Abstract
ATP-gated purinergic P2X4 receptors (P2X4Rs) are the most alcohol-sensitive P2XR subtype. We recently reported that ivermectin (IVM), an antiparasitic used in animals and humans, antagonized ethanol inhibition of P2X4Rs. Furthermore, IVM reduced ethanol intake in mice. The first molecular model of the rat P2X4R, built onto the X-ray crystal structure of zebrafish P2X4R, revealed an action pocket for both ethanol and IVM formed by Asp331, Met336 in TM2 and Trp46, and Trp50 in TM1 segments. The role of Asp331 and Met336 was experimentally confirmed. The present study tested the hypothesis that Trp46 plays a role in ethanol and IVM modulation of P2X4Rs. Trp46 was mutated to residues with different physicochemical properties and the resultant mutants tested for ethanol and IVM responses using Xenopus oocyte expression system and two-electrode voltage clamp. Nonaromatic substitutions at position 46 reduced ethanol inhibition at higher concentrations and switched IVM potentiation to inhibition. Simultaneous substitution of alanine at positions Trp46 and Met336 also resulted in similar changes in ethanol and IVM responses. Furthermore, a new molecular model based on the open pore conformation of zebrafish P2X4R suggested a role for Tyr42 that was further supported experimentally. Our previous and current findings, combined with our preliminary evidence of increased ethanol consumption in P2X4R knockout mice, suggest that the ethanol and IVM action pocket in P2X4Rs formed by positions 42, 46, 331, and 336 presents a potential target for medication development for alcohol use disorders.
Keywords: ATP-gated P2X4 receptors, Ethanol and ivermectin sensitivity, Site-directed mutagenesis, Xenopus oocytes, Whole-cell two-electrode voltage clamp, Position 46, GluCl, Molecular model, Homology model, Zebrafish
Introduction
Alcohol use disorders (AUDs) have a staggering socioeconomic impact, yet current therapeutic strategies to treat AUDs remain a serious unmet medical need [1]. Presently, mechanism(s) of ethanol action that cause and modulate its effects are not well understood. This information is critical for development of effective medications aimed to prevent or treat AUDs.
ATP-gated P2X receptors (P2XRs) are a superfamily of cation-permeable ligand-gated ion channels (LGICs) [2, 3]. Currently, seven P2XRs subunits have been identified (P2X1–P2X7) that form homo- or heterotrimeric channels. Each P2X subunit consists of two transmembrane (TM) segments, a large extracellular domain (ectodomain), and intracellular amino (N) and carboxy (C) terminals (for review, see [3, 4]). A previous 3.1-Å resolution closed-state crystal structure of zebrafish P2X4Rs verified the trimeric construction of P2XRs [5], which was confirmed by recent crystal structures of zebrafish P2X4Rs in the presence or absence of ATP at 2.8 and 2.9-Å resolution. The later models revealed a previously unseen ATP-binding motif and an open pore conformation [6].
P2X4Rs are the most abundant P2XR subtype expressed in the brain. This includes key brain regions implicated in the reinforcing properties of alcohol and other drugs, such as the striatum [7–10]. P2X4Rs are the most alcohol-sensitive P2XR subtype known [11]. In vitro, they are inhibited by a broad range of ethanol concentrations beginning well below the concentrations considered legally intoxicating in the United States (0.08 % or 17 mM) [11–17]. Evidence from genetic and genomic studies [18, 19] suggests that these receptors play a role in regulating ethanol intake.
Ivermectin (IVM), an antiparasitic drug used in humans and animals, is a selective positive modulator of P2X4Rs [20, 21]. We have shown that IVM antagonizes ethanol-mediated inhibition of P2X4Rs in vitro [12]. Moreover, IVM significantly reduced ethanol intake and preference in mouse models of ethanol self-administration [22]. These findings suggest that IVM's anti-alcohol effects result, at least in part, from blocking the action of ethanol on P2X4Rs.
The sites of ethanol and IVM action in P2X4Rs are not known. Previous investigations suggest that the interface between the ectodomain and the TM segments in P2X4Rs contains targets for ethanol and IVM action or modulation [17, 23, 24]. We have identified residues Asp331 and Met336 in the ectodomain–TM2 segment interface to be important for ethanol modulation, whereas residues Asn338, Ser341, Gly342, Leu346, Gly347, Ala349, and Ile356 in the TM2 segment were found to play a role in IVM modulation of the receptor [25, 26]. Furthermore, we found that Met336 at the ectodomain–TM2 interface is a common site of action for both ethanol and IVM [12]. Interestingly, these sites are closely located to lateral fenestrations found in P2X4Rs that were shown to be sites for allosteric modulation of the receptor [27], including by IVM [28].
The molecular model built on the zebrafish P2X4R structure in the closed state [5] revealed a common action pocket for ethanol and IVM formed by residues at the ectodomain–TM2 segment interface: Asp331 and Met336, and TM1 segment residues Trp46 and Trp50. While the role for Asp331 and Met336 in TM2 as putative sites of ethanol action were supported experimentally, as well as with the model [14], the roles of Trp46 and Trp50 have not been fully tested experimentally. Our initial investigation demonstrated that mutating Trp46, but not Trp50, at the interface of the TM1 segment and the ectodomain to alanine significantly altered ethanol effect compared to that of WT P2X4Rs [14].
The present study focused on Trp46 and tested the hypothesis that this is a site for ethanol and IVM action. We mutated Trp46 to residues with different physicochemical properties and tested the effects of these substitutions on the sensitivity of the receptor to the agonist and the effects of ethanol and IVM. We have also tested effects of ethanol and IVM on the Trp46–Met336 double alanine mutant. The results support the hypothesis that position 46 is important for ethanol and IVM action. These data led to a new model of the site of ethanol and IVM action on P2X4Rs.
Materials and methods
Materials
Adenosine 5′-triphosphate disodium salt (ATP), ethanol (95 %, USP), collagenase, and IVM (ivermectin B1A) were purchased from Sigma-Aldrich (St. Louis, MO, USA). Stock solution of IVM (10 mM) were made in dimethyl sulfoxide and kept at −20 °C. All other chemicals were of reagent grade.
Isolation of Xenopus laevis oocytes and cRNA injections
Xenopus oocytes were isolated from X. laevis (Nasco, CA, USA) and maintained as described previously [11, 13]. Stages V and VI oocytes were selected, rinsed, and stored in incubation medium containing (in millimolars), NaCl 96, KCl 2, MgCl2 1, CaCl2 1, HEPES 5, theophylline 0.6, pyruvic acid 2.5, with 1 % horse serum, and 0.05 mg/ml gentamycin. The following day, oocytes were injected (32 nl) cytosolically with capped ribonucleic acid (cRNA) encoding rat P2X4 receptor (20 ng per oocyte). The injections were performed with Nanoject II Nanoliter injection system (Drummond Scientific, Broomall, PA, USA). The injected oocytes were stored in incubation medium, in Petri dishes (VWR, San Dimas, CA, USA) at 18 °C and usually expressed WT and mutant P2X4Rs on day 2 postinjection. Oocytes were used in electrophysiological recordings for 3–7 days after cRNA injection. Water-injected and noninjected oocytes were used as negative controls during the experiments. Defolliculated oocytes do not possess native P1 or P2 receptors that might otherwise complicate the analysis of agonist activity [29, 30].
Site-directed mutagenesis
Mutations were generated in vitro by using QuikChange II XL Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA, USA). Primers were purchased from Integrated DNA Technologies (Coralville, IA, USA). Mutations were sequence verified (DNA Core Facility, University of Southern California, Los Angeles, CA, USA).
cRNA synthesis
The DNA of wild type (WT) or mutant receptors was linearized and transcribed using the mMESSAGE mMACHINE kit (Ambion, Austin, TX, USA) to result in cRNA, which was stored at −70 °C until injection.
Whole-cell voltage clamp recording
WT and mutant receptors were screened using an eight-channel, computer-controlled automated two-electrode voltage clamp system, OpusXpress (Molecular Devices, Union City, CA, USA). Oocytes were placed in eight recording chambers (volume 200 μl), superfused with P2X buffer solution (in millimolars) (NaCl 110, KCl 2.5, BaCl2 1.8, HEPES 10, pH 7.5) at a speed of 3–4 ml/min, and impaled with two-glass electrodes filled with 3 M KCl (0.5 to 2 MΩ). The membrane potential was held at −70 mV, and currents were sampled at 5 kHz and filtered at 1 kHz. The currents acquired were stored on the computer for offline analysis using Axon pCLAMP 9 software (Axon Instruments, Union City, CA, USA).
Experimental procedures
All experiments were performed at room temperature (20–23 °C). Ethanol and/or IVM were co-applied with ATP for 20 s. Submaximal ATP concentrations that ranged from EC5 to EC10 were used in these studies that are designated as EC10 throughout the manuscript. We have previously shown that ethanol inhibition of ATP function on P2X4Rs is more robust and reliable when tested in the presence of low concentrations of ATP (typically EC5–20) [11, 13]. ATP-activated EC10 responses were measured before and after each ethanol application to take into account possible shifts in the baseline current values. Ethanol or IVM did not affect the resting membrane currents in oocytes expressing P2X4Rs in the absence of agonists or in uninjected oocytes. To generate ATP concentration curves, ATP in the range of 0.05–100 μM were applied for 20 s followed by 15-min washout for higher ATP and 5–10 min for lower ATP concentrations.
Molecular modeling
We built a homology model of rat P2X4R by threading the primary sequence of rat P2X4R onto the recent X-ray structure of the zebrafish P2X4R [12]. Briefly, we used the crystal structure of the open conformation of zebrafish P2X4R-A (PDB ID 4DW1) [6] as a template for the homology models of the rat P2X4R. The homology was high (identity 62.3 %, similarity 79.3 %) and, after editing the C- and N-terminal segments to fit the template, it was possible to align the sequences without gaps. We threaded the rat sequence onto the backbone atoms of the template using the Modeler module of Discovery Studio 3.5 (DS 3.5; Accelrys, San Diego, CA, USA), essentially as previously described for GABAA and glycine receptors models [31]. Then, we used the autorotamer module of Discovery Studio to optimize the positions of the side chains, while the backbone atoms were tethered. We assigned the CHARMM force field to all atoms and optimized the resulting model by tethering the backbone atoms with a harmonic restraint of 10 kcal/A2 and then optimized to a gradient of 0.0001 kcal/A. Before adding the IVM ligand, as described below, we relaxed the initial model with molecular dynamics for 100,000 1-fs steps at 300 K. Because our focus was on the fit of IVM into a putative binding site, we used harmonic restraints on the backbone atoms instead of embedding the P2X4 model in an explicit membrane.
We noted that the recent X-ray structure of the pentameric glutamate chloride channel (GluCL, PDP ID 3RHW) [32] contains five IVM molecules positioned between each of the five subunits and within the membrane, but near the membrane–water interface. We expected that this orientation of IVM in GluCl would present the moieties known to be important to binding toward the P2X4R structure and should make a good starting point for modeling. However, since GluCl is a pentamer and P2X4R is a trimer, we were not able to use typical superposition algorithms to align the subunits. Instead, we chose one pair of subunits from the GluCl X-ray structure with an IVM molecule between them at the subunit interface. Then, we manually superimposed this pair of subunits with IVM onto our new model of rat P2X4R. First, we moved the two GluCl subunits horizontally in the plane of the membrane to line up the channel pores. Then, we moved the two GluCl subunits vertically along the ion pore axis to get the IVM molecule between two rat P2X4R subunits and between Trp46, Trp50, and Met336. The vertical movement continued until the interface between the helices and the extracellular domains of both models lined up. Then, the pair of GluCl subunits was removed, but the IVM molecule remained in place to preserve the orientation of IVM with respect to the membrane and the model interface that was described in the GluCl X-ray structure [32].
We expected that the dissimilarity between the nearly parallel alpha helices of GluCl compared to the sharply angled helices in the model of P2X4R would cause some initial van der Walls overlaps with unfavorable energy. Therefore, we assigned the CHARMM force field to all atoms and optimized the resulting model of IVM in P2X4R with 100,000 1-fs steps of molecular dynamics at 300 K with a harmonic restraint of 10 kcal / (mol A2) on all atoms. After the major “bumps” in the model were removed, the harmonic restraint on IVM was removed but a restraint of 10 kcal / (mol A2) was imposed on only the backbone atoms of the P2X4R model. After 5,000,000 2-fs steps (10 ns) of molecular dynamics at 300 K, the IVM remained positioned in the intersubunit site between the amino acid residues Trp46, Trp50 of subunit 1, Met336 of subunit 2, and the ectodomain.
This model revealed that IVM made two H bonds to the phenolic OH of Tyr 42 in P2X4. We tested this new interaction in two ways: (1) computationally, by “mutating” the model described above to have Y42F and then reproducing the previously described optimization and 5,000,000 2-fs steps of molecular dynamics and (2), experimentally, by making, expressing, and testing the Y42F mutant in oocytes. We tested the hypothesis that the mutation would reduce the affinity of IVM for the binding site and change its position in the site.
Data analyses
Results are presented as a percentage change in ATP EC10-activated currents (nanoamperes) after normalizing them with the response obtained with agonist alone. All results are expressed as mean ± SEM. Data were obtained from oocytes from at least two different frogs.The n refers to the number of different oocytes tested. Significant differences were determined by either paired or unpaired Student's t test, as appropriate. Prism (GraphPad Software, San Diego, CA, USA) was used to perform statistical analyses and curve fitting. The ATP concentration response data were fitted to a concentration–response curve using the logistic equation: I / Imax = 100 * [drug]n / ([drug]n + (EC50)n), where I/Imax is the percentage of the maximum obtainable response, EC50 is the concentration producing a half-maximal response and n is the Hill coefficient (nH). Statistical significance was defined as P < 0.05.
We used linear regression analyses (Prism) to test for possible relationships between the responses to ethanol, IVM, and physicochemical properties (molecular volume, molecular weight, hydrophobicity, and hydropathy) of the substituted residues taking into consideration both the mean and variance of the data. Molecular volumes were calculated using Peptide Property Calculator (Northwestern University Medical School). Hydrophobicity values were assigned to amino acid residues using the Wimley–White whole-residue hydrophobicity scales [33]. Hydropathy values were assigned using the Kyte–Doolittle hydropathy scale[34].
Results
Agonist sensitivity of WT and position 46 mutant P2X4Rs
WT and all mutant receptors yielded measurable ATP-gated currents (see Imax values, Table 1). The EC50 value for the WT P2X4Rs was similar to EC50 values reported by others [15, 35, 36]. There were statistically significant differences in EC50 and Hill slopes for some of the mutant receptors versus WT (Table 1). Position 46 is not located near the proposed ATP binding site [6], suggesting that the changes in agonist properties are not likely the result of changes in ATP binding.
Table 1.
I max, EC50, and Hill slope values for ATP concentration–response curves of WT and W46 mutant P2X4Rs. I max represents peak currents generated by 100 μM ATP. EC50 and Hill slope values were determined from ATP concentration–response curves. Data shown are mean ± SEM, n = 4–11
| Receptor | ATP | ||
|---|---|---|---|
| I max | EC50 (μM) | Hill slope | |
| WT P2X4 | 4,163 ± 1,504 | 6.05 ± 0.53 | 1.57 ± 0.22 |
| W46Y | 3,078 ± 816 | 3.77 ± 0.98 | 1.82 ± 0.40 |
| W46F | 4,847 ± 1,152 | 4.13 ± 1.12 | 2.08 ± 0.62 |
| W46D | 2,059 ± 373 | 1.24 ± 0.31* | 1.09 ± 0.11 |
| W46K | 6,523 ± 962 | 1.97 ± 0.29* | 0.75 ± 0.12* |
| W46N | 3,711 ± 717 | 1.37 ± 0.45* | 0.64 ± 0.07* |
| W46L | 2,118 ± 241 | 3.59 ± 1.20 | 1.18 ± 0.27 |
| W46V | 2,394 ± 192 | 1.85 ± 0.21* | 1.06 ± 0.15 |
| W46A | 2,593 ± 363 | 3.18 ± 0.52 | 0.97 ± 0.07* |
* P < 0.05 compared to the EC50 values of WT P2X4R
Aromaticity and molecular volume of the residue at position 46 affect ethanol sensitivity
Consistent with our previous findings [12, 14], ethanol (10–200 mM) inhibited ATP-induced currents in WT P2X4Rs in a reversible and concentration-dependent manner (Fig. 1a, b). Substituting the naturally occurring aromatic tryptophan residue at position 46 (Trp), with other aromatic residues (Tyr or Phe), did not significantly alter ethanol inhibition compared to WT at all ethanol concentrations (Fig. 1, Groups WT, Y, and F). In contrast, nonaromatic mutations significantly decreased sensitivity to higher ethanol concentrations (100 and 200 mM) with hydrophilic substitutions (Asp, Lys, and Asn) producing a 20–25 % reduction from WT (Fig. 1, Groups D, K, and N) and hydrophobic substitutions (Leu and Val) producing a much greater reduction (50 %) in response to ethanol (Groups L and V). Interestingly, the smallest, hydrophobic nonaromatic substitution, alanine, further reduced ethanol response at 100 mM and essentially eliminated it at 200 mM ethanol. These findings suggest that the physicochemical characteristics of the residue at position 46 play a key role in determining the P2X4R sensitivity to ethanol.
Fig. 1.
Ethanol concentration–response curves for position 46 mutant P2X4Rs. (a) Concentrations in the range of 10–200 mM were used. There were no significant differences among the mutants with respect to sensitivity to low ethanol concentrations (10, 25, and 50 mM) compared to WT P2X4R. However, at 100 and 200 mM ethanol, all mutations, with the exception of the aromatic residues (Y, F), significantly altered the response to ethanol (P < 0.05 compared to ethanol effects in the WT P2X4R). Data are expressed as mean + SEM, n = 6–28. The curve for WT is shown as an interrupted line. Boxes represent groups of receptors with similar ethanol responses. The following ATP EC10 values (in μM) were used for studies in Figs. 2–4: WT 0.9, Tyr 0.9, Phe 0.8, Asp 0.15, Lys 0.25, Asn 0.15, Leu 0.25, Val 0.30, and Ala 0.30. (b) Representative tracings of ATP-induced currents in the absence and presence of 100 mM ethanol for WT, W46N, and W46A mutant P2X4Rs. ATP EC10 for each receptor was used
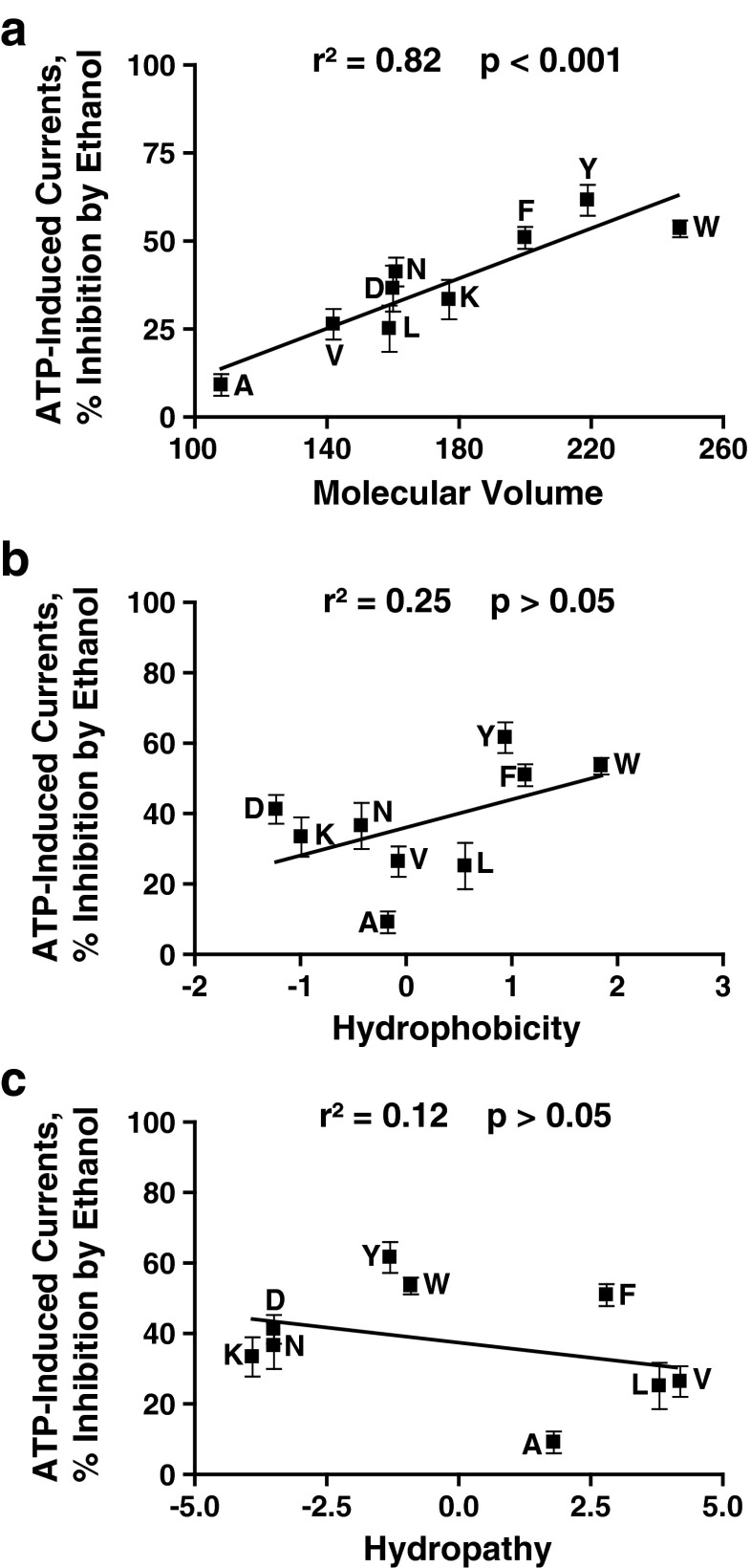
We used linear regression analyses to test this possibility by determining the relationships between the responses to 100 mM ethanol and the physicochemical properties of the residue at position 46. As shown in Fig. 2a, there was a significant direct relationship between the molecular volume of the residue at position 46 and the ethanol sensitivity. We also found a similar significant direct correlation for the molecular weight and ethanol response (r2 = 0.73; p < 0.01). There was no significant correlation between the hydrophobicity or hydropathy and the magnitude of ethanol response (Fig. 2b and c, respectively).
Fig. 2.
Correlation analyses of physicochemical properties of the residues at position 46 and ethanol sensitivity. Ethanol inhibition significantly correlates with the molecular volume of the residue at position 46. There is no significant correlation between hydrophobicity, hydropathy, and ethanol response. (a) Molecular volume, (b) hydrophobicity, and (c) hydropathy were correlated against sensitivity to 100 mM ethanol
Since some mutations at position 46 significantly affected the EC50 values, we used correlational analysis to evaluate if mutant-induced changes in the agonist sensitivity of the receptor influenced the magnitude of ethanol response. This analysis found no correlation between the EC50 values for the ATP concentration responses and ethanol sensitivity (r2 = 0.29; p > 0.05), indicating that changes in agonist properties do not account for altered ethanol response.
Several conclusions can be drawn from these findings: (1) physicochemical parameters influence the magnitude or threshold of ethanol inhibition at concentrations above 50 mM, (2) aromaticity is required for maximal ethanol response, (3) the magnitude of ethanol inhibition increases with the size of the substitution at position 46, and (4) hydrophobicity and hydropathy do not significantly affect ethanol sensitivity.
Aromaticity, molecular volume, and hydrophobicity of the residue at position 46 affect IVM sensitivity
Our previous findings demonstrated that 0.5 μM concentration of IVM elicited small but significant potentiation in P2X4Rs and was able to completely antagonize ethanol effects at intoxicating concentrations in WT receptors [20]. In addition, our previous work in mice demonstrated that low doses of IVM were able to cause reduction in ethanol consumption [22], suggesting that 0.5 μM concentration falls within therapeutically relevant doses in regard to IVM's anti-alcohol effects. For these reasons, we focused to investigate the effects of 0.5 μM IVM.
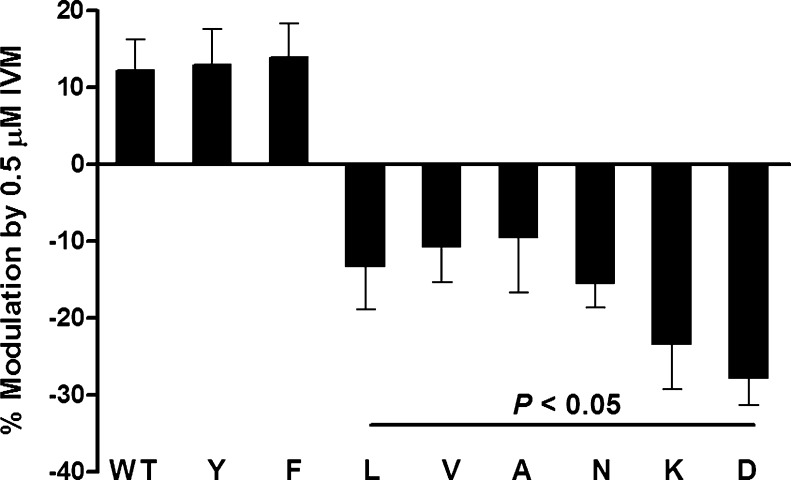
As previously reported [20], 0.5 μM IVM potentiated ATP-activated currents in WT P2X4Rs (Fig. 3). Substituting the naturally occurring aromatic tryptophan at position 46 with another aromatic residue, Tyr or Phe, did not significantly affect IVM potentiation of ATP-gated currents versus WT (Fig. 3). In contrast, substitution with nonaromatic amino acids (Leu, Val, Ala, Asn, Lys, or Asp) switched the potentiating effect of IVM to inhibition (Fig. 3).
Fig. 3.
Effects of 0.5 μM IVM on WT and mutant P2X4Rs. Application of 0.5 μM IVM potentiated the ATP-activated currents in P2X4Rs. Aromatic W46 substitution with other aromatic residues, W46Y or W46F, did not affect the direction or the degree of IVM effect compared to the WT receptor. Substituting W46 with nonaromatic residues (W46L, W46V, W46A, W46N, W46K, and W46D) switched IVM potentiation to inhibition. Data are expressed as mean ± SEM, n = 6–18. P < 0.05 compared to WT
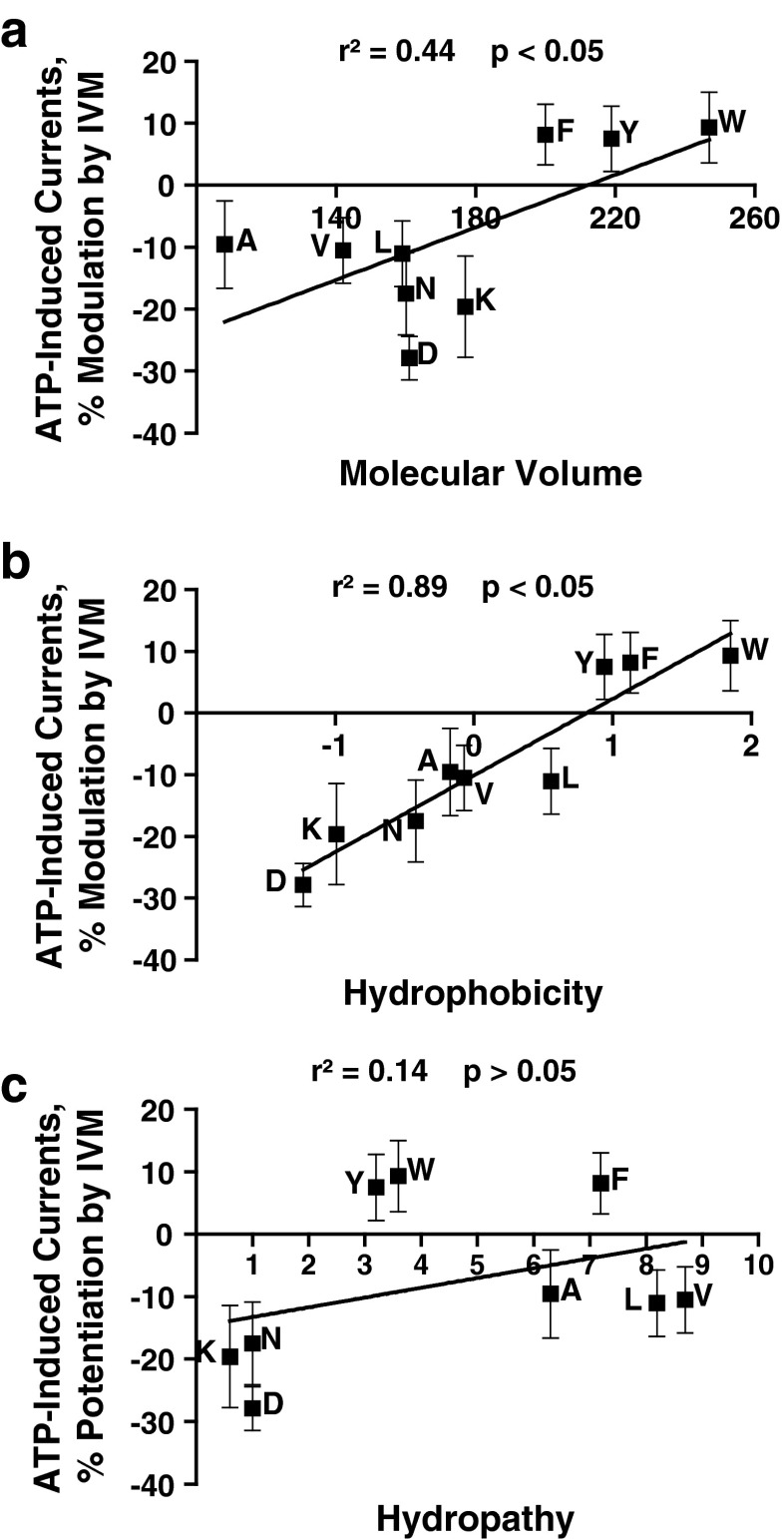
Linear regression analyses were used to test for possible relationships between the responses to 0.5 μM IVM and the physicochemical properties of the residue at position 46. The analyses revealed a statistically significant correlation between the response to 0.5-μM IVM and the molecular volume (Fig. 4a) or the molecular weight (r2 = 0.47, p < 0.05). There was a significant direct relationship between hydrophobicity and IVM potentiation (Fig. 4b). This contrasts with ethanol, in which there was no correlation (Fig. 2b). As with ethanol, hydropathy did not significantly correlate with the magnitude of IVM response (Fig. 4c).
Fig. 4.
Correlation analyses of physicochemical properties of the residues at position 46 and response to 0.5 μM IVM. IVM effects significantly correlate with the molecular volume and hydrophobicity of the residue at position 46. There is no significant correlation between hydropathy and IVM response. (a) Molecular volume, (b) hydrophobicity, and (c) hydropathy were correlated against sensitivity to 0.5 μM IVM
Overall, the distribution of the data suggests two separate groups with respect to IVM responses: aromatic and nonaromatic. It appears that the presence or absence of an aromatic group, rather than the volume of the residue at position 46, is the key determinant of whether IVM causes potentiation or inhibition. In addition, the presence of a charged residue at position 46 further increased the degree of IVM inhibition over the other nonaromatic residues.
In contrast to ethanol, there was a correlation between the EC50 values for ATP-induced currents and IVM response (r2 = 0.75; p < 0.01). Interestingly, removing the aromatic residues from the analysis eliminated the correlation (r2 = 0.51; p > 0.05), which further supports the concept that the presence of an aromatic residue at position 46 is key to modulation by IVM.
Effects of ethanol and IVM in W46A–M336A double mutant P2X4Rs
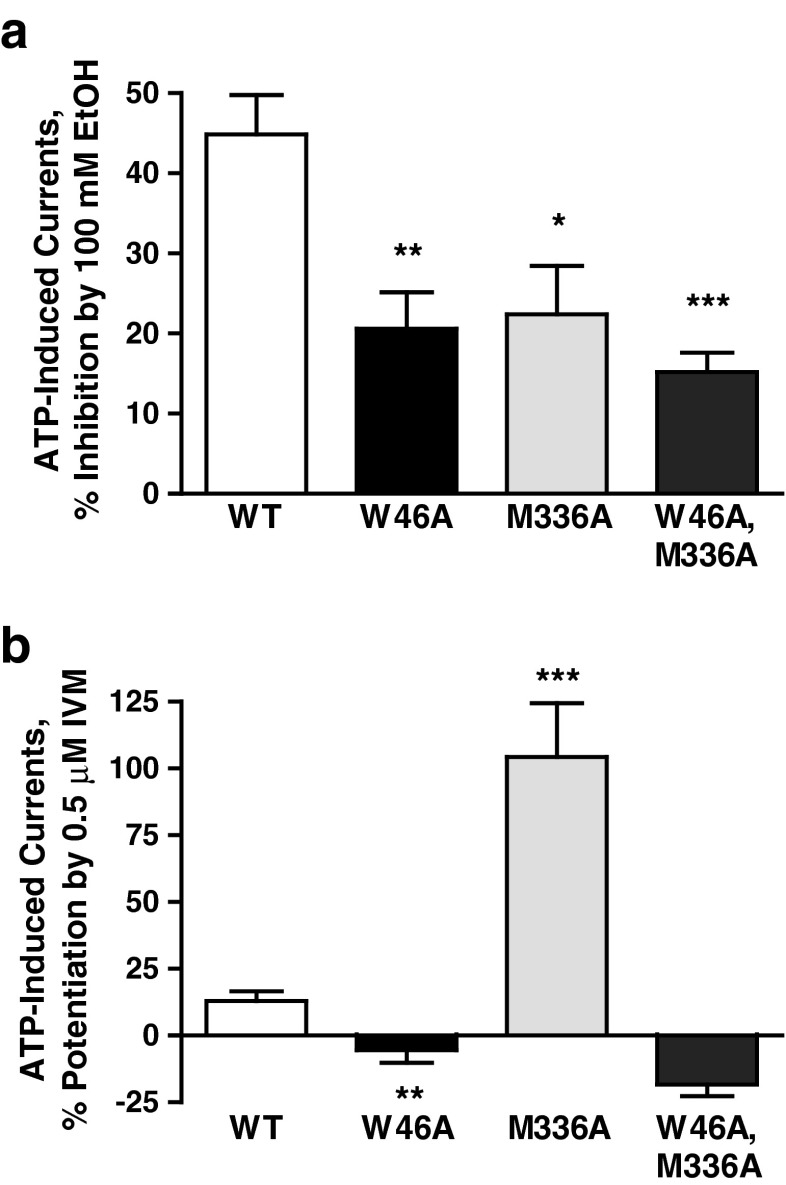
Our previous molecular model suggested that the interaction of Met336 with Trp46 is part of an ethanol/IVM binding pocket [12]. To further test this notion, we investigated the effects of both drugs in a W46A–M336A double mutant receptor. We tested the effect of 100 mM ethanol, since involvement of Trp46 was demonstrated at higher ethanol concentrations. Ethanol response was significantly reduced in the single mutants (W46A and M336A) as well as the double mutant (W46A–M336A, Fig. 5a). The extent of reduction was similar in all the mutants. The response to 0.5 μM IVM was significantly reduced in W46A and dramatically increased in M336A mutant P2X4Rs. Interestingly, simultaneous alanine substitutions at both positions 46 and 336 (W46A–M336A) switched the effect of IVM from potentiation to inhibition (Fig. 5b). The latter was not significantly different from that of W46A. These findings add support for the contention that position 46 plays a critical role in IVM effect and that interaction between the residues at positions 46 and 336 are important in modulating the effects of ethanol and IVM.
Fig. 5.
Comparison of the effects of 100 mM ethanol and 0.5 μM IVM in WT, W46A, M336A, and W46A–M336 mutant P2X4Rs. (a) Response to ethanol is significantly reduced in all the mutants compared to that of the WT. (b) Substitution of alanine for Met336 caused a drastic increase in the IVM potentiation compared to the effect in WT P2X4Rs. However, mutating both Trp46 and Met336 to alanine reversed IVM potentiation to inhibition in W46A–M336A double mutant that was not significantly different than the effect in W46A mutant receptor. Data are expressed as mean ± SEM, n = 4–8. *P < 0.05, **P < 0.01, ***P < 0.001 compared to WT
Molecular model of P2X4Rs focused on position 46 interactions with ethanol and IVM
To visualize potential interactions of Trp46 and other key residues with IVM, we generated a model based on the recent structure of zebrafish P2X4R in the open conformation [6]. We positioned IVM into that model by combining the known position of IVM in a pentameric GluCl receptor [32] with our new model of trimeric P2X4R (Fig. 6a–d). We started with the long axis of the IVM molecule essentially parallel to and in proximity of the extracellular membrane surface, as in GluCl [32]. We, then, compared the position of IVM in this model with the orientation of IVM in the alpha-helical bundle described previously [37]. In that model the spiroketal and benzofuran rings are near Trp46 and Trp50 while the disaccharide is pointing along the long axis of the alpha helices. Consequently, many assumptions and approximations are involved in the resulting model of an IVM binding site. Nevertheless, the combined model was stable during 5,000,000 2-fs steps of molecular dynamics with no restraints applied to the IVM molecule, which remained positioned in the intersubunit site between the amino acid residues Tyr42, Trp46, and Trp50 of subunit 1, Met336 of subunit 2, and the ectodomain (Fig. 6b). This stability implies that the site shown in Fig. 6b is a reasonable one.
Fig. 6.
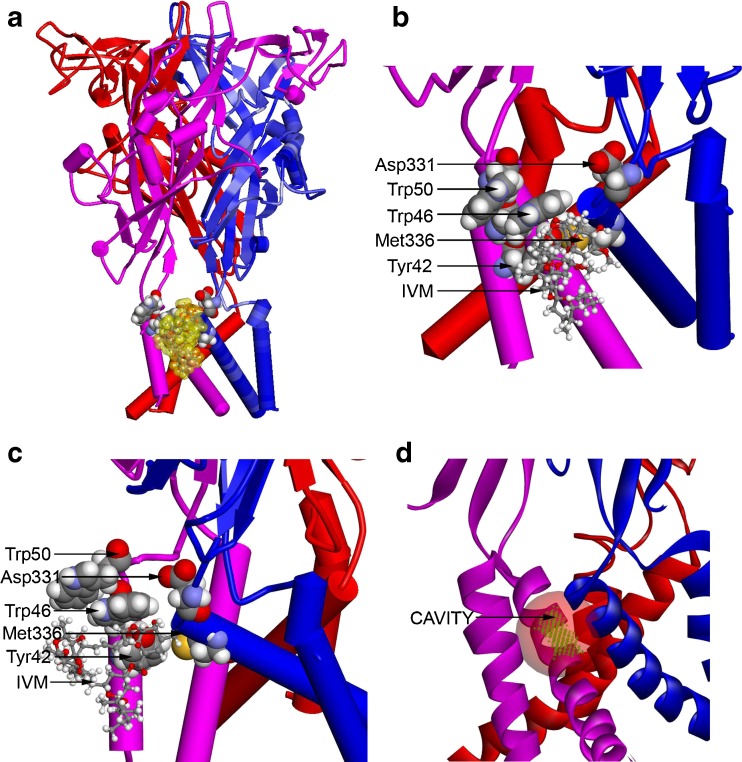
Homology model of rat P2X4R. (a) A homology model of rat P2X4R was built by threading the rat primary sequence onto the recent X-ray structure of the zebrafish P2X4R in the open conformation (PDB ID 4DW1) as described in “Materials and methods”. In this model, the P2X4R subunits are rendered as blue, pink, and red cartoon structures. The residues important for IVM binding are rendered with a van der Waals surface (carbon, oxygen, nitrogen, and hydrogen are colored gray, red, blue, and white), whereas the IVM is rendered in ball and stick and then enveloped with a transparent van der Walls surface (yellow). (b) A detailed view of the region surrounding the IVM molecule in Fig. 6a showing the relationship of residues important for IVM and ethanol binding. As viewed from the plane of the membrane and looking toward the ion pore, the IVM is between two subunits and within the membrane, but near the interface between the extracellular and transmembrane domains. The initial position of the IVM molecule was parallel to the plane of the membrane, as it is in the X-ray structure of GluCl. (c) A view of the region surrounding the IVM molecule in Fig. 6b, but rotated 90° in order to show the relationship of residues important for IVM and ethanol binding. The view is from the plane of the membrane but tangential to the ion pore, which is aligned with the right edge of the figure. The IVM molecule is between Trp46 and Met336; it also forms an H bond with Tyr42. (d) The IVM molecule was removed from the model in Fig. 6a and a cavity-finding algorithm was used to detect internal and external cavities. The module found 39 cavities using a 0.5-Å3 grid. The most relevant cavity site (to be compared to the IVM surface in Fig. 6a) is shown as a green grid enclosed within a transparent red sphere. It is located between two P2X4R subunits and is near the extracellular domain. In this figure, the P2X4R subunits are rendered as blue, pink, and red ribbons to better reveal the cavity
Hydrogen bond (H bond) and Pi–Pi monitors in Discovery Studio 3.5 were applied to IVM and the highlighted residues in Fig. 6b. A strong H bond was formed between the phenolic hydrogen of Tyr42 and the hydroxyl oxygen (O10) of IVM. Although not an H bond, H72 of IVM was in close contact (2.4 Å center to center) with the sulfur atom of Met336. In contrast, the closest hydrogen atom of Trp42 (HH2) was 6 Å away from the nearest oxygen atom of Asp331, suggesting a lack of interaction in this conformation.
The Pi–Pi interactions were very interesting; the aromaticring of Trp46 was sandwiched between Trp50 on the extracellular side and Tyr42 on the intracellular side (Fig. 6b and c). In addition, the axes perpendicular to the aromatic rings of Trp46 and Trp50 are offset in a way that optimizes Pi stacking (the N–N distance was 5.2 Å). A clear Pi–Pi interaction was detected between the aromatic rings of Trp46 and Trp50 as well as between the aromatic ring of Trp46 and the phenolic hydrogen of Tyr42. In this conformation, the position of Trp50 can directly influence the orientation of Trp46 and, indirectly, the orientation of Tyr42. Finally, we formed a van der Waals surface around IVM in this position, so we could relate it to four of the residues known to be important for IVM binding. The surface representation fit well within the intersubunit space. The cavity-finding algorithm in the “Define and Edit Binding Site” module of DS 3.5 detected 39 cavities within the rat P2X4R model, the most relevant of which (transparent red sphere in Fig. 6d) corresponds to the location of the van der Waals surface shown in Fig. 6a.
Altered responses of Y42F mutant to ethanol and IVM support findings from the molecular model
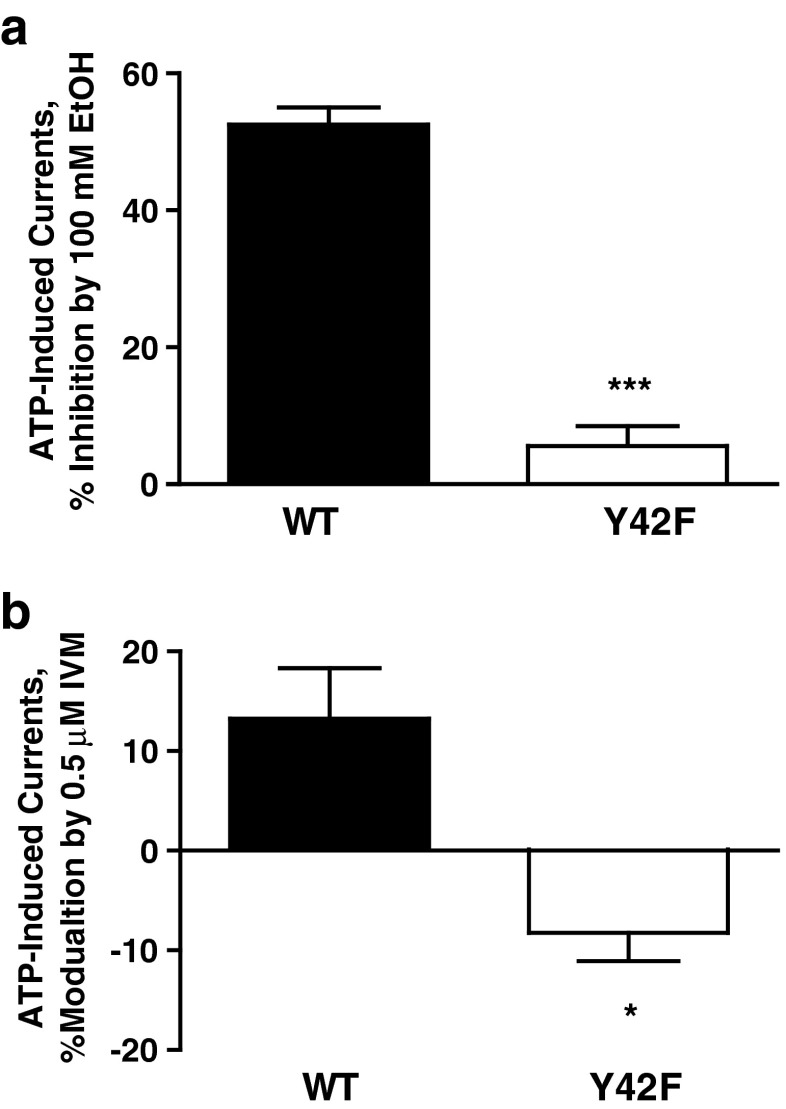
The model of P2X4 with IVM shown in Fig. 6 revealed two H bonds, one between the H20 hydrogen of IVM and the OH of Y42 and a second between O10 of IVM and the phenolic (HH) hydrogen of Y42. We tested the involvement of Tyr42, and in particular the phenolic OH group, in modulation of ethanol and IVM responses suggested by the molecular model. Mutating the tyrosine residue at position 42 to phenylalanine that lacks a phenolic OH group resulted in reduced sensitivity to 100 mM ethanol (Fig. 7a). Interestingly, this mutation switched the potentiation response of IVM to inhibition (Fig. 7b) similarly to the nonaromatic substitutions at position 46 (Fig. 3). In addition, the agonist properties of Y42F were not significantly different from WT P2X4Rs (ATP EC50 = 4.3 ± 1.23 μM versus 6.06 ± 0.53 μM for WT). This supports the notion derived from the molecular model that the phenolic hydroxyl group of Tyr42 is involved in the modulation of IVM and ethanol in P2X4Rs.
Fig. 7.
Effects of ethanol and IVM in Y42F mutant P2X4Rs. Mutating Tyr42 to Phe resulted in significantly reduced sensitivity to 100 mM ethanol (a) and reversal of IVM potentiation to inhibition (b). Data are expressed as mean ± SEM, n = 4–7. *P < 0.05, **P < 0.01 compared to WT
Discussion
The present study tested the hypothesis that Trp46 is a site of ethanol and IVM action. In support of the hypothesis, we found that position 46: (1) is important for ethanol action at higher concentrations, (2) modulates the response of IVM, and (3) interactions with other critical residues play an important role for the formation of the putative ethanol/IVM pocket. Taken in the context of prior studies, these findings support the notion that there are multiple sites of interaction for ethanol and IVM in P2X4Rs. Finally, to help visualize and interpret the role of position 46, we present a new homology model of the rat P2X4R.
The present findings suggest that position 46 in the TM1 segment of P2X4Rs is a site for ethanol action. We found that Trp46 substitutions differentially altered sensitivity of P2X4Rs to ethanol. That is, aromatic substitutions at position 46 did not change ethanol inhibition, whereas nonaromatic mutations reduced the sensitivity of the receptor to ethanol in a concentration-specific manner. The reduction of ethanol inhibition was found at higher concentrations, i.e., 100 and 200 mM. These results add position 46 to the TM2 ectodomain positions 331 and 336 [14] as a putative site of ethanol action/modulation. As with Trp46, Asp331 mutations affected ethanol sensitivity at higher ethanol concentrations (>50 mM), whereas Met336 mutations were sensitive to lower, intoxicating ethanol concentrations (10 mM) [14]. The new findings may be relevant to chronic alcohol abusers whose ethanol concentrations generally reach levels as high as 50 mM and above. These findings also support the notion that there are multiple sites for ethanol action in P2X4Rs—high affinity (such as position 336) that are sensitive to lower intoxicating concentrations experienced by most social drinkers and low affinity (such as positions 46 and 331) that show responses at the higher concentrations achieved by chronic alcoholics.
The physicochemical properties of amino acid substitutions at position 46 appeared to play a key role in determining the effect of the mutation on ethanol sensitivity. Correlation analyses showed a significant direct association between molecular volume/molecular weight and ethanol sensitivity. In support of this, substitution to alanine, a small-sized residue, eliminated ethanol response to 200 mM ethanol. Importantly, the presence of an aromatic ring at position 46 appeared to be required for the maximal response to ethanol seen in WT, W46Y, and W46F mutant receptors. Neither hydrophobicity nor hydropathy was important for the ethanol response.
The physicochemical requirements at position 46 for ethanol sensitivity differ from previous findings for positions 331 and 336, where hydropathy and polarity were correlated with ethanol sensitivity [14]. Together, these findings add a new line of evidence to support the notion that there are multiple sites of ethanol action/modulation in P2X4Rs. In addition, these results suggest that there are structural and functional differences between the different sites upon which ethanol acts. The concept of multiple sites of ethanol action with different properties is similar to findings in other LGICs [38–43].
IVM is a hydrophobic molecule that is known to bind at the lipid–protein interfaces and to act on multiple sites within the TM segments of P2X4Rs [25, 26]. Prior studies, using site-directed mutagenesis, suggested that positions at the TM1–ectodomain interface, i.e., Val47, Trp50, and Val60, play important roles in IVM regulation of the channel function [26]. The current study adds evidence that positions 42 and 46, in TM1 segment, are also sites of IVM action.
Moreover, the present findings provide new evidence indicating that Trp46 plays a critical role in determining the response of the receptor to IVM. These studies found a significant correlation between the sensitivity of the receptor to IVM and hydrophobicity, as well as size, of the substituted residue. Of potential greater importance, the presence or absence of an aromatic group at position 46 played a pivotal role in determining whether IVM causes potentiation or inhibition of the receptor at 0.5 μM IVM. In support of this, nonaromatic substitutions at position 46 switched the response of IVM from potentiation to inhibition. In contrast, aromatic substitutions at position 46 did not significantly affect the direction or magnitude of the response of the receptor to IVM. The importance of the presence of an aromatic residue at position 46 for the IVM response is further supported by the findings of the correlational analyses indicating significant relationship between the EC50 values for the ATP concentration responses of WT and mutant receptors and IVM modulation. These findings suggest that the aromatic ring at position 46 may interact with the IVM molecule through a cation–Pi interaction [44]. Once this interaction is disrupted, the IVM molecule may exert its action on a different site, which causes the switch of the response from potentiation to inhibition. Similar to the findings on ethanol, these findings add more evidence to the notion that there are multiple sites for IVM action in P2X4Rs.
Our previous model revealed a pocket formed by residues at the ectodomain–TM2 segment interface, Asp331 and Met336, and TM1 segment residues Trp46 and Trp50. Furthermore, the model suggested that Trp46 and Met336 side chains interact, further implicating the importance of Trp46 in ethanol and IVM modulation. The present findings from the double mutant W46A–M336A P2X4Rs suggest an important role of the position 46 interaction with Met336 in ethanol and IVM action. Similar to the effect in the single mutants (W46A and M336A), response to ethanol was reduced in the double mutant receptor. Importantly, findings in the double mutant supported the notion that Trp46 plays a critical role in IVM response. This is based on the finding that simultaneous alanine mutation of Trp46 and Met336 resulted in a switch in IVM response from potentiation to inhibition, an effect seen with nonaromatic mutants at position 46.
Collectively, these findings demonstrating the importance of an aromatic residue at position 46 in determining the magnitude of the response to ethanol and the direction of the response to IVM led to a new molecular model of the P2X4R. This model revealed potential sites for ethanol and IVM action. The model replicated the vertical and lateral positions of IVM in the X-ray structure of the pentameric glutamate-gated chloride ion channel GluCl [32]. The van der Waals surface of IVM molecule fit well within one of the 39 cavities found in P2X4R model. Molecular dynamic simulations demonstrated that the IVM molecule was stable in that position. The position of IVM in this model is consistent with prior mutagenesis data on P2X4R and adds support for a pocket within P2X4Rs that includes positions 42 and 46 (TM1) and positions 331 and 336 (TM2) [12].
The present model provides insight into possible mechanism by which the presence and absence of aromatic residues at position 46 in our mutagenesis switched the response of P2X4Rs to IVM. Specifically, the model suggests that Trp46 and Met336 are part of the same pocket and that the two residues face each other [12]. This positioning opens the potential for the aromatic rings and sulfur groups to interact in a variety of ways, including hydrogen bonding to the aryl hydrogens, SH–Pi interactions, electrostatic, or hydrophobic interactions [45]. In addition, interactions of the sulfur atom of methionine with multiple aromatic groups can produce “hot spots” in protein–protein interactions [46]. These interactions could contribute to the stabilization of the protein [45]. Our mutagenesis findings with the single and double mutations at positions 46 and 336 support this conclusion.
The sites identified in our previous [12] and the present study are likely located close to fenestrations that were recently described at the membrane interface of P2X4Rs [27, 28]. These “lateral portals” are closed in the closed state of the receptor and open to approximately 8 Å in the open state [27], thus providing sufficient size for IVM binding. The latter stabilizes the open state of the receptor. Therefore, it is reasonable to speculate that changing the physicochemical properties of the residue at positions 42, 46, or other important sites may disrupt the interactions with other critical residues described above, thus changing the physical structure of this site and resulting in the switch in the response to IVM or altering ethanol response.
Aromatic residues located within the upper part of the TM1 segment of P2X4Rs, such as Tyr42, Trp46, and Trp50, have been suggested to play a role in the three-dimensional organization of the receptor [26]. The current model demonstrated that Trp46 is “sandwiched” between Tyr42 and Trp50. Previous mutagenesis studies have shown that Trp50 does not affect ethanol response [14]. Moreover, the disruption of the Pi–Pi interaction between the aromatic rings of Trp46 and Trp50 does not seem to alter the response to ethanol when Trp50 residue is mutated. On the other hand, the model showed Pi–Pi interactions between the aromatic ring of Trp46 and the phenolic hydrogen of Tyr42 as well as H bond formation between the phenolic OH group of Tyr42 and the hydroxyl oxygen of IVM. Substituting Tyr42 for Phe, which lacks the hydroxyl group on the aromatic ring, significantly affected the proposed interactions that altered ethanol and IVM responses. These findings suggest the importance of Tyr42 in the modulation of the two drugs.
As described in the “Introduction”, IVM antagonizes ethanol inhibition of P2X4Rs and reduces ethanol intake in mice [22]. Further studies with P2X4 knockout mice [47], site-directed mutations, and molecular modeling link this anti-alcohol effect of IVM, at least in part, to its action on P2X4Rs, specifically through interference with ethanol acting on residues in the transmembrane domain [12]. The present study supports the hypothesis that Trp46 also plays a role in ethanol and/or IVM modulation of P2X4Rs and suggests that positions 42, 46, 50, 331, and 336 form an action pocket for both ethanol and IVM. In the light of these findings, combined with our preliminary study demonstrating increased ethanol consumption in P2X4 knockout mice, this pocket represents a potential target for medication development for AUD.
Acknowledgments
We would like to thank Miriam Fine, Sylvia Nguyen, and Angeline Chen for technical and laboratorial assistance. This work was supported, in part, by research grants NIAAA/NIH F31 AA017029 (M.P.), KO1 AA017243 (L.A.), AA03972 (R.L.A.), AA013922 (D.L.D.), and AA013378 (J.R.T.) and the USC School of Pharmacy. This work was conducted as partial fulfillment of the requirements for the Ph.D. degree in Pharmaceutical Sciences, University of Southern California (M.P.).
References
- 1.Bouchery EE, Harwood HJ, Sacks JJ, Simon CJ, Brewer RD. Economic costs of excessive alcohol consumption in the US, 2006. Am J Prev Med. 2011;41(5):516–524. doi: 10.1016/j.amepre.2011.06.045. [DOI] [PubMed] [Google Scholar]
- 2.Khakh BS, Burnstock G, Kennedy C, King BF, North RA, Seguela P, Voigt M, Humphrey PA. International union of pharmacology. XXIV. Current status of the nomenclature and properties of P2X receptors and their subunits. Pharmacol. Rev. 2001;53:107–118. [PubMed] [Google Scholar]
- 3.North RA. Molecular physiology of P2X receptors. Physiol. Rev. 2002;82:1013–1067. doi: 10.1152/physrev.00015.2002. [DOI] [PubMed] [Google Scholar]
- 4.Burnstock G. Unresolved issues and controversies in purinergic signalling. J Physiol Online. 2008;586(14):3307–3312. doi: 10.1113/jphysiol.2008.155903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kawate T, Michel JC, Birdsong WT, Gouaux E. Crystal structure of the ATP-gated P2X4 ion channel in the closed state. Nature. 2009;460(7255):592–598. doi: 10.1038/nature08198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hattori M, Gouaux E. Molecular mechanism of ATP binding and ion channel activation in P2X receptors. Nature. 2012;485(7397):207–212. doi: 10.1038/nature11010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buell G, Lewis C, Collo G, North RA, Suprenant A. An antagonist insensitive P2X receptor expressed in epithelia and brain. EMBO J. 1996;15(1):55–62. [PMC free article] [PubMed] [Google Scholar]
- 8.Soto F, Garcia-Guzman M, Gomez-Hernandez JM, Hollmann M, Karschin C, Stuhmer W. P2X4: an ATP-activated ionotropic receptor clonned from rat brain. Proc Natl Acad Sci USA. 1996;93:3684–3688. doi: 10.1073/pnas.93.8.3684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Amadio S, Montilli C, Picconi B, Calabrei PCV. Mapping P2X and P2Y receptor proteins in striatum and substantia nigra: an immunohistological study. Purinergic Signal. 2007;3:389–398. doi: 10.1007/s11302-007-9069-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Krügel U, Kittner H, Franke H, Illes P. Purinergic modulation of neuronal activity in the mesolimbic dopaminergic system in vivo. Synapse. 2003;47:134–142. doi: 10.1002/syn.10162. [DOI] [PubMed] [Google Scholar]
- 11.Davies DL, Kochegarov AA, Kuo ST, Kulkarni AA, Woodward JJ, King BF, Alkana RL. Ethanol differentially affects ATP-gated P2X(3) and P2X(4) receptor subtypes expressed in Xenopus oocytes. Neuropharmacol. 2005;49:243–253. doi: 10.1016/j.neuropharm.2005.03.015. [DOI] [PubMed] [Google Scholar]
- 12.Asatryan L, Popova M, Perkins DI, Trudell JR, Alkana RL, Davies DL. Ivermectin antagonizes ethanol inhibition in P2X4 receptors. J. Pharmacol. Exp. Ther. 2010;334:720–728. doi: 10.1124/jpet.110.167908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Davies DL, Machu TK, Guo Y, Alkana RL. Ethanol sensitivity in ATP-gated P2X receptors is subunit dependent. Alcohol. Clin. Exp. Res. 2002;26:773–778. doi: 10.1111/j.1530-0277.2002.tb02604.x. [DOI] [PubMed] [Google Scholar]
- 14.Popova M, Asatryan L, Ostrovskaya O, Wyatt RL, Li K, Alkana RL, Davies DL. A point mutation in the ectodomain-transmembrane 2 interface eliminates the inhibitory effects of ethanol in P2X4 receptors. J Neurochem. 2010;112:307–317. doi: 10.1111/j.1471-4159.2009.06460.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xiong K, Peoples RW, Montgomery JP, Chiang Y, Stewart RR, Weight FF, Li C. Differential modulation by copper and zinc of P2X2 and P2X4 receptor function. J Neurophysiol. 1999;81:2088–2094. doi: 10.1152/jn.1999.81.5.2088. [DOI] [PubMed] [Google Scholar]
- 16.Xiong K, Li C, Weight FF. Inhibition by ethanol of rat P2X4 receptors expressed in Xenopus oocytes. Br. J. Pharmacol. 2000;130:1394–1398. doi: 10.1038/sj.bjp.0703439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xiong K, Hu XQ, Stewart RR, Weight FF, Li C. The mechanism by which ethanol inhibits rat P2X4 receptors is altered by mutation of histidine 241. Br J Pharmacol. 2005;145:576–586. doi: 10.1038/sj.bjp.0706192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kimpel MW, Strother WN, McClintick JN, Carr LG, Liang T, Edenberg HJ, McBride WJ. Functional gene expression differences between inbred alcohol-preferring and -non-preferring rats in five brain regions. Alcohol. 2007;41(2):95–132. doi: 10.1016/j.alcohol.2007.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tabakoff B, Saba L, Printz M, Flodman P, Hodgkinson C, Goldman D, Koob G, Richardson H, Kechris K, Bell RL, Hübner N, Heinig M, Pravenec M, Mangion M, Legault L, Dongier M, Conigrave KM, Whitfield JB, Saunders JB, Grant B, Hoffman PL. Genetical genomic determinants of alcohol consumption in rats and humans BMC. Biology. 2009;7:70. doi: 10.1186/1741-7007-7-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Khakh BS, Proctor WR, Dunwiddie TV, Labarca C, Lester HA. Allosteric control of gating and kinetics at P2X4 receptor channels. J. Neurosci. 1999;19:7289–7299. doi: 10.1523/JNEUROSCI.19-17-07289.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Norenberg W, Sobottka H, Hempel C, Plotz T, Fischer W, Schmalzing G, Schaefer M. Positive allosteric modulation by ivermectin of human but not murine P2X7 receptors. Br. J. Pharmacol. 2012;167(1):48–66. doi: 10.1111/j.1476-5381.2012.01987.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yardley MM, Wyatt L, Khoja S, Asatryan L, Ramaker MJ, Finn DA, Alkana RL, Huynh N, Louie SG, Petasis NA, Bortolato M, Davies DL (2012) Ivermectin reduces alcohol intake and preference in mice. Neuropharmacol 63. doi:10.1016/j.neuropharm.2012.03.014 [DOI] [PMC free article] [PubMed]
- 23.Asatryan L, Popova M, Woodward JJ, King BF, Alkana RL, Davies DL. Roles of ectodomain and transmembrane regions in ethanol and agonist action in purinergic P2X2 and P2X3 receptors. Neuropharmacol. 2008;55(5):835–843. doi: 10.1016/j.neuropharm.2008.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yi CL, Liu YW, Xiong KM, Stewart RR, Peoples RW, Tian X, Zhou L, Ai YX, Li ZW, Wang QW, Li CY. Conserved extracellular cysteines differentially regulate the inhibitory effect of ethanol in rat P2X4 receptors. Biochem. Biophys. Res. Commun. 2009;381:102–106. doi: 10.1016/j.bbrc.2009.02.018. [DOI] [PubMed] [Google Scholar]
- 25.Silberberg SD, Li M, Swartz KJ. Ivermectin interaction with transmembrane helices reveals widespread rearrangements during opening of P2X receptor channels. Neuron. 2007;54(2):263–274. doi: 10.1016/j.neuron.2007.03.020. [DOI] [PubMed] [Google Scholar]
- 26.Jelinkova I, Vavra V, Jindrichova M, Obsil T, Zemkova HW, Zemkova H, Stojilkovic SS. Identification of P2X(4) receptor transmembrane residues contributing to channel gating and interaction with ivermectin. Pflugers Arch. 2008;456:939–950. doi: 10.1007/s00424-008-0450-4. [DOI] [PubMed] [Google Scholar]
- 27.Jiang R, Taly A, Grutter T. Moving through the gate in ATP-activated P2X receptors. Trends Biochem Sci. 2013;38(1):20–29. doi: 10.1016/j.tibs.2012.10.006. [DOI] [PubMed] [Google Scholar]
- 28.Samways DS, Khakh BS, Egan TM. Allosteric modulation of Ca2+ flux in ligand-gated cation channel (P2X4) by actions on lateral portals. J. Biol. Chem. 2012;287(10):7594–7602. doi: 10.1074/jbc.M111.322461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.King BF, Pintor J, Wang S, Ziganshin AU, Ziganshina LE, Burnstock G. A novel P1 purinoceptor activates an outward K + current in follicular oocytes of Xenopus laevis. J. Pharmacol. Exp. Ther. 1996;276:93–100. [PubMed] [Google Scholar]
- 30.King BF, Wang S, Burnstock G. P2 purinoceptor-activated inward currents in follicular oocytes of Xenopus laveis. J Physiol (Lond) 1996;494:17–28. doi: 10.1113/jphysiol.1996.sp021472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Perkins DI, Trudell JR, Crawford DK, Asatryan L, Alkana RL, Davies DL. Loop 2 structure in glycine and GABAA receptors plays a key role in determining ethanol sensitivity. J. Biol. Chem. 2009;284:27304–27314. doi: 10.1074/jbc.M109.023598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hibbs RE, Gouaux E. Principles of activation and permeation in an anion-selective Cys-loop receptor. Nature. 2011;474(7349):54–60. doi: 10.1038/nature10139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wimley WC, White SH. Experimentally determined hydrophobicity scale for proteins at membrane interfaces. Nat. Struct. Biol. 1996;3(10):842–848. doi: 10.1038/nsb1096-842. [DOI] [PubMed] [Google Scholar]
- 34.Kyte J, Doolittle RF. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 35.Wildman SS, King BF, Burnstock G. Modulation of ATP-responses at recombinant rP2X4 receptors by extracellular pH and zinc. Br J Pharmacol. 1999;126:762–768. doi: 10.1038/sj.bjp.0702325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jelinkova I, Yan Z, Liang Z, Moonat S, Teisinger J, Stojilkovic SS, Zemkova H. Identification of P2X4 receptor-specific residues contributing to the ivermectin effects on channel deactivation. Biochem. Biophys. Res. Commun. 2006;349:619–625. doi: 10.1016/j.bbrc.2006.08.084. [DOI] [PubMed] [Google Scholar]
- 37.Sattelle DB, Buckingham SD, Akamatsu M, Matsuda K, Pienaar I, Jones AK, Sattelle BM, Almond A, Blundell CD. Comparative pharmacology and computational modelling yield insights into allosteric modulation of human [alpha]7 nicotinic acetylcholine receptors. Biochem. Pharmacol. 2009;78(7):836–843. doi: 10.1016/j.bcp.2009.06.020. [DOI] [PubMed] [Google Scholar]
- 38.Borghese CM, Henderson LA, Bleck V, Trudell JR, Harris RA. Sites of excitatory and inhibitory actions of alcohols on neuronal alpha2beta4 nicotinic acetylcholine receptors. J. Pharmacol. Exp. Ther. 2003;307:42–52. doi: 10.1124/jpet.102.053710. [DOI] [PubMed] [Google Scholar]
- 39.Crawford DK, Trudell JR, Bertaccini E, Li KX, Davies DL, Alkana RL. Evidence that ethanol acts on a target in Loop 2 of the extracellular domain of alpha1 glycine receptors. J. Neurochem. 2007;102(6):2097–2109. doi: 10.1111/j.1471-4159.2007.04680.x. [DOI] [PubMed] [Google Scholar]
- 40.Mihic SJ, Ye Q, Wick MJ, Koltchine VV, Krasowski MD, Finn SE, Mascia MP, Valenzuela CF, Hanson KK, Greenblatt EP, Harris RA, Harrison NL. Sites of alcohol and volatile anaesthetic action on GABA A and glycine receptors. Nature. 1997;389:385–389. doi: 10.1038/38738. [DOI] [PubMed] [Google Scholar]
- 41.Perkins DI, Trudell JR, Crawford DK, Alkana RL, Davies DL. Targets for ethanol action and antagonism in loop 2 of the extracellular domain of glycine receptors. J. Neurochem. 2008;106:1337–1349. doi: 10.1111/j.1471-4159.2008.05476.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang J, Lester HA, Dougherty DA. Establishing an ion pair interaction in the homomeric rho1 gamma-aminobutyric acid type A receptor that contributes to the gating pathway. J. Biol. Chem. 2007;282(36):26210–26216. doi: 10.1074/jbc.M702314200. [DOI] [PubMed] [Google Scholar]
- 43.Ye Q, Koltchine VV, Mihic SJ, Mascia MP, Wick MJ, Finn SE, Harrison NL, Harris RA. Enhancement of glycine receptor function by ethanol is inversely correlated with molecular volume at position à267. J. Biol. Chem. 1998;273:3314–3319. doi: 10.1074/jbc.273.6.3314. [DOI] [PubMed] [Google Scholar]
- 44.Dougherty DA. Cation–pi interactions in chemistry and biology: a new view of benzene, Phe, Tyr, and Trp. Science. 1996;271(5246):163–168. doi: 10.1126/science.271.5246.163. [DOI] [PubMed] [Google Scholar]
- 45.Tatko CD, Waters ML. Investigation of the nature of the methionine–pi interaction in beta-hairpin peptide model systems. Protein Sci. 2004;13(9):2515–2522. doi: 10.1110/ps.04820104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ma B, Nussinov R. Trp/Met/Phe hot spots in protein–protein interactions: potential targets in drug design. Curr Topics Medicin Chem. 2007;7(10):999–1005. doi: 10.2174/156802607780906717. [DOI] [PubMed] [Google Scholar]
- 47.Bortolato M, Yardley M, Khoja S, Godar SC, Asatryan L, Finn DA, Huynh N, Alkana RL, Louie SG, Davies DL (2013) Pharmacological insights into the role of P2X4 receptors in behavioral regulation: lessons from ivermectin. Int J Neuropsychopharmacol 16(5):1059–1070. doi:10.1017/S1461145712000909 [DOI] [PMC free article] [PubMed]