Abstract
The androgen receptor (AR) has critical functions as a transcription factor in both normal and cancer cells, but the specific mechanisms that regulate its nuclear localization are not well defined. We found that an AR mutation commonly reported in prostate cancer generates an androgen-independent gain of function for nuclear import. The substitution, Thr877Ala, is within the ligand-binding domain, but the nuclear import gain of function is mediated by the bipartite nuclear localization signal (NLS) spanning the DNA-binding domain (DBD) and hinge region. Bipartite NLS activity depends on the structure provided by the DBD, and protein interactions with the bipartite NLS are repressed by the hinge region. The bipartite NLS is recognized by importin 7, a nuclear import receptor for several proteins. Importin 7 binding to AR, however, inhibits import by shielding the bipartite NLS. Androgen binding relieves the inhibition by inducing a switch that promotes exchange of importin 7 for karyopherin alpha import receptors. Importin 7 contributes to the regulation of AR import by restraining import until androgen is detected in the cytoplasm.
INTRODUCTION
Nuclear import of proteins is mediated by cis-acting nuclear localization signals (NLSs) that usually contain either one or two clusters of basic amino acids (1). Import signals similar to the monopartite NLS in simian virus 40 (SV40) (PKKKRRV) and the bipartite NLS in nucleoplasmin (KRPAATKKAGQAKKKK) have been identified in hundreds of proteins and provide the specificity for import through interactions with nuclear import receptors (1). Three steps common to all NLS and transport receptor-mediated pathways are (i) receptor recognition of the NLS, (ii) translocation of the NLS-receptor complex through the nuclear pore complex (NPC), and (iii) dissociation of the NLS-receptor complex in the nucleoplasm (2). After release of the NLS-containing protein into the nucleoplasm, import receptors are exported to the cytoplasm for a new round of protein import.
The import receptors that recognize NLSs in proteins are phylogenetically conserved. Within a species, import receptors are either members of the importin β superfamily, which also includes structurally related export receptors (3) or the importin-α family of import receptors (4). The latter are termed karyopherin α (KPNA) proteins. Importin β superfamily members that mediate import are 95- to 125-kDa polypeptides composed of HEAT repeats, which were first recognized in Huntingtin, elongation factor 3, the PR65/A subunit of protein phosphatase 2A, and the lipid kinase TOR and can bind directly to NLSs. Importin β proteins then undergo transient binding to nucleoporins during translocation through the NPC (5). Certain HEAT repeats within importin β family members form a domain that can bind RanGTP, an interaction that promotes NLS cargo release from these receptors within the nucleus (6). KPNA proteins are encoded by seven genes in humans (7). They are polypeptides of ∼60 kDa in size that are composed of Armadillo (ARM) repeats, which bind directly to NLSs. KPNA proteins also interact with importin β, which mediates translocation through the NPC and facilitates NLS cargo release through its interaction with RanGTP (8). KPNA proteins, therefore, function as molecular adaptors between NLS containing cargos and importin β.
NLS recognition and nuclear translocation are generally thought to occur immediately after protein translation in the cytoplasm. Some nuclear proteins, however, accumulate in the cytoplasm after translation and await cues to signal nuclear entry. The steroid hormones androgen and cortisol bind and stimulate nuclear entry and transcriptional activation of the androgen receptor (AR) and glucocorticoid receptor (GR), respectively (9, 10). The ability of AR and GR to selectively bind cognate ligand and directly regulate gene expression makes these receptors important transducers of steroid hormones. Hormones and nonhormone ligands specific for nuclear receptors bind to the ligand-binding domains (LBDs), which rely on chaperone proteins such as Hsp90 to maintain a LBD conformation that is competent for ligand binding. Ligand binding promotes changes in LBD structure that are accompanied by Hsp90 release and nuclear translocation (11). The correlation between hormone-binding, chaperone release, and nuclear import of the steroid receptor complex has led to the view that molecular chaperones could play key roles in regulating the activity of NLSs.
Defining AR import and export mechanisms will lay the groundwork for understanding if AR trafficking is altered in prostate cancer and whether the AR transport mechanisms might be exploited therapeutically. Here, we report that a single amino acid substitution in AR observed in prostate cancers (T877A) induces androgen-independent import of AR. The effect of T877A, which is in the LBD, is communicated to the bipartite NLS in the DNA-binding domain (DBD)–hinge region. Thus, the T877A gain of function for nuclear import is relayed between two structural domains in AR. Using biochemical and cell-based assays, we show that importin 7, previously known only as an import factor, binds directly to the bipartite NLS of unliganded AR and inhibits import by competing with KPNA import receptors. Ligand binding releases importin 7 from AR, permits engagement of the NLS with a KPNA receptor, and results in AR import into the nucleus. Our data suggest that importin 7 functions as a molecular chaperone that inhibits AR import and that androgen promotes nuclear import by inducing a switch from importin 7 to KPNA binding. Our data are consistent with a model based on cytoplasmic retention mediated by masking of the AR NLS.
MATERIALS AND METHODS
Plasmid construction.
Flag-AR(T877A), Flag-AR(K633E), and Flag-AR(K633E, T877A), were constructed using QuikChange II site-directed mutagenesis kit (Stratagene) with Flag-AR(WT), which has been described (12). AR ΔAF1 cDNA fragment containing amino acids 503 to 919 was cloned into vector pcDNA3 (Invitrogen) (13). AR ΔLBD cDNA fragment containing amino acids 1 to 710 was cloned into vector PCI-Neo (Promega) (14). Flag-AR with C-terminal fusion of c-Abl kinase NES was described previously (15), and T877A mutation was introduced by site-directed mutagenesis. Full-length AR, AR K633E and AF1 (1–565), and LBD (710–920) were cloned into pCMVTNT vector (Promega) for in vitro translation. The GST-DBD-hinge plasmid was kindly provided by Daniel Gioeli (University of Virginia). Other glutathione S-transferase (GST)-tagged proteins were generated by cloning the respective cDNAs into pGEX4T-1. Human importin 7 cDNA was cloned into pKH3 vector (a gift from Ian Macara, Vanderbilt University) to generate three-hemagglutinin (3×HA)-tagged importin 7. N-terminal HA-tagged human KPNA1-7 cDNAs were cloned into pCMVTNT vector. Plasmids encoding nuclear receptors with N-terminal green fluorescent protein (GFP) fusions were kindly provided by Gordon Hager (National Institutes of Health).
Antibodies.
A polyclonal antibody against the AR NLS was produced in rabbits by using standard methods (14). In brief, a peptide containing a portion of the AR NLS bipartite sequence (CYEAGMTLGARKLKK) was coupled to keyhole limpet hemocyanin through an N-terminal cysteine. The peptide conjugate was injected into rabbits (Cocalico), and immune serum was purified using immobilized peptide. AR21 (against the AR N terminus) and AR hinge antibodies were previously described (14). Commercial antibodies to HA (mouse monoclonal antibody 16B12 [Covance]; rabbit polyclonal antibody SC-805 [Santa Cruz]), KPNA4 (SC-81364 [Santa Cruz]), importin β (antibody 610560 [BD]), and importin 7 (IM6-3131 [IMGenex]) were also used in the study.
Tissue culture and cell transfection.
PC3, Cos7, HeLa, and LNCaP cells were purchased from the American Type Culture Collection. LAPC4 cells were kindly provided by Charles Sawyers (Memorial Sloan-Kettering Cancer Center). Transient transfections were performed in PC3 and Cos7 cells using Fugene 6 (Roche) according to the manufacturer's protocols. The following media (Gibco/Invitrogen) were used: RPMI 1640 supplemented with 5% fetal bovine serum (FBS; Invitrogen or Atlanta Biologicals) for PC3 cells, Dulbecco modified Eagle medium (DMEM) supplemented with 10% FBS (Invitrogen) for Cos7 cells, DMEM supplemented with 5% FBS and 5% NCS for HeLa cells, T Medium supplemented with 5% FBS for LNCaP cells, and Iscove modified Dulbecco medium supplemented with 10% FBS for LAPC4 cells. For experiments involving androgen treatment, cells were transferred to phenol red-free medium supplemented with 5% charcoal-stripped serum at least 24 h prior to addition of the synthetic androgen R1881 (1 nM) or ethanol vehicle (0.01%).
IF microscopy.
Cos7 cells were plated directly on glass coverslips, whereas PC3 and LNCaP cells were plated on glass coverslips precoated with 10 μg of poly-d-lysine (Sigma-Aldrich)/ml. Cells pretreated with either vehicle or 1 nM R1881 (Perkin-Elmer) were fixed using 4% formaldehyde. Immunofluorescence (IF) microscopy was performed according to standard methods with antibody AR21, AR hinge, and anti-HA monoclonal antibody. Nuclei were counterstained with DAPI (4′,6′-diamidino-2-phenylindole). Wide-field fluorescence microscopy was performed using a Nikon Eclipse E800 equipped with a Hamamatsu C4742-95 charge-coupled device camera. Fluorescence measurements of digital images were performed using Openlab 5.5 (Improvision) or ImageJ (16), and the P values were calculated using the TTEST function of Microsoft Excel 2007. Unless indicated, the nuclear to cytoplasmic ratio of AR was measured for 50 to 100 cells per condition.
Digitonin-permeabilized cell import assay.
Nuclear import assays in digitonin-permeabilized cells were performed essentially as described previously (17). In brief, HeLa cells were seeded onto coverslips 24 h before use. Cells were washed three times with transport buffer (20 mM HEPES [pH 7.4], 110 mM potassium acetate, 2 mM magnesium acetate, 0.5 mM EGTA) containing 2 mM dithiothreitol (DTT) and 1-μg/ml portions (each) of leupeptin, pepstatin, and aprotinin and then permeabilized with 0.005% digitonin for 5 min at 25°C. Import reactions contained 1.2 μg of transport ligands (GST-GFP fusion proteins), and an energy regenerating system (5 mg of bovine serum albumin [BSA]/ml, 80 U of creatine phosphokinase/ml, 1.6 mg of creatine phosphate/ml, 1 mM ATP, 1 mM GTP) diluted in transport buffer. The reaction was carried out at 30°C for 30 min and terminated by transferring coverslips to ice-cold transport buffer. After additional washes in transport buffer, the coverslips were fixed, stained with DAPI, and imaged by fluorescence microscopy. The levels of nuclear fluorescence in randomly selected fields were determined in ≥100 cells per condition.
Silver staining and immunoblotting.
Proteins were resolved by SDS-PAGE and detected by silver staining or Coomassie blue staining or by immunoblotting and enhanced chemiluminescence with the antibodies indicated in the figures. For silver staining, SDS-PAGE gels were fixed in 50% methanol and 10% acetic acid overnight. The fixed gels were washed extensively, incubated with sodium thiosulfate solution (2 mM) for 90 s, and then incubated with 1.8 mg of silver nitrate/ml for 25 min. Gels were transferred to developing solution (20 mg of K2CO3/ml, 0.002% formaldehyde, 0.08 mM sodium thiosulfate) until protein bands were visible. Development was stopped in 10% acetic acid. For quantitative immunoblotting, blots were detected with fluorescently labeled secondary antibodies and quantified by using an Odyssey infrared imaging system (LI-COR, Lincoln, NE).
Protein expression and binding assays.
GST fusion proteins were expressed in BL21(DE3) bacteria, lysed by using a French press, and purified by standard methods (18). In brief, GST-tagged proteins were isolated using glutathione beads, eluted in 50 mM Tris-Cl (pH 8.0) containing 10 mM glutathione, and dialyzed into phosphate-buffered saline (PBS). His-tagged proteins (importin β, importin 7, KPNA4, and Ran) were expressed and purified using a Talon metal affinity resin (Clontech). Recombinant proteins were dispensed as single use aliquots, flash frozen in liquid N2, and stored at −80°C. Purified histone H1 was purchased from Calbiochem. In vitro transcription-translation assays with 35S-labeled methionine were performed using the TNT-coupled system (Promega). For experiments involving MgCl2 elutions, glutathione beads containing the indicated GST fusion proteins were incubated with 2 ml of reticulocyte lysate (RL) containing 1 mM DTT, 1 μg (each) of leupeptin, pepstatin, and aprotinin/ml for 4 h at 4°C. After three PBS washes, bound proteins were eluted with a gradient of MgCl2 (0.05 to 1.6 M). Proteins were precipitated with methanol and analyzed by SDS-PAGE. Amino acid substitutions that interfere with NLS function have been published (19). We tested the effect of charge reversal on six different amino acids (single mutants: R617E, K618E, R629E, K630E, K632E, and K633E) in the bipartite NLS using a GST-DBD-NLS fusion protein and [35S]methionine-labeled importin 7 binding as the readout. Each of these substitutions reduced importin 7 binding to GST-DBD-NLS (data available upon request).
Receptor competition binding to the AR bipartite NLS was performed using GST-DBD-NLS immobilized on beads (1 μg/μl beads). Recombinant His-tagged KPNA4 and importin 7 were thawed on ice and clarified by using an air-driven ultracentrifuge (200,000 × g) for 10 min. KPNA4 was added (1 μM final) to a series of binding reactions that contained 0, 0.075, 0.125, 0.25, 0.5, and 1.0 μM importin 7. The samples were mixed end over end for 3 h at 4°C and washed five times with 0.5 ml of wash buffer (25 mM Tris [pH 7.5], 50 mM NaCl, 5 mM MgCl2, 0.1 mM EDTA, 0.1% NP-40, 1 mM DTT, protease inhibitors, 10 μM BSA). Beads were eluted with 15 μl of 2× SDS sample buffer and analyzed by immunoblotting for KPNA4 and importin 7.
Enzyme-linked immunosorbent assay (ELISA).
High protein-binding 96-well dishes (Thermo Scientific) were coated with 280 ng of GST recombinant protein per well in 0.05 M sodium carbonate buffer (pH 9.6). Wells were blocked with 1% BSA for 1 h and incubated with dilutions of either an anti-GST monoclonal antibody or an anti-AR NLS antibody. Washes were performed using PBS containing 0.05% Tween 20. Wells were subsequently incubated with horseradish peroxidase-conjugated secondary antibodies and developed with substrate (Sigma Fast OPD tablet), and the absorbance was measured at 492 nm. The absorbance values were normalized to the signal obtained by using an anti-GST monoclonal antibody to correct for the amount of protein immobilized in each well. Half-maximal binding of the anti-AR NLS antibody was determined by curve fitting with OriginPro7.5 data analysis and graphing software (Origin Lab).
Immunoprecipitation.
Cos7 cells were transfected with plasmids using Fugene-6 transfection reagent (Roche). At 24 h posttransfection, cells were treated with 1 nM R1881 for 30 min and lysed in 0.5 ml of immunoprecipitation (IP) buffer (20 mM Tris-HCl [pH 7.5], 150 mM NaCl, 1% NP-40, 1 mM EDTA, 1 mM EGTA, 2 mM DTT, protease inhibitors, and phosphatase inhibitor cocktail [Sigma]). Lysates were sonicated and clarified by centrifugation. Samples were subjected to IP for 3 h at 4°C using HA- or Flag antibody-conjugated agarose beads (Sigma). Complexes were washed three times with IP buffer, solubilized in SDS gel sample buffer, and analyzed by electrophoresis and immunoblotting.
Transcription assays.
AR transcription was measured using the dual-luciferase reporter system and plotted as firefly/Renilla as described previously (13). The firefly luciferase reporter gene used to measure AR activity is based on the 5-kb promoters for prostate-specific antigen (PSA). Reactions were prepared in triplicate, the results were analyzed by using the Student t test, and the data presented are representative of at least three experiments. Small interfering RNA duplex (siRNA) against the human importin 7 mRNA target sequence of 5′-CAGAUAGAUAACACCUGCC-3′ was synthesized by Integrated DNA Technologies. Control siRNA was purchased from Invitrogen. Lipofectamine RNAiMAX (Invitrogen) reagent was used to transfect siRNA into LAPC4 cells. Endogenous transcript levels of two AR-regulated genes (KLK2 and PSA) were measured by real-time PCR. Reactions were prepared in duplicate, the results were analyzed using the Student t test, and the data are representative of at least two experiments. In brief, cells were treated with ligands for 18 h, and RNA was prepared by using an RNeasy minikit (Qiagen), including a DNase treatment step. For each sample, 1 μg of RNA was reverse transcribed using iScript cDNA synthesis kit and amplified using the IQ SYBR green PCR master mix in a MyiQ instrument (all from Bio-Rad). The relative mRNA levels were normalized to β-glucuronidase (GUS) and actin mRNA levels in the same samples by the 2−ΔΔCT method. The forward and reverse primers used for real-time PCR were as follows: KLK2, 5′-CACAGCTGCCCATTGCCTAAAGAA-3′ and 5′-GGCCTGTGTCTTCAGGCTCAAA-3′; PSA, 5′-TGGTGCATTACCGGAAAGTGGATCA-3′ and 5′-GCTTGAGTCTTGGCCTGGTCATTTC-3′; and importin 7, 5′-GCAGTGTTGAGACAAAATTCAGA-3′ and 5′-AACACCATTATCGAGGCCC-3′.
RESULTS
The T877A substitution is permissive for the androgen-independent import of AR.
In a variety of cell types, including the prostate cancer line PC3, ectopically expressed AR assembles with chaperones into a cytoplasmic complex but can undergo androgen-stimulated import into the nucleus (Fig. 1A, middle row). In contrast, endogenous AR in the human prostate cancer line LNCaP is predominantly nuclear in the absence of androgens (Fig. 1A, top row). Because these assays were performed in cells grown in charcoal- and dextran-treated FBS (which depletes steroids), this observation suggests that AR in LNCaP cells can undergo androgen-independent import. Although this could involve a difference in the expression level of a factor that mediates AR import (see Fig. 5), we first considered an explanation based on the DNA mutation in exon 8 of the AR gene that results in a threonine to alanine substitution (T877A) in the LBD (20). We engineered the T877A substitution in AR and introduced it into AR-negative PC3 cells. T877A AR localized to the nucleus of PC3 cells without androgen treatment (Fig. 1A, bottom row). T877A AR nuclear localization contrasted that of wild-type (WT) AR, which was mostly cytoplasmic in the absence of androgen (Fig. 1A, middle row). The T877A substitution, which occurs in advanced prostate cancer (21) and broadens the ligand-binding specificity of AR (20), results in a gain of function for androgen-independent nuclear localization.
Fig 1.
The nuclear import gain of function provided by T877A is mediated by the bipartite NLS in AR. (A) Localization of endogenous AR (T877A) in LNCaP cells and transfected AR (WT and T877A) in PC3 cells. Cells were treated with vehicle (0.1% ethanol) or synthetic androgen (1 nM R1881) for 2 h and processed for IF microscopy. (B) Immunoblotting AR from nuclear (N) and cytoplasmic (C) fractions from LNCaP cells with or without R1881. To separate nuclear and cytoplasmic fractions, cell pellets were permeabilized on ice for 6 min in the same buffer described in a digitonin-permeabilized cell import assay. After a brief centrifugation step, the supernatants were treated as cytoplasmic fractions, and the pellets were treated as nuclear fractions. (C) Immunoblotting WT and T877A forms of AR in nuclear and cytoplasmic fractions from transfected PC3 cells ± R1881. (D) Localization of WT AR and AR containing LBD and NLS mutations in PC3 cells with or without R1881. (E) Nuclear/cytoplasmic (N/C) ratios of AR proteins with or without R1881. The data are presented as means ± the standard deviations (SD). *, P < 0.01.
Fig 5.
Importin 7 can negatively regulate AR and GR import. (A) Effect of importin 7 cotransfection on AR localization, assayed with or without R1881. Wild-type AR was coexpressed with empty vector (left panels) or HA-importin 7 (middle and right panels) in Cos7 cells. Cells were treated with 1 nM R1881 for 30 min (lower panels). The samples were examined by IF microscopy. (B) N/C ratios of AR (from panel A) were quantified and plotted by box plot. **, P < 0.001. (C) AR interaction with importin 7. Cos7 cells were cotransfected with Flag-tagged AR (Flag-AR) and HA-tagged importin 7 (HA-Imp7). After the cells were treated with 1 nM R1881 for 30 min, AR complexes were isolated by IP using anti-Flag–agarose beads, and immunoblotted for AR and HA. (D) Importin 7 binds to AR via the bipartite NLS, and complexes formed in vitro can be dissociated by androgen. WT and K633E mutant AR were translated as [35S]methionine-labeled proteins and used in GST-importin 7 fusion protein binding assays. (E) GFP-tagged AR, GR, and ERβ coexpressed with empty vector (first column) or HA-tagged importin 7 (middle and left panels) in Cos7 cells and visualized by IF microscopy.
The T877A gain of function requires the bipartite NLS.
We used mutational analysis to investigate how the T877A substitution promotes AR localization to the nucleus. Because the LBD is necessary to maintain androgen-free AR in the cytoplasm (Fig. 1D and E; ARΔLBD), we hypothesized that the T877A substitution in the LBD altered an interdomain interaction that normally impinges on the bipartite NLS within the DBD-hinge when AR is not bound by androgen (10, 19). Point mutations in the bipartite NLS have been shown by the Wilson group to inhibit AR import (19). We found that single point mutations that reverse the charge of any of the basic residues of the bipartite NLS reduced function (data available upon request). We used K618E and K633E for our experiments, which correspond to the first and second elements of the bipartite NLS. The K633E substitution prevented the nuclear import gain of function provided by T877A (Fig. 1D and E; P < 0.01). Androgen treatment induced nuclear import of each of the forms of AR (WT, T877A, K633E, and T877A/K633E), presumably through the NLS that resides within the LBD. This result suggests the single and double substitutions do not impair LBD folding. Because the bipartite NLS (i) mediates androgen-independent AR import in the context of the T877A variant and (ii) helps dictate the nuclear/cytoplasmic ratio of AR in the presence of androgen, we set out to characterize the bipartite NLS in more detail.
The bipartite NLS of AR is necessary but not sufficient for nuclear import.
We used a digitonin-permeabilized cell assay to characterize the import pathway utilized by the bipartite NLS of AR. The bipartite NLS in AR contains two clusters of positively charged residues, amino acids 617 to 618 in the second α-helix of the DBD and amino acids 629 to 633 in a region of undefined structure (Fig. 2A, PDB ID 1R4I [22]). Recombinant GST-GFP-DBD-NLS (human AR amino acids 556 to 633) and GST-GFP (control protein) were used as import substrates, and nuclear accumulation quantified ≥100 cells per condition using fluorescence microscopy. We found that GST-GFP-DBD-NLS was imported into the nuclei of digitonin-permeabilized cells (Fig. 2B). Import was reduced significantly in reactions performed on ice (0°C), as well as in the absence of an energy-regenerating system (−ERS) (Fig. 2B and C). Import was reduced dramatically by the presence of a 20-fold excess of the NLS-containing protein histone H1 (Fig. 2D). The DBD-NLS import pathway is therefore temperature sensitive, energy dependent, and saturable. The GST-GFP control protein was efficiently excluded from the nucleus under all reaction conditions (Fig. 2C), and GST-GFP-DBD-NLS containing a lysine-to-glutamate substitution (K618E) in the first element of the bipartite NLS was not imported (Fig. 2E). This indicates nuclear import of the GST-GFP-DBD-NLS reporter protein is strictly dependent on a functional NLS. Of note, a GST-GFP fusion protein containing the bipartite NLS (amino acids 617 to 633) without the DBD was not imported (Fig. 2F). Because the bipartite NLS is necessary but not sufficient for import, the structure contributed by the DBD appears to be critical for NLS activity.
Fig 2.
DBD structure is important for activity of the bipartite NLS in AR. (A) Ribbon model of the AR DBD. Amino acids that comprise the first element of the bipartite NLS (R617, K618) are shown as a wire-frame diagram. The protein sequence of the second element and the hinge region is shown but was not part of the crystal structure. The model was generated with PyMOL using the AR DBD structure (1R4I) solved by the Gewirth laboratory (22). (B) Fluorescence images (GFP) showing nuclear import of GST-GFP-DBD-NLS reconstituted in the digitonin-permeabilized cell import assay. The assay used HeLa cells and conditions indicated in the figure, including with (+) or without (−) an energy-regenerating system (ERS). (C) Quantitation of nuclear import in the digitonin-permeabilized cell import assay. Nuclear levels of GST-GFP-DBD-NLS and the control protein GST-GFP were measured by microscopy (n = number of cells measured). (D) Quantitation of GST-GFP-DBD-NLS import in the digitonin-permeabilized cell import assay. Including histone H1 in the reaction inhibits GST-GFP-DBD-NLS import. (E) Mutation of a lysine (K618E) in the first element of the bipartite NLS of AR inhibits nuclear import in vitro. Nuclear import of GST-GFP-DBD-NLS containing the K618E mutation was analyzed in the permeabilized cell import assay, quantified, and compared to the WT protein. (F) The bipartite NLS of AR requires the DBD for import activity. Sequence encoding AR amino acids 617 to 633 were engineered as a GST-GFP fusion protein and compared to GST-GFP-DBD-NLS for import activity in the permeabilized cell import assay. (G) Reticulocyte lysate (RL) has opposite effects on import of AR DBD-NLS and SV40 NLS. RL addition to the digitonin-permeabilized cell import assay inhibited nuclear import of GST-GFP-DBD-NLS and stimulated nuclear import of GST-GFP-SV40 NLS. **, P < 0.001.
RL inhibits import mediated by the AR bipartite NLS.
Reticulocyte lysate (RL) is a rich source of transport factors and stimulates nuclear import in permeabilized cell assays (23). RL addition to the permeabilized cell assay, however, reduced import of GST-GFP-DBD-NLS by ∼80% (Fig. 2G). Under the same reaction conditions, RL addition increased by 2-fold the nuclear import of a GST-GFP protein containing the SV40 NLS (Fig. 2G). The fact that RL has opposite effects on import mediated by the DBD-NLS and SV40-NLS suggests there are signal-specific differences in these two import pathways.
Transport factors from RL bind the bipartite NLS.
To identify the factors involved in regulating import mediated by the AR bipartite NLS, we used GST-DBD-NLS to isolate binding proteins from RL. In brief, glutathione beads containing GST-DBD-NLS and GST were incubated with RL, washed, eluted with a step gradient of MgCl2, and analyzed by SDS-PAGE and silver staining. Two of the polypeptides that eluted from GST-DBD-NLS beads with 50 to 100 mM MgCl2 had molecular masses (∼95 and 120 kDa) that were similar to those of importin β superfamily members (Fig. 3A). Immunoblotting revealed the two proteins were importin β and importin 7 (Fig. 3B). To determine whether importin β and importin 7 from RL bind specifically to the bipartite NLS, we performed the experiment with GST-DBD-NLS K633E, a substitution that causes a loss of function in the DBD-NLS. By immunoblotting, importin β and importin 7 from RL bound to the WT protein, but neither protein bound to GST-DBD-NLS K633E (Fig. 3C). Thus, importin β and importin 7 binding is specific for a functional import signal with the AR DBD-NLS.
Fig 3.
Importin 7 and importin β bind the bipartite NLS of AR. (A and B) Isolation of importin 7 and importin β from RL using GST-DBD-NLS immobilized on glutathione beads. The beads were eluted with a step gradient of MgCl2, and the fractions were precipitated and analyzed by SDS-PAGE and silver staining (A) and by immunoblotting (B) with antibodies to importin 7 and importin β. Two arrows in panel A indicate the positions of importin 7 (upper) and importin β (lower), respectively. (C) Importin 7 and importin β binding to the DBD-NLS requires a functional NLS. WT (left panels) and mutant (K633E) forms of GST-DBD-NLS (right panels) were used to enrich for binding factors from RL. The beads were eluted with MgCl2 and analyzed by SDS-PAGE and immunoblotting.
Importin 7 binding to the bipartite NLS is specific and inhibited by the hinge region.
We analyzed in more detail the specificity of importin 7 binding to the bipartite NLS in AR. We used 35S-labeled importin 7 and a set of GST-DBD-NLS proteins immobilized on glutathione beads. Importin 7 binding required both elements of the bipartite NLS, since mutation of the first element (R617E or K618E) or deletion of the second element (ΔE2; amino acids 629 to 633) eliminated binding (Fig. 4A). Importin 7 also failed to bind to the isolated bipartite NLS (Fig. 4B, lane 2), suggesting that DBD structure is critical for the interaction.
Fig 4.
The hinge region in AR represses importin 7 binding to the NLS. (A) Importin 7 binding to the AR bipartite NLS. Importin 7 was translated as [35S]methionine-labeled proteins and used in binding assays with GST fusion proteins immobilized on glutathione beads. (B) The DBD and hinge regions affect importin 7 binding to the bipartite NLS. Binding assays (see panel A) were performed in duplicate, and bound fractions were measured by densitometry of X-ray films. (C) Quantitation of importin 7 binding to the bipartite NLS in AR. Interactions between importin 7 with the bipartite AR NLS requires the DBD, since only background levels of binding are observed with GST-NLS (amino acids 617 to 633). The hinge region strongly represses importin 7 binding the bipartite AR NLS. The data are presented as means ± the SD. *, P < 0.01. (D) Immunoblotting with an anti-AR NLS antibody. Affinity-purified antibody was used to probe the indicated GST fusion proteins. A duplicate gel was stained with Coomassie blue. (E) The hinge region reduces anti-AR NLS antibody binding measured by ELISA. GST-DBD-NLS and GST-DBD-NLS-hinge fusion proteins were adsorbed to microtiter wells and incubated with a dilution series of antibody. (F) Inhibitory effect of the hinge region on nuclear import. Nuclear import of GST-GFP-DBD-NLS and GST-GFP-DBD-NLS-hinge was measured in digitonin-permeabilized HeLa cells. **, P < 0.001.
Including the 38 amino acids hinge region (amino acids 634 to 671) on the C-terminal side of the DBD-NLS caused a large reduction in importin 7 binding (Fig. 4B and C), showing that the hinge can negatively regulate importin 7 binding to the AR NLS. To explore how the hinge inhibits importin 7 binding, we generated an antibody to a segment within the bipartite NLS (AR amino acids 619 to 633) for the purpose of testing whether the hinge alters NLS accessibility. By immunoblotting (Fig. 4D), the anti-NLS antibody bound to DBD-NLS-hinge (lane 1, AR amino acids 548 to 671), DBD-NLS (lane 2, AR amino acids 548 to 633), DBD-NLS lacking the second element of the NLS (lane 3, AR amino acids 548 to 628), and AR amino acids 617 to 633 (lane 4). These data, together with the sequence of the peptide used to generate and purify the antibody (see Materials and Methods), places the epitope within amino acids 619 to 628 of AR. The anti-NLS antibody does not recognize the SV40 NLS (lane 5), and the antibody can immunoprecipitate full-length AR from cell extracts (data not shown).
The anti-NLS antibody was used to probe for NLS accessibility in the GST-DBD-NLS-hinge (amino acids 556 to 675) and GST-DBD-NLS (amino acids 556 to 633) proteins by ELISA. Equivalent amounts of GST-DBD-NLS and GST-DBD-NLS-hinge proteins were adsorbed to microtiter wells, based on detection with an anti-GST antibody. We determined that anti-NLS antibody binding to GST-DBD-NLS-hinge protein was 3.6-fold lower than binding to GST-DBD-NLS (Fig. 4E). This result suggests the presence of the hinge region reduces protein interaction with the bipartite NLS. This could occur if the hinge reduces NLS accessibility to the antibody or alters the structure in a manner that affects the epitope. To determine whether the hinge affects nuclear import mediated by the DBD-NLS, we compared the levels of nuclear localization of GST-GFP-DBD-NLS and GST-DBD-NLS-hinge in a digitonin-permeabilized cell assay. The hinge reduced nuclear import mediated by the AR NLS by ∼50% (Fig. 4F). Thus, the hinge negatively affects antibody and importin 7 binding to the NLS and nuclear localization mediated by the bipartite NLS.
Importin 7 inhibits AR import, and the inhibition is reversed by androgen addition.
Specific binding of importin 7 to the bipartite NLS in AR, together with its role as an import receptor for histone H1 (24), suggested that importin 7 functions as an AR import factor. We examined the effects of importin 7 on AR localization in Cos7 cells, where, in contrast to PC3 cells, AR is predominantly nuclear under standard culture conditions (Fig. 5A and B). Cotransfecting importin 7 (HA-Imp7) caused a pronounced shift of AR from the nucleus to the cytoplasm, a distribution that was reversed by androgen addition (Fig. 5A and B). Importin 7 binding to AR was observed by coimmunoprecipitation, and the interaction was significantly reduced by treating cells with androgen (Fig. 5C). Thus, importin 7 retains ligand-free AR in the cytoplasm, and release from retention is correlated with androgen-induced dissociation of AR from importin 7. Androgen also reduced 35S-labeled AR binding to GST-importin 7 immobilized on glutathione beads (Fig. 5D, +R1881). Androgen-induced AR release from importin 7 can therefore occur without AR-induced gene expression.
A low level of AR binding to importin 7 persists in the presence of R1881, similar to the level of importin 7 binding to AR that contains an amino acid substitution in the second element of the bipartite NLS (Fig. 5D, K633E). Since this amino acid substitution abrogates importin 7 binding to the bipartite NLS (Fig. 3), the data suggest that AR might contain a second importin 7 binding site, which was proposed for GR by the Yamamoto group (25).
To determine whether importin 7 can reduce nuclear import of other nuclear receptors, we cotransfected importin 7 with GFP-tagged AR, GR, ERα and ERβ. Importin 7 retained GFP-AR and GFP-GR in the cytoplasm, without a noticeable effect on GFP-ERβ (Fig. 5E) and GFP-ERα (data not shown). This indicates the cytoplasmic retention mediated by importin 7 is a feature that does not extend to all steroid receptors.
AR domains involved in cytoplasmic retention.
To identify the AR domains involved in the importin 7-mediated retention mechanism, we cotransfected AR truncation mutants lacking the N- and C-terminal domains (ΔAF1 and ΔLBD, respectively) with importin 7 and quantified the localizations of the AR proteins by IF microscopy. Deleting the AF1 reduced the extent of cytoplasmic retention, and deleting the LBD essentially abolished retention by importin 7 (Fig. 6A and B). In binding experiments, importin 7 displayed strong interactions with full-length AR and with the isolated LBD, both of which were reduced slightly by the addition of R1881 in vitro (Fig. 6C). Importin 7 binding to the AF1 was extremely low and probably represents background in this assay (Fig. 6C). The binding data (Fig. 5D; Fig. 6A to C) suggest that importin 7 likely contacts two sites within AR, the bipartite NLS and the LBD. The data also suggest that the AF1 contributes indirectly to cytoplasmic retention by importin 7 (Fig. 6A), which could involve the interdomain interaction between the AF1 and the LBD (26, 27).
Fig 6.
AR domains that contribute to cytoplasmic retention by importin 7. (A) Full-length WT AR, AR ΔAF1, and AR ΔLBD were coexpressed with empty vector or HA-tagged importin 7 in Cos7 cells. Cells were stained with antibody against AR hinge (14) and processed for IF microscopy. (B) N/C ratios of AR (from panel A) were quantified and displayed as box plots. **, P < 0.001. (C) Full-length, AF1, and LBD domains of AR were translated as [35S]methionine-labeled proteins and used in GST-importin 7 fusion protein binding assays with or without R1881. (D) WT AR and T877A AR was coexpressed with empty vector or HA-tagged importin 7 in Cos7 cells in the absence of androgen. (E) N/C ratios of AR (from panel D) were quantified and displayed as box plots. **, P < 0.001.
We tested whether the T877A gain of function that increases AR nuclear localization (Fig. 1) reflects escape from cytoplasmic retention. Ectopic expression of importin 7 induced similar fold changes in the nuclear/cytoplasmic distribution of T877A and WT forms of AR, respectively (Fig. 6D and E). By coimmunoprecipitation, importin 7 showed comparable levels of binding to WT and T877A forms of AR (data not shown). Thus, while importin 7 exerts an affect on the localization of both forms of AR, it does not fully overcome the nuclear import gain of function provided by the T877A substitution.
Importin 7 reduces AR transcriptional activity.
To test whether importin 7 affects AR activity as a transcription factor, we assayed for potential effects on transactivation of the PSA promoter using a reporter gene (28). Ectopic expression of importin 7 had a pronounced inhibitory effect on androgen-induced transcription (Fig. 7A). The strong inhibition of AR activity occurs in the presence of androgen, which promotes release of importin 7 (Fig. 5). This suggests that the androgen-independent importin 7 binding reduces AR activity or that importin 7 affects the localization or activity of other factors that regulate AR. Reducing importin 7 levels with siRNA in LAPC4 cells increased transcription of two endogenous genes, PSA and KLK2 (Fig. 7B).
Fig 7.
Importin 7 negatively regulates AR-dependent transcription. (A) AR transcription activity was measured by using a PSA-luciferase reporter gene in Cos7 cells. WT AR and T877A AR was cotransfected with empty vector or HA-importin 7. Cells were treated with 1 nM R1881 for 18 h prior to harvest and luciferase assay. F/R, firefly/Renilla. (B) Real-time PCR to measure endogenous androgen-responsive genes in LAPC4 cells. LAPC4 cells were transfected with siRNA against importin 7 with or without 1 nM R1881 for 18 h before RNA isolation. The transcript levels of PSA, KLK2, and importin 7 were measured by real-time PCR.
Importin 7 and KPNA proteins display reciprocal binding to AR.
Androgen stimulates importin 7 release from AR under conditions where AR translocates into the nucleus (Fig. 5A to D). Release of importin 7 from AR in response to androgen binding could represent a step that occurs prior to import, since its release can be observed in RL in the absence of a nucleus (Fig. 5D). This consideration led us to test whether other transport factors undergo androgen-induced recruitment to AR, which could in turn mediate AR import. We tested the seven KPNA isoforms for AR binding in the absence and presence of androgen by coimmunoprecipitation. All seven isoforms bound to AR, with six of the seven isoforms (KPNA2 to -7) showing androgen-induced binding (Fig. 8A). Thus, androgen treatment of cells decreases importin 7 binding and increases KPNA binding to AR. The fact that KPNA proteins and importin 7 both bind specifically to NLS suggested these factors compete for binding the DBD-NLS on AR. We tested this hypothesis using an in vitro binding assay with GST-DBD-NLS immobilized on beads and importin 7 and KPNA4 added in solution, both as purified, recombinant proteins. Importin 7 and KPNA4 added separately each bound to the AR DBD-NLS (Fig. 8B, lanes 3 and 4). Including importin 7 in reactions that contained a constant amount of KPNA4 resulted in a dose-dependent reduction in binding of the latter (Fig. 8B, lanes 5 to 9). This indicates that importin 7 and KPNA4 compete for the DBD-NLS on AR. These data, together with the fact that androgen has opposite effects on importin 7 and KPNA binding to AR, suggest that androgen binding to AR induces a switch from importin 7 to KPNA binding. This switch in protein interactions is correlated with AR release from cytoplasmic retention and stimulation of nuclear import.
Fig 8.
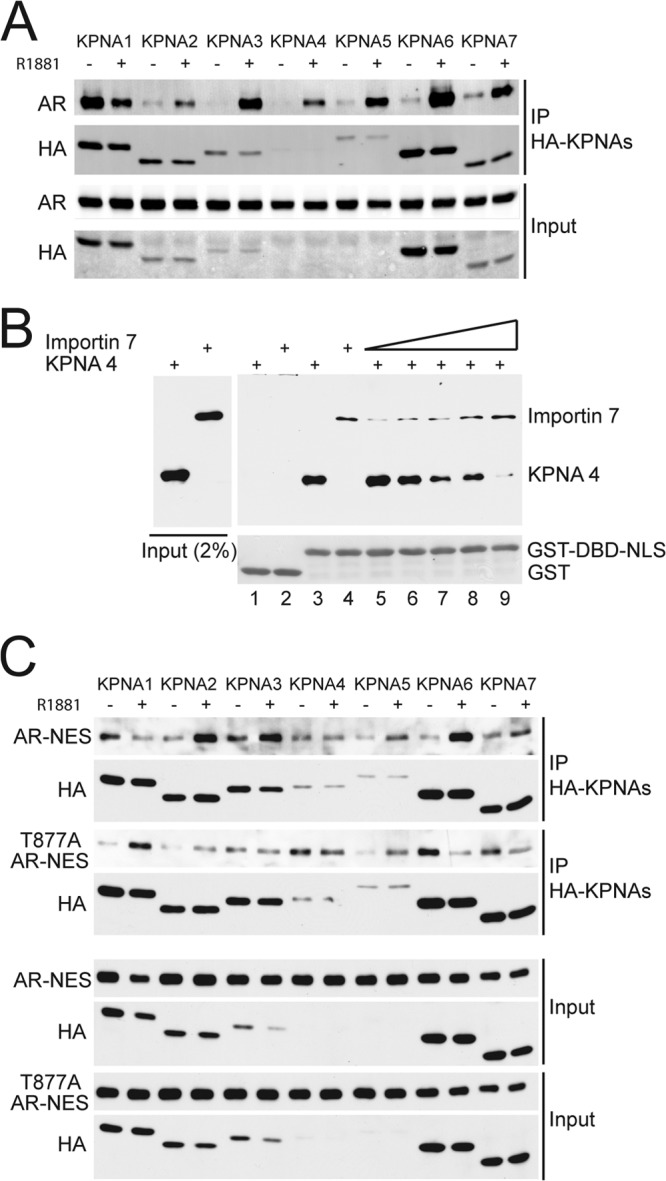
Importin 7 and KPNA proteins compete for binding AR. (A) AR interactions with KPNA proteins in response to androgen treatment. Cos7 cells were cotransfected with AR and HA-tagged KPNAs (HA-KPNAs). After the cells were treated with 1 nM R1881 for 30 min, protein complexes were isolated using HA-agarose beads and immunoblotted for AR and HA. (B) Importin 7 and KPNA4 can compete for binding to AR NLS. Recombinant importin 7 and KPNA4 (each His-tagged) were combined with GST-DBD-NLS immobilized on glutathione beads, and bound fractions were examined by immunoblotting. Increasing amounts of importin 7 were used to compete with a constant amount of KPNA4. (C) Comparison of WT and T877A mutant AR binding to KPNAs. Nuclear export signal (NES) fusions of WT and T877A mutant were cotransfected with HA-KPNAs in Cos7 cells. Immunoprecipitation and immunoblotting were performed using the same methods as panel A.
We also compared KPNA binding to WT and T877A forms of AR to determine whether the androgen-independent import of T877A is associated with changes in import factor interactions. Since KPNA binding to AR is a cytoplasmic reaction, we used NES fusions of WT and T877A to force cytoplasmic localization of both forms of AR. Although all KPNA family members were capable of binding the NES fusions with WT and T877A forms of AR (Fig. 8C), there were changes in the relative levels of KPNA binding in the absence and presence of androgen. Androgen reduced KPNA1 binding to WT AR, but induced binding to T877A AR. Likewise, androgen induced KPNA6 binding to WT AR but reduced binding to T877A AR. Given the complexity of the changes in these interactions, we cannot specifically link the nuclear import gain of function of T877A AR import to increased binding to a particular KPNA family member. We can say that in the context of this assay the T877A substitution can alter the binding interaction between AR and several KPNA proteins, but whether this is the basis of the nuclear import observed by IF microscopy remains a point of speculation.
DISCUSSION
Ligand binding to steroid hormone receptors, including AR, has long been known to induce a “transformation” associated with changes in receptor structure, cellular localization, and transcriptional activity (18, 29). From crystallographic and biochemical studies on nuclear receptors, the fundamental effect of ligand binding at the structural level is repositioning of helix 12 in the LBD, which helps create a favorable binding surface for factors that promote transcription (30, 31). Ligand binding also regulates interactions between the LBD and the N-terminal AF1 domain of AR (26, 27, 32). Although androgen induces AR import and even affects AR mobility within the nucleus (33, 34), exactly how ligand induces nuclear translocation has remained one of the least understood aspects of AR biology.
A topic of inquiry since the pioneering observations by Yamamoto and coworkers on GR (9), the textbook model of ligand-induced translocation (35) posits that nuclear receptors such as GR are retained in the cytoplasm by chaperones, including Hsp90, that contact the LBD. Some features of the chaperone pathway have been described in detail, including how chaperones and cochaperones regulate a cycle of ligand binding, dissociation, and rebinding (36). The textbook model does not articulate how ligand-free nuclear receptors are retained in the cytoplasm, how ligand binding turns “on” the activity of the bipartite NLS, and why certain nuclear receptors, including ER, can undergo efficient translocation in the ligand-free state.
Using transfection-based approaches, we found that the T877A substitution in the LBD of AR generates a nuclear import gain of function (Fig. 1). Given the central role of the LBD in regulating nuclear receptor localization, we anticipated the T877A substitution in AR would enhance the so-called “weak” NLS activity encoded within the three-dimensional structure of the LBD (37, 38). This prediction proved to be incorrect, since the enhancement of nuclear import by the T877A substitution was instead mediated by the bipartite NLS in the DBD-hinge region of AR. Since the LBD has a strong inhibitory effect on AR import, the data suggest that the T877A substitution weakens the repressive effect of the LBD on nuclear import of androgen-free AR. Thus, interdomain interactions, already known to be relevant to the transactivation function of AR (26, 27, 39) and detected by FRET in both the WT and T877A forms of AR (40), can play a role in regulating nuclear import as well. A combination of protein modeling and mutagenesis was used by the Claussens group to show that interdomain communication in AR also occurs between the LBD and DBD (39), and this type of interaction likely occurs in other nuclear receptors as well (41). Whether the T877A substitution stabilizes a conformation that is similar to an androgen-induced state in WT AR or whether this AR variant adopts a unique structure remain an open question. The T877A substitution appears to induce features that affect binding to some, but not all, KPNA family members.
Because the T877A substitution modulated AR import via the bipartite NLS, we focused on the transport pathway utilized by this import signal. We obtained formal evidence that both elements of the bipartite NLS are necessary for full import activity, which is consistent with the observations made originally by the Wilson and Brinkmann groups (10, 19). One of the new findings in our study was that DBD structure was critical for the activity of the AR bipartite NLS. Since the first element of the NLS falls within an alpha helix in the DBD, the DBD probably provides a structural context that helps orient the first element of the bipartite NLS. Although another group concluded that AR uses a monopartite NLS, the contribution of the first element was probably missed because synthetic peptides lacking the DBD were utilized (42).
The most surprising finding in our study was that importin 7 functions as a cytoplasmic retention factor for AR by binding to the bipartite NLS (Fig. 9). Given that importin 7 has been shown to be an import factor for ribosomal proteins, histone H1, HIV-1 reverse transcription complex, hypoxia-inducible factor 1α, EZI, and extracellular signal-regulated kinase (24, 43–48) and the fact that importin 7 binding to the AR bipartite NLS was reduced by the same amino acid substitutions that disrupt AR import, our expectation was that importin 7 would function as an AR import factor. This is clearly not the case, since importin 7 promotes strong cytoplasmic retention of AR. Using isolated domains of AR, we found that importin 7 was also capable of binding to the LBD and that the LBD was required for importin 7 to retain AR in the cytoplasm. There are at least three interpretations of these results, which are not mutually exclusive: (i) importin 7 binding to the LBD mediates cytoplasmic retention; (ii) the LBD modulates the conformation of the bipartite NLS, such that it can be recognized by importin 7; and (iii) in the context of full-length AR, a single molecule of importin 7 contacts both the bipartite NLS and the LBD (Fig. 9). We note that ectopic expression of importin 7 does not inhibit binding of radiolabeled androgen to AR (data available upon request), this suggests that importin 7 does not inhibit AR import by interfering with the ligand binding cycle.
Fig 9.

Summary of the proposed AR import mechanism. After its translation in the cytoplasm, AR associates with importin 7, which shields the bipartite NLS and causes cytoplasmic retention. Androgen binding induces a switch in protein structure that results in importin 7 dissociation and binding of KPNA/importin β receptors. The AR/KPNA/importin β complex translocates into the nucleus where it is disassembled via RanGTP binding to importin β. The switch is presumed to involve a change in structure that increases importin 7 dissociation from AR, but it could also reflect a change that is necessary for or enhances KPNA binding.
In cultured cells, androgen treatment relieves cytoplasmic retention and allows AR to translocate into the nucleus. In protein interaction experiments with AR-importin 7 complexes assembled in vitro, androgen treatment promotes importin 7 release from AR. Androgen-induced release of importin 7 from AR, does not, therefore, require transcription, and it does not require entry into a nucleus. Because importin 7 and KPNA proteins compete for NLS binding, the import mechanism likely involves release of importin 7, exposure of the bipartite NLS to KPNA import receptors, and formation of an AR/KPNA/importin β complex that undergoes nuclear translocation (Fig. 9). We propose that the effect of androgen binding is to induce a switch that shifts the preference from importin 7 to KPNA binding. After translocation into the nucleus, RanGTP binding to the AR/KPNA/importin β complex will promote complex disassembly and release AR for genomic action (49). Our data suggest that multiple KPNA family members could act as import factors for AR; the specific KPNA proteins utilized for AR import would depend on multiple considerations, including relative expression levels of KPNA proteins, and competition with other NLS cargoes. It is interesting that KPNA2, one of the receptors that can recognize AR, displays elevated expression in prostate cancer and is an independent adverse predictor of biochemical recurrence after radical prostatectomy (50).
The ability of importin 7 to recognize a functional, bipartite NLS in AR is a key property of import receptors and it explains how it can shield the AR NLS from interactions with KPNA import receptors. Nuclear transport receptors, including importin 7, were previously shown to reduce aggregation of proteins with clusters of basic charge in vitro, the first indication that they can have a chaperone-like function. In the course of mediating cytoplasmic retention of AR, importin 7 might also reduce aggregation between AR molecules (51).
To our knowledge, our data provide the first evidence that competitive binding between two transport factors can limit nuclear import of an NLS-containing protein. In the absence of androgen, importin 7 binding to the bipartite NLS can dominate and cause cytoplasmic localization of AR. Ligand binding induces a switch in AR that changes the binding preference from importin 7 to a KPNA receptor. Implicit in the model are structural features in AR induced by ligand binding that reduce importin 7 binding to the bipartite NLS, but it is equally possible the switch simply increases AR affinity for KPNA proteins. It is plausible that the hinge region is part of the switch, since it displays a strong inhibitory effect on factor binding to the bipartite NLS. Further analysis of the androgen-induced switch should provide insight into these questions.
ACKNOWLEDGMENTS
These studies were supported by grant P01 CA104106 from the National Cancer Institute.
We thank D. Gioeli, I. Macara, C. Sawyers, G. Hager, and K. Yamamoto for the generous gifts of reagents.
Footnotes
Published ahead of print 7 October 2013
REFERENCES
- 1. Lange A, Mills RE, Lange CJ, Stewart M, Devine SE, Corbett AH. 2007. Classical nuclear localization signals: definition, function, and interaction with importin alpha. J. Biol. Chem. 282:5101–5105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Pemberton LF, Paschal BM. 2005. Mechanisms of receptor-mediated nuclear import and nuclear export. Traffic 6:187–198 [DOI] [PubMed] [Google Scholar]
- 3. Harel A, Forbes DJ. 2004. Importin beta: conducting a much larger cellular symphony. Mol. Cell 16:319–330 [DOI] [PubMed] [Google Scholar]
- 4. Goldfarb DS, Corbett AH, Mason DA, Harreman MT, Adam SA. 2004. Importin alpha: a multipurpose nuclear-transport receptor. Trends Cell Biol. 14:505–514 [DOI] [PubMed] [Google Scholar]
- 5. Gorlich D, Vogel F, Mills AD, Hartmann E, Laskey RA. 1995. Distinct functions for the two importin subunits in nuclear protein import. Nature 377:246–248 [DOI] [PubMed] [Google Scholar]
- 6. Gorlich D, Pante N, Kutay U, Aebi U, Bischoff FR. 1996. Identification of different roles for RanGDP and RanGTP in nuclear protein import. EMBO J. 15:5584–5594 [PMC free article] [PubMed] [Google Scholar]
- 7. Kelley JB, Talley AM, Spencer A, Gioeli D, Paschal BM. 2010. Karyopherin α7 (KPNA7), a divergent member of the importin α family of nuclear import receptors. BMC Cell Biol. 11:63–2121-11-63. 10.1186/1471-2121-11-63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gorlich D, Henklein P, Laskey RA, Hartmann E. 1996. A 41-amino-acid motif in importin-alpha confers binding to importin-beta and hence transit into the nucleus. EMBO J. 15:1810–1817 [PMC free article] [PubMed] [Google Scholar]
- 9. Picard D, Yamamoto KR. 1987. Two signals mediate hormone-dependent nuclear localization of the glucocorticoid receptor. EMBO J. 6:3333–3340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Jenster G, Trapman J, Brinkmann AO. 1993. Nuclear import of the human androgen receptor. Biochem. J. 293(Pt 3):761–768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Prescott J, Coetzee GA. 2006. Molecular chaperones throughout the life cycle of the androgen receptor. Cancer Lett. 231:12–19 [DOI] [PubMed] [Google Scholar]
- 12. Gioeli D, Ficarro SB, Kwiek JJ, Aaronson D, Hancock M, Catling AD, White FM, Christian RE, Settlage RE, Shabanowitz J, Hunt DF, Weber MJ. 2002. Androgen receptor phosphorylation: regulation and identification of the phosphorylation sites. J. Biol. Chem. 277:29304–29314 [DOI] [PubMed] [Google Scholar]
- 13. Yang CS, Vitto MJ, Busby SA, Garcia BA, Kesler CT, Gioeli D, Shabanowitz J, Hunt DF, Rundell K, Brautigan DL, Paschal BM. 2005. Simian virus 40 small t antigen mediates conformation-dependent transfer of protein phosphatase 2A onto the androgen receptor. Mol. Cell. Biol. 25:1298–1308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yang CS, Xin HW, Kelley JB, Spencer A, Brautigan DL, Paschal BM. 2007. Ligand binding to the androgen receptor induces conformational changes that regulate phosphatase interactions. Mol. Cell. Biol. 27:3390–3404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kesler CT, Gioeli D, Conaway MR, Weber MJ, Paschal BM. 2007. Subcellular localization modulates activation function 1 domain phosphorylation in the androgen receptor. Mol. Endocrinol. 21:2071–2084 [DOI] [PubMed] [Google Scholar]
- 16. Schneider CA, Rasband WS, Eliceiri KW. 2012. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9:671–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Adam SA, Sterne-Marr R, Gerace L. 1991. In vitro nuclear protein import using permeabilized mammalian cells. Methods Cell Biol. 35:469–482 [DOI] [PubMed] [Google Scholar]
- 18. Shank LC, Paschal BM. 2005. Nuclear transport of steroid hormone receptors. Crit. Rev. Eukaryot. Gene Expr. 15:49–73 [DOI] [PubMed] [Google Scholar]
- 19. Zhou ZX, Sar M, Simental JA, Lane MV, Wilson EM. 1994. A ligand-dependent bipartite nuclear targeting signal in the human androgen receptor. Requirement for the DNA-binding domain and modulation by NH2-terminal and carboxyl-terminal sequences. J. Biol. Chem. 269:13115–13123 [PubMed] [Google Scholar]
- 20. Veldscholte J, Ris-Stalpers C, Kuiper GG, Jenster G, Berrevoets C, Claassen E, van Rooij HC, Trapman J, Brinkmann AO, Mulder E. 1990. A mutation in the ligand binding domain of the androgen receptor of human LNCaP cells affects steroid binding characteristics and response to anti-androgens. Biochem. Biophys. Res. Commun. 173:534–540 [DOI] [PubMed] [Google Scholar]
- 21. Taplin ME, Bubley GJ, Shuster TD, Frantz ME, Spooner AE, Ogata GK, Keer HN, Balk SP. 1995. Mutation of the androgen-receptor gene in metastatic androgen-independent prostate cancer. N. Engl. J. Med. 332:1393–1398 [DOI] [PubMed] [Google Scholar]
- 22. Shaffer PL, Jivan A, Dollins DE, Claessens F, Gewirth DT. 2004. Structural basis of androgen receptor binding to selective androgen response elements. Proc. Natl. Acad. Sci. U. S. A. 101:4758–4763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kehlenbach RH, Paschal BM. 2006. Analysis of nuclear protein import and export in digitonin-permeabilized cells, p 267–275 In Celis JE. (ed), Cell biology, 3rd ed. Academic Press, Burlington, VT [Google Scholar]
- 24. Jakel S, Albig W, Kutay U, Bischoff FR, Schwamborn K, Doenecke D, Gorlich D. 1999. The importin beta/importin 7 heterodimer is a functional nuclear import receptor for histone H1. EMBO J. 18:2411–2423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Freedman ND, Yamamoto KR. 2004. Importin 7 and importin alpha/importin beta are nuclear import receptors for the glucocorticoid receptor. Mol. Biol. Cell 15:2276–2286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. He B, Kemppainen JA, Voegel JJ, Gronemeyer H, Wilson EM. 1999. Activation function 2 in the human androgen receptor ligand binding domain mediates interdomain communication with the NH2-terminal domain. J. Biol. Chem. 274:37219–37225 [DOI] [PubMed] [Google Scholar]
- 27. He B, Bowen NT, Minges JT, Wilson EM. 2001. Androgen-induced NH2- and COOH-terminal interaction inhibits p160 coactivator recruitment by activation function 2. J. Biol. Chem. 276:42293–42301 [DOI] [PubMed] [Google Scholar]
- 28. Shang Y, Myers M, Brown M. 2002. Formation of the androgen receptor transcription complex. Mol. Cell 9:601–610 [DOI] [PubMed] [Google Scholar]
- 29. Kumar R, McEwan IJ. 2012. Allosteric modulators of steroid hormone receptors: structural dynamics and gene regulation. Endocrinol. Rev. 33:271–299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Weatherman RV, Fletterick RJ, Scanlan TS. 1999. Nuclear-receptor ligands and ligand-binding domains. Annu. Rev. Biochem. 68:559–581 [DOI] [PubMed] [Google Scholar]
- 31. Hur E, Pfaff SJ, Payne ES, Gron H, Buehrer BM, Fletterick RJ. 2004. Recognition and accommodation at the androgen receptor coactivator binding interface. PLoS Biol. 2:E274. 10.1371/journal.pbio.0020274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. van Royen ME, Cunha SM, Brink MC, Mattern KA, Nigg AL, Dubbink HJ, Verschure PJ, Trapman J, Houtsmuller AB. 2007. Compartmentalization of androgen receptor protein-protein interactions in living cells. J. Cell Biol. 177:63–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Marcelli M, Stenoien DL, Szafran AT, Simeoni S, Agoulnik IU, Weigel NL, Moran T, Mikic I, Price JH, Mancini MA. 2006. Quantifying effects of ligands on androgen receptor nuclear translocation, intranuclear dynamics, and solubility. J. Cell. Biochem. 98:770–788 [DOI] [PubMed] [Google Scholar]
- 34. Georget V, Lobaccaro JM, Terouanne B, Mangeat P, Nicolas JC, Sultan C. 1997. Trafficking of the androgen receptor in living cells with fused green fluorescent protein-androgen receptor. Mol. Cell. Endocrinol. 129:17–26 [DOI] [PubMed] [Google Scholar]
- 35. Lodish HF. 2000. Molecular cell biology. W.H. Freeman/Macmillan, New York, NY [Google Scholar]
- 36. Pratt WB, Toft DO. 1997. Steroid receptor interactions with heat shock protein and immunophilin chaperones. Endocrinol. Rev. 18:306–360 [DOI] [PubMed] [Google Scholar]
- 37. Poukka H, Karvonen U, Yoshikawa N, Tanaka H, Palvimo JJ, Janne OA. 2000. The RING finger protein SNURF modulates nuclear trafficking of the androgen receptor. J. Cell Sci. 113(Pt 17):2991–3001 [DOI] [PubMed] [Google Scholar]
- 38. Jenster G, van der Korput JA, Trapman J, Brinkmann AO. 1992. Functional domains of the human androgen receptor. J. Steroid Biochem. Mol. Biol. 41:671–675 [DOI] [PubMed] [Google Scholar]
- 39. Helsen C, Dubois V, Verfaillie A, Young J, Trekels M, Vancraenenbroeck R, De Maeyer M, Claessens F. 2012. Evidence for DNA-binding domain–ligand-binding domain communications in the androgen receptor. Mol. Cell. Biol. 32:3033–3043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Schaufele F, Carbonell X, Guerbadot M, Borngraeber S, Chapman MS, Ma AA, Miner JN, Diamond MI. 2005. The structural basis of androgen receptor activation: intramolecular and intermolecular amino-carboxy interactions. Proc. Natl. Acad. Sci. U. S. A. 102:9802–9807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Chandra V, Huang P, Hamuro Y, Raghuram S, Wang Y, Burris TP, Rastinejad F. 2008. Structure of the intact PPAR-γ-RXR-nuclear receptor complex on DNA. Nature 456:350–356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Cutress ML, Whitaker HC, Mills IG, Stewart M, Neal DE. 2008. Structural basis for the nuclear import of the human androgen receptor. J. Cell Sci. 121:957–968 [DOI] [PubMed] [Google Scholar]
- 43. Jakel S, Gorlich D. 1998. Importin beta, transportin, RanBP5 and RanBP7 mediate nuclear import of ribosomal proteins in mammalian cells. EMBO J. 17:4491–4502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Bauerle M, Doenecke D, Albig W. 2002. The requirement of H1 histones for a heterodimeric nuclear import receptor. J. Biol. Chem. 277:32480–32489 [DOI] [PubMed] [Google Scholar]
- 45. Chachami G, Paraskeva E, Mingot JM, Braliou GG, Gorlich D, Simos G. 2009. Transport of hypoxia-inducible factor HIF-1α into the nucleus involves importins 4 and 7. Biochem. Biophys. Res. Commun. 390:235–240 [DOI] [PubMed] [Google Scholar]
- 46. Chuderland D, Konson A, Seger R. 2008. Identification and characterization of a general nuclear translocation signal in signaling proteins. Mol. Cell 31:850–861 [DOI] [PubMed] [Google Scholar]
- 47. Fassati A, Gorlich D, Harrison I, Zaytseva L, Mingot JM. 2003. Nuclear import of HIV-1 intracellular reverse transcription complexes is mediated by importin 7. EMBO J. 22:3675–3685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Saijou E, Itoh T, Kim KW, Iemura S, Natsume T, Miyajima A. 2007. Nucleocytoplasmic shuttling of the zinc finger protein EZI is mediated by importin-7-dependent nuclear import and CRM1-independent export mechanisms. J. Biol. Chem. 282:32327–32337 [DOI] [PubMed] [Google Scholar]
- 49. Kaku N, Matsuda K, Tsujimura A, Kawata M. 2008. Characterization of nuclear import of the domain-specific androgen receptor in association with the importin alpha/beta and Ran-guanosine 5′-triphosphate systems. Endocrinology 149:3960–3969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Mortezavi A, Hermanns T, Seifert HH, Baumgartner MK, Provenzano M, Sulser T, Burger M, Montani M, Ikenberg K, Hofstadter F, Hartmann A, Jaggi R, Moch H, Kristiansen G, Wild PJ. 2011. KPNA2 expression is an independent adverse predictor of biochemical recurrence after radical prostatectomy. Clin. Cancer Res. 17:1111–1121 [DOI] [PubMed] [Google Scholar]
- 51. Jakel S, Mingot JM, Schwarzmaier P, Hartmann E, Gorlich D. 2002. Importins fulfill a dual function as nuclear import receptors and cytoplasmic chaperones for exposed basic domains. EMBO J. 21:377–386 [DOI] [PMC free article] [PubMed] [Google Scholar]









