Abstract
The influenza M2 protein forms an acid-activated and drug-sensitive proton channel in the virus envelope that is important for the virus lifecycle. The functional properties and high-resolution structures of this proton channel have been extensively studied to understand the mechanisms of proton conduction and drug inhibition. We review biochemical and electrophysiological studies of M2 and discuss how high-resolution structures have transformed our understanding of this proton channel. Comparison of structures obtained in different membrane-mimetic solvents and under different pH using X-ray crystallography, solution NMR, and solid-state NMR spectroscopy revealed how the M2 structure depends on the environment and showed that the pharmacologically relevant drug-binding site lies in the transmembrane (TM) pore. Competing models of proton conduction have been evaluated using biochemical experiments, high-resolution structural methods, and computational modeling. These results are converging to a model in which a histidine residue in the TM domain mediates proton relay with water, aided by microsecond conformational dynamics of the imidazole ring. These mechanistic insights are guiding the design of new inhibitors that target drug-resistant M2 variants and may be relevant for other proton channels.
Keywords: solid-state NMR, magic angle spinning, drug inhibition, membrane protein structure determination, protein dynamics
Introduction
The M2 protein of the influenza A virus was first discovered as the protein target of the anti-influenza drug, amantadine.1 This modular protein serves multiple functions, which are localized in different domains of the short 97-residue sequence. Its highly conserved N-terminal 23 residues are located on the outside of the virus and assist M2 incorporation into the virion.2 Following this region is a transmembrane (TM) helix (Fig. 1) that serves as a tetramerization and proton-conducting domain. Conduction of protons into the virion acidifies the virus after endocytosis and initiates viral uncoating.6,7 Residues 46–60 form the next module, which helps induce membrane curvature and membrane scission to release newly assembled viruses from the host cell.8 Finally, the C-terminal tail of the protein interacts with the matrix protein M1 and is essential for virus packaging and budding.9 In some strains of the influenza A virus, M2 is also important for equilibrating the pH of the lumen of the Golgi apparatus with the cytoplasm, preventing premature conformational change of the viral hemagglutinin.10 The M2 protein has been extensively studied because of its medical importance, particularly following the emergence of widespread resistance to M2-blocking drugs amantadine and rimantadine.11 The small size and modular structure of M2 also make it attractive for biophysical studies.
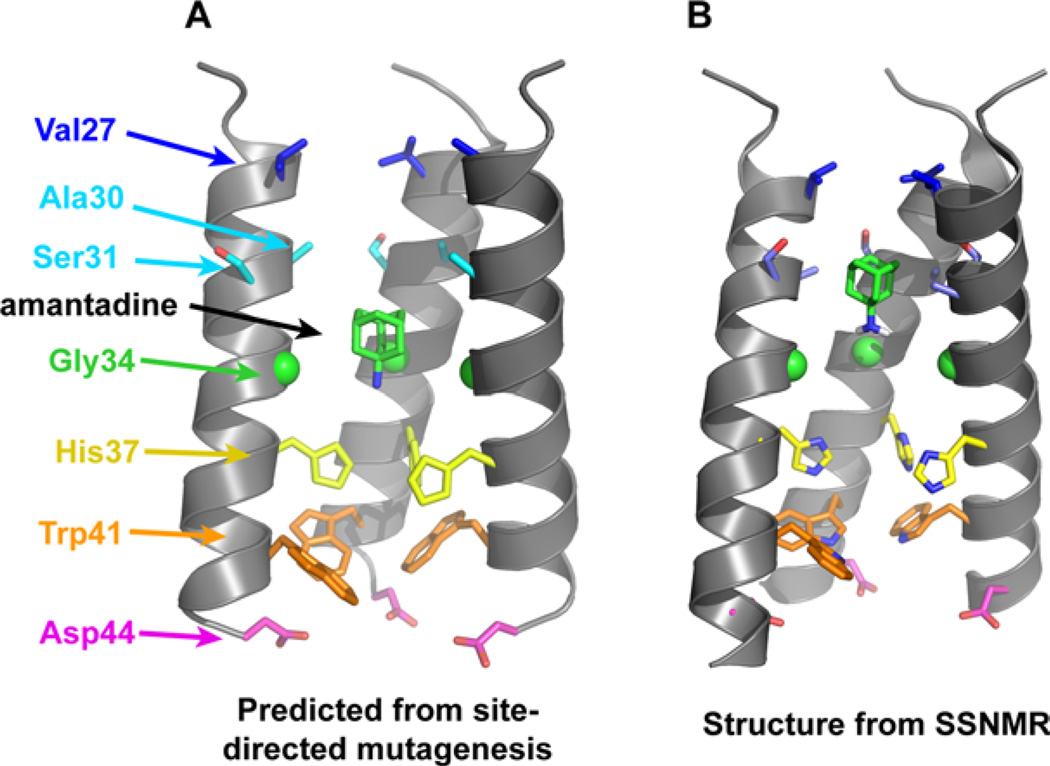
Figure 1.
(A) Functional model of the TM domain of the M2 tetramer, showing the positions of crucial side chains and the drug amantadine. The model was obtained from cysteine scanning mutagenesis.3,4 (B) High-resolution structure of the amantadine-bound M2TM in DMPC bilayers obtained from SSNMR (PDB: 2KQT).5 The overall shape of the tetrameric bundle from the functional model is in excellent agreement with the high-resolution structure; however, specific differences exist such as the helix tilt angle and the conformations of several side chains (e.g., Ser31 and Trp41). In both images, the “front” helix has been removed for clarity.
This review will focus primarily on the structure and function of the TM domain of M2 (M2TM), which contains the proton-conducting residue, histidine 37 (His37),12 and the channel-gating residue, tryptophan 41 (Trp41).13 The TM domain reproduces most of the electrophysiological, pharmacological, and biophysical features of the full-length protein, such as low-pH activated proton conductivity, amantadine sensitivity of the proton current, and tetramerization of the protein.14–16 Since its discovery and until 2008, the mechanism of proton conduction was extensively studied by electrophysiology, site-directed mutagenesis, and molecular dynamics (MD) simulations.3 However, while these studies established the overall topology and approximate location of key side chains, no high-resolution structures were available. In 2008, this situation changed dramatically with the publication of solution NMR and crystallographic structures.17,18 These were followed by solid-state NMR (SSNMR) structures of phospholipid-bilayer-bound M2.5,19 In this review, we first summarize our understanding of M2’s mechanism of proton conduction and inhibition based on biochemical and electrophysiological studies. We then discuss how high-resolution structures have transformed our understanding of this proton channel. At times, high-resolution structures can give misleading results if their conclusions are not supported by functional data. Thus, to distinguish mechanistic conclusions based on biochemical data versus mechanistic insights from high-resolution structures, we review these subjects in separate sections. Because structures are emerging from several experimental methods, we also provide a brief technical description of the advantages as well as underlying uncertainties of the different methods.
Finally, we point to remaining areas of disagreement and open questions for future studies. Progress in the area of M2 has been punctuated by competing models of proton conduction and the site of pharmacological inhibition. Because these competing models were well defined and articulated, they stimulated new and more discriminating experiments and interpretation. The first debate concerned the proton conduction mechanism. An early model envisioned a continuous aqueous channel that was gated by pH (shutter mechanism),20 versus the currently accepted model in which protons diffuse along a water wire until reaching His37, where they are then “shuttled” by His37 through alternate protonation and deprotonation events. A second debate focused on whether drugs inhibited the channel by binding to a site within the aqueous pore or on the surface of the protein. This debate stimulated extensive functional experiments21–25 and SSNMR investigations.5,26,27 The physiologically important site was eventually shown by SSNMR to lie in the pore, highlighting the utility of this atomic resolution spectroscopy for structure determination of membrane proteins in near-native lipid bilayers.
The primary remaining debate in the literature centers on the mechanism by which charge is stabilized in the His37 tetrad when it is protonated. One early model focused on a low-barrier hydrogen bond (LBHB) between His37 residues, which was motivated by the observation of a high pKa for the first two protonation steps.28 A more recent study, however, suggested that the His37 cluster is less basic than originally anticipated.29 Magic-angle spinning (MAS) SSNMR measurements of the His37 conformation and dynamics,30 combined with crystal structure information,31 indicate that His37 residues interact with each other indirectly via water molecules in the dominant structure at equilibrium. We present a model for conduction and inhibition that combines our understanding of the pharmacology and the full ensemble of structural and functional studies now accumulated for this proton channel.
Conduction properties of M2
The rate and pH dependence of proton conductance through M2 appear to be evolutionarily tuned to provide enough conductances to mediate acidification of the virion when encapsulated in the endosome, but not excessive or nonselective conductance that could cause toxicity to the cell before it produces viral progeny. M2 conducts protons ~ 105–106 times more effectively than other ions such as sodium.32–35 The selectivity, however, is not absolute, and K+ ions, which are at a much higher concentration than protons, are slowly conducted out of the virus as the inside of the virus is acidified.36 This small K+ conductance prevents the formation of a large electrical gradient that would otherwise counter proton flux before acidification is complete. Classically, the conductance of a channel is described by its radius, length, ion selectivity, and most importantly, the concentration of ions on the two sides of the bilayer. Typically, the rate of an ion’s conductance scales linearly with its concentration on a given side of the bilayer, and a net flux is observed only at asymmetric ion concentration or in the presence of a TM electrical potential. The proton conductance of M2 is relatively small compared with K+ or Na+ channels, largely because the concentration of protons is very low (10−5M at pH 5) relative to alkali metal ions (0.15M) in a cell. Near neutrality, M2 conducts protons at a rate near what is expected for a channel formed from a tetramer of α-helices;37 a second-order rate constant of ~ 107–108M−1 s−1 can be computed for the net diffusion of ions into and through the channel. However, as the pH is reduced below pH 6, the conductance fails to increase linearly and instead levels off, which is reminiscent of the Michaelis–Menton behavior seen in transporters. This saturation behavior has also been observed in studies of M2 reconstituted in phospholipid vesicles.36,38
Therefore, M2 was proposed to work by a mechanism in which His37, the only ionizable residue in the TM domain, served to shuttle protons through the channel.3,34 Saturation was proposed to occur at low pH as one or more His37 residues became fully protonated. The conduction curve showed a midpoint near pH 6,33,39 which was taken to be the pKa of the conducting His37. The leveling seen at low pH is indicative of the rate-limiting step switching from diffusion into the channel (at high pH) to another step at low pH. This step was assigned to proton dissociation from His37, given the known dissociation rate of histidines in proteins. Histidines with a pKa near 6 have proton dissociation rates of ~ 104 s−1 at room temperature.40 In a channel with a constricted diameter like that of M2, the rate is expected to be one to two orders of magnitude slower, 102–103 s−1.37 This value is in good agreement with the experimentally measured maximal conductance of M2, which is about 10–1000 s−1.
The His37 shuttle hypothesis was further refined following the report that the first two protonation steps of this residue occurred with a pKa near 8.2.28 This value has considerable uncertainty because it was measured in dimyristoylphosphatidylcholine (DMPC)/dimyristoylphosphatidylglycerol (DMPG) bilayers at a temperature where residual peptide motion remained to broaden the 15N spectra. More recently, the pKa’s of the His37 tetrad were determined more accurately using samples reconstituted in a membrane whose lipid composition more closely resembles that of a virus, and the first two protonation steps were resolved, with pKa’s of 7.6 and 6.8.29 Both studies found that binding of the third proton occurred near the midpoint of the pH/ current curve, suggesting that conduction occurs when the His37 tetrad cycles between the +2 and +3 states. In some variants, however, there is a secondary conductance near neutral pH that requires consideration of a low-level of conductance for the +1 to +2 cycle. More recent data suggest that the +3 to +4 cycle might also lead to productive proton flux at pH <5 (Chunlong Ma, Lawrence Pinto, personal communication).
Natural and artificial sequence variants of M2
While influenza A virus is a relatively fast evolving virus that constantly mutates and shuffles its genome, the M2 protein is largely conserved compared with other proteins encoded by the genome. There is an extensive record of sequence variation in M2 from viruses dating back to 1918, which shows particularly little variation in the TM region. From a practical perspective, it is important to understand why the protein is so conserved and how it tolerates the few observed mutations to pore-lining residues, since this information can guide the design of new drugs and anticipate potential sources of new resistance. From a more fundamental perspective, both natural and artificial mutants of M2 provide valuable information about the residues involved in proton conduction and inhibition. Fortunately, the tetrameric structure of M2 is simple that it was possible to predict the probable location of these mutations even before the availability of high-resolution structures.
The first models for the structures of the TM region of M2 arose from Cys-scanning mutagenesis and unrestrained MD simulations.4,20,41 These models guided the design and interpretation of electrophysiological experiments for over a decade, until crystallographic and NMR structures became available. These early models3 captured the overall shape of the pore seen in subsequent high-resolution structures [Fig. 1(A)]. Starting at the exterior of the virus, the N-terminal half of the sequence forms a water-filled pore lined by Val27, Ala30, Ser31, and Gly34. These sites are frequently mutated in amantadine-resistant mutants. The pore is interrupted by His37 and Trp41, which project to the center of the channel. Asp44 defines the C-terminal end of the pore. Below, we will first discuss mutations to the N-terminal pore, which is important for diffusion of protons to His37, followed by key residues His37, Trp41, and Asp44, which are important for charge storage and proton transfer.
The N-terminal aqueous pore
Early studies of the mechanism of amantadine resistance focused on the selection of viruses that could replicate in cell culture in the presence of the pore-blocking drug amantadine. Mutations that cause resistance occurred at Val27, Ala30, Ser31, and Gly34, which line the N-terminal aqueous pore.1,42 A subset of these mutations are found in infected patients following treatment with amantadine,43 and reverse-engineered viruses harboring various pore-lining mutations are able to replicate in vitro and in a mouse model.44 However, many of these mutations give rise to somewhat attenuated viruses that are less transmissible than wild-type (WT) and tend to revert in the absence of drug pressure.42,45 Indeed, large-scale sequencing of transmissible viruses from 1918 to 2010 showed that mutations to pore-lining residues are allowed only within the first turn of the TM helix at positions 26, 27, and 31 [Fig. 1(A)]. S31N has long been the dominant amantadine-resistant mutation in M2,46–49 accounting for 98–100% of the transmissible amantadine-resistant H1N1, H5N1, and H3N2 strains isolated from humans, birds, and swine in the last decade.11,50–60 V27A and L26F are less frequent and generally have been found in nonpandemic amantadine-resistant H1N1.48,61,62
Extensive studies of point mutations to the pore-lining residues of M2 have been carried out to probe the conductance mechanism and to identify additional sites that might impart amantadine-resistance.21,63 A surprisingly large number of mutants in the N-terminal aqueous pore retained the ability to conduct protons selectively over other ions, although the magnitude and pH dependence of their conductance varied. Functional channels were generally observed so long as the mutation did not disrupt the tetrameric structure of the channel64 or introduce a large hydrophobic residue that could block the aqueous pathway leading to His37. While these “functional” mutations gave proton-selective channels, they differed from WT in the magnitude of their proton conduction and the shape of their pH–current curves. Only a few mutations—V27A, S31N, and L26F—had properties very similar to WT. These are also the same mutants that comprise more than 99.9% of reported resistance in transmissible viruses. The stringency of sequence conservation in M2 reflects tight functional constraints of the pore-lining residues, where a single mutation to a monomer causes four changes within a very constricted area of the tetrameric pore.
The proton-selective and gating residues: His37, Trp41, and Asp44
Mutations to the invariant residue, His37, increase the conductance of the channel and eliminate its stringent proton selectivity.12,32,65 Interestingly, the proton selectivity of H37G can be restored by adding exogenous imidazole, which presumably binds to the site normally occupied by His.65
Mutations to Trp41 have identified this residue as the “proton gate.”13 Most strains of influenza have M2 proteins that conduct protons asymmetrically—when pHout is less than pHin, there is a robust inward proton flux, which is much greater than the outward current observed when the situation is reversed. This asymmetry is lost when Trp41 is replaced with other side chains except Tyr, which also has an electron-rich aromatic ring capable of stabilizing cation-π interactions.66,67 These data suggest that Trp41’s side chain is a gate that can block diffusion of protons from the inside but not from the outside of the virus. So long as this gate is closed, protons cannot rapidly access His37 from the inside of the virus, explaining why outward flux is slow under low pHin and high pHout. However, protons coming from the outside can access the His37 tetrad, allowing it to reach a threshold protonation state, which is now known to be the +3 state. This leads to opening of the Trp41 gate and inward proton flux.
Asp44, the final residue in the conduction path of the channel, influences the proton conductance in the pH range 5–7. The Rostock virus has the more conducting variant, D44N.68,69 This virus has a particularly acid-labile hemagglutinin, which M2 protects by preventing the acidification of the late Golgi. Other substitutions at residue 44 also lead to larger proton flux, suggesting that Asp44 helps to stabilize Trp41 in the closed form, and disruption of this interaction results in enhancement of the proton flux.
High-resolution structures of M2
High-resolution structures elevated mechanistic understanding of the M2 protein to the atomic level. All three major high-resolution techniques, X-ray crystallography, solution NMR, and SSNMR have been used to determine the structure of M2. Because of the differing sample requirements of these techniques, the structures were solved in different membrane-mimetic solvents, making M2 a rare case for understanding the conformational dependence of membrane proteins on the environment.70,71 So far, the high-resolution structures of M2 include three crystal structures of the TM domain in the detergent octyl glucoside from pH 5.3 to pH 7.5;18,31 solution NMR structures of influenza A M2(18–60)17 and an AM2-BM2 chimera in dihexanoylphosphatidylcholine (DHPC) micelles at pH 7.5; SSNMR orientational structure of M2(22–62) in dioleyolphosphatidylcholine (DOPC)/dioleoylphosphatidylethanolamine (DOPE) bilayers at pH 7.519 and M2(22–46) in DMPC bilayers at both acidic and neutral pH;74 and MAS SSNMR structure of M2(22–46) in DMPC bilayers5 and dilauroylphosphati-dylcholine (DLPC) bilayers26 at neutral pH. Except for the M2(22–62) orientational structure, the other SSNMR structures were solved in the presence of the antiviral drug amantadine.
It is important to examine the type and number of experimental restraints and the underlying assumptions for the various M2 structures. For the crystal structures, the crystals diffracted to 3.5–1.65 Å, the highest resolution of which allowed the detection of bound water molecules in the pore31 whereas the lowest resolution could not pinpoint the direction of the polar amine of amantadine.18 For the solution NMR structure of M2(18–60), the most important restraints were 27 intermonomer and drug-protein NOEs, 23 side chain dihedral angles, and 27 residual N–H dipolar couplings.17 The SSNMR experiments and structural constraints fall into two categories. Oriented-membrane SSNMR experiments involve glass-plate-aligned or bicelle-aligned membranes that are kept static during data acquisition.75,76 15N-1H dipolar couplings, which reflect the N–H bond orientations and in turn the helix orientation from the bilayer normal, are measured from 2D correlation spectra. In the solid state, the 15N-1 H dipolar couplings exhibit the full range of possible values, compared with the much smaller range in weakly aligned molecules in solution;77 thus, the SSNMR extracted N–H dipolar couplings are more sensitive to protein orientation. Compared with oriented-membrane SSNMR, MAS NMR experiments utilize unoriented hydrated proteoliposomes that are spun fast around an axis tilted by 54.7° from the magnetic field. MAS SSNMR gives a wide variety of structural information, including interatomic distances, torsion angles, rotamer structure, oligomeric number, molecular motion, and chemical structure.75,78–82 The inputs for the MAS SSNMR structure of amantadine-bound M2(22–46)5 [Fig. 1(B)] included drug-protein distances, side chain rotamers,83,84 chemical shifts,85 intermonomer distances,84 as well as N–H bond orientations.74 In general, SSNMR structures have fewer constraints than solution NMR and crystal structures, but these sparse constraints are measured in the more native environment of lipid bilayers and usually have high precision and accuracy.
Although various environmental factors such as pH, membrane-mimetic solvent, and drug differed among the various high-resolution studies, the overall shape of the TM domain is largely preserved (Fig. 2). All structures showed four helices assembled into a left-handed parallel bundle, with a tilt angle of 30°–35° for the N-terminal half of the helix in most cases. The only exception is the DHPC-micelle bound solution NMR structure, which showed a much smaller tilt angle of ~ 15°.17 The tilt angle of the C-terminal half of the helix differs by about 10° between the low-pH crystal structure and the high-pH SSNMR structures. All structures agreed on the relative positions of pore-facing versus lipid-facing side chains; however, key differences exist about the rotameric states of His37 and Trp41.17,30,31,84 It is worth noting that side chain conformations cannot be obtained from oriented-membrane SSNMR experiments, as they only measure backbone N–H bond orientations. The His37 and Trp41 rotamers in the recent oriented-membrane SSNMR structure of M2(22–62) were based on computational modeling.19
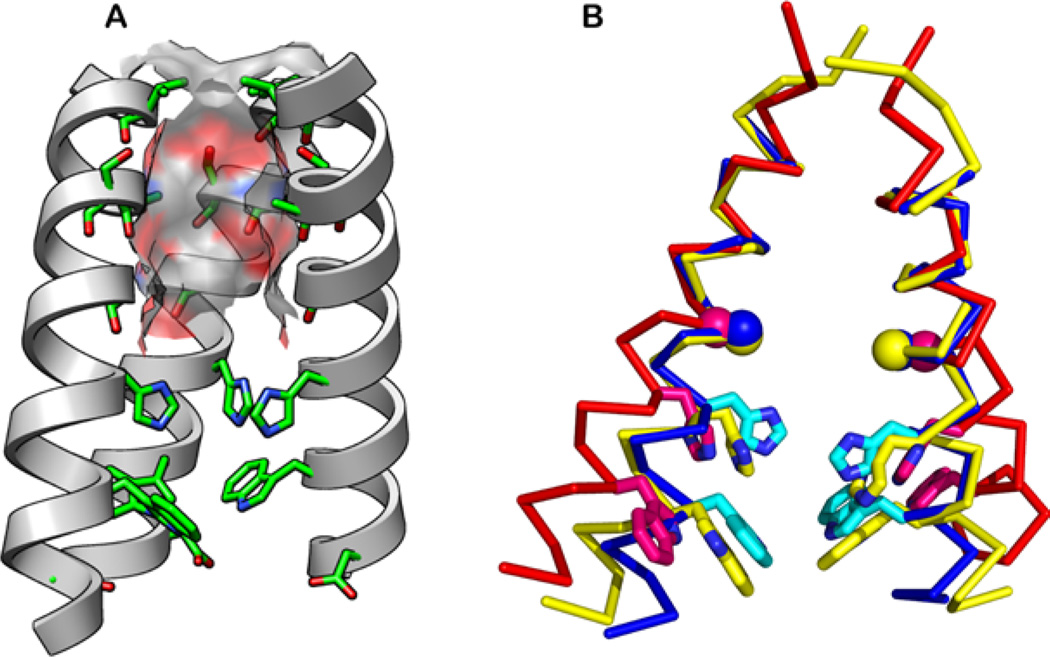
Figure 2.
Structures of the TM domain of M2. (A) The N-terminal pore is lined by the hydroxyl of Ser31 and backbone carbonyl groups (pictured structure is the 1.65Å crystal structure at pH 6.5, PDB: 3LBW). The molecular surface of the channel is color-coded with the oxygen atoms in red, carbon in gray, and nitrogen in blue. The “front” helix has been removed for clarity. (B) Superimposed solid-state NMR structure at pH 7.5 (2L0J, yellow), the 1.65-Å crystal structure at pH 6.5 (3LBW, blue), and the 3.5-Å crystal structure at pH 5.3 (3C9J, red). His37 and Trp41 side chains (sticks) and Gly34 Cα (ball) are shown.
Compared with the TM domain, much larger structural variations are seen for the cytoplasmic helix between the solution NMR structure17 and the oriented-membrane SSNMR structure.19 In DHPC micelles, the four cytoplasmic helices form a helical bundle that is well separated from the TM domain and from the putative micelle surface by a dynamic loop.17 In contrast, in DOPC/DOPE bilayers, this cytoplasmic helix is tightly connected to the TM domain by a rigid Leu46–Phe47 turn19 and lies at the membrane-water interface. The interfacial location is consistent with earlier H/D exchange data showing very slow exchange for the hydrophobic residues in this domain.86
Mechanism of proton transfer from high-resolution structures
The availability of the high-resolution structures has provided atomic-level understanding of the structural and dynamic basis of proton conduction. With these structural data, we can now address how protons are transported through the aqueous pore to the His37 tetrad, how charge is stabilized in the His37 cluster, and how protons are released into the virus interior.
The N-terminal aqueous pore
The structures of the TM domain show that the pore is replete with H-bond acceptors but deficient in H-bond donors. The pore is lined by small side chains (Val27, Ala30, Ser31, and Gly34) that allow substantial exposure of the main chain atoms to the pore. Carbonyl groups in helices are able to accept two hydrogen bonds, one from the main-chain amide NH and one from solvent water or a Ser/Thr located one turn away, whereas the amide NH group can donate only a single hydrogen bond. The walls of the N-terminal aqueous pore are therefore positioned to accept, but not donate, hydrogen bonds. In the neutral state of the tetramer, the His37 tetrad has the lone pair of its Nδδ projecting toward the pore, ready to accept a hydrogen bond [Figs. 1(B) and 2(A)]. By contrast, water is at its lowest energy state when forming equal numbers of H-bond donors and acceptors. Thus, water in the channel experiences a deficit of H-bond donors when His37 is neutral, which provides a strong driving force for binding H-bond donors, such as ammonium groups in drugs or hydronium ion-like species during proton conduction. This provides a rationale for the elevated pKa of the first two protonation events of the His37 tetrad, which introduces H-bond donors into the system.
Dynamic, water-mediated stabilization of charge in the His37 tetrad
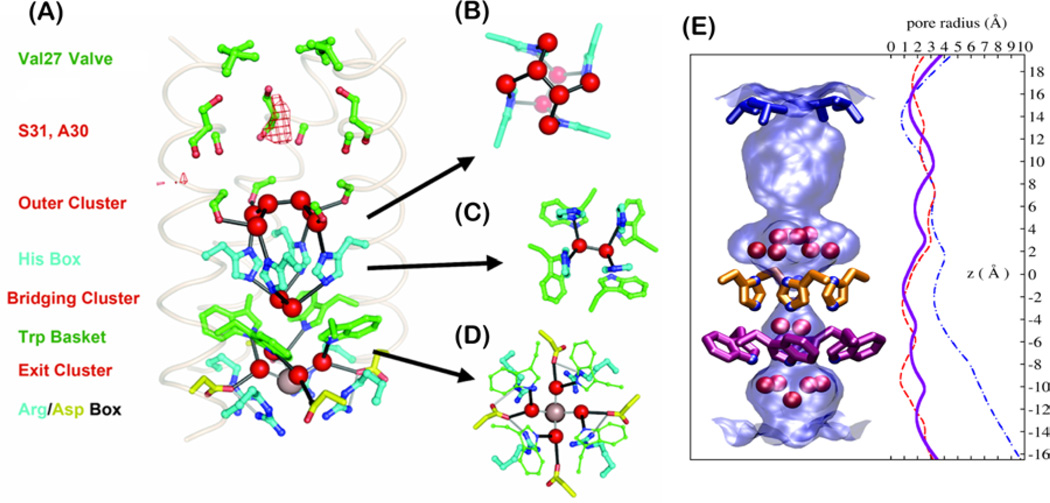
The 1.65 Å crystal structure at pH 6.531 provides a very high-resolution snapshot of the protein at intermediate pH where it appears to be in the +2 state (Fig. 3). The structure shows residues essential for conduction interspersed with layers of well-ordered water molecules. The N-terminal portion of the channel shows a region of diffuse density, suggestive of disordered water molecules [mesh in Fig. 3(A)]. Below this region, three layers of water clusters are found between pore-facing Gly34, His37, Trp41, and Asp44 to form a continuous pathway for proton conduction. The four side chains of His37 are packed into a box-like structure. The imidazoles are not connected by direct hydrogen bonds but by highly structured water molecules above and below, supporting the His37-water model for proton shuttling. Two bridging water molecules are seen between His37 and Trp41. As the channel is roughly in the +2 state at pH 6.5, these bridging water molecules are hypothesized to delocalize the excess protons, thus reducing the energy barrier for proton storage in the hydrophobic part of the lipid membrane. The crystal structure was obtained at cryogenic temperatures where water is frozen. Complementary SSNMR30,87 and two-dimensional IR experiments88 carried out at ambient temperature, as well as MD simulations,89,90 were also reported and showed that the water molecules near His37 are in fact dynamic. Even water molecules between His37 and Trp41 are found to be dynamic based on 2D 15N-1H correlation SSNMR spectra.30 These dynamic water molecules likely solvate the Cu2+ that was recently determined to bind within this His-Trp aromatic cage.91 Two-dimensional MAS SSNMR spectra showed that water interaction with M2 is pH dependent: the water–protein cross-peaks build up faster at low pH than at high pH, and the different buildup rates indicate that the water-exposed surface area of the tetramer is ~ 33% larger at low pH (open state) than at high pH (closed state).87 Thus, the channel pore widens at low pH, which is consistent with the observed increase of the helix tilt angle at low pH.18,31,73,74 Taken together, these results depict a highly solvated His37 tetrad in the multiply charged states.
Figure 3.
(A) The proton conduction pathway seen in a 1.65-Å resolution crystal structure including three clusters of crystallographic waters. (B–D) A second perspective of the outer (B), bridging (C), and exit (D) clusters viewed normal to the membrane plane. (E) The surface of the pore (light blue shading) is shown along with the crystallographic waters (red spheres). Val27, His37, and Trp41 residues are rendered in blue, orange, and magenta, respectively. Pore radius profiles are plotted for the high-resolution crystal structure (3LBW, blue solid line), low-pH (3C9J, blue dash-dotted line), and amantadine-inhibited (red dashed line) structures.
SSNMR experiments yielded three conclusive pieces of evidence that His37 rapidly exchanges protons with the surrounding water molecules. First, N spectra showed that the imidazolium nitrogens interchange between the unprotonated and protonated states, manifested as 15N intensities halfway between the limiting frequencies of the N and NH peaks. The linewidths of the 15N exchange peaks indicate an exchange rate of 4.5 × 105 s−1. This N <–> NH exchange rate is about two orders of magnitude larger than the proton conduction rate, indicating that most exchange events do not result in conduction of protons into the virus, but rather occur between multiple His37 residues through the intervening water (vide infra). The larger His-water proton exchange rate compared with the proton conduction rate supports the delocalization of the protons over the His37 tetrad and the surrounding water. The 15N exchange peaks are the highest between pH 5 and 6, the pH range of the endosome, where the +2 and +3 states dominate.29 Second, as the 15N exchange peaks do not in themselves indicate that the exchange partner of His37 is water, the chemical shifts of the imidazole HN protons were recently measured, and were found to lie at the water frequency at high temperature.92 This observation indicates that the proton-exchange partner of His37 is water, rather than another histidine. Third, the N–H bond lengths of the imidazole rings were found to be elongated (1.11 Å) from the covalent bond length at low pH,30 whereas at high pH, the Nε2-H bond of neutral His37 remains short and strongly covalent (1.03 Å). Thus, the low-pH imidazolium forms hydrogen bonds with water molecules.
The microsecond His37-water proton exchange is accompanied by reorientation of the imidazolium rings at acidic pH.30 In a cholesterol-containing complex lipid membrane that immobilizes the backbone of the TM helix, the imidazolium side chains show motionally averaged dipolar couplings. The reduced order parameters, which depend on the reorientation angles of motion, indicate that the imidazolium reorients around the Cβ-Cγ bond by ~ 45°. This motion points the unprotonated nitrogen to the acidic N-terminal side and the protonated nitrogen to the high-pH C-terminal side to facilitate proton transfer with water. The ring reorientation occurs at least 105 times per second, and the energy barrier was measured to be at least 60 kJ mol−1 based on the temperature dependence of the dipolar couplings. This minimum barrier is consistent with the energy barrier of 50–120 kJ mol−193,94, for 180° ring flips of imidazole model compounds. Moreover, the minimum energy barrier is consistent with the proton conduction barrier of ~ 100 kJ mol−1 estimated from temperature-dependent single-channel proton conductivities,95 indicating that the conformational change of the His37 side chain constitutes the highest energy barrier in the conduction process.
A low-barrier hydrogen bond between His37 residues?
While the above experimental data indicate that the His37 forms hydrogen bonds with water, a second model suggests that His37 instead hydrogen bonds directly to one another, through a LBHB between Nδ1 of a neutral imidazole and Nε2—H of a cationic imidazolium in the +2 state of the channel.19,28 The primary rationale for this model is the early observation of a high pKa (8.2) for the first two protonation steps of the His37 tetrad, suggesting high basicity of the His residues and prompting the need to explain how the excess protons are stabilized before conduction occurs. However, the measured phenomenological pKa values not only reflect the intrinsic basicity of the side chains but also include a statistical factor due to the number of energetically degenerate permutations for arranging the protons in a tetramer. In particular, the first protonation can occur at any of the four histidines; thus, each His37 is in fact less protonatable than the tetrad pKa suggests. The statistical factor for the first pKa is log(4) = 0.6, so that the basicity of the individual His37 decreases to 7.6, which is within one standard deviation of the average pKa (6.6 ± 1.0) for partially or fully buried histidines in proteins. For the first tetrad pKa of 7.6 measured in the virus-mimetic membrane, the modified value would be 7.0, even closer to the average histidine pKa in proteins. We suggest that the stabilizing interactions of water molecules polarized by amide carbonyls, cation-π interactions between His37 and Trp41, and electrostatic interactions from Asp44, explain the modest increase in the first pKa of His37.
While these energetic and statistical considerations largely remove the need for proposing a His–His LBHB, experimental data also indicate an absence of LBHB. The original 15N MAS NMR spectra lack specific peaks that are required for LBHB.28 The orientational NMR structure of M2(22–62) containing the LBHB was computed without experimental side chain constraints,19 and the putative LBHB was used as a starting distance restraint to enforce the expected geometry during MD simulations. The measured His37 rotamer was trans–trans for both χ1 and χ2,30 which places the Nδ1-Nε2 vector parallel to the channel axis, thus making it impossible to establish an Nε2-H…Nδ1 hydrogen bond. Finally, in the +2 state proposed to contain the LBHB, no imidazole-imidazolium 13C-13C cross-peaks were observed in 2D spectra,29 indicating that the His37 rings are not closely packed. Therefore, all experimental evidence so far indicates that the LBHB-stabilized dimer model, while interesting, does not represent the dominant equilibrium structure of M2 in the +2 charged state.
Release of protons into the virus interior
The final step in proton conduction involves release of protons from the His37 cluster into the viral interior. This step occurs through the transient formation of a conformational form that “opens” the Trp gate and then closes upon diffusion of proton into the interior of the virus. Solution and SSNMR spectra show that M2 becomes increasingly dynamic17,30,97,98 and the pore increasingly hydrated87,88 with decreasing pH, supporting a conformational exchange model in which the helices move apart as the degree of protonation increases. As proton flux down the electrochemical gradient is a dynamic process in which the protein cycles between different protonation states, protein conformational changes can be expected for proton conduction. Examination of the multiple high-resolution structures of M2 suggests possible main chain motions that might assist proton movement out of the channel. The aligned structures in Figure 2(B) show a trend for the C-terminus of the channel to dilate with decreasing pH. For example, solution NMR, SSNMR, and X-ray structures in the protonation states between 0 and +2 show a closed-off C-terminus, whereas the crystal structure at more acidic pH shows a more open C-terminus. In the most dilated structure, the TM helix is straight, causing the helices to diverge beyond a common point of closest approach near the N-terminus, resulting in dilation near the C-terminus. This C-terminal dilation is consistent with the increased hydration and dynamics of the protein at low pH. In the less dilated structures, a slight bend near Gly34 is present, which keeps the helices close together at the C-terminus. It has been suggested that the dilation in the low pH structure of the M2 bundle is a crystallographic artifact arising from packing between bundles, but recent crystal structures from lipidic cubic phases confirmed this structure in crystals devoid of interbundle contacts (unpublished results).
Asymmetric bundles of the TM domain have been observed18 that have hybrid characteristics between the fully dilated and more restricted bundles. Thus, only a single helix might transiently change conformations to release a proton. SSNMR spectra, both oriented98 and MAS,97 showed the existence of multiple conformations for key residues such as His37, Gly34, and Val27. The exact nature of these conformations and the rates of interconversion between them are not yet well understood and require further studies. So far, the larger line broadening of the low-pH spectra suggests small-amplitude conformational dynamics on the microsecond timescale, whereas large-amplitude motion between very different states, which should exhibit large chemical shift differences, have not been detected.
In addition to the backbone structural differences at different pH, significant variations in the Trp41 side chain conformation were observed from different structures. In the rimantadine complex solved by solution NMR at pH 7.5,17 the six-membered benzenoid rings of the indole point to the center of the pore, and pack tightly together to occlude the C-terminal end of the channel [Fig. 4(A)]. This rotamer (t-105, with χ1 at the trans conformation and a χ2 of ca. −120°) was obtained from residual Nε1-Hε1 dipolar coupling and NOEs.17 In comparison, the pH 6.5 crystal structure shows a more open Trp gate31 due to a t90 rotamer (χ2 = +80°, which is ca. 180° flipped from the t-105 rotamer). This projects the polar five-membered pyrrole ring into the channel [Fig. 4(B)]. This t90 rotamer was also found from 19F-19F distances measured between Hζ3-fluorinated Trp41 using SSNMR.84 However, these 19F SSNMR experiments were carried out at high pH, thus contradicting the solution NMR result. This discrepancy could result from the different solvents (detergents vs. lipid bilayers) used in the solution and SSNMR samples or from the different lengths of the M2 construct, thus additional experiments are necessary to clarify the Trp41 rotameric state. Even with the same t90 rotamer, the crystal structure solved at pH 5.3 shows larger separations among the indole rings than at pH 6.5 due to the backbone dilation [Fig. 2(B)]. These changes support the original model that Trp41 acts as a gate that moves out of the way when His37 reaches a critical protonation state, thus allowing protons to enter the virus.
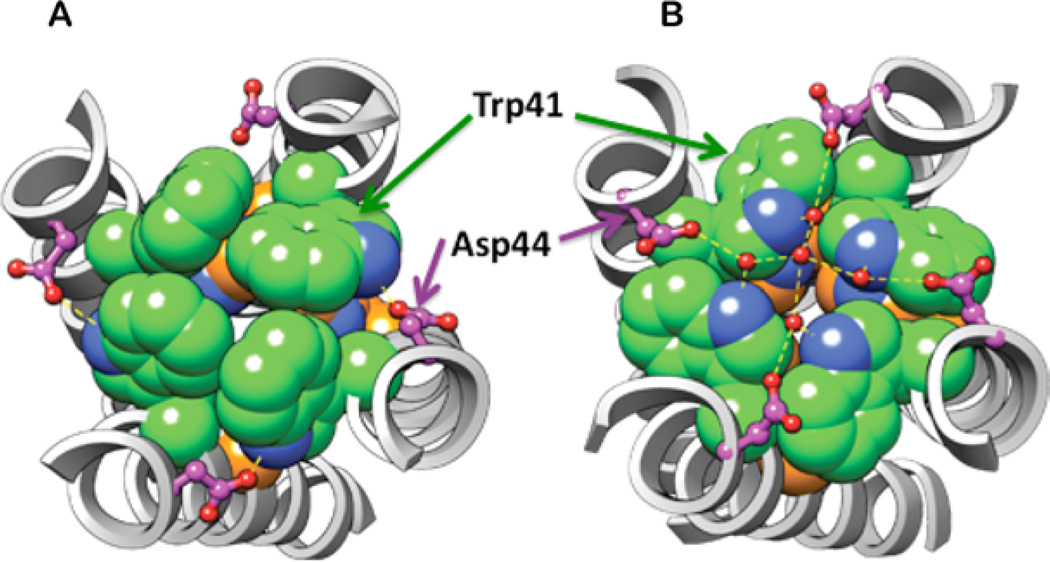
Figure 4.
The Trp41 rotamer and Trp41-Asp44 contact in the (A) high pH, drug-bound solution NMR structure (2RLF), and (B) the pH 6.5 X-ray crystal structure (3LBW). A larger pore radius at Trp41 is found in B due to the 180° χ2 angle change. The Trp41 side chains are shown in spheres for Trp41 (green for C, blue for N), and in ball-and-stick for Asp44 (pink C, blue O). His37 is shown as spheres with orange for C, blue for N. His37’s side chain is almost fully occluded by Trp41 in (A); in (B), it has slightly more accessibility. Crystallographically defined water molecules are shown in small red spheres in B, with hydrogen bonds shown in dashed lines.
Asp44 appears to play a critical role in stabilizing the Trp41 indole in distinct conformations. In the high-pH solution NMR structure, Asp44 is positioned to interact directly with the indole proton in about half of the members of the structural ensemble.17 In the pH 6.5 crystal structure, the Asp44 side chains interact indirectly with Trp41 via a small cluster of water molecules.31 Thus, Asp44 may contribute to gating by stabilizing the Trp41 gate in the fully closed and/or partially closed states until a critical number of protons accumulate on the His37 tetrad. In addition, the negative charge of Asp44 might stabilize the positive charge accumulated on the His37 tetrad. These conclusions are consistent with electrophysiological data showing that Asp44 mutations resulted in proton-selective and amantadine-sensitive channels with enhanced conductance in the physiological pH range,21,68,69 indicating that His37, Trp41, and Asp44 all interact to regulate both the pH dependence of conductance and channel gating.
Drug-binding site and inhibition mechanism from high-resolution structures
As described above, mutagenesis and whole-cell electrophysiology have long suggested the N-terminal pore to be the drug-binding region, as amantadine-resistant mutations such as V27A, A30T, S31N, and G34E21,99 are all located in this region. Thus, it was a major surprise when the solution NMR structure of M2(18–60) showed rimantadine NOEs to lipid-facing residues between Leu40 and Arg45 near the C-terminus of the TM domain, but no NOEs to the N-terminal pore residues.17 In contrast, the crystal structure of M2TM at pH 5.3 found electron densities of the right shape and size for the drug in the pore, surrounded by Val27, Ala30, Ser31, and Gly34.18 Subsequent structural and functional experiments showed that the pore site was the physiologically relevant site, whereas the surface site is nonspecific and has low affinity, and much was learned in the process of resolving this controversy.
Because both solution NMR and crystal structures were obtained using samples reconstituted in detergent micelles, which is an imperfect mimic of lipid bilayers, it was important to show how the drug bound to M2 in a true bilayer environment by SSNMR. Measurement of 13C-2H distances between 13C-labeled M2TM and perdeuterated amantadine in lipid bilayers revealed how the drug concentration affected the binding site.5 At the stoichiometric ratio of one drug per tetramer, only N-terminal residues (Val27, Ser31, and Gly34) showed REDOR dipolar dephasing by the deuterium spins. Only when excess amantadine was added did Asp44 in the surface site show dipolar dephasing. Thus, the first equivalent of drug binds to the N-terminal pore, while excess drugs bind to the surface site with lower affinity. As 2H-quadrupolar splitting is sensitive to molecular motion and orientation, the 2H-spectra of the perdeuterated amantadine revealed the orientation of the drug at the two binding sites. The Ser31-proximal drug is mostly upright, with the threefold molecular axis parallel to the channel axis [Fig. 1(B)], whereas the Asp44-bound drug is tilted by 37° or 80° from the membrane normal. The same tilted orientation is also adopted by lipid-bound drug in the absence of the protein; thus, the surface binding site results from nonspecific association of excess drugs from the lipid membrane. Quantitative analysis of the 13C-2H REDOR data resulted in six distance constraints between the perdeuterated adamantane cage and the Val27 side chain, Ser31, and Gly34. These protein-drug distances provided the crucial constraints for a high-resolution structure that utilized only bilayer-based SSNMR data [Fig. 1(B)].5
While these data clearly showed that the drug bound to the N-terminal pore in lipid bilayers with high affinity, there remained the possibility that the results were skewed by the fact that the TM construct lacked the cytoplasmic amphipathic helix. This motivated a series of more biological experiments using truncations and site-specific mutations, which ultimately showed that the cytoplasmic helix was unimportant for channel function but very important for stabilizing membrane curvature during virus budding.8,16,24 The results suggested that under conditions used for structure determination, the cytoplasmic helix was actually destabilizing the TM conformation required for drug binding. Supporting this conclusion, Chou and coworkers solved a structure of a chimeric protein consisting of the N-terminal region of M2 and the C-terminal helical region of a homologue of the influenza B virus, in which rimantadine was observed to bind in the pore only.72 The same pore-binding site was found in the longer M2 construct in phospholipid bilayers, as long as the membrane does not contain cholesterol and sphingomyelin.100 These results reveal a complex interaction between the cytoplasmic helix and the membrane-mimetic solvent. 31P NMR spectra and lipid–protein correlation MAS experiments indicate that the amphipathic helix, cholesterol, and sphingomyelin together promote an isotropic membrane domain with diameters less than ~ 30 nm.101 This high curvature likely perturbed the TM helix assembly, which in turn interfered with drug binding to the pore. A correlation is found between high membrane curvature and less drug binding to the pore. Thus, the amphipathic helix exerts an allosteric effect on the drug-competent conformation of the TM helical bundle.
Mutations that place hydrophobic residues in the water-filled pore of the channel physically occlude the channel and drastically decrease its conductance. In a similar manner, the adamantane moiety of amantadine fits into the channel with excellent geometric complementarity, doubtlessly contributing to blocking of protons. Drug binding causes a cascade of structural and dynamical changes to the channel. Water–protein cross-peak significantly decreased in intensity, indicating channel dehydration.87 The unprotonated 15N peak of His37 persisted to lower pH, indicating decrease of the His37 pKa.28 The 15N exchange peak at low pH was lost upon drug binding. Backbone chemical shifts of the low-pH protein adopt high-pH values. Finally, imidazole dipolar couplings revert to rigid-limit values, indicating immobilization of the His37 side chain.28 Thus, drug binding dehydrates the channel, which prevents protonation and chemical exchange of the His37, which in turn prevents backbone dilation, thus stopping side chain reorientation. All these effects abolish proton relay into the virus.
Design of broader-spectrum drugs with high affinity
There is an urgent need for new drugs that inhibit drug-resistant mutants, particularly S31N. The design of new drugs has been greatly advanced by the above structural studies as well as numerous computational studies that probe both the location and the driving force for binding.21,25,31,90,102–110 Amantadine and rimantadine have amphiphilic structures with a polar amine head and an apolar adamantyl or adamantylethyl group. Structure–activity relationships have shown that a variety of apolar substituents can replace the adamantyl substituent, and that a cationic primary ammonium group is optimal for high-affinity binding, as tertiary amines, alcohols, and other neutral groups tend to have lower affinity (secondary amines can be tolerated in some cases).111 The effectiveness of primary amines suggested that the charged amine (ammonium) group might mimic hydronium ions, formed as protons percolate through the outer pore to His37. Indeed, MD simulations of amantadine in the channel showed that, on average, its ammonium group was hydrated by four water molecules in a square planar array, and this hydrate was further stabilized by hydrogen-bonding to four carbonyl groups from Ala30.25,90,107
Thus, we can now understand the affinity of amantadine for the channel in terms of the properties of carbonyl groups on the surface of helices. About half of the solvent-accessible carbonyls in helices have an additional hydrogen bond to water. They can receive hydrogen bonds to waters to stabilize hydrated ammonium or hydronium ions. However, they can be easily dehydrated to hydrophobically stabilize the binding of an adamantyl substituent. The apolar isopropyl group of Val27 caps the site, resulting in geometric and physiochemical complementarity with amantadine and rimantadine in the WT.
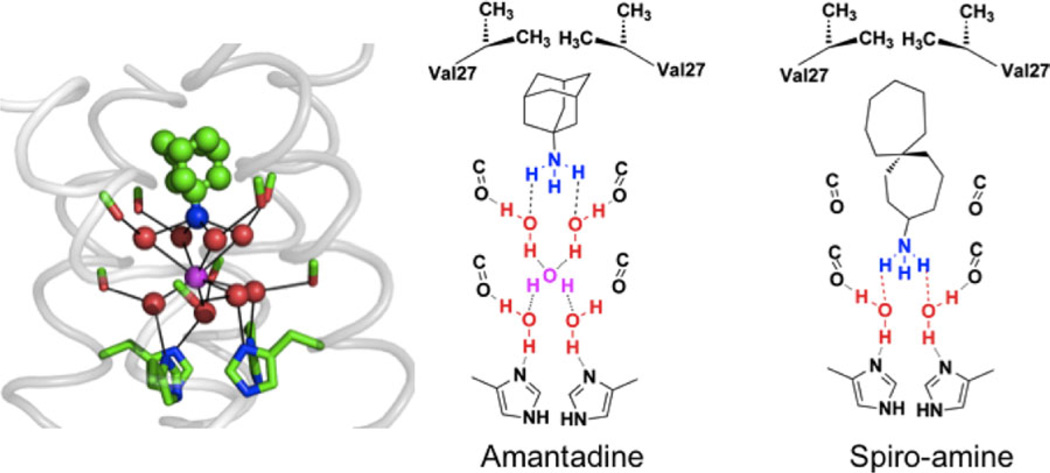
MD simulations of other complexes indicated the presence of additional sites capable of stabilizing ammonium or hydronium one turn up in the helix from Ala30, at a site formed by the carbonyl of Val27 and the hydroxyl of Ser31, or one turn down where a set of four water molecules are strongly hydrogen-bonded by the carbonyl groups of Gly34 and the imidazoles of His37. By targeting the ammonium group to the lower putative hydronium-binding site deeper in the pocket (Fig. 5), it was possible to engineer spiro-compounds with greater affinity for WT M2.107
Figure 5.
Snapshot from a simulation of amantadine with WT (far left). Water molecules (red) associate with carbonyl groups (green/red sticks) in a square planar array. This array of water molecules can stabilize the bound ammonium group of amantadine (green and blue bound drug) or a centrally located water molecule (magenta). The remaining panels show vertical slices of the channel in schematic form, showing how a longer inhibitor than amantadine places its ammonium group deeper in the channel and displaces more water molecules.
For drug-resistant mutants, MD simulations suggested that V27A and L26F had wider openings near the top of the channel, leading to a poorer fit and decreased hydrophobic burial of the adamantyl group of amantadine.107 The water structure in the central and lower portion of the channel, however, appeared to be retained. This led to the design of spiro-bicyclic and spiro-adamantane amines107 that targeted not only WT, but also V27A and L26F mutants with IC50s similar to or better than that of amantadine inhibition of the WT channel. In MD simulations, these drugs shifted upward in V27A, to allow their alkyl groups to fill the larger cavity near the channel entrance; their ammonium groups occupy the upper site in V27A but the lower aqueous site in WT. SSNMR data confirmed that the drugs bound directly to the targeted site.107 The potencies of these inhibitors were further demonstrated in binding and plaque reduction assays. These results demonstrate the power of MD simulations to probe the mechanism of drug binding and to guide design of inhibitors of targets that had previously appeared to be undruggable.
In addition to adamantane drugs, other inhibitors of M2 have been studied functionally, and some of these inhibitors were characterized structurally. Among divalent metal ions, Cu2+ displayed significant inhibitory effects, with an equilibrium dissociation constant of ~ 2 µM4 (cf. amantadine’s IC50 of ~ 16 µM22). Cu2+ inhibition exhibits similar functional features as amantadine: it is sensitive to pH and applied voltage, is competitive with the hydrophobic drug BL-1743,112 and inhibits both inward and outward currents.4 SSNMR studies of Cu2+ -bound M2TM using paramagnetic relaxation enhancement effects showed that the Cu2+ binding site is His37 Nε2, between His37 and Trp41, which explains the relatively slow dissociation of the Cu2+ ion.91 Therefore, compared with amantadine, Cu2+ directly targets the heart of the proton-conducting His37 instead of the N-terminal pore-facing residues, which suggests new routes for inhibitor design to target the S31N mutant.
Acknowledgment
The authors thank the many students and postdoctoral fellows in the two groups who have contributed to this work.
Grant sponsor: NIH; Grant numbers: GM088204 (M.H.); GM56423, GM54616, and AI74571 (W.F.D.); Grant sponsor: NSF; Grant number: MCB0543473 (M.H.).
Footnotes
Conflict of interest: WFD was the scientific founder of InfluMedix.
References
- 1.Hay AJ, Wolstenholme AJ, Skehel JJ, Smith MH. The molecular basis of the specific anti-influenza action of amantadine. EMBO J. 1985;4:3021–3024. doi: 10.1002/j.1460-2075.1985.tb04038.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Park EK, Castrucci MR, Portner A, Kawaoka Y. The M2 ectodomain is important for its incorporation into influenza A virions. J Virol. 1998;72:2449–2455. doi: 10.1128/jvi.72.3.2449-2455.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pinto LH, Dieckmann GR, Gandhi CS, Papworth CG, Braman J, Shaughnessy MA, Lear JD, Lamb RA, DeGrado WF. A functionally defined model for the M2 proton channel of influenza A virus suggests a mechanism for its ion selectivity. Proc Natl Acad Sci USA. 1997;94:11301–11306. doi: 10.1073/pnas.94.21.11301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gandhi CS, Shuck K, Lear JD, Dieckmann GR, DeGrado WF, Lamb RA, Pinto LH. Cu(II) inhibition of the proton translocation machinery of the influenza A virus M2 protein. J Biol Chem. 1999;274:5474–5482. doi: 10.1074/jbc.274.9.5474. [DOI] [PubMed] [Google Scholar]
- 5.Cady SD, Schmidt-Rohr K, Wang J, Soto CS, DeGrado WF, Hong M. Structure of the amantadine binding site of influenza M2 proton channels in lipid bilayers. Nature. 2010;463:689–692. doi: 10.1038/nature08722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pinto LH, Holsinger LJ, Lamb RA. Influenza virus M2 protein has ion channel activity. Cell. 1992;69:517–528. doi: 10.1016/0092-8674(92)90452-i. [DOI] [PubMed] [Google Scholar]
- 7.Pinto LH, Lamb RA. The M2 proton channels of influenza A and B viruses. J Biol Chem. 2006;281:8997–9000. doi: 10.1074/jbc.R500020200. [DOI] [PubMed] [Google Scholar]
- 8.Rossman JS, Jing X, Leser GP, Lamb RA. Influenza virus M2 protein mediates ESCRT-independent membrane scission. Cell. 2010;142:902–913. doi: 10.1016/j.cell.2010.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McCown MF, Pekosz A. Distinct domains of the influenza a virus M2 protein cytoplasmic tail mediate binding to the M1 protein and facilitate infectious virus production. J Virol. 2006;80:8178–8189. doi: 10.1128/JVI.00627-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ciampor F, Cmarko D, Cmarková J, Závodská E. Influenza virus M2 protein and haemagglutinin conformation changes during intracellular transport. Acta Virol. 1995;39:171–181. [PubMed] [Google Scholar]
- 11.Bright RA, Medina MJ, Xu X, Perez-Oronoz G, Wallis TR, Davis XM, Povinelli L, Cox NJ, Klimov AI. Incidence of adamantane resistance among influenza A (H3N2) viruses isolated worldwide from 1994 to 2005: a cause for concern. Lancet. 2005;366:1175–1181. doi: 10.1016/S0140-6736(05)67338-2. [DOI] [PubMed] [Google Scholar]
- 12.Wang C, Lamb RA, Pinto LH. Activation of the M2 ion channel of influenza virus: a role for the transmembrane domain histidine residue. Biophys J. 1995;69:1363–1371. doi: 10.1016/S0006-3495(95)80003-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tang Y, Zaitseva F, Lamb RA, Pinto LH. The gate of the influenza virus M2 proton channel is formed by a single tryptophan residue. J Biol Chem. 2002;277:39880–39886. doi: 10.1074/jbc.M206582200. [DOI] [PubMed] [Google Scholar]
- 14.Kochendoerfer GG, Salom D, Lear JD, Wilk-Orescan R, Kent SB, DeGrado WF. Total chemical synthesis of the integral membrane protein influenza A virus M2: role of its C-terminal domain in tetramer assembly. Biochemistry. 1999;38:11905–11913. doi: 10.1021/bi990720m. [DOI] [PubMed] [Google Scholar]
- 15.Salom D, Hill BR, Lear JD, DeGrado WF. pH-dependent tetramerization and amantadine binding of the transmembrane helix of M2 from the influenza A virus. Biochemistry. 2000;39:14160–14170. doi: 10.1021/bi001799u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ma C, Polishchuk AL, Ohigashi Y, Stouffer AL, Schón A, Magavern E, Jing X, Lear JD, Freire E, Lamb RA, DeGrado WF, Pinto LH. Identification of the functional core of the influenza A virus A/M2 proton-selective ion channel. Proc Natl Acad Sci USA. 2009;106:12283–12288. doi: 10.1073/pnas.0905726106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schnell JR, Chou JJ. Structure and mechanism of the M2 proton channel of influenza A virus. Nature. 2008;451:591–595. doi: 10.1038/nature06531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stouffer AL, Acharya R, Salom D, Levine AS, Di Costanzo L, Soto CS, Tereshko V, Nanda V, Stayrook S, DeGrado WF. Structural basis for the function and inhibition of an influenza virus proton channel. Nature. 2008;451:596–599. doi: 10.1038/nature06528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sharma M, Yi M, Dong H, Qin H, Peterson E, Busath D, Zhou HX, Cross TA. Atomistic mechanism of the influenza A proton channel from a structure solved in a lipid bilayer. Science. 2010;330:509–512. doi: 10.1126/science.1191750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sansom MSP, Kerr ID, Smith GR, Son HS. The influenza A virus M2 channel: a molecular modeling and simulation study. Virology. 1997;233:163–173. doi: 10.1006/viro.1997.8578. [DOI] [PubMed] [Google Scholar]
- 21.Balannik V, Carnevale V, Fiorin G, Levine BG, Lamb RA, Klein ML, Degrado WF, Pinto LH. Functional studies and modeling of pore-lining residue mutants of the influenza a virus M2 ion channel. Biochemistry. 2010;49:696–708. doi: 10.1021/bi901799k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jing X, Ma C, Ohigashi Y, Oliveira FA, Jardetzky TS, Pinto LH, Lamb RA. Functional studies indicate amantadine binds to the pore of the influenza A virus M2 proton-selective ion channel. Proc Natl Acad Sci USA. 2008;105:10967–10972. doi: 10.1073/pnas.0804958105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ohigashi Y, Ma C, Jing X, Balannick V, Pinto LH, Lamb RA. An amantadine-sensitive chimeric BM2 ion channel of influenza B virus has implications for the mechanism of drug inhibition. Proc Natl Acad Sci USA. 2009;106:18775–18779. doi: 10.1073/pnas.0910584106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rosenberg MR, Casarotto MG. Coexistence of two adamantane binding sites in the influenza A M2 ion channel. Proc Natl Acad Sci USA. 2010;107:13866–13871. doi: 10.1073/pnas.1002051107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Khurana E, Devane RH, Dal Peraro M, Klein ML. Computational study of drug binding to the membrane-bound tetrameric M2 peptide bundle from influenza A virus. Biochim Biophys Acta. 2011;2010:530–537. doi: 10.1016/j.bbamem.2010.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cady SD, Mishanina TV, Hong M. Structure of amantadine-bound M2 transmembrane peptide of influenza A in lipid bilayers from magic-angle-spinning solid-state NMR: the role of Ser31 in amantadine binding. J Mol Biol. 2009;385:1127–1141. doi: 10.1016/j.jmb.2008.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cady SD, Wang J, Wu Y, DeGrado WF, Hong M. Specific binding of adamantane drugs and direction of their polar amines in the pore of the influenza M2 transmembrane domain in lipid bilayers and dodecyl-phosphocholine micelles determined by NMR spectroscopy. J Am Chem Soc. 2011;133:4274–4284. doi: 10.1021/ja102581n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hu J, Fu R, Nishimura K, Zhang L, Zhou HX, Busath DD, Vijayvergiya V, Cross TA. Histidines, heart of the hydrogen ion channel from influenza A virus: toward an understanding of conductance and proton selectivity. Proc Natl Acad Sci USA. 2006;103:6865–6870. doi: 10.1073/pnas.0601944103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hu F, Schmidt-Rohr K, Hong M. NMR detection of pH-dependent histidine-water proton exchange reveals the conduction mechanism of a transmem-brane proton channel. J Am Chem Soc. 2012;134:3703–3713. doi: 10.1021/ja2081185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hu F, Luo W, Hong M. Mechanisms of proton conduction and gating by influenza M2 proton channels from solid-state NMR. Science. 2010;330:505–508. doi: 10.1126/science.1191714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Acharya A, Carnevale V, Fiorin G, Levine BG, Polishchuk A, Balannick V, Samish I, Lamb RA, Pinto LH, DeGrado WF, Klein ML. Structural mechanism of proton transport through the influenza A M2 protein. Proc Natl Acad Sci USA. 2010;107:15075–15080. doi: 10.1073/pnas.1007071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shimbo K, Brassard DL, Lamb RA, Pinto LH. Ion selectivity and activation of the M2 ion channel of influenza virus. Biophys J. 1996;70:1335–1346. doi: 10.1016/S0006-3495(96)79690-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mould JA, Drury JE, Frings SM, Kaupp UB, Pekosz A, Lamb RA, Pinto LH. Permeation and activation of the M2 ion channel of influenza A virus. J Biol Chem. 2000;275:31038–31050. doi: 10.1074/jbc.M003663200. [DOI] [PubMed] [Google Scholar]
- 34.Mould JA, Li HC, Dudlak CS, Lear JD, Pekosz A, Lamb RA, Pinto LH. Mechanism for proton conduction of the M(2) ion channel of influenza A virus. J Biol Chem. 2000;275:8592–8599. doi: 10.1074/jbc.275.12.8592. [DOI] [PubMed] [Google Scholar]
- 35.Mustafa M, Henderson DJ, Busath DD. Free-energy profiles for ions in the influenza M2-TMD channel. Proteins. 2009;76:794–807. doi: 10.1002/prot.22376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Leiding T, Wang J, Martinsson J, DeGrado WF, Arsköld SP. Proton and cation transport activity of the M2 proton channel from influenza A virus. Proc Natl Acad Sci USA. 2010;107:15409–15414. doi: 10.1073/pnas.1009997107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Decoursey TE. Voltage-gated proton channels and other proton transfer pathways. Physiol Rev. 2003;83:475–579. doi: 10.1152/physrev.00028.2002. [DOI] [PubMed] [Google Scholar]
- 38.Pielak RM, Chou JJ. Kinetic analysis of the M2 proton conduction of the influenza virus. J Am Chem Soc. 2010;132:17695–17697. doi: 10.1021/ja108458u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chizhmakov IV, Geraghty FM, Ogden DC, Hayhurst A, Antoniou M, Hay AJ. Selective proton permeability and pH regulation of the influenza virus M2 channel expressed in mouse erythroleukaemia cells. J Physiol. 1996;494:329–336. doi: 10.1113/jphysiol.1996.sp021495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Markley JL. Observation of histidine residues in proteins by means of nuclear magnetic resonance spectroscopy. Acc Chem Res. 1975;8:70–80. [Google Scholar]
- 41.Forrest LR, DeGrado WF, Dieckmann GR, Sansom MS. Two models of the influenza A M2 channel domain: verification by comparison. Fold Des. 1998;3:443–448. doi: 10.1016/S1359-0278(98)00061-3. [DOI] [PubMed] [Google Scholar]
- 42.Grambas S, Bennett MS, Hay AJ. Influence of amantadine resistance mutations on the pH regulatory function of the M2 protein of influenza A viruses. Virology. 1992;191:541–549. doi: 10.1016/0042-6822(92)90229-i. [DOI] [PubMed] [Google Scholar]
- 43.Shiraishi K, Mitamura K, Sakai-Tagawa Y, Goto H, Sugaya N, Kawaoka Y. High frequency of resistant viruses harboring different mutations in amantadine-treated children with influenza. J Infect Dis. 2003;188:57–61. doi: 10.1086/375799. [DOI] [PubMed] [Google Scholar]
- 44.Abed Y, Goyette N, Boivin G. Generation and characterization of recombinant influenza A (H1N1) viruses harboring amantadine resistance mutations. Antimicrob Agents Chemother. 2005;49:556–559. doi: 10.1128/AAC.49.2.556-559.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Suzuki H, Saito R, Masuda H, Oshitani H, Sato M, Sato I. Emergence of amantadine-resistant influenza A viruses: epidemiological study. J Infect Chemother. 2003;9:195–200. doi: 10.1007/s10156-003-0262-6. [DOI] [PubMed] [Google Scholar]
- 46.Deyde V, Garten R, Sheu T, Smith C, Myrick A, Barnes J, Xu X, Shaw M, Klimov A, Gubareva L. Genomic events underlying the changes in ada-mantane resistance among influenza A(H3N2) viruses during 2006–2008. Influenza Other Respi Viruses. 2009;3:297–314. doi: 10.1111/j.1750-2659.2009.00103.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Deyde VM, Xu X, Bright RA, Shaw M, Smith CB, Zhang Y, Shu Y, Gubareva LV, Cox NJ, Klimov AI. Surveillance of resistance to adamantanes among influenza A(H3N2) and A(H1N1) viruses isolated worldwide. J Infect Dis. 2007;196:249–257. doi: 10.1086/518936. [DOI] [PubMed] [Google Scholar]
- 48.Krumbholz A, Schmidtke M, Bergmann S, Motzke S, Bauer K, Stech J, Durrwald R, Wutzler P, Zell R. High prevalence of amantadine resistance among circulating European porcine influenza A viruses. J Gen Virol. 2009;90:900–908. doi: 10.1099/vir.2008.007260-0. [DOI] [PubMed] [Google Scholar]
- 49.Simonsen L, Viboud C, Grenfell BT, Dushoff J, Jennings L, Smit M, Macken C, Hata M, Gog J, Miller MA, Holmes EC. The genesis and spread of reassortment human influenza A/H3N2 viruses conferring adamantane resistance. Mol Biol Evol. 2007;24:1811–1820. doi: 10.1093/molbev/msm103. [DOI] [PubMed] [Google Scholar]
- 50.Bright RA, Shay DK, Shu B, Cox NJ, Klimov AI. Adamantane resistance among influenza A viruses isolated early during the 2005–2006 influenza season in the United States. JAMA. 2006;295:891–894. doi: 10.1001/jama.295.8.joc60020. [DOI] [PubMed] [Google Scholar]
- 51.Salter A, Ni Laoi B, Crowley B. Emergence and phylogenetic analysis of amantadine-resistant influenza a subtype H3N2 viruses in Dublin, Ireland, over six seasons from 2003/2004 to 2008/2009. Intervirology. 2011;54:305–315. doi: 10.1159/000321358. [DOI] [PubMed] [Google Scholar]
- 52.Tosh C, Murugkar HV, Nagarajan S, Tripathi S, Katare M, Jain R, Khandia R, Syed Z, Behera P, Patil S, Kulkarni DD, Dubey SC. Emergence of amantadine-resistant avian influenza H5N1 virus in India. Virus Genes. 2011;42:10–15. doi: 10.1007/s11262-010-0534-z. [DOI] [PubMed] [Google Scholar]
- 53.Ujike M, Ejima M, Anraku A, Shimabukuro K, Obuchi M, Kishida N, Hong X, Takashita E, Fujisaki S, Yamashita K, Horikawa H, Kato Y, Oguchi A, Fujita N, Tashiro M, Odagiri T. Monitoring and characterization of oseltamivir-resistant pandemic (H1N1) 2009 virus, Japan, 2009–2010. Emerg Infect Dis. 2011;17:470–479. doi: 10.3201/eid1703.101188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zaraket H, Saito R, Suzuki Y, Baranovich T, Dapat C, Caperig-Dapat I, Suzuki H. Genetic makeup of amantadine-resistant and oseltamivir-resistant human influenza A/H1N1 viruses. J Clin Microbiol. 2010;48:1085–1092. doi: 10.1128/JCM.01532-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kiselev OI, Komissarov AB, Stukova MA, Buzitskaia Zh V, Pisareva MM, Elpaeva EA, Danilenko DM, Konovalova NI, Gudkova TM, Grigor’eva VA, Smirnova TS, Slita AV, Romanovskaia-Roman’ko EA, Tsybalova LM, Sominina AA, Eropkin M, Grudinin MP. [The 2009 pandemic influenza in Russia. I. Diagnosis and molecular biological characteristics of the virus] Vopr Virusol. 2011;56:17–21. [PubMed] [Google Scholar]
- 56.Puzelli S, Facchini M, Di Martino A, Fabiani C, Lackenby A, Zambon M, Donatelli I. Evaluation of the antiviral drug susceptibility of influenza viruses in Italy from 2004/05 to 2009/10 epidemics and from the recent 2009 pandemic. Antiviral Res. 2011;90:205–212. doi: 10.1016/j.antiviral.2011.04.003. [DOI] [PubMed] [Google Scholar]
- 57.Zhou J, Zou L, Zhang X, Liao J, Ni H, Hou N, Wang Y, Li H, Wu J, Jonges M, Meijer A, Koopmans M, Ke C. Adamantane- and oseltamivir-resistant seasonal A (H1N1) and pandemic A (H1N1) 2009 influenza viruses in Guangdong, China, during 2008 and 2009. J Clin Microbiol. 2011;49:2651–2655. doi: 10.1128/JCM.00535-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dapat C, Suzuki Y, Kon M, Tamura T, Saito R, Dapat IC, Yamazaki O, Odagiri T, Fujisaki S, Suzuki H. Phylogenetic analysis of an off-seasonal influenza virus A (H3N2) in Niigata, Japan, 2010. Japan J Infect Dis. 2011;64:237–241. [PubMed] [Google Scholar]
- 59.Qu Y, Zhang R, Cui P, Song G, Duan Z, Lei F. Evolutionary genomics of the pandemic 2009 H1N1 influenza viruses (pH1N 1v) Virol J. 2011;8:250. doi: 10.1186/1743-422X-8-250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gubareva LV, Trujillo AA, Okomo-Adhiambo M, Mishin VP, Deyde VM, Sleeman K, Nguyen HT, Sheu TG, Garten RJ, Shaw MW, Fry AM, Klimov AI. Comprehensive assessment of 2009 pandemic influenza A (H1N1) virus drug susceptibility in vitro. Anti-vir Ther. 2010;15:1151–1159. doi: 10.3851/IMP1678. [DOI] [PubMed] [Google Scholar]
- 61.Furuse Y, Suzuki A, Kamigaki T, Shimizu M, Fuji N, Oshitani H. Reversion of influenza A (H3N2) virus from amantadine resistant to amantadine sensitive by further reassortment in Japan during the 2006-to-2007 influenza season. J Clin Microbiol. 2009;47:841–844. doi: 10.1128/JCM.01622-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schmidtke M, Zell R, Bauer K, Krumbholz A, Schrader C, Suess J, Wutzler P. Amantadine resistance among porcine H1N1, H1N2, and H3N2 influenza A viruses isolated in Germany between 1981 and 2001. Intervirology. 2006;49:286–293. doi: 10.1159/000094244. [DOI] [PubMed] [Google Scholar]
- 63.Stouffer AL, Ma C, Cristian L, Ohigashi Y, Lamb RA, Lear JD, Pinto LH, DeGrado WF. The interplay of functional tuning, drug resistance, and thermodynamic stability in the evolution of the M2 proton channel from the influenza Avirus. Structure. 2008:1067–1076. doi: 10.1016/j.str.2008.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Howard KP, Lear JD, DeGrado WF. Sequence determinants of the energetics of folding of a trans-membrane four-helix-bundle protein. Proc Natl Acad Sci USA. 2002;99:8568–8572. doi: 10.1073/pnas.132266099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Venkataraman P, Lamb RA, Pinto LH. Chemical rescue of histidine selectivity filter mutants of the M2 ion channel of influenza A virus. J Biol Chem. 2005;280:21463–21472. doi: 10.1074/jbc.M412406200. [DOI] [PubMed] [Google Scholar]
- 66.Okada A, Miura T, Takeuchi H. Protonation of histidine and histidine-tryptophan interaction in the activation of the M2 ion channel from influenza A virus. Biochemistry. 2001;40:6053–6060. doi: 10.1021/bi0028441. [DOI] [PubMed] [Google Scholar]
- 67.Takeuchi H, Okada A, Miura T. Roles of the histidine and tryptophan side chains in the M2 proton channel from influenza A virus. FEBS Lett. 2003;552:35–38. doi: 10.1016/s0014-5793(03)00781-6. [DOI] [PubMed] [Google Scholar]
- 68.Betakova T, Ciampor F, Hay AJ. Influence of residue 44 on the activity of the M2 proton channel of influenza A virus. J Gen Virol. 2005;86:181–184. doi: 10.1099/vir.0.80358-0. [DOI] [PubMed] [Google Scholar]
- 69.Chizhmakov IV, Ogden DC, Geraghty FM, Hayhurst A, Skinner A, Betakova T, Hay AJ. Differences in conductance of M2 proton channels of two influenza viruses at low and high pH. J Physiol. 2003;546:427–438. doi: 10.1113/jphysiol.2002.028910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cady SD, Luo WB, Hu F, Hong M. Structure and function of the influenza M2 proton channel. Biochemistry. 2009;48:7356–7364. doi: 10.1021/bi9008837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cross TA, Sharma M, Yi M, Zhou HX. Influence of solubilizing environments on membrane protein structures. Trends Biochem Sci. 2010 doi: 10.1016/j.tibs.2010.07.005. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Pielak RM, Oxenoid K, Chou JJ. Structural investigation of rimantadine inhibition of the AM2-BM2 chimera channel of influenza viruses. Structure. 2011;19:1655–1663. doi: 10.1016/j.str.2011.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wang J, Kim S, Kovacs F, Cross TA. Structure of the transmembrane region of the M2 protein H+ channel. Protein Sci. 2001;10:2241–2250. doi: 10.1110/ps.17901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hu J, Asbury T, Achuthan S, Li C, Bertram R, Quine JR, Fu R, Cross TA. Backbone structure of the amantadine-blocked trans-membrane domain M2 proton channel from Influenza A virus. Biophys J. 2007;92:4335–4343. doi: 10.1529/biophysj.106.090183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hong M. Oligomeric structure, dynamics, and orientation of membrane proteins from solid-state NMR. Structure. 2006;14:1731–1740. doi: 10.1016/j.str.2006.10.002. [DOI] [PubMed] [Google Scholar]
- 76.Opella SJ, Marassi FM. Structure determination of membrane proteins by NMR spectroscopy. Chem Rev. 2004;104:3587–3606. doi: 10.1021/cr0304121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tjandra N, Bax A. Direct measurement of distances and angles in biomolecules by NMR in a dilute liquid crystalline medium. Science. 1997;278:1111–1114. doi: 10.1126/science.278.5340.1111. [DOI] [PubMed] [Google Scholar]
- 78.Hong M. Solid-state NMR studies of the structure, dynamics, and assembly of beta-sheet membrane peptides and alpha-helical membrane proteins with antibiotic activities. Acc Chem Res. 2006;39:176–183. doi: 10.1021/ar040037e. [DOI] [PubMed] [Google Scholar]
- 79.Hong M, Zhang Y, Hu F. Membrane protein structure and dynamics from NMR spectroscopy. Annu Rev Phys Chem. 2012;63:1–24. doi: 10.1146/annurev-physchem-032511-143731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Luca S, Heise H, Baldus M. High-resolution solid-state NMR applied to polypeptides and membrane proteins. Acc Chem Res. 2003;36:858–865. doi: 10.1021/ar020232y. [DOI] [PubMed] [Google Scholar]
- 81.McDermott AE. Structure and dynamics of membrane proteins by magic angle spinning solid-state NMR. Annu Rev Biophys. 2009;38:385–403. doi: 10.1146/annurev.biophys.050708.133719. [DOI] [PubMed] [Google Scholar]
- 82.Tycko R. Solid-state NMR studies of amyloid fibril structure. Annu Rev Phys Chem. 2011;62:279–299. doi: 10.1146/annurev-physchem-032210-103539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hong M, Mishanina TV, Cady SD. Accurate measurement of methyl 13C chemical shifts by solid-state NMR for the determination of protein sidechain conformation: the influenza M2 transmembrane peptide as an example. J Am Chem Soc. 2009;131:7806–7816. doi: 10.1021/ja901550q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Luo W, Mani R, Hong M. Sidechain conformation and gating of the M2 transmembrane peptide proton channel of influenza A virus from solid-state NMR. J Phys Chem. 2007;111:10825–10832. doi: 10.1021/jp073823k. [DOI] [PubMed] [Google Scholar]
- 85.Cady SD, Hong M. Amantadine-induced conformational and dynamical changes of the influenza M2 transmembrane proton channel. Proc Natl Acad Sci USA. 2008;105:1483–1488. doi: 10.1073/pnas.0711500105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Tian C, Gao PF, Pinto LH, Lamb RA, Cross TA. Initial structural and dynamic characterization of the M2 protein transmembrane and amphipathic helices in lipid bilayers. Prot Sci. 2003;12:2597–2605. doi: 10.1110/ps.03168503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Luo W, Hong M. Conformational changes of an ion channel membrane protein detected through water-protein interactions using solid-state NMR spectroscopy. J Am Chem Soc. 2010;132:2378–2384. doi: 10.1021/ja9096219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ghosh A, Qiu J, DeGrado WF, Hochstrasser RM. Tidal surge in the M2 proton channel, sensed by 2D IR spectroscopy. Proc Natl Acad Sci USA. 2011;108:6115–6120. doi: 10.1073/pnas.1103027108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Khurana E, Dal Peraro M, DeVane R, Vemparala S, DeGrado WF, Klein ML. Molecular dynamics calculations suggest a conduction mechanism for the M2 proton channel from influenza A virus. Proc Natl Acad Sci USA. 2009;106:1069–1074. doi: 10.1073/pnas.0811720106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Carnevale V, Fiorin G, Levine BG, Degrado WF, Klein ML. Multiple proton confinement in the M2 channel from the influenza A virus. J Phys Chem C Nanomater Interfaces. 2010;114:20856–20863. doi: 10.1021/jp107431g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Su Y, Hu F, Hong M. Paramagnetic Cu(II) for probing membrane protein structure and function: inhibition mechanism of the influenza M2 proton channel. J Am Chem Soc. 2012;134:8693–8702. doi: 10.1021/ja3026328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Hong M, Fritzsching KJ, Williams J. Hydrogen-bonding partner of the proton-conducting histidine in the influenza M2 proton channel revealed from 1H chemical shifts. J Am Chem Soc. 2012 doi: 10.1021/ja307453v. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Fischbach I, Spiess HW, Saalwachter K, Goward GR. Solid state NMR spectroscopic investigations of model compounds for imidazole-based proton conductors. J Phys Chem. 2004;108:18500–18508. [Google Scholar]
- 94.Goward GR, Saalwächter K, Fischbach I, Spiess HW. Reorientation phenomena in imidazolium methyl sulfonate as probed by advanced solid-state NMR. Solid State Nucl Magn Reson. 2003;24:150–162. doi: 10.1016/S0926-2040(03)00043-2. [DOI] [PubMed] [Google Scholar]
- 95.Lin TI, Schroeder C. Definitive assignment of proton selectivity and attoampere unitary current to the M2 ion channel protein of influenza A virus. J Virol. 2001;75:3647–3656. doi: 10.1128/JVI.75.8.3647-3656.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Edgcomb SP, Murphy KP. Variability in the pKa of histidine side-chains correlates with burial within proteins. Proteins. 2002;49:1–6. doi: 10.1002/prot.10177. [DOI] [PubMed] [Google Scholar]
- 97.Hu F, Luo W, Cady SD, Hong M. Conformational plasticity of the influenza A M2 transmembrane peptide in lipid bilayers under varying pH, drug binding and membrane thickness. Biochim Biophys Acta. 2011;1808:415–423. doi: 10.1016/j.bbamem.2010.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Li C, Qin H, Gao FP, Cross TA. Solid-state NMR characterization of conformational plasticity within the transmembrane domain of the influenza A M2 proton channel. Biochim Biophys Acta. 2007;1768:3162–3170. doi: 10.1016/j.bbamem.2007.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Wang C, Takeuchi K, Pinto LH, Lamb RA. Ion channel activity of influenza A virus M2 protein: characterization of the amantadine block. J Virol. 1993;67:5585–5594. doi: 10.1128/jvi.67.9.5585-5594.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Cady SD, Wang T, Hong M. Membrane-dependent effects of a cytoplasmic helix on the structure and drug binding of the influenza virus M2 protein. J Am Chem Soc. 2011;133:11572–11579. doi: 10.1021/ja202051n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Wang T, Cady SD, Hong M. NMR determination of protein partitioning into membrane domains with different curvatures and application to the influenza M2 peptide. Biophys J. 2012;102:787–794. doi: 10.1016/j.bpj.2012.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Chuang GY, Kozakov D, Brenke R, Beglov D, Guarnieri F, Vajda S. Binding hot spots and amantadine orientation in the influenza a virus M2 proton channel. Biophys J. 2009;97:2846–2853. doi: 10.1016/j.bpj.2009.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Laohpongspaisan C, Rungrotmongkol T, Intharathep P, Malaisree M, Decha P, Aruksakunwong O, Sompornpisut P, Hannongbua S. Why amantadine loses its function in influenza m2 mutants: MD simulations. J Chem Inf Model. 2009;49:847–852. doi: 10.1021/ci800267a. [DOI] [PubMed] [Google Scholar]
- 104.Kozakov D, Chuang GY, Beglov D, Vajda S. Where does amantadine bind to the influenza virus M2 proton channel? Trends Biochem Sci. 2010;35:471–475. doi: 10.1016/j.tibs.2010.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Phongphanphanee S, Rungrotmongkol T, Yoshida N, Hannongbua S, Hirata F. Proton transport through the influenza A M2 channel: three-dimensional reference interaction site model study. J Am Chem Soc. 2010;132:9782–9788. doi: 10.1021/ja1027293. [DOI] [PubMed] [Google Scholar]
- 106.Astrahan P, Flitman-Tene R, Bennett ER, Krugliak M, Gilon C, Arkin IT. Quantitative analysis of influenza M2 channel blockers. Biochim Biophys Acta. 2010 doi: 10.1016/j.bbamem.2010.08.021. PMID: 20831860 {Medline} [DOI] [PubMed] [Google Scholar]
- 107.Wang J, Ma C, Fiorin G, Carnevale V, Wang T, Hu F, Lamb RA, Pinto LH, Hong M, Klein ML, DeGrado WF. Molecular dynamics simulation directed rational design of inhibitors targeting drug-resistant mutants of influenza A virus M2. J Am Chem Soc. 2011;133:12834–12841. doi: 10.1021/ja204969m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Wang J, Qiu JX, Soto CS, DeGrado WF. Structural and dynamic mechanisms for the function and inhibition of the M2 proton channel from influenza A virus. Curr Opin Struct Biol. 2011;21:68–80. doi: 10.1016/j.sbi.2010.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Gu RX, Liu LA, Wei DQ, Du JG, Liu L, Liu H. Free energy calculations on the two drug binding sites in the M2 proton channel. J Am Chem Soc. 2011;133:10817–10825. doi: 10.1021/ja1114198. [DOI] [PubMed] [Google Scholar]
- 110.Rungrotmongkol T, Yotmanee P, Nunthaboot N, Han-nongbua S. Computational studies of influenza A virus at three important targets: hemagglutinin, neuraminidase and M2 protein. Curr Pharm Des. 2011;17:1720–1739. doi: 10.2174/138161211796355083. [DOI] [PubMed] [Google Scholar]
- 111.Wang J, Cady SD, Balannik V, Pinto LH, DeGrado WF, Hong M. Discovery of spiro-piperidine inhibitors and their modulation of the dynamics of the M2 proton channel from influenza A virus. J Am Chem Soc. 2009;131:8066–8076. doi: 10.1021/ja900063s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Tu Q, Pinto LH, Luo G, Shaughnessy MA, Mullaney D, Kurtz S, Krystal M, Lamb RA. Characterization of inhibition of M2 ion channel activity by BL-1743, an inhibitor of influenza A virus. J Virol. 1996;70:4246–4252. doi: 10.1128/jvi.70.7.4246-4252.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]