ABSTRACT
The establishment and maintenance of cell polarity is an essential property governing organismal homeostasis, and loss of polarity is a common feature of cancer cells. The ability of epithelial cells to establish apical-basal polarity depends on intracellular signals generated from polarity proteins, such as the Par-1 family of proteins, as well as extracellular signals generated through cell contacts with the extracellular matrix (ECM). The Par-1 family has a well-established role in regulating cell–cell contacts in the form of tight junctions by phosphorylating Par-3. In addition, Par-1 has been shown to impact on cell–ECM interactions by regulating laminin receptor localization and laminin deposition on the basal surface of epithelial cells. Laminins are major structural and signaling components of basement membrane (BM), a sheet of specialized ECM underlying epithelia. In this study, we identify RNF41, an E3 ubiquitin ligase, as a novel Par-1b (also known as MARK2) effector in the cell–ECM pathway. Par-1b binds to and phosphorylates RNF41 on serine 254. Phosphorylation of RNF41 by Par-1b is required for epithelial cells to localize laminin-111 receptors to their basolateral surfaces and to properly anchor to laminin-111. In addition, phosphorylation of RNF41 is required for epithelial cells to establish apical-basal polarity. Our data suggests that phosphorylation of RNF41 by Par-1b regulates basolateral membrane targeting of laminin-111 receptors, thereby facilitating cell anchorage to laminin-111 and ultimately forming the cell–ECM contacts required for epithelial cells to establish apical-basal cell polarity.
KEY WORDS: Polarity, Laminin, Extracellular matrix, Par-1, Par-1b, MARK2, RNF41, NRDP1, Ubiquitin ligase
INTRODUCTION
Epithelial cell polarity depends on extrinsic cues from the extracellular matrix (ECM) as well as intrinsic cues from polarity proteins including Par-1 (Rodriguez-Boulan and Nelson, 1989). Par-1 is a serine/threonine protein kinase and is encoded by one of six partitioning-defective (Par) genes that were first identified in a genetic screen conducted in Caenorhabditis elegans (C. elegans) as regulators of asymmetric cell division during early embryonic development (Kemphues et al., 1988): par-1, par-2, par-3 (ASIP in mammals), par-4 (LKB1 or STK11 in mammals), par-5 (14-3-3 in mammals) and par-6. Orthologs of each of these genes have been identified except for par-2. Par-2 contains a RING finger and it colocalizes with Par-1 to the posterior pole of the C. elegans zygote and cooperates with other Par gene products to establish polarity during early embryonic development.
In epithelial cells, the establishment of apical-basal polarity is dependent on the antagonistic relationship between Par-1 and the Par-3–Par-6–atypical protein kinase C (aPKC) complex. The Par-3–Par-6–aPKC complex localizes to tight junctions and the integrity of this complex is required for maintaining polarity (Etienne-Manneville and Hall, 2003 ; Joberty et al., 2000). Tight junctions help to maintain cell polarity by preventing the lateral diffusion of integral membrane proteins between the apical and basolateral surfaces. Par-1 is excluded from tight junctions and instead localizes to basolateral membranes (Böhm et al., 1997). Par-1 phosphorylates Par-3 to regulate its association with tight junctions (Benton and St Johnston, 2003; Hurd et al., 2003), whereas aPKC in complex with Par-3–Par-6 phosphorylates Par-1 on T595 to keep it away from tight junctions (Chen et al., 2006; Hurov et al., 2004; Suzuki et al., 2004). In mammals there are four members of the Par-1 family: Par-1a (C-TAK1 or MARK3), Par-1b (EMK or MARK2), Par-1c (MARK1) and Par-1d (MARKL1 or MARK4) (Drewes et al., 1997; Espinosa and Navarro, 1998; Hurov et al., 2001; Inglis et al., 1993; Kato et al., 2001; Müller et al., 2001; Peng et al., 1998).
In addition to cell–cell contacts, in the form of tight junctions, cell–ECM interactions provide additional spatial cues required for epithelial cell polarity (Ekblom, 1989). The three-dimensional (3D) microenvironment is crucial for the ability of breast epithelial cells to form polarized differentiated acinar structures in culture (Emerman and Pitelka, 1977). In addition, the ability of the ECM to direct formation of apical domains requires laminin assembly on the basal cell surface (O'Brien et al., 2002). Laminins are major components of the basement membrane (BM), a sheet of specialized ECM underlying epithelia, and play critical roles during morphogenesis and tissue organization in vivo. Epithelial-cell–laminin interactions influence cellular responses, such as adhesion, polarity and survival (Li et al., 2003) and disregulated cell–laminin interactions are implicated in cancer progression (Patarroyo et al., 2002). Two examples of BM laminins are laminin-111 and laminin-332, and deposition and/or formation of matrices composed of these laminins is regulated by cell-surface receptors, including integrins and the dystroglycan (DG) complex. Previous studies have implicated Par-1b in ECM-directed regulation of epithelial cell polarity and have shown that Par-1b is involved in laminin deposition through the DG complex and its actin-regulatory substrate IRSp53 (Cohen et al., 2004; Cohen et al., 2011; Masuda-Hirata et al., 2009; Yamashita et al., 2010).
A previous proteomic screen employing tandem affinity purification (TAP) followed by tandem mass spectrometry identified a complex of proteins that associate with one of the human Par-1 orthologs (Par-1d/MARK4) (Brajenovic et al., 2004). This screen identified both known (aPKC and 14-3-3) and novel Par-1d-interacting proteins (Brajenovic et al., 2004). RNF41, a RING finger E3 ubiquitin ligase was identified as one component of the Par-1d complex. RNF41, also known as NRDP1 (neuregulin receptor degradation protein-1), contains an N-terminal RING domain, two zinc-finger domains (B-Boxes), a coiled-coil domain, and a C-terminal domain that binds to various substrates (Abdullah et al., 2001; Bouyain and Leahy, 2007; Diamonti et al., 2002; Qiu and Goldberg, 2002). RNF41 has been shown to regulate steady-state levels of ErbB3 and ErbB4, and RNF41 associates with ErbB3 independently of receptor stimulation (Diamonti et al., 2002; Qiu and Goldberg, 2002) to promote ligand-independent ErbB3 ubiquitylation and degradation (Qiu and Goldberg, 2002). RNF41 also regulates the interleukin-3 and erythropoietin cytokine receptors (Jing et al., 2008) by controlling receptor degradation and shedding in a ligand-independent manner (Wauman et al., 2011). RNF41 regulates toll-like receptor (TLR)-mediated responses through ubiquitylation of MyD88 and TBK1 (Singh et al., 2009) and has been implicated in the ubiquitin-mediated proteolysis of two ubiquitin ligases, BRUCE and PARKIN (Qiu et al., 2004; Zhong et al., 2005). Although several targets of RNF41 have been identified, very little is known regarding how RNF41 is regulated except that it is subject to auto-ubiquitylation and proteolysis and that this is counterbalanced by the activity of USP8, a deubiquitylating enzyme (Cao et al., 2007; Wu et al., 2004).
In this study, we identified RNF41 as a novel Par-1b substrate and identified a role for RNF41 in regulating epithelial cell polarity. We show that phosphorylation of RNF41 on S254 by Par-1b is increased as breast epithelial cells polarize in 3D cultures and that the ability of breast epithelial cells to deposit laminin-111, to localize laminin-111 receptors on the BM and to form polarized spheroids in vitro requires phosphorylation of RNF41 by Par-1b.
RESULTS
RNF41 co-precipitates with Par-1b and is required for MCF10A cells to polarize in 3D cultures
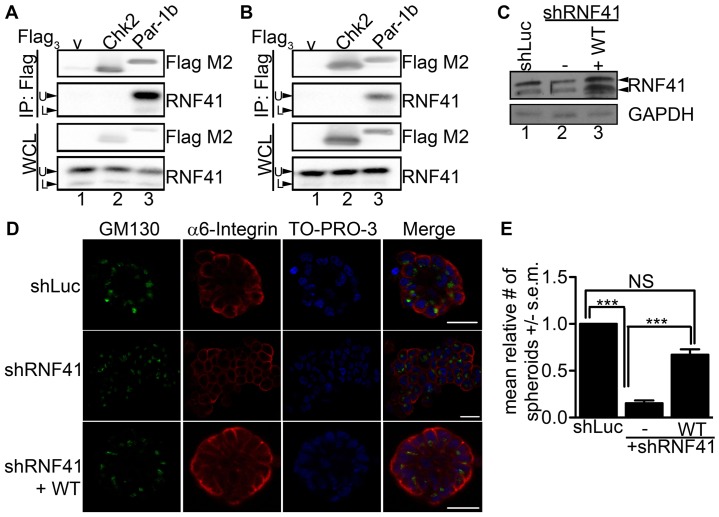
A previous proteomic screen employing tandem affinity purification (TAP) followed by tandem mass spectrometry identified RNF41 in a complex of proteins that associate with Par-1d (MARK4) (Brajenovic et al., 2004). To determine whether RNF41 interacted with a second human Par-1 ortholog (Par-1b, MARK2), HeLa and HEK 293T cells were transfected with control plasmid (V), plasmid encoding human Flag-tagged CHK2 as a negative control, or plasmid encoding Flag-tagged Par-1b. Lysates were prepared and incubated with Flag–agarose followed by western blotting. As seen in Fig. 1A,B, endogenous RNF41 was detected in Par-1b (lane 3) but not in control (lanes 1, 2) precipitates. Two electrophoretic forms of RNF41 were detected in HeLa (Fig. 1A) and HEK293T (Fig. 1B) cells, a predominant 41 kDa form (U) and a minor 34 kDa form (L), and this is likely due to differential splicing.
Fig. 1.
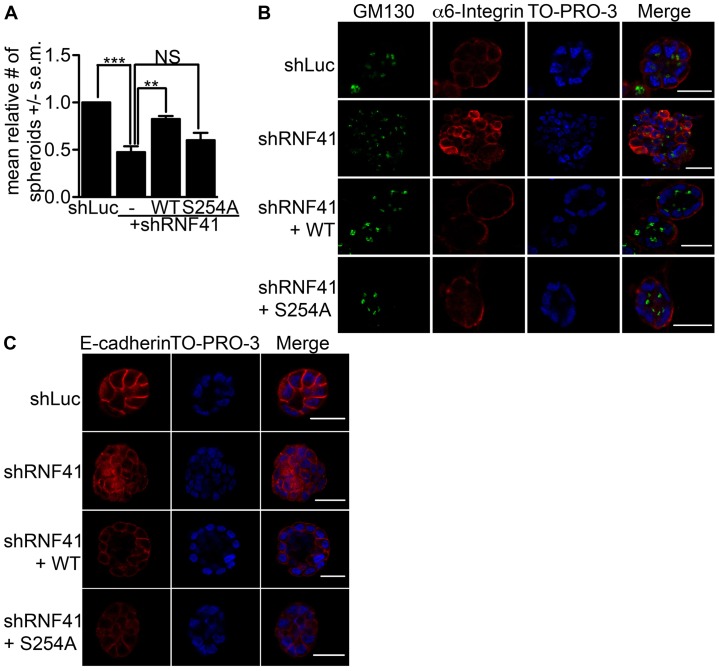
RNF41 binds to Par-1b and is necessary for epithelial cell polarity. HeLa (A) and HEK293T (B) cells were transfected with control plasmid (lane 1) or with plasmids encoding Flag3–Chk2 (lane 2) or Flag3–Par-1b (lane 3). Cells were harvested 24 hours later and lysates were resolved directly by SDS-PAGE or were incubated with anti-Flag agarose prior to SDS-PAGE. Western blotting was performed with the indicated antibodies. Arrows indicate the two electrophoretic forms of RNF41 that are designated upper (U) and lower (L). A representative image from n = 3 is shown. (C) MCF10A cells expressing shRNA specific for luciferase (shLuc), shRNA specific for sequences within the 3′UTR of RNF41 (shRNF41#1, labeled –) or shRNF41#1 plus C-terminally tagged wild-type (WT) RNF41 (labeled +WT) were cultured in a 3D matrix of collagen I and harvested after 20 days. Lysates were resolved by SDS-PAGE and analyzed by western blotting with the indicated antibodies. A representative image from n = 3 is shown. (D) Representative immunofluorescent confocal images of 3D colonies from 20-day cultures stained with antibody against α6 integrin to detect basolateral membranes, and GM130 to detect Golgi at the apical surface. 3D colonies were visualized with secondary antibodies conjugated to Alexa Fluor 594 (red) and 488 (green), respectively, and To-Pro-3 to detect nuclei (blue). A serial immunofluorescent confocal cross section of a representative colony is shown. Scale bars: 20 µm. The data presented is representative of the 500 colonies that we observed in each of three independent experiments (total, 1500 colonies). (E) The number of colonies that formed acini-like spheroids was determined. 500 colonies were counted in each of three independent experiments (total, 1500 colonies). The data are presented as the mean (±s.e.m.) relative number of acini-like spheroids. ***P<0.001 (one-way ANOVA with a Tukey's multiple comparison post test); NS, not significantly different.
Next, knockdown experiments were conducted in MCF10A cells to determine whether RNF41, like Par-1b, is required for epithelial cell polarity (Böhm et al., 1997; Cohen et al., 2007; Cox et al., 2001a; Doerflinger et al., 2003; Ducharme et al., 2006). MCF10A is a spontaneously immortalized but untransformed mammary epithelial cell line. When grown in 3D cultures, MCF10A cells form polarized acini-like spheroids (Debnath et al., 2003). We employed a lentiviral vector that concurrently knocks down and rescues expression of a target gene in the same cell (Feng et al., 2010). Lentiviral vectors encoding shRNA specific for luciferase (shLuc) as a control or shRNA specific for sequences within the 3′UTR of RNF41 (shRNF41) were generated. Two independent shRNAs were used throughout our studies (Fig. 1; supplementary material Fig. S1). In addition, a vector simultaneously expressing shRNF41 and shRNA-resistant RNF41 tagged with a C-terminal Flag epitope followed by six histidines (FlagHis6) was generated. In this way, we could determine whether phenotypes observed upon knockdown of RNF41 could be rescued by expression of wild-type (WT) RNF41. The maximum knockdown of RNF41 achievable in MCF10A cells was ∼50% and in the experiment shown in Fig. 1C, shRNA treatment resulted in a 54% knockdown (lane 2). The finding that only a 50% knockdown of RNF41 is achievable might indicate that RNF41 is essential for cell viability.
As seen in Fig. 1D, MCF10A cells expressing control shRNA (shLuc) formed normal acinar architecture along with a hollow lumen over the 20-day culture period in collagen I matrix. Serial confocal cross sections of an acinus were stained with To-Pro-3 to detect nuclei, as well as antibodies against α6 integrin, to detect basolateral membranes, and GM130, to detect Golgi directed towards the apical surface. Staining confirmed that MCF10A cells expressing control shRNA polarized under these 3D culture conditions and that the establishment of cell polarity was accompanied by loss of proliferation as judged by negative Ki67 staining (supplementary material Fig. S2). Despite achieving only a 54% knockdown of RNF41, dramatic effects were observed when MCF10A cells were cultured in collagen I matrix. As seen in Fig. 1D, cells expressing RNF41 shRNA had a abnormal architecture after growing for 20 days under 3D culture conditions and these cells were not able to establish apical-basal polarity. In addition, RNF41-deficient cells continued to proliferate under 3D culture conditions resulting in ∼4-fold more RNF41-deficient cells compared with control cells by 20 days (supplementary material Fig. S2). Importantly, the polarity defect was partially rescued when WT RNF41 was expressed in MCF10A cells knocked down for RNF41 (Fig. 1D,E) suggesting a role for RNF41 in establishing apical-basal cell polarity in mammary epithelial cells. Similar observations were made in MCF10A cells engineered to express a second hairpin specific to the 3′UTR of RNF41 (supplementary material Fig. S1).
Par-1b phosphorylates RNF41 on S254 in vitro and RNF41 is phosphorylated on S254 in vivo
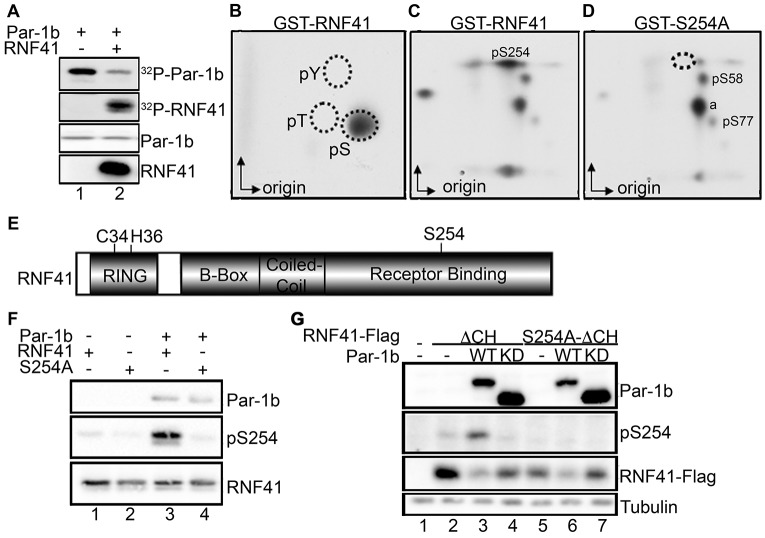
To determine whether RNF41 is a substrate of Par-1b, kinase assays were performed in vitro in conjunction with two-dimensional tryptic phosphopeptide mapping. RNF41 was phosphorylated by Par-1b in vitro (Fig. 2A), and this occurred on one or more serine residues (Fig. 2B). Mapping studies revealed several phosphopeptides, and substitution of alanine for serine at position 254 resulted in the disappearance of a single phosphopeptide (Fig. 2C,D), suggesting that Par-1b phosphorylates RNF41 on S254 in vitro. To confirm that Par-1b phosphorylates RNF41 on S254 in vitro and that RNF41 is phosphorylated on S254 in vivo, a phosphospecific antibody recognizing RNF41 only when it is phosphorylated on S254 was generated. As seen in Fig. 2F, recognition of RNF41 by the pS254 antibody was evident when RNF41-WT (lane 3) but not RNF41-S254A (lane 4) was phosphorylated by Par-1b in vitro. RNF41 consists of an N-terminal RING domain followed by a B box, a coiled-coil domain, and a C-terminal region that mediates interactions between RNF41 and ErbB3 (the receptor-binding domain) (Fig. 2E). Mutation of two residues within the RING domain [cysteine (C) 34 and histidine (H) 36] ablates the E3 ubiquitin ligase activity of RNF41 (Qiu and Goldberg, 2002). Owing to the ability of RNF41 to auto-ubiquitylate and promote its own proteosomal degradation, we generated a ligase-deficient mutant of RNF41 encoding serine for C34 and glutamine for H36 (denoted ΔCH) in order to produce adequate levels of RNF41 in vivo. As seen in Fig. 2G, the pS254 antibody recognized ectopic RNF41 (lane 2) and phosphorylation of S254 increased when RNF41 was co-produced with WT Par-1b in vivo (lane 3). By contrast, phosphorylation of RNF41 on S254 was reduced in cells expressing kinase-inactive Par-1b (lane 4). Mutation of S254 eliminated recognition of RNF41 by the phosphospecific antibody under similar conditions (lanes 5–7). These results demonstrate that the antibody is specific for RNF41 when it is phosphorylated on S254 and that ectopically expressed RNF41 is phosphorylated on S254 in vivo. Furthermore, the ability of kinase-active Par-1b to enhance S254 phosphorylation and kinase-inactive Par-1b to decrease S254 phosphorylation suggests that phosphorylation of RNF41 on S254 is regulated in vivo by Par-1b. Par-1b also phosphorylated RNF41 on serines 58 and 77 in vitro. We were unable to generate antibodies specific for these sites and therefore did not pursue their functional relevance. The identity of phosphopeptide a, shown in Fig. 2D, has not been determined, but preliminary evidence suggests it is derived from the GST tag.
Fig. 2.
Par-1b phosphorylates RNF41. (A–D) Bacterially produced GST–Par-1b was incubated in the absence or presence of bacterially produced GST–RNF41 and kinase assays were performed in vitro. Radiolabeled reaction products were resolved by SDS-PAGE (A) and phosphotryptic peptides were subjected to phosphoamino acid analysis (PAA analysis) (B). (C,D) Kinase assays were also performed in the presence of GST–Par-1b and GST–RNF41 (WT or S254A) followed by two-dimensional phosphopeptide mapping. a, unidentified phosphopeptide. Arrows indicate the direction of electrophoresis (x-axis) and chromatography (y-axis). A representative image from n = 4 is shown. (E) Schematic representation of RNF41 protein. Protein domains are indicated, C34 and H36 are required for the E3 ligase activity of RNF41 and S254 is phosphorylated by Par-1. (F) Kinase assays were performed in vitro with GST–RNF41 WT (lanes 1 and 3) or GST–S254A (lanes 2 and 4) in the presence (lanes 3 and 4) or absence (lanes 1 and 2) of GST–Par-1b. Proteins were resolved by SDS-PAGE and detected by western blotting using the indicated antibodies. A representative image from n = 3 is shown. (G) Lysates prepared from mock-transfected HEK293T cells (lane 1) or from HEK293T cells expressing RNF41–FlagΔCH (lanes 2–4) or RNF41–FlagS254A-ΔCH (lanes 5–7), with Flag3–Par-1b WT (lanes 3 and 6) or Flag3–Par-1b KD (lanes 4 and 7) were resolved by SDS-PAGE and immunoblotted with the indicated antibodies. A representative image from n = 3 is shown. KD, kinase dead.
Phosphorylation of RNF41 on serine 254 is required for apical-basal polarity
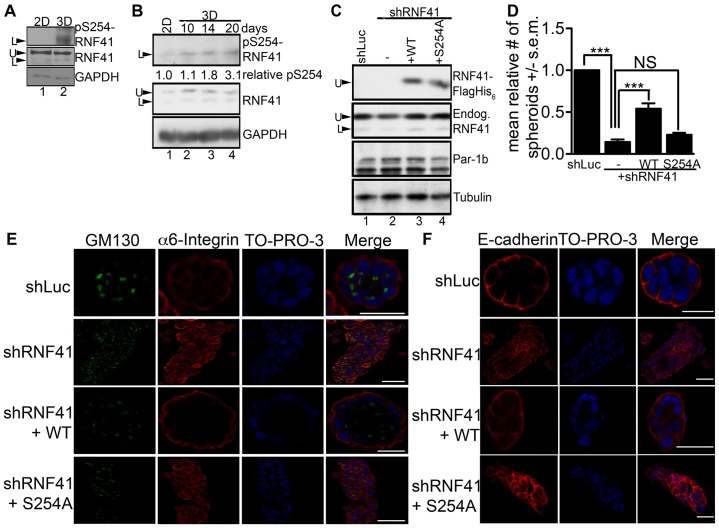
Phosphorylation of endogenous RNF41 on S254 was barely detectable when cells were grown in two-dimensional (2D) cultures but increased 3- to 5-fold after 20 days of growing MCF10A cells in collagen I matrix (Fig. 3A,B). Increased phosphorylation of S254 was seen as early as after 10 days of culturing under 3D conditions (Fig. 3B). Two electrophoretic forms of RNF41 were detected in MCF10A cells and the faster migrating (L) form was phosphorylated on S254 under 3D culture conditions. Knockdown–rescue experiments were conducted to determine whether phosphorylation of RNF41 on S254 was required for MCF10A cells to polarize into acini-like spheroids in 3D cultures. In the experiment shown in Fig. 3C, shRNA treatment resulted in a 48% knockdown of RNF41 (lane 2) and expression of WT RNF41 (lane 3) or the S254A mutant (lane 4) restored RNF41 levels in knockdown cells to approximately those detected for endogenous RNF41 in control cells (lane 1). Cells expressing RNF41 shRNA formed abnormal architecture (Fig. 3D), were unable to establish apical-basal polarity (Fig. 3E) and their adherens junctions lacked organization (Fig. 3F). Partial rescue of colony architecture, adherens junction formation and polarity was observed when WT RNF41, but not the S254A mutant, was expressed in MCF10A cells knocked down for RNF41 (Fig. 3D–F; Table 1). These results indicate that phosphorylation of RNF41 by Par-1b is required for the establishment of apical-basal polarity and suggests that RNF41 acts downstream of Par-1b in the polarity pathway.
Fig. 3.
Phosphorylation of RNF41 by Par-1 is required for epithelial cells to establish polarity. (A) MCF10A cells were cultured on plastic (2D) or in collagen I matrix (3D) and spheroids were harvested after 20 days. Cell lysates were resolved by SDS-PAGE and immunoblotted with the indicated antibodies, n = 3. The slower (U) and faster (L) electrophoretic forms of RNF41 are indicated. (B) MCF10A cells were cultured on plastic (2D) or in collagen I matrix (3D) and spheroids were harvested after 10, 14 and 20 days. Cell lysates were resolved by SDS-PAGE and immunoblotted with the indicated antibodies, n = 3. Relative levels of pS254 RNF41 were determined from western blotting by using the ImageJ program and are indicated below the western blot. (C) MCF10A stable cells expressing control shRNA (shLuc), or shRNA specific for RNF41 (shRNF41#1) alone (labeled −) or in the presence of tagged (FlagHis6) WT RNF41 (+ WT) or the tagged S254A mutant (+ S254A) were cultured in collagen I matrix and harvested after 20 days. Lysates were resolved directly by SDS-PAGE or were first incubated with anti-Flag agarose to isolate tagged RNF41 (top panel, indicated by U), followed by western blotting. (D) Quantification of the ratio of round acini-like spheroids generated for the cells described in C and cultured in a 3D collagen I matrix for 20 days. The number of colonies that formed acini-like spheroids was determined. 500 colonies were counted in each of three independent experiments (total, 1500 colonies). The data are presented as the mean (±s.e.m.) relative number of acini-like spheroids. ***P<0.001 (one-way ANOVA with a Tukey's multiple comparison post test); NS, not significantly different. (E,F) Representative immunofluorescent confocal images of 3D colonies from 20 day cultures stained with antibodies against α6 integrin to detect basolateral membranes, GM130 to detect Golgi at the apical surface and E-cadherin to detect adherens junctions. 3D colonies were visualized with secondary antibodies conjugated to Alexa Fluor 594 [red, α6 integrin (E) and E-cadherin (F)] and 488 [green, GM130 (E)], and To-Pro-3 to detect nuclei (blue). A serial immunofluorescent confocal cross section of a representative colony is shown. Scale bars: 20 µm. The data presented is representative of the 500 colonies that we observed in each of three independent experiments (total, 1500 colonies).
Table 1. Summary of rescue experiments.
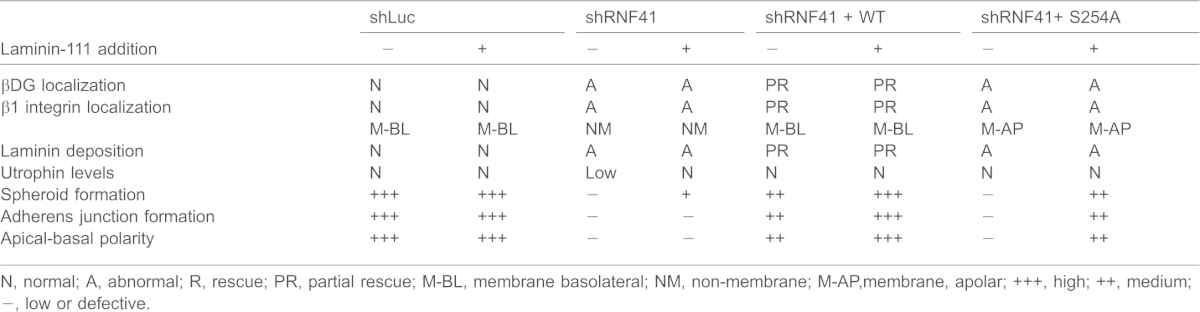
N, normal; A, abnormal; R, rescue; PR, partial rescue; M-BL, membrane basolateral; NM, non-membrane; M-AP,membrane, apolar; +++, high; ++, medium; −, low or defective.
Phosphorylation of RNF41 on S254 is required for proper laminin-111 receptor localization and laminin-111 deposition
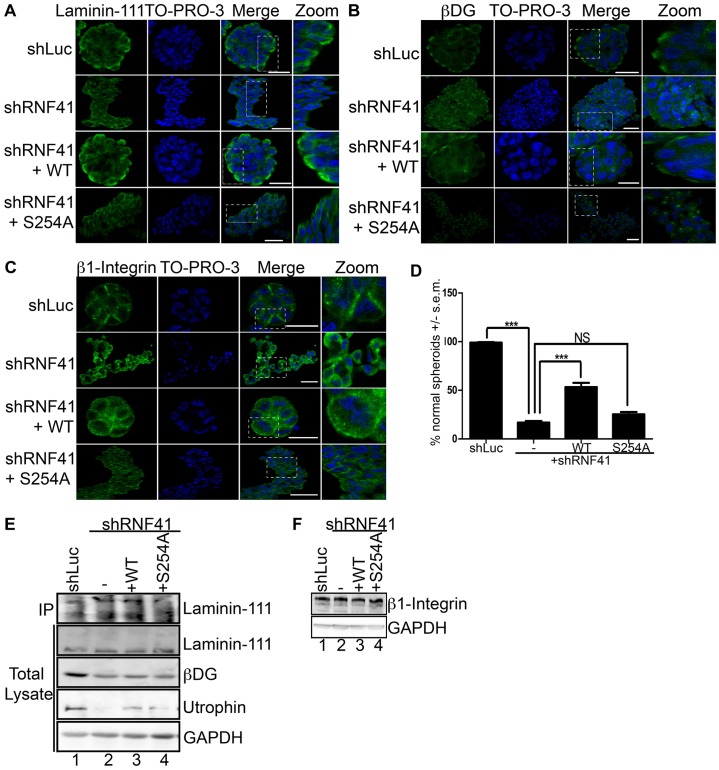
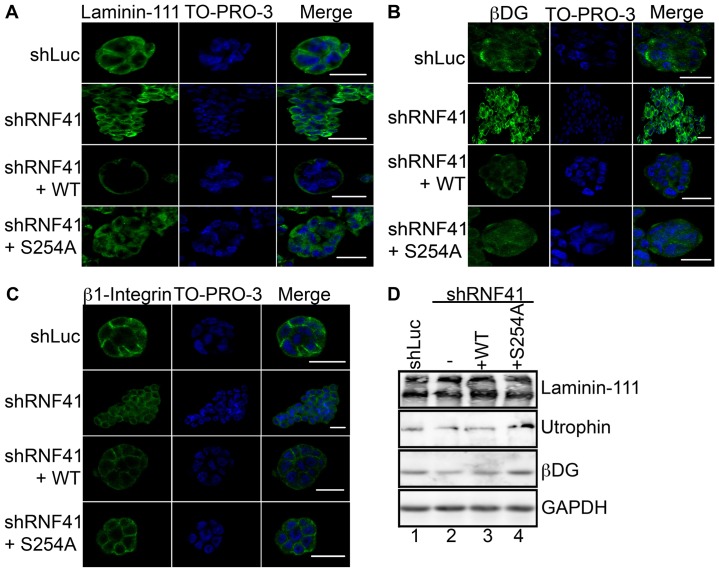
Par-1b kinase activity has been shown to be required in epithelial cells for proper basolateral membrane localization of dystroglycan (DG), a laminin-111 receptor, and for laminin-111 deposition on the basal surface of epithelial cells (Masuda-Hirata et al., 2009). Therefore, we monitored the effects of RNF41 knockdown on these processes using the MCF10A 3D culture system. MCF10A cells expressing control shRNA were capable of depositing laminin-111 when grown in collagen I matrix (Fig. 4A; Table 1) and formed normal acini-like spheroids (Fig. 1D). Although RNF41-deficient cells were able to secrete laminin-111 (Fig. 4E), its deposition was defective in these cells (Fig. 4A). Expression of WT RNF41 but not the S254A mutant partially rescued laminin-111 deposition, suggesting that phosphorylated RNF41 is required for laminin-111 deposition (Fig. 4A; Table 1).
Fig. 4.
Phosphorylation of RNF41 by Par-1b is required for laminin-111 deposition and polarized localization of laminin-111 receptors. (A–C) MCF10A cells (control, knockdown and rescue cells, as described in Fig. 3C) were cultured in a collagen I matrix and on day 20, colonies were stained with antibodies against laminin-111 (A), βDG (B), or β1 integrin (C). Colonies were visualized with secondary antibody conjugated to Alexa Fluor 488 (green) and To-Pro-3 to detect nuclei (blue). A serial immunofluorescent confocal cross section of a representative colony is shown. Scale bars: 20 µm. The data presented is representative of the 500 colonies that were observed in each of three independent experiments, for a total of 1500 colonies. (D) Percentage of normal spheroids. The percentage of MCF10A cells in panels A–C that formed normal acini-like spheroids was determined. 500 colonies were counted in each of three independent experiments for a total of 1500 colonies. The data are presented as the mean (±s.e.m.) percentage of normal spheroids. ***P<0.001 (one-way ANOVA with a Tukey's multiple comparison post test); NS, not significantly different. (E,F) Lysates from 3D colonies were resolved by SDS-PAGE and analyzed by western blotting with antibodies specific for laminin-111, βDG, utrophin, β1 integrin or GAPDH. In addition, laminin-111 was immunoprecipitated from the culture medium and immunoprecipitates were analyzed by western blotting (IP, panel E). A representative image from n = 3 is shown.
Receptors for laminin-111 include the α3β1 integrin and the DG complex, which consists of utrophin, αDG and βDG (Ekblom et al., 1998; Li et al., 2003; Ojakian and Schwimmer, 1994; Schoenenberger et al., 1994). These receptors reside on the basolateral surface of epithelial cells where they anchor laminin-111. Loss of the DG complex in mammary epithelial cells impairs laminin anchoring and the establishment of apical-basal polarity (Weir et al., 2006). MCF10A cells knocked down for RNF41 mislocalized laminin-111 receptors (Fig. 4B–D; Table 1). Partial rescue of basal-lateral localization of receptors was observed when WT RNF41 but not the S254A mutant was expressed in these cells (Fig. 4B,C). Membrane localization of β1 integrin was partially restored when the S254A mutant was expressed in RNF41 knockdown cells (Fig. 4C and Table 1). By contrast, the βDG subunit localized to discrete intracellular foci in S254A-expressing cells (Fig. 4B). Mislocalization of βDG, and β1 integrin, as well as failure to anchor laminin was observed after 14 days of culture under 3D conditions (supplementary material Fig. S3), suggesting that receptor mislocalization and subsequent failure to anchor laminin-111 might be the cause of abnormal spheroid formation. Levels of utrophin (Fig. 4E) but not β1 integrin (Fig. 4F) were severely diminished in MCF10A cells knocked down for RNF41. Utrophin levels were partially restored when either WT RNF41 or the S254A mutant were expressed in knockdown cells (Fig. 4E). However, even though utrophin levels were partially restored, rescue of laminin deposition, proper laminin-111 receptor localization and apical-basal cell polarity were not observed in these cells (Table 1). Additionally, although a decrease in βDG receptor levels was observed in cells knocked down for RNF41, βDG receptor levels were not rescued when RNF41 expression was restored (Fig. 4E). Thus, decreased βDG receptor levels might be an indirect effect of RNF41 loss. Taken together, these results suggest that a threshold of RNF41 protein is important in regulating utrophin levels, whereas phosphorylation of RNF41 on S254 is required for regulating proper laminin-111 receptor localization, laminin deposition and apical-basal cell polarity. The inability of RNF41-deficient and S254A-expressing cells to properly localize laminin-111 receptors might explain why laminin-111 deposition is defective and, therefore, why MCF10A cells fail to establish apical-basal polarity.
Laminin-111 rescues apical-basal polarity in S254A-expressing MCF10A cells
Next, we asked whether any of the defects observed in RNF41-defective MCF10A cells could be rescued by the addition of exogenous laminin-111 to the collagen I matrix (Figs 5, 6). Exogenous laminin-111 caused an increase in the number of spherical, acinar-like structures in MCF10A cells knocked down for RNF41 (47.5% as compared with 14.4% without laminin-111), and this was also true for cells expressing the S254A mutant (60% as compared with 22.9% without laminin-111) (Fig. 3D and Fig. 5A). Experiments shown in Fig. 3C–F and Fig. 5 were performed without and with the addition of exogenous laminin-111 in parallel. Although an increase in the number of spherical, acinar-like structures was observed in RNF41-deficient cells when exogenous laminin-111 was provided (Fig. 5A), neither polarity (Fig. 5B) nor adherens junction formation (Fig. 5C) were rescued in these cells. In addition, exogenous laminin did not rescue laminin-111 deposition (Fig. 6A) or laminin-111 receptor localization (Fig. 6B,C) in cells knocked down for RNF41. However, utrophin levels were restored when MCF10A cells were cultured in collagen I matrix supplemented with laminin-111 (Fig. 6D). Interestingly, exogenous laminin enhanced the incomplete rescue of all phenotypes in knockdown cells expressing WT RNF41, but only enhanced the rescue of spheroid formation, adherens junction formation and apical-basal polarity in S254A-expressing cells (Figs 5, 6; Table 1). Proper localization of βDG and laminin-111 deposition were not observed in S254A-expressing cells and although β1 integrin localized to membranes in mutant-expressing cells, it was not restricted to basolateral membranes.
Fig. 5.
Ability of exogenous laminin-111 to rescue adherens junction formation and polarity in RNF41-deficient MCF10A cells. (A) Control, knockdown and rescued MCF10A cells, as described in Fig. 3C, were cultured in collagen I matrix supplemented with exogenous laminin-111 for 20 days. The number of colonies that formed round acini-like spheroids was determined. 500 colonies were counted in each of three independent experiments (total, 1500 colonies). The data are presented as the mean (±s.e.m.) relative number of acini-like spheroids. **P<0.01; ***P<0.001 (one-way ANOVA with a Tukey's multiple comparison post test); NS, not significantly different. (B,C) Day 20 3D colonies were stained with antibodies against α6 integrin to detect basolateral membranes, GM130 to detect Golgi at the apical surface and E-cadherin to detect adherens junctions. 3D colonies were visualized with secondary antibodies conjugated to Alexa Fluor 594 [red, α6 integrin (B) and E-cadherin (C)] and 488 [green, GM130 (B)], and To-Pro-3 to detect nuclei (blue). A serial immunofluorescent confocal cross section of a representative colony is shown. Scale bars: 20 µm. The data presented is representative of the 500 colonies that were observed in three independent experiments (total, 1500 colonies).
Fig. 6.
Ability of exogenous laminin-111 to rescue laminin-111 deposition and polarized localization of laminin-111 receptors in RNF41-deficient MCF10A cells. (A–C) Control, knockdown and rescued MCF10A colonies, as shown in Fig. 5, were stained with antibodies against laminin-111 (A), βDG (B) or β1 integrin (C). 3D colonies were visualized with secondary antibody conjugated to Alexa Fluor 488 (green) and To-Pro-3 to detect nuclei (blue). A serial immunofluorescent confocal cross section of a representative colony is shown. Scale bars: 20 µm. The data presented is representative of the 500 colonies that were observed in each of three independent experiments (total, 1500 colonies). (D) Cells were also harvested for western blotting with antibodies specific for the indicated proteins. A representative image from n = 3 is shown.
DISCUSSION
In this study, RNF41 was identified as a novel substrate of the Par-1b polarity kinase and we discovered a role for RNF41 in regulating epithelial cell polarity. The Par-1 protein kinase regulates cell polarity in worms, flies, frogs and mammals (Böhm et al., 1997; Cohen et al., 2004; Cohen and Müsch, 2003; Cox et al., 2001b; Müller et al., 2001; Ossipova et al., 2005; Shulman et al., 2000; Sun et al., 2001; Tomancak et al., 2000). In order to identify Par-1-binding proteins, mammalian Par-1 orthologs had previously been used as bait in a TAP screen (Brajenovic et al., 2004). The RING finger containing E3 ubiquitin ligase RNF41/NRDP1 was found in Par-1d-containing complexes, and here we demonstrate that RNF41 also binds to Par-1b. Mammalian orthologs have been identified for five out of six of the C. elegans Par genes with the exception of par-2. par-2 encodes a RING-finger-containing protein that co-localizes with Par-1 in the C. elegans zygote. Par-2 and RNF41 share similarity in both their RING finger domains (53% sequence similarity) and C-terminal domains (63% similarity in N-terminal region) leaving open the interesting possibility that RNF41 is a Par-2 ortholog.
In the early C. elegans embryo, par-1 is among several partitioning-defective genes whose mutation disrupts asymmetric cell division, blastomere cell fate, localization of P granules and mitotic spindle orientation (Kemphues et al., 1988; Kirby et al., 1990). Some of the mutations are within the kinase domain of Par-1, indicating kinase activity is necessary for its polarity function (Guo and Kemphues, 1995). Of the reported Par-1 substrates, several are known to play a role in regulating polarity, including Par-3, utrophin and IRSp53. Par-3, in complex with Par-6 and aPKC, localizes to tight junctions, and the integrity of this complex is required for maintaining polarity (Etienne-Manneville and Hall, 2003; Joberty et al., 2000; Tabuse et al., 1998). Par-1 phosphorylates Par-3 to exclude it from tight junctions and this in turn disrupts epithelial cell polarity (Benton and St Johnston, 2003; Hurd et al., 2003). In addition, Par-1 phosphorylates utrophin to promote its interactions with DG (Yamashita et al., 2010) and phosphorylates IRSp53 to block interactions between IRS53 and its effector proteins (Cohen et al., 2011). Here we identified a novel substrate of Par-1 (RNF41) that also plays a role in regulating cell polarity. Par-1 phosphorylates RNF41 on S254, a site that is conserved in mammals and frogs. However, S254 is not conserved in flies or the C. elegans Par-2 protein, suggesting that Par-1-mediated phosphorylation of RNF41 is not a conserved mode of polarity signaling. MCF10A cells knocked down for RNF41 failed to polarize when grown in collagen I matrix. Interestingly, 50% knockdown of RNF41 was sufficient to prevent MCF10A cells from forming polarized acini-like spheroids in 3D culture. The establishment of cell polarity was accompanied by a loss of cell proliferation in control cells. By contrast, MCF10A cells deficient in RNF41 failed to polarize and continued to proliferate. Polarity was rescued by expressing WT RNF41 in knockdown cells. Phosphorylation of RNF41 on S254 was observed to increase as MCF10A cells polarized in 3D cultures, and expression of the S254A mutant of RNF41 failed to rescue polarity defects of RNF41-deficient MCF10A cells. These results indicate that phosphorylation of RNF41 on S254 is necessary for establishing apical-basal polarity and place RNF41 downstream of Par-1 in this pathway (Fig. 7).
Fig. 7.
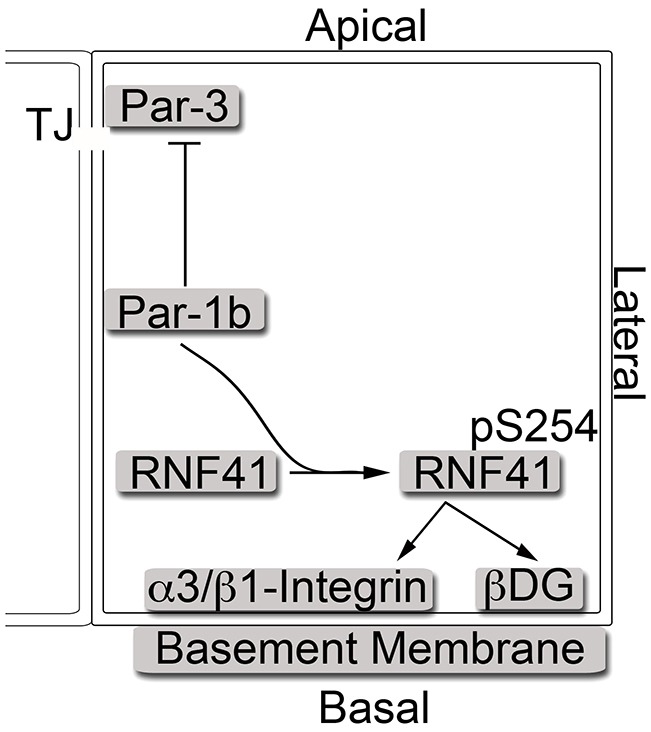
The Par-1–RNF41 pathway regulates epithelial cell polarity through regulation of laminin receptors. The Par-1 protein kinase regulates cell–cell communication through its regulation of tight junctions (TJ) and also regulates ECM–cell communications by regulating laminin-111 anchoring. A key substrate in the cell–cell communication pathway is Par-3. Here, we show that RNF41 is a key substrate in the cell–ECM pathway. The Par-1b protein kinase phosphorylates RNF41 on S254. Phosphorylated RNF41 regulates the polar localization of laminin-111 receptors, including α3β1 integrin and the DG complex. This facilitates laminin-111 anchoring to the basal cell surface. The establishment of epithelial-cell–laminin interactions facilitates cell–ECM communication, which is a requirement for epithelial cells to establish apical-basal cell polarity.
The BM provides spatial cues necessary for establishing epithelial cell polarity (Ekblom, 1989; Li et al., 2003). We demonstrate that MCF10A cells deficient in RNF41 mislocalize laminin-111 receptors (the DG complex and the α3β1 integrin) and fail to anchor laminin-111 on their basal surfaces. These phenotypes are similar to those reported in Par-1b-deficient MDCK cells (Masuda-Hirata et al., 2009). Specifically, Par-1b has been shown to interact with laminin-111 receptors in cultured MDCK cells, and Par-1b-deficient MDCK cells fail to localize the DG complex properly and are defective in anchoring laminins on their basal cell surface. It has been shown that the DG complex is important in the initial binding of laminin-111, where it serves to concentrate laminin-111 on the cell surface (Henry et al., 2001). Laminin-111 binding and activation of DG receptors, in turn, increases interactions between β1 integrin and laminin-111 (Driss et al., 2006) and β1 integrin is responsible for the polymerization of the concentrated laminin-111 on the cell surface (Henry et al., 2001). Thus, by regulating DG receptor localization, RNF41 might promote cell–ECM communication, a prerequisite for epithelial cell polarity.
The addition of exogenous laminin-111 to 3D cultures of RNF41-deficient MCF10A cells partially rescued spheroid formation (Fig. 5A) but not apical-basal polarity (Fig. 5B), adherens junction formation (Fig. 5C), laminin-111 deposition (Fig. 6A) or laminin-111 receptor localization (Fig. 6B,C). Interesting, exogenous laminin-111 potentiated the rescue of all phenotypes in RNF41-deficient cells that had been engineered to re-express WT RNF41. By contrast, exogenous laminin-111 potentiated the rescue of only a subset of phenotypes in RNF41-deficient cells that has been engineered to re-express the S254A mutant of RNF41. These included spheroid formation, apical-basal polarity and adherens junction formation. How does exogenous laminin-111 rescue cell polarity in S254A-expressing cells if laminin-111 deposition and laminin-111 receptor localization are defective? Interestingly, β1 integrin localized to membranes in S254A-expressing cells, although it was not restricted to basolateral membranes. Thus, in the presence of excess laminin-111, cells might bypass the requirement for the DG receptor to concentrate laminin-111 on their cell surface in order to promote interactions between laminin-111 and α3β1 integrin. In this case, signaling through α3β1 integrin receptors might be sufficient to rescue polarity (Fig. 7).
Epithelial cell polarity is controlled by three conserved polarity protein complexes termed the Par, Crumbs and Scibble complexes, and deregulation of members of all three complexes contribute to the onset of progression of human tumors (Ellenbroek et al., 2012). The Par complex consists of aPKC, Par-3 and Par-6, and members of this complex have been reported to have tumor suppressive potential (Par-3, Par-6) as well as tumor-promoting activities (aPKC, Par-3 and Par-6) (Ellenbroek et al., 2012). Deregulation of Par-1 has not been directly linked to oncogenesis; however, its downstream effector RNF41 has. RNF41 is an E3 ubiquitin ligase that regulates steady state levels of ErbB3 (Diamonti et al., 2002). ErbB3 is overexpressed in a subset of breast cancers and the ErbB2–ErbB3 heterodimer has been shown to function as an oncogenic unit to drive breast tumor cell proliferation (Holbro et al., 2003). There is a correlation between decreased levels of RNF41 and increased levels of ErbB3 in a transgenic mouse model of ErbB2-induced mammary tumorigenesis and in a panel of primary breast tumors (Yen et al., 2006). It has been assumed that elevated levels of active ErbB3 signaling is the major contributing factor driving tumorigenesis under these conditions. Our results uncover a more general role for RNF41 – that of regulating apical-basal cell polarity – and demonstrate that phosphorylation of RNF41 on S254 by Par-1 is required for the polarity function of RNF41. The establishment of cell polarity enables cells to sense and transmit spatiotemporal responses to cues that arise from neighboring cells and the surrounding microenvironment (St Johnston and Ahringer, 2010). When polarity is disrupted, cells might override signals that inhibit proliferation. Indeed, loss of RNF41 not only disrupted cell polarity but also allowed cells to continue to proliferate under 3D culture conditions. A similar correlation between loss of polarity and enhanced proliferative capacity was made in neuroepithelial cells deficient in DG function (Schröder et al., 2007). Thus, disruption of cell polarity associated with hyperproliferation might contribute to the tumorigenic phenotypes observed in tumors with RNF41 deficiency.
MATERIALS AND METHODS
Cell culture
HeLa cells were routinely maintained in DMEM (Gibco-BRL) supplemented with 10% bovine growth serum (HyClone), 100 units per ml of penicillin and streptomycin, and 1 mM L-glutamine. HEK293T cells were grown in the presence of 10% FBS (HyClone). MCF10A cells were maintained in DMEM/F12 (Invitrogen) supplemented with 5% horse serum (Invitrogen), 20 ng/ml epidermal growth factor (EGF) (PeproTech), 0.5 mg/ml hydrocortisone (Sigma) 100 ng/ml Cholera toxin (Sigma), 10 µg/ml insulin (Sigma), and 100 units per ml of penicillin and streptomycin.
Cloning and mutagenesis
GST–RNF41 was cloned by PCR from the pCDNA3.1-RNF41-Flag plasmid (Diamonti et al., 2002) and ligation into the pGex2T vector (GE Healthcare). The pFLRU6RNF41shRNA-cmv-MCS-FH plasmid was generated by PCR. First, PCR of the U6 promoter from the pFLRU6Luc-cmv-MCS-FH plasmid was performed followed by PCR of the RNF41shRNA sequence targeted against the 3′UTR of RNF41 [5′-GCCACATTGTTGGCAATTTAA-3′ (shRNA #1) or 5′-GGTAAGTAGTGAGGCCGTTAA-3′ (shRNA #2)]. The final PCR was performed with the U6 promoter and the RNF41 shRNA template generated in the previous step, in order to create a U6–RNF41shRNA fusion. The fusion PCR product was ligated into the XbaI and XhoI restriction sites of the pFLRcmv-MCS-FH plasmid. The pFLRU6RNF41shRNA-cmvRNF41-FlagHis6-FH (WT and S254A) plasmids were generated by PCR of RNF41 (WT and S254A) from the appropriate pCDNA3.1-RNF41-Flag plasmid and inserted into the pFLRU6RNF41shRNA-cmv-MCS-FH plasmid. The RNF41-Flag C34S/H36Q (ΔCH), RNF41-Flag C34S/H36Q/S254A, and GST-RNF41 S254A mutants were generated by using the QuickChange XL Site-Directed Mutagenesis Kit (Stratagene). The sequences of all constructs were verified by sequencing.
Plasmids
Plasmids encoding Flag3–CMV-10-Par-1b, Flag3–CMV-10-Par-1b (D193N) (KD), GST–Par-1b (Hurov et al., 2004) and pCDNA3.1-RNF41-Flag (Diamonti et al., 2002) have been described previously. The pFLRU6Luc-CMV-MCS-FH and pFLRCMV-MCS-FH plasmids were a generous gift from Greg Longmore (Washington University in St. Louis, MO) (Feng et al., 2010).
Transient transfections
Transient transfections were performed using Lipofectamine 2000 reagent according to the recommendations of the manufacturer (Invitrogen). Cells were harvested 24 hours post transfection.
MCF10A-derived cell lines
HEK293T cells were plated at 8×105 cells in a 6-cm dish the day before transfection. The next day, cells were transfected with a mixture of 1 µg viral DNA encoding the gene of interest and 1 µg pHR'8.2deltaR packaging plasmid at a ratio of 8∶1 with the pCMV-VSV-G envelope plasmid, using Mirus LTR1 transfection reagent (Mirus). MCF10A cells were plated at 1×106 cells in a 10-cm dish the day before infection with the lentivirus. After HEK293T cells were transfected with viral DNA for 48 hours, target MCF10A cells were infected in the presence of 10 µg/ml protamine sulfate for 4 hours and then the virus was removed and the medium replaced. Infected cells were allowed to recover for 24 hours and then were re-infected with lentivirus. Infected cells were trypsinized and plated in medium containing 1 µg/ml puromycin 48 hours after the first infection with lentivirus. RNF41 protein expression of selected cells was measured by western blot.
Antibodies and western blotting
Antibodies specific for RNF41 (Bethyl), Flag M2 (Sigma), α-tubulin (Sigma), utrophin (Leica) and GAPDH (IMGENEX) were purchased. Par-1b was detected with ascites made from a monoclonal antibody that has been previously described (Hurov et al., 2001). The antibody specific for RNF41 phosphorylated on serine 254 was generated by immunizing rabbits with the phosphopeptide ENAHER-pS-WPQGLATC, coupled to keyhole limpet hemocyanin (KLH). Par-1b fusion protein was precipitated with anti-Flag M2 antibody-agarose affinity gel (Sigma) and detected by western blotting with anti-Par-1b ascites. The RNF41–FlagHis6 fusion protein was precipitated with anti-Flag M2 antibody-agarose affinity gel (Sigma) and detected by western blotting with anti-Flag M2 monoclonal antibody (Sigma). Indirect immunofluorescence was performed with antibodies against GM130 (BD Biosciences), α6 integrin (Millipore), E-cadherin (BD Biosciences), laminin-111 (Sigma), βDG (Leica), β1 integrin (Millipore) and Ki67 (Cell Signaling Technology) antibodies.
Immunoprecipitation
HeLa and HEK293T cells were transfected with the indicated plasmids using Lipofectamine 2000 reagent (Invitrogen) at 75% confluence. Cells were grown for 24 hours before washing in ice-cold phosphate-buffered saline (PBS) once and harvesting in ice-cold mammalian cell lysis buffer (MCLB, 50 mM Tris-HCl pH 8.0, 100 mM NaCl, 2 mM DTT, 5 mM EDTA, 0.5% NP-40, 1 µm microcystin, 1 mM sodium orthovanadate, 2 mM PMSF, 0.15 U/ml aprotinin, 20 µm leupeptin, and 20 µm pepstatin). A total of 1 mg of total cell lysate was used for the indicated immunoprecipitations (IP). M2 Flag-agarose (Sigma) (1∶1 slurry in MCLB, 10 µl) was used to immunoprecipitate Flag3–Par-1b and RNF41-FlagHis6 proteins for 1 hour at 4°C. Immunoprecipitations were washed six times with ice-cold MCLB before boiling in SDS-PAGE sample buffer and loading on an SDS gel.
Secreted laminin-111 was immunoprecipitated from 3D culture medium 20 days after seeding in matrix. SDS and Triton X-100 were added to the culture medium so that the final percentage was 0.1% and 0.01% respectively. The medium was then incubated at 70°C for 10 minutes and then incubated with anti-laminin-111 antibody rocking at 4°C overnight. Protein A/G agarose (Santa Cruz Biotechnology) (1∶1 slurry in MCLB, 10 µl) was added and samples rocked at 4°C for 1 hour. Immunoprecipitations were washed six times with ice-cold MCLB before boiling in SDS-PAGE sample buffer and loading onto an SDS gel.
Expression and purification of proteins in bacteria
JM109 cells were transformed with a plasmid encoding GST–RNF41. Cultures were grown at 37°C to an A600 of 0.6, and isopropyl-1-thio-β-D-galactopyranoside (IPTG) was added to a final concentration of 0.5 mM. After growing for an additional 4 hours at 30°C, cells were pelleted by centrifugation. Cell pellets were washed with PBS buffer and resuspended in STE (100 mM NaCl, 10 mM Tris-HCl pH 8.0 and 1 mM EDTA) supplemented with 2 mM phenylmethylsulfonyl fluoride (PMSF), 0.15 units/ml aprotinin, 20 µM leupeptin, 20 µM pepstatin (referred to as 1× protease inhibitors), and 1.0 mg/ml lysozyme. After rocking at 4°C for 20 minutes, Sarkosyl was added to a final concentration of 1.5%, and lysis was accomplished by sonication. Lysates were clarified by centrifugation, and Triton X-l00 was added to a final concentration of 2%. Proteins were precipitated with GSH agarose (Sigma) and washed twice in STE, twice in LiCl buffer (0.5 M LiCl, 50 mM Tris-HCl pH 8.0), and twice in 50 mM Tris-HCl, pH 7.4. GST fusion proteins were eluted with 20 mM glutathione in 50 mM Tris-HCl (pH 7.4), and the concentration of each GST fusion protein was estimated by comparison to known BSA standards after SDS-PAGE and Coomassie Blue staining.
To express GST–Par-1b in bacteria, the E. coli strain JM109 was transformed with GST–Par-1b, and cultures were grown at 37°C until reaching an A600 of ∼0.6. Cells were induced with 100 µM IPTG, grown at 28°C for 11 hours, collected by centrifugation and resuspended in STE buffer supplemented with 1× protease inhibitors and 1.0 mg/ml lysozyme. After rocking at 4°C for 20 minutes, cells were lysed by sonication and centrifuged at 3000 g for 10 minutes. Proteins were precipitated with GSH agarose and washed as described above. Finally, precipitated proteins were washed with incomplete kinase buffer (50 mM Tris-HCl, pH 7.5, 12.5 mM MgCl2, 1 mM DTT).
Kinase assays
Recombinant GST–Par-1b bound to GSH agarose was incubated with 5 µg of recombinant GST–RNF41 in complete kinase buffer (50 mM Tris-HCl pH 7.5, 12.5 mM MgCl2, 1 mM DTT, 440 µM ATP and 7 µCi γ[32P]ATP [>4000 Ci/mmol]) at 37°C for 30 minutes. Samples were boiled, resolved by SDS-PAGE, and transferred onto nitrocellulose.
Mapping studies
The nitrocellulose containing radiolabeled GST–RNF41 was blocked with 0.5% polyvinylpyrrolidone (PVP-40) in 100 mM acetic acid for 30 minutes at 37°C, followed by washing six times with water. Radiolabeled GST–RNF41 was digested in a solution containing 0.2 mg of trypsin (Worthington) per ml in 50 mM ammonium bicarbonate. Tryptic phosphopeptides were separated in the first dimension by thin layer chromatography at pH 1.9 and in the second dimension by ascending chromatography in a buffer consisting of n-butanol:pyridine:acetic acid:water in a ratio of 75:50:15:60. For PAA analysis, washed tryptic labeled peptides were boiled in 6 M hydrochloric acid for 90 minutes, followed by washing six times with water (van der Geer and Hunter, 1994).
3D cultures
Each well of a 6-well plate was coated with 500 µl of a mixture of collagen type 1 (BD Biosciences), 1 M sodium hydroxide, horse serum (Invitrogen), 5× DMEM (Sigma) and sterile water with a final concentration of collagen type 1 at 2.4 mg/ml. The collagen coating was allowed to polymerize in a tissue culture incubator for 30 minutes at 37°C. MCF10A cells or transduced MCF10A cells selected with puromycin were trypsinized and 7.49×104 cells were added to mixture consisting of collagen type 1, 1 M sodium hydroxide, horse serum, 5× DMEM and sterile water mixture (1.0:0.02:0.08:0.3:0.06 ratio) on ice. The cell–collagen mixture was gently mixed so that cells were in suspension and 1 ml of the mixture was added to each of three wells of the six-well plate. The cell–collagen mixture was allowed to polymerize in a tissue culture incubator for 1 hour at 37°C and then 2 ml of medium with or without 1 µg/ml puromycin was added to each well. Cells were harvested after 8, 10, 14 or 20 days in culture. Cells were harvested by pooling each piece of polymerized collagen and cells from each of the three wells and incubating them in 20 mg/ml Collagenase A (Roche) in PBS for 5 minutes at 37°C. The cells were washed three times with ice-cold PBS and either spotted onto a glass slide for analysis by indirect immunofluorescence microscopy or lysed in ice-cold MCLB followed by western blotting.
For cells cultured in collagen I matrix supplemented with ectopic laminin-111, 3D cultures were generated as described above except a final concentration of 1.3 µg/ml purified laminin-111 (BD Biosciences) was added to the cell mixture before it was allowed to polymerize.
Determining cell number
MCF10A cells cultured in 3D collagen I matrix for 20 days were isolated and suspended in 20 mg/ml Collagenase A (Roche) in PBS for 5 minutes at 37°C to obtain single cells. Cells were then counted using a hemocytometer. Experiments were repeated three times.
Indirect immunofluorescence microscopy
MCF10A cells grown in 3D culture were spotted onto microscope slides and fixed with 4% paraformaldehyde (PFA) in PBS for 20 minutes at room temperature. Slides were washed two times with room temperature PBS and three times (10 minutes each wash) at room temperature in 100 mM glycine in PBS. Fixed cells were blocked in IF blocking solution [10% normal goat or donkey serum, 0.2% Triton X-100, 0.1% BSA (radioimmunoassay grade, Sigma) and 0.05% Tween 20 in PBS, pH 7.4] for 1.5 hours at room temperature in a humidified chamber. Fixed cells were stained with primary antibody diluted in IF buffer (IF blocking solution without 10% normal goat or donkey serum) for 2 hours at room temperature in a humidified chamber. Slides were washed three times for 20 minutes in IF buffer in a Coplin jar at room temperature with gentle shaking. Cells were then incubated with a secondary antibody (conjugated to Alexa Fluor 488 or Alexa Fluor 594; Molecular Probes) diluted in IF buffer for 45 minutes at room temperature in a humidified chamber. Slides were washed once with IF buffer for 20 minutes and then two times with PBS for 10 minutes at room temperature with gentle shaking. Finally, cells were stained with 1 µM TO-PRO-3 iodide (Molecular Probes) for 10 minutes at room temperature in a humidified chamber, slides were washed once with PBS for 10 minutes with gentle shaking and were mounted with Prolong Antifade mounting medium (Molecular Probes). Images were obtained using an Olympus FV-500 confocal microscope with a 60× water objective. Images were processed using the Olympus FLUOVIEW Ver.2.1a Viewer software.
Supplementary Material
Acknowledgments
We thank Greg Longmore, Robert Mercer, Jeff Minor, Tracy Adair-Kirk and Robert Mecham for providing helpful discussion and recommendations throughout this study. We thank Emily Powell and Anuarg Agarwal for thoughtful discussions and help with manuscript editing. Emily Powell is also thanked for help in establishing 3D culture conditions. We thank Kermit Carraway for pcDNA3.1 RNF41-Flag and Greg Longmore for pFLRU6Luc-CMV-MCS-FH and pFLRCMV-MCS-FH. Dennis Oakley at the Bakewell Neuroimaging Laboratory is thanked for assistance with confocal microscopy.
Footnotes
Competing interests
The authors declare no competing interests.
Author contributions
K.T.L. performed the experiments; K.T.L. and H.P.-W. conceived the project, analyzed data, and wrote the manuscript.
Funding
This work was supported, in part, by the Bakewell Neuroimaging Core, supported in part by the Bakewell Family Foundation and National Institutes of Health Neuroscience Blueprint Interdisciplinary Center Core Grant [grant number P30 NS057105 to Washington University]. K.T.L. was supported, in part, by the Cancer Biology Pathway program administered through the Siteman Cancer Center at Washington University which is supported, in part, by an NCI Cancer Center Support Grant [grant number P30 CA91842]. H.P.-W. is a Research Professor of the American Cancer Society. Deposited in PMC for release after 12 months.
Supplementary material available online at http://jcs.biologists.org/lookup/suppl/doi:10.1242/jcs.129148/-/DC1
References
- Abdullah J. M., Li X., Nachtman R. G., Jurecic R. (2001). FLRF, a novel evolutionarily conserved RING finger gene, is differentially expressed in mouse fetal and adult hematopoietic stem cells and progenitors. Blood Cells Mol. Dis. 27, 320–333 10.1006/bcmd.2001.0390 [DOI] [PubMed] [Google Scholar]
- Benton R., St Johnston D. (2003). Drosophila PAR-1 and 14-3-3 inhibit Bazooka/PAR-3 to establish complementary cortical domains in polarized cells. Cell 115, 691–704 10.1016/S0092--8674(03)00938--3 [DOI] [PubMed] [Google Scholar]
- Böhm H., Brinkman V., Drab M., Henske A., Kurzchalia T. V. (1997). Mammalian Homologues of C-elegans polarization gene product PAR-1 are asymmetrically localized in epithelial cells and may influence their polarity. Curr. Biol. 7, 603–606 10.1016/S0960--9822(06)00260--0 [DOI] [PubMed] [Google Scholar]
- Bouyain S., Leahy D. J. (2007). Structure-based mutagenesis of the substrate-recognition domain of Nrdp1/FLRF identifies the binding site for the receptor tyrosine kinase ErbB3. Protein Sci. 16, 654–661 10.1110/ps.062700307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brajenovic M., Joberty G., Küster B., Bouwmeester T., Drewes G. (2004). Comprehensive proteomic analysis of human Par protein complexes reveals an interconnected protein network. J. Biol. Chem. 279, 12804–12811 10.1074/jbc.M312171200 [DOI] [PubMed] [Google Scholar]
- Cao Z., Wu X., Yen L., Sweeney C., Carraway K. L., III (2007). Neuregulin-induced ErbB3 downregulation is mediated by a protein stability cascade involving the E3 ubiquitin ligase Nrdp1. Mol. Cell. Biol. 27, 2180–2188 10.1128/MCB.01245--06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y. M., Wang Q. J., Hu H. S., Yu P. C., Zhu J., Drewes G., Piwnica-Worms H., Luo Z. G. (2006). Microtubule affinity-regulating kinase 2 functions downstream of the PAR-3/PAR-6/atypical PKC complex in regulating hippocampal neuronal polarity. Proc. Natl. Acad. Sci. USA 103, 8534–8539 10.1073/pnas.0509955103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen D., Müsch A. (2003). Apical surface formation in MDCK cells: regulation by the serine/threonine kinase EMK1. Methods 30, 269–276 10.1016/S1046--2023(03)00033--1 [DOI] [PubMed] [Google Scholar]
- Cohen D., Brennwald P. J., Rodriguez-Boulan E., Müsch A. (2004). Mammalian PAR-1 determines epithelial lumen polarity by organizing the microtubule cytoskeleton. J. Cell Biol. 164, 717–727 10.1083/jcb.200308104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen D., Tian Y., Müsch A. (2007). Par1b promotes hepatic-type lumen polarity in Madin Darby canine kidney cells via myosin II- and E-cadherin-dependent signaling. Mol. Biol. Cell 18, 2203–2215 10.1091/mbc.E07--02--0095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen D., Fernandez D., Lázaro-Diéguez F., Müsch A. (2011). The serine/threonine kinase Par1b regulates epithelial lumen polarity via IRSp53-mediated cell-ECM signaling. J. Cell Biol. 192, 525–540 10.1083/jcb.201007002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox D. N., Lu B., Sun T. Q., Williams L. T., Jan Y. N. (2001a). Drosophila par-1 is required for oocyte differentiation and microtubule organization. Curr. Biol. 11, 75–87 10.1016/S0960--9822(01)00027--6 [DOI] [PubMed] [Google Scholar]
- Cox D. N., Seyfried S. A., Jan L. Y., Jan Y. N. (2001b). Bazooka and atypical protein kinase C are required to regulate oocyte differentiation in the Drosophila ovary. Proc. Natl. Acad. Sci. USA 98, 14475–14480 10.1073/pnas.261565198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debnath J., Muthuswamy S. K., Brugge J. S. (2003). Morphogenesis and oncogenesis of MCF-10A mammary epithelial acini grown in three-dimensional basement membrane cultures. Methods 30, 256–268 10.1016/S1046--2023(03)00032--X [DOI] [PubMed] [Google Scholar]
- Diamonti A. J., Guy P. M., Ivanof C., Wong K., Sweeney C., Carraway K. L., III (2002). An RBCC protein implicated in maintenance of steady-state neuregulin receptor levels. Proc. Natl. Acad. Sci. USA 99, 2866–2871 10.1073/pnas.052709799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doerflinger H., Benton R., Shulman J. M., St Johnston D. (2003). The role of PAR-1 in regulating the polarised microtubule cytoskeleton in the Drosophila follicular epithelium. Development 130, 3965–3975 10.1242/dev.00616 [DOI] [PubMed] [Google Scholar]
- Drewes G., Ebneth A., Preuss U., Mandelkow E-M., Mandelkow E. (1997). MARK, a novel family of protein kinases that phosphorylate microtubule-associated proteins and trigger microtubule disruption. Cell 89, 297–308 10.1016/S0092--8674(00)80208--1 [DOI] [PubMed] [Google Scholar]
- Driss A., Charrier L., Yan Y., Nduati V., Sitaraman S., Merlin D. (2006). Dystroglycan receptor is involved in integrin activation in intestinal epithelia. Am. J. Physiol. 290, G1228–G1242 10.1152/ajpgi.00378.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ducharme N. A., Hales C. M., Lapierre L. A., Ham A. J., Oztan A., Apodaca G., Goldenring J. R. (2006). MARK2/EMK1/Par-1Balpha phosphorylation of Rab11-family interacting protein 2 is necessary for the timely establishment of polarity in Madin-Darby canine kidney cells. Mol. Biol. Cell 17, 3625–3637 10.1091/mbc.E05--08--0736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekblom P. (1989). Developmentally regulated conversion of mesenchyme to epithelium. FASEB J. 3, 2141–2150 [DOI] [PubMed] [Google Scholar]
- Ekblom M., Falk M., Salmivirta K., Durbeej M., Ekblom P. (1998). Laminin isoforms and epithelial development. Ann. N. Y. Acad. Sci. 857, 194–211 10.1111/j.1749--6632.1998.tb10117.x [DOI] [PubMed] [Google Scholar]
- Ellenbroek S. I., Iden S., Collard J. G. (2012). Cell polarity proteins and cancer. Semin. Cancer Biol. 22, 208–215 10.1016/j.semcancer.2012.02.012 [DOI] [PubMed] [Google Scholar]
- Emerman J. T., Pitelka D. R. (1977). Maintenance and induction of morphological differentiation in dissociated mammary epithelium on floating collagen membranes. In Vitro 13, 316–328 10.1007/BF02616178 [DOI] [PubMed] [Google Scholar]
- Espinosa L., Navarro E. (1998). Human serine/threonine protein kinase EMK1: genomic structure and cDNA cloning of isoforms produced by alternative splicing. Cytogenet. Cell Genet. 81, 278–282 10.1159/000015046 [DOI] [PubMed] [Google Scholar]
- Etienne-Manneville S., Hall A. (2003). Cell polarity: Par6, aPKC and cytoskeletal crosstalk. Curr. Opin. Cell Biol. 15, 67–72 10.1016/S0955--0674(02)00005--4 [DOI] [PubMed] [Google Scholar]
- Feng Y., Nie L., Thakur M. D., Su Q., Chi Z., Zhao Y., Longmore G. D. (2010). A multifunctional lentiviral-based gene knockdown with concurrent rescue that controls for off-target effects of RNAi. Genomics Proteomics Bioinformatics 8, 238–245 10.1016/S1672--0229(10)60025--3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo S., Kemphues K. J. (1995). par-1, a gene required for establishing polarity in C. elegans embryos, encodes a putative Ser/Thr kinase that is asymmetrically distributed. Cell 81, 611–620 10.1016/0092--8674(95)90082--9 [DOI] [PubMed] [Google Scholar]
- Henry M. D., Satz J. S., Brakebusch C., Costell M., Gustafsson E., Fässler R., Campbell K. P. (2001). Distinct roles for dystroglycan, beta1 integrin and perlecan in cell surface laminin organization. J. Cell Sci. 114, 1137–1144 [DOI] [PubMed] [Google Scholar]
- Holbro T., Beerli R. R., Maurer F., Koziczak M., Barbas C. F., III and Hynes N. E. (2003). The ErbB2/ErbB3 heterodimer functions as an oncogenic unit: ErbB2 requires ErbB3 to drive breast tumor cell proliferation. Proc. Natl. Acad. Sci. USA 100, 8933–8938 10.1073/pnas.1537685100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurd T. W., Fan S., Liu C. J., Kweon H. K., Hakansson K., Margolis B. (2003). Phosphorylation-dependent binding of 14-3-3 to the polarity protein Par3 regulates cell polarity in mammalian epithelia. Curr. Biol. 13, 2082–2090 10.1016/j.cub.2003.11.020 [DOI] [PubMed] [Google Scholar]
- Hurov J. B., Stappenbeck T. S., Zmasek C. M., Ranganath S. H., White L. S., Russell J. H., Chan A. C., Murphy K. M., Piwnica-Worms H. (2001). Immune cell dysfunction and autoimmune disease in mice lacking the EMK1/Par-l protein kinase. Mol. Cell. Biol. 21, 3206–3219 10.1128/MCB.21.9.3206--3219.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurov J. B., Watkins J. L., Piwnica-Worms H. (2004). Atypical PKC phosphorylates PAR-1 kinases to regulate localization and activity. Curr. Biol. 14, 736–741 10.1016/j.cub.2004.04.007 [DOI] [PubMed] [Google Scholar]
- Inglis J. D., Lee M., Hill R. E. (1993). Emk, a protein kinase with homologs in yeast maps to mouse chromosome 19. Mamm. Genome 4, 401–403 10.1007/BF00360595 [DOI] [PubMed] [Google Scholar]
- Jing X., Infante J., Nachtman R. G., Jurecic R. (2008). E3 ligase FLRF (Rnf41) regulates differentiation of hematopoietic progenitors by governing steady-state levels of cytokine and retinoic acid receptors. Exp. Hematol. 36, 1110–1120 10.1016/j.exphem.2008.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joberty G., Petersen C., Gao L., Macara I. G. (2000). The cell-polarity protein Par6 links Par3 and atypical protein kinase C to Cdc42. Nat. Cell Biol. 2, 531–539 10.1038/35019573 [DOI] [PubMed] [Google Scholar]
- Kato T., Satoh S., Okabe H., Kitahara O., Ono K., Kihara C., Tanaka T., Tsunoda T., Yamaoka Y., Nakamura Y. et al. (2001). Isolation of a novel human gene, MARKL1, homologous to MARK3 and its involvement in hepatocellular carcinogenesis. Neoplasia 3, 4–9 10.1038/sj.neo.7900132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemphues K. J., Priess J. R., Morton D. G., Cheng N. S. (1988). Identification of genes required for cytoplasmic localization in early C. elegans embryos. Cell 52, 311–320 10.1016/S0092--8674(88)80024--2 [DOI] [PubMed] [Google Scholar]
- Kirby C., Kusch M., Kemphues K. (1990). Mutations in the par genes of Caenorhabditis elegans affect cytoplasmic reorganization during the first cell cycle. Dev. Biol. 142, 203–215 10.1016/0012--1606(90)90164--E [DOI] [PubMed] [Google Scholar]
- Li S., Edgar D., Fässler R., Wadsworth W., Yurchenco P. D. (2003). The role of laminin in embryonic cell polarization and tissue organization. Dev. Cell 4, 613–624 10.1016/S1534--5807(03)00128--X [DOI] [PubMed] [Google Scholar]
- Masuda-Hirata M., Suzuki A., Amano Y., Yamashita K., Ide M., Yamanaka T., Sakai M., Imamura M., Ohno S. (2009). Intracellular polarity protein PAR-1 regulates extracellular laminin assembly by regulating the dystroglycan complex. Genes Cells 14, 835–850 10.1111/j.1365--2443.2009.01315.x [DOI] [PubMed] [Google Scholar]
- Müller J., Ory S., Copeland T., Piwnica-Worms H., Morrison D. K. (2001). C-TAK1 regulates Ras signaling by phosphorylating the MAPK scaffold, KSR1. Mol. Cell 8, 983–993 10.1016/S1097--2765(01)00383--5 [DOI] [PubMed] [Google Scholar]
- O'Brien L. E., Zegers M. M., Mostov K. E. (2002). Opinion: Building epithelial architecture: insights from three-dimensional culture models. Nat. Rev. Mol. Cell Biol. 3, 531–537 10.1038/nrm859 [DOI] [PubMed] [Google Scholar]
- Ojakian G. K., Schwimmer R. (1994). Regulation of epithelial cell surface polarity reversal by beta 1 integrins. J. Cell Sci. 107, 561–576 [PubMed] [Google Scholar]
- Ossipova O., Dhawan S., Sokol S., Green J. B. (2005). Distinct PAR-1 proteins function in different branches of Wnt signaling during vertebrate development. Dev. Cell 8, 829–841 10.1016/j.devcel.2005.04.011 [DOI] [PubMed] [Google Scholar]
- Patarroyo M., Tryggvason K., Virtanen I. (2002). Laminin isoforms in tumor invasion, angiogenesis and metastasis. Semin. Cancer Biol. 12, 197–207 10.1016/S1044--579X(02)00023--8 [DOI] [PubMed] [Google Scholar]
- Peng C-Y., Graves P. R., Ogg S., Thoma R. S., Byrnes M. J., 3rd, Wu Z., Stephenson M. T., Piwnica-Worms H. (1998). C-TAK1 protein kinase phosphorylates human Cdc25C on serine 216 and promotes 14-3-3 protein binding. Cell Growth Differ. 9, 197–208 [PubMed] [Google Scholar]
- Qiu X. B., Goldberg A. L. (2002). Nrdp1/FLRF is a ubiquitin ligase promoting ubiquitination and degradation of the epidermal growth factor receptor family member, ErbB3. Proc. Natl. Acad. Sci. USA 99, 14843–14848 10.1073/pnas.232580999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu X. B., Markant S. L., Yuan J., Goldberg A. L. (2004). Nrdp1-mediated degradation of the gigantic IAP, BRUCE, is a novel pathway for triggering apoptosis. EMBO J. 23, 800–810 10.1038/sj.emboj.7600075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Boulan E., Nelson W. J. (1989). Morphogenesis of the polarized epithelial cell phenotype. Science 245, 718–725 10.1126/science.2672330 [DOI] [PubMed] [Google Scholar]
- Schoenenberger C. A., Zuk A., Zinkl G. M., Kendall D., Matlin K. S. (1994). Integrin expression and localization in normal MDCK cells and transformed MDCK cells lacking apical polarity. J. Cell Sci. 107, 527–541 [DOI] [PubMed] [Google Scholar]
- Schröder J. E., Tegeler M. R., Grosshans U., Porten E., Blank M., Lee J., Esapa C., Blake D. J., Kröger S. (2007). Dystroglycan regulates structure, proliferation and differentiation of neuroepithelial cells in the developing vertebrate CNS. Dev. Biol. 307, 62–78 10.1016/j.ydbio.2007.04.020 [DOI] [PubMed] [Google Scholar]
- Shulman J. M., Benton R., St Johnston D. (2000). The Drosophila homolog of C. elegans PAR-1 organizes the oocyte cytoskeleton and directs oskar mRNA localization to the posterior pole. Cell 101, 377–388 10.1016/S0092--8674(00)80848--X [DOI] [PubMed] [Google Scholar]
- Singh R., Kaushik S., Wang Y., Xiang Y., Novak I., Komatsu M., Tanaka K., Cuervo A. M., Czaja M. J. (2009). Autophagy regulates lipid metabolism. Nature 458, 1131–1135 10.1038/nature07976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- St Johnston D., Ahringer J. (2010). Cell polarity in eggs and epithelia: parallels and diversity. Cell 141, 757–774 10.1016/j.cell.2010.05.011 [DOI] [PubMed] [Google Scholar]
- Sun T. Q., Lu B., Feng J. J., Reinhard C., Jan Y. N., Fantl W. J., Williams L. T. (2001). PAR-1 is a Dishevelled-associated kinase and a positive regulator of Wnt signalling. Nat. Cell Biol. 3, 628–636 10.1038/35083016 [DOI] [PubMed] [Google Scholar]
- Suzuki A., Hirata M., Kamimura K., Maniwa R., Yamanaka T., Mizuno K., Kishikawa M., Hirose H., Amano Y., Izumi N. et al. (2004). aPKC acts upstream of PAR-1b in both the establishment and maintenance of mammalian epithelial polarity. Curr. Biol. 14, 1425–1435 10.1016/j.cub.2004.08.021 [DOI] [PubMed] [Google Scholar]
- Tabuse Y., Izumi Y., Piano F., Kemphues K. J., Miwa J., Ohno S. (1998). Atypical protein kinase C cooperates with PAR-3 to establish embryonic polarity in Caenorhabditis elegans. Development 125, 3607–3614 [DOI] [PubMed] [Google Scholar]
- Tomancak P., Piano F., Riechmann V., Gunsalus K. C., Kemphues K. J., Ephrussi A. (2000). A Drosophila melanogaster homologue of Caenorhabditis elegans par-1 acts at an early step in embryonic-axis formation. Nat. Cell Biol. 2, 458–460 10.1038/35017101 [DOI] [PubMed] [Google Scholar]
- van der Geer P., Hunter T. (1994). Phosphopeptide mapping and phosphoamino acid analysis by electrophoresis and chromatography on thin-layer cellulose plates. Electrophoresis 15, 544–554 10.1002/elps.1150150173 [DOI] [PubMed] [Google Scholar]
- Watkins J. L., Lewandowski K. T., Meek S. E., Storz P., Toker A., Piwnica-Worms H. (2008). Phosphorylation of the Par-1 polarity kinase by protein kinase D regulates 14-3-3 binding and membrane association. Proc. Natl. Acad. Sci. USA 105, 18378–18383 10.1073/pnas.0809661105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wauman J., De Ceuninck L., Vanderroost N., Lievens S., Tavernier J. (2011). RNF41 (Nrdp1) controls type 1 cytokine receptor degradation and ectodomain shedding. J. Cell Sci. 124, 921–932 10.1242/jcs.078055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weir M. L., Oppizzi M. L., Henry M. D., Onishi A., Campbell K. P., Bissell M. J., Muschler J. L. (2006). Dystroglycan loss disrupts polarity and beta-casein induction in mammary epithelial cells by perturbing laminin anchoring. J. Cell Sci. 119, 4047–4058 10.1242/jcs.03103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X., Yen L., Irwin L., Sweeney C., Carraway K. L., III (2004). Stabilization of the E3 ubiquitin ligase Nrdp1 by the deubiquitinating enzyme USP8. Mol. Cell. Biol. 24, 7748–7757 10.1128/MCB.24.17.7748--7757.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita K., Suzuki A., Satoh Y., Ide M., Amano Y., Masuda-Hirata M., Hayashi Y. K., Hamada K., Ogata K., Ohno S. (2010). The 8th and 9th tandem spectrin-like repeats of utrophin cooperatively form a functional unit to interact with polarity-regulating kinase PAR-1b. Biochem. Biophys. Res. Commun. 391, 812–817 10.1016/j.bbrc.2009.11.144 [DOI] [PubMed] [Google Scholar]
- Yen L., Cao Z., Wu X., Ingalla E. R., Baron C., Young L. J., Gregg J. P., Cardiff R. D., Borowsky A. D., Sweeney C. et al. (2006). Loss of Nrdp1 enhances ErbB2/ErbB3-dependent breast tumor cell growth. Cancer Res. 66, 11279–11286 10.1158/0008--5472.CAN--06--2319 [DOI] [PubMed] [Google Scholar]
- Zhong L., Tan Y., Zhou A., Yu Q., Zhou J. (2005). RING finger ubiquitin-protein isopeptide ligase Nrdp1/FLRF regulates parkin stability and activity. J. Biol. Chem. 280, 9425–9430 10.1074/jbc.M408955200 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.