Abstract
The genes for all cytoplasmic and potentially all mitochondrial aminoacyl-tRNA synthetases (aaRSs) were identified, and all those tested by RNA interference were found to be essential for the growth of Trypanosoma brucei. Some of these enzymes were localized to the cytoplasm or mitochondrion, but most were dually localized to both cellular compartments. Cytoplasmic T. brucei aaRSs were organized in a multiprotein complex in both bloodstream and procyclic forms. The multiple aminoacyl-tRNA synthetase (MARS) complex contained at least six aaRS enzymes and three additional non-aaRS proteins. Steady-state kinetic studies showed that association in the MARS complex enhances tRNA-aminoacylation efficiency, which is in part dependent on a MARS complex-associated protein (MCP), named MCP2, that binds tRNAs and increases their aminoacylation by the complex. Conditional repression of MCP2 in T. brucei bloodstream forms resulted in reduced parasite growth and infectivity in mice. Thus, association in a MARS complex enhances tRNA-aminoacylation and contributes to parasite fitness. The MARS complex may be part of a cellular regulatory system and a target for drug development.
INTRODUCTION
Aminoacyl-tRNA synthetases (aaRSs) are ubiquitous enzymes that charge specific tRNAs with their cognate amino acids and thus contribute to accurate mRNA translation during protein synthesis (1, 2). In eukaryotes, these enzymes are organized in a multiprotein complex called the multiple aminoacyl-tRNA synthetase (MARS) complex (3–6). A MARS complex that is composed of nine cytoplasmic aaRSs and three accessory proteins, p38, p43, and p18 (also called aminoacyl-tRNA synthetase-interacting multifunctional proteins 1, 2, and 3, respectively), has been characterized in mammalian cells (6, 7). In this complex, methionyl-tRNA synthetase (MetRS), isoleucyl-tRNA synthetase (IleRS), leucyl-tRNA synthetase (LeuRS), and the fused Glu/prolyl-tRNA synthetase (Glu/ProRS) associate with p18, forming subcomplex I (3, 6), and arginyl-tRNA synthetase (ArgRS) and (GlnRS) associate with p43, forming subcomplex II (3, 6, 8). Protein p38 bridges both subcomplexes by interacting with Glu/ProRS and p43 and also interacts with both lysyl-tRNA synthetase (LysRS) and aspartyl-tRNA synthetase (AspRS) (3, 6, 8, 9).
The associations between aaRSs are in most cases mediated by accessory domains that are often at their N or C termini (6, 8, 10, 11). For example, the MARS complex in Caenorhabditis elegans lacks the protein p43, but a p43-like domain is at the C terminus of MetRS. This sequence has a leucine zipper (LZ) domain and a tRNA-binding domain (TRBD), which interacts with the p38 ortholog and other aaRSs (12). Saccharomyces cerevisiae, on the other hand, contains an ortholog of p43, called aminoacyl-tRNA synthetase cofactor 1 protein (Arc1p). Arc1p is part of a well-characterized ternary complex which includes MetRS and glutamyl-tRNA synthetase (GluRS) (11, 13–15). Yeast lacks p38, but six other aaRSs, including threonyl-tRNA synthetase (ThrRS), seryl-tRNA synthetase (SerRS), phenylalanyl-tRNA synthetase (PheRS), and glycyl-tRNA synthetase (GlyRS), have been identified in the MARS complex (16). Interestingly, this complex is also part of the translasome, a multicomplex machinery that is involved in protein synthesis and degradation (16).
The organization of aaRSs in MARS complexes in eukaryotes from yeast to humans appears to improve the efficiency of tRNA-aminoacylation and result in tRNA channeling, which avoids tRNA diffusion in the cytoplasm and enhances its recycling and, hence, translation efficiency (15, 17, 18). Importantly, the MARS complex is also involved in cell signaling and transcriptional and translational control (19–21). The early branching of Trypanosoma brucei within the eukaryotic lineage provides an opportunity to gain insight into the evolutionary diversification of the MARS complex and how this relates to the physiology of different organisms. In addition, T. brucei is a protozoan pathogen that causes human African trypanosomiasis (also known as African sleeping sickness), a lethal disease that is endemic in 36 sub-Saharan countries in Africa (22, 23). Thus, analysis of aaRSs and their association and function within a complex may reveal useful targets for drug development, given their central role in protein synthesis and possibly other cellular regulatory functions.
Little is known about aaRSs in T. brucei and the related trypanosomatid parasites Leishmania spp. and T. cruzi. Few aaRSs have been characterized in T. brucei, and most of the genetic studies have been conducted on the insect stage of the parasite (24–31). Although in T. brucei translation takes place in both the cytoplasm and the mitochondria, only 24 genes have been identified in the genome as potentially encoding aaRSs (32). A few of these genes have been shown to encode both cytoplasmic and mitochondrial enzymes (25–27, 30), and dual cytoplasmic and mitochondrial localization has been shown to result from alternative trans-splicing of IleRS mRNA (29). How these parasites aminoacylate all the tRNAs that are needed for cytoplasmic and mitochondrial translation with this limited set of genes remains to be determined. It is also unknown whether the aaRSs in trypanosomatids associate in multiprotein complexes and, if so, what physiological or regulatory roles this association in complexes might play.
We show here that T. brucei has genes encoding aaRSs to charge all 20 aminoacyl-tRNAs required for protein synthesis and that all tested aaRSs are essential for parasite growth. Some of these enzymes were localized to the cytoplasm or mitochondrion, but most were dually localized to both cellular compartments. We found that cytoplasmic T. brucei aaRSs are organized in a multiprotein complex, which contains at least six aaRSs and three associated proteins. Steady-state kinetic studies show that association in the MARS complex enhances tRNA-aminoacylation efficiency, which in part results from a MARS complex-associated protein (MCP), MCP2, that binds tRNAs and increases their aminoacylation by the complex. Conditional repression of MCP2 results in reduced parasite growth and infectivity in mice. Thus, association in a MARS complex enhances tRNA-aminoacylation and contributes to parasite fitness.
MATERIALS AND METHODS
Cell growth.
T. brucei single-marker Lister 427 (SM427) bloodstream forms were grown in vitro at 37°C in Hirumi modified Iscove's medium 9 (HMI-9) supplemented with 10% (vol/vol) fetal bovine serum (FBS) in the presence of 2 μg/ml of G418. T. brucei 29.13 procyclic forms were grown in vitro at 27°C in semidefined medium 79 (SDM-79) containing hemin (7.5 mg/ml) and 10% (vol/vol) FBS in the presence of G418 (15 μg/ml) and hygromycin (25 μg/ml).
Generation of tandem affinity purification (TAP)-tagged and 3V5-tagged T. brucei cell lines. (i) Generation of pLEW100-3v5 vector and 3V5-tagged aaRS plasmids.
The 3v5 sequences were amplified from the pT73v5 vector (a gift from Marilyn Parsons) by PCR with Phusion high-fidelity DNA polymerase (Thermo Scientific) using specific primers (Table 1). The sequences were cloned in the pLEW100 vector (33) using the vector restriction sites HindIII and BamHI and the PCR product restriction sites HindIII and BglII with T4 DNA ligase (New England BioLabs). The final vector has a 3V5 tag followed by two stop codon sequences (TAGTAA). The 3V5 tag is preceded by a multiple-cloning site containing HindIII, XhoI, and BamHI. This vector was used for generation of 3V5-tagged proteins. DNA sequences were amplified by PCR using specific primers (Table 1) and cloned into the pLEW100-3v5 vector with T4 DNA ligase (New England BioLabs).
Table 1.
Plasmids used in this study with the GeneDB accession number and primers used for each gene cloned
| Primer no. | Plasmid used in this study | Gene or GeneDB accession no. of sequence cloned | Sequencea |
|
|---|---|---|---|---|
| Forward primer | Reverse primer | |||
| 1 | pLEW100-3v5 | 3v5 tag | CCCGGTCTCAAGCTTCTCGAGGGATCCGGTAAGCCTATCCCTAACCCTCTC | CCCGGTCTCAGATCTTTACTACGTGCTATCAAGACCGAGG |
| 2 | pLEW100-MetRS-3v5 | Tb927.10.1500 | CCCAAGCTTATGGCTCTAAAGCTGCTTTCAGA | CCCAGATCTTGTACTCTTTGTATTCTCTGTTGAGCG |
| 3 | pLEW100-ValRS-3v5 | Tb927.6.4480 | CCCGTCGACATGAAACAACTTGCTCCCCAG | CCCAGATCTGAGTAAACTCTTCATCTTTGTTAAACCCTC |
| 4 | pLEW100-SerRS-3v5 | Tb927.11.7170 | CCCGGTCTCAAGCTTCATATGGTGCTTGATATACAGCTGTTTC | CCCGGTCTCCTCGAGCTCCCCCTTGTCGGGT |
| 5 | pLEW100-GlyRS-3v5 | Tb927.11.9640 | CCCGGTCTCAAGCTTATGAGCACCGGGCTGC | CCCGGTCTCGGATCCCGTTGCGGCTGCCTG |
| 6 | pLEW100-CisRS-3v5 | Tb927.6.950 | CCCGGTCTCAAGCTTCATATGAAGGAAAGTGACGGTTTGTTAT | CCCGGTCTCCTCGAGTGATACTTCGGTGCTTTTTAAGCT |
| 7 | pLEW100-GluRS-3v5 | Tb927.6.4590 | CCCGGTCTCAAGCTTATGCTCCGGTTAGCTAACAAGTTTA | CCCGGTCTCGGATCCGACAGCTTGTTCTTTGTTCTGACTTT |
| 8 | pLEW100-MCP2-3v5 | Tb927.8.5330 | CCCGGTCTCAAGCTTATGGAAGGTCCTTTCCTCAGC | CCCGGTCTCGGATCCTGAAACGCTACCGTTGGG |
| 9 | pT7-IleRS-3v5 | Tb927.10.9190 | CACAAGCTCCTAGGAAGCTTCATGAATCGATGCTACTCC | CTCTAGGGATCCCTCGAGCGATTCACCAGCCGACGG |
| 10 | pT7-IleRS-3v5 ΔM1-G66 | Tb927.10.9190 | CACAAGCTCCTAGGAAGCTTCATGACTGGACCACTACAA | CTCTAGGGATCCCTCGAGCGATTCACCAGCCGACGG |
| 11 | pT7-GFP | GFP | CCCGGTCTCCTCGAGATGGTGAGCAAGGGCGAGGAGCTG | CCCGGTCTCGGATCCGGTACCCTACTTGTACAGCTCGTCCATGCC |
| 12 | pT7-IleRSM1-G66-GFP | Tb927.10.9190 | CCCGGTCTCAAGCTTATGAATCGATGCTACTCCCTCCTT | GCCCTTGCTCACCATCGTGGTTTTACTCGC (reverse #12.1) and CCCGGTCTCGGATCCCTACTTGTACAGCTCGTCCATGCC (reverse #12.2) |
| 13 | pT7-ProRS-3v5 | Tb927.10.12890 | CCCCGGTCTCAAGCTTATGCTGATGGCAACATGTATC | CCCCGGTCTCGGATCCATAGCTTCTCCCAAATAAAACCCA |
| 14 | PT7-ProRS-3v5 ΔM1-G46 | Tb927.10.12890 | CCCGGTCTCAAGCTTATGTCAGCAAGTAATTGT | CCCCGGTCTCGGATCCATAGCTTCTCCCAAATAAAACCCA |
| 15 | pLEW79-MH-TAP-MetRS | Tb927.10.1500 | CCCAAGCTTATGGCTCTAAAGCTGCTTTCAGA | CCCAGATCTTGTACTCTTTGTATTCTCTGTTGAGCG |
| 16 | pLEW79-MH-TAP-ProRS | Tb927.10.12890 | CCCGGTCTCAAGCTTATGTCAGCAAGTAATTGT | CCCCGGTCTCGGATCCATAGCTTCTCCCAAATAAAACCCA |
| 17 | pLEW79-MH-TAP-MCP2 | Tb927.8.5330 | CCCGGTCTCAAGCTTATGGAAGGTCCTTTCCTCAGC | CCCGGTCTCGGATCCTGAAACGCTACCGTTGGG |
| 18 | pLEW100-NTAP-IleRS | Tb927.10.9190 | CCCAAGCTTATGACTGGACCACTACAAAACTTTG | CCCAGATCTTTACGATTCACCAGCCGAC |
| 19 | pLEW100-NTAP | N-terminal TAP | ACTTCAATTACACCAAAAAGTAAAATTCACATGCACGATGAAGCCGT (forward #19.1) | CCCAGATCTTCATTAGGATCCCTCGAGAAGCTTAAGTGCCCCGGAGGAT (reverse #19.1) |
| ATAGCGGCCGCTATCGAT (forward #19.2) | GTGAATTTTACTTTTTGGTGTAATTGAAGT (reverse #19.2) | |||
| 20 | pET29-MCP2 | Tb927.8.5330 | CCCCATATGGAAGGTCCTTTCCTC | CCCCTCGAGTGAAACGCTACCGTTG |
| 21 | pQuadra-AlaRS | Tb927.6.700 | ATACCAATGTGATGGGGAGGCAATACTGTGGTCGT | ATACCATAGAGTTGGTGCGCCTTGCAGTATAGATG |
| 22 | pTrypRNAiGate-ProRS | Tb927.10.12890 | TTTCTTGATCACTATGCGGCTG | AAACCCTTAAGCTCCCAGTGGT |
| 23 | pTrypRNAiGate-LysRS | Tb927.8.1600 | ACGCATAATCCGGAGTTCAC | CTGAGCCGAAGGTCTTTACG |
| 24 | pTrypRNAiGate-LeuRS | Tb927.11.3730 | CGGAGGAGCTGTTGAAGAAC | CTCGGAGTAGGCACCAGAAG |
Restriction sites are shown underlined.
(ii) Generation of TAP-tagged aaRSs and pLEW-NTAP vector.
The genes of interest were amplified by PCR with Phusion high-fidelity DNA polymerase (Thermo Scientific) using specific primers (Table 1) and cloned into pLEW79MH-TAP (for C-terminal TAP tagging) (34). For construction of the pLEW100-NTAP vector, the N-terminal TAP tag was amplified by PCR from pLEW79-NTAP (34) without the mitochondrion-targeting sequence with Phusion high-fidelity DNA polymerase (Thermo Scientific) using primers forward #19.1 and reverse #19.1 (Table 1). A second PCR product was obtained by PCR from the pLEW100 vector amplified by primers forward #19.2 and reverse #19.2 and corresponds to the sequence of pLEW100 from the NotI site to the sequence adjacent to the HindIII site. Both fragments were fused by PCR using primers forward #19.1 and reverse #19.2. The fused fragment was cloned into pLEW100. For this final cloning, the pLEW100 vector was digested with NotI and BamHI, the PCR fused fragment was digested with NotI and BglII, and both fragments were ligated with T4 DNA ligase (New England BioLabs). The final vector contains the TAP tag (protein A sequence, tobacco etch virus [TEV] protease site, and calmodulin binding peptide [CBP] sequence), followed by a multiple-cloning site containing HindIII, XhoI, and BamHI (which was added to primer reverse #19.1) The pLEW100-NTAP was used for cloning IleRS. IleRS was amplified by PCR from T. brucei SM427 genomic DNA with specific primers (Table 1) and cloned in pLEW100-NTAP with T4 DNA ligase.
(iii) pT7IleRS-GFP constructs.
For the pT7IleRS-GFP constructs, the green fluorescent protein (GFP) sequence was amplified with Phusion high-fidelity DNA polymerase (Thermo Scientific) and specific primers (Table 1). The GFP template was obtained from the pT7GFP vector (a gift from Marilyn Parsons). For pT7-IleRSM1-G66-GFP, the fragment of DNA corresponding to the N terminus of IleRS (methionine 1 [M1] to glycine 66 [G66], nucleotides 1 to 198) was amplified by PCR with Phusion high-fidelity DNA polymerase (Thermo Scientific) and the corresponding forward primer and reverse primer #12.1 (Table 1). The PCR fragment was fused to GFP using the same forward primer and reverse primer #12.2 by PCR. The final fragment was cloned in the pT7 vector using HindIII and BamHI. The plasmids obtained were digested with NotI, and 10 μg of DNA was used to transfect bloodstream or procyclic forms T. brucei, as previously described (35). Parasites transfected with pLEW100- and pLEW79-derived vectors were selected with phleomycin (5 μg/ml for SM427 bloodstream forms and 2.5 μg/ml for 29.13 procyclic forms), whereas parasites transfected with pT7-derived vectors were selected with 0.1 μg/ml of puromycin. Selection also contained antibiotics needed for maintaining the parental cell line.
TAP-tagged cell lines.
The aaRS genes of interest were cloned into TAP-tagged expression plasmids, which generate a fusion protein of the aaRS enzyme with the TAP tag, which is composed of a protein A domain separated from a CBP sequence by a TEV protease cleavage site (36). Plasmids for expression of TAP-tagged aminoacyl-tRNA synthetases were constructed using the pLEW79-MH-TAP (36) and pLEW100-NTAP vectors (Table 1). The open reading frames of interest were PCR amplified from T. brucei strain Lister 427 genomic DNA, digested with restriction enzymes, and cloned in the HindIII-BamHI sites of the plasmids. Plasmids were linearized with NotI and transfected into procyclic- or bloodstream-form T. brucei. Phleomycin-resistant clones were selected with 5 μg/ml of phleomycin, and the clones were screened for tetracycline-regulated expression.
Purification of TAP-tagged complexes.
TAP-tagged proteins were expressed by induction with tetracycline (50 to 100 ng/ml). After 48 h, 3 liters of T. brucei bloodstream forms growing at 1.2 × 106 to 1.6 × 106 cells/ml was harvested by centrifugation. For procyclic cells, 1 liter of culture growing at 2.0 × 107 cells/ml was used. TAP-tagged complexes were purified from the cell lysate as described previously (36). Briefly, the harvested cells were lysed with 1% Triton X-100 in 50 mM Tris, 150 mM NaCl, 0.2% NP-40, pH 7.4, with EDTA-free protease inhibitor cocktail (Roche). Cell lysate was centrifuged at 10,000 × g for 10 min at 4°C, and the cleared lysate was incubated with protein A-Sepharose 4 fast flow medium (Pharmacia) overnight with rotation at 4°C. The protein-resin mix was washed with 100 ml of wash buffer (50 mM Tris, 150 mM NaCl, 0.2% NP-40), followed by washing in 10 ml of TEV protease buffer (50 mM Tris, 150 mM NaCl, 0.2% NP-40, 1 mM dithiothreitol [DTT]). The mix was incubated with 100 U of ac TEV protease (Life Technologies) in 1 ml of TEV buffer at 4°C for 2 h. Afterward, the eluted fractions (1.5 ml of TEV buffer) were diluted in binding buffer (50 mM Tris, 150 mM NaCl, 0.2% NP-40, 1 mM CaCl2, 10 mM β-mercaptoethanol, 1 mM Mg acetate, 1 mM imidazole) and incubated with CBP-agarose (Pharmacia). The mix was incubated for 1 h with rotation at 4°C and then washed in the same binding buffer. Fractions were eluted with elution buffer (50 mM Tris, 150 mM NaCl, 0.2% NP-40, 10 mM β-mercaptoethanol, 1 mM Mg acetate, 1 mM imidazole, 2 mM EGTA). Eluted fractions were stored in aliquots at −80°C until use.
Mass spectrometry analysis of cell and mitochondrial lysates.
Lysates of T. brucei procyclic forms were prepared with 1% Triton X-100 in 50 mM Tris, 150 mM NaCl, 0.2% NP-40, pH 7.4, with EDTA-free protease inhibitor cocktail (Roche). Cell lysate was centrifuged at 10,000 × g for 10 min at 4°C, and the cleared lysate was prepared for mass spectrometry (2 experiments) by gel-based approaches as described previously (37). Isolation of mitochondria from T. brucei procyclic forms and mass spectrometry (4 experiments) analysis were performed as previously described (37, 38).
Protein identification.
Samples were prepared for mass spectrometry by gel-based approaches as described previously (37). Briefly, proteins in gel pieces were reduced with 10 mM DTT at 56°C for 30 min, alkylated with 55 mM iodoacetamide at room temperature for 45 min, and digested with sequencing-grade modified trypsin (Promega, Madison, WI) at 37°C overnight. The resulting peptides were extracted with 5% formic acid in 50% acetonitrile. Mass spectrometry analyses were performed at the Fred Hutchinson Cancer Research Center facilities using an LTQ or LTQ-Orbitrap mass spectrometer. Peptide and protein identifications was performed using the X!-Tandem and Mascot programs. Using Mascot, the LTQ mass spectrometry results were analyzed with a peptide mass tolerance of ±0.5 Da, a fragment mass tolerance of ±0.5 Da, and average mass values; and for the LTQ Orbitrap mass spectrometry results, a peptide mass tolerance of ±10 ppm, a fragment mass tolerance of ±0.5 Da, and monoisotopic mass values of C13 equal to 2 were used. Analyses with X!-Tandem used a fragment monoisotopic mass error of ±0.5 Da. Data were analyzed against the data in the T. brucei v4.0 predicted protein sequence database, which contains 9,211 protein entries and, additionally, 18 predicted protein sequences from mitochondrion-encoded edited and unedited RNAs and bovine serum albumin (BSA), immunoglobulin heavy and light chain, and keratin sequences. The data set presented here includes only data for the doubly tryptic peptides that have a minimum peptide identification probability of 0.9. Peptides containing more than one missed trypsin cleavage site in the sequence were excluded.
Western blotting, native PAGE, and glycerol gradient analyses.
Western blotting and native PAGE were performed as described previously (37). For glycerol gradient sedimentation, T. brucei bloodstream forms expressing 3V5-tagged proteins were grown at 1.2 × 106 to 1.6 × 106 parasites/ml in 400 ml of medium with 100 ng/ml of tetracycline. Cells were harvested, and lysates were obtained with 1% Triton X-100 in 50 mM Tris–150 mM NaCl with EDTA-free protease inhibitor cocktail (Roche). Volumes of 500 μl of lysate were loaded on 10 to 30% continuous glycerol gradients, as previously described (36), and centrifuged for 8 h at 100,000 × g in a Beckman ultracentrifuge. Twenty-two fractions of 500 μl each were collected from the top, and 30 μl from each fraction was analyzed by Western blotting.
Cloning, expression, and purification of recombinant protein.
The DNA sequence of the gene Tb927.8.5330 was amplified by PCR using specific primers (Table 1) and cloned into the pET29a+ vector (Novagen) using NdeI and XhoI restriction sites with a C-terminal 6× His tag. The constructs were used to transform the Escherichia coli Rosetta 2(DE3)/pLysS strain (Novagen), and protein expression was induced with 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside). E. coli was grown in LB medium and harvested, and the lysate was prepared with the BugBuster reagent (Novagen). Proteins were purified using nickel (Ni2+)-magnetic beads (Millipore), dialyzed against 50 mM Tris, 140 mM NaCl, and kept at 4°C (with 0.05% sodium azide) until use.
RNA binding assays and quantitative RT-PCR.
The RNA binding assay used was adapted from a published method (39). Briefly, 100 μl of T. brucei bloodstream-form total RNA (250 ng/μl) was mixed with or without recombinant MCP2 (rMCP2) containing a 6× His tag at the C terminus in 50 mM HEPES, 140 mM NaCl buffer with 40 U RNase inhibitor (Life Technologies). The protein-RNA mixes were incubated at room temperature for 2 h with rotation. Afterwards, 15 μl of Ni2+-magnetic beads (Millipore) was added and the mixture was incubated with rotation for an additional 1 h at room temperature. Then, the mixes were washed 5 times with 1 ml of phosphate-buffered saline (PBS) and the RNA-protein-magnetic beads mixes were resuspended in 30 μl of diethyl pyrocarbonate (DEPC)-treated water. Random hexamer primers (Life Technologies) were added to the RNA-protein-bead mix and heated for 5 min at 70°C, followed by incubation in ice for 5 min. The mixes were used for cDNA synthesis using a TaqMan cDNA synthesis kit (Life Technologies) according to the manufacturer's instructions. The cDNA was used for quantitative PCRs (qPCRs) using EvaGreen qPCR mix (Bio-Rad), and reactions were performed in a real-time PCR 7500 apparatus (Applied Biosystems) with specific primers to amplify tRNAMet (Tb927.4.3294; forward primer TAATACGACTCACTATAGGGCGAGCGTGGCGC and reverse primer TGGTGCGATCGGTGAGGCT), tRNAGln (Tb927.8.2852; forward primer TAATACGACTCACTATAGGGCTCCCATAGTGTAGCGGTTATC and reverse primer TGGCACTCCCACCTGGACTCG), tRNATyr (Tb927.4.1219; forward primer TAATACGACTCACTATAGGGCTTCTGTAGCTCAATTGGTAGAGCATG and reverse primer TGGTCCTTCCGGCCGGA), tRNASer (Tb927.4.296; forward primer TAATACGACTCACTATAGGGTCACCATACCCAAGTGGTTACG and reverse primer TGGCGTCACCAGCAGGATTCG), tRNAIle (Tb927.8.2851; forward primer TAATACGACTCACTATAGGGCTCCTATAGCTCAGTCGGTTAGAG and reverse primer TGGTGCTCCCAACAGGGGTC), tRNAPro (Tb927.4.1195; forward primer TAATACGACTCACTATAGGGGCCGTTTGGTCTAGTGGCA and reverse primer TGGGGGCCGTTGCGGG), 18S rRNA (Tb927.2.1452; forward primer CGGAATGGCACCACAAGAC and reverse primer TGGTAAAGTTCCCCGTGTTGA), and 5S rRNA (Tb927.8.1381; forward primer CATACTTGGCCGAATGCAC and reverse primer GTACAACACCCCGGGTTCC). The threshold cycle (CT) values for each pulled-down RNA fraction (PDRNA) were normalized to the input RNA CT values (IPRNA) for the same qPCR assay using the following formula: ΔCT for normalized PDRNA = CT for PDRNA − [CT for IPRNA − (log2 IDF)], where IDF is the input dilution factor, which corresponds to the dilution of the input RNA before cDNA synthesis. The percentage of the input was calculated according to the formula 2(−ΔCT for normalized PDRNA) using the average of the normalized PDRNA values for the three experiments.
For qPCR analysis of gene knockdown, T. brucei cell lines were harvested at room temperature (10 min, 1,300 × g) and RNA was isolated using the TRIzol reagent (Life Technologies cDNAs were prepared using TaqMan reverse transcriptase reagents (Applied Biosystems) according to the manufacturer's instructions. Reverse transcription-PCR (RT-PCR) analysis was carried out with EvaGreen reagents (Bio-Rad) according to the manufacturer's instructions with ABI Prism 7000 software (Applied Biosystems) using primers specific for LysRS (Tb927.8.1600; forward primer CGTAAAGACCTTCGGCTCAG and reverse primer ACGCAGTTGCCTTTCAAACT), ProRS (Tb927.10.12890; forward primer ATTTGAGGTCCCTTGCATTGG and reverse primer CGTTTTCATGGCGTTCTGTA), LeuRS (Tb927.11.3730; forward primer ATATTCAGCAACACCATTA and reverse primer ACTCGTTAGACAACTCAT), alanyl-tRNA synthetase (AlaRS; Tb927.6.700; forward primer TGGGTGACAGCGCGATT and reverse primer TGATGCTCCCGTCAATTTCTT), telomere reverse transcriptase (TERT; Tb927.11.10190; forward primer GAGCGTGTGACTTCCGAAGG and reverse primer AGGAACTGTCACGGAGTTTGC), and β-tubulin (Tb927.1.2390; forward primer TTCCGCACCCTGAAACTGA and reverse primer TGACGCCGGACACAACAG). Relative amounts of RNA template in the preparations were calculated using the ΔΔCT method (40), and LinRegPCR software (41) was used for calculating primer efficiency. Parallel amplifications minus the reverse transcription step revealed only insignificant contamination with genomic DNA.
Aminoacylation assays.
Aminoacylation assays were performed as previously described (42). Briefly, reactions were performed in aminoacylation buffer (30 mM HEPES buffer, 140 mM NaCl, 30 mM KCl, 40 mM MgCl2) with 1 mM DTT, 200 μM ATP, 2 U/ml inorganic pyrophosphatase (PPiase; Sigma-Aldrich), 10 mM l-methionine (l-Met; Sigma-Aldrich), and 8 μM tRNAMet at 37°C, and 23 nM MetRS-TAP-tagged protein was purified. The T. brucei tRNAMet (GeneDB accession number Tb927.4.3294) was produced by in vitro transcription as previously described (42). Briefly, in vitro transcription with a MEGAScript T7 polymerase kit (Ambion; Life Technologies) was used for producing tRNAs from a tRNA template (the PCR product contained a T7 promoter, tRNA sequence, and a CCA sequence [42]). Reaction mixtures were extracted with phenol-chloroform-isoamyl alcohol (25:24:1, vol/vol; Sigma-Aldrich), and tRNAs were precipitated with isopropanol (Sigma-Aldrich). tRNAs were resuspended in DEPC-treated water and folded by heating at 70°C for 5 min, followed by slow cooling. The eluate of MetRS-TAP washed with 600 mM NaCl was quantified using a bicinchoninic acid protein assay (Pierce). Quantification of MetRS from the eluate of MetRS-TAP washed with 140 mM NaCl (which retained the complex) was done by comparing the intensity of the MetRS protein to that of the previously quantified MetRS-TAP washed with 600 mM NaCl using SDS-PAGE and Western blotting. Different dilutions of both purifications were separated by SDS-PAGE and stained with Imperial stain (Pierce) or transferred to an Immobilon-FL polyvinylidene difluoride membrane (Millipore) and probed with rabbit polyclonal anti-CBP antibodies (GenScript). Finally, the MetRS-TAP washed with 140 mM NaCl (MetRS complex preparation) was diluted to obtain equal amounts of MetRS in relation to the quantified preparation (MetRS washed with 600 mM NaCl). Figure 4b shows a final comparison of both preparations tested by SDS-PAGE and Western blotting. Variations of the aminoacylation assay conditions are described below. The aminoacylation reactions (total volume, 50 μl to 100 μl each) were performed in clear, flat-bottom 96-well plates (Costar 96-well standard microplates), and the reaction mixtures were incubated for 30 min at 37°C. The reactions were stopped by addition of 100 μl of malachite green (Echelon Biosciences) and developed for 30 min at room temperature. Absorbances were then measured at 620 nm using a Spectramax M2 reader (Molecular Devices). Time course assays in the presence of recombinant MCP2 were performed as described above, except that 10-μl aliquots were withdrawn at times of 0, 2.5, 5, 10, and 30 min, incubated with 10 mM EDTA, and developed with malachite green. The recombinant protein was added to the reaction mixtures at concentrations of 12 and 48 nM. Complex-associated MetRS (MetRS complex washed with 140 mM NaCl) was used at 8 nM, whereas 23 nM was used for dissociated MetRS (MetRS washed with 600 mM NaCl). For enzymological studies, reactions were performed in a 50-μl volume with MetRS-TAP-tagged purifications (washed with 140 mM or 600 mM, both at 23 nM in relation to MetRS), l-Met (0.1, 0.5, 1, 5, and 20 mM), 8 μM tRNAmet, 2 U/ml PPiase, 200 μM ATP, and 1 mM DTT in aminoacylation buffer, for calculating the Km in relation to l-Met. For calculating the Km in relation to tRNAMet, we performed reactions with tRNAMet at 0.1, 0.5, 1, 5, and 25 μM and l-Met at 10 mM. The reaction mixtures were incubated at 37°C; at 2.5, 5, and 10 min, 15-μl aliquots were withdrawn and mixed with 10 mM EDTA on ice to stop the reactions. One hundred microliters of malachite green solution was added, and the absorbance was measured at 620 nm. Reaction velocities were calculated with data collected at the linear phase of the reaction using GraphPad Prism software (v5). Velocities were plotted against the amino acid concentrations, and Michaelis-Menten constants and kcat values were calculated using GraphPad Prism software (v5).
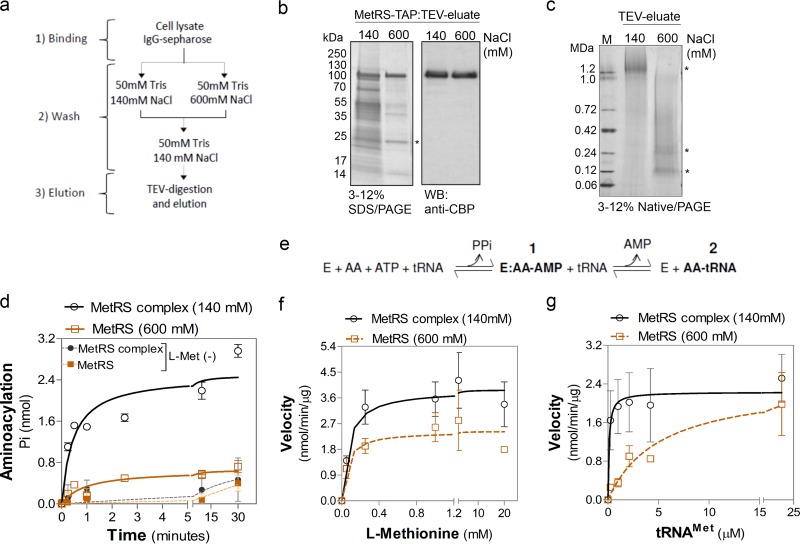
Fig 4.
tRNA-aminoacylation is more efficient in a multienzyme complex in T. brucei. (a) Scheme use for purification of the T. brucei procyclic form as a TAP-tagged MetRS complex and as a complex-dissociated enzyme. (b) SDS-PAGE (4 to 20%) analysis of TEV eluates of MetRS-TAP after 140 mM and 600 mM NaCl washes. Results of Imperial staining of SDS-polyacrylamide gels (left) or Western blotting with polyclonal antibodies against CBP (right) are shown. Asterisk, TEV protease. (c) Native PAGE (3 to 12%) of the TEV eluates after Imperial staining. Top asterisk, MetRS complex; bottom and middle asterisks, bands with sizes similar to those of MetRS-TAP and its dimer, respectively. Lane M, molecular mass markers. (d) Time course of tRNAMet aminoacylation by TEV eluates containing the MetRS complex (after a 140 mM NaCl wash) or dissociated MetRS (after a 600 mM NaCl wash). Reactions were performed with l-Met and tRNAMet as the substrates. Reactions without l-Met [l-Met(−)] are shown as a control. The data show the averages of three experiments performed in duplicate ± SD. (e) Diagram of tRNA-aminoacylation where the amino acid was (i) first activated, creating an enzyme (E):aminoacyladenylate (AA)-AMP intermediate via ATP hydrolysis and PPi release, and (ii) then transferred to tRNA, forming the aminoacyl-tRNA (AA-tRNA), releasing AMP. (f and g) Aminoacylation kinetics of MetRS complex and dissociated MetRS as a function of l-Met concentration (f) or tRNAMet concentration (g) with the calculated kinetic values shown in Table 6. Data in panel g are the averages of three experiments ± SDs, and those in panel f are the averages of four experiments ± SDs.
Generation of conditional-null cells and growth curves.
T. brucei SM427 was transfected with the pLEW100-MCP2-3V5 vector by electroporation, and cells were selected by resistance to phleomycin. Expression of the MCP2-3V5 protein was checked by Western blotting, and the cell line obtained was used for transfection with constructs targeting the endogenous alleles using a PCR-based method (43). Cell lines were selected by resistance to blasticidin and puromycin. For growth curves, 25-cm2 cell culture flasks were seeded with 5.0 × 104 parasites/ml in 10 ml of HMI-9 medium supplemented with 10% FBS, 2.0 μg/ml G418, and 2.5 μg/ml of phleomycin with or without tetracycline (0.25 μg/ml). Parasites were counted daily using a cell counter (Beckman) and diluted daily to 5.0 × 104 parasites/ml in new medium, and the procedure was repeated for 10 consecutive days.
Generation of RNAi cell lines.
T. brucei bloodstream-form SM427 was transfected with vectors for RNA interference (RNAi) and selected with the antibiotics G418 (2 μg/ml) and phleomycin (5 μg/ml). RNAi cell lines were generated for genes for the following: LysRS (Tb927.8.1600), ProRS (Tb927.10.12890), LeuRS (Tb927.11.3730), and AlaRS (Tb927.6.700). The PCR products were generated using high-fidelity platinum Taq DNA polymerase (Life Technologies) (see Table S1 in the supplemental material for the primer sequences). AlaRS PCR products were cloned in the pQuadra vector for RNAi, as previously described (35). The LysRS, LeuRS, and ProRS PCR products were cloned in the vector pTrypRNAiGate, as previously described (44). Briefly, PCR products were incubated with the pCR8/GW/TOPO vector (supplied ready to use) for 5 min at room temperature, transformed into Top10 competent cells, and plated on spectinomycin (100 μg/ml)-containing LB plates as described by the manufacturer (Life Technologies). Plasmid DNAs were isolated from positive transformants and sequenced to verify the correct orientation of the genes between the attL1 and attL2 sites. They were then used in the LR recombination reaction with the pTrypRNAiGate destination vector. For growth curve analysis, RNAi was induced with 1 μg/ml of tetracycline. Parasites were counted daily using a cell counter (Beckman) and diluted daily to 5.0 × 104 parasites/ml in new medium, and the procedure was repeated for from 5 to 10 consecutive days.
Immunofluorescence assay.
Expression of a 3V5-tagged protein in the T. brucei bloodstream form was performed by induction with 0.5 μg/ml of tetracycline. Cells growing at mid-log phase were fixed with 4% paraformaldehyde in PBS, adhered in a poly-l-lysine-treated 2-mm cover glass (Fisher), and permeabilized with 0.2% Triton X-100 in PBS. Cells were blocked with 3% BSA (or 5% FBS) diluted in PBS. Then, they were labeled with fluorescein isothiocyanate (FITC)-conjugated anti-V5 monoclonal antibody (Life Technologies) diluted 1:500 in PBS–3% BSA. Mitochondrial staining was performed by treating the cells with 0.01 μg/ml MitoTracker (Life Technologies) (1 μl of a 100-μg/ml stock diluted in 10 ml of HMI-9 medium with 10% FBS), and the cells were incubated for 30 min at 37°C in 5% CO2 before cell fixation. DNA staining was performed with DAPI (4′,6-diamidino-2-phenylindole; 1 μg/ml diluted in PBS). Cells were analyzed with a Deltavision fluorescence microscope (Olympus IX70).
Infection of mice with an MCP2 conditional-null line.
The T. brucei MCP2 conditional-null line growing at mid-log phase was used to infect BALB/cAnNHsd mice (male; age, 6 to 8 weeks; Harlan Laboratories). Parasites (1.0 × 104) in 200 μl of HMI-9 medium were injected intraperitoneally. Doxycycline at 200 μg/ml and 5% sucrose were added to the mouse drinking water 24 h before infection to induce MCP2 expression (the treated drinking water was replaced daily). Mice in which MCP2 expression was not induced received drinking water containing 5% sucrose only. Parasitemia was monitored daily starting at day 2 postinfection by testing blood obtained by tail prick, and parasites were counted under a light microscope using a Neubauer chamber. Mice with parasitemias higher than 1.0 × 108 parasites/ml of blood were euthanized. All procedures were performed in the vivarium of the Seattle Biomedical Research Institute in compliance with the laws and institutional guidelines (approval number IACUC KS-01).
Sequence analysis and data presentation.
Amino acid sequences were obtained from the NCBI, TriTrypDB, or GeneDB database. Amino acid sequences were analyzed using Geneious Pro (v5.5.5) software (Biomatters Ltd.). Mass spectrometry data were visualized using the Scaffold (v3.6.4) program (Proteome Software Inc.), and intensity maps were generated using the Java Treeview program (v1.1.6r2; Alok).
Statistical analysis.
All data are shown as means ± standard deviations (SDs). Comparisons among groups were done by two-tailed Student's t test for repeated measures using GraphPad Prism (v5.00) for Windows (GraphPad Software). P values of <0.05 with confidence intervals of 95% were considered statistically significant, unless otherwise specified.
RESULTS
Aminoacyl-tRNA synthetases: genes, subcellular location, and essentiality.
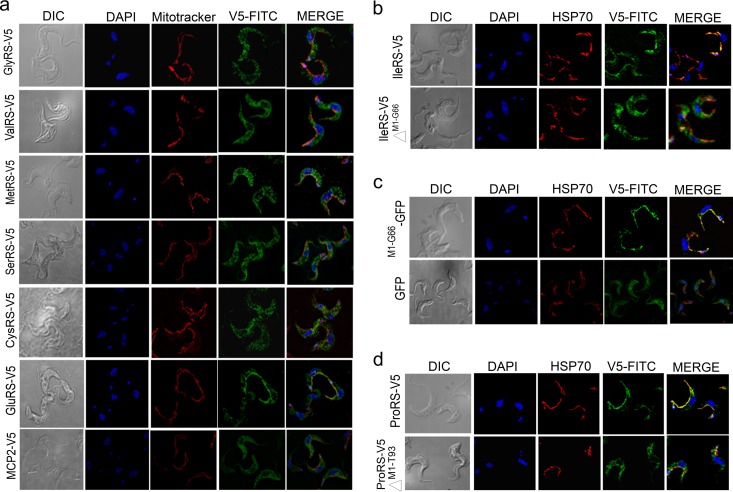
T. brucei was found to have 24 nuclear genes that encode aaRSs (Table 2). Sequence analysis revealed that they are highly conserved between T. cruzi and Leishmania spp. (Table 3), and phylogenetic comparisons showed that most of them have greater predicted amino acid sequence similarity to eukaryotic than prokaryotic aaRSs (Table 3; see Fig. S1 in the supplemental material). We determined the subcellular locations of aaRSs in T. brucei. These are summarized in Table 2, along with the results for others. We generated cell lines that express tetracycline-regulatable ectopic 3V5-tagged copies of some of these genes in bloodstream forms. Immunofluorescence staining localized six of them predominantly to the cytoplasm, namely, valyl-tRNA synthetase (ValRS), cysteinyl-tRNA synthetase (CysRS), MetRS, SerRS, ThrRS, and GlyRS (Fig. 1a and Table 2). Alternative mRNA trans-splicing has previously been shown for other mRNAs in T. brucei, i.e., those which encode IleRS, ProRS, GluRS, GlnRS, and asparaginyl-tRNA synthetase (AsnRS) (45, 46). This results in two mRNA isoforms that either retain or lack the N-terminal mitochondrial targeting sequence (MTS), and dual cytoplasmic and mitochondrial localization was confirmed for IleRS in procyclic forms (29). We expressed the spliced variants of IleRS, ProRS, and GluRS with a 3V5 tag in bloodstream forms and showed dual cytoplasmic and mitochondrial localization by immunofluorescence staining (Fig. 1b to d and Table 2). In addition, mass spectrometry analysis of purified mitochondria and whole-cell lysates of procyclic forms confirmed the mitochondrial localization of tryptophanyl-tRNA synthetase (TrpRS), AspRS, LysRS, ProRS, IleRS, GluRS, GlnRS, and AsnRS and identified seven aaRSs which are potentially dually localized, CysRS, GlyRS, LeuRS, MetRS, PheRS-β, ThrRS, and ValRS (Table 2). Other aaRSs were identified in mitochondrial preparations with low spectral counts, i.e., AlaRS, ArgRS, LysRS, PheRS-α, SerRS, and tyrosyl-tRNA synthetase (TyrRS), and these may be of low abundance in the mitochondrion or possibly adventitious contaminants (Table 2).
Table 2.
T. brucei aaRS enzyme classification, subcellular localization, and essentialitya
| aaRS no. | aaRS | GeneDB accession no. | Subclass | IFA (reference) | Mass spectrometry countb |
Localization | Growth defect (reference)d | |
|---|---|---|---|---|---|---|---|---|
| WC | MP | |||||||
| 1 | AlaRS | Tb927.6.700 | IIc | − | 23 | 3 | C/M | Yes |
| 2 | ArgRS | Tb927.11.1990 | Ic | − | 23 | 2 | C/M | − |
| 3 | AsnRS | Tb927.4.2310 | IIb | − | 10 | 3 | C/M | − |
| 4 | AspRS-2 | Tb927.10.1260 | IIb | M (26) | 0 | 8 | M | Yes (26) |
| 5 | AspRS-1 | Tb927.6.1880 | IIb | C (26 | 12 | 0 | C | Yes (26) |
| 6 | CysRS | Tb927.6.950 | Ia | C | 9 | 27 | C/M | − |
| 7 | GlnRS | Tb927.9.5210 | Ic | C/M (30)c | 14 | 0 | C/M | Yes (30) |
| 8 | GluRS | Tb927.6.4590 | Ic | C/M (30)c | 10 | 8 | C/M | Yes (30) |
| 9 | GlyRS | Tb927.11.9640 | IIc | C | 25 | 13 | C/M | − |
| 10 | HisRS | Tb927.6.2060 | IIa | − | 8 | 0 | C | − |
| 11 | IleRS | Tb927.10.9190 | Ia | C/M (29) | 31 | 70 | C/M | Yes (24, 29) |
| 12 | LeuRS | Tb927.11.3730 | Ia | − | 28 | 13 | C/M | Yes |
| 13 | LysRS-1 | Tb927.8.1600 | IIb | C (27) | 13 | 4 | C/M | Yes (27) |
| 14 | LysRS-2 | Tb927.6.1510 | IIb | M (27) | 0 | 11 | M | Yes (27) |
| 15 | MetRS | Tb927.10.1500 | Ia | C | 22 | 19 | C/M | Yes (31) |
| 16 | PheRS-α | Tb927.11.14120 | IIc | − | 8 | 4 | C/M | − |
| 17 | PheRS-β | Tb927.11.2360 | IIc | − | 10 | 13 | C/M | − |
| 18 | ProRS | Tb927.10.12890 | IIa | C/M | 28 | 11 | C/M | Yes |
| 19 | SerRS | Tb927.11.7170 | IIa | C | 11 | 3 | C/M | Yes (28) |
| 20 | ThrRS | Tb927.5.1090 | IIa | C | 23 | 16 | C/M | Yes (Kalidas et al.) |
| 21 | TrpRS-1 | Tb927.3.5580 | Ib | C (25) | 0 | 0 | C | Yes (25) |
| 22 | TrpRS-2 | Tb927.8.2240 | Ib | M (25) | 0 | 39 | M | Yes (25, 70) |
| 23 | TyrRS | Tb927.7.3620 | Ib | − | 6 | 2 | C/M | − |
| 24 | ValRS | Tb927.6.4480 | Ia | C | 27 | 20 | C/M | − |
The enzyme classification is based on sequence and motif similarity (1, 2). The immunofluorescence assay (IFA) and growth defect results are reported here (Fig. 1 and 2) and elsewhere, as indicated by the citations in parentheses or as reported by S. Kalidas, I. Cestari, K. Stuart, and M. Phillips (unpublished data). Abbreviations: C, cytoplasmic localization; M, mitochondrial localization; C/M, dual localization; C/M, lower-confidence dual localization; −, not determined.
Total spectral counts for each aaRS identified by mass spectrometry of whole-cell (WC) or purified mitochondrial (MP) lysates. The mass spectrometry analyses were performed on procyclic forms.
GlnRS and GluRS were localized by Western analysis of cytosolic and mitochondrial subcellular fractions (30).
Table 3.
Amino acid sequence similarities between aaRSs of T. brucei and various other speciesa
| aaRS no. | GeneDB accession no. | GeneDB annotation | % amino acid sequence similarity |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Trypanosoma cruzi | Leishmania major |
Homo sapiens |
Saccharomyces cerevisiae |
E. coli | Methanobrevibacter smithii | |||||
| Cytoplasmic | Mitochondrial | Cytoplasmic | Mitochondrial | |||||||
| 1 | Tb927.10.1500 | MetRS | 70.9 | 59.5 | 15.7 | 36.2 | 12.6 | 30.7 | 20.9 | 21.1 |
| 2 | Tb927.6.4480 | ValRS | 80.7 | 73.9 | 45 | 43.6 | 43.9 | 36.1 | 24.1 | |
| 3 | Tb927.6.950 | CysRS | 70.8 | 64.1 | 37.8 | 26.5 | 23.1 | 22.2 | ||
| 4 | Tb927.11.9640 | GlyRS | 79.5 | 67 | 43.7 | 41.7 | 41.7 | 9.8/9.3 | 34.8 | |
| 5 | Tb927.11.7170 | SerRS | 78.8 | 73.6 | 49 | 21.8 | 43.6 | 25.4 | 28.3 | 38.1 |
| 6 | Tb927.5.1090 | ThrRS | 78.2 | 68.8 | 46.5 | 39.3 | 43.9 | 29.1 | 31.6 | 14.4 |
| 7 | Tb927.6.2060 | HisRS | 78.4 | 67.5 | 24.8 | 21.8 | 24.4 | 23.1 | 21.5 | 26.2 |
| 8 | Tb927.11.3730 | LeuRS | 76.1 | 63.4 | 39.4 | 12 | 37.5 | 12.3 | 12.4 | 24 |
| 9 | Tb927.6.700 | AlaRS | 74.1 | 65.9 | 34.8 | 20.1 | 19.4 | 17.9 | 31.2 | 16.5 |
| 10 | Tb927.7.3620 | TyrRS | 68.8 | 60.7 | 20 | 6.4 | 19.2 | 6.4 | 26 | |
| 11 | Tb927.11.1990 | ArgRS | 75.5 | 68.1 | 40.5 | 34.9 | 42.2 | 42.2 | 33.3 | 15.9 |
| 12 | Tb927.11.14120 | PheRS-α | 75.6 | 61.4 | 44.6 | 7 | 41.1 | 6.9 | 22.1 | 27 |
| 13 | Tb927.11.2360 | PheRS-β | 76 | 63.4 | 39.1 | 6.1 | 35.5 | 6.9 | 11.5 | 20.2 |
| 14 | Tb927.10.9190 | IleRS | 76.4 | 64.5 | 47.9 | 19.7 | 47.6 | 17.7 | 21.4 | 26 |
| 15 | Tb927.10.12890 | ProRS | 68.9 | 64.9 | 32.5 | 7.2 | 45.6 | 9.3 | 10.3 | 30.2 |
| 16 | Tb927.6.4590 | GluRS | 77.5 | 67.2 | 38.3 | 14.3 | 43.5 | 14.5 | 15 | |
| 17 | Tb927.9.5210 | GlnRS | 82.5 | 74.6 | 31.7 | 32.4 | 40.7 | 20.9 | ||
| 18 | Tb927.4.2310 | AsnRS | 69 | 51.8 | 34.4 | 22.7 | 34.4 | 23.2 | 23.9 | 25.7b |
| 19 | Tb927.3.5580 | TrpRS-1 | 85.1 | 78.4 | 55.3 | 13.2 | 47.2 | 12.3 | 23.7 | |
| 20 | Tb927.8.2240 | TrpRS-2 | 53.3 | 47.7 | 33.3 | 10.5 | 34.5 | 10.9 | 20 | |
| 21 | Tb927.8.1600 | LysRS-1 | 74.7 | 68.7 | 45.6 | 48.4 | 44.8 | 26.1 | 37.3 | 7.4 |
| 22 | Tb927.6.1510 | LysRS-2 | 66.4 | 31.4 | 30.7 | 32.4 | 33.2 | 25.3 | 32.7 | 9.5 |
| 23 | Tb927.6.1880 | AspSR-1 | 79.3 | 69.7 | 43.3 | 15.8 | 39.4 | 12.2 | 15.5 | 26.8 |
| 24 | Tb927.10.1260 | AspRS-2 | 54.5 | 42.9 | 31.4 | 16.2 | 31 | 14.2 | 15.9 | 23.9 |
Sequence identification numbers are listed in Fig. S1 in the supplemental material.
The sequence was compared to the AsnRS sequence of Archaeoglobus veneficus SNP6 (GI: 327315644).
Fig 1.
Subcellular localization analysis of T. brucei aaRSs in bloodstream forms expressing C-terminal 3V5-tagged aaRS (except as indicated). The cells were fixed in 4% paraformaldehyde and stained with FITC-conjugated anti-V5 monoclonal antibody (V5-FITC), mitochondria were stained with Texas Red-conjugated anti-HSP70 monoclonal antibody (HSP70) or MitoTracker, and DNA was stained with DAPI or by GFP fluorescence. The images were then merged, as indicated. Differential interference contrast (DIC) images are also shown. (a) Parasites expressing 3V5-tagged proteins, as indicated. GlnRS has a predicted MTS from M1 to I51 and a trans-splicing site at nucleotide 80 (45), which would remove most of the MTS. Expression of the longer variant resulted in proteins localized in both the cytoplasm and mitochondrion. MCP2 is not an aaRS. (b) IleRS splicing variants. See reference 45 for alternative splicing information. The longer variant, IleRS-3V5 (IleRS-V5), starts from M1 and contains a predicted MTS from M1 to G66, and the shorter variant, IleRS-3V5 with the deletion of amino acids M1 to G66 (IleRS-V5 ΔM1-G66), starts at M67 and lacks the predicted MTS. (c) The IleRS MTS (M1 to G66) was cloned in frame with GFP (M1-G66-GFP) and is sufficient to target GFP to the mitochondrion. Parasites expressing GFP alone were used as a control. (d) ProRS splicing variants. See reference 45 for alternative splicing information. The longer variant, ProRS-3V5 (ProRS-V5), starts with M1 and contains a predicted MTS, and the shorter splicing variant, ProRS-3V5 with a deletion from amino acids M1 to T93 (ProRS-V5 ΔM1-T93), starts at M94 and has the predicted MTS region from M1 to T93 removed.
Overall, T. brucei has 24 aaRS genes for all 20 amino acids (PheRS has two subunits) needed for protein synthesis, as follows: (i) 3 genes encode cytoplasmic AspRS-1, histidyl-tRNA synthetase (HisRS), and TrpRS-1, (ii) 3 genes encode mitochondrial TrpRS-2, AspRS-2, and LysRS-2 (25–27), (iii) 5 genes encode alternatively trans-spliced mRNAs (45, 46), and we confirmed their dual cytoplasmic and mitochondrial localization, and (iv) 13 genes encode cytoplasmic enzymes, of which 7 also appear to have a dual localization and the remaining 6 may also have a dual localization (Table 2).
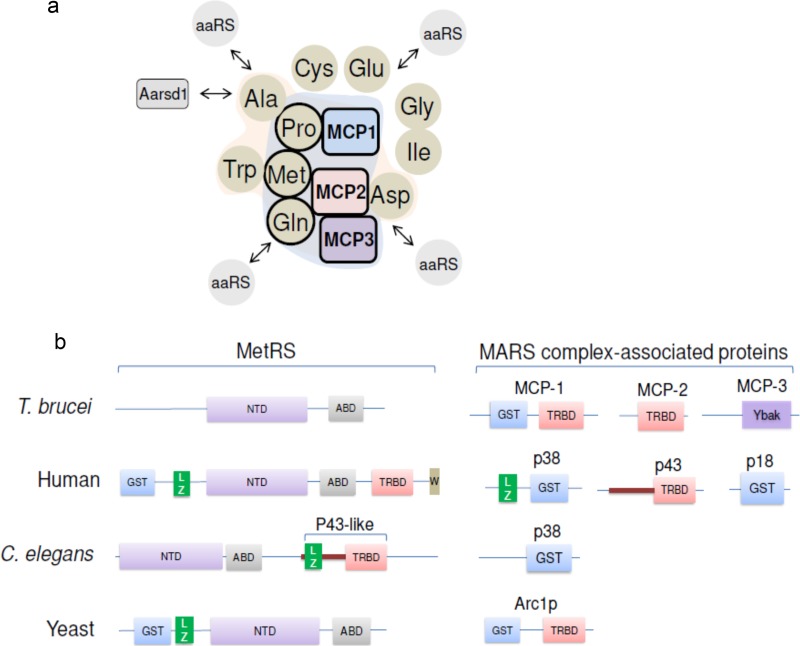
A search of the T. brucei genome for sequences that encode aaRS domains identified two genes that had been annotated as hypothetical proteins: Tb927.9.6650, which has 49% similarity to the alanyl-tRNA editing protein (Aarsd1) of Mus musculus, and Tb927.10.1250, which has 64% similarity to the Arabidopsis thaliana YbaK/prolyl-tRNA synthetase-associated domain-containing protein (Table 4). These proteins are involved in aminoacylation editing activity, i.e., deacylation of misaminoacylated tRNAs (47–49), and they may have a similar function in T. brucei. The gene Tb927.8.5330 has been annotated as encoding tyrosyl/methionyl-tRNA synthetase. However, it appears to not have an anticodon-binding domain or a nucleotidyltransferase domain. On the other hand, it has 54% similarity to the Aquifex aeolicus tRNA-binding protein 111 (Trbp111) (Table 4), a cofactor that increases aaRS activity but which is not an enzyme per se (50, 51). The gene Tb927.7.2400 has been annotated as tyrosyl-tRNA synthetase; however, it also does not have nucleotidyltransferase or anticodon-binding domains. It has a glutathione S-transferase (GST) and a TRBD and has 39% identity to the yeast protein Arc1p, which is a MARS complex-associated protein that also acts as a cofactor for efficient tRNA-aminoacylation (11). Based on this and results presented below, we suggest reannotating Tb927.7.2400 as MARS complex-associated protein 1 (MCP1), Tb927.8.5330 as MCP2, and Tb927.10.1250 as MCP3 (Table 4).
Table 4.
T. brucei genes encoding aaRS-related proteins, their homologs, and description of homolog function
| New name | GeneDB accession no. | GeneDB annotation | Homolog (% amino acid identity to T. brucei) | Function |
|---|---|---|---|---|
| MCP1 | Tb927.7.2400 | Tyrosyl-tRNA synthetase, putative | Saccharomyces cerevisiae aaRS cofactor 1 protein (Arc1p) (39) | Arc1p forms a ternary complex with MetRS and GluRS in yeast; it binds tRNAs and increases their aminoacylation by these enzymes |
| MCP2 | Tb927.8.5330 | Tyrosyl/methionyl-tRNA synthetase, putative | Aquifex aeolicus tRNA-binding protein (Trbp111) (54) | Trbp111 binds to tRNAs and improves theirs aminoacylation by aaRS enzymes |
| MCP3 | Tb927.10.1250 | Hypothetical protein | Arabidopsis thaliana YbaK/aaRS-associated domain-containing protein (64) | Ybak proteins deacylate misaminoacylated Cys-tRNAPro and Ala-tRNAPro |
| TbAarsd1 | Tb927.9.6650 | Hypothetical protein | Mus musculus alanyl-tRNA editing protein Aarsd1 (49) | Aarsd1 protein (also known as AlaXp) deacylates misaminoacylated Ser-tRNAAla and Gly-tRNAAla |
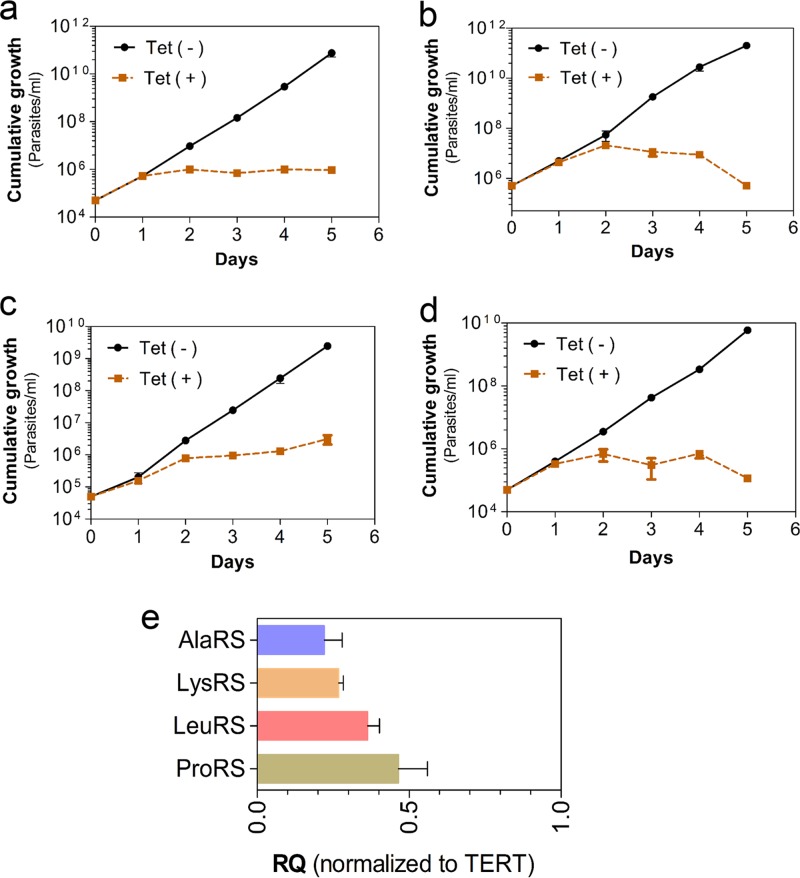
Knockdown of expression of the genes encoding LeuRS, ProRS, LysRS, and AlaRS in T. brucei bloodstream forms in vitro by RNA interference (RNAi) resulted in parasite growth arrest, which indicates that each of these genes is essential for parasite growth (Fig. 2 and Table 2). Indeed, RNAi knockdown of all aaRS genes tested to date resulted in growth inhibition of either procyclic or bloodstream forms in vitro (Table 2) and also prevented infection of mice (24). Overall, T. brucei has all aaRS genes needed to aminoacylate the 20 aminoacyl-tRNAs required for protein synthesis. Most of the aaRSs localize to the cytoplasm, and some localize to the mitochondrion or both compartments, but all tested genes are essential for the growth of procyclic and bloodstream forms.
Fig 2.
Growth curves of T. brucei bloodstream forms upon RNAi knockdown with 1 μg/ml of tetracycline (Tet). (a) AlaRS (Tb927.6.700); (b) ProRS (Tb927.10.12890); (c) LeuRS (Tb927.11.3730); (d) LysRS-1 (Tb927.8.1600). Data from panels a to d are the averages of at least three experiments ± SDs. (e) Quantitative RT-PCR analysis of T. brucei bloodstream-form RNA collected 24 h after tetracycline induction of RNAi from the cell lines for which the results are shown in panels a to d. Data were normalized with telomerase reverse transcriptase (TERT) relative to those for the noninduced matched controls and are the averages of three experiments ± SDs. RQ, relative quantification.
T. brucei aaRSs are organized in a multiprotein complex.
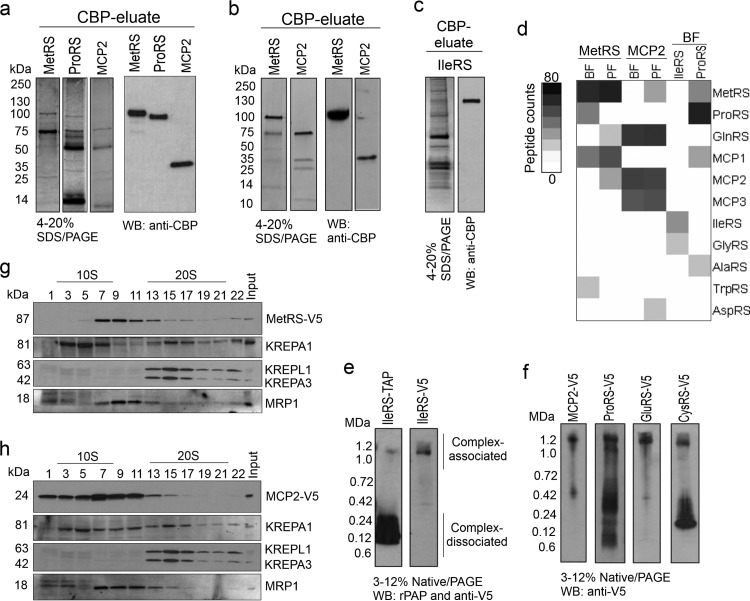
Since aaRSs are organized as a multiprotein complex in mammalian cells and yeast (6, 7, 11), we sought to determine whether T. brucei aaRSs are also associated in a complex. We performed tandem affinity purification and mass spectrometry to identify aaRS-interacting proteins. N- or C-terminal TAP-tagged MetRS, MCP2, and cytoplasmic isoforms of the ProRS and IleRS proteins were expressed in T. brucei bloodstream forms, and TAP-tagged MetRS and MCP2 were also expressed in procyclic forms. The TAP tag used has a protein A domain separated from a CBP domain by a TEV protease cleavage site. The tagged proteins were purified under native conditions by affinity chromatography using IgG-Sepharose and CBP-Sepharose with an intervening TEV protease cleavage step to remove the protein A. Analysis of the CBP eluates by SDS-PAGE and Western blotting confirmed the purification of the tagged proteins and revealed several copurified proteins (Fig. 3a to c).
Fig 3.
T. brucei aminoacyl-tRNA synthetase multiprotein complex. (a to c) TAP-tagged purified aaRS examined by SDS-PAGE (4 to 20%) and Western blotting (WB) in which the gels and blots were probed with polyclonal antibodies to anti-CBP. (a) MetRS, ProRS (cytoplasmic; the MTS was removed), and MCP2 from bloodstream forms (BF) (SYPRO Ruby stain). (b) MetRS and MCP2 from procyclic forms (PF) (Imperial stain). (c) IleRS (cytoplasmic; the MTS was removed) from bloodstream forms (silver stain). (d) Intensity map of peptide spectral counts of TAP-tagged proteins that were purified from bloodstream forms and procyclic forms and identified by mass spectrometry. The data summarize the results of two independent TAP tag purifications and mass spectrometry analyses (Table 5). Table S1 in the supplemental material gives a list of all identified proteins. (e) Native PAGE (3 to 12%; Life Technologies) and Western blotting of T. brucei bloodstream forms expressing IleRS-TAP or IleRS-3V5 tags. Protein expression was induced with 100 ng/ml of tetracycline. Protein lysates from 1.0 × 107 parasites per lane were analyzed. The TAP tag was detected with the rPAP reagent (Sigma-Aldrich), and the 3V5 tag was detected with anti-V5 monoclonal antibody. rPAP, rabbit peroxidase antiperoxidase (f) Native PAGE (3 to 12%; Life Technologies) and Western blotting of T. brucei bloodstream forms expressing 3V5-tagged MCP2, ProRS, GluRS, and CysRS. Protein lysates from 1.0 × 107 parasites were analyzed per lane, and the 3V5 tag was detected with anti-V5 monoclonal antibody. Note that GluRS-3V5 and CysRS-3V5 were not identified by tandem affinity purification and mass spectrometry in experiments whose results are shown in Fig. 1d, likely due to weak affinity to the complex or interference of their interaction with tagged proteins or because they were not directly interacting with tagged proteins/complexes. (g) Western blot analysis of glycerol gradient (10 to 30%) fractions of lysate from bloodstream forms expressing 3V5-tagged MetRS probed with anti-V5 monoclonal antibody and then stripped and reprobed with monoclonal antibodies against editosome proteins KREPA1, KREPA3, and KREPL1 (36) and MRP1 (55). (h) Similar to panel g, but with lysate from T. brucei bloodstream forms expressing 3V5-tagged MCP2.
Mass spectrometry analysis of the MetRS-TAP identified interactions between MetRS, ProRS, MCP1, and TrpRS in bloodstream forms but MCP1, GlnRS, and MCP2 in procyclic forms (Fig. 3d and Table 5). ProRS-TAP confirmed the interactions between MetRS, ProRS, and MCP1 that were observed with MetRS-TAP and also copurified AlaRS. In addition, MCP2-TAP copurified GlnRS and MCP3 in both parasite life stages, and in procyclic forms it also copurified MetRS and AspRS. IleRS-TAP copurified GlyRS. Several purifications of IleRS with either an N- or C-terminal TAP tag resulted in low yields, which likely reflects the interference of the TAP tag with the IleRS association with its binding partners, since native PAGE and Western blotting of IleRS that had only a 3V5 tag revealed more protein associated with a high-molecular-mass complex (∼1.2 MDa) than when it was TAP tagged (Fig. 3e). The 3V5-tagged proteins MCP2 and ProRS also migrated in an ∼1.2-MDa complex by native PAGE and Western blotting, and similar results were observed with CysRS and GluRS (Fig. 3f), which indicates that the latter aaRSs may also interact with a protein complex. Overall, TAP tagging and mass spectrometry revealed associations between six aaRSs, MetRS, ProRS, GlnRS, AlaRS, TrpRS, and AspRS, and three other proteins that we suggest naming MCP1, MCP2, and MCP3 (Fig. 3d and Table 5), suggesting that they may form a complex. Whether the other aaRSs analyzed by native PAGE interact with this complex or other complexes is still unknown. The differences in interactions between bloodstream and procyclic forms likely reflect experimental variations due to sample preparation, the fewer cells used for bloodstream-form purifications, and mass spectrometry limitations rather than biological differences. Several other proteins were also identified by mass spectrometry of the affinity-purified material, including chaperones, cytoskeletal proteins, and components of the translation apparatus, such as ribosomal proteins and initiation and elongation factors (see Table S1 in the supplemental material). Although these proteins have been shown to interact with aaRSs in other organisms (3, 16, 52), additional experiments are needed to assess the specificity of the interaction, since their high abundance in cells suggests that some may be adventitious contaminants.
Table 5.
Unique peptide count and percent amino acid coverage of proteins in the MARS complex identified by TAP and mass spectrometry
| GeneDB accession no. or tag | GeneDB annotation | New name | Length (no. of amino acids) | Unique peptide count (% amino acid coverage)a |
|||||
|---|---|---|---|---|---|---|---|---|---|
| BF_MetRS-TAP | PF_MetRS-TAP | BF_MCP2-TAP | PF_MCP2-TAP | BF_IleRS-TAP | BF_ProRS-TAP | ||||
| Tb927.10.1500 | Methionyl-tRNA synthetase | Same | 773 | 53 (64.29) | 76 (81.11) | 2 (3.62) | 4 (6.99) | ||
| Tb927.10.12890 | Bifunctional aaRS | ProRS | 808 | 5 (10.15) | 73 (71.78) | ||||
| Tb927.9.5210 | Glutaminyl-tRNA synthetase | Same | 627 | 1 (3.19) | 36 (49.92 | 41 (54.55) | |||
| Tb927.7.2400 | Tyrosyl-tRNA synthetase | MCP1 | 425 | 6 (13.18) | 17 (44.00) | 2 (3.06) | |||
| Tb927.8.5330 | Tyrosyl/methionyl-tRNA synthetase | MCP2 | 223 | 2 (14.35) | 25 (87.44) | 20 (78.92) | |||
| Tb927.10.1250 | Hypothetical protein, conserved | MCP3 | 261 | 14 (61.69) | 17 (52.49) | ||||
| Tb927.10.9190 | Isoleucyl-tRNA synthetase | Same | 1143 | 3 (4.2) | |||||
| Tb927.11.9640 | Glycyl-tRNA synthetase | Same | 624 | 1 (1.92) | |||||
| Tb927.6.700 | Alanyl-tRNA synthetase | Same | 966 | 1 (2.28) | |||||
| Tb927.8.2240 | Tryptophanyl-tRNA synthetase | Same | 473 | 1 (1.90) | |||||
| Tb927.6.1880 | Aspartyl-tRNA synthetase | Same | 560 | 1 (2.68) | |||||
| CBP tagb | 26 | 2 (5.95) | 3 (7.03) | 1 (5.95) | 4 (8.1) | 2 (5.95) | |||
The data show the total unique peptide counts from two independent purification and mass spectrometry experiments for PF_MetRS-TAP, BF_ProRS-TAP, and BF_MCP2-TAP, three experiments for BF_MetRS-TAP, and one experiment for PF_MCP2-TAP and BF_IleRS-TAP. Peptides that were identified more than once in two or more experiments were only counted once. BF, bloodstream forms; PF, procyclic forms.
The CBP tag is part of the TAP tag and remains associated with the tagged protein after TEV protease digestion.
Glycerol gradient sedimentation analysis of T. brucei bloodstream-form lysates from cell lines expressing 3V5-tagged MetRS or MCP2 revealed a main peak for both between fractions 7 and 11 (Fig. 3g and h). Because the proteins of the ∼20-Svedberg (20S) editosome (53) and the lower-S-unit mitochondrial RNA-binding 1 (MRB1) complex (54, 55) sediment between ∼10S and 20S, we compared their sedimentation to the sedimentation of 3V5-tagged MetRS and MCP2, which primarily sediment between 10S and 20S (Fig. 3g and h). These proteins were also detected above and below the 10S to 20S region, which suggests that they may occur in larger protein associations perhaps via transient or weak interactions. Taken together, these data indicate that T. brucei aaRSs exist in a multiprotein complex that contains aaRS enzymes and non-aaRS proteins.
The MARS complex enhances the efficiency of tRNA-aminoacylation.
To assess the function of the MARS complex in T. brucei, we purified the MetRS-TAP complex and analyzed the effect of its dissociation on tRNA-aminoacylation. The complexes were enriched from lysates of procyclic forms expressing TAP-tagged MetRS by incubation with IgG-Sepharose (Fig. 4a). The bound complexes were initially washed either with low-salt buffer, i.e., buffer containing 140 mM NaCl, or with high-salt buffer that contained 600 mM NaCl in order to disrupt interactions between proteins. The bound complexes were rewashed with 140 mM NaCl to reconstitute the salt concentrations and eluted with TEV protease for enzymatic studies. Similar amounts of MetRS were purified under both conditions, but the high-salt wash resulted in substantial disruption of the MARS complex, as assessed by SDS-PAGE and Western blotting (Fig. 4b). A high-salt wash does not affect MetRS folding or activity (42, 56). Neither salt concentration affected the interaction of the tagged protein with IgG-Sepharose (Fig. 4b). Native PAGE analysis of TEV eluates that had been washed with salt at the lower concentration (140 mM NaCl) revealed a complex migrating at about 1.2 MDa (Fig. 4c), similar to the migration of the 3V5-tagged complexes when analyzed from cell lysates (Fig. 3e and f). In contrast, TEV eluates that had been washed with salt at the higher concentration (600 mM NaCl) migrated at a lower molecular mass, similar to the mass of a monomeric MetRS with a CBP tag (∼90 kDa) or its dimer (∼180 kDa) (56) (Fig. 4c).
We performed steady-state kinetic assays to compare the MetRS aminoacylation rates (using specific substrates l-Met and tRNAMet) with material from both purifications and found that the aminoacylation rate was about 6-fold higher with the complex (the MetRS complex washed with 140 mM NaCl) compared to the dissociated enzyme (MetRS washed with 600 mM NaCl) (Fig. 4d). Assays performed in the absence of l-Met showed negligible levels of product, indicating MetRS specific activity (Fig. 4d). The aminoacylation reaction occurs in two steps: (i) activation of the amino acid via ATP hydrolysis, forming an enzyme-aminoacyladenylate intermediate and releasing inorganic pyrophosphate (PPi), followed by (ii) the transfer of the amino acid to the tRNA, releasing AMP (Fig. 4e). In order to identify how the MARS complex was affecting aminoacylation, we performed steady-state kinetic measurements to determine the Km and kcat of MetRS both in the complex and in its dissociated form in relation to the l-Met or tRNAMet substrate. The MetRS Km and kcat in relation to l-Met was similar whether MetRS was in the complex (MetRS complex washed with 140 mM NaCl) or was a dissociated enzyme (MetRS washed with 600 mM NaCl) (Fig. 4f and Table 6). In contrast, the Km for tRNAMet was about 50-fold lower for MetRS when in the complex (0.1 μM) than when in the dissociated form (5.2 μM), resulting in a 46-fold higher enzyme efficiency when in the complex (Fig. 4g and Table 6). These results indicate that the formation of the complex results in an increased efficiency of aminoacylation at the second step of the reaction, i.e., the transfer of the amino acid to the tRNA, by reducing its Km toward the tRNA.
Table 6.
Steady-state kinetic parameters of MetRS and MetRS complex for l-methionine and tRNAMet
| Substrate and MetRS form | Km (mol/liter) | Vmax (nmol/min/μg) | kcat (s−1) | Efficiency [(mol/liter)−1 s−1] |
|---|---|---|---|---|
| l-Methionine | ||||
| MetRS complex | 7.0 × 10−5 ± 3.8 × 10−5 | 3.9 ± 0.3 | 10 ± 0.8 | 1.4 × 105 ± 2.0 × 104 |
| MetRS | 5.4 × 10−5 ± 4.2 × 10−5 | 2.4 ± 0.3 | 6.3 ± 0.7 | 1.2 × 105 ± 1.6 × 104 |
| tRNAMet | ||||
| MetRS complex | 1.0 × 10−7 ± 1.3 × 10−7 | 2.2 ± 0.3 | 5.8 ± 0.8 | 5.6 × 107 ± 6.0 × 106 |
| MetRS | 5.2 × 10−6 ± 2.9 × 10−6 | 2.4 ± 0.5 | 6.3 ± 1.4 | 1.2 × 106 ± 4.8 × 105 |
MCP2 enhances tRNA-aminoacylation by the MARS complex.
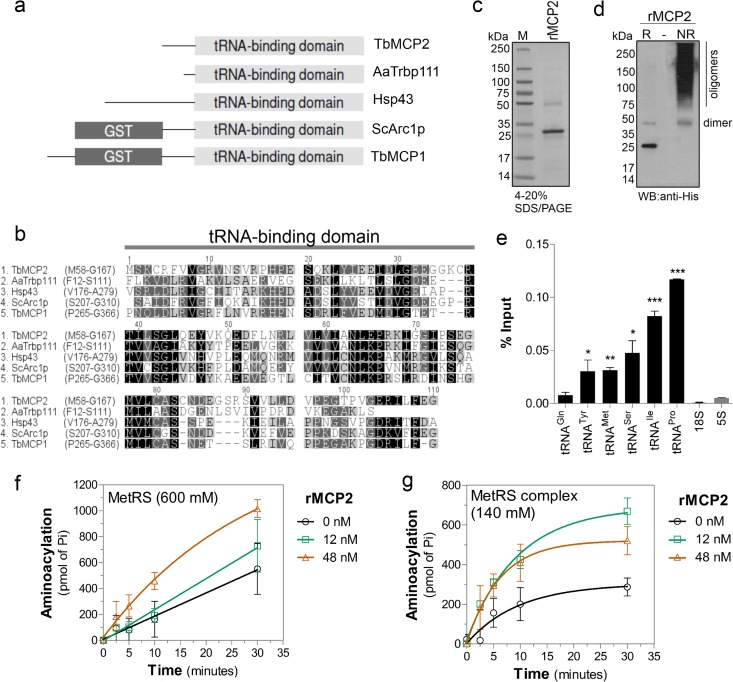
Because our data suggested that formation of the MARS complex enhances the efficiency of aminoacylation at the tRNA ligation step, we explored whether components of the MARS complex could directly bind tRNA since this might explain the increase in catalytic efficiency. Amino acid sequence analysis of MCP2 identified a tRNA-binding domain (from amino acids M58 to G167) that is highly similar to the bacterial protein Trbp111 (54%) (Fig. 5a and b). Trbp111 has been shown to specifically interact with tRNAs and increase their aminoacylation (50, 51). The amino acid sequence also has similarity to the C terminus of Arc1p (55%; from amino acids S207 to G346), which is involved in tRNA binding (11, 15) (Fig. 5a and b). Arc1p interacts with MetRS and GluRS through its N terminus and increases their aminoacylation rates in yeast (11, 15). Therefore, we reasoned that MCP2 might bind to tRNAs and facilitate their aminoacylation when associated with the complex. We cloned the mcp2 gene with a C-terminal 6× His tag and expressed and purified it from E. coli (Fig. 5c). Recombinant MCP2 formed a dimer and, possibly, oligomers when analyzed under nonreducing conditions (Fig. 5d), similar to Trbp111 (50). rMCP2 was incubated with T. brucei total RNA, and the bound RNAs were analyzed by quantitative RT-PCR after protein affinity purification using Ni2+-magnetic beads. The RT-PCR showed that several tRNAs were bound to rMCP2, whereas highly abundant 18S rRNA and 5S rRNA did not (Fig. 5e), suggesting selectivity to tRNAs.
Fig 5.
MCP2 enhances tRNA-aminoacylation by the MARS complex. (a) Diagram of domains conserved in proteins associated with aaRS complexes, including Tb927.7.2400 (T. brucei MCP1 [TbMCP1]) and Tb927.8.5330 (T. brucei MCP2 [TbMCP2]), Saccharomyces cerevisiae Arc1p (ScArc1p; GI: 1620460), Homo sapiens Hsp43 (GI: 85700432), and Aquifex aeolicus Trbp111 (AaTrbp111; GI: 34810888), showing the conserved TRBD and GST domains. (b) Amino acid sequence alignment of the TRBDs of the proteins shown in panel a (amino acid positions are shown). (c) SDS-PAGE (4 to 20%) of rMCP2 purified from E. coli (Imperial stain). Lane M, molecular mass markers. (d) Western blot with anti-His tag antibodies of T. brucei rMCP2 run under reducing conditions (lane R; Laemmli running buffer with 5% β-mercaptoethanol and boiling) or nonreducing conditions (lane NR; Laemmli running buffer without β-mercaptoethanol) in 4 to 20% SDS-polyacrylamide gels. (e) Detection of tRNAs from the binding assay with T. brucei total RNA and rMCP2 analyzed by quantitative RT-PCR. The data show the averages of three independent experiments ± SDs; significance tests were performed by unpaired t test comparing tRNA to 5S rRNA data. ***, P < 0.0001; **, P < 0.005; *, P < 0.05. Aminoacylation kinetics of dissociated MetRS (washed with 600 mM NaCl) (f) or the MetRS complex (washed with 140 mM NaCl [Fig. 4]) (g) upon addition of rMCP2. The averages of three independent experiments ± SDs are shown.
To analyze whether MCP2 was responsible for increasing the aminoacylation efficiency of MetRS, we performed steady-state kinetic analysis with both complex-associated and dissociated MetRS in the presence and absence of rMCP2. The aminoacylation rates of MetRS (MetRS washed with 600 mM NaCl) increased in a dose-dependent manner in the presence of rMCP2 (Fig. 5f). Increased aminoacylation rates were also observed when rMCP2 was added to the MARS complex purification (MetRS complex washed with 140 mM NaCl) (Fig. 5g). Lower molar ratios of 1.5:1 for rMCP2 to MetRS (see the results for 12 nM rMCP2 in Fig. 5g) were sufficient for the optimum activity of MetRS in complex, likely because the purified complex contains the native protein, limiting the association of additional recombinant protein to the complex. The increase in molar ratios to 6:1 (48 nM rMCP2) did not increase the rate of aminoacylation in the complex (Fig. 5g), but it did with the dissociated MetRS enzyme (Fig. 5f). This suggests that multiple molecules of MCP2 are associated with the complex. Together, these results indicate that MCP2 interacts with tRNAs and facilitates their aminoacylation by the enzymes on the MARS complex, perhaps by inducing an enzyme or a tRNA conformational fit acting as a cofactor for aminoacylation.
Conditional repression of MCP2 expression results in reduced parasite fitness.
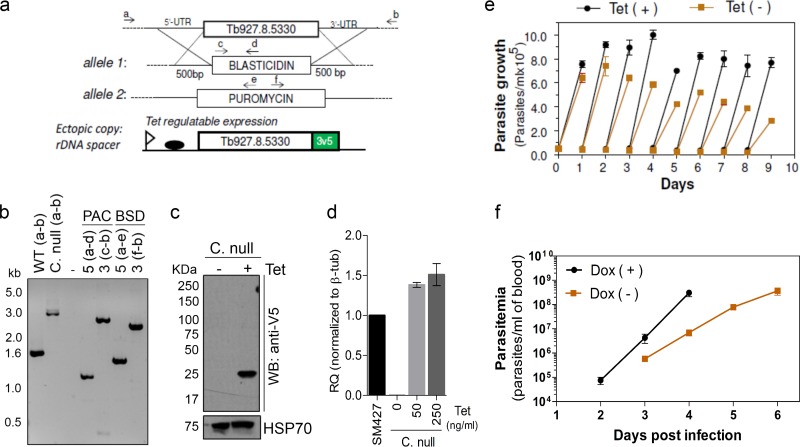
Since the T. brucei MCP2 is a component of the MARS complex and contributes to efficient tRNA-aminoacylation, we explored whether its depletion from the cell would affect aminoacylation in vivo and impact parasite growth and infection in a mouse model. We tested this possibility by generating conditional-null cell lines for the mcp2 gene. A tetracycline-regulated ectopic copy of the mcp2 gene containing a 3V5 tag at the C terminus was inserted at the ribosomal DNA (rDNA) spacer of T. brucei bloodstream-form single-marker 427. Using this cell line, we replaced both endogenous alleles encoding MCP2 with drug resistance markers (Fig. 6a). PCR genotyping analysis of the conditional-null cells confirmed the replacement of endogenous alleles (Fig. 6a and b). Expression of the mcp2 gene was assessed by Western blotting, confirming that it was conditionally expressed upon tetracycline induction (Fig. 6c). Quantitative RT-PCR analysis showed that the level of the tetracycline-induced ectopic expression of MCP2 was similar to that of the parental (SM427) cell line (Fig. 6d). Immunofluorescence analysis also revealed a cytoplasmic distribution of MCP2-3V5 (Fig. 1a), consistent with its association with the MARS complex. Conditional repression of this gene in T. brucei bloodstream forms by withdrawal of tetracycline resulted in slow parasite growth (Fig. 6e), which may be due to the inefficient formation of aminoacyl-tRNAs in vivo. To determine if MCP2 depletion would affect parasite infectivity in vivo, we infected mice with the MCP2 conditional-null line and analyzed the parasitemia upon induction or repression of mcp2 expression in vivo. Doxycycline (a stable tetracycline analog) was added to the mouse drinking water 24 h before infection and was maintained in the drinking water throughout the course of infection. T. brucei parasitemia in doxycycline-treated mice was initially detected at day 2 postinfection and increased rapidly, reaching ∼3.0 × 108 parasites/ml of blood on day 4 postinfection (Fig. 6f), which resembles the parasitemia achieved during infection by the wild-type parasite (24). However, in mice that did not receive doxycycline, the parasitemia was delayed, and it rose more slowly in untreated mice than in doxycycline-treated mice, suggesting that mcp2 expression is required for efficient parasite infection (Fig. 6f). The requirement of T. brucei MCP2 for optimal parasite growth and infectivity in mouse probably reflects its role in increasing the efficiency of tRNA-aminoacylation, perhaps by recruiting tRNAs and other accessory proteins to the complex. This suggests that assembly of a MARS complex may also contribute to parasite fitness.
Fig 6.
The MARS complex in T. brucei contributes to parasite fitness. (a) Diagram of a conditional-null T. brucei MCP2 cell line in which the endogenous alleles were replaced with blasticidin (BSD-Ty1-HSVTK) and puromycin (PAC-Ty1-HSVTK) resistance markers and a tetracycline (Tet)-regulatable ectopic 3V5-tagged allele was inserted into the rDNA spacer. UTR, untranslated region. (b) TBE-agarose gel (1%) of PCR products from the conditional-null (C. null) cells diagrammed in panel a (see Materials and Methods for details). WT, wild type. (c) Western blot analysis (10% SDS-PAGE) of lysates of conditional-null cells diagrammed in panel a after 2 days of growth in the absence (lane −) or presence (lane +) of 250 ng/ml tetracycline using anti-V5 monoclonal antibodies. The blot was reprobed with monoclonal antibody MAb78 for HSP70 as a loading control. (d) Quantitative real-time PCR analysis of MCP2 mRNA in SM427 or conditional-null cells grown for 24 h without tetracycline or in the presence of 50 ng/ml or 250 ng/ml. β-Tubulin (β-tub) was used as an internal control. (e) Growth curve of conditional-null cells in presence or absence of 250 ng/ml of tetracycline. Results are averages of three experiments ± SDs. (f) Parasitemia in mice infected with the T. brucei MCP2 conditional-null line. Experiments were performed in the absence (−) or presence (+) of doxycycline (Dox). Mice received doxycycline (200 μg/ml) in drinking water starting 24 h before infection, and the doxycycline was maintained in the drinking water throughout infection (replaced daily). The data show the averages of three independent experiments ± SDs.
DISCUSSION
We show here that T. brucei has genes that encode aaRS enzymes to aminoacylate all 20 amino acids needed for protein synthesis, and all analyzed genes were essential for parasite growth. We identified the cytoplasmic, mitochondrial, and dually localized aaRSs. We also found that cytoplasmic aaRSs are organized in a multiprotein complex which contains about six aaRSs and three associated proteins. The MARS complex provides for efficient tRNA-aminoacylation and contributes to parasite fitness.
Analysis of the T. brucei genome revealed 24 nuclear genes annotated as aaRSs, of which 2 encode the two subunits of one enzyme (PheRS), but no aaRS gene has been identified in the mitochondrial genome (38, 57). Analysis of the TriTryp (T. brucei, T. cruzi, and Leishmania) genome database did not identify any additional genes with a predicted aaRS domain. It did show that all 24 genes are highly conserved among the species in the TriTryp database and, although to a lesser extent, among other organisms, but they were more similar to genes for eukaryotic than prokaryotic enzymes, as observed for Leishmania (58). It is unlikely that such genes are gaps in all three databases, and thus, these 24 genes probably represent all aaRS genes in these organisms.
Localization studies indicate that these genes encode three aaRSs (AspRS-1, HisRS, and TrpRS-1) that are primarily, if not exclusively, cytoplasmic and three (AspRS-2, LysRS-2, and TrpRS-2) that are primarily, if not exclusively, mitochondrial, as reported by others (25–27) and confirmed here. The remaining 13 aaRSs are or may be localized both in the cytoplasm and in the mitochondrion (Table 2). Most of the annotated aaRSs had been identified in the mitochondrial proteome of T. brucei procyclic forms (38). Five of the dually localized aaRSs are encoded by alternatively trans-spliced transcripts (45, 46), which provides for the joint localization, as reported by others (29, 30) and confirmed here. Dual localization of these enzymes may be due to (i) alternative splicing of mRNAs that were not detected due to their low abundance, (ii) alternative start codons that could result in addition or removal of MTSs, as seen in other eukaryotes (59, 60), (iii) protein processing that could expose or remove an MTS (61), or (iv) other mechanisms, such as cotranslocation of proteins into the mitochondrion.
These 24 genes appear to account for all 20 amino acids that are needed for protein synthesis in both compartments. Mass spectrometry did not detect HisRS peptides in purified mitochondria, perhaps due to low abundance or issues related to the limitations of mass spectrometry. The dual localization of aaRSs is in agreement with all mitochondrial tRNAs in T. brucei being imported from the cytoplasm (62–64), although it is also conceivable that charged tRNAs are also imported.
Several of the T. brucei aaRSs occur in a multiprotein complex along with non-aaRS proteins. Mass spectrometry of TAP-tagged and purified complexes and native gel analysis identified the association of about six aaRS enzymes and three related proteins. However, other aaRS enzymes may also interact with this complex, as detected by native PAGE and Western blotting of 3V5-tagged aaRSs. The complex sediments at somewhat over 10S in a glycerol gradient and migrates with a molecular mass of ∼1.2 MDa in native polyacrylamide gels. These are similar to the findings for the ∼1.5-MDa human MARS complex, which has at least 9 aaRS enzymes and 3 associated proteins (3). Since we did not detect all aaRSs in a complex, it may be that the identified complex represents the more stably associated proteins. Association of other aaRSs may also have been affected by the TAP tag (as observed for IleRS-TAP), they may be less stably associated with the complex, or perhaps they only transiently interact with the MARS complex (Fig. 7a). For example, the proteins CysRS and GluRS were detected migrating in an ∼1.2-MDa complex in native PAGE and Western blotting experiments, but they were not detected by TAP tagging and mass spectrometry. We also do not rule out the possibility that they interact with other components of the MARS complex that we have not yet identified or the existence of multiple subcomplexes.
Fig 7.
The T. brucei MARS complex and the distribution of protein domains among MetRS and MARS-associated proteins. (a) Diagram of proteins associated in the T. brucei MARS complex. Interactions are indicated on the basis of the TAP tagging and mass spectrometry data. Circles, aaRSs; squares, associated non-aaRS proteins. Proteins with the most peptide counts (Fig. 3d) are outlined in bold lines and inside the blue background shading. Proteins with lower peptide counts (AlaRS, TrpRS, and AspRS) are inside the magenta background shading. Whether the proteins IleRS, GlyRS, CysRS, and GluRS interact with the identified complex is unknown; however, they were added to the diagram due to their similar migration in the native gel. The arrows associated with aaRSs indicate potential dynamic interactions of proteins with the MARS complex. TbAarsd1 is indicated. (b) Arrangement of the various protein domains in MetRSs and MARS-associated proteins in different species. NTD, nucleotidyltransferase domain; ABD, anticodon-binding domain; LZ, leucine zipper motif; GST, glutathione S-transferase domain; TRBD, tRNA-binding domain; Ybak, T. brucei domain with homology to prokaryote Ybak domain; W, WHEP domain.
The T. brucei MARS complex has general similarities with MARS complexes identified in other eukaryotes, including humans, C. elegans, and yeast, namely, the association of several aaRSs and proteins that have domains that function in protein interaction, tRNA binding, and/or prevention of misaminoacylation (Fig. 7a and b). Depending on the organism, these domains may be fused to an aaRS or exist in separate proteins that are associated with the complex. For example, GST, TRBD, and the LZ domain are part of the MetRS of humans (Fig. 7b); however, they are also in the p38 and p43 complex-associated proteins in humans and in MCP1 and MCP2 in T. brucei. Thus, it is possible that these domains confer an advantage for tRNA-aminoacylation in these organisms, as they appear as part of the enzymes in higher eukaryotes. In general, the MARS complexes in different organisms do not contain the same set of aaRSs, which may reflect the differences in overall protein synthesis and its regulation among them. However, protein and RNA interaction domains are conserved, although they are differently distributed within the enzymes and associated proteins.
The consequence of aaRS organization into a MARS complex is efficient formation of aminoacyl-tRNAs and their channeling to ribosomes, which contributes to translation efficiency by avoiding diffusion of charged tRNAs in the cytoplasm and permits tRNA reutilization (17, 18). We demonstrated this greater aminoacylation efficiency in the T. brucei MARS complex and showed that the increased efficiency is at the second step of the reaction, i.e., the ligation of the amino acid to the tRNA. Conditional repression of T. brucei MCP2 resulted in slower growth of T. brucei and reduced infectivity in a mouse model, suggesting that the MARS complex enhances aminoacylation rates in vivo. This enhanced efficiency was evident as a decrease in the MetRS Km for tRNAMet upon dissociation of the complex (Fig. 4f and g and Table 6). Partial reconstitution of aminoacylation activity was also achieved upon the addition of rMCP2 to MetRS or to the purified complex. Thus, MCP2 may act as a cofactor for tRNA-aminoacylation. MCP2 may induce a conformational fit to MetRS and/or tRNAMet which facilitates tRNA recognition and thus enhances aminoacylation. MCP2 was previously annotated as Tyr/MetRS, but it is unlikely to function as an aaRS since it has no nucleotidyltransferase or anticodon-binding domain. In addition, we did not detect tRNAMet- or tRNATyr-aminoacylation activity with rMCP2 (not shown). MCP2 is likely a tRNA-binding protein on the basis of its similarity to the TRBD of A. aeolicus Trb111 protein and to related domains in eukaryotic tRNA-binding proteins (50, 51). MCP2 oligomerizes and interacts with tRNAs, as does A. aeolicus Trb111 and other accessory proteins (14, 50, 51), such as yeast Arc1p, which also has a TRBD (11, 15). Thus, MCP2 is a component of the T. brucei MARS complex, but unlike in C. elegans, plants, and humans, it is not found as an accessory domain of the MetRS (Fig. 7b) (10, 12, 65). Since MCP2 binds to other tRNAs, it may also contribute to aminoacylation by other aaRS enzymes and perhaps to tRNA recycling by recruiting and delivering tRNAs to the complex, as suggested for Arc1p (15). On the basis of its association with the MARS complex and interaction with tRNAs, perhaps via its TRBD, it may function as a structural MARS component and as a cofactor for aminoacylation. Similarly, MCP1 was previously annotated as TyrRS and also has no nucleotidyltransferase or anticodon-binding domain. It also may not function as an aaRS, although we have not analyzed its aminoacylation activity. MCP1 has a GST and a TRBD, the presence of which implies protein and RNA interaction roles, and thus, it may also function as structural protein and/or as a cofactor for aminoacylation. MCP1 has sequence similarity to yeast Arc1p, which interacts with MetRS and GluRS and improves their aminoacylation rates (11, 14, 15). Thus, MCP1 may have a similar function in the T. brucei MARS complex.
Some proteins in the T. brucei MARS complex likely function to ensure accurate aminoacylation, as in other organisms. MCP3, which is part of the MARS complex, has sequence similarity to the Ybak protein (47, 49) and to Ybak domains of prokaryote ProRS (66). Ybak functions to prevent misaminoacylation by deacylating mischarged tRNAs (67). In addition, bioinformatic analysis identified the T. brucei Aarsd1 (TbAarsd1), the homolog of mouse Aarsd1, although it was not found in the MARS complex. TbAarsd1 may be involved in preventing misaminoacylation, similar to its homologs (68, 69). The distribution of these domains in separate proteins that may or may not be stably associated with the MARS complex or be part of the aaRS enzyme in different species likely reflects their different needs for aminoacylation quality control.
AaRSs are attractive drug targets in parasites because of their essentiality for protein synthesis. The structural differences between host enzymes and those of protozoan parasites, which diverged early in eukaryotic evolution, may be exploited to identify drug leads. Both cytoplasmic and mitochondrial translation are essential for T. brucei bloodstream and procyclic forms (27, 70, 71), and the dually localized enzymes may be especially attractive since their inhibition may block protein synthesis in both cellular compartments. For example, we showed that inhibition of IleRS, which is dually localized in the cytoplasm and mitochondrion, cures mice infected with T. brucei (24).
In summary, we show here that T. brucei aaRSs are essential for survival, and most of them are eukaryote-like and localize to the cytoplasm. They form a multiprotein complex that is required for efficient tRNA-aminoacylation and which contributes to parasite fitness. This complex has accessory proteins that contain RNA- and protein-interacting domains, including MCP2, which binds to tRNAs and contributes to their efficient aminoacylation by the MARS complex.
Supplementary Material
ACKNOWLEDGMENTS
We are very grateful to Lindsay N. Carpp for revision of the manuscript and Christina McCormick and Anna Sokolov for administrative support.
This work was supported by the National Institutes of Health (grant 1R01AI078962 to K.S. and M.A.P.) and the Welch Foundation (grant I-1257 to M.A.P.). M.A.P. holds the Beatrice and Miguel Elias Distinguished Chair in Biomedical Science and the Carolyn R. Bacon Professorship in Medical Science and Education. I.C. was awarded a National Institutes of Health fellowship (5T32AI007509-12).
Footnotes
Published ahead of print 14 October 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/MCB.00711-13.
REFERENCES
- 1. Ibba M, Soll D. 2000. Aminoacyl-tRNA synthesis. Annu. Rev. Biochem. 69:617–650 [DOI] [PubMed] [Google Scholar]
- 2. Perona JJ, Hadd A. 2012. Structural diversity and protein engineering of the aminoacyl-tRNA synthetases. Biochemistry 51:8705–8729 [DOI] [PubMed] [Google Scholar]
- 3. Kaminska M, Havrylenko S, Decottignies P, Gillet S, Le Marechal P, Negrutskii B, Mirande M. 2009. Dissection of the structural organization of the aminoacyl-tRNA synthetase complex. J. Biol. Chem. 284:6053–6060 [DOI] [PubMed] [Google Scholar]
- 4. Mirande M, Kellermann O, Waller JP. 1982. Macromolecular complexes from sheep and rabbit containing seven aminoacyl-tRNA synthetases. II. Structural characterization of the polypeptide components and immunological identification of the methionyl-tRNA synthetase subunit. J. Biol. Chem. 257:11049–11055 [PubMed] [Google Scholar]
- 5. Quevillon S, Robinson JC, Berthonneau E, Siatecka M, Mirande M. 1999. Macromolecular assemblage of aminoacyl-tRNA synthetases: identification of protein-protein interactions and characterization of a core protein. J. Mol. Biol. 285:183–195 [DOI] [PubMed] [Google Scholar]
- 6. Rho SB, Kim MJ, Lee JS, Seol W, Motegi H, Kim S, Shiba K. 1999. Genetic dissection of protein-protein interactions in multi-tRNA synthetase complex. Proc. Natl. Acad. Sci. U. S. A. 96:4488–4493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kaminska M, Havrylenko S, Decottignies P, Le Marechal P, Negrutskii B, Mirande M. 2009. Dynamic organization of aminoacyl-tRNA synthetase complexes in the cytoplasm of human cells. J. Biol. Chem. 284:13746–13754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Robinson JC, Kerjan P, Mirande M. 2000. Macromolecular assemblage of aminoacyl-tRNA synthetases: quantitative analysis of protein-protein interactions and mechanism of complex assembly. J. Mol. Biol. 304:983–994 [DOI] [PubMed] [Google Scholar]
- 9. Kim JY, Kang YS, Lee JW, Kim HJ, Ahn YH, Park H, Ko YG, Kim S. 2002. p38 is essential for the assembly and stability of macromolecular tRNA synthetase complex: implications for its physiological significance. Proc. Natl. Acad. Sci. U. S. A. 99:7912–7916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Havrylenko S, Legouis R, Negrutskii B, Mirande M. 2010. Methionyl-tRNA synthetase from Caenorhabditis elegans: a specific multidomain organization for convergent functional evolution. Protein Sci. 19:2475–2484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Simos G, Segref A, Fasiolo F, Hellmuth K, Shevchenko A, Mann M, Hurt EC. 1996. The yeast protein Arc1p binds to tRNA and functions as a cofactor for the methionyl- and glutamyl-tRNA synthetases. EMBO J. 15:5437–5448 [PMC free article] [PubMed] [Google Scholar]
- 12. Havrylenko S, Legouis R, Negrutskii B, Mirande M. 2011. Caenorhabditis elegans evolves a new architecture for the multi-aminoacyl-tRNA synthetase complex. J. Biol. Chem. 286:28476–28487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Deinert K, Fasiolo F, Hurt EC, Simos G. 2001. Arc1p organizes the yeast aminoacyl-tRNA synthetase complex and stabilizes its interaction with the cognate tRNAs. J. Biol. Chem. 276:6000–6008 [DOI] [PubMed] [Google Scholar]
- 14. Simader H, Hothorn M, Kohler C, Basquin J, Simos G, Suck D. 2006. Structural basis of yeast aminoacyl-tRNA synthetase complex formation revealed by crystal structures of two binary sub-complexes. Nucleic Acids Res. 34:3968–3979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Simos G, Sauer A, Fasiolo F, Hurt EC. 1998. A conserved domain within Arc1p delivers tRNA to aminoacyl-tRNA synthetases. Mol. Cell 1:235–242 [DOI] [PubMed] [Google Scholar]
- 16. Sha Z, Brill LM, Cabrera R, Kleifeld O, Scheliga JS, Glickman MH, Chang EC, Wolf DA. 2009. The eIF3 interactome reveals the translasome, a supercomplex linking protein synthesis and degradation machineries. Mol. Cell 36:141–152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cannarozzi G, Schraudolph NN, Faty M, von Rohr P, Friberg MT, Roth AC, Gonnet P, Gonnet G, Barral Y. 2010. A role for codon order in translation dynamics. Cell 141:355–367 [DOI] [PubMed] [Google Scholar]
- 18. Negrutskii BS, Deutscher MP. 1991. Channeling of aminoacyl-tRNA for protein synthesis in vivo. Proc. Natl. Acad. Sci. U. S. A. 88:4991–4995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Han JM, Jeong SJ, Park MC, Kim G, Kwon NH, Kim HK, Ha SH, Ryu SH, Kim S. 2012. Leucyl-tRNA synthetase is an intracellular leucine sensor for the mTORC1-signaling pathway. Cell 149:410–424 [DOI] [PubMed] [Google Scholar]
- 20. Han JM, Park BJ, Park SG, Oh YS, Choi SJ, Lee SW, Hwang SK, Chang SH, Cho MH, Kim S. 2008. AIMP2/p38, the scaffold for the multi-tRNA synthetase complex, responds to genotoxic stresses via p53. Proc. Natl. Acad. Sci. U. S. A. 105:11206–11211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ofir-Birin Y, Fang P, Bennett SP, Zhang HM, Wang J, Rachmin I, Shapiro R, Song J, Dagan A, Pozo J, Kim S, Marshall AG, Schimmel P, Yang XL, Nechushtan H, Razin E, Guo M. 2013. Structural switch of lysyl-tRNA synthetase between translation and transcription. Mol. Cell 49:30–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Brun R, Blum J, Chappuis F, Burri C. 2010. Human African trypanosomiasis. Lancet 375:148–159 [DOI] [PubMed] [Google Scholar]
- 23. Simarro PP, Cecchi G, Franco JR, Paone M, Diarra A, Ruiz-Postigo JA, Fevre EM, Mattioli RC, Jannin JG. 2012. Estimating and mapping the population at risk of sleeping sickness. PLoS Negl. Trop. Dis. 6:e1859. 10.1371/journal.pntd.0001859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Cestari I, Stuart K. 2013. Inhibition of isoleucyl-tRNA synthetase as a potential treatment for human African trypanosomiasis. J. Biol. Chem. 288:14256–14263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Charriere F, Helgadottir S, Horn EK, Soll D, Schneider A. 2006. Dual targeting of a single tRNA(Trp) requires two different tryptophanyl-tRNA synthetases in Trypanosoma brucei. Proc. Natl. Acad. Sci. U. S. A. 103:6847–6852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Charriere F, O'Donoghue P, Helgadottir S, Marechal-Drouard L, Cristodero M, Horn EK, Soll D, Schneider A. 2009. Dual targeting of a tRNAAsp requires two different aspartyl-tRNA synthetases in Trypanosoma brucei. J. Biol. Chem. 284:16210–16217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Espanol Y, Thut D, Schneider A, Ribas de Pouplana L. 2009. A mechanism for functional segregation of mitochondrial and cytosolic genetic codes. Proc. Natl. Acad. Sci. U. S. A. 106:19420–19425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Geslain R, Aeby E, Guitart T, Jones TE, Castro de Moura M, Charriere F, Schneider A, Ribas de Pouplana L. 2006. Trypanosoma seryl-tRNA synthetase is a metazoan-like enzyme with high affinity for tRNASec. J. Biol. Chem. 281:38217–38225 [DOI] [PubMed] [Google Scholar]
- 29. Rettig J, Wang Y, Schneider A, Ochsenreiter T. 2012. Dual targeting of isoleucyl-tRNA synthetase in Trypanosoma brucei is mediated through alternative trans-splicing. Nucleic Acids Res. 40:1299–1306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Rinehart J, Horn EK, Wei D, Soll D, Schneider A. 2004. Non-canonical eukaryotic glutaminyl- and glutamyl-tRNA synthetases form mitochondrial aminoacyl-tRNA in Trypanosoma brucei. J. Biol. Chem. 279:1161–1166 [DOI] [PubMed] [Google Scholar]
- 31. Shibata S, Gillespie JR, Kelley AM, Napuli AJ, Zhang Z, Kovzun KV, Pefley RM, Lam J, Zucker FH, Van Voorhis WC, Merritt EA, Hol WG, Verlinde CL, Fan E, Buckner FS. 2011. Selective inhibitors of methionyl-tRNA synthetase have potent activity against Trypanosoma brucei infection in mice. Antimicrob. Agents Chemother. 55:1982–1989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Berriman M, Ghedin E, Hertz-Fowler C, Blandin G, Renauld H, Bartholomeu DC, Lennard NJ, Caler E, Hamlin NE, Haas B, Bohme U, Hannick L, Aslett MA, Shallom J, Marcello L, Hou L, Wickstead B, Alsmark UC, Arrowsmith C, Atkin RJ, Barron AJ, Bringaud F, Brooks K, Carrington M, Cherevach I, Chillingworth TJ, Churcher C, Clark LN, Corton CH, Cronin A, Davies RM, Doggett J, Djikeng A, Feldblyum T, Field MC, Fraser A, Goodhead I, Hance Z, Harper D, Harris BR, Hauser H, Hostetler J, Ivens A, Jagels K, Johnson D, Johnson J, Jones K, Kerhornou AX, Koo H, Larke N, et al. 2005. The genome of the African trypanosome Trypanosoma brucei. Science 309:416–422 [DOI] [PubMed] [Google Scholar]
- 33. Wirtz E, Leal S, Ochatt C, Cross GA. 1999. A tightly regulated inducible expression system for conditional gene knock-outs and dominant-negative genetics in Trypanosoma brucei. Mol. Biochem. Parasitol. 99:89–101 [DOI] [PubMed] [Google Scholar]
- 34. Carnes J, Soares CZ, Wickham C, Stuart K. 2011. Endonuclease associations with three distinct editosomes in Trypanosoma brucei. J. Biol. Chem. 286:19320–19330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Inoue M, Nakamura Y, Yasuda K, Yasaka N, Hara T, Schnaufer A, Stuart K, Fukuma T. 2005. The 14-3-3 proteins of Trypanosoma brucei function in motility, cytokinesis, and cell cycle. J. Biol. Chem. 280:14085–14096 [DOI] [PubMed] [Google Scholar]
- 36. Carnes J, Schnaufer A, McDermott SM, Domingo G, Proff R, Steinberg AG, Kurtz I, Stuart K. 2012. Mutational analysis of Trypanosoma brucei editosome proteins KREPB4 and KREPB5 reveals domains critical for function. RNA 18:1897–1909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Acestor N, Zikova A, Dalley RA, Anupama A, Panigrahi AK, Stuart KD. 2011. Trypanosoma brucei mitochondrial respiratome: composition and organization in procyclic form. Mol. Cell. Proteomics 10:M110.006908. 10.1074/mcp.M110.006908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Panigrahi AK, Ogata Y, Zikova A, Anupama A, Dalley RA, Acestor N, Myler PJ, Stuart KD. 2009. A comprehensive analysis of Trypanosoma brucei mitochondrial proteome. Proteomics 9:434–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Keene JD, Komisarow JM, Friedersdorf MB. 2006. RIP-Chip: the isolation and identification of mRNAs, microRNAs and protein components of ribonucleoprotein complexes from cell extracts. Nat. Protoc. 1:302–307 [DOI] [PubMed] [Google Scholar]
- 40. Ingham DJ, Beer S, Money S, Hansen G. 2001. Quantitative real-time PCR assay for determining transgene copy number in transformed plants. Biotechniques 31:132–134, 136–140 [DOI] [PubMed] [Google Scholar]
- 41. Batsch A, Noetel A, Fork C, Urban A, Lazic D, Lucas T, Pietsch J, Lazar A, Schomig E, Grundemann D. 2008. Simultaneous fitting of real-time PCR data with efficiency of amplification modeled as Gaussian function of target fluorescence. BMC Bioinformatics 9:95. 10.1186/1471-2105-9-95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Cestari I, Stuart K. 2013. A spectrophotometric assay for quantitative measurement of aminoacyl-tRNA synthetase activity. J. Biomol. Screen. 18:490–497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Merritt C, Stuart K. 2013. Identification of essential and non-essential protein kinases by a fusion PCR method for efficient production of transgenic Trypanosoma brucei. Mol. Biochem. Parasitol. 190:44–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kalidas S, Li Q, Phillips MA. 2011. A Gateway(R) compatible vector for gene silencing in bloodstream form Trypanosoma brucei. Mol. Biochem. Parasitol. 178:51–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Nilsson D, Gunasekera K, Mani J, Osteras M, Farinelli L, Baerlocher L, Roditi I, Ochsenreiter T. 2010. Spliced leader trapping reveals widespread alternative splicing patterns in the highly dynamic transcriptome of Trypanosoma brucei. PLoS Pathog. 6:e1001037. 10.1371/journal.ppat.1001037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Siegel TN, Hekstra DR, Wang X, Dewell S, Cross GA. 2010. Genome-wide analysis of mRNA abundance in two life-cycle stages of Trypanosoma brucei and identification of splicing and polyadenylation sites. Nucleic Acids Res. 38:4946–4957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. An S, Musier-Forsyth K. 2005. Cys-tRNA(Pro) editing by Haemophilus influenzae YbaK via a novel synthetase.YbaK.tRNA ternary complex. J. Biol. Chem. 280:34465–34472 [DOI] [PubMed] [Google Scholar]
- 48. Chong YE, Yang XL, Schimmel P. 2008. Natural homolog of tRNA synthetase editing domain rescues conditional lethality caused by mistranslation. J. Biol. Chem. 283:30073–30078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Ruan B, Soll D. 2005. The bacterial YbaK protein is a Cys-tRNAPro and Cys-tRNA Cys deacylase. J. Biol. Chem. 280:25887–25891 [DOI] [PubMed] [Google Scholar]
- 50. Morales AJ, Swairjo MA, Schimmel P. 1999. Structure-specific tRNA-binding protein from the extreme thermophile Aquifex aeolicus. EMBO J. 18:3475–3483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Nomanbhoy T, Morales AJ, Abraham AT, Vortler CS, Giege R, Schimmel P. 2001. Simultaneous binding of two proteins to opposite sides of a single transfer RNA. Nat. Struct. Biol. 8:344–348 [DOI] [PubMed] [Google Scholar]
- 52. Raina M, Elgamal S, Santangelo TJ, Ibba M. 2012. Association of a multi-synthetase complex with translating ribosomes in the archaeon Thermococcus kodakarensis. FEBS Lett. 586:2232–2238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Schnaufer A, Ernst NL, Palazzo SS, O'Rear J, Salavati R, Stuart K. 2003. Separate insertion and deletion subcomplexes of the Trypanosoma brucei RNA editing complex. Mol. Cell 12:307–319 [DOI] [PubMed] [Google Scholar]
- 54. Acestor N, Panigrahi AK, Carnes J, Zikova A, Stuart KD. 2009. The MRB1 complex functions in kinetoplastid RNA processing. RNA 15:277–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Panigrahi AK, Zikova A, Dalley RA, Acestor N, Ogata Y, Anupama A, Myler PJ, Stuart KD. 2008. Mitochondrial complexes in Trypanosoma brucei: a novel complex and a unique oxidoreductase complex. Mol. Cell. Proteomics 7:534–545 [DOI] [PubMed] [Google Scholar]
- 56. Koh CY, Kim JE, Shibata S, Ranade RM, Yu M, Liu J, Gillespie JR, Buckner FS, Verlinde CL, Fan E, Hol WG. 2012. Distinct states of methionyl-tRNA synthetase indicate inhibitor binding by conformational selection. Structure 20:1681–1691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Sloof P, de Haan A, Eier W, van Iersel M, Boel E, van Steeg H, Benne R. 1992. The nucleotide sequence of the variable region in Trypanosoma brucei completes the sequence analysis of the maxicircle component of mitochondrial kinetoplast DNA. Mol. Biochem. Parasitol. 56:289–299 [DOI] [PubMed] [Google Scholar]
- 58. Gowri VS, Ghosh I, Sharma A, Madhubala R. 2012. Unusual domain architecture of aminoacyl tRNA synthetases and their paralogs from Leishmania major. BMC Genomics 13:621. 10.1186/1471-2164-13-621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Chang KJ, Lin G, Men LC, Wang CC. 2006. Redundancy of non-AUG initiators. A clever mechanism to enhance the efficiency of translation in yeast. J. Biol. Chem. 281:7775–7783 [DOI] [PubMed] [Google Scholar]
- 60. Chen SJ, Lin G, Chang KJ, Yeh LS, Wang CC. 2008. Translational efficiency of a non-AUG initiation codon is significantly affected by its sequence context in yeast. J. Biol. Chem. 283:3173–3180 [DOI] [PubMed] [Google Scholar]
- 61. Dias J, Octobre G, Kobbi L, Comisso M, Flisiak S, Mirande M. 2102. Activation of human mitochondrial lysyl-tRNA synthetase upon maturation of its premitochondrial precursor. Biochemistry 51:909–916 [DOI] [PubMed] [Google Scholar]
- 62. Seidman D, Johnson D, Gerbasi V, Golden D, Orlando R, Hajduk S. 2012. Mitochondrial membrane complex that contains proteins necessary for tRNA import in Trypanosoma brucei. J. Biol. Chem. 287:8892–8903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Tan TH, Pach R, Crausaz A, Ivens A, Schneider A. 2002. tRNAs in Trypanosoma brucei: genomic organization, expression, and mitochondrial import. Mol. Cell. Biol. 22:3707–3717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Tschopp F, Charriere F, Schneider A. 2011. In vivo study in Trypanosoma brucei links mitochondrial transfer RNA import to mitochondrial protein import. EMBO Rep. 12:825–832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Kaminska M, Deniziak M, Kerjan P, Barciszewski J, Mirande M. 2000. A recurrent general RNA binding domain appended to plant methionyl-tRNA synthetase acts as a cis-acting cofactor for aminoacylation. EMBO J. 19:6908–6917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Kumar S, Das M, Hadad CM, Musier-Forsyth K. 2012. Substrate and enzyme functional groups contribute to translational quality control by bacterial prolyl-tRNA synthetase. J. Phys. Chem. B 116:6991–6999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Kumar S, Das M, Hadad CM, Musier-Forsyth K. 2012. Substrate specificity of bacterial prolyl-tRNA synthetase editing domain is controlled by a tunable hydrophobic pocket. J. Biol. Chem. 287:3175–3184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Beebe K, Ribas De Pouplana L, Schimmel P. 2003. Elucidation of tRNA-dependent editing by a class II tRNA synthetase and significance for cell viability. EMBO J. 22:668–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Nawaz MH, Merriman E, Yang XL, Schimmel P. 2011. p23H implicated as cis/trans regulator of AlaXp-directed editing for mammalian cell homeostasis. Proc. Natl. Acad. Sci. U. S. A. 108:2723–2728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Cristodero M, Seebeck T, Schneider A. 2010. Mitochondrial translation is essential in bloodstream forms of Trypanosoma brucei. Mol. Microbiol. 78:757–769 [DOI] [PubMed] [Google Scholar]
- 71. Zikova A, Panigrahi AK, Dalley RA, Acestor N, Anupama A, Ogata Y, Myler PJ, Stuart K. 2008. Trypanosoma brucei mitochondrial ribosomes: affinity purification and component identification by mass spectrometry. Mol. Cell. Proteomics 7:1286–1296 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.