage variation among HN and F genes are determined by mutational bias, ENC values of each HN and F gene were plotted against its corresponding GC3s. Genes, whose codon choice is constrained only by a G+C mutational bias, will lie on or just below the curve of the predicted values. All of the spots lie below the expected curve as shown in Fig. 2. In addition, a significantly positive correlation between GC3s and ENC values was observed. The results indicated that the codon usage bias in these HN and F genes are greatly influenced by the G+C mutation bias.
Fig. 2.
Effective number of codons used in each gene plotted against the GC3s. The continuous curve plots the relationship between GC3s and ENC in the absence of selection. All of spots lie below the expected curve both in HN and F genes
Furthermore, the GC index was used to calculate the overall GC content in the ORF, while the index GC3s was used to calculate the fraction of GC nucleotides at the synonymous third codon position (excluding Met, Trp, and the termination codons). We analyzed the correlation between the first or second axis values in COA and GC3s values of each gene. The first axis value in COA of each selected gene, which contains most of the variation in synonymous codon usage bias between these HN and F genes, is not correlated with the GC composition in third codon position. The second axis in the COA of each gene is also not correlated with GC3s. This analysis indicated that most of the codon usage bias among different ORFs is not related to the nucleotide composition. Therefore, the compositional constraint is not the main determinant of the variation in synonymous codon usage among different HN and F ORFs. At the amino acid level, the general average hydrophobicity score (GRAVY) and the frequency of aromatic amino acids (Aromo) in the putative gene product were also analyzed. As showed in Fig. 2, all of the actual ENC values are significantly lower than the expected ones, which indicated mutational bias is the main factor determining codon usage in HN gene. In order to test whether selection pressure contributes to the codon usage variation among HN and F gene, we performed a correlation analysis to evaluate whether GRAVY and Aromaticity values were related to first two axes of COA. Our results showed that only Aromaticity was correlated with axis 1 in F gene (Table 1). However, GRAVY was correlated with both axis 1 and axis 2, and Aromaticity was correlated with axis 2 in HN gene (Table 2), indicating that the degree of hydrophobicity and the frequency of aromatic amino acids (Phe, Tyr, Trp) were also associated with the codon usage variation in HN gene.
Table 1.
Summary of correlation analysis between the first two axes in COA and GC3s, GRAVY, or Aromaticity in the selected 69 NDV HN genes
| GRAVY | Aromaticity | GC3s | |
|---|---|---|---|
| Axis 1 | |||
| r | −0.434** | −0.357** | −0.111 |
| P | ≤0.01 | 0.003 | 0.365 |
| Axis 2 | |||
| r | −0.213** | 0.064 | −0.007 |
| P | 0.079 | 0.599 | 0.953 |
* P value ≤ 0.05; ** P value ≤ 0.01
Table 2.
Summary of correlation analysis between the first two axes in COA and GC3s, GRAVY, or Aromaticity in the selected 69 NDV F genes
| GRAVY | Aromaticity | GC3s | |
|---|---|---|---|
| Axis 1 | |||
| r | 0.234 | −0.518** | −0.119 |
| P | 0.053 | ≤0.01 | 0.329 |
| Axis 2 | |||
| r | −0.185 | −0.224 | 0.018 |
| P | 0.129 | 0.064 | 0.883 |
* P value ≤ 0.05; ** P value ≤ 0.01
In our report, the synonymous codon usage biases in NDV HN and F genes were analyzed, and we found that both NDV HN and F genes had low codon usage bias. Mutational pressure rather selection pressure is the main factor determining the codon usage biases. In addition, aromaticity and hydrophobicity could be partially accounting for the codon usage variation.
Electronic supplementary material
Acknowledgments
This work is supported by National Natural Science Foundation of China (NSFC, Grant no 31200121, 31300145 and 31310103018), and Natural Science Foundation of Heilongjiang Province of China (C201322).
References
- 1.Duret L, Mouchiroud D. Expression pattern and, surprisingly, gene length shape codon usage in Caenorhabditis, Drosophila, Arabidopsis. Proc Natl Acad Sci USA. 1999;96:4482–4487. doi: 10.1073/pnas.96.8.4482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Francino HP, Ochman H. Isochores result from mutation not selection. Nature. 1999;400:30–31. doi: 10.1038/21804. [DOI] [PubMed] [Google Scholar]
- 3.Gouy M, Gautier C. Codon usage in bacteria: correlation with gene expressivity. Nucleic Acids Res. 1982;10:7055–7074. doi: 10.1093/nar/10.22.7055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Grantham R, Gautier C, Gouy M, Jacobzone M, Mercier R. Codon catalog usage is a genome strategy modulated for gene expressivity. Nucleic Acids Res. 1981;9:r43–r74. doi: 10.1093/nar/9.1.213-b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Grantham R, Gautier C, Gouy M, Mercier R, Pave A. Codon catalog usage and the genome hypothesis. Nucleic Acids Res. 1980;8:R49–R62. doi: 10.1093/nar/8.1.197-c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gu WJ, Zhou T, Ma JM, Sun X, Lu ZH. Analysis of synonymous codon usage in SARS Coronavirus and other viruses in the Nidovirales. Virus Res. 2004;101:155–161. doi: 10.1016/j.virusres.2004.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ikemura T. Codon usage and tRNA content in unicellular and multicellular organisms. Mol Biol Evol. 1985;2:13–34. doi: 10.1093/oxfordjournals.molbev.a040335. [DOI] [PubMed] [Google Scholar]
- 8.Ikemura T. Correlation between the abundance of Escherichia coli transfer RNAs and the occurrence of the respective codons in its protein genes: a proposal for a synonymous codon choice that is optimal for the E. coli translational system. J Mol Biol. 1981;151:389–409. doi: 10.1016/0022-2836(81)90003-6. [DOI] [PubMed] [Google Scholar]
- 9.Jang J, Hong SH, Choi D, Choi KS, Kang S, Kim IH. Overexpression of Newcastle disease virus (NDV) V protein enhances NDV production kinetics in chicken embryo fibroblasts. Appl Microbiol Biotechnol. 2010;85:1509–1520. doi: 10.1007/s00253-009-2189-z. [DOI] [PubMed] [Google Scholar]
- 10.Karlin S, Burge C. Dinucleotide relative abundance extremes: a genomic signature. Trends Genet. 1995;11:283–290. doi: 10.1016/S0168-9525(00)89076-9. [DOI] [PubMed] [Google Scholar]
- 11.Karlin S, Mrazek J. What drives codon choices in human genes? J Mol Biol. 1996;262:459–472. doi: 10.1006/jmbi.1996.0528. [DOI] [PubMed] [Google Scholar]
- 12.King DJ, Seal BS. Biological and molecular characterization of Newcastle disease virus (NDV) field isolates with comparisons to reference NDV strains. Avian Dis. 1998;42:507–516. doi: 10.2307/1592677. [DOI] [PubMed] [Google Scholar]
- 13.Lesnik T, Solomovici J, Deana A, Ehrlich R, Reiss C. Ribosome traffic in E. coli and regulation of gene expression. J Theor Biol. 2000;202:175–185. doi: 10.1006/jtbi.1999.1047. [DOI] [PubMed] [Google Scholar]
- 14.Lloyd AT, Sharp PM. Evolution of codon usage patterns: the extent and nature of divergence between candida and albicans and Saccharomyces cerevisiae. Nucleic Acids Res. 1992;20:5289–5295. doi: 10.1093/nar/20.20.5289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ma JM, Zhou T, Gu WJ, Sun X, Lu ZH. Cluster analysis of the codon use frequency of MHC genes from different species. Biosystems. 2002;65:199–207. doi: 10.1016/S0303-2647(02)00016-3. [DOI] [PubMed] [Google Scholar]
- 16.Makkay AM, Krell PJ, Nagy E. Antibody detection-based differential ELISA for NDV-infected or vaccinated chickens versus NDV HN-subunit vaccinated chickens. Vet Microbiol. 1999;66:209–222. doi: 10.1016/S0378-1135(99)00016-4. [DOI] [PubMed] [Google Scholar]
- 17.Marin A, Bertranpetit J, Oliver JL, Medina JR. Variation in G+C-content and codon choice: differences among synonymous codon groups in vertebrate genes. Nucleic Acids Res. 1989;17:6181–6189. doi: 10.1093/nar/17.15.6181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moriyama EN, Powell JR. Synonymous substitution rates in Drosophila: mitochondrial versus nuclear genes. J Mol Evol. 1997;45:378–391. doi: 10.1007/PL00006243. [DOI] [PubMed] [Google Scholar]
- 19.Shackelton LA, Parrish CR, Holmes EC. Evolutionary basis of codon usage and nucleotide composition bias in vertebrate DNA viruses. J Mol Evol. 2006;62:551–563. doi: 10.1007/s00239-005-0221-1. [DOI] [PubMed] [Google Scholar]
- 20.Sharp PM, Tuohy TMF, Mosurski KR. Codon usage in yeast: cluster-analysis clearly differentiates highly and lowly expressed genes. Nucleic Acids Res. 1986;14:5125–5143. doi: 10.1093/nar/14.13.5125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tao P, Dai L, Luo MC, Tang FQ, Tien P, Pan ZS. Analysis of synonymous codon usage in classical swine fever virus. Virus Genes. 2009;38:104–112. doi: 10.1007/s11262-008-0296-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vijay DP, Vijayarani K, Kumanan K. Cloning and expression of fusion (F) protein gene of Newcastle disease virus (NDV) Indian J Anim Sci. 2012;82:132–134. [Google Scholar]
- 23.Wang M, Liu YS, Zhou JH, Chen HT, Ma LN, Ding YZ, Liu WQ, Gu YX, Zhang J. Analysis of codon usage in Newcastle disease virus. Virus Genes. 2011;42:245–253. doi: 10.1007/s11262-011-0574-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wright F. The effective number of codons used in a gene. Gene. 1990;87:23–29. doi: 10.1016/0378-1119(90)90491-9. [DOI] [PubMed] [Google Scholar]
- 25.Zhou T, Gu WJ, Ma JM, Sun X, Lu ZH. Analysis of synonymous codon usage in H5N1 virus and other influenza A viruses. Biosystems. 2005;81:77–86. doi: 10.1016/j.biosystems.2005.03.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.