Abstract
A recombinant plasmid expressing the VP6 inner capsid coding gene of grass carp reovirus (GCRV) was constructed and expressed in a Ctenopharyngodon idellus kidney (CIK) cell line and grass carps. The VP6 gene was amplified by RT-PCR, cloned into a pEGFP-N1 eukaryotic expression vector and transfected into CIK cells. Results from enhanced green fluorescent protein (EGFP) experiments and flow cytometry showed highest protein expression at 48 h. The immunoreactivity of fusion protein was confirmed using an indirect immunofluorescent assay. The specific binding between the fusion protein and polyclonal mouse GCRV VP6-specific antiserum indicated that the fusion protein was translated in vitro and had good immunogenicity. An antiviral activity assay showed that the virus titer was 100-fold lower in the GCRV VP6 expressed cells than in the pEGFP-N1 transfected cells. The expression levels of three immune genes in the head kidney of grass carps injected with the recombinant plasmid were used. Mx, TLR3 and IgM mRNA expression increased sharply at the 1st and 15th days post-injection (dpi). Specific antibodies were detected 30 days after vaccination. Neutralizing titers of the antibodies in vaccinated fish detected ranged from 160 to 320. Intramuscular injection of grass carps with 1 μg of pEGFP-N1-VP6 was found to provide strong protection against GCRV. These results suggested that the VP6 gene was a good candidate for the design of GCRV-DNA vaccines and to investigate the use of cytokines as co-stimulatory molecules.
Keywords: Grass carp reovirus, GCRV VP6, Recombinant plasmid, Vaccine, Immunity
Introduction
The grass carp reovirus (GCRV), which causes hemorrhage in about 85 % of the fingerling and yearling grass carp population, is the first aquatic virus isolated and characterized in China [5]. As a member of the genus aquareovirus and family reoviridea [15], GCRV has a multilayered spherical structure and encloses a genome consisting of eleven segments of dsRNA [11, 19]. It has been recognized that GCRV is the most pathogenic among all aquareovirus isolates reported to date [1, 13]. Therefore, GCRV provides a good model system to study aquareovirus replication and pathogenesis and such studies also have significance in the fish farming agriculture [6]. There are various vaccines to prevent GCRV, but none are effective [7, 27]. An inactivated vaccine was applied as the main method to prevent GCRV, but this kind of vaccine has subtype specificity, which limits its application [23].
DNA vaccines, compared to the traditional inactivated vaccine, have several practical and immunological advantages that make them attractive to the aquaculture industry. The early success of DNA vaccines in animal models was encouraging, but fish was unique in many aspects. Findings using other classes of vertebrate, namely mammals and birds, do not necessarily apply to aquatic animals [20]. However, more recent studies with reporter genes showed that fish cells efficiently expressed foreign proteins encoded by eukaryotic expression vectors [3, 9]. Therefore, DNA vaccine may be a better choice for vaccine construction against GCRV. The protein GCRV VP6 encoded by the segment 8 (S8) is similar to the σ2 protein in mammalian orthoreovirus (MRV) [6]. A recent report showed GCRV VP6 is oval shaped with eight major helices [28]. This protein binds on the outer surface of the VP3 inner shell at two instead of three positions as seen in orthoreoviruses [12, 26]. Fang et al. [7] successfully constructed a co-expression vector and obtained high-level co-expression of GCRV VP6 and enhanced green fluorescence protein (EGFP) in a baculovirus expression system, which provide useful evidence for establishing a stable system for the structural proteins of GCRV expression. Recently, a baculovirus transfer vector with dual promoters of GCRV VP6 (pFastBac-FA-VP6-ph-VP6) was also constructed and the oral vaccination of this vector could evoke antibody response in grass carp against GCRV [10, 24].
In this paper, we report the co-expression of GCRV VP6 and EGFP in a Ctenopharyngodon idellus kidney (CIK) cell line. Moreover, GCRV VP6 protein expressed in CIK cells was detected with an immunofluorescent assay (IFA), and the fluorescent cells were quantified by flow cytometry and observed by fluorescence microscopy. The cytopathic effect (CPE) assay was used to determine the antiviral activity against GCRV induced by pEGFP-N1-VP6 transfection in CIK cells. In addition, the effect of a DNA vaccine expressing the GCRV VP6 on the kinetics of Mx [18], TLR3 [17] or IgM [21] mRNA expression was assayed in the head kidney of grass carp. Specific antibodies were detected on the 30th day after vaccination. GCRV challenge trials were also carried out in grass carp, and the effect of vaccination on mortality was evaluated. This work will help us to understand the interactions of GCRV with its host cell and development of an effective vaccine against this virus.
Materials and methods
Virus and cells
Hubei grass carp disease reovirus (grass carp reovirus strain 104, GCRV104, CCTCC NO: V201217) was isolated from Jingzhou, Hubei Province in China by our laboratory [4]. The CIK cell line, established by Zuo et al. [29], was used for the propagation of GCRV in transfection experiments and for immunofluorescence studies. Cells were grown at 28 °C in MEM medium (Sigma, USA) supplemented with 100 IU/ml penicillin G (Sigma, USA), 100 mg/ml streptomycin (Sigma, USA), 2 mmol/L l-glutamine and 10 % FBS (Hangzhou, China).
Cloning of GCRV VP6 gene
Total RNA was extracted using Trizol reagent (Invitrogen, USA) according to the manufacturer’s instructions. The concentration and purity of the obtained RNA were measured spectrophotometrically (Eppendorf, Germany). The primers were designed from the coding gene fragment of GCRV VP6 (the accession no. JN967636). Two PCR primers contained specific restriction enzyme digestion sites (BamHI and HindIII) on the multiple cloning sites in the expression vector pEGFP-N1. First strand cDNA was synthesized with Superscript II reverse transcriptase (Invitrogen, USA). Briefly, RNA (5 μg) was incubated with 1 μL of a GCRV VP6 sense primer S8-F (Table 1) and 1 μL of dNTP mix (10 mmol/L) for 5 min at 65 °C. Subsequently, 4 μL of 5× first strand buffer and 2 μL DTT (0.1 mol/L) was added, mixed and left for 2 min at 42 °C. Finally, 1 μL of Superscript II reverse transcriptase was added and the mixture was incubated at 42 °C for 50 min. The reaction was stopped by heating at 70 °C for 15 min. cDNA fragment encoding 1257 bp of GCRV104 VP6 gene was amplified by PCR using S8-F and S8-R primers. The reaction was carried out in 95 °C for 5 min, 30 cycles of 94 °C for 45 s, 55 °C for 45 s, 72 °C for 1 min and 72 °C for 10 min. The AAG CTT ATG GCA sequence was incorporated into the S8-F primer because the expression vector used has no ATG initiation code.
Table 1.
Primers and probes used in this study for standard and RT-PCR analyses
| Gene | Accession no. | Name | Primers | Product size(bp) | Cycles | Cycling conditions |
|---|---|---|---|---|---|---|
| GCRV VP6 | HM234682 | S8-F | 5′-CCCAAGCTTATGGCACGTGTGGTT TATG-3′ | 1,257 | 1× | 94 °C 5 min |
| S8-R | 5′-CGCGGATCCGTGGTTACGCGGGTCA-3′ | 30× | 94 °C 45 s, 56 °C 45 s, 72 °C 1 min | |||
| Mx | AY395698 | Mx-F | 5′-ACGCTGTCCTCTGGTATTGA-3′ | 178 | 1× | 50 °C 2 min, 95 °C 10 min |
| Mx-R | 5′-CATGACTGATCCCTTCTCCC-3′ | 40× | 94 °C 15 s, 60 °C 1 min | |||
| TLR3 | DQ864497 | TLR3-F | 5′-TCTGTATCTTCGTCGTGG-3′ | 102 | 1× | 50 °C 2 min, 95 °C 10 min |
| TLR3-R | 5′-TAATGCTGAGCGGAGTTT-3′ | 40× | 94 °C 15 s, 60 °C 1 min | |||
| IgM | DQ417927 | IgM-F | 5′-AACCATCTCCGCCGAAGTCAA-3′ | 177 | 1× | 50 °C 2 min, 95 °C 10 min |
| IgM-R | 5′-CACTCCCAAACGCTGGATACTG-3′ | 40× | 94 °C 15 s, 60 °C 1 min | |||
| Actin | M25013 | Actin-F | 5′-GATGATGAAATTGCCGCACTG-3′ | 151 | 1× | 50 °C 2 min, 95 °C 10 min |
| Actin-R | 5′-TGGTCAGCCCGAAACTATC-3′ | 40× | 94 °C 15 s, 60 °C 1 min |
PCR product was visualized on an ethidium bromide stained 1.2 % agarose gel, which was recovered and purified with the Wizard SV Gel and PCR Clean Up System (Promega, USA). The purified PCR product was sequenced prior to and after cloning into the pEGFP-N1 (Clontech, USA) vector to verify correct insertion. The construct was designated as pEGFP-N1-VP6 and transformed into chemically competent Escherichia coli TOP10 (Invitrogen, USA). The positive clone was screened in selective plate of ampicillin and then incubated at 37 °C overnight with shaking. Recombinant plasmid DNA was isolated with the Endo-free Plasmid Mini Kit (Omega, USA) following the manufacturer’s instructions and the concentration was measured in a spectrophotometer before the recombinant plasmid was aliquoted and conserved at −20 °C.
Characterization of pEGFP-N1-VP6 in vitro
In order to characterize the pEGFP-N1-VP6 clone in vitro, CIK cells were transfected with pEGFP-N1-VP6 using Lipofectamine 2000 (Invitrogen, USA). The Lipofectamine 2000 transfection reagent was used at a ratio of 20 μl Lipofectamine 2000: 8 μg recombinant plasmid DNA in accordance with the manufacturer’s instructions. Some transfected cells were examined at different times (day 1, 2, 3 and 4) for the presence of GCRV VP6 using fluorescence microscopy (Leica, DMIL LED). Meanwhile, at the times (12, 24, 36, 48, 72 and 96 h), transfected cells were measured using a flow cytometry (Beck Man, Cell LabQuanta™ SC) with a fixed 488 nm argon laser through the FL-1 detection channel to count the EGFP-positive cells. To obtain a stable transfected cell, which was carried out in MEM growth medium supplemented with 10 % FBS and 400 μg/mL G-418 (Sigma, USA). The medium containing G418 was changed every 2–3 days to compensate for loss of selection pressure. After 30 days, surviving cells were transferred to cell culture plates to obtain a cell line, which stably expressed GCRV VP6. Meanwhile, stable cell line was detected by fluorescence microscopy and flow cytometry.
GCRV VP6 expressed in CIK cells was detected with an IFA, and the fluorescent cells were observed by fluorescence microscopy. Briefly, after incubation of transfected and non-transfected CIK, cells were fixed with 3.7 % formaldehyde in PBS for 15 min and permeabilized with 0.1 % Triton X-100 in PBS for 1 min. The cells were washed with PBS and incubated with a polyclonal mouse antiserum against GCRV (prepared in our laboratory) for 30 min at room temperature. After washing with PBS, the cells were treated with a Cy3-labelled goat anti-mouse IgG antibody (Jiangsu, China) and examined by fluorescence microscopy (Nikon, Japan) [2].
A CPE assay was used to determine the antiviral activity against GCRV induced by pEGFP-N1-VP6 transfected CIK cells. This assay measures the percentage of cells against virus-induced lysis by calculating the proportion of cells that survive virus infection. Standard GCRV serum (TCID50 = 108.4) was diluted into nine concentrations, ranging from 10−1 to 10−9. Monolayers of the transfected cells (96 wells/sample) were infected with GCRV at various concentrations and pEGFP-N1 (mock plasmid) transfected cells were used as controls. The plates were incubated at 28 °C with GCRV until a viral CPE developed in the viral controls and the plates were then processed to evaluate percentage of CPE in monolayer. The antiviral activity was titrated on CIK cells by the TCID50 method [14].
Characterization of pEGFP-N1-VP6 in vivo
Seven-month-old grass carps (mean weight 10 g) were kindly provided by our experimental base, a facility with no history of viral disease. Fish were kept under a 12/12 h light/dark regime at 25 °C in 350 L closed re-circulating water tanks. The fish were fed daily with a diet of commercial pellets.
Fish were placed in a temperature controlled (25 °C) 80 L aquaria with dechlorinated water, and they were acclimatized for 1 week. In each group, 15 grass carps were t injected intramuscularly below the dorsal fin with one of the following [2]: (1) 1 μg of pEGFP-N1 diluted in 100 μL PBS; (2) 1 μg of pEGFP-N1-VP6 vaccine diluted in 100 μL PBS. Three fish from each group were sacrificed at 1, 2, 7, 15 and 30 dpi. Head kidney was removed aseptically and processed to isolate the total RNA in order to determine the expression of Mx, TLR3 and IgM mRNA by real-time PCR.
Amplifications were carried out in a Rotor-Gene Q Real-Time PCR Detection System (QIAGEN, Germany), using primers designed to amplify the grass carp β-actin (as an endogenous control), Mx, TLR3 and IgM genes (Table 1). Primers designed to amplify the GCRV VP6 gene were also used.
RT-PCR amplification was performed in a final volume of 20 and 1 μL of cDNA was added to the following mix in an individual PCR tube: 10 μL of iScript one-step RT-PCR supermix (Bio Rad), 1 μL of iScript one-step RT-PCR reverse transcriptase (Bio Rad), 7 μL of dH2O and 1 μL of a 20× mix containing the forward primer (18 μmol/L) and reverse primer (18 μmol/L). Fish injected with mock plasmids were used as the calibrator group for Mx, TLR3 and IgM. All samples were analyzed in triplicate and the results were expressed as a relative fold increase. The data were analyzed using the Rotor-Gene Q series Software and the relative quantification (comparative method) was calculated using ΔΔCt method [16]. All samples were normalized to the ΔCt value of a calibrator sample to obtain a ΔΔCt value (ΔCttarget − ΔΔCtcalibrator). The final relative expression was calculated using the following formula: 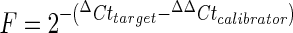 [16].
[16].
The value of the target normalized to the β-actin was expressed as 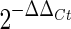 , where ΔCt was determined by subtracting the average β-actin Ct value from the average target Ct. The standard deviation of the difference was calculated from the standard deviation of the target and β-actin values, and ΔΔCt = ΔCt of samples of target gene −ΔCt of the calibrator.
, where ΔCt was determined by subtracting the average β-actin Ct value from the average target Ct. The standard deviation of the difference was calculated from the standard deviation of the target and β-actin values, and ΔΔCt = ΔCt of samples of target gene −ΔCt of the calibrator.
Immunization and detection of neutralizing antibodies
The pEGFP-N1-VP6 plasmid (1 μg diluted in 100 μL of PBS) was injected intramuscularly at the base of the dorsal fin of fish. Control fish were injected with the empty plasmid vector or with PBS. Treatment groups were placed in 80 L aquaria and maintained as specified above. After 30 days, fish blood was collected by caudal puncture using plastic disposable syringes with a 25-gauge needle (BD, USA). The blood sample was left at 4 °C overnight, centrifuged for 10 min at 1,600×g and the sera thereby isolated was stored at −70 °C until analysis for antibody production or the presence of virus-specific antibodies. A neutralization test (NT) was used to evaluate the humoral immune response against GCRV after DNA immunization. Briefly, constant amounts of GCRV were mixed with two-fold serial dilutions of fish sera, and the mixtures were then assayed in triplicate on CIK cells. After incubation for 3 days, the cell cultures were fixed and stained with crystal violet solution (2 % in ethanol). The titre of a serum was determined as the reciprocal of the serum dilution that reduced the viral infectivity (TCID50 mL−1) by approximately 50 % when compared to a negative control.
Challenge test
In each group, forty grass carps were injected intramuscularly below the dorsal fin with one of the following: (1) 1 μg of pEGFP-N1 diluted in 100 μL PBS; (2) 1 μg of pEGFP-N1-VP6 vaccine diluted in 100 μL PBS; (3) 100 μL PBS. At 30 days after vaccination, each fish was injected intramuscularly with 4.0 × 103 TCID50 of GCRV strain 104 in 100 μL PBS [4]. Mortalities were recorded daily for 14 days.
Results
Recombinant plasmid construction
The PCR product containing the 1257 bp segment of the GCRV VP6 gene was amplified from extracted RNA from GCRV infected CIK cells. The PCR amplified fragment and pEGFP-N1 were digested with BamHI and HindIII, respectively, and then ligated by T4 DNA ligase. Positive recombinant plasmid pEGFP-N1-VP6 was identified by restriction enzyme digestion, PCR amplification and sequencing (data not shown). The gene had inserted into the correct orientation and its sequence was identical to the GCRV VP6 reference strain (the accession no. JN967636).
Characterization of pEGFP-N1-VP6 in vitro
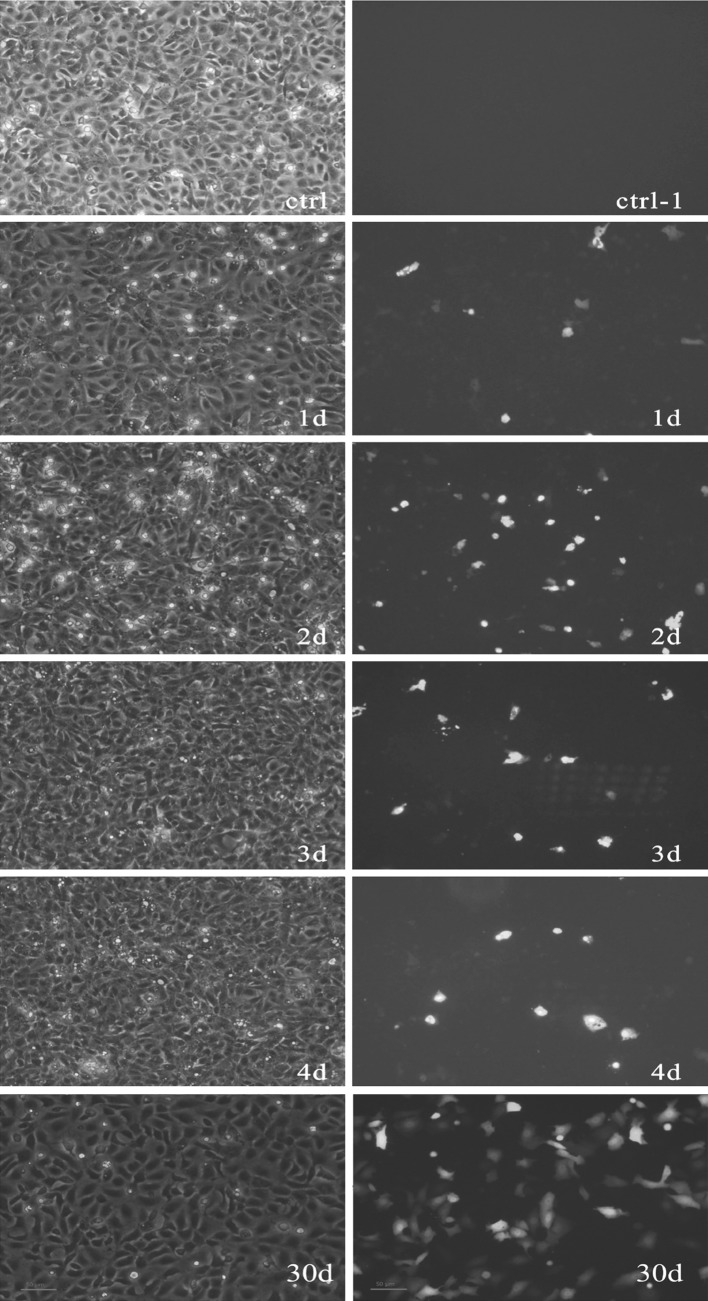
CIK cells were transfected with pEGFP-N1-VP6 using Lipofectamine 2000 and detected by fluorescent microscopy (Fig. 1). The 2nd day after the cells were transfected with pEGFP-N1-VP6, the percentage of fluorescent cells was approximately 40 %, and thereafter there were gradually fewer cells detected. When the CIK cells transfected with pEGFP-N1-VP6 were screened by G418 in 1 month, about 90 % fluorescent CIK cells were detected by fluorescent microscopy, which reflects the expression of GCRV VP6 in a large proportion of cells.
Fig. 1.
pEGFP-N1-VP6 expressed in CIK cells. Images were taken with white light (left) and fluorescent light (right). CIK cells expressing green fluorescent protein on different days (day 1, 2, 3 and 4) post transfection with pEGFP-N1-VP6 were examined by microscopy and fluorescent microscopy (control–4d) and the CIK cells transformed with pEGFP-N1-VP6 were screened by G418 in 1 month (30d)
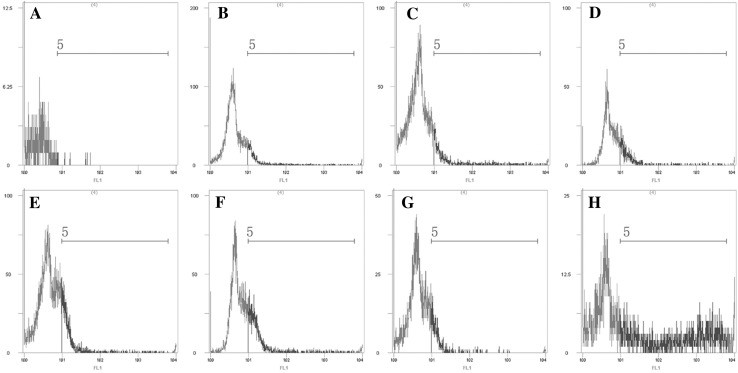
In addition, EGFP was used in flow cytometry to quantitatively analyze the interactions between the fusion protein and CIK cells. As shown in Fig. 2b–g, blue wave lines were evident in the cells transfected with pEGFP-N1-VP6, while nearly no fluorescence was detected in untransfected cells (Fig. 2a). The expression of EGFP was detected at 12 h (about 3.6 % of cells) after transfection, reached 12 % of cells at 48 h, and then gradually became weaker. The same result was observed with fluorescent microscope and analysis with flow cytometry. When the CIK cells transfected with the pEGFP-N1-VP6 were screened by G418 after 1 month, the expression of GCRV VP6 and EGFP fusion protein was detected in nearly all CIK cells (Fig. 2h).
Fig. 2.
Flow cytometry evaluation of Ctenopharyngodon idellus kidney cells expressing pEGFP-N1-VP6. Untransfected cells (a); transfected cells 12, 24, 36, 48, 72 and 96 h post-transfection (b–g), respectively; transfected cells are screened by G418 in 1 month (h)
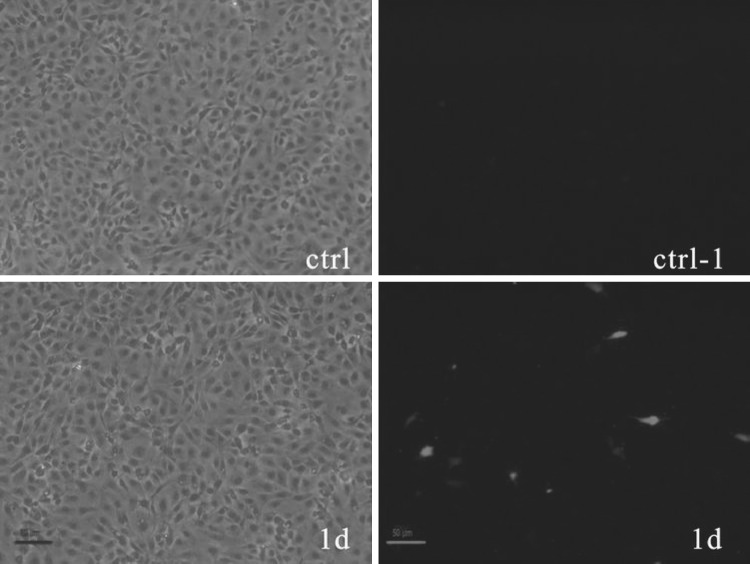
GCRV VP6 expression was assessed by IFA using a polyclonal antiserum raised against the GCRV VP6. As shown in Fig. 3, red fluorescence was evident in the cytoplasm of cells transfected with pEGFP-N1-VP6 (Fig. 31d), while no fluorescence was detected in untransfected cells (Fig. 3ctrl).
Fig. 3.
Immunofluorescent staining of pEGFP-N1-VP6 infected CIK cells. Images were taken with white light (left) and fluorescent light (right). The CIK cells transformed with pEGFP-N1-VP6 were first detected as positive by immunofluorescence on day 1 post-transfection. There was no fluorescence in uninfected CIK cells
Antiviral activity assay
Standard GCRV serum (TCID50 = 108.4) was diluted into nine concentrations, ranging from 10−1 to 10−9, and was inoculated into the transfected CIK cells of each group, respectively. The TCID50 of GCRV VP6 expressing cells and pEGFP-N1 transfected cells was 105.3 and and 107.2, respectively. These results were confirmed in the virus yield reduction assay, in which the virus titer was 100-fold lower in the GCRV VP6 expressing cells than in pEGFP-N1 transfected cells. All theses results suggested that GCRV VP6 has a direct or indirect antiviral effect against GCRV in vitro, although the plasmid also induces some protection against these viruses.
Real-time PCR analysis to study the expression changes of TLR3, Mx and IgM genes after injection with pEGFP-N1-VP6
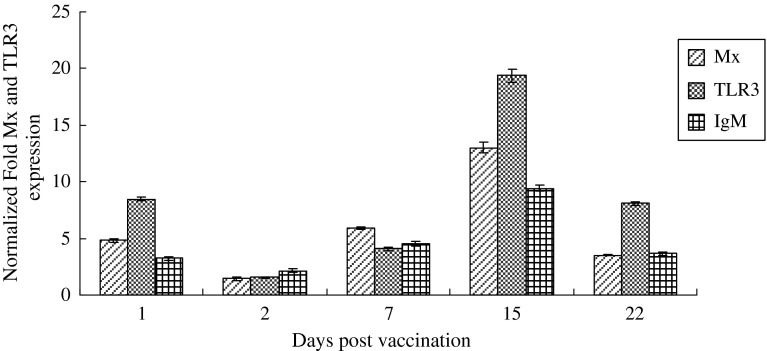
The effect of the DNA vaccine expressing the GCRV VP6 on the kinetics of Mx, TLR3 and IgM mRNA expression was assayed in the head kidney of grass carp. Fish injected with pEGFP-N1 were used as negative controls. In the head kidney of fish injected with pEGFP-N1-VP6, TLR3 expression was significantly up-regulated to 8-fold on the 1st dpi when compared with the controls. The TLR3 expression decreased to basal levels on the 2nd day and then progressively increased until reaching strong expression on the 15th day (20-fold). Similar changes were also observed for Mx and IgM gene expressions (Fig. 4). In addition, the permanence of GCRV VP6 expression at the 30th dpi was checked and confirmed by RT-PCR.
Fig. 4.
Real-time PCR analysis to study the changes in TLR3, Mx and GCRV VP6 gene expression after injection of pEGFP-N1-VP6. Expression of Mx, TLR3 and IgM genes in grass carp receiving an intramuscular injection (1 μg in 50 μL PBS) of empty plasmid or pEGFP-N1-VP6. On day 1, 2, 3, 15, and 22, fish (n = 3 fish/group) were sampled and head kidney tissues were pooled and processed to evaluate the gene expression by real-time PCR
Levels of GCRV specific antibodies
The ability of the VP6 protein expressed in the vaccinated fish to induce specific anti-GCRV antibodies was examined in a neutralizing assay with serum samples from fish 30 days post-vaccination. The level of anti-GCRV-neutralizing antibodies is shown in Table 2. No antibodies were detected in the PBS or pEGFP-N1 infected fish group, while in the pEGFP-N1-VP6 vaccinated fish group the neutralizing titres of the antibodies detected ranged from 160 to 320.
Table 2.
GCRV neutralizing titres in sera from fish injected with pEGFP-N1-VP6
| pEGFP-N1-VP6 vaccined fish | pEGFP-N1 infected fish | PBS infected fish | |||
|---|---|---|---|---|---|
| Fish | Titre | Fish | Titre | Fish | Titre |
| V1 | 320 | N1 | 5 | P1 | 5 |
| V2 | 320 | N2 | 5 | P2 | 10 |
| V3 | 160 | N3 | 10 | P3 | 5 |
| V4 | 320 | N4 | 10 | P4 | 5 |
| V5 | 160 | N5 | 5 | P5 | 5 |
Sera were obtained from individual fish at 30 days after DNA injection. V, N and P are pEGFP-N1-VP6 vaccinated fish, pEGFP-N1 injected fish and PBS injected fish, respectively
Protection of grass carp from GCRV challenge
Fish vaccinated with the pEGFP-N1-VP6 DNA vaccine were able to survive better than the controls after the GCRV challenge. Fourteen days after the GCRV challenge, cumulative mortalities of the fish injected with 1 μg of pEGFP-N1-VP6 were about 15 %, while the cumulative mortalities of the control groups were 90 % (Table 3).
Table 3.
Cumulative percentage mortality (%) and calculated RPS values following challenge with GCRV in experimental groups of DNA vaccinated fish by i.m. injection
| Injected plasmid | No. of injected virus: 4.0 × 103 TCID50/fish | |
|---|---|---|
| Mortalities death/total | RPSa | |
| pEGFP-N1-VP6 | 15 % (6/40) | 83.33 |
| pEGFP-N1 | 90 % (36/40) | 0 |
| PBS | 90 % (36/40) | – |
a RSP Relative percentage survival = {1 − [% mortality (vaccinated)/% mortality (control)]} × 100
Discussion
The GCRV, which is considered to be the most pathogenic aquareovirus, can cause a large number of deaths in grass carp and adversely affect the progression of fresh water aquaculture [5]. Although some experimental vaccines for GCRV have been developed, only few have been commercialized, and the protective effect against GCRV demonstrated in laboratory trials are not consistent with field observations. This may be due to the fact that in the water the fish may be exposed to several other pathogens in addition to GCRV. Every year, many grass carp farms and hatcheries suffer severe economic losses, due to GCRV outbreaks. It has been speculated that the high impact of GCRV, despite the availability of the vaccine in China, could be due to the poor antigenic nature of the GCRV antigens produced in different expression systems [8]. There is a great necessity for new and improved vaccines for early vaccination of grass carps before they naturally get infected with GCRV. In this sense, DNA vaccines are promising tools since they have proved extremely effective for grass carps. In the present study, the DNA vaccine was designed according to the coding sequence of GCRV VP6 gene. The GCRV VP6 comes from GCRV strain 104, which is a new species of the genus Aquareovirus and is distantly related to any known species within this genus [4, 22, 25]. We evaluated its processing in vitro and its in vivo effects on grass carps immune response by induction of immune gene expression. This current study has important significance in the development of a DNA vaccine for GCRV.
First, we used a cell-free expression system to investigate the proteins expressed by pEGFP-N1-VP6. We successfully constructed a co-expression vector and obtained co-expression protein of GCRV VP6 and EGFP in CIK cells. Through fluorescence microscopy and flow cytometry, we confirmed the expression of the fusion protein at the highest level 48 h after pEGFP-N1-VP6 transfection of CIK cells. A CIK cell line constitutively expressing GCRV VP6 was established and selected in growth medium with the G-418, and the cells showed a direct or indirect antiviral effect against GCRV in vitro. The specific binding between the fusion protein and polyclonal mouse GCRV VP6-specific antiserum indicated that the protein had been translated in vitro and showed specific immunogenicity. In addition, our results demonstrated that expression of the GCRV VP6 in a cell line induced an antiviral state that was active against GCRV. Although there was a decrease in cell survival as the infective titer of the inoculum increased, the protection against virus-induced lysis in GCRV VP6 expressing cells was more effective. This may indicate a synergy of GCRV VP6 and viral infections in the activation of antiviral states. The virus showed a 100-fold reduction in its yield and its CPE was reduced when this established cell line was infected. Our results show that a non-specific antiviral response was produced in GCRV VP6-transfected cells, as GCRV was similarly inhibited. However, the mock plasmid also induced some protection against viral infections, but to a lesser extent than the plasmid encoding GCRV VP6. The most possible reason is that the high concentrated antibiotic G418 screening used in CIK cells might reduce the cells’ susceptibility to GCRV infection.
In vivo, the recombinant plasmid encoding the GCRV VP6 was used as a DNA vaccine and it produced an early up-regulation of cytokine-related gene expression, including Mx, TLR3 and IgM in head kidneys of grass carps. Our results show there is a strong increase in TLR3 mRNA expression at 1 and 15 dpi, as well as that of other cytokines such as Mx and IgM in grass carps after vaccination. Mx, TLR3 and IgM genes are markers of the non-specific innate immune responses to viruses, and their involvement in antiviral defenses has been documented extensively. Injected pEGFP-N1-VP6 could enhance the immunity of fish. The presence of GCRV-neutralizing antibodies was demonstrated on the 30th day post-vaccination in most of the fish tested. Anti-GCRV-neutralizing antibodies were found in serum from five grass carps injected with the pEGFP-N1-VP6 vaccine, while no neutralization occurred with the sera from the control fish. The time when mortality was first observed after GCRV challenge was also different between the vaccinated and control groups. Almost all control fish exhibited acute mortality starting on the 8th day after challenge, following which low mortalities were observed until the termination of the experiment. The vaccinated grass garps, however, had lower mortality rates than that of the controls throughout the challenge period. In line with this observation, these results indicated that anti-GCRV-neutralizing antibodies play a crucial role in mounting a protective immune response against viral diseases in fish. The present immunization experimental results are in accordance with previous studies [10, 24], which proved the good immune efficacy of a GCRV (873 strain) VP6 DNA vaccine.
In conclusion, a recombinant plasmid encoding the GCRV VP6 was constructed and its antiviral activity was studied in vitro by establishing CIK cells that constitutively expressed GCRV VP6. The usefulness of the DNA vaccine of GCRV VP6 in studying the early immune responses was evaluated. The results showed that GCRV VP6 expression in vitro inhibits the virus-induced cytopathic effect to some extent, as well as the viral yield of GCRV. In vivo, the vaccine induced innate immunity responses with a more than 10-fold increase in Mx, TLR3 and IgM expression levels at 15 days post vaccination. VP6 expressed in grass carp, induced specific immune response and protected fish against viral challenge infection. Future work will focus on the characterization of GCRV VP6 and co-expression of other GCRV structural proteins in vitro and in vivo.
Acknowledgments
This work was supported by the earmarked fund for China Agriculture Research System (CARS-46).
Footnotes
Yong Zhou and Yu-ding Fan have contributed equally to this paper.
References
- 1.Ahne W. Viral infectious of aquatic animals with special reference to Asian aquaculture. Annu Rev Fish Dis. 1994;4:375–388. doi: 10.1016/0959-8030(94)90036-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ana IH, Sara IPP, Sylvia RS. In vitro and in vivo immune responses induced by a DNA vaccine encoding the VP2 gene of the infectious pancreatic necrosis virus. Fish Shellfish Immunol. 2009;27:120–129. doi: 10.1016/j.fsi.2008.11.021. [DOI] [PubMed] [Google Scholar]
- 3.Dhar AK, Bowers RM, Rowe CG, Allnutt FC. Expression of a foreign epitope on infectious pancreatic necrosis virus VP2 capsid protein subviral particle (SVP) and immunogenicity in rainbow trout. Antivir Res. 2010;85:525–531. doi: 10.1016/j.antiviral.2009.12.009. [DOI] [PubMed] [Google Scholar]
- 4.Fan YD, Rao SJ, Zeng LB, Ma J, Zhou Y, Xu J, Zhang H. Identification and genomic characterization of a novel fish reovirus, Hubei grass carp disease reovirus, isolated in 2009 in China. J Gen Virol. 2013;94:2266–2277. doi: 10.1099/vir.0.054767-0. [DOI] [PubMed] [Google Scholar]
- 5.Fang Q, Ke LH, Cai YQ. Growth characteristics and high titer culture of grass carp hemorrhage virus (GCHV)-873 in vitro. Virol Sin. 1989;4:315–319. [Google Scholar]
- 6.Fang Q, Shah S, Liang Y, Zhou ZH. 3D reconstruction and capsid protein characterization of grass carp reovirus. Sci China C Life Sci. 2005;48:593–600. doi: 10.1360/062004-105. [DOI] [PubMed] [Google Scholar]
- 7.Fang Q, Seng EK, Dai W. Construction and co-expression of grass carp reovirus VP6 protein and enhanced green fluorescence protein in the insect cells. Virol Sin. 2007;22(5):397–404. doi: 10.1007/s12250-007-0038-8. [DOI] [Google Scholar]
- 8.He Y, Xu H, Yang Q. The use of an in vitro microneutralization assay to evaluate the potential of recombinant VP5 protein as an antigen for vaccinating against Grass carp reovirus. Virol J. 2011;8(1):132–138. doi: 10.1186/1743-422X-8-132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Heppell J, Davis HL. Application of DNA vaccine technology to aquaculture. Adv Drug Deliv Rev. 2000;43:29–43. doi: 10.1016/S0169-409X(00)00075-2. [DOI] [PubMed] [Google Scholar]
- 10.Liu L, Xu SY, Li JH. Evaluation of immune efficacy of GCRV vp6 DNA vaccine. J Fish Sci China. 2012;19(5):841–847. doi: 10.3724/SP.J.1118.2012.00841. [DOI] [Google Scholar]
- 11.Lupiani B, Hetrick FM, Samal SK. Genetic analysis of aquareoviruses using RNA–RNA blot hybridization. Virology. 1993;197:475–479. doi: 10.1006/viro.1993.1616. [DOI] [PubMed] [Google Scholar]
- 12.Nason EL, Samal SK, Venkataram PBV. Trypsin induced structural transformation in aquareovirus. J Virol. 2000;74:6546–6555. doi: 10.1128/JVI.74.14.6546-6555.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rangel AA, Rockemann DD, Hetrick FM, Samal SK. Identification of grass carp hemorrhage virus as a new genogroup of aquareovirus. J Gen Virol. 1999;80:2399–2402. doi: 10.1099/0022-1317-80-9-2399. [DOI] [PubMed] [Google Scholar]
- 14.Reed LJ, Nuench H. A simple method of estimating fifty per cent endpoints. The Am J Hyg. 1938;27(3):493–497. [Google Scholar]
- 15.Regenmortel MHV, Fauquet CM, Bishop DHL. Herpesvirus family. In virus taxonomy: classification and nomenclature of viruses, 7th ICTV report. San Diego: Academic Press; 2000. pp. 395–480. [Google Scholar]
- 16.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative CT method. Nat Protoc. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 17.Su JG, Jang SH, Yang CR. Genomic organization and expression analysis of Toll-like receptor 3 in grass carp (Ctenopharyngodon idella) Fish Shellfish Immunol. 2009;27:433–439. doi: 10.1016/j.fsi.2009.06.009. [DOI] [PubMed] [Google Scholar]
- 18.Su JG, Yang CR, Zhu ZY. Enhanced grass carp reovirus resistance of Mx-transgenic rare minnow (Gobiocypris rarus) Fish Shellfish Immunol. 2009;26:828–835. doi: 10.1016/j.fsi.2008.12.007. [DOI] [PubMed] [Google Scholar]
- 19.Subramanian K, Hetrick FM, Samal SK. Identification of a new genogroup of aquareovirus by RNA–RNA hybridization. J Gen Virol. 1997;78(6):1385–1388. doi: 10.1099/0022-1317-78-6-1385. [DOI] [PubMed] [Google Scholar]
- 20.Tom CT, Jarl B, Roy AD. What happens to the DNA vaccine in fish? A review of current knowledge. Fish Shellfish Immunol. 2008;25:1–18. doi: 10.1016/j.fsi.2008.03.007. [DOI] [PubMed] [Google Scholar]
- 21.Wang XX, Sun BJ, Chang MX, Nie P. The sequence and expression of the immunoglobulin M heavy chain cDNA of Ctenopharyngodon idellus. J Fish China. 2008;32:13–20. [Google Scholar]
- 22.Wang Q, Zeng WW, Liu C. Complete genome sequence of a reovirus isolated from grass carp, indicating different genotypes of GCRV in China. J Virol. 2012;86(22):12466. doi: 10.1128/JVI.02333-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xu SY, Li HL, Deng GC. The preparation and immune effect of attenuated live vaccine obtained through cell culture for hemorrhage of grass carp. J Fish China. 1994;18(2):110–116. [Google Scholar]
- 24.Xue RY, Liu L, Cao GL, Xu SY, Li JH, Zou Y, Chen H, Gong CL. Oral vaccination of BacFish-vp6 against grass carp reovirus evoking antibody response in grass carp. Fish Shellfish Immunol. 2013;34:348–355. doi: 10.1016/j.fsi.2012.11.024. [DOI] [PubMed] [Google Scholar]
- 25.Ye X, Tian YY, Deng GC. Complete genomic sequence of a reovirus isolated from grass carp in China. Virus Res. 2012;163(1):275–283. doi: 10.1016/j.virusres.2011.10.014. [DOI] [PubMed] [Google Scholar]
- 26.Zhang X, Tang J, Walker SB, O’Hara D, Nibert ML, Duncan R, Baker TS. Structure of avian orthoreovirus virion by electron cryomicroscopy and image reconstruction. Virology. 2005;343:25–35. doi: 10.1016/j.virol.2005.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang LL, Shen JY, Lei CF, Li XM, Fang Q. High level expression of grass carp reovirus VP7 protein in prokaryotic cells. Virol Sin. 2008;23:51–56. doi: 10.1007/s12250-008-2921-3. [DOI] [Google Scholar]
- 28.Zhang X, Lei J, Fang Q, Zhou ZH. 3.3A° Cryo-EM structure of a nonenveloped virus reveals a priming mechanism for cell entry. Cell. 2010;141:472–484. doi: 10.1016/j.cell.2010.03.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zuo WG, Qian HX, Xu YF. A cell line derived from the kidney of grass carp (Ctenopharyngodon idellus) J Fish China. 1983;10:11–17. [Google Scholar]