Abstract
Heat shock proteins (HSPs), which are members of the chaperone family of proteins, are essential factors for cellular responses to environmental stressors, such as hyperthermia, and are antiapoptotic. The transcription of HSPs is mainly controlled by heat shock transcription factor 1 (HSF1). In response to environmental stress, HSF1 forms a trimer, undergoes hyperphosphorylation, and is translocated to the nucleus. In this study, we show that upon heat shock treatment of cells, a WW domain-containing propyl-isomerase, PIN1, is able to colocalize to and associate with phospho-HSF1 at Ser326 in the nucleus via its WW domain. This interaction is required for the DNA-binding activity of HSF1 and is consistent with the lower induction of HSPs in PIN1-deficient cells. This function of PIN1 is further demonstrated by in vivo refolding and survival assays, which have shown that PIN1-deficient cells are temperature sensitive and develop apoptosis upon exposure to an environmental challenge. Moreover, the reduced levels of HSPs in PIN1-deficient cells resulted in less efficient refolding of denatured proteins. Based on our results, we propose a novel role for PIN1 whereby it acts as a stress sensor regulating HSF1 activity in response to stress on multiple levels through the transcriptional activation of stress response elements in embryonic fibroblast cells, tumor cells, and neurons.
INTRODUCTION
Exposure of cells to environmental stress factors such as heat shock, heavy metals, and proteasome inhibition causes the induction of heat shock proteins (HSPs), which have been shown to have cytoprotective functions (1). HSP induction is regulated at the transcriptional level by heat shock factor 1 (HSF1), which recognizes the heat shock element (HSE) in the promoter of hsp genes (2). Under normal conditions, HSF1 is present as a monomer and localizes primarily to the cytoplasm. Upon the induction of stress via methods such as hyperthermia, proteasome inhibition by MG132 treatment and heavy metal treatment, HSF1 trimerizes and translocates to the nucleus (3, 4). In addition, rigorous mechanisms controlling HSF1 activation have been reported. For example, HSP70 and HSP90 stably associate with HSF1 under normal conditions, thereby preventing HSF1 activation (5). In cells exposed to heat, hyperphosphorylation of HSF1 has been observed (6–8), but the role of phosphorylation has remained controversial. For instance, Holmberg et al. demonstrated that calcium-/calmodulin-dependent protein kinase II (CaMKII) enhances both the level of in vivo Ser230 phosphorylation and transactivation of HSF1 (8). However, Ser303 is a target for robust, heat-inducible phosphorylation, corresponding to the inducible HSF1 sumoylation (9). The small ubiquitin-like modifier (SUMO) modification maintains HSF1 in its inactive form (10, 11). Guettouche et al. have described in detail the phosphorylation status of HSF1 in stressed cells and have systematically identified the phospho-residues involved in activation of downstream factors (6). The majority of these newly identified phosphorylation sites, such as Ser292, Ser326, Ser314, and Ser363, are serine residues located adjacent to proline residues. However, the significance of phosphorylation of these new sites remains to be elucidated.
Phosphorylation-dependent isomerization has recently been characterized as a posttranslational modification step that controls protein activity and conformation. This modification is catalyzed by a propyl-isomerase, PIN1 (peptidyl-prolyl cis-trans isomerase NIMA-interacting 1), that specifically recognizes phosphoserine/threonine-proline motifs via the WW domain at its amino terminus. The carboxy-terminal peptidyl-prolyl isomerase (PPIase) domain of PIN1 catalyzes the isomerization of the peptide bond, resulting in a conformational change in the substrate. PIN1 regulates various protein functions, including protein stability, transcriptional activity, catalytic activity, protein-protein interactions, and subcellular localization (12–15). Moreover, PIN1 has been reported to protect cells from a variety of stress stimuli. For example, Akiyama et al. have demonstrated that PIN1 protects mice from severe lipopolysaccharide-induced inflammation (16). Other studies have shown that PIN1 increases cell survival by preventing cell death induced by oxidative stress or DNA damage (17). However, the role of PIN1 in heat shock-induced HSF1 activation or HSP expression is still unclear.
A number of PIN1-regulated proteins are transcription factors, suggesting that PIN1 might also play a role in the regulation of HSF1. In the present study, we investigated the role of PIN1 in HSF1 activation after hyperthermia stress. We demonstrated that heat shock stress promotes phosphorylation-dependent association of PIN1 with HSF1 in the nucleus. Our results suggest that PIN1 is essential for the efficient interaction of HSF1 with DNA. Furthermore, PIN1 deficiency leads to the attenuation of HSP expression, protein refolding ability and cell viability. The involvement of PIN1 in heat shock-induced HSF1 activation suggests a model in which PIN1 binding to phosphorylated Ser326 of HSF1 is required for PIN1 to isomerize the phospho-Ser326-Pro327 bond to the trans-conformation, which enhances the DNA-binding activity of HSF1. Taken together, this study describes a novel function for PIN1 as an important mediator of HSF1 transcriptional regulation. Our results suggest that the PIN1-mediated modulation of HSP expression may be helpful for further understanding the diverse roles of PIN1.
MATERIALS AND METHODS
Cell culture and transfection.
Human HeLa, MCF7, and mouse embryonic fibroblast (MEF) cells were grown in Dulbecco modified Eagle medium (DMEM) supplemented with 10% fetal calf serum (FCS), 2 mM l-glutamine, and antibiotics (penicillin and streptomycin) in a humidified 5% CO2 chamber at 37°C. For all experiments, HeLa and MCF7 cells were seeded at 3 × 106 cells per 10-cm plate. Heat shock treatment was performed at 43°C in a water bath. In some experiments, cells were allowed to recover at 37°C. For efficient transfection, Lipofectamine 2000 (Invitrogen, Carlsbad, CA) was used according to the manufacturer's instructions.
Primary cortical neuron purification and culture.
Primary cortical neurons were prepared from embryonic brains (embryonic days 16 to 18) of rats or mice. The meninges were removed, and the cortical neurons were separated by mechanical dissociation and mild trypsinization. The cells were plated at a density of 5 × 106 cells/10-cm dish on poly-d-lysine-precoated plates. Neurobasal medium (Invitrogen, Karlsruhe, Germany) supplemented with 1.2 mM glutamine and 2% (vol/vol) B27 supplement (Invitrogen, Karlsruhe, Germany [20 ml/liter]) was used as the culture medium. After 48 h, neurons were treated with 5 μM cytosine arabinoside for another 48 h to inhibit non-neuronal cell growth. After 6 to 7 days of in vitro culture, the neurons were used for experiments.
Plasmid DNA.
The Flag-HSF1 plasmid was purchased from Addgene (plasmid 32537). The HSP70B-Luc plasmid was kindly provided by Barry Trink (Division of Head and Neck Cancer Research). The mouse PIN1 expression plasmid was purchased from Origene. The 3HSE-Luc reporter plasmid was constructed by the insertion of three repeats of the heat shock element (HSE) into the pGL3-Basic vector.
Genotyping.
The PIN1 heterozygous knockout mouse model was kindly provided by Anthony Means (Department of Pharmacology and Cancer Biology, Duke University Medical Center) (18). Mouse genotyping was performed by PCR, using the primers 5′-TTAATGGAAGGTGCGTAGGGTGCT-3′ and 5′-CCATTTGAGGATGCGTCGTTTGCT-3′ for the wild-type Pin1 allele and the primers 5′-GAACAAGATGGATTGCACGCAGGT-3′ and 5′-ATGTTTCGCTTGGTGCTCGAATGG-3′ for the disrupted allele. A total of 50 ng of genomic DNA were used in the PCR, with a program of one cycle at 95°C for 90 s; 26 cycles at 95°C for 30 s, 59.7°C for 30 s, and 65°C for 3 min; and one cycle at 65°C for 10 min. The PCR products were separated on a 1% agarose gel, stained with ethidium bromide, and imaged.
Generating PIN1 knockdown cell lines.
Lentiviruses for generating stable control and PIN1 knockdown cell lines in MCF7 and HeLa cells were prepared using PIN1-specific and scrambled shRNA constructs. Cell lines were selected by subculturing cells in medium containing 10 μg of puromycin/ml, and the knockdown efficiency was validated by Western blot analysis with the anti-PIN1 antibody.
Western blotting.
Cells were disrupted in 2× sample buffer (0.1 M Tris-HCl [pH 6.8], 4% sodium dodecyl sulfate [SDS], 20% glycerol, 2% β-mercaptoethanol), boiled for 10 min, centrifuged, placed on ice for 5 min, stored at −80°C, and then separated using 8 to 15% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). The proteins were then transferred onto polyvinylidene difluoride transfer membranes (Millipore, Lincoln Park, NJ). The membranes were blocked with 5% bovine serum albumin in TBST buffer (10 mM Tris-HCl [pH 7.4], 150 mM NaCl, 0.5% Tween 20) at 4°C for 2 h or overnight and were incubated for 2 h at room temperature with specific primary antibodies (monoclonal anti-PIN1 [Santa Cruz Biotechnology, Santa Cruz, CA], anti-HSF1 [Santa Cruz Biotechnology, Santa Cruz, CA], and monoclonal antiactin [Sigma, St. Louis, MO]). The membranes were washed with TBST six times for 10 min each. Immunoreactive proteins were detected using horseradish peroxidase-conjugated secondary antibodies, and the membrane was washed with TBST six times for 10 min each during the detection process. The protein signal was detected by an enhanced chemiluminescence system (ECL Plus; Perkin-Elmer Life and Analytical Sciences, Inc., Waltham, MA).
Immunoprecipitation and glutathione S-transferase (GST) pulldown assays.
For immunoprecipitation assays, the cells were washed twice with phosphate-buffered saline (PBS) at the indicated times and lysed in ice-cold radioimmunoprecipitation assay (RIPA) buffer (100 mM HEPES [pH 7.4], 150 mM NaCl, 2 mM EDTA, 0.5% Tween 20, 0.1% Triton X-100, 1 mM dithiothreitol, 50 μg of AEBSF [4-(2-aminoethyl)-benzenesulfonyl fluoride]/ml, 10 μg of leupeptin/ml, 10 μg of aprotinin/ml, 5 mM NaF, and 1 mM Na3VO4). Anti-PIN1 or anti-HSF1 antibody was incubated with protein G-Sepharose beads for 1 h at 4°C, and the cell lysates were then incubated with the protein G-Sepharose beads for 2 h at 4°C. The immune complexes were washed three times with RIPA buffer, eluted by boiling in 2× SDS sample buffer, and subjected to Western blot analysis.
For glutathione S-transferase (GST) pulldown assays, GST or GST-PIN1 wild-type and mutant proteins expressed in BL21(DE3) cells were adsorbed to glutathione-agarose beads (Sigma) for 1 h after three washes with PBS. The beads were then washed three times with the same buffer, boiled in sample buffer, and subjected to Western blot analysis.
Immunofluorescence assays.
Cells cotransfected with pcDNA3-HSF1-flag and PIN1-DsRed plasmids were grown on coverslips, washed in PBS, fixed with 3.7% paraformaldehyde in PBS, and permeabilized with 10% Triton X-100. After permeabilization, the cells were washed with PBS and incubated with mouse anti-HA monoclonal antibody (Santa Cruz Biotechnology) at 4°C overnight. After a rinsing step with PBS, the cells were incubated with goat anti-rat IgG fluorescence-activated cell sorting (Santa Cruz Biotechnology). After final washes with PBS, the cells were stained with DAPI (4′,6′-diamidino-2-phenylindole), and the localization of fluorescently labeled proteins was visualized using a Zeiss laser confocal microscope (LSM510; Zeiss, Inc., Oberkochen, Germany).
DNA affinity precipitation assay (DAPA) and electrophoretic mobility shift assay (EMSA).
Nuclear extracts were prepared using the NE-PER nuclear extraction kit (Pierce). The oligonucleotide containing the HSE sequence corresponding to the sequence within the HSP70 promoter and the mutant oligonucleotides were individually biotinylated at the 5′ terminus and annealed with their complementary strands. Biotinylated probes were incubated with streptavidin-agarose beads (GE Healthcare) for 1 h, and the cell lysates were incubated with the protein G-Sepharose beads either for 2 h or overnight at 4°C. The protein-DNA complexes with streptavidin-agarose beads were washed five times with binding buffer containing 0.5% (wt/vol) Nonidet P-40. The streptavidin-precipitated DNA-protein complexes were boiled in sample buffer (2×) and resolved by SDS–10% PAGE, which was then followed by Western blot detection with specific antibodies.
A total of 20 μg of nuclear protein extract was used for each sample in the EMSA experiment. Oligonucleotides were end labeled with [γ-32P]ATP using the T4 polynucleotide kinase (NEB). The binding reactions were carried out with 20 μg of nuclear extract or whole extract and 50,000 cpm of oligonucleotide in a 25-μl reaction volume containing binding buffer and 2.5 μg of poly(dI-dC) at room temperature for 30 min. For supershift analysis, antibodies were added to the reaction mixture on ice for 20 min prior to the addition of radiolabeled probes. The binding reactions were resolved on a 3% nondenaturing polyacrylamide gel at 100 mA for 2 h at 4°C in 1× TBE (0.089 M Tris-borate, 0.089 M boric acid, and 0.002 M EDTA). Subsequently, the gels were dried and exposed to a phosphor screen before visualizing using phosphorimager.
Promoter activity assay.
At 48 h after transfection with pGL3 empty vector or the pGL3-HSP70B plasmids, the cells were washed twice with PBS and lysed in passive lysis buffer (Promega). Cell lysates were centrifuged at 16,500 × g for 1 min at 4°C, and the supernatant was collected. Firefly and Renilla luciferase activities were measured by using a luminometer (Victor), and the relative luciferase activity was calculated as the ratio of the firefly luciferase intensity and the Renilla luciferase intensity.
Cytosolic total RNA extraction and quantitative reverse transcription-PCR (RT-PCR).
Total cellular RNA was isolated using TRIzol reagent (Gibco), and the contaminant genomic DNA was further digested with RNase-free DNase I (Promega). A total of 2 μg of total RNA and 0.5 μg of oligo(dT) were added in a sterile RNase-free microcentrifuge tube and heated at 70°C for 5 min to melt secondary structures within the template. A mixture containing 5 μl of Moloney murine leukemia virus 5× reaction buffer (Promega), 1.25 μl of 10 mM deoxynucleoside triphosphates, 1 μl of recombinant RNasin inhibitor (Promega), M-MLV reverse transcriptase (RTase) (Promega), and nuclease-free water was added to yield a final reaction volume of 25 μl. The reverse transcription reaction was performed at 42°C for 1 h in a water bath.
Chromatin immunoprecipitation assay.
Cells (107) were cross-linked with 1% formaldehyde in PBS for 10 min at 30°C. The treated cells were washed two times with ice-cold PBS and lysed in lysis buffer. The chromatin was fragmented by sonication to an average size of 1 kb and centrifuged at 16,000 × g at 4°C for 10 min. The supernatant was diluted 5-fold with 1× binding buffer (500 mM NaCl, 20 mM Tris-HCl [pH 7.9]) containing protease inhibitors and mixed well with 50 μl of salmon sperm DNA-saturated nickel resin slurry (GE Healthcare) for 1 h. The resin was precipitated and washed two times with 1 ml of binding buffer and two times with 1 ml of wash buffer (50 mM imidazole, 0.5 M NaCl, 20 mM Tris-HCl [pH 7.9]). Protein-DNA complexes were separated from the His-binding resin by using two washes of 100 μl of elution buffer (1 M imidazole, 0.5 M NaCl, 20 mM Tris-HCl [pH 7.9]). The protein-DNA cross-links were reversed by heating the samples at 65°C overnight, and the proteins were digested with proteinase K (0.5 mg/ml) at 55°C for 1 h. The DNA was purified by phenol-chloroform extraction and ethanol precipitation and was analyzed by PCR amplification with HSP70-specific primers.
LDH assay.
The cells were subjected to heat shock treatment and allowed to recover at 37°C for 10 h. After recovery, the supernatant was collected for lactate dehydrogenase (LDH) measurement according to the manufacturer's instructions (Abcam).
TUNEL assay.
The apoptosis rates were determined by the TUNEL (terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling) method according to the manufacturer's suggested protocol (R&D Systems).
In vivo refolding assay.
The cells were cotransfected with pTet-off and pTRE-Luc plasmids (Clontech). At 12 h posttransfection, doxycycline was added to culture medium to turn off luciferase production. Transfected cells were subjected to heat shock treatment for 30 min and allowed to recover for 10 h. After being washed twice with PBS, the cells were lysed in passive lysis buffer (Promega) and centrifuged at 16,500 × g for 1 min at 4°C; the supernatant was collected. The luciferase activity was measured by a luminometer (Victor), and the relative activity was calculated as the ratio of the firefly luciferase luminescence intensity (obtained from the assay for firefly luciferase) and the protein concentration of the sample.
RESULTS
Phosphorylation-dependent association of PIN1 with HSF1.
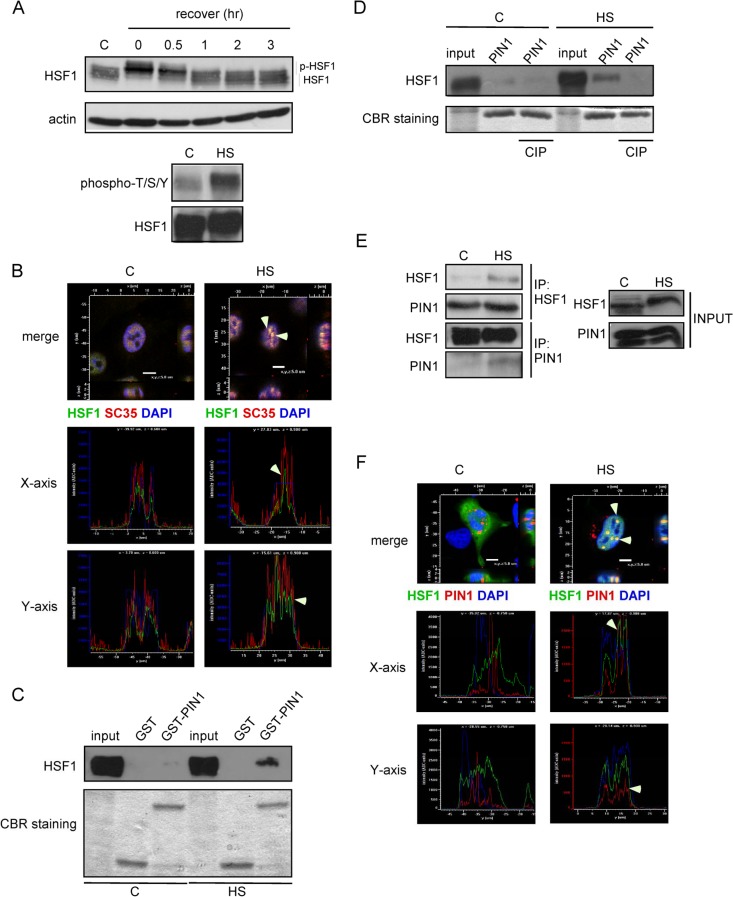
Previous studies have shown that hyperthermia induces the phosphorylation of HSF1 and the formation of stress granules in cells (19, 20). In an attempt to confirm these observations, HeLa cells were subjected to heat shock and were allowed to recover at 37°C for different increments of time, as indicated. Indeed, after a 1-h heat shock treatment, the hyperphosphorylated HSF1 had reduced mobility, which gradually disappeared upon the recovery of the cells at 37°C (Fig. 1A, upper panel). We used the phospho-Ser/Thr specific HSF1 antibody to confirm the phosphorylation status of HSF1. As shown in Fig. 1A, phosphorylation was significantly induced upon hyperthermia treatment (lower panel, Fig. 1A). Previously, Jolly et al. reported that activation of the heat shock response correlates with a rapid relocalization of HSF1 to nuclear structures called nuclear stress granules (20, 21). Consistent with this observation, we observed nuclear stress granules only upon heat shock at 43°C in HeLa cells. The colocalization of the spliceosome marker, SC35, with nuclear stress granules was confirmed by codetection of HSF1 (Fig. 1B). Guettouche et al. have shown that 12 serine residues can be phosphorylated in response to hyperthermia (6). Many of these serine residues, such as Ser292 and Ser326, present a consensus motif (phospho-Thr/Ser-Pro) for PIN1 binding.
Fig 1.
Heat shock stress promotes the association of PIN1 and HSF1. (A) (Top) Heat shock-induced hyperphosphorylation of HSF1 in HeLa cells. The slower mobility of HSF1 in heat shock-treated cells is due to inducible phosphorylation. (Bottom) HSF1 was immunoprecipitated and separated by SDS-PAGE. The phosphorylation of HSF1 was examined by using anti-phospho-Ser/Thr and anti-HSF1 antibodies. (B) SC35 (red) and HSF1 (green) were detected by immunofluorescence either in untreated or heat shock-treated HeLa cells. (C) Purified GST and GST-tagged PIN1 recombinant proteins were incubated with untreated or heat shock-treated HeLa cell lysates. Equivalent loading of the proteins was confirmed by Coomassie blue staining. Western blot analysis with anti-HSF1 antibody was used to confirm PIN1 and HSF1 interaction. (D) The cell lysates were treated with calf alkaline phosphatase (CIP) prior to GST-pulldown assays and immunoblotting for HSF1 to identify the interaction. (E) Untreated and heat shock-treated cell extracts were subject to immunoprecipitation and Western blotting with anti-HSF1 or anti-PIN1 antibodies. (F) At 24 h after transfection with Flag-HSF1 and PIN1-RFP expression plasmids, HeLa cells were exposed to heat shock for 1 h. The cells were then fixed with 3.7% formaldehyde, permeabilized, incubated with mouse anti-Flag primary and rhodamine-conjugated secondary antibodies, and observed under a Zeiss LSM510 confocal microscope.
To explore whether PIN1 and HSF1 associate upon hyperthermia stress, GST-pulldown assays were performed. Extracts of HeLa cells transiently transfected with Flag-tagged HSF1 were incubated with GST or GST-fused PIN1 proteins. After extensive washes, the bound proteins were eluted, separated by SDS-PAGE and stained with Coomassie blue. As shown in Fig. 1C, a strong band that was recognized by the anti-Flag antibody was retained only in cells subjected to hyperthermia treatment. Interestingly, this interaction was abolished when cell extracts was pretreated with the alkaline phosphatase CIP (Fig. 1D). This finding is consistent with previous reports demonstrating that the interaction of PIN1 with its substrates is usually phosphorylation dependent (22, 23). To further define the role of PIN1 in HSF1 regulation, immunoprecipitation was carried out to examine the direct association of HSF1 with PIN1. Indeed, endogenous HSF1 was pulled down by anti-PIN1 antibody in hyperthermia-treated HeLa cells, and the ectopically expressed HA-tagged PIN1 was immunoprecipitated by anti-HSF1 antibody (Fig. 1E). We also analyzed the cellular localization of PIN1 and HSF1. At 2 days after transfection with Flag-HSF1 and PIN1-RFP expression plasmids, the cells were subject to heat shock. Upon exposure to heat shock, the overexpressed HSF1 formed visible stress granules in the nucleus, and consistent with our hypothesis, PIN1 colocalized with HSF1 in the nucleus after hyperthermia treatment (Fig. 1F).
PIN1 regulates the interact of HSF1 with DNA.
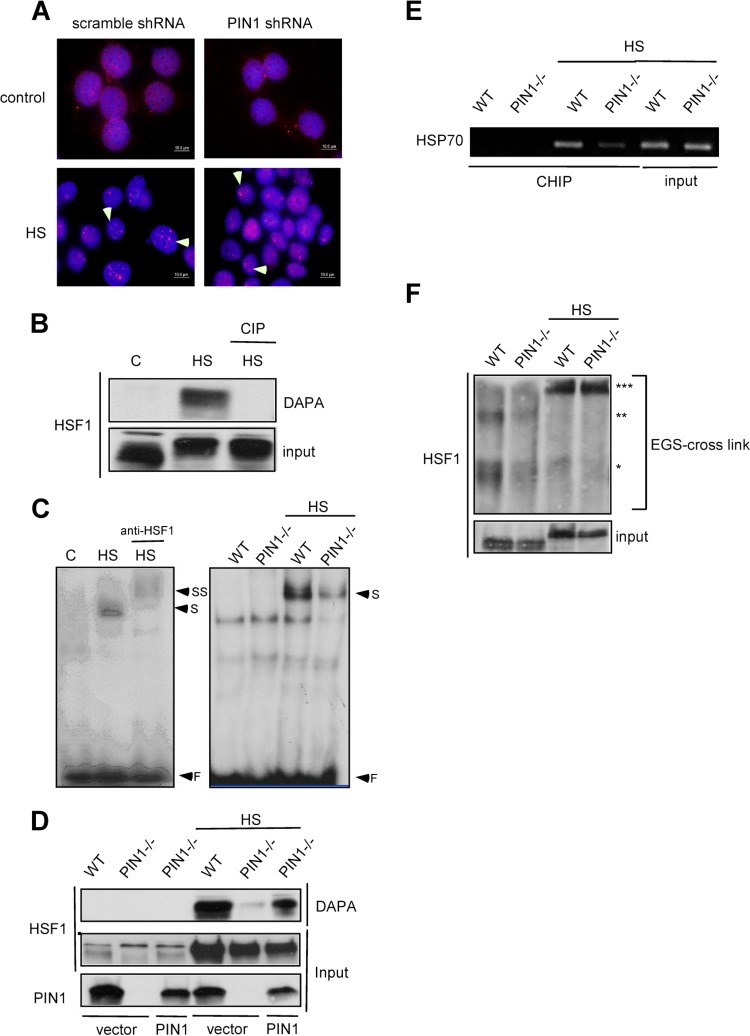
We have demonstrated that PIN1 associates with HSF1 upon heat shock treatment (Fig. 1). Next, we wanted to examine whether PIN1 is critical to the formation of nuclear stress granules. Scrambled and PIN1-specific shRNA lentiviral vectors were constructed to infect MCF7 cells. In Fig. 2A anti-HSF1 antibody staining revealed the predominantly nuclear localization of HSF1 in untreated MCF7 cells, although some cytoplasmic localization was visible. The exposure of MCF7 cells to heat shock resulted in 3 to 15 brightly stained nuclear HSF1 granules of various sizes in every cell. However, the stress granules were visible in both scrambled and PIN1 shRNA-infected cells after heat shock.
Fig 2.
PIN1 is required for HSF1 DNA-binding activity. (A) MCF7 cells were infected by scrambled-shRNA or PIN1-shRNA lentivirus constructs. HSF1 was detected by immunofluorescence in control or heat shock-treated MCF7 control shRNA-treated and PIN1-shRNA-treated cells. (B) To determine the DNA-binding activity levels, cell lysates were treated with calf alkaline phosphatase (CIP) prior to DNA-affinity precipitation assay (DAPA) and immunoblotting for HSF1. (C) (Left) Supershift analysis shows that the induced DNA-protein complexes contain HSF1. (Right) EMSA analysis of whole-cell extracts from wild-type and PIN1−/− MEF cells treated with 43°C heat shock. (D) The PIN1−/− MEF cells were transfected with mouse PIN1 DNA. The nuclear extracts of wild type, PIN1−/− MEFs and PIN1-expressing PIN1−/− MEFs were incubated with the probe and immunoblotted for HSF1. (E) In vivo chromatin immunoprecipitation analysis of HSF1-DNA-binding activity in wild-type and PIN1−/− MEF cells treated with 43°C heat shock. (F) Total protein extracts of MEFs were evaluated for HSF1 multimerization by EGS cross-linking, SDS-PAGE, and immunoblotting with an HSF1-specific antibody. *, **, and *** on the right indicate the expected migration of the HSF1 monomer, dimer, and trimer, respectively.
Under normal conditions, HSF1 is present in a non-DNA binding, monomeric state. Upon hyperthermia, HSF1 localizes to the nucleus and trimerizes to associate with DNA via the HSE in the promoter region of HSPs. The phosphorylation-dependent interaction of PIN1 and HSF1 has also been demonstrated. Next, we wanted to examine whether phosphorylation is required for HSF1-DNA association. Heat shock-treated HeLa cell lysates were incubated with biotin-labeled probes, and the presence of HSF1 in the HSF1-HSE complex was confirmed by DAPA. Interestingly, CIP treatment completely abolished the DNA-binding activity of HSF1, suggesting that PIN1 may play a role in HSF1-DNA association (Fig. 2B). Next, electromobility shift assays (EMSAs) and DAPAs were performed to examine whether PIN1 is involved in HSF1 regulation. DAPA analysis showed that upon heat shock treatment, the DNA-binding activity of HSF1 was attenuated in PIN1−/− cells compared to wild-type cells (Fig. 2C). This result was further confirmed by the isotope-based EMSA experiment (Fig. 2D). Next, chromatin immunoprecipitation assay was performed to confirm in vivo HSF1-DNA binding. Consistent with our hypothesis, the DNA-binding activity of HSF1 was decreased in heat shock-treated PIN1−/− cells (Fig. 2E) compared to wild-type controls. Taken together, our results indicate that PIN1 may play a role in the regulation of HSF1 and DNA interactions. Xia and Voellmy have demonstrated that HSF1 trimerization is a critical step for its DNA-binding activity (24); therefore, we hypothesized that PIN1 interferes with HSF1 trimerization. However, when the extracts of heat shock-treated cells were treated with the cross-linking agent EGS, HSF1 was still shown to trimerize in both wild-type and PIN1-deficient cells (Fig. 2F).
PIN1 is required for the induction of heat shock-induced HSPs.
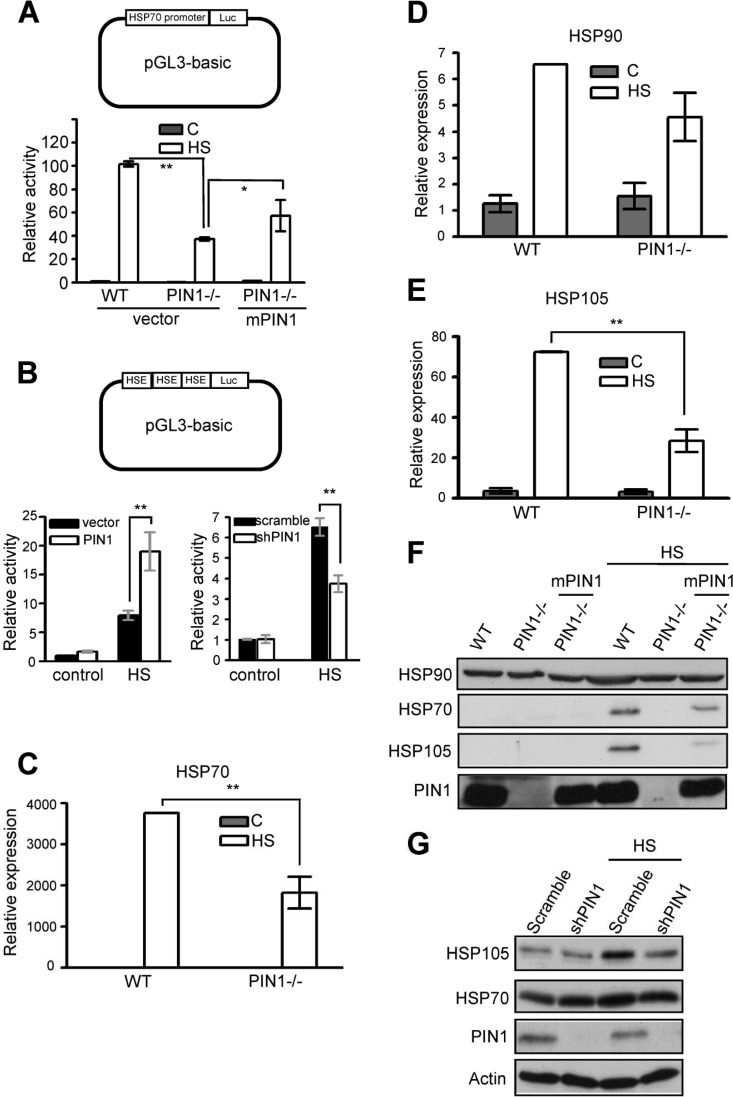
The HSF1-regulated transcription of molecular chaperones mRNAs (including heat shock proteins HSP70, HSP90, and HSP105) is among the most prevalent responses to environmental challenges. Because HSP70 is predominantly regulated at the transcriptional level by nuclear proteins such as HSF1, we performed reporter gene assays using a reporter plasmid containing HSP70 promoter to address the effect of PIN1 on HSF1 regulation (25). HSP70 promoter activity can potentially be induced after hyperthermia treatment in wild-type MEF cells. Interestingly, in heat shock-treated PIN1-deficient cells, the HSP70 promoter activity was attenuated and exogenous PIN1 was able to partially restore HSF1 activity (Fig. 3A). Next, a luciferase plasmid containing three copies of HSE was used to confirm that PIN1 is indispensable for HSF1-dependent transcriptional activity. Our results showed that PIN1 overexpression enhances HSF1 activity compared to vector only controls (Fig. 3B, left panel). Similarly, downregulation of PIN1 leads to the attenuation of HSF1-driven luciferase activity (Fig. 3B, right panel). Furthermore, the expression of HSF1-dependent genes, such as Hsp70, Hsp90, and Hsp105, was assessed by real-time PCR and Western blot analysis. A heat-induced increase in HSP mRNA levels was readily detected in wild-type cells at 3 h after heat shock treatment (Fig. 3A). Consistent with the reporter assay, the induction of HSP70 and HSP105 was reduced in PIN1−/− cells (Fig. 3C to E). Moreover, the downregulation of the HSP gene was confirmed at the protein level in MEF cells (Fig. 3F). To confirm the role of PIN1 in HSF1 regulation, a lentivirus was used to knock down PIN1 in MCF7 cells. As shown in Fig. 3G, knocking down PIN1 in MCF7 cells resulted in a lower level of HSP gene expression compared to controls. These results strongly suggested a critical, nonredundant role for PIN1 in HSF1 activation.
Fig 3.
PIN1 is required for HSF1-dependent HSP gene expression. (A) Schematic of the HSP70B promoter-driven luciferase construct. Wild type MEFs, PIN1−/− MEFs, and PIN1-expressing-PIN1−/− MEFs were exposed to heat shock and analyzed by luciferase assay. (B) Schematic of the luciferase construct containing three repeats of the HSE (3HSE). In the left panel, HeLa cells cotransfected with PIN1-HA and 3HSE constructs were exposed to heat shock and analyzed by luciferase assay. In the right panel, HeLa cells cotransfected with the PIN1 shRNA plasmid and 3HSE constructs were subjected to heat shock and analyzed by luciferase assay. Wild-type and PIN1−/− MEFs were heat shock treated and allowed to recover for 3 h. Cytoplasmic RNA was extracted and analyzed by quantitative RT-PCR. Comparative HSP gene expression profiles for HSP70 (C), HSP90 (D), and HSP105 (E) are shown. (F) Wild-type and PIN1−/− MEFs were subjected to heat shock treatment and allowed to recover for 10 h. Cells were lysed with RIPA buffer and analyzed by Western blotting. (G) Heat-induced HSP expression in MCF7 cells treated with scramble-shRNA or shPIN1 (PIN1 shRNA). Cells were exposed to heat shock treatment for 1 h at 43°C as indicated (HS) and analyzed by Western blotting after 10 h of recovery at 37°C. An actin Western blot serves as the loading control.
PIN1 recognizes phospho-Ser326 of HSF1 via the WW domain.
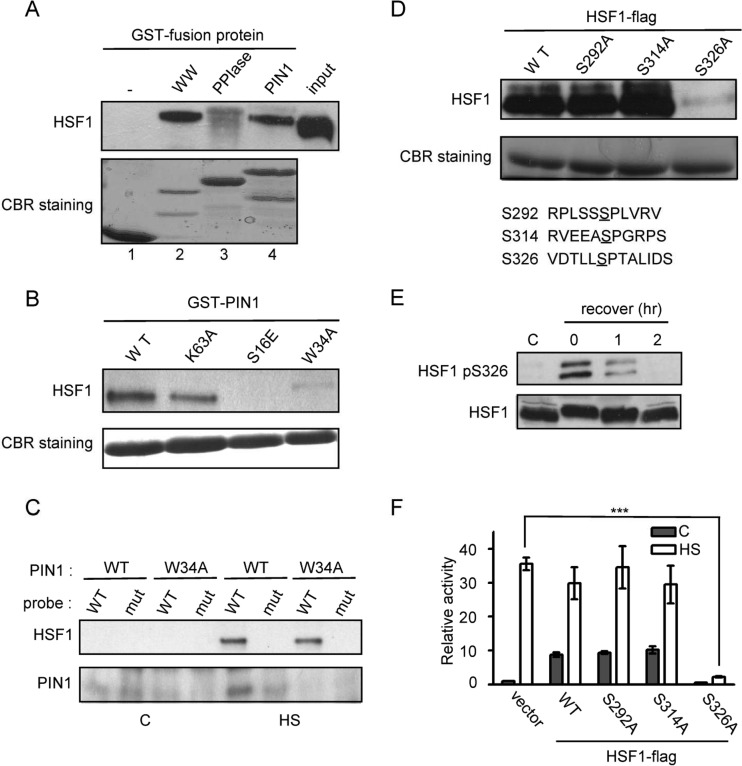
To determine the motif responsible for the direct interaction of PIN1 with HSF1, the two PIN1 functional domains, WW and PPIase, were expressed as glutathione S-transferase (GST) fusion proteins in Escherichia coli and were affinity purified using glutathione beads. Flag-HSF1-expressing HeLa cell extracts were incubated with the GST-PIN1 fusion proteins. After an extensive number of washes, the bound proteins were eluted, separated by SDS-PAGE, and stained with Coomassie blue. As shown in Fig. 4A, the intense band that was recognized by the anti-Flag antibody was retained only by the GST-PIN1 and GST-WW affinity matrices (lanes 2, 4). Furthermore, in contrast to wild-type PIN1, both of the WW-domain PIN1 mutants (PIN1W34A or PIN1S16E) failed to pull down HSF1 (Fig. 4B). These results indicate that PIN1 associates with HSF1 via the WW domain. Therefore, we hypothesized that PIN1 is involved in the HSF1-DNA complex. At 24 h after transfection with wild-type or mutant PIN1 (W34A), the HeLa cells were subjected to heat shock. Upon stress induction, the biotin-labeled probe containing the wild-type HSE sequences pulled down HSF1 and PIN1 but not mutant PIN1 (Fig. 4C). However, the probe containing the mutant HSE sequences was not able to pull down the HSF1-PIN1 complex (Fig. 4C).
Fig 4.
PIN1 associates with phospho-Ser326 of HSF1 via the WW domain (A) Each GST-tagged PIN1 and PIN1 mutant construct was incubated in binding buffer with cell lysates containing Flag-HSF1. Equivalent loading of proteins was confirmed by Coomassie blue staining. Western blot analysis with anti-HSF1 was used to identify the interaction domain. (B) GST pulldown assay with GST-fused PIN1 and PIN1 mutant proteins (S16E, W34A, and K63A) was used to confirm the binding with HSF1. (C) The cell lysates of PIN1- or W34A PIN1-expressing cells were incubated with a biotin-labeled HSP70 probe. HSF1 and PIN1 were detected by Western blot analysis. (D) HeLa cell lysates containing ectopically expressed HSF1 were incubated with GST-PIN1 in binding buffer. Equivalent loading of proteins was confirmed by Coomassie blue staining. Western blot analysis with anti-HSF1 antibody was used to verify the interacting residue. (E) HeLa cells were subjected to heat shock treatment, allowed to recover, and analyzed by Western blotting with antibodies against pHSF1-Ser326, HSF1, and actin. (F) HeLa cells were transfected with HSP70B promoter-driven luciferase reporter and HSF1 plasmids. The cells were subjected to heat shock treatment and were analyzed by luciferase assay.
Next, we wanted to determine the specific PIN1-interacting site in HSF1. As demonstrated in Fig. 3, the heat-induced HSP expression was abolished in PIN1- deficient cells, suggesting that heat shock-induced HSP expression is PIN1 dependent. Moreover, using mutagenesis and functional studies, Guettouche et al. showed that after hyperthermia stress, 12 serine residues on HSF1 can be phosphorylated but only phosphorylation Ser326 is critical for HSF1 activation (6). We hypothesized that PIN1 associates with the Ser326-Pro motif via its WW domain to induce isomerization. To evaluate the importance of Ser326 in PIN1-HSF1 interactions, we examined the effect of an alanine substitution for HSF1-Ser326 on the PIN1 association. Interestingly, the HSF1 and PIN1 interaction was diminished when Ser326 was mutated to alanine (Fig. 4D). We next examined the effect of HSF1-Ser326 phosphorylation in vivo in HeLa cells subjected to heat stress. The cells were exposed to heat shock at 43°C for 1 h and were then assayed for HSF1-Ser326 phosphorylation using a phosphor-specific antibody. Ser326 phosphorylation was detected immediately after heat shock, persisted for ∼1 h, and gradually decreased after the cells were allowed to recover (Fig. 4E). To examine the effects of alanine substitution of HSF1-Ser326 on HSP70 promoter activity, wild-type and alanine substituted HSF1 expression plasmids (Ser292A, Ser314A, and Ser326A substitution mutants) were cotransfected with an HSP70 promoter-driven luciferase reporter plasmid and analyzed 24 h posttransfection. Heat-induced activation of the HSP70 promoter was reduced by 80% in cells expressing the Ser326A HSF1 mutant compared to the cells expressing wild-type, Ser292A, or Ser314A HSF1 (Fig. 4F).
Lack of heat resistance in PIN1-deficient cells.
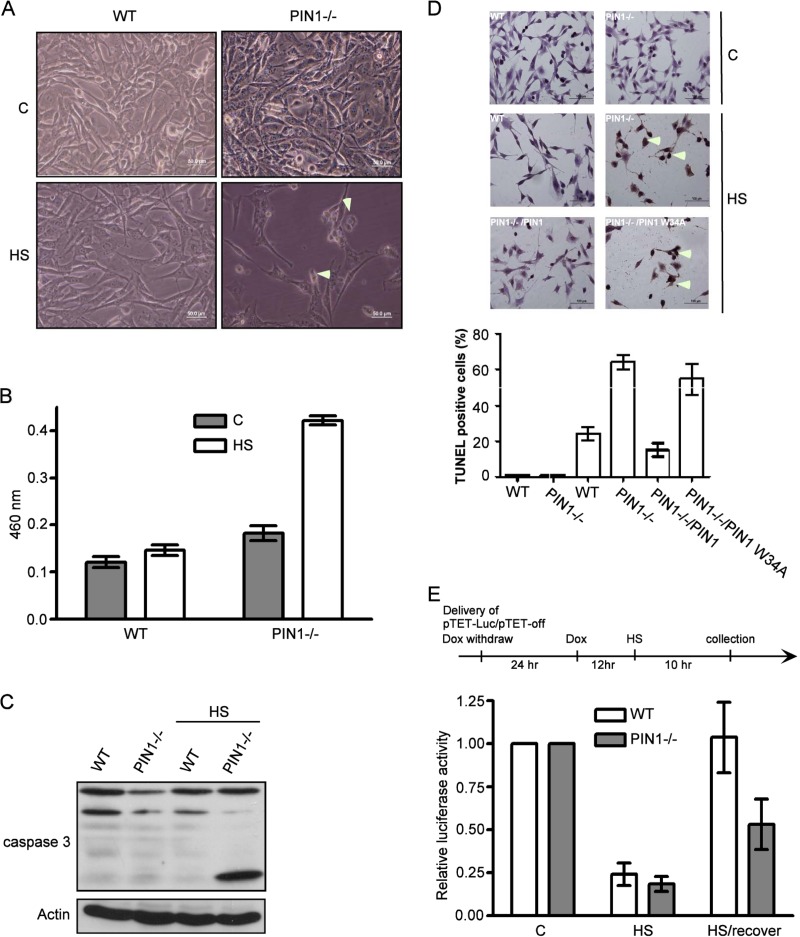
We have demonstrated the inefficient heat-induced HSP activation in PIN1-deficient cells (Fig. 3). Recently, several studies have demonstrated that activation of the heat shock response (HSR), specifically the elevation of HSP70 levels, can protect cells from protein damage-induced stress such as during hyperthermia and proteasome inhibition. We hypothesized that PIN1-deficient cells are less resistant to hyperthermia stress compared to wild-type cells. To test this hypothesis, wild-type and PIN1-deficient cells were subjected to 1 h heat treatment and allowed to recover for 12 h. The cellular morphology was then observed under a microscope. In Fig. 5A, the morphology of heat-treated wild-type cells is similar to that of untreated control cells, whereas heat-treated PIN1-deficient cells show shrinkage. Next, we performed the LDH assay to evaluate heat shock-induced toxicity in cells and found that the LDH level was elevated in heat shock-treated PIN1-deficient cells (Fig. 5B). Furthermore, the cleavage of caspase-3 was increased in heat shock-treated PIN1-deficient cells (Fig. 5C). In order to precisely observe the apoptotic effect, TUNEL assay was applied. In Fig. 5D, the TUNEL signal of heat-treated PIN1−/− cells is higher than wild-type cells. Interestingly, the ectopic expression of wild-type PIN1 but not PIN1 W34A is capable of decreasing the amount of TUNEL stained cells. Next, we hypothesized that increased toxicity in heat shock-treated PIN1-deficient cells could be due to the reduced ability to refold or degrade the denatured proteins in the cell. This finding was confirmed by the in vivo refolding assay, which evaluates the refolding activity of HSPs in cells. Wild-type and PIN1 knockout cells were transfected with a tetracycline-control (Tet-off) luciferase plasmid, and after 24 h, doxycycline was added to switch off the luciferase expression. These cells were then subjected to heat shock and allowed to recover for 10 h at 37°C. The results showed that 1 h after heat shock treatment but without recovery, the luciferase activity was diminished both in wild-type and in PIN1 knockout cells. Interestingly, although luciferase activity was completely restored during recovery in wild-type cells, the restoration was much less efficient in PIN1-deficient cells (Fig. 5E).
Fig 5.
Lack of heat resistance in PIN1-deficient cells. (A) Morphological changes in wild-type and PIN1−/− MEFs before and after heat shock treatment. (B) Estimation of LDH levels in the culture medium of wild-type and PIN1−/− MEFs before and after heat shock treatment. (C) Wild-type and PIN1−/− MEF lysates were separated by SDS-PAGE and were analyzed by Western blotting to detect PIN1, actin, and caspase-3. (D) Wild-type and PIN1−/− MEF cells were heated, fixed, and stained with TUNEL reagent. The images were taken by microscope. (E) Wild-type and PIN1−/− MEFs were transfected with pTet-off and pTRE-Luc plasmids and subjected to heat shock treatment. Transfected cell lysates were analyzed by luciferase assay.
Impaired responses to thermal challenge in PIN1-deficient neurons.
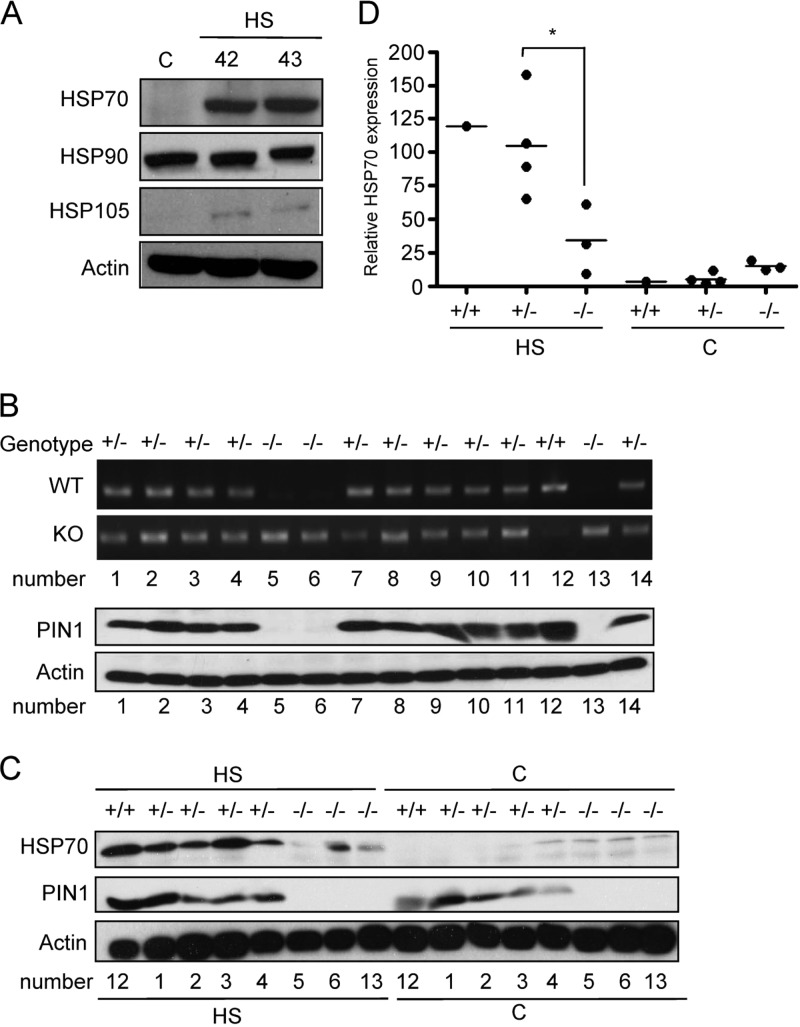
Neurons are particularly vulnerable to the detrimental effects of misfolded and/or aggregated proteins because they are postmitotic and cannot dilute potentially toxic species through cell division. Therefore, misfolded proteins accumulate in neurons during aging (26), and it has been reported that PIN1 is abundant in the nucleus and cytoplasm of neurons (27, 28). To test the hypothesis that PIN1 plays a role in protecting neurons from protein damage-induced stress, we first confirmed that heat stress induces HSP gene expression in neurons. Primary neurons of embryonic stage day 18 of rats were purified and cultured. At DIV8, primary neurons were exposed to heat shock and allowed to recover for 10 h. As shown in Fig. 6A, HSP70 and HSP105 were induced in response to hyperthermia stress. To determine the role of PIN1 in neurons, one pair of PIN1 heterozygous mice were mated. Cortical neurons were purified from embryonic mice and genotyped using specific primers. Figure 6B shows that one wild-type mice, 10 PIN1 heterozygous knockout mice and three homozygous knockout mice were obtained. Consistent with our hypothesis, the PIN1 expression was lower in homozygous knockout mice than in wild-type mice. Next, protein extracts of heat shock-treated wild-type, heterozygous, and homozygous knockout cultured neurons at DIV8 were analyzed by Western blotting. The results showed that the expression of HSP70 was attenuated in homozygous knockout neurons after hyperthermia treatment (Fig. 6C and D).
Fig 6.
PIN1 is required for heat resistance and heat-induced HSP gene expression in neurons. (A) Primary cortical neurons at DIV8 were subjected to heat shock treatment and allowed to recover for 10 h. HSP expression was evaluated by Western blotting. (B) The neurons of fetal mice were purified and cultured. Genotyping and Western blotting analyses were used to characterize the genotypes of the mice used. (C) The neurons obtained from wild-type, PIN1+/−, and PIN1−/− mice were exposed to heat shock treatment and allowed to recover in the incubator for 10 h. Whole-cell protein extracts were analyzed by Western blotting with anti-HSP70, anti-PIN1, and antiactin antibodies. (D) Quantification of the HSP70 expression in panel C.
Taken together, these results suggest that PIN1 and HSF1 directly interact and that PIN1 synergizes with HSF1 to induce HSP expression in tumor cells and primary neurons.
DISCUSSION
The cellular response to stress includes not only the induction of heat shock proteins that assist the cell in protecting against cellular stress but also cell cycle arrest, which gives the cell an opportunity to repair the damage. There are many HSF1 binding proteins that regulate HSF1 activity, such as DAXX, CHIP, and HSP90 (29–31). In the present study, we provide evidence that hyperthermia stress induces the phosphorylation-dependent nuclear association of PIN1 and HSF1 and that PIN1 is indispensable for the DNA-binding activity of HSF1. Upon its interaction with HSF1, PIN1 may catalyze the cis/trans isomerization of HSF1, enhancing its DNA-binding activity. Moreover, damaged proteins were refolded or produced at reduced levels after HSP gene induction, suggesting that PIN1 functions as a stress sensor.
HSF1 transcriptionally regulates heat shock genes to protect cells against environmental stress. The activation of HSF1 is a component of a complex mechanism that includes posttranslational modifications, trimerization, nuclear localization, and DNA binding. Sumoylation, phosphorylation, and acetylation have been reported as posttranslational modifications that are involved in this process (8–10, 32). Specifically, phosphorylation seems to play a vital role in the regulation of HSF1. For example, Holmberg et al. have reported that calmodulin kinase II phosphorylates HSF1 at Ser230 and promotes the transcriptional activity of HSF1 (8). In addition, the mTOR-mediated phosphorylation of HSF1 at Ser326 is critical for HSF1 activity (33). Our results show that both the PIN1-binding and DNA-binding activities of HSF1 are phosphorylation dependent (Figs. 1 and 2B). Moreover, PIN1 recognizes the Ser326-Pro motif in HSF1 and modulates its DNA-binding activity. We hypothesize that in addition to associating with HSF1, PIN1 also brings about HSF1 isomerization, which alters the three-dimensional structure of HSF1 leading to the upregulation of HSF1 activity. Interestingly, PIN1 does not recognize other Ser-Pro motifs (Ser292-Pro and Ser314-Pro), suggesting that neighboring sequences may also interfere with the binding affinity of PIN1 and its substrate. This study proposes a novel concept that the isomerization of HSF1 is a critical step in its activation.
PIN1 has previously been shown to be a cell cycle regulator (34) and to restore the function of Alzheimer-associated phosphorylated tau and APP proteins (28, 35). Similar to HSF1, PIN1 seems to be beneficial for the cell because it is often required for mechanisms that are indispensable for protecting the cell. HSF1 is a transcription factor that regulates the expression of heat shock proteins. HSF1 is activated in cells upon exposure to environmental stressors such as heat shock, proteosomal inhibition and heavy metal treatment. The results of our study clearly demonstrate that PIN1 collaborates with HSF1 and resists stress via the upregulation of heat shock proteins. The hallmark of the heat shock treatment of cells is the formation of nuclear stress granules (20, 21). We found that PIN1 colocalizes with HSF1 in stress granules and promotes HSF1 assembly into the stress granules in HeLa cells (Fig. 1F). However, PIN1 knockdown does not reduce the formation of stress granules. Therefore, the exact role of PIN1 in the formation of the stress granules is unknown.
A few HSF1 binding partners that regulate HSF1 DNA-binding activity have been reported previously (32, 36). Westerheide et al. demonstrated that a protein deacetylase called sirtuin 1 (SIRT1) can deacetylate HSF1 by decreasing the negative charge on HSF1 and its DNA-binding activity. In this study, although we have shown that PIN1 regulates HSF1 activity, the precise mechanism by which PIN1 enhances HSF1 DNA-binding activity is still unknown. SIRT1 was shown to assist HSF1 in regulating life span and resisting aging (32). Interestingly, PIN1 has been reported to attenuate tauopathy in Alzheimer's disease, which is a disease of aging (28). On the basis of previous reports, we can conclude that PIN1 may promote HSF1 binding to DNA via the enhancement of HSF1 and SIRT1 interactions. In addition to its role in SIRT1 regulation, the role of HSF1 in HSP gene expression in aging has been demonstrated previously (37). Chiang et al. showed that HSF1 is indispensable for the modulation of longevity in Caenorhabditis elegans. Taken together, these results suggest that PIN1 is involved in the regulation of life span.
Heat shock proteins are extremely important in postmitotic cells such as neurons. Neurons cannot undergo cell division to dilute the denatured proteins resulting from environmental stress, and heat shock proteins are therefore required for refolding or eliminating the impaired proteins in neurons. The role of PIN1 in neuronal cells has not been evaluated completely. In the present study, we showed that heat shock proteins are robustly induced after hyperthermia treatment in neurons cultured in vitro. To our surprise, HSP gene expression was attenuated in PIN1-deficient neurons after heat shock treatment, indicating that PIN1 is indispensable for heat shock-induced HSP gene expression. Moreover, both HSP110/105 and PIN1 deficiency have been shown to result in age-dependent tau hyperphosphorylation and the early accumulation of insoluble amyloid proteins (28, 35, 38). The accumulation of hyperphosphorylated tau and insoluble amyloid proteins is a pathological consequence of Alzheimer's disease. Consequently, the downregulation of HSP110/105 and other HSP genes due to PIN1 deficiency could contribute to Alzheimer disease progression.
In conclusion, our study demonstrates that PIN1 physically interacts with HSF1 via its WW domain in a phosphorylation-dependent manner and enhances HSF1 DNA-binding activity. Furthermore, we showed that PIN1 is critical for heat shock-induced HSF1 activation and HSP gene expression in embryonic fibroblasts, tumor cells, and primary neurons. HSPs are produced to repair environmental stress-induced protein damage. Thus, we present a novel mechanism for the regulation of HSF1 by PIN1.
ACKNOWLEDGMENTS
We thank the medical imaging core facility of the Clinical Medicine Research Center in National Cheng Kung University Hospital for assisting with image processing.
This work received financial support from the National Science Council (NSC102-2325-B-006-011-), the National Research Program for Biopharmaceuticals (DOH102-TD-PB-111-TM027), and the National Health Research Institutes (NHRI-EX102-10152SI) in Taiwan.
Footnotes
Published ahead of print 14 October 2013
REFERENCES
- 1. Sharp FR, Massa SM, Swanson RA. 1999. Heat-shock protein protection. Trends Neurosci. 22:97–99 [DOI] [PubMed] [Google Scholar]
- 2. Morimoto RI. 1998. Regulation of the heat shock transcriptional response: cross talk between a family of heat shock factors, molecular chaperones, and negative regulators. Genes Dev. 12:3788–3796 [DOI] [PubMed] [Google Scholar]
- 3. Kim SA, Yoon JH, Lee SH, Ahn SG. 2005. Polo-like kinase 1 phosphorylates heat shock transcription factor 1 and mediates its nuclear translocation during heat stress. J. Biol. Chem. 280:12653–12657 [DOI] [PubMed] [Google Scholar]
- 4. Flick KE, Gonzalez L, Jr, Harrison CJ, Nelson HC. 1994. Yeast heat shock transcription factor contains a flexible linker between the DNA-binding and trimerization domains: implications for DNA binding by trimeric proteins. J. Biol. Chem. 269:12475–12481 [PubMed] [Google Scholar]
- 5. Shi Y, Mosser DD, Morimoto RI. 1998. Molecular chaperones as HSF1-specific transcriptional repressors. Genes Dev. 12:654–666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Guettouche T, Boellmann F, Lane WS, Voellmy R. 2005. Analysis of phosphorylation of human heat shock factor 1 in cells experiencing a stress. BMC Biochem. 6:4. 10.1186/1471-2091-6-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Murshid A, Chou SD, Prince T, Zhang Y, Bharti A, Calderwood SK. 2010. Protein kinase A binds and activates heat shock factor 1. PLoS One 5:e13830. 10.1371/journal.pone.0013830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Holmberg CI, Hietakangas V, Mikhailov A, Rantanen JO, Kallio M, Meinander A, Hellman J, Morrice N, MacKintosh C, Morimoto RI. Eriksson JE, Sistonen L. 2001. Phosphorylation of serine 230 promotes inducible transcriptional activity of heat shock factor 1. EMBO J. 20:3800–3810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hietakangas V, Ahlskog JK, Jakobsson AM, Hellesuo M, Sahlberg NM, Holmberg CI, Mikhailov A, Palvimo JJ, Pirkkala L, Sistonen L. 2003. Phosphorylation of serine 303 is a prerequisite for the stress-inducible SUMO modification of heat shock factor 1. Mol. Cell. Biol. 23:2953–2968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hietakangas V, Anckar J, Blomster HA, Fujimoto M, Palvimo JJ, Nakai A, Sistonen L. 2006. PDSM, a motif for phosphorylation-dependent SUMO modification. Proc. Natl. Acad. Sci. U. S. A. 103:45–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Anckar J, Hietakangas V, Denessiouk K, Thiele DJ, Johnson MS, Sistonen L. 2006. Inhibition of DNA binding by differential sumoylation of heat shock factors. Mol. Cell. Biol. 26:955–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Tsang N, Wu YH, Lim JS, Wee Ong C, Salto-Tellez M, Ito K, Ito Y, Chen LF. 2012. Prolyl isomerase Pin1 downregulates tumor suppressor RUNX3 in breast cancer. Oncogene 32:1488–1496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Shen ZJ, Braun RK, Hu J, Xie Q, Chu H, Love RB, Stodola LA, Rosenthal LA, Szakaly RJ, Sorkness RL, Malter JS. 2012. Pin1 protein regulates Smad protein signaling and pulmonary fibrosis. J. Biol. Chem. 287:23294–23305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wulf G, Finn G, Suizu F, Lu KP. 2005. Phosphorylation-specific prolyl isomerization: is there an underlying theme? Nat. Cell Biol. 7:435–441 [DOI] [PubMed] [Google Scholar]
- 15. Liou YC, Zhou XZ, Lu KP. 2011. Prolyl isomerase Pin1 as a molecular switch to determine the fate of phosphoproteins. Trends Biochem. Sci. 36:501–514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Akiyama H, Misawa T, Ono M, Uchida C, Uchida T. 2011. Prolyl isomerase pin1 protects mice from endotoxin shock. PLoS One 6:e14656. 10.1371/journal.pone.0014656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ryo A, Hirai A, Nishi M, Liou YC, Perrem K, Lin SC, Hirano H, Lee SW, Aoki I. 2007. A suppressive role of the prolyl isomerase Pin1 in cellular apoptosis mediated by the death-associated protein Daxx. J. Biol. Chem. 282:36671–36681 [DOI] [PubMed] [Google Scholar]
- 18. Atchison FW, Capel B, Means AR. 2003. Pin1 regulates the timing of mammalian primordial germ cell proliferation. Development 130:3579–3586 [DOI] [PubMed] [Google Scholar]
- 19. Cotto J., Fox S, Morimoto R. 1997. HSF1 granules: a novel stress-induced nuclear compartment of human cells. J. Cell Sci. 110(Pt 23):2925–2934 [DOI] [PubMed] [Google Scholar]
- 20. Jolly C, Metz A, Govin J, Vigneron M, Turner BM, Khochbin S, Vourc'h C. 2004. Stress-induced transcription of satellite III repeats. J. Cell Biol. 164:25–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Jolly C, Konecny L, Grady DL, Kutskova YA, Cotto JJ, Morimoto RI, Vourc'h C. 2002. In vivo binding of active heat shock transcription factor 1 to human chromosome 9 heterochromatin during stress. J. Cell Biol. 156:775–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ranganathan R, Lu KP, Hunter T, Noel JP. 1997. Structural and functional analysis of the mitotic rotamase Pin1 suggests substrate recognition is phosphorylation dependent. Cell 89:875–886 [DOI] [PubMed] [Google Scholar]
- 23. Lu PJ, Zhou XZ, Shen M, Lu KP. 1999. Function of WW domains as phosphoserine- or phosphothreonine-binding modules. Science 283:1325–1328 [DOI] [PubMed] [Google Scholar]
- 24. Xia W, Voellmy R. 1997. Hyperphosphorylation of heat shock transcription factor 1 is correlated with transcriptional competence and slow dissociation of active factor trimers. J. Biol. Chem. 272:4094–4102 [DOI] [PubMed] [Google Scholar]
- 25. Pirkkala L, Nykanen P, Sistonen L. 2001. Roles of the heat shock transcription factors in regulation of the heat shock response and beyond. FASEB J. 15:1118–1131 [DOI] [PubMed] [Google Scholar]
- 26. Muchowski PJ, Wacker JL. 2005. Modulation of neurodegeneration by molecular chaperones. Nat. Rev. Neurosci. 6:11–22 [DOI] [PubMed] [Google Scholar]
- 27. Becker EB, Bonni A. 2006. Pin1 mediates neural-specific activation of the mitochondrial apoptotic machinery. Neuron 49:655–662 [DOI] [PubMed] [Google Scholar]
- 28. Lu PJ, Wulf G, Zhou XZ, Davies P, Lu KP. 1999. The prolyl isomerase Pin1 restores the function of Alzheimer-associated phosphorylated tau protein. Nature 399:784–788 [DOI] [PubMed] [Google Scholar]
- 29. Dai Q, Zhang C, Wu Y, McDonough H, Whaley RA, Godfrey V, Li HH, Madamanchi N, Xu W, Neckers L, Cyr D, Patterson C. 2003. CHIP activates HSF1 and confers protection against apoptosis and cellular stress. EMBO J. 22:5446–5458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Boellmann F, Guettouche T, Guo Y, Fenna M, Mnayer L, Voellmy R. 2004. DAXX interacts with heat shock factor 1 during stress activation and enhances its transcriptional activity. Proc. Natl. Acad. Sci. U. S. A. 101:4100–4105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ali A, Bharadwaj S, O'Carroll R, Ovsenek N. 1998. HSP90 interacts with and regulates the activity of heat shock factor 1 in Xenopus oocytes. Mol. Cell. Biol. 18:4949–4960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Westerheide SD, Anckar J, Stevens SM, Jr, Sistonen L, Morimoto RI. 2009. Stress-inducible regulation of heat shock factor 1 by the deacetylase SIRT1. Science 323:1063–1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Chou SD, Prince T, Gong J, Calderwood SK. 2012. mTOR is essential for the proteotoxic stress response, HSF1 activation and heat shock protein synthesis. PLoS One 7:e39679. 10.1371/journal.pone.0039679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lu KP, Hanes SD, Hunter T. 1996. A human peptidyl-prolyl isomerase essential for regulation of mitosis. Nature 380:544–547 [DOI] [PubMed] [Google Scholar]
- 35. Pastorino L, Sun A, Lu PJ, Zhou XZ, Balastik M, Finn G, Wulf G, Lim J, Li SH, Li X, Xia W, Nicholson LK, Lu KP. 2006. The prolyl isomerase Pin1 regulates amyloid precursor protein processing and amyloid-beta production. Nature 440:528–534 [DOI] [PubMed] [Google Scholar]
- 36. Raynes R, Pombier KM, Nguyen K, Brunquell J, Mendez JE, Westerheide SD. 2013. The SIRT1 modulators AROS and DBC1 regulate HSF1 activity and the heat shock response. PLoS One 8:e54364. 10.1371/journal.pone.0054364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Chiang WC, Ching TT, Lee HC, Mousigian C, Hsu AL. 2012. HSF-1 regulators DDL-1/2 link insulin-like signaling to heat-shock responses and modulation of longevity. Cell 148:322–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Eroglu B, Moskophidis D, Mivechi NF. 2010. Loss of Hsp110 leads to age-dependent tau hyperphosphorylation and early accumulation of insoluble amyloid beta. Mol. Cell. Biol. 30:4626–4643 [DOI] [PMC free article] [PubMed] [Google Scholar]