Abstract
The P2X7 receptor is a trimeric ATP-gated cation channel important in health and disease. We have observed that the specific phospholipase D (PLD)1 antagonist, CAY10593 impairs P2X7-induced shedding of the ‘low affinity’ IgE receptor, CD23. The current study investigated the mode of action of this compound on P2X7 activation. Measurements of ATP-induced ethidium+ uptake revealed that CAY10593 impaired P2X7-induced pore formation in human RPMI 8226 B cells, P2X7-transfected HEK-293 cells and peripheral blood mononuclear cells. Concentration response curves demonstrated that CAY10593 impaired P2X7-induced pore formation in RPMI 8226 cells more potently than the PLD2 antagonist CAY10594 and the non-specific PLD antagonist halopemide. Electrophysiology measurements demonstrated that CAY10593 also inhibited P2X7-induced inward currents. Notably, RT-PCR demonstrated that PLD1 was absent in RPMI 8226 cells, while choline-Cl medium or 1-butanol, which block PLD stimulation and signalling respectively did not impair P2X7 activation in these cells. This data indicates that CAY10593 impairs human P2X7 independently of PLD1 stimulation and highlights the importance of ensuring that compounds used in signalling studies downstream of P2X7 activation do not affect the receptor itself.
Keywords: P2X7 receptor, Phospholipase D, CD23, B cell, T cell, Monocyte
Introduction
P2X7 is a trimeric ligand-gated cation (Ca2+, Na+ and K+) channel present on various cell types and plays important roles in many disease states, including inflammatory, immune, neoplastic, musculoskeletal and neurological disorders [1]. Prolonged exposure of P2X7 to extracellular ATP opens a second permeability state or pore that allows the uptake of organic cations including fluorescent dyes such as ethidium+ [2]. Whether this second permeability state is attributed to intrinsic channel dilation [3], the pannexin-1 channel [4] or an alternate but unknown uptake pathway [5–7] remains controversial. Moreover, our understanding of this permeability state is further complicated with some [8–10] but not other [11–13] studies showing that P2X7-induced dye uptake involves the p38 mitogen-activated protein kinase. Regardless of the true identity of the P2X7 pore and the mechanism by which it opens, P2X7 activation stimulates several intracellular signalling pathways to induce various cellular events including inflammatory mediator release, reactive oxygen and nitrogen species formation, and cell proliferation or death [14, 15]. P2X7 activation also induces the shedding of cell-surface molecules including the ‘low-affinity’ IgE receptor, CD23 [16–19]. However, the intracellular signalling pathways that mediate this process are unknown.
Phospholipase D (PLD) catalyses the hydrolysis of phosphatidylcholine to phosphatidic acid and choline, which subsequently participate in various cellular events [20]. Two isoforms of mammalian PLD have been described, PLD1 and PLD2 [20]. P2X7 activation can stimulate PLD in B cells [21, 22] and macrophages [23, 24]. P2X7-induced PLD stimulation in macrophages plays a role in the killing of intracellular mycobacteria [25, 26] and the generation of microvesicles capable of further macrophage activation [27]. In contrast, the role of P2X7-induced PLD stimulation in B cells remains unknown.
We have previously shown that human RPMI 8226 B cells express P2X7, and that activation of P2X7 on these cells induces pore formation and the shedding of CD23 [17, 28, 29]. During our preliminary investigations of the possible intracellular signalling enzymes involved in P2X7-induced CD23 shedding from RPMI 8226 cells we observed that the 1-(piperidin-4-yl)-1H-benzo[d]imidazol-2(3H)-one analogue and specific PLD1 inhibitor, CAY10593 [30], significantly impaired P2X7-induced CD23 shedding. Therefore, the aim of this study was to investigate the mode of action by which CAY10593 impairs P2X7-induced CD23 shedding. Measurements of P2X7-induced pore formation and channel activity, and PLD analysis by RT-PCR demonstrated that CAY10593 impairs human P2X7 independently of PLD1.
Materials and methods
Reagents
RPMI-1640 medium (containing 10 mM HEPES), ATP, poly-d-lysine, imipramine and diphenyleneiodonium were from Sigma Chemical Company (St. Louis, MO). Foetal bovine serum was from Bovogen Biologicals (East Keilor, Australia) or Lonza (Basal, Switzerland). DMEM:F12 medium (containing 10 mM HEPES), GlutaMAX, l-glutamine, penicillin/streptomycin and G418 were from Invitrogen (Grand Island, NY). Ficoll-PaqueTM PLUS was from GE Healthcare Bio-Sciences AB (Uppsala, Sweden). Dimethyl sulphoxide (DMSO) and ethidium bromide were from Amresco (Solon, OH). AZ10606120, AZ11645373, A-438079 and SB216763 were from Tocris Bioscience (Ellisville, MO). Rottlerin, LY294002, SB202190, SB203580, U0126 and SP600125 were from Merck Chemicals (Darmstadt, Germany). AG-126, GF109203X, D609, Fasudil, Y-27632 and AACOCF3 were from Enzo Life Sciences (Bristol, UK). CAY10593, CAY10594 and halopemide were from the Cayman Chemical Company (Ann Arbor, MI). Phycoerythrin (PE) or allophycocyanin (APC)-conjugated murine anti-human CD23 (clone EBVCS2), isotype control (clone P3.6.2.8.1), and APC-conjugated murine anti-human CD19 (clone HIB19) monoclonal antibodies (mAb) were from eBioscience (San Diego, CA). Primers were from GeneWorks (Hindmarsh, Australia).
Cells
Human RPMI 8226 multiple myeloma B cells and human A431 skin epithelial carcinoma cells (European Collection of Cell Cultures, Porton Down, UK) were maintained in complete RPMI-1640 medium (RPMI-1640 medium containing 10 % foetal bovine serum and 2 mM GlutaMAX) at 37 °C and 95 % air/5 % CO2. HEK-293 cells (American Type Culture Collection, Rockville, MD) were maintained in complete DMEM/F12 medium (DMEM/F12 medium containing 10 % foetal bovine serum, 100 U/ml penicillin, 100 μg/ml streptomycin and 2 mM l-glutamine). A stable HEK-293 cell line expressing human P2X7 was established by clonal dilution and kept under G418 selection medium. Experiments with human blood were approved by the University of Wollongong Human Ethics Committee. Peripheral blood was collected into VACUETTE® lithium heparin tubes (Greiner Bio-One, Frickenhausen, Germany) and diluted with an equal volume of phosphate-buffered saline. Peripheral blood mononuclear cells (PBMCs) were separated by density gradient centrifugation over Ficoll-PaqueTM PLUS (560×g for 20 min) and washed once in phosphate-buffered saline (450×g for 10 min).
Measurement of P2X7-induced pore formation by flow cytometry
P2X7-induced pore formation in RPMI 8226 cells or PBMCs was assessed by flow cytometric measurements of ATP-induced ethidium+ uptake as described [17]. Briefly, cells suspended in NaCl medium (145 mM NaCl, 5 mM KCl, 5 mM glucose, 10 mM HEPES, pH 7.4) (1 × 106 cells/ml) were pre-incubated in the absence or presence of antagonist (as indicated), and then with 25 μM ethidium+ in the absence or presence of ATP (as indicated) for 5 min at 37 °C. In some experiments, cells were suspended in choline-Cl medium (150 mM choline-Cl, 5 mM KCl, 5 mM glucose, 10 mM HEPES, pH 7.4). Incubations were stopped by addition of an equal volume of ice-cold MgCl2 medium (NaCl medium containing 20 mM MgCl2) and centrifugation (300×g for 5 min). Cells were washed once with NaCl medium. PBMCs were also incubated with APC-conjugated anti-human CD19 mAb and washed once with NaCl medium. The mean fluorescence intensity (MFI) of ethidium+ uptake was determined using a LSR II flow cytometer (BD, San Jose, CA) (using a 575/26-nm band-pass filter) and FlowJo software (Tree Star, Ashland, OR). For PBMCs, lymphocytes and monocytes were gated by forward and side scatter and CD19 expression was detected using a 660/20 band-pass filter.
Measurement of P2X7-induced CD23 shedding by flow cytometry
P2X7-induced CD23 shedding from RPMI 8226 cells was indirectly assessed by flow cytometric measurements of ATP-induced loss of cell-surface CD23 as described [17]. Briefly, cells suspended in NaCl medium (1 × 106 cells/ml) were pre-incubated in the absence or presence of antagonist (as indicated), and then in the absence or presence of 1 mM ATP for 7 min at 37 °C. In some experiments, cells were suspended in either choline-Cl medium, KCl medium (150 mM KCl, 5 mM glucose, 10 mM HEPES, pH 7.4), or NaCl medium containing either 0.1 mM EGTA or 50 μM BAPTA-AM for 5 min, and then in the absence or presence of 1 mM ATP for 7 min at 37 °C. All ATP incubations were stopped by addition of an equal volume of ice-cold MgCl2 medium and centrifugation (300×g for 5 min). Cells were washed once with NaCl medium and incubated with PE or APC-conjugated anti-human CD23 or isotype-control mAb. The MFI of cell-surface CD23 expression was determined using flow cytometry (using a 575/26 or 660/20 nm band-pass filter for PE or APC respectively) and FlowJo software.
Detection of PLD mRNA expression by RT-PCR
Total RNA was isolated from cells using the RNeasy Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. Primer pairs (forward and reverse, respectively) to PLD1 were: 5′-TCATGTGTCATCCACCGTCT-3′ and 5′-GGCGTGGAGTACCTGTCAAT-3′, and PLD2 [30] were: 5′-GGCGATGAGATTGTGGACA-3′ and 5′-CTGGAAGAAGTCATCACAGA-3′. PCR amplification was performed using the MyTaq One-Step RT-PCR Kit (Bioline, Sydney, Australia) according to the manufacturer’s instructions. PCR cycling conditions were 45 °C for 20 min, 94 °C for 2 min, 30 cycles of 94 °C for 30 s, 54 °C (PLD1 primer pair) or 57 °C (PLD2 primer pair), and 72 °C for 1 min, and a final step of 72 °C for 5 min. Products were separated on a 2 % agarose gel and visualised using ethidium bromide staining.
Measurement of P2X7 channel activity by electrophysiology
P2X7 channel activity in P2X7-transfected HEK-293 cells was assessed by electrophysiological measurements of ATP-induced currents as described [31]. Briefly, whole-cell patch-clamp recordings were performed at room temperature using an EPC10 amplifier and Patchmaster acquisition software (HEKA, Lambrecht, Germany). ATP and CAY10593 were delivered using the RSC-160 fast-flow system (Bio-Logic Science Instruments, Claix, France). Membrane potential was clamped at −60 mV in all experiments. External solution was 145 mM NaCl, 5 mM KCl, 2 mM CaCl2, 1 mM MgCl2, 13 mM glucose, 10 mM HEPES, and internal solution was 145 mM NaCl, 10 mM HEPES, 10 mM EGTA. Both solutions were adjusted to pH 7.3 with 5 M NaOH and were 300–310 mOsm/L.
Measurement of P2X7-induced pore formation using a fluorescent plate reader
Ethidium+ uptake assays on human P2X7-transfected HEK-293 cells were performed using an Optima FLUOSTAR plate reader (BMG Labtech, Ortenberg, Germany). Cells (5 × 104 cells/well) were incubated overnight in a 96-well poly-d-lysine coated plate. Ethidium+ (25 μM) was added in low divalent solution (145 mM NaCl, 5 mM KCl, 0.2 mM CaCl2, 13 mM glucose, 10 mM HEPES, pH 7.3). Cells were pre-incubated with CAY10593 for 15 min at 37 °C before measurements started. ATP was injected after 40 s measurements commenced. Fluorescence was measured using a 485 nm excitation filter and a 520 nm emission filter block. Gain was set at the beginning of the experiment to 30 % required value and fluorescence measurements were taken every 10 s.
Presentation of data and statistics
Data is presented as mean ± SD. Differences between treatments were compared using the unpaired Student’s t test using Prism 5 (Windows version 5.01; GraphPad Software, San Diego, CA) with P < 0.05 considered significant. Concentration response curves of log(agonist) vs. response, or log(inhibitor) vs. response were fitted using the least squares (ordinary) fit method using Prism 5.
Results
P2X7 antagonists inhibit ATP-induced pore formation in a concentration-dependent manner
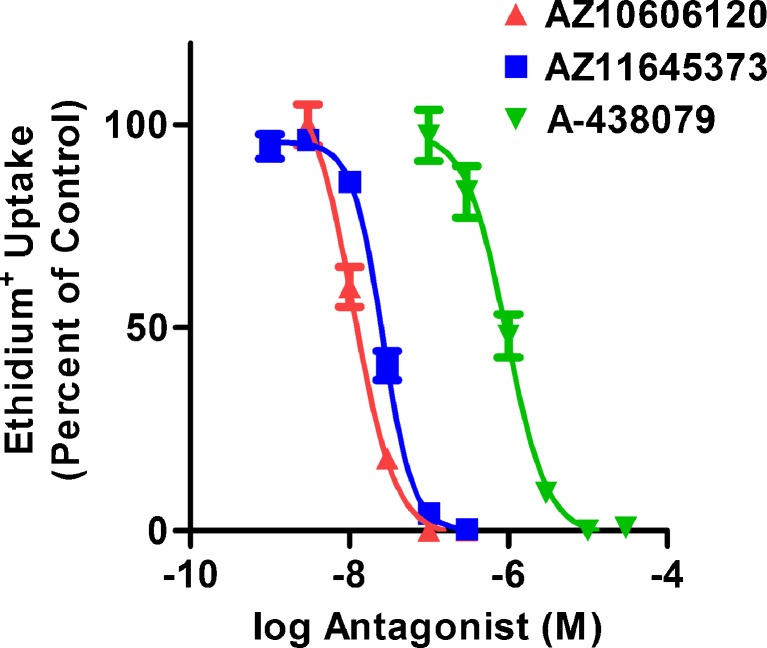
Specific P2X7 antagonists including AZ10606120 [32], AZ11645373 [33] and A-438079 [34] have been characterised using cells expressing recombinant P2X7, however these antagonists have been far less studied on cells expressing endogenous (or native) P2X7. RPMI 8226 cells have been previously shown to express endogenous P2X7 [17, 28, 29]. Therefore, to test these specific P2X7 antagonists on endogenously expressed P2X7, RPMI 8226 cells were pre-incubated in the absence or presence of increasing concentrations of AZ10606120, AZ11645373 or A-438079, and the ATP-induced ethidium+ uptake (pore formation) was measured by flow cytometry. AZ10606120, AZ11645373 and A-438079 inhibited 1 mM ATP-induced ethidium+ uptake in a concentration-dependent manner, with maximal inhibition occurring at 100 nM, 300 nM and 10 μM and with an IC50 of 11 ± 1 nM, 27 ± 3 nM and 900 ± 100 nM, respectively (Fig. 1).
Fig. 1.
P2X7 antagonists impair ATP-induced ethidium+ uptake into RPMI 8226 cells. RPMI 8226 cells in NaCl medium were pre-incubated at 37 °C for 15 min in the absence or presence of varying concentrations of antagonist (as indicated). Cells were then incubated with 25 μM ethidium+ in the absence or presence of 1 mM ATP at 37 °C for 5 min. Incubations were stopped by addition of MgCl2 medium and centrifugation, and the mean fluorescence intensity (MFI) of ethidium+ uptake determined by flow cytometry. Results are the mean percent of ATP-induced ethidium+ uptake in the absence of antagonist ± SD (triplicate data from one experiment for each antagonist)
Changes in intracellular cation concentrations are not essential for P2X7-induced CD23 shedding
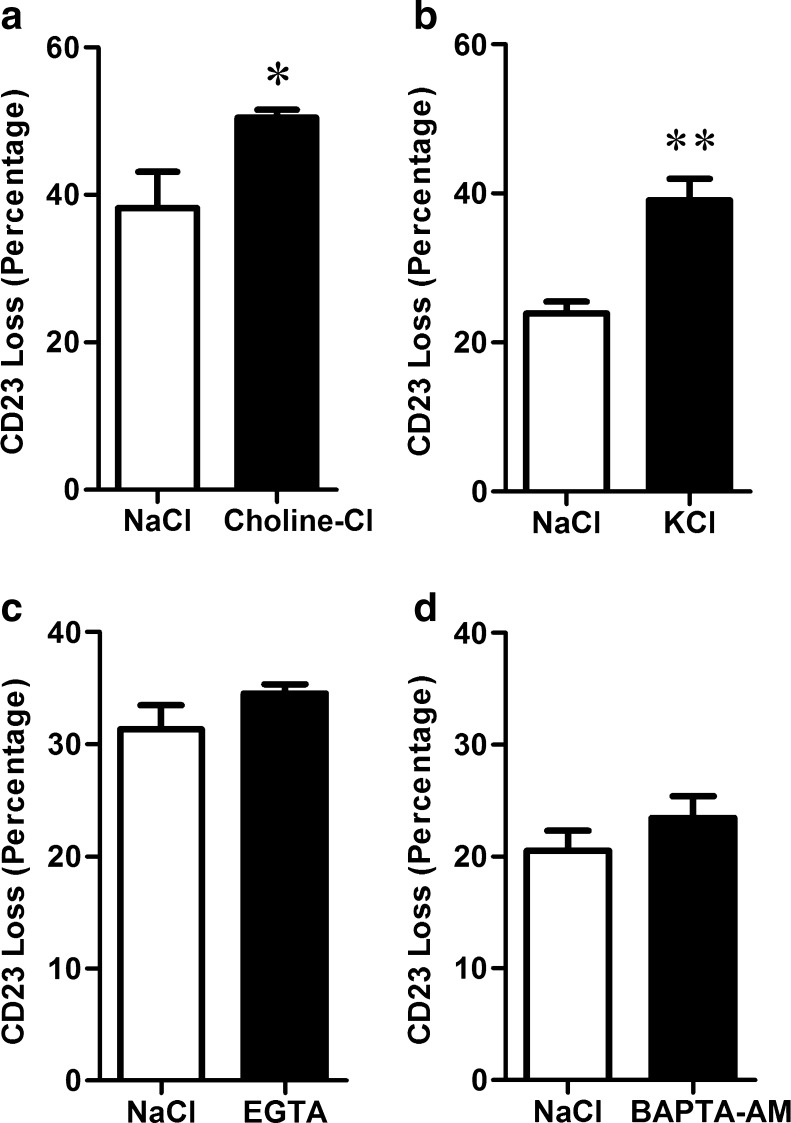
To assess a potential role for changes in intracellular cation concentrations in P2X7-induced CD23 shedding, ATP-induced CD23 shedding from RPMI 8226 cells was compared between cells suspended in NaCl medium (control) to cells suspended in either choline-Cl medium, KCl medium, or in NaCl medium containing EGTA or BAPTA-AM, which prevent Na+ influx, K+ efflux, Ca2+ influx or intracellular Ca2+ increases, respectively. ATP-induced CD23 shedding was indirectly assessed by measuring the loss of cell-surface CD23 using an anti-CD23 mAb and flow cytometry following 7 min incubation with 1 mM ATP, which is approximate to the t1/2 for this process [17, 29]. As previously observed [17, 29], ATP induced CD23 shedding from RPMI 8226 cells in NaCl medium (Fig. 2). ATP-induced CD23 shedding was potentiated from cells suspended in either choline-Cl or KCl medium compared to cells in NaCl medium (Fig. 2a, b). In contrast, ATP-induced CD23 shedding was similar from cells suspended in NaCl medium containing 0.1 mM EGTA or 50 μM BAPTA-AM compared to cells in NaCl medium (Fig. 2c, d).
Fig. 2.
ATP-induced CD23 shedding from RPMI 8226 cells is not prevented by changes in intracellular cation concentrations. a–d Cells in NaCl medium, in a choline-Cl or b KCl medium, or in NaCl medium containing c 0.1 mM EGTA or d 50 μM BAPTA-AM were pre-incubated at 37 °C for a–c 5 min or d 30 min. a–d Cells were then incubated in the absence or presence of 1 mM ATP at 37 °C for 7 min. Incubations were stopped by addition of MgCl2 medium and centrifugation. Cells were labelled with PE-conjugated anti-CD23 or isotype-control mAb, and the mean fluorescence intensity (MFI) of cell-surface CD23 expression determined by flow cytometry. Results are the mean percent of ATP-induced CD23 loss ± SD (triplicate data from one experiment for each comparison); *P < 0.05 and **P < 0.01 compared to NaCl medium
PLD antagonists inhibit P2X7-induced CD23 shedding
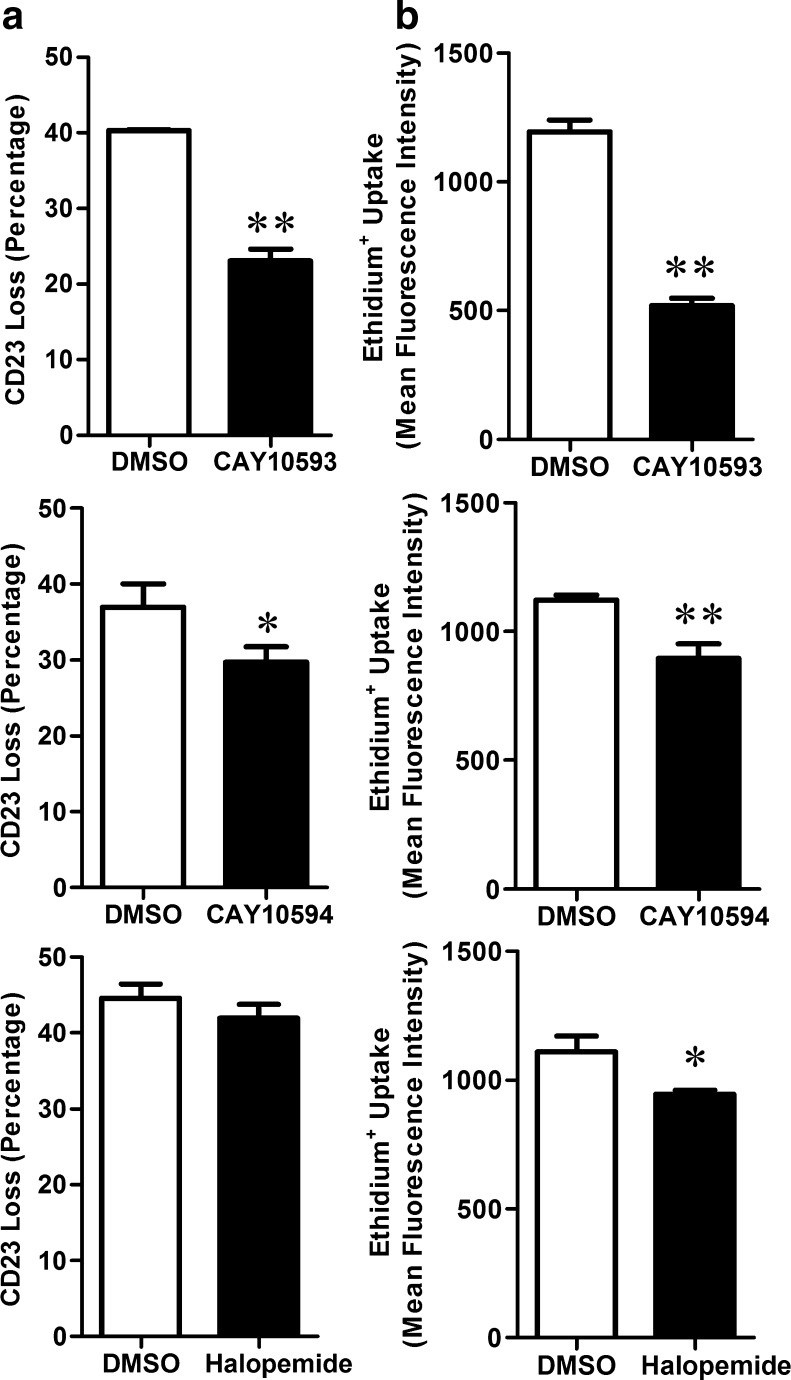
To determine the involvement of intracellular signalling pathways in P2X7-induced CD23 shedding, cells were pre-incubated in the presence of various enzyme antagonists (as listed in the “Materials and methods”) or their corresponding diluent control, and the ATP-induced CD23 shedding assessed as above. Antagonist concentrations were based on previously used concentrations, and AZ10606120 was included as a positive control. As expected, 100 nM AZ10606120 significantly inhibited ATP-induced shedding of CD23 by 88 ± 9 % (n=3). In contrast, most of the enzyme antagonists failed to significantly impair ATP-induced CD23 shedding (results not shown). Of note, the PLD1 antagonist CAY10593 and to a lesser extent the PLD2 antagonist CAY10594 (both at 10 μM) significantly impaired ATP-induced CD23 shedding (Fig. 3a). The non-selective PLD antagonist and structural analogue of CAY10593, halopemide (10 μM) however had no significant effect on ATP-induced CD23 shedding (Fig. 3a). In the absence of ATP, no antagonist significantly altered cell-surface expression of CD23 compared to control-treated cells (results not shown).
Fig. 3.
PLD antagonists impair ATP-induced CD23 shedding and ethidium+ uptake in RPMI 8226 cells. a, b Cells in NaCl medium were pre-incubated at 37 °C for 15 min in the presence of DMSO, or 10 μM CAY10593, CAY10594 or halopemide. a Cells were then incubated in the absence or presence of 1 mM ATP at 37 °C for 7 min. Incubations were stopped by addition of MgCl2 medium and centrifugation. Cells were then labelled with PE-conjugated anti-CD23 or isotype-control mAb, and the mean fluorescence intensity (MFI) of cell-surface CD23 expression determined by flow cytometry. b Cells were then incubated with 25 μM ethidium+ in the absence or presence of 1 mM ATP at 37 °C for 5 min. Incubations were stopped by addition of MgCl2 medium and centrifugation, and the mean fluorescence intensity (MFI) of ethidium+ uptake determined by flow cytometry. Results are the a mean percent of ATP-induced CD23 loss ± SD and b mean ATP-induced ethidium+ uptake ± SD (triplicate data from one experiment for each antagonist); *P < 0.05 and **P < 0.01 compared to DMSO
PLD antagonists inhibit ATP-induced ethidium+ uptake in a concentration-dependent manner
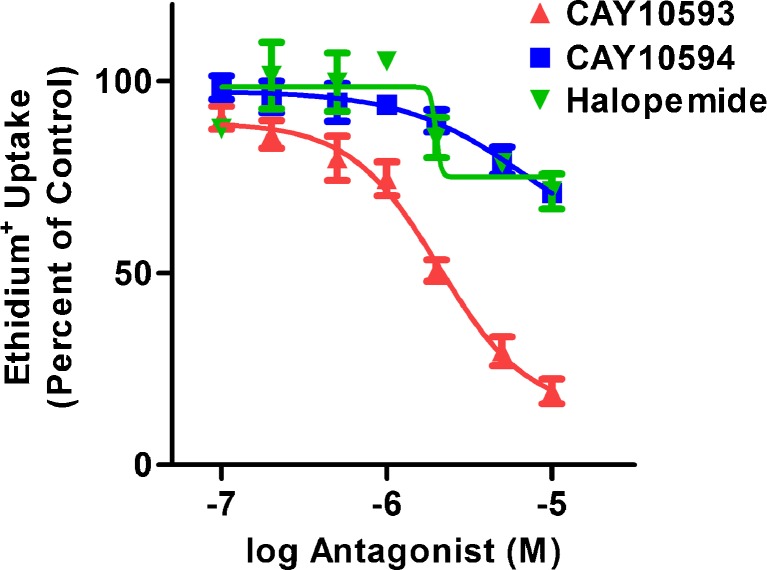
The above data indicates that the PLD1 antagonist, CAY10593, can inhibit P2X7-induced CD23 shedding. Therefore, the current study investigated the mode of action by which this compound impaired P2X7-induced CD23 shedding. The PLD2 antagonist CAY10594 and the non-selective PLD antagonist halopemide were studied as a comparison. To investigate whether CAY10593 impaired ATP-induced CD23 shedding by blocking P2X7 activation, RPMI 8226 cells were pre-incubated in the presence of DMSO (diluent control), CAY10593, CAY10594 or halopemide (each at 10 μM), and the ATP-induced ethidium+ uptake was measured. The PLD antagonists significantly inhibited ATP-induced ethidium+ uptake by 56 ± 4 %, 20 ± 6 % and 15 ± 5 % respectively compared to ATP-induced ethidium+ uptake in cells pre-incubated with DMSO (Fig. 3b). In the absence of ATP, these antagonists did not significantly alter ethidium+ uptake compared to DMSO-treated cells (results not shown).
The above data suggests that the inhibitory action of CAY10593 and CAY10594 on P2X7-induced CD23 shedding is due to impaired P2X7 activation. Therefore, to further characterise the effect of the above three PLD antagonists on P2X7 activation, cells were pre-incubated in the presence of increasing concentrations of each antagonist and the ATP-induced ethidium+ uptake measured, using 120 μM ATP which is approximate to the EC50 for ATP in this process [17, 28]. CAY10593 inhibited ATP-induced ethidium+ uptake in a concentration-dependent manner, with maximal inhibition occurring near 10 μM and an IC50 of 2.0 ± 0.5 μM (Fig. 4). Inhibition of ATP-induced ethidium+ uptake by CAY10594 and halopemide was less than 29 % on average at the highest concentration used (10 μM), and IC50 values for these antagonists could not be reliably determined due to this low amount of inhibition (Fig. 4). Thus, the mode of action by which CAY10593 impairs P2X7 was studied further below.
Fig. 4.
CAY10593, CAY10594 and halopemide impair ATP-induced ethidium+ uptake into RPMI 8226 cells in a concentration-dependent manner. Cells in NaCl medium were pre-incubated at 37 °C for 15 min in the presence of DMSO or varying concentrations of antagonist (as indicated). Cells were then incubated with 25 μM ethidium+ in the absence or presence of 120 μM ATP at 37 °C for 5 min. Incubations were stopped by addition of MgCl2 medium and centrifugation, and the mean fluorescence intensity (MFI) of ethidium+ uptake determined by flow cytometry. Results are the mean percent of ATP-induced ethidium+ uptake in the absence of compound ± SD (triplicate data from one experiment for each antagonist)
CAY10593 inhibits P2X7-induced pore formation in a non-competitive-like manner
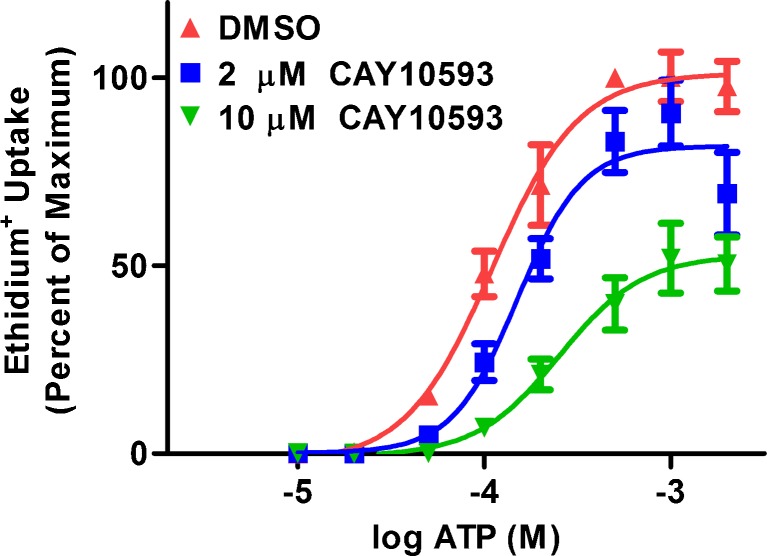
To determine whether CAY10593 inhibits P2X7-induced pore formation in a competitive or non-competitive manner, RPMI 8226 cells were pre-incubated in the presence of DMSO, or 2 or 10 μM CAY10593, and then the ethidium+ uptake was measured in the presence of increasing concentrations of ATP. In the absence of CAY10593, ATP induced ethidium+ uptake in a concentration-dependent manner with maximal uptake occurring at 0.5 mM ATP and with an EC50 of 116 ± 31 μM (Fig. 5). In the presence of 2 μM CAY10593, the mean maximum ATP response was reduced by 17 % and with a slight increase in the EC50 to 154 ± 26 μM (Fig. 5). In the presence of 10 μM CAY10593, the mean maximum ATP response was reduced by 60 % and with a larger increase in the EC50 to 256 ± 22 μM (Fig. 5).
Fig. 5.
CAY10593 impairs ATP-induced ethidium+ uptake into RPMI 8226 cells in a non-competitive-like manner. RPMI 8226 cells in NaCl medium were pre-incubated at 37 °C for 15 min in the presence of DMSO, or 2 μM or 10 μM CAY10593. Cells were then incubated with 25 μM ethidium+ in the absence or presence of varying concentrations of ATP at 37 °C for 5 min. Incubations were stopped by addition of MgCl2 medium and centrifugation, and the mean fluorescence intensity (MFI) of ethidium+ uptake determined by flow cytometry. Results are the mean percent of maximum ATP (0.5 mM)-induced ethidium+ uptake ± SD (data from three independent experiments)
PLD signalling is not required for P2X7-induced pore formation
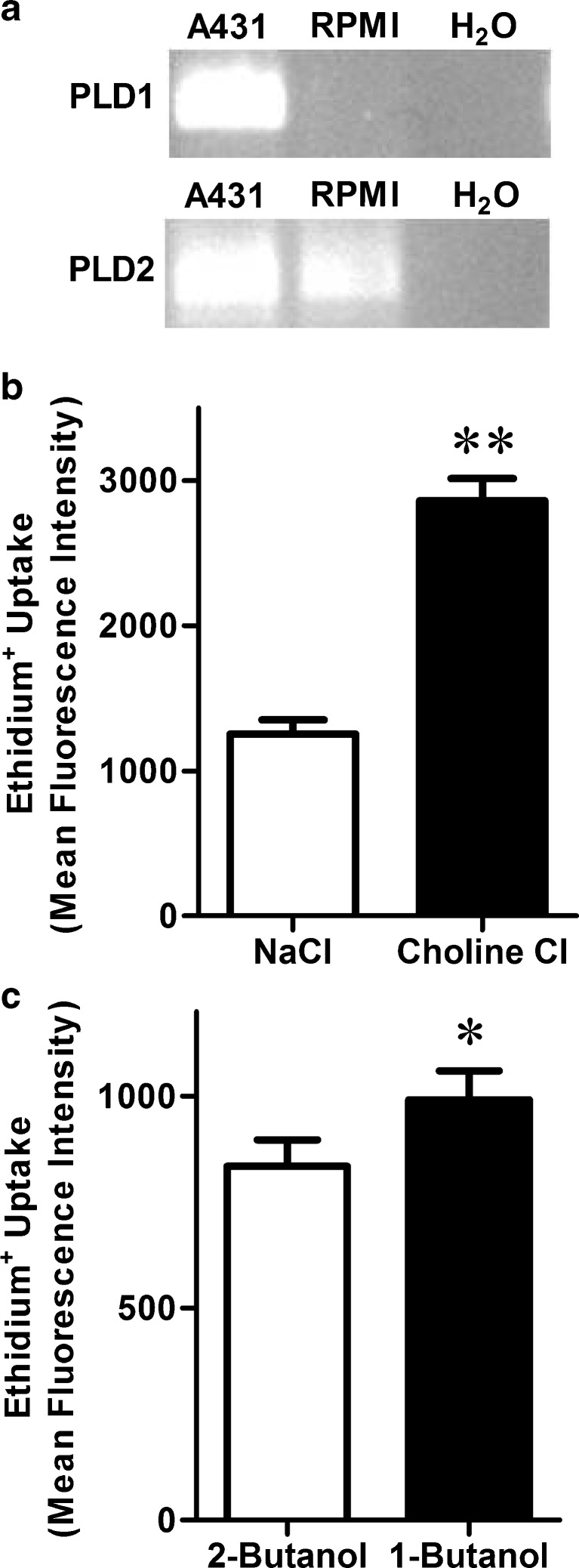
Collectively, the above results show that the PLD1 specific antagonist CAY10593 can impair P2X7-induced pore formation. Therefore, to determine whether this effect is due to inhibition of PLD1 or direct inhibition of P2X7 itself, a series of experiments were performed. First, the presence of PLD1 and PLD2 in RPMI 8226 cells was examined by RT-PCR. The human skin epithelial carcinoma cell line, A431, which expresses both PLD isoforms [35], was used as a positive control. RT-PCR revealed the presence of PLD1 in A431 cells but not in RPMI 8226 cells, despite the presence of PLD2 in both cell lines (Fig. 6a). Choline-Cl medium has previously been shown to prevent P2X7-induced PLD stimulation [24, 36]. Therefore RPMI 8226 cells were suspended in either choline-Cl or NaCl medium and the ATP-induced ethidium+ uptake measured. ATP-induced ethidium+ uptake was potentiated in cells incubated in choline-Cl compared to NaCl medium (Fig. 6b). In the presence of a primary alcohol, PLD catalyses a transphosphatidylation reaction to form a phosphatidyl alcohol product, which does not serve as a substrate for PLD-mediated signal transduction [37]. Therefore, RPMI 8226 cells were pre-incubated in the presence of the primary alcohol, 0.27 % (v/v) 1-butanol, or the secondary alcohol, 0.27 % (v/v) 2-butanol as a negative control, and the ATP-induced ethidium+ uptake measured. ATP-induced ethidium+ uptake into cells treated with 1-butanol was also increased compared to cells treated with 2-butanol (Fig. 6c). In the absence of ATP, ethidium+ uptake into cells in choline-Cl or NaCl media in the presence of either alcohol was similar (results not shown).
Fig. 6.
PLD1 is not required for ATP-induced ethidium+ uptake into RPMI 8226 cells. a RNA was isolated from A431 (positive control) and RPMI 8226 cells, and then analysed by RT-PCR using primers for PLD1 and PLD2. RNA substituted with H2O was used as a negative control. PCR products were visualised by agarose gel electrophoresis and ethidium bromide (representative result from three independent experiments is shown). b, c RPMI 8226 cells were pre-incubated at 37 °C for 5 min in b choline-Cl or NaCl medium, or c NaCl medium in the presence of 0.27 % (v/v) 2-butanol (negative control) or 1-butanol. b, c Cells were then incubated with 25 μM ethidium+ in the absence or presence of 1 mM ATP at 37 °C for 5 min. Incubations were stopped by addition of MgCl2 medium and centrifugation, and the mean fluorescence intensity (MFI) of ethidium+ uptake determined by flow cytometry. Results are the mean ATP-induced ethidium+ uptake ± SD (triplicate data from one experiment for each comparison); *P < 0.05 and **P < 0.01 compared to corresponding control
CAY10593 inhibits P2X7-induced channel activation and pore formation in P2X7-transfected HEK-293 cells
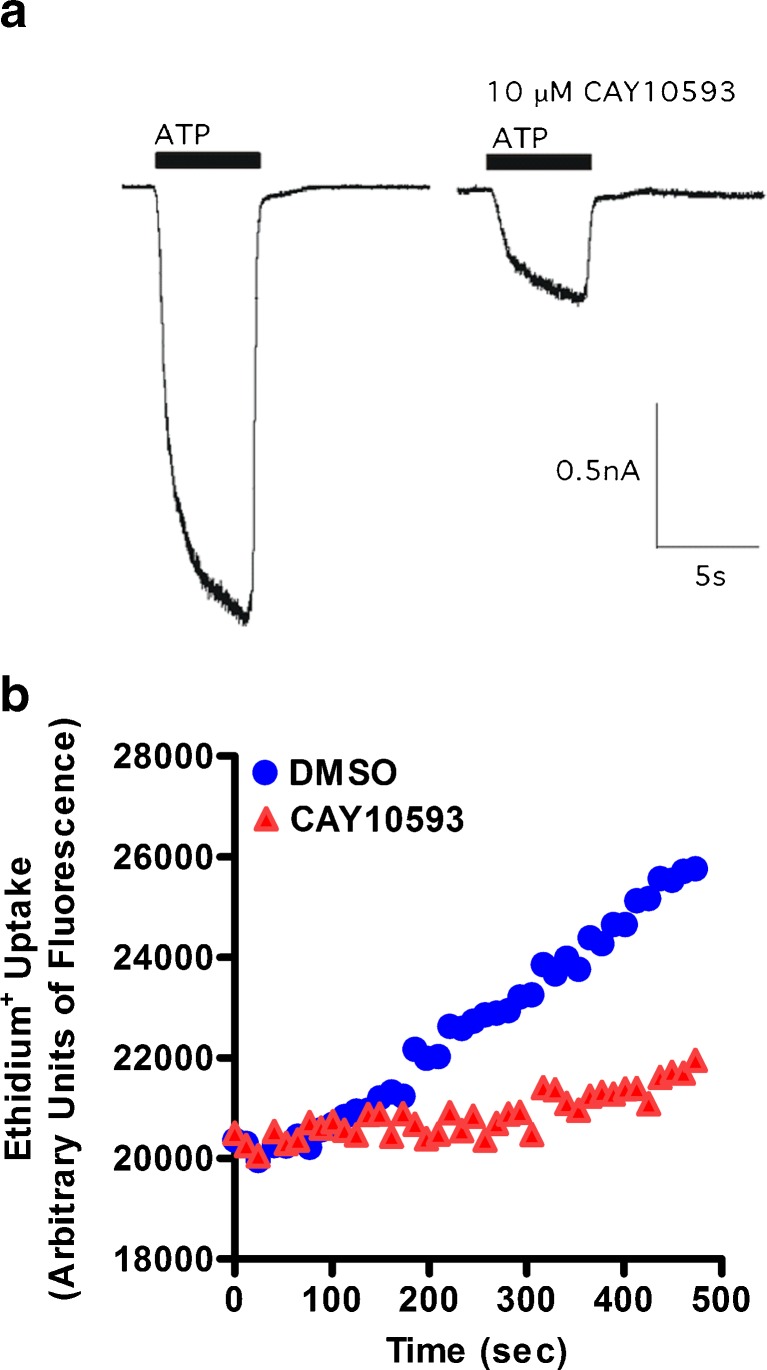
The above results indicate that PLD signalling is not required for P2X7-induced pore formation and that CAY10593 directly impairs P2X7. Therefore, the effect of CAY10593 on P2X7 channel activity in human P2X7-transfected HEK-293 cells was assessed by electrophysiology. In the absence of 10 μM CAY10593, ATP induced an inward current typical of P2X7 (Fig. 7a). Removal of extracellular ATP, and subsequent 3–5 min incubation with 10 μM CAY10593 reduced the ATP-induced inward current to 29.5 ± 2.6 % of control (Fig. 7a). To confirm that CAY10593 impairs P2X7 pore formation in these cells, P2X7-transfected HEK-293 cells were pre-incubated with CAY10593 for 15 min at 37 °C and ATP-induced ethidium+ uptake was measured. CAY10593 impaired ATP-induced ethidium+ uptake into P2X7-transfected HEK-293 cells by 72 % (Fig. 7b).
Fig. 7.
CAY10593 impairs ATP-induced inward currents and pore formation in human P2X7-transfected HEK-293 cells. a Inward currents were elicited using 1 mM ATP in divalent NaCl solution. ATP was added for 5 s (denoted by black bar) before and after treatment with 10 μM CAY10593 (3–5 min). ATP was added in the continued presence of CAY10593 (single representative trace of four to seven cells is shown). b P2X7-transfected HEK-293 cells in low divalent NaCl solution containing 25 μM ethidium+ were pre-incubated in the presence of DMSO or CAY10593 for 15 min at 37 °C. ATP (1 mM) was injected after 40 s. Fluorescence was measured every 10 s using a plate reader (data from one experiment)
CAY10593 inhibits P2X7-induced pore formation in human peripheral blood mononuclear cells
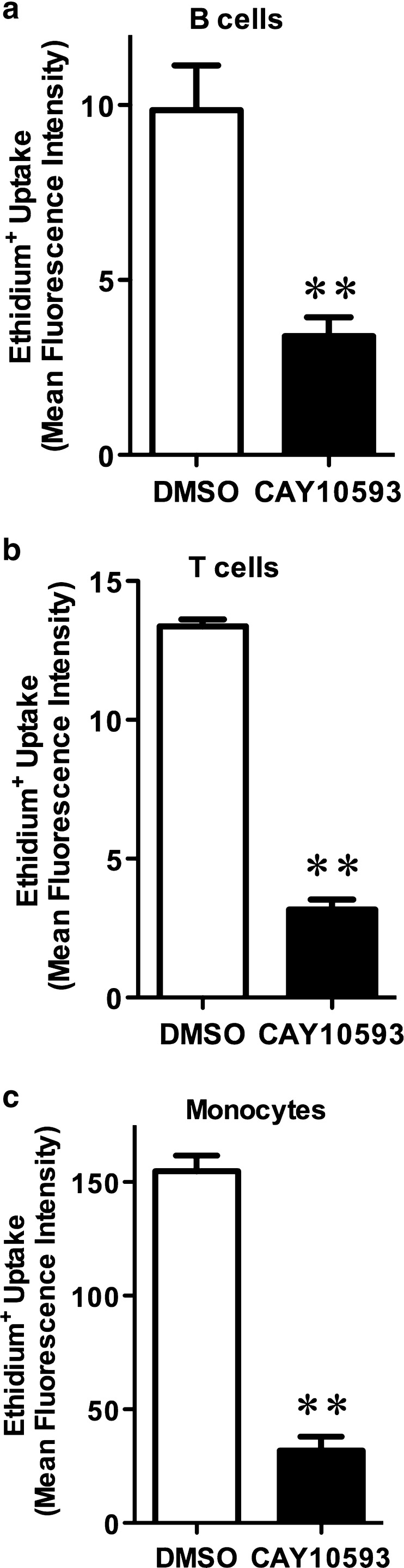
To determine if CAY10593 inhibits P2X7-induced pore formation in primary cells, PBMCs from a human donor were pre-incubated in the presence of DMSO or 10 μM CAY10593 and ethidium+ uptake was measured in the absence or presence of ATP. CAY10593 inhibited ATP-induced ethidium+ uptake into B cells, T cells and monocytes by 66 ± 5 %, 76 ± 3 % and 80 ± 4 % respectively (Fig. 8). In the absence of ATP, CAY10593 did not significantly alter ethidium+ uptake compared to DMSO-treated cells (results not shown). Similar amounts of inhibition of ATP-induced ethidium+ uptake by CAY10593 were observed in B cells, T cells and monocytes from a second donor (results not shown).
Fig. 8.
CAY10593 impairs ATP-induced ethidium+ uptake in primary human peripheral blood mononuclear cells (PBMCs). PBMCs in NaCl medium were pre-incubated for 15 min in the presence of DMSO or 10 μM CAY10593. Cells were then incubated with 25 μM ethidium+ in the absence or presence of 1 mM ATP at 37 °C for 5 min. Incubations were stopped by addition of MgCl2 medium and centrifugation, and washed once with NaCl medium. Cells were then labelled with APC-conjugated anti-CD19. The mean fluorescence intensity (MFI) of ethidium+ uptake into a B cells (CD19+) b T cells (CD19−) and (c) monocytes was determined by flow cytometry. Results are the mean ATP-induced ethidium+ uptake ± SD (triplicate data from one donor; similar amounts of inhibition were observed in with triplicate data from a second donor, results not shown); **P < 0.01 compared to corresponding control
Discussion
The current study demonstrates that the PLD1 antagonist, CAY10593 impairs human P2X7 activation. Our preliminary investigations demonstrated that CAY10593 impaired ATP-induced CD23 shedding. Moreover, CAY10593 impaired ATP-induced ethidium+ uptake into RPMI 8226 cells in a concentration-dependent manner, and with an IC50 similar to that of the P2X7 antagonist, A-438079 as observed previously [34] and in this study. Unlike A-438079, however, which blocks P2X7 in a competitive manner [34], CAY10593 impaired P2X7 in a non-competitive-like manner. CAY10593 also impaired ATP-induced ethidium+ uptake into P2X7-transfected HEK-293 cells, and primary human B cells, T cells and monocytes. In contrast, CAY10593 failed to affect ATP-induced ethidium+ uptake into murine erythroleukemia cells (unpublished observations), which like RPMI 8226 cells also express endogenous P2X7 [12, 38], indicating that CAY10593 does not act on murine P2X7.
The current study also shows that CAY10593 impairs P2X7 activation independently of PLD1 stimulation. First, CAY10593 was far more effective at inhibiting P2X7-induced pore formation than halopemide, which both impair cellular PLD1 with similar efficacies [30]. Second, the IC50 value of CAY10593 for inhibition of P2X7-induced pore formation was two logs greater than that observed for the inhibition of phorbol 12-myristate-induced PLD1 stimulation in the non-small-cell lung cancer cell line, Calu-1 [30] (2 μM vs. 11 nM, respectively). Third, PLD1 mRNA was absent in RPMI 8226 cells, indicating that these cells do not contain this PLD isoform. Fourth, the primary alcohol, 1-butanol, which prevents PLD-mediated signalling [37], did not impair P2X7-induced pore formation. In fact, 1-butanol potentiated this process, similar to a previous study in which incubation with 1-butanol increased P2X7-induced pore formation and cytolysis in murine macrophages compared to controls [39]. The authors of this previous study concluded that phosphatidic acid production, resulting from PLD stimulation, delays P2X7-induced pore formation and cytolysis. Thus, it is possible that PLD-induced phosphatidic acid production may also delay P2X7-induced pore formation in human cells. Fifth and similar to the effect of 1-butanol, P2X7-induced pore formation and CD23 shedding were not impaired in choline-Cl medium, which prevents P2X7-induced PLD stimulation in murine macrophages [24] and human chronic lymphocytic leukemic lymphocytes [36]. Sixth, CAY10593 also impaired P2X7 channel activity, which is not directly linked to the activation of intracellular signalling molecules [40]. Finally, it is unlikely that CAY10593 impairs P2X7 activity via a PLD2-dependent mechanism despite the presence of PLD2 mRNA in RPMI 8226 cells; the PLD2 inhibitor, CAY10594, was far less effective at impairing ATP-induced ethidium+ uptake and CD23 shedding compared to CAY10593. Combined this data highlights the importance of ensuring that antagonists used in intracellular signalling studies downstream of P2X7 activation do not directly affect P2X7 itself. In this regard, CAY10593 has been used at 50 μM to support a role for PLD in the generation of P2X7-induced microvesicles capable of activating macrophages [27]. However, our data suggests that CAY10593 may have also acted on P2X7 itself in this previous study. Conversely, our data in combination with that of Scott and colleagues [30] indicates that the use of CAY10593 at nanomolar concentrations will be of potential value in determining if PLD1 is involved in signalling pathways downstream of P2X7 activation.
As noted above, CAY10593 impaired P2X7-induced pore formation more efficaciously than CAY10594 or halopemide. Moreover, CAY10593 blocked P2X7-induced CD23 shedding to a greater extent than CAY10594 or halopemide. CAY10593 was originally synthesised via the modification of the 1-(piperidin-4-yl)-1H-benzo[d]imidazol-2(3H)-one analogue halopemide, but unlike halopemide, CAY10593 contains a chiral (S)-methyl group which prompts PLD1 preferring pharmacology [30]. CAY10594, which has a 1-phenyl-1,3,8-triazaspiro[4,5]decan-4-one scaffold instead of a 1-(piperidin-4-yl)-1H-benzo[d]imidazol-2(3H)-one scaffold, also lacks a chiral (S)-methyl group [30]. Therefore, this structural group may be of importance in the interaction of CAY10593 with P2X7. The scaffold or structural groups of CAY10593 may provide useful leads in the development of new P2X7 antagonists. In this regard, the study of analogues of the P2X7 antagonist KN-62, which is also an inhibitor of Ca2+/calmodulin-dependent protein kinase II, has provided valuable insight into moieties which interact with P2X7 [41].
Broad spectrum metalloprotease antagonists have implicated a role for metalloproteases in P2X7-induced CD23 shedding from chronic lymphocytic leukemic B-lymphocytes [16] and RPMI 8226 cells [17]. More recently, we have shown that P2X7-induced CD23 shedding is mediated by ADAM10 [29]. However, it is unknown if P2X7-induced CD23 shedding results from changes in intracellular cation concentrations, stimulation of signalling pathways downstream of P2X7 activation or by a direct physical interaction of P2X7 itself. The current study reports data exploring the first two of these potential mechanisms. In this study, we show that P2X7-induced CD23 shedding does not require changes in intracellular Na+, K+ or Ca2+ concentrations. Changes in intracellular cation concentrations are crucial for some P2X7-mediated downstream processes. For example, P2X7-induced IL-1β processing and release is dependent on K+ efflux from human monocytes [42, 43], murine and human macrophages [44–46] and murine microglia [47]. Moreover, P2X7-induced secretion of IL-1β is dependent on the influx of extracellular Ca2+ and a sustained increase in intracellular Ca2+ in human monocytes [42, 48], murine macrophages and P2X7-transfected HEK-293 cells [48]. Finally, P2X7-induced rapid phosphatidylserine exposure on murine thymocytes is dependent on Na+ influx [49]. In contrast to these studies, we found that neither K+ efflux, Na+ influx, Ca2+ influx nor an increase in intracellular Ca2+ is essential for P2X7-induced CD23 shedding from RPMI 8226 cells. Of note, choline-Cl and KCl medium potentiated ATP-induced CD23 shedding compared to NaCl medium, which is likely due to the omission of extracellular Na+, a cation known to inhibit P2X7 activity [50, 51], as well as the possible inhibition of phosphatidic acid production by choline as discussed above.
The current study also reports that several signalling molecules downstream of P2X7 are unlikely to be involved in P2X7-induced CD23 shedding. However, it should be noted that the compounds used to target these enzymes were only used at a single concentration, and the efficacy of these compounds was not verified by relevant enzymatic assays. Thus, the involvement of these signalling pathways in P2X7-induced CD23 shedding cannot be definitively excluded. Future studies, using large antagonist libraries and high throughput assays may prove a more useful approach to identify the potential signalling pathway mediating P2X7-induced CD23 shedding, rather than the candidate approach applied in this study.
In conclusion, this study demonstrates the PLD1 antagonist, CAY10593, impairs human P2X7 independently of PLD1 stimulation. This study highlights the importance of ensuring that antagonists used in intracellular signalling studies downstream of P2X7 activation do not directly affect P2X7 itself. Moreover, this study suggests that CAY10593 may be of value in future studies of P2X7-induced PLD1 stimulation when used at nanomolar concentrations, as well as a lead compound in the development of novel P2X7 antagonists.
Acknowledgments
We gratefully acknowledge Marie Ranson for helpful advice and critically reviewing the manuscript, and Vanessa Sluyter and the staff of Illawarra Health and Medical Research Institute for excellent technical assistance.
References
- 1.Sluyter R, Stokes L. Significance of P2X7 receptor variants to human health and disease. Recent Pat DNA Gene Seq. 2011;5:41–54. doi: 10.2174/187221511794839219. [DOI] [PubMed] [Google Scholar]
- 2.Wiley JS, Gargett CE, Zhang W, Snook MB, Jamieson GP. Partial agonists and antagonists reveal a second permeability state of human lymphocyte P2Z/P2X7 channel. Am J Physiol. 1998;275:C1224–C1231. doi: 10.1152/ajpcell.1998.275.5.C1224. [DOI] [PubMed] [Google Scholar]
- 3.Yan Z, Khadra A, Li S, Tomic M, Sherman A, et al. Experimental characterization and mathematical modeling of P2X7 receptor channel gating. J Neurosci. 2010;30:14213–14224. doi: 10.1523/JNEUROSCI.2390-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pelegrin P, Surprenant A. Pannexin-1 mediates large pore formation and interleukin-1β release by the ATP-gated P2X7 receptor. EMBO J. 2006;25:5071–5082. doi: 10.1038/sj.emboj.7601378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Marques-da-Silva C, Chaves MM, Castro NG, Coutinho-Silva R, Guimaraes MZ. Colchicine inhibits cationic dye uptake induced by ATP in P2X2 and P2X7 receptor-expressing cells: implications for its therapeutic action. Br J Pharmacol. 2011;163:912–926. doi: 10.1111/j.1476-5381.2011.01254.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schachter J, Motta AP, de Souza Zamorano A, da Silva-Souza HA, Guimaraes MZ, et al. ATP-induced P2X7-associated uptake of large molecules involves distinct mechanisms for cations and anions in macrophages. J Cell Sci. 2008;121:3261–3270. doi: 10.1242/jcs.029991. [DOI] [PubMed] [Google Scholar]
- 7.Cankurtaran-Sayar S, Sayar K, Ugur M. P2X7 receptor activates multiple selective dye-permeation pathways in RAW 264.7 and human embryonic kidney 293 cells. Mol Pharmacol. 2009;76:1323–1332. doi: 10.1124/mol.109.059923. [DOI] [PubMed] [Google Scholar]
- 8.Donnelly-Roberts DL, Namovic MT, Faltynek CR, Jarvis MF. Mitogen-activated protein kinase and caspase signaling pathways are required for P2X7 receptor (P2X7R)-induced pore formation in human THP-1 cells. J Pharmacol Exp Ther. 2004;308:1053–1061. doi: 10.1124/jpet.103.059600. [DOI] [PubMed] [Google Scholar]
- 9.Faria RX, Defarias FP, Alves LA. Are second messengers crucial for opening the pore associated with P2X7 receptor? Am J Physiol Cell Physiol. 2005;288:C260–C271. doi: 10.1152/ajpcell.00215.2004. [DOI] [PubMed] [Google Scholar]
- 10.Bianco F, Perrotta C, Novellino L, Francolini M, Riganti L, et al. Acid sphingomyelinase activity triggers microparticle release from glial cells. EMBO J. 2009;28:1043–1054. doi: 10.1038/emboj.2009.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.da Cruz CM, Ventura AL, Schachter J, Costa-Junior HM, da Silva Souza HA, et al. Activation of ERK1/2 by extracellular nucleotides in macrophages is mediated by multiple P2 receptors independently of P2X7-associated pore or channel formation. Br J Pharmacol. 2006;147:324–334. doi: 10.1038/sj.bjp.0706559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang B, Sluyter R. P2X7 receptor activation induces reactive oxygen species formation in erythroid cells. Purinergic Signal. 2013;9:101–112. doi: 10.1007/s11302-012-9335-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Michel AD, Thompson KM, Simon J, Boyfield I, Fonfria E, et al. Species and response dependent differences in the effects of MAPK inhibitors on P2X7 receptor function. Br J Pharmacol. 2006;149:948–957. doi: 10.1038/sj.bjp.0706938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lenertz LY, Gavala ML, Zhu Y, Bertics PJ. Transcriptional control mechanisms associated with the nucleotide receptor P2X7, a critical regulator of immunologic, osteogenic, and neurologic functions. Immunol Res. 2011;50:22–38. doi: 10.1007/s12026-011-8203-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wiley JS, Sluyter R, Gu BJ, Stokes L, Fuller SJ. The human P2X7 receptor and its role in innate immunity. Tissue Antigens. 2011;78:321–332. doi: 10.1111/j.1399-0039.2011.01780.x. [DOI] [PubMed] [Google Scholar]
- 16.Gu B, Bendall LJ, Wiley JS. Adenosine triphosphate-induced shedding of CD23 and L-selectin (CD62L) from lymphocytes is mediated by the same receptor but different metalloproteases. Blood. 1998;92:946–951. [PubMed] [Google Scholar]
- 17.Farrell AW, Gadeock S, Pupovac A, Wang B, Jalilian I, et al. P2X7 receptor activation induces cell death and CD23 shedding in human RPMI 8226 multiple myeloma cells. Biochim Biophys Acta. 2010;1800:1173–1182. doi: 10.1016/j.bbagen.2010.07.001. [DOI] [PubMed] [Google Scholar]
- 18.Sluyter R, Wiley JS. Extracellular adenosine 5'-triphosphate induces a loss of CD23 from human dendritic cells via activation of P2X7 receptors. Int Immunol. 2002;14:1415–1421. doi: 10.1093/intimm/dxf111. [DOI] [PubMed] [Google Scholar]
- 19.Georgiou JG, Skarratt KK, Fuller SJ, Martin CJ, Christopherson RI, et al. Human epidermal and monocyte-derived Langerhans cells express functional P2X receptors. J Invest Dermatol. 2005;125:482–490. doi: 10.1111/j.0022-202X.2005.23835.x. [DOI] [PubMed] [Google Scholar]
- 20.McDermott M, Wakelam MJ, Morris AJ. Phospholipase D. Biochem Cell Biol. 2004;82:225–253. doi: 10.1139/o03-079. [DOI] [PubMed] [Google Scholar]
- 21.Gargett CE, Cornish EJ, Wiley JS. Phospholipase D activation by P2Z-purinoceptor agonists in human lymphocytes is dependent on bivalent cation influx. Biochem J. 1996;313:529–535. doi: 10.1042/bj3130529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shemon AN, Sluyter R, Wiley JS. Rottlerin inhibits P2X7 receptor-stimulated phospholipase D activity in chronic lymphocytic leukaemia B-lymphocytes. Immunol Cell Biol. 2007;85:68–72. doi: 10.1038/sj.icb.7100005. [DOI] [PubMed] [Google Scholar]
- 23.Humphreys BD, Dubyak GR. Induction of the P2z/P2X7 nucleotide receptor and associated phospholipase D activity by lipopolysaccharide and IFN-γ in the human THP-1 monocytic cell line. J Immunol. 1996;157:5627–5637. [PubMed] [Google Scholar]
- 24.el-Moatassim C, Dubyak GR. Dissociation of the pore-forming and phospholipase D activities stimulated via P2z purinergic receptors in BAC1.2F5 macrophages. Product inhibition of phospholipase D enzyme activity. J Biol Chem. 1993;268:15571–15578. [PubMed] [Google Scholar]
- 25.Kusner DJ, Adams J. ATP-induced killing of virulent Mycobacterium tuberculosis within human macrophages requires phospholipase D. J Immunol. 2000;164:379–388. doi: 10.4049/jimmunol.164.1.379. [DOI] [PubMed] [Google Scholar]
- 26.Fairbairn IP, Stober CB, Kumararatne DS, Lammas DA. ATP-mediated killing of intracellular mycobacteria by macrophages is a P2X7-dependent process inducing bacterial death by phagosome-lysosome fusion. J Immunol. 2001;167:3300–3307. doi: 10.4049/jimmunol.167.6.3300. [DOI] [PubMed] [Google Scholar]
- 27.Thomas LM, Salter RD. Activation of macrophages by P2X7-induced microvesicles from myeloid cells is mediated by phospholipids and is partially dependent on TLR4. J Immunol. 2010;185:3740–3749. doi: 10.4049/jimmunol.1001231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gadeock S, Pupovac A, Sluyter V, Spildrejorde M, Sluyter R. P2X7 receptor activation mediates organic cation uptake into human myeloid leukaemic KG-1 cells. Purinergic Signal. 2012;8:669–676. doi: 10.1007/s11302-012-9320-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pupovac A, Foster CM, Sluyter R. Human P2X7 receptor activation induces the rapid shedding of CXCL16. Biochem Biophys Res Commun. 2013;432:626–631. doi: 10.1016/j.bbrc.2013.01.134. [DOI] [PubMed] [Google Scholar]
- 30.Scott SA, Selvy PE, Buck JR, Cho HP, Criswell TL, et al. Design of isoform-selective phospholipase D inhibitors that modulate cancer cell invasiveness. Nat Chem Biol. 2009;5:108–117. doi: 10.1038/nchembio.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stokes L, Fuller SJ, Sluyter R, Skarratt KK, Gu BJ, et al. Two haplotypes of the P2X7 receptor containing the Ala-348 to Thr polymorphism exhibit a gain-of-function effect and enhanced interleukin-1β secretion. FASEB J. 2010;24:2916–2927. doi: 10.1096/fj.09-150862. [DOI] [PubMed] [Google Scholar]
- 32.Michel AD, Chambers LJ, Walter DS. Negative and positive allosteric modulators of the P2X7 receptor. Br J Pharmacol. 2008;153:737–750. doi: 10.1038/sj.bjp.0707625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stokes L, Jiang LH, Alcaraz L, Bent J, Bowers K, et al. Characterization of a selective and potent antagonist of human P2X7 receptors, AZ11645373. Br J Pharmacol. 2006;149:880–887. doi: 10.1038/sj.bjp.0706933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nelson DW, Gregg RJ, Kort ME, Perez-Medrano A, Voight EA, et al. Structure-activity relationship studies on a series of novel, substituted 1-benzyl-5-phenyltetrazole P2X7 antagonists. J Med Chem. 2006;49:3659–3666. doi: 10.1021/jm051202e. [DOI] [PubMed] [Google Scholar]
- 35.Min DS, Ahn BH, Jo YH. Differential tyrosine phosphorylation of phospholipase D isozymes by hydrogen peroxide and the epidermal growth factor in A431 epidermoid carcinoma cells. Mol Cells. 2001;11:369–378. [PubMed] [Google Scholar]
- 36.Fernando KC, Gargett CE, Wiley JS. Activation of the P2Z/P2X7 receptor in human lymphocytes produces a delayed permeability lesion: involvement of phospholipase D. Arch Biochem Biophys. 1999;362:197–202. doi: 10.1006/abbi.1998.1045. [DOI] [PubMed] [Google Scholar]
- 37.Morris AJ, Frohman MA, Engebrecht J. Measurement of phospholipase D activity. Anal Biochem. 1997;252:1–9. doi: 10.1006/abio.1997.2299. [DOI] [PubMed] [Google Scholar]
- 38.Constantinescu P, Wang B, Kovacevic K, Jalilian I, Bosman GJ, et al. P2X7 receptor activation induces cell death and microparticle release in murine erythroleukemia cells. Biochim Biophys Acta. 2010;1798:1797–1804. doi: 10.1016/j.bbamem.2010.06.002. [DOI] [PubMed] [Google Scholar]
- 39.Le Stunff H, Raymond MN. P2X7 receptor-mediated phosphatidic acid production delays ATP-induced pore opening and cytolysis of RAW 264.7 macrophages. Cell Signal. 2007;19:1909–1918. doi: 10.1016/j.cellsig.2007.04.015. [DOI] [PubMed] [Google Scholar]
- 40.Pelegrin P. Many ways to dilate the P2X7 receptor pore. Br J Pharmacol. 2011;163:908–911. doi: 10.1111/j.1476-5381.2011.01325.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Romagnoli R, Baraldi PG, Di Virgilio F. Recent progress in the discovery of antagonists acting at P2X7 receptor. Expert Opin Ther Patents. 2005;15:271–287. doi: 10.1517/13543776.15.3.271. [DOI] [Google Scholar]
- 42.Andrei C, Margiocco P, Poggi A, Lotti LV, Torrisi MR, et al. Phospholipases C and A2 control lysosome-mediated IL-1β secretion: Implications for inflammatory processes. Proc Natl Acad Sci U S A. 2004;101:9745–9750. doi: 10.1073/pnas.0308558101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sluyter R, Shemon AN, Wiley JS. Glu496 to Ala polymorphism in the P2X7 receptor impairs ATP-induced IL-1β release from human monocytes. J Immunol. 2004;172:3399–3405. doi: 10.4049/jimmunol.172.6.3399. [DOI] [PubMed] [Google Scholar]
- 44.Kahlenberg JM, Dubyak GR. Mechanisms of caspase-1 activation by P2X7 receptor-mediated K+ release. Am J Physiol Cell Physiol. 2004;286:C1100–C1108. doi: 10.1152/ajpcell.00494.2003. [DOI] [PubMed] [Google Scholar]
- 45.Perregaux D, Gabel CA. Interleukin-1β maturation and release in response to ATP and nigericin. Evidence that potassium depletion mediated by these agents is a necessary and common feature of their activity. J Biol Chem. 1994;269:15195–15203. [PubMed] [Google Scholar]
- 46.Ferrari D, Chiozzi P, Falzoni S, Dal Susino M, Melchiorri L, et al. Extracellular ATP triggers IL-1β release by activating the purinergic P2Z receptor of human macrophages. J Immunol. 1997;159:1451–1458. [PubMed] [Google Scholar]
- 47.Sanz JM, Di Virgilio F. Kinetics and mechanism of ATP-dependent IL-1β release from microglial cells. J Immunol. 2000;164:4893–4898. doi: 10.4049/jimmunol.164.9.4893. [DOI] [PubMed] [Google Scholar]
- 48.Gudipaty L, Munetz J, Verhoef PA, Dubyak GR. Essential role for Ca2+ in regulation of IL-1β secretion by P2X7 nucleotide receptor in monocytes, macrophages, and HEK-293 cells. Am J Physiol Cell Physiol. 2003;285:C286–C299. doi: 10.1152/ajpcell.00070.2003. [DOI] [PubMed] [Google Scholar]
- 49.Courageot MP, Lepine S, Hours M, Giraud F, Sulpice JC. Involvement of sodium in early phosphatidylserine exposure and phospholipid scrambling induced by P2X7 purinoceptor activation in thymocytes. J Biol Chem. 2004;279:21815–21823. doi: 10.1074/jbc.M401426200. [DOI] [PubMed] [Google Scholar]
- 50.Michel AD, Chessell IP, Humphrey PP. Ionic effects on human recombinant P2X7 receptor function. Naunyn Schmiedebergs Arch Pharmacol. 1999;359:102–109. doi: 10.1007/PL00005328. [DOI] [PubMed] [Google Scholar]
- 51.Wiley JS, Chen R, Wiley MJ, Jamieson GP. The ATP4− receptor-operated ion channel of human lymphocytes: inhibition of ion fluxes by amiloride analogs and by extracellular sodium ions. Arch Biochem Biophys. 1992;292:411–418. doi: 10.1016/0003-9861(92)90010-T. [DOI] [PubMed] [Google Scholar]