Abstract
The yeast shuttling serine/arginine-rich protein Npl3 is required for the export of mRNAs and pre-60S ribosomal subunits from the nucleus to the cytoplasm. Here, we report a novel function of Npl3 in translation initiation. A mutation in its C terminus that prevents its dimerization (npl3Δ100) is lethal to cells and leads to translational defects, as shown by [35S]methionine incorporation assays and a hypersensitivity to the translational inhibitor cycloheximide. Moreover, this Npl3 mutant shows halfmers in polysomal profiles that are indicative of defects in monosome formation. Strikingly, the loss of the ability of Npl3 to dimerize does not affect mRNA and pre-60S export. In fact, the mRNA and rRNA binding capacities of npl3Δ100 and wild-type Npl3 are similar. Intriguingly, overexpression of the dimerization domain of Npl3 disturbs dimer formation and results in a dominant-negative effect, reflected in growth defects and a halfmer formation phenotype. In addition, we found specific genetic interactions with the ribosomal subunit joining factors Rpl10 and eukaryotic translation initiation factor 5B/Fun12 and detected a substantially decreased binding of npl3Δ100 to the Rpl10-containing complex. These findings indicate an essential novel function for Npl3 in the cytoplasm, which supports monosome formation for translation initiation.
INTRODUCTION
Serine/arginine-rich (SR) proteins are well known for their function in pre-mRNA splicing. In addition, several members of this family, among others, facilitate the export of the mature mRNAs from the nucleus to the cytoplasm by recruitment of the export receptor heterodimer TAP-p15 (yeast Mex67-Mtr2) to the mRNAs (1–3). Although the function of the SR proteins in splicing does not seem to be conserved in Saccharomyces cerevisiae, the function of the shuttling SR proteins (human SRSF1, SRSF3, and SRSF7 and yeast Npl3, Gbp2, and Hrb1) in mRNA export is conserved (4). All of them contain RNA recognition motifs (RRMs) and a domain rich in repeating arginines and serines (the SR domain) (5). In addition to its function in mRNA export, Npl3 has recently been shown to mediate the export of large ribosomal subunits from the nucleus to the cytoplasm (6). In contrast to mRNA export, Npl3 directly mediates the contact between the ribosomal 25S rRNA and the nuclear pore complex to export pre-60S ribosomal subunits (6). However, mutations in the RRMs lead to export defects of both transport cargoes, mRNA and pre-60S particles. In addition to the RRMs, Npl3 contains an N-terminal domain of unknown function and a C-terminal domain, which consists of repeated motifs of arginine, glycine, and glycine (RGG) and serine and arginine (SR), that is important for its nuclear import mediated by the karyopherin Mtr10 (7).
Interestingly, contrary to other transport factors, the nucleocytoplasmic shuttling SR proteins remain bound to their cargoes in the cytoplasm and can be detected on early polyribosomes (4, 8). Npl3 has been shown to repress translation when it is defective in dissociation from the mRNA (8). In addition, in vitro experiments of RGG-motif containing proteins, including Npl3, suggested that these proteins have repressor functions by interacting with eIF4G to prevent translation (9), but because these interaction studies have been obtained with purified proteins in vitro, the in vivo situation remains still unclear. Moreover, an active role of Npl3 in translation in vivo has not been shown yet. Further indications for a potential involvement of SR proteins in translation were obtained by the association of the mammalian shuttling SR protein and Npl3 homolog SF2/ASF (SFRS1) with polyribosomes and its stimulatory activity on the phosphorylation of the eIF4-binding protein 4E-BP1, which led to an enhancement of translation of a subset of mRNAs (10). Thus, all findings hint to a potential involvement of shuttling SR proteins in translation. However, their exact in vivo function is currently rather nebulous.
Upon arrival of the mRNA in the cytoplasm, the small ribosomal subunit (40S) binds via translation initiation factors to the mRNA, which awaits translation in a closed loop formation. This 43S initiation complex scans the 5′ untranslated region (5′UTR) to identify the (AUG) start codon, where it forms a 48S complex that awaits the 60S ribosomal subunit for subunit joining (11).
Prior to the association of the 60S particle with the mRNA-bound small ribosomal subunit, different cytoplasmic maturation steps of the pre-60S particle occur that involve the removal of assembly factors and the loading of ribosomal protein components, most of which are catalyzed by ATPases or GTPases (12). One of the last events is the release of Tif6, which is a prerequisite for the Lsg1 mediated release of the transport factor Nmd3 (12). GTP hydrolysis on Lsg1 was suggested to drive a conformational change in the pre-60S particle that culminates in the accommodation of Rpl10 into the complex and the release of Nmd3, preparing the 60S subunit for joining the 40S subunit on the mRNA (13). Lastly, the eukaryotic initiation factor 5B (eIF5B/Fun12) associates with the initiation complex and stimulates subunit joining when bound to GTP. Upon 80S monosome formation GTP hydrolysis finally occurs and is required for the release of eIF5B and eIF1A, suggesting that GTP hydrolysis by eIF5B serves as a final checkpoint for correct 80S ribosome assembly (14).
Given that Npl3 transports both, mRNAs and pre-60S ribosomal subunits, we investigated the possibility that Npl3 has a role in bringing together both RNPs in the cytoplasm for translation, in particular because Npl3 has been found in association with early polysomes (8). By analyses of systematic C-terminal truncations of Npl3, we generated mutants that are neither affected in mRNA export, nor in pre-60S export, but have severe defects in monosome formation important for translation initiation, correlating with the fact that they are not able to dimerize. Moreover, we uncover physical and genetic interactions with other ribosomal subunit joining factors and finally show that the dimerization-defective mutant of Npl3 in contrast to its wild-type counterpart does not interact with the ribosomal subunit joining factor Rpl10, which is required for proper monosome formation. Together, our findings support a function of an SR protein in translation initiation to ensure proper formation of the monosomes, which uncovers an unexpected role of this SR protein in connecting nuclear and cytoplasmic steps in gene expression.
MATERIALS AND METHODS
Plasmids and yeast strains.
All of the yeast strains and plasmids used in the present study are listed in Tables S1 and S2, respectively, in the supplemental material.
Construction of RPL10 plasmids.
The sequences for the 5′UTR and the open reading frame (ORF) of RPL10 were amplified by PCR using the primers HK500 and HK501. The fragment was inserted into the SacII-XhoI sites of pHK12 resulting in a C-terminal green fluorescent protein (GFP)-tagged RPL10 (pHK747). The point mutation in rpl10(G161D) in pHK756 was inserted by site-directed mutagenesis of pHK747 using the primer set HK522 and HK523. pHK758 was created as follows. RPL10 was amplified with the primers HK500 and HK501 and inserted into the SacII-XhoI sites of pPS1525. By site-directed mutagenesis, the glycine residue at position 161 was exchanged with an aspartate residue resulting in rpl10(G161D). pHK1291 was made by cutting the coding sequence for GFP out of pHK758 with XhoI-BamHI and the subsequent insertion of a 323-bp terminator sequence of NUF2. The resulting construct was subcloned via SacII-BamHI sites into pRS313. The vector pHK1302 was created by amplifying RPL10 with HK503 and HK504 and inserted into the XhoI-BamHI sites of pHK104.
Construction of vectors for yeast two-hybrid analyses.
The XmaI-BamHI flanked NPL3 ORF from pHK501 was cloned into pHK391 resulting in pHK763 (bait plasmid). The prey plasmid was generated by amplification of NPL3 with the primer set HK181 and HK518 from pHK25, subcloned via TOPO TA cloning kit (Invitrogen) from which the final 1.2-kb NdeI-BamHI fragment was inserted into pHK392, resulting in pHK764. Stepwise truncations of NPL3 were created as follows: PCR fragments amplified with the primer pairs HK578-HK609 (npl3Δ25), HK578-HK610 (npl3Δ50), HK578-HK611 (npl3Δ75), HK578-HK612 (npl3Δ100), and HK578-HK613 (npl3Δ125) were inserted into the XbaI-BamHI sites of pHK764, resulting in pHK829-833.
Construction of NPL3 vectors.
Galactose-inducible GFP-C-npl3 was amplified with appropriate primers and inserted into the BamHI site of pHK230. pHK598 was constructed by inserting the NdeI fragment from pHK195 after filling the overhangs into the SmaI site of Yep351-ADE3. pHK777 was made by amplification of 6×Myc with HK553 and HK554 and insertion into the StuI sites of pHK418 in frame with NPL3. Stepwise truncations of the 3′ part of NPL3 were obtained as follows: NdeI-BamHI fragments of the vectors pHK830 (npl3Δ50), pHK831 (npl3Δ75), pHK832 (npl3Δ100), or pHK833 (npl3Δ125) were treated with Klenow fragment and cloned into StuI-NsiI sites (Klenow treated) of pHK418 resulting in pHK835-838. GFP-NES-C-npl3 (pHK1227) was constructed by inserting the PCR fragment (HK582 and HK680) into the XbaI-HindIII sites of pHK230. The NLS-npl3Δ100 (pHK1274) was constructed in two steps. At first, amplification of the NLS sequence from pHK12 with HK1083 and HK1084 was performed, and the fragment was inserted into the StuI-NsiI site of pHK418. Second, npl3Δ100 was amplified with HK1085 and HK1086 and fused with the NLS sequence via the NsiI site. Plasmids pHK1303 and pHK1304 were made by amplification of fragments with HK615 and HK530 (NPL3) or HK615 and HK612 (npl3Δ100) and insertion into the XhoI-BamHI sites of pHK883. pHK1305 was constructed by PCR with HK1220 and HK612 from the template pHK1276 and inserted into the XhoI-BamHI sites of pHK883. For the GFP-split constructs, the N domain (HK644 and HK645) or the C domain (HK646 and HK647; pHK1322) of eGFP was amplified and fused to NPL3 in pHK418 via the StuI sites. To create pHK1321, the N-GFP-NPL3 fragment was further cut with PvuII and inserted into the same sites of pHK88.
Construction of GST-NPL3 for heterologous expression.
NPL3 was amplified with the primers HK617 and HK686. The resulting fragment was inserted into the BamHI-XhoI sites of pHK119, creating pHK1276.
Purification of 6×His-Npl3.
The purification of recombinant 6×His-Npl3 was performed as described previously (6).
In vitro binding studies.
Both recombinant proteins 6×His-Npl3 (encoded by pHK845) and GST-Npl3 (encoded by pHK1276) were expressed in Escherichia coli BL21DE3. For the affinity pull down of GST-Npl3 via batch purification, glutathione-Sepharose 4B (GE Healthcare) was used. First, the Sepharose was blocked with lysate from bacteria for 4 h at 4°C. The Sepharose (15 μl of 50% slurry) was then incubated with bacterial lysate containing GST-Npl3 in binding buffer (5% glycerin, 100 mM NaCl, 2 mM MgCl2, 20 mM HEPES, 0,14% [vol/vol] 2-β-mercaptoethanol, 1 mM phenylmethylsulfonyl fluoride [PMSF]), to which 45 μg of purified 6×His-Npl3 was also added. As a negative control, bacterial lysate containing GST plus 6×His-Npl3 was incubated. To all lysates, RNase A at a final concentration of 200 μg/ml was added. After agitation for 2 h at 4°C, the Sepharose was washed six times with binding buffer before the addition of sodium dodecyl sulfate (SDS) sample buffer for the subsequent SDS-PAGE.
Yeast two-hybrid analyses.
Dimerization studies of Npl3 were performed by using a Matchmaker two-hybrid system (Clontech). The ORF of the NPL3 gene was fused to the DNA-binding domain or transcription-activating domain of GAL4 in the vectors pGBKT7 DNA-BD and pGADT7-Rec AD, respectively. Truncations of NPL3 were made and expressed from the pGADT7-Rec AD.
GFP fluorescence.
All light microscopy studies were performed as described previously (6).
In situ poly(A)+ RNA hybridization.
Localization of poly(A)+ RNA by in situ hybridization was performed as described previously (6).
[35S]methionine incorporation assay.
Log-phase yeast cells were grown in medium lacking methionine, collected by centrifugation, and resuspended in 1 ml of medium. After preincubation at either 25 or 37°C for 10 or 30 min, 200 μCi of [35S]methionine was added for 30 min at the same temperature. The cells were washed twice before they were lysed in SDS sample buffer (80 mM Tris-HCl, 2.5% [wt/vol] SDS, 10% [vol/vol] glycerol, 5% [vol/vol] 2-mercaptoethanol, 1% [wt/vol] bromphenol blue). The samples were separated by using 10 to 15% (wt/vol) SDS-PAGE, and the amounts of newly synthesized proteins were determined by using a PhosphorImager system FLA-3000 (Fuji) and quantified using Image Gauge 3.1 software.
Extraction of total RNA.
Total RNA was extracted with an RNeasy minikit (Qiagen) according to the manufacturer's instructions. Purified RNA was either analyzed for rRNA integrity by using Experion (Bio-Rad) or further subjected to dot blot analyses.
Sucrose density gradient fractionation.
Polysome analyses were essentially carried out as described previously (15). Yeast cells were grown at 25 or 30°C overnight in 100 to 200 ml of the appropriate medium to 1 × 107 to 2 × 107 cells/ml. Where indicated, cells were shifted to 37°C before cycloheximide was added to a final concentration of 100 μg/ml. The cultures were incubated on ice for 10 min in the presence of cycloheximide prior to centrifugation. Cell pellets were lysed in lysis buffer (20 mM Tris-HCl [pH 7.5], 20 mM KCl, 5 mM MgCl2, 100 μg of cycloheximide/ml, 12 mM β-mercaptoethanol) with two cell volumes of glass beads by beating with the FastPrep-24 (MP Biomedical). Lysates were clarified for 10 min at 13,000 × g and 15 optical density at 260 nm (OD260) units were loaded onto linear 12-ml 7 to 47% (wt/vol) sucrose gradients (20 mM Tris [pH 7.5], 20 mM KCl, 5 mM MgCl2). When we analyzed the amounts of ribosomal subunits, 7 OD260 units of the lysate were preincubated with 100 mM EDTA on ice before they were loaded onto the gradient. After centrifugation at 40,000 rpm for 2.5 to 3.5 h in a Beckman SW40 rotor, the gradient was fractionated, and the absorbance at 254 nm was measured.
Coimmunoprecipitation experiments.
Cells were grown at 25°C to a density of 2 × 107 to 3 × 107 cells/ml in the appropriate medium. Where indicated, a temperature shift to 37°C was performed before the cells were lysed in PBSKMT buffer (137 mM NaCl, 5.7 mM KCl, 10 mM Na2HPO4, 2 mM KH2PO4, 2.5 mM MgCl2, 0.1% [vol/vol] Triton X-100) and glass beads by beating the samples using a FastPrep-24 (MP Biomedicals). Yeast protease inhibitor cocktail (Sigma-Aldrich) and EDTA-free protease inhibitor (Roche; one pill dissolved in 2 ml of distilled water) were added (5 μl of each inhibitor per 100-μl cell pellet). The crude extracts were centrifuged at 13,000 × g until the supernatant was clear. Depending on the immunoprecipitation experiment, lysates were incubated either with 2 μg of antibody (anti-GFP or anti-myc antibody [Santa Cruz]) and 10 μl of glutathione-Sepharose in a 50% slurry (Amersham Biosciences) or 10 μl of a 50% slurry of GFP-Trap_A (ChromoTek) for 3 to 4 h at 4°C. Where indicated, crude extracts were split and treated with or without RNase A (200 μg/ml). Beads were washed five times with 1 ml of PBSKMT buffer and mixed with SDS sample buffer for SDS-PAGE.
Coimmunoprecipitation of RNA.
Yeast cells were grown at 25°C in selective medium to a density of 2 × 107 to 3 × 107 cells/ml. The cells were collected by centrifugation, washed with 1× Tris-buffered saline and resuspended in one cell volume of RNA immunoprecipitation (RNA-IP) buffer (25 mM Tris-HCl [pH 7.5], 100 mM KCl, 0.2% [vol/vol] Triton X-100, 0.2 mM PMSF, 5 mM dithiothreitol). One cell volume of glass beads, RiboLock RNase inhibitor (Fermentas), and protease inhibitor cocktails (from Sigma-Aldrich and Roche) were added. The cells were lysed with a FastPrep-24 instrument (MP Biomedicals) with the following parameters: 20 s and a rotation speed of 6 m/s. For DNase I digestion, 1 ml of lysate was incubated with 10 U of DNase I (Fermentas) at 30°C for 30 min. Lysates were incubated at 4°C for 3 h by incubating the lysates with 10 μl (50% slurry) of GFP-Trap_A (Chromotek). The beads were washed five times with RNA-IP buffer and then split into two fractions. One fraction contained one-tenth of the beads to control the pulldown efficiency via Western blot analysis. The other fraction was treated with proteinase K at 37°C for 30 min. The RNA was purified via phenol-chloroform extraction, followed by ethanol precipitation. Finally, the RNA was eluted in 10 μl of diethyl pyrocarbonate-treated double-distilled H2O.
Dot blot analyses.
For the dot blot experiments, equal amounts of total RNA or eluted RNA from RNA coimmunoprecipitations were transferred onto a Hybond N+ nylon membrane (GE Healthcare) and UV cross linked (254 nm, 120,000 μJ/cm2). Afterward, the membrane was prehybridized for 4 h in prehybridization buffer (0.5 M sodium phosphate buffer [pH 7.5], 7% SDS, 1 mM EDTA) and hybridized overnight in prehybridization buffer, to which 32P-labeled oligo(dT)50 probe was added. The membrane was washed three times in 0.04 M sodium phosphate (pH 7.2) and 0.1% SDS for 10 min each time. The signal intensity was quantified using the Fuji PhosphoImager FLA-3000. The dot blot analyses were performed at least three times.
Quantitative reverse transcription-PCR.
To analyze the amount of coprecipitated 25S rRNA, 250 to 500 ng of the eluted RNA was reverse transcribed with random hexamer primers and Maxima reverse transcriptase (Thermo Scientific) according to the manufacturer's protocol. The PCRs were carried out by using GoTaq qPCR Master Mix (Promega), the primers HK653 and HK654 (each at 0.1 μM), and a Rotor-Gene machine (Qiagen) for 45 cycles at a 61°C annealing temperature. ΔCT values were calculated from the eluate of the negative control. The binding of the 25S rRNA mutant protein was compared to the binding of the wild-type protein.
Software used for quantification.
Signals derived from the [35S]methionine incorporation assay were quantified by using ImageJ, Western blot analyses were quantified by using Bio1D software (Peqlab), and the signals from dot blot experiments were analyzed by using PhosphoImager FLA-3000 software (Fuji).
RESULTS
A functional dimerization domain of Npl3 is essential for cell viability.
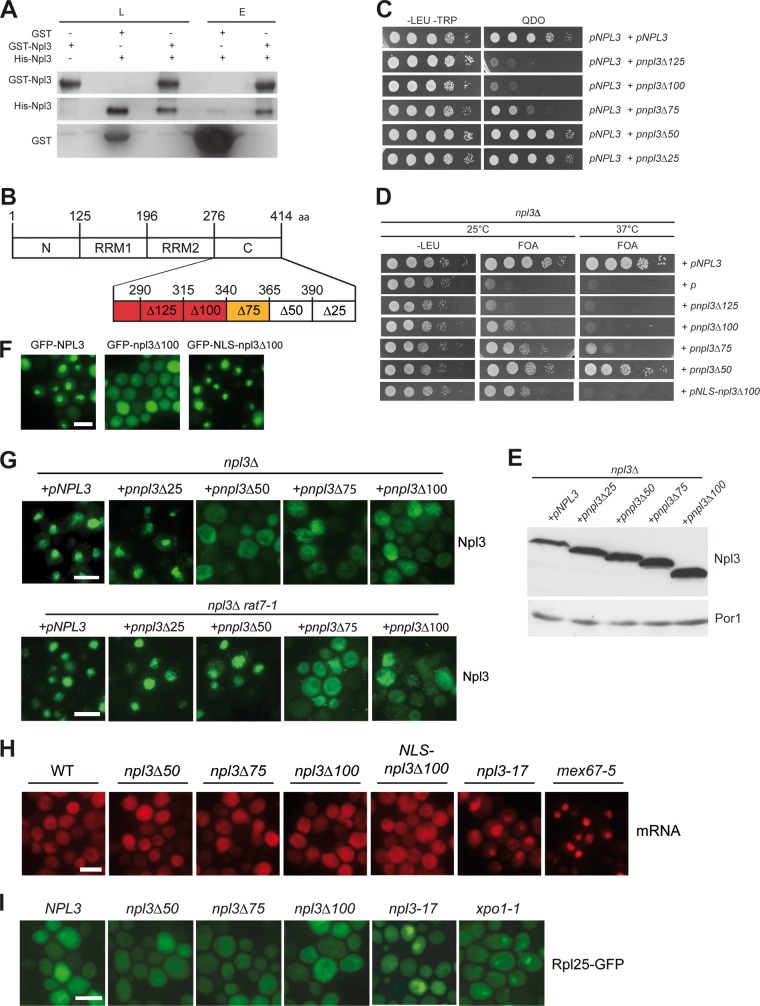
Npl3 can dimerize in vivo (16); however, it is unknown whether this interaction is direct or whether additional factors are required. Thus, we investigated the Npl3-Npl3 interaction in vitro and found that Npl3 can dimerize (Fig. 1A). Thus, we sought to identify a mutant defective in dimerization by constructing systematic truncations. First, we identified the C-terminal domain to be responsible for dimerization (see Fig. S1 in the supplemental material). We then mapped the full dimerization domain by two hybrid analyses to amino acids 276 to 364 and the minimal dimerization domain to amino acids 276 to 339, which is located in the C-terminal SR domain (Fig. 1B and C). Interestingly, only NPL3 mutants that possess full dimerization activity support growth in an NPL3 deletion strain (Fig. 1D), showing that this domain is essential for survival. The growth defects of npl3Δ100 cells are not caused by differences in the expression level of the truncated constructs, as shown in Western blot analyses (Fig. 1E). Similar protein levels, for example, were detected for npl3Δ50 and npl3Δ100, which have different phenotypes. The mitochondrial protein Por1 served as a loading control.
Fig 1.
The dimerization domain of Npl3 is essential for survival but is not crucial for mRNA or pre-60S export. (A) In vitro dimerization of Npl3. Co-affinity precipitations of recombinantly expressed GST-Npl3 with His-Npl3 are shown in the presence of bacterial lysate and RNase A in Western blots. L, lysate; E, eluate. (B) Domain organization of the 414-amino-acid (aa) protein Npl3. N, N-terminal domain; RRM, RNA recognition motif; C, C-terminal domain. Domains necessary (red) or supportive (yellow) for dimerization are also indicated. Deletion mutants used in the present study are indicated by the “Δ” symbol. (C) Two-hybrid analyses reveal a minimal dimerization domain reaching from amino acids 276 to 339 and the full dimerization domain to amino acids 276 to 364. Deletions of the C terminus of Npl3 in 25-amino-acid residue steps were created and cells expressing the truncated versions of Npl3 in combination with wild-type NPL3 were tested for growth on quadruple-dropout (QDO) plates. (D) The C-terminal domain of Npl3 is essential for survival. Truncated versions of NPL3 were subcloned and investigated for the complementation of an npl3 knockout strain. Since the deleted C terminus also lacks the Mtr10 import receptor interaction domain, a nuclear localization signal (NLS) was fused to npl3Δ100 (bottom), allowing Mtr10-independent import. Strains were spotted onto −LEU and 5-fluoorotic acid (FOA) plates on which cells were selected that have lost the covering NPL3 gene on a URA3 vector. (E) The expression of the C-terminal truncation mutants of Npl3 is similar. Western blot analyses of the truncated proteins are shown. Por1 served as a loading control. (F) GFP-NLS-npl3Δ100 is localized to the nucleus. Localization of the indicated GFP-tagged Npl3 variants is shown in a wild-type strain. Scale bars (F, G, H, and I), 5 μm. (G) The nuclear import of npl3Δ100 is inhibited. The localization of wild-type Npl3 and the indicated truncated versions of the protein are shown in npl3Δ and the double-mutant npl3Δ rat7-1 upon a temperature shift to 37°C for 1 h. Antibodies against Npl3 were used for immunofluorescence studies. (H) Dimerization-defective mutants have no mRNA export defects. In situ hybridizations with Cy3-labeled oligo(dT) probes were performed in log-phase wild-type cells, the mex67-5 and npl3-17 mRNA export factor mutants and the indicated truncated npl3 mutant strains shifted to 37°C for 30 min. (I) Dimerization-defective mutants have no pre-60S export defects. The localization of Rpl25-GFP was determined in the wild type, the indicated npl3 mutants, and the 60S export receptor mutant xpo1-1 after a shift to 37°C for 30 min.
Since the dimerization domain also comprises the Mtr10 import receptor-binding domain (7), we added a different nuclear localization signal (NLS), which is recognized by the importin α/β pathway, to direct the protein into the nucleus. Importantly, although this fusion protein, GFP-NLS-npl3Δ100, is properly localized to the nucleus (Fig. 1F), it is still defective in growth (Fig. 1D), indicating that the altered intracellular localization is not responsible for the growth defects. However, we cannot exclude that the dimerization domain contributes to the binding to other proteins as well; the dimerization of Npl3 seems to be important for cell growth. Furthermore, immunofluorescence studies revealed that increasing C-terminal deletions lead to a defect in the proper nuclear steady-state localization of the proteins (npl3Δ50, npl3Δ75, and npl3Δ100) and to defects in the nucleocytoplasmic shuttling (npl3Δ75 and npl3Δ100), as determined in the rat7-1 mRNA export mutant, which is defective in the nuclear porin Nup159 (Fig. 1G).
Strikingly, dimerization-defective mutants of Npl3 have neither detectable mRNA export defects (Fig. 1H) nor apparent defects in the export of 60S and 40S ribosomal subunits (Fig. 1I and data not shown), indicating that the dimerization domain might not be required for the transport functions of Npl3. Further, we found that the dimerization-defective mutants have no obvious defects in mRNA synthesis, rRNA synthesis, or the integrity of the rRNA (data not shown).
The Npl3 dimerization domain is essential for translation.
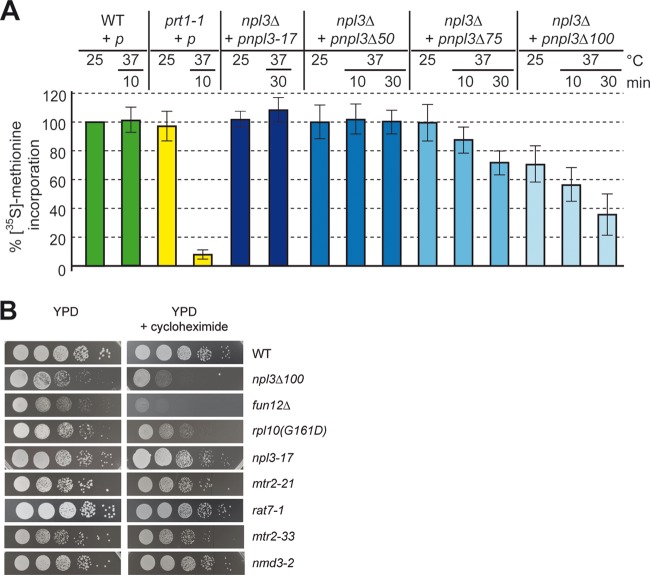
Since truncations of the dimerization domain in Npl3 do not affect the known functions of Npl3, we explored the possibility that Npl3 is involved in translation. Therefore, we analyzed the de novo synthesis of proteins by [35S]methionine incorporation assays, in which the cells were incubated with [35S]methionine at 25 or 37°C upon preincubations at these temperatures (Fig. 2A). Significantly, translation was reduced to <40% after a 30-min shift of the cells to 37°C in npl3Δ100 cells and to 75% in npl3Δ75 cells but was unaffected in the npl3Δ50 cells. The observed translational defect is specific for Npl3 dimerization mutants, because the npl3-17 mutant, which exhibits strong mRNA export and 60S export defects (Fig. 1H and I) (6), has no visible defects in translation (Fig. 2A).
Fig 2.
Npl3 is required for translation. (A) npl3Δ100 is defective in general protein synthesis. The wild type, the translation initiation mutant prt1-1, and the indicated npl3 mutants were grown to log phase in medium lacking methionine. Strains were split and either retained at 25°C or shifted to 37°C for the indicated times before protein synthesis was measured by [35S]methionine incorporation and quantified relative to the wild type (10 different experiments were quantified, and the standard deviations are shown). (B) The dimerization-defective npl3Δ100 mutant is hypersensitive to the translation inhibitor cycloheximide. Strains are shown on yeast extract-peptone-dextrose (YPD) plates and YPD plates containing 0.04 μg of cycloheximide/ml upon incubation at 25°C for 3 days.
Interestingly, we found that mutations in the C-terminal domain in vivo lead to a decrease in translation (Fig. 2A), suggesting that full-length Npl3 in the cellular context enhances translation. This impact of Npl3 in translation is also in accordance with a hypersensitivity of npl3Δ100 cells to the translational inhibitor cycloheximide, similar to other mutants defective in translation such as the fun12Δ (eIF5B) mutant and the rpl10(G161D) 60S ribosomal protein mutant. In contrast, the npl3-17 mutant and two other mRNA export mutants, the mtr2-21 and rat7-1 mutants, as well as mutants in pre-60S export, the mtr2-33 and nmd3-2 mutants, are not affected by treatment with cycloheximide (Fig. 2B), indicating that defects in mRNA or pre-60S export do not generally result in a hypersensitivity to cycloheximide. Interestingly, the hypersensitivity to cycloheximide is also visible for the other C-terminal truncations of Npl3, suggesting that the C terminus of the protein is rather important for this effect (see Fig. S2 in the supplemental material). Taken together, these in vivo data support an essential novel function of Npl3 in enhancing translation.
Mutation of the dimerization domain of Npl3 results in monosome formation defects.
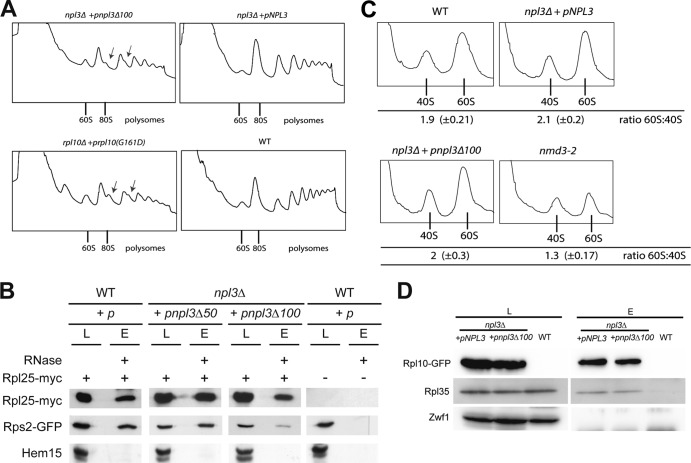
The nature of the translational defect in the dimerization-defective mutant of Npl3 was further investigated by polysomal profile analyses. Compared to the wild type, npl3Δ100 cells show decreased amount of polysomes and, most strikingly, the formation of halfmers, which is a typical feature of mutants that are defective in monosome formation and reflects a delayed formation of the 80S ribosome on mRNAs (Fig. 3A). The formation of halfmers was comparable to that seen in rpl10(G161D) mutant cells known to have defects in monosome formation (17) (Fig. 3A). To further confirm the 80S monosome formation defects in npl3Δ100 cells, we performed coimmunoprecipitation experiments with Rpl25, a protein of the large ribosomal subunit, and Rps2, a component of the small subunit. As a control, we used antibodies against the mitochondrial protein Hem15. Although the association of both subunits is similar in wild-type (100% ± 30%) and npl3Δ50 (91% ± 18%) cells, this interaction is significantly diminished to 26% ± 8% in npl3Δ100 cells (Fig. 3B), indicating that Npl3 dimerization is indeed required for proper 80S formation.
Fig 3.
Npl3 is required for ribosomal subunit joining. (A) The npl3Δ100 mutant is defective in the formation of proper mono- and polysomes. Log-phase wild-type, rpl10(G161D), and npl3Δ100 cells were subjected to sucrose density gradient centrifugation (7 to 47%) upon a shift to 37°C for 30 min. The positions of 60S, the 80S monosomes, and polysomes in the profiles (A254) are indicated. Arrows indicate halfmers. (B) The dimerization of Npl3 is important for the proper interaction of the 40S and 60S ribosomal subunits. Coimmunoprecipitations of Rps2-GFP with Rpl25-myc, indicative for proper 80S formation, were performed in the indicated strains. Western blots including the negative control protein Hem15 are shown. L, lysate; E, eluate. (C) The subunit joining defects in the npl3Δ100 mutant are not due to decreased amounts of ribosomal subunits. Lysates were incubated with EDTA, and the ratio of 60S to 40S ribosomal subunit was determined by sucrose density analyses (7 to 47%) in wild-type, npl3Δ100, and nmd3-2 cells upon a shift to 37°C for 30 min. The ratios of the peak areas of both subunits were calculated from six independent experiments. (D) The incorporation of Rpl10 into the cytoplasmic 60S particle is functional in npl3Δ100 cells. Coimmunoprecipitations of Rpl35 with Rpl10-GFP were compared in wild-type and npl3Δ100 cells in the presence of RNase A. The results are shown in a Western blot. Zwf1 served as a negative control.
Possible causes for a delayed attachment of the 60S subunit to the mRNA-bound 40S subunit and thus halfmer formation are (i) defects in the nuclear pre-60S maturation, (ii) pre-60S export defects, (iii) defects in the cytoplasmic 60S maturation, or (iv) impaired translation initiation. Maturation and export defects are associated with a net decrease in free 60S, because fewer 60S particles are present in the cell. This is reflected in a reduced 60S peak in the polysome profile. In contrast, for defects in the actual joining of the subunits, there should be no dramatic decrease in free 60S versus 40S subunits (18). Thus, a decreased amount of 60S particles in polysome profile experiments can be determined by setting the peak areas of both subunits into relation. We analyzed the ratio of 60S ribosomal subunits to 40S ribosomal subunits (the 60S/40S ratio) in polysomal profile experiments of wild-type cells and found the expected value of ∼2:1 (i.e., 2.0). Similarly, polysome profiles of npl3Δ100 cells show the same ratio. This indicates that npl3Δ100 has no visible defects in the 60S maturation and transport of the large subunit. In contrast, the export defective nmd3-2 mutant clearly shows a decreased 60S/40S ratio of 1.3 in polysome profiles compared to the wild type (Fig. 3C).
Upon arrival of the pre-60S ribosomal subunit in the cytoplasm, sequential protein dissociation and association steps occur that finally lead to the mature, Rpl10-containing 60S particle (12). Since earlier steps of the maturation pathway are a prerequisite for the later steps, we examined whether the levels of the Rpl10-containing 60S subunit were comparable in npl3Δ100 and wild-type cells and found no significant differences, indicating that the earlier steps are not impaired in the npl3Δ100 mutant (Fig. 3D). The glucose-6-phosphate dehydrogenase Zwf1 served as a negative control. This indicates that there are no detectable defects in the cytoplasmic 60S maturation that could lead to monosome formation defects and, in fact, supports a more direct role for Npl3 at the actual step of ribosomal subunit joining.
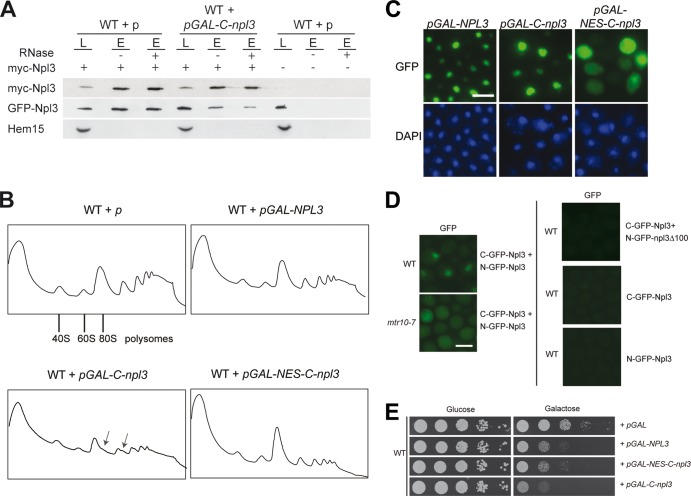
Overexpression of the Npl3 dimerization domain triggers halfmer formation.
An alternative approach to the dimerization mutant to assess the functional importance of dimerization would be to impair dimerization by overexpression of the C terminal domain of Npl3, because it might interfere with the dimer formation of the full-length protein. Such overexpression could have a dominant effect that would clarify the requirement for the Npl3 dimerization in vivo. This dominant-negative effect was indeed visible in vivo (Fig. 4A). Importantly, an excess of the dimerization domain also results in halfmer formation defects (Fig. 4B). The dimerization domain of Npl3 overlaps partially with the Mtr10 import receptor-binding domain (7), which is responsible for the nuclear localization (Fig. 4C). However, the essential novel function of Npl3 that requires dimerization is suggested to occur in the cytoplasm during translation initiation. Therefore, we fused a nuclear export signal (NES) to the C-terminal domain of Npl3 (NES-C-npl3) to enrich the protein in the cytoplasm (Fig. 4C). Strikingly, the presence of an NES resulted in a decrease in the polysomes similar to the overexpression of wild-type NPL3; however, the halfmer formation was no longer visible (Fig. 4B). This is not caused by different expression levels of the constructs, as shown in Fig. S3 in the supplemental material. These data could indicate that dimerization of Npl3 might occur in the nucleus and only there can be disturbed. If so, one would expect to detect Npl3 dimerization also in the nucleus. To analyze this, we used the fluorescence complementation system (19) and fused Npl3 to the N-terminal or the C-terminal part of GFP, so that fluorescent GFP is only generated upon its dimerization. As shown in Fig. 4D, a nuclear signal is visible in wild-type cells, which reflects the steady-state localization of Npl3 and confirms that Npl3 dimers are already detectable in the nucleus. To show that Npl3 dimers are also present in the cytoplasm, we expressed the split-GFP versions of Npl3 in the mtr10-7 mutant that is defective in the nuclear import of Npl3 and found a clear cytoplasmic GFP signal upon a temperature shift to the nonpermissive temperature (Fig. 4D). Moreover, as expected, the combination of Npl3 with npl3Δ100 in the split-GFP system did not lead to any signal, which is due to the defect of npl3Δ100 in dimerization (Fig. 4D). Thus, Npl3 dimers are present in both compartments: the nucleus and the cytoplasm. However, preventing a nuclear dwell time of the C domain by forcing it out of the nucleus via an NES prohibits halfmer formation (Fig. 4B), suggesting that dimer formation requires the nuclear compartment to occur. This is also reflected in the cellular growth rate. Although overexpression of the Npl3 dimerization domain is highly toxic, expression of the C domain fused to an NES only leads to a milder growth defect (Fig. 4E).
Fig 4.
Overexpression of the Npl3 dimerization domain is toxic and leads to halfmer formation. (A) Expression of the C-terminal domain of Npl3 inhibits the dimerization of the full-length proteins. Coimmunoprecipitations of myc-Npl3 and GFP-Npl3 were performed in the presence or absence of the C-terminal domain of Npl3. Western blot analyses, including the negative control protein Hem15, are shown. L, lysate; E, eluate. (B) Overexpression of the C-terminal domain of Npl3 disturbs proper monosome formation. Polysomal profiles of wild-type cells carrying the indicated plasmids are shown. Arrows indicate halfmers. (C) The C-terminal domain of Npl3 is localized to the nucleus. The localization of full-length Npl3 and the C-terminal domain of Npl3 tagged with GFP is shown with or without an NES in wild-type cells. Scale bars (C and D), 5 μm. (D) Npl3 dimerization occurs in the nucleus and in the cytoplasm. Split-GFP domains were used to detect the dimerized Npl3 proteins in vivo in the wild type and the mtr10-7 import receptor mutant. C-GFP-Npl3 and N-GFP-npl3Δ100 or the single (C- or N-GFP)-Npl3 proteins were used as negative controls. (E) Overexpression of the C-terminal domain of Npl3 in the nucleus severely inhibits cellular growth. Serial dilutions of wild-type strains carrying the indicated galactose-inducible vectors are shown on −URA glucose or galactose plates after incubation at 25°C for 3 days.
Genetic evidence for a function of Npl3 in monosome formation.
Monosome formation during the initiation phase of translation occurs on the exported looped mRNA at the AUG start codon. The 43S initiation complex scans the mRNA and, upon recognition of the AUG, it forms a 48S complex that awaits the 60S ribosomal subunit for subunit joining (11). Prior to the association of the 60S particle, different cytoplasmic maturation steps of the exported pre-60S particle occur. These steps include the removal of assembly factors and the loading of certain ribosomal proteins. A final step is the exchange of the transport factor Nmd3 with Rpl10 (12).
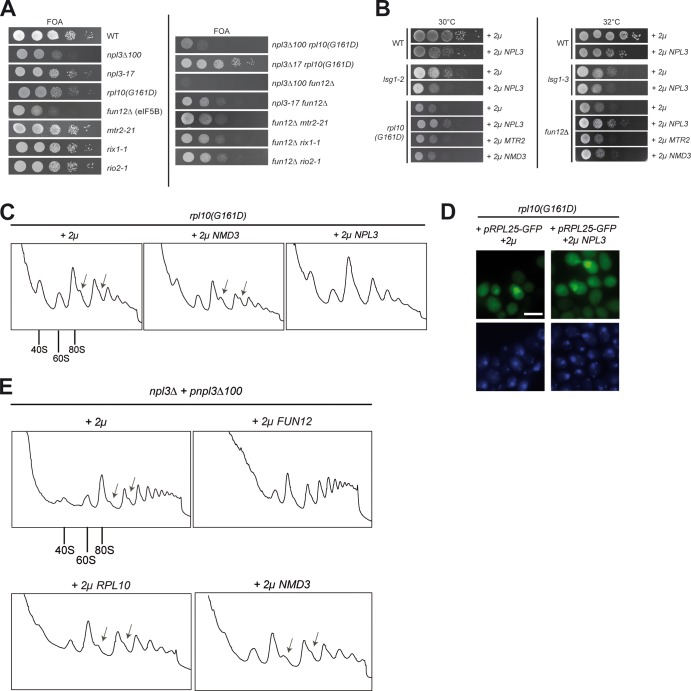
The final ribosomal subunit joining is assisted by eIF5B/Fun12 (14). To study the novel Npl3 function in the context of the other two subunit joining factors, we investigated genetic interactions of npl3Δ100 with mutants of RPL10 and FUN12 and found that their combination significantly impacts the ability of these cells to grow, uncovering their joined activities in the same process (Fig. 5A). Although the growth of the npl3Δ100 rpl10(G161D) double mutant is severely inhibited at 25°C, the npl3Δ100 fun12Δ double mutation is synthetically lethal. Importantly, mutations that lead to mRNA export defects, both in NPL3 (npl3-17) and in MTR2 (mtr2-21), or to ribosome biogenesis defects (rix1-1 and rio2-1) show no synthetic growth interaction with subunit joining factor mutants. To investigate whether the presence of excess Npl3 can compensate the growth defects of mutants involved in ribosomal subunit joining, which would uncover a more direct and specific coordinated function in the same process, we performed high-copy-number suppression experiments. Although overexpression of NPL3 is slightly toxic to wild-type cells, it modestly suppresses the growth defects of fun12Δ and rpl10(G161D) mutants but not of the lsg1 cytoplasmic 60S maturation-defective mutant, further supporting the notion for a direct involvement of Npl3 in monosome formation (Fig. 5B). The specificity of the suppression by NPL3 was shown by overexpression of the pre-60S export factor genes MTR2 and/or NMD3. Both factors are known suppressors of pre-60S export defects (6); however, 2μ NMD3 and MTR2 are not able to reduce the growth defects of rpl10(G161D) or fun12Δ cells (Fig. 5B), indicating that they have no direct function in subunit joining. This is further supported by the fact that both Mtr2 and Nmd3 are not present in polysomes: Nmd3 is exchanged upon entry of Rpl10 (13), and the heterodimer Mex67-Mtr2 dissociates prior to translation (8).
Fig 5.
Genetic interactions support the novel function of Npl3 in monosome formation. (A) The dimerization-defective mutant of NPL3 shows synthetic growth defects or is synthetically lethal in combination with mutants involved in ribosomal subunit joining. Serial dilutions of the wild type and the indicated single and double mutants were spotted onto FOA plates, followed by incubation for 3 days at 25°C. (B) High-copy-number NPL3 is toxic in wild-type cells but supports growth in ribosomal subunit joining factor mutants. Serial dilutions of wild-type, lsg1-2, lsg1-3, rpl10(G161D), and fun12Δ cells, containing either empty vector, 2μ NPL3, MTR2, or NMD3 were spotted in serial dilution onto −LEU plates, followed by incubation for 3 days at the indicated temperatures. (C) High-copy-number NPL3 suppresses the halfmer formation in the rpl10(G161D) ribosomal subunit joining factor mutant. Log-phase rpl10(G161D) cells grown at 30°C carrying either an empty vector (2μ) or 2μ NPL3 or 2μ NMD3 plasmids were subjected to sucrose density gradient centrifugation experiments (7 to 47%). The polysomal profiles (A254) are shown. In panels C and E, arrows indicate the halfmers. (D) High-copy-number NPL3 does not suppress the pre-60S export defects in rpl10(G161D) cells. The Rpl25-GFP localization is shown in the rpl10(G161D) mutant without (2μ) or with high-copy-number NPL3. The DNA was stained with DAPI (4′,6′-diamidino-2-phenylindole). Scale bar, 5 μm. (E) The halfmer formation in npl3Δ100 cells is suppressed by high-copy-number FUN12 (eIF5B). Polysomal profiles of npl3Δ100 cells are shown in the presence of the indicated overexpression plasmids upon growth at the semipermissive temperature of 30°C.
The suppression of the slow-growth phenotype of rpl10(G161D) prompted us to investigate whether high-copy-number NPL3 would also rescue the halfmer formation defects of this mutant. Indeed, although using the same expression vector for both, high-copy-number NMD3 did not affect the formation of halfmers, but NPL3 clearly suppressed this phenotype in the rpl10(G161D) mutant under semipermissive growth conditions (Fig. 5C). RPL10 mutants have been shown to also possess, in addition to the ribosomal subunit joining phenotype, defects in the export of the pre-60S ribosomal subunit from the nucleus to the cytoplasm, which are due to defects in maturation (20). Interestingly, in contrast to the suppression effect of high-copy-number NPL3 on proper monosome formation, the overexpression of NPL3 could not suppress the export/maturation defects of rpl10(G161D) cells (Fig. 5D), supporting a shared function of Npl3 and Rpl10 in monosome formation.
In contrast, when we did the experiment the other way around and overexpressed FUN12 or RPL10 in npl3Δ100 cells, we detected a suppression of the halfmer phenotype only in the presence of high-copy-number FUN12 but not of RPL10 or the pre-60S export factor gene NMD3 (Fig. 5E). This might be due to the fact that the cellular occurrence of Fun12 is rather low (approximately six times less than Npl3). Elevating the level of this enzyme might be beneficial for the final potentially rate-limiting step at the border of translation initiation and elongation. In contrast, the overexpression of RPL10 did not suppress the npl3Δ100 phenotype because it is most likely not possible to incorporate additional copies of Rpl10 into the 60S particle since Rpl10 is a structural component. All genetic experiments support an involvement of Npl3 in ribosomal subunit joining, where it might act together with Rpl10 and eIF5B/Fun12 in connecting both subunits to build an mRNA-associated monosome, prepared for translation.
Npl3 lacking the dimerization domain does not interact with Rpl10 anymore.
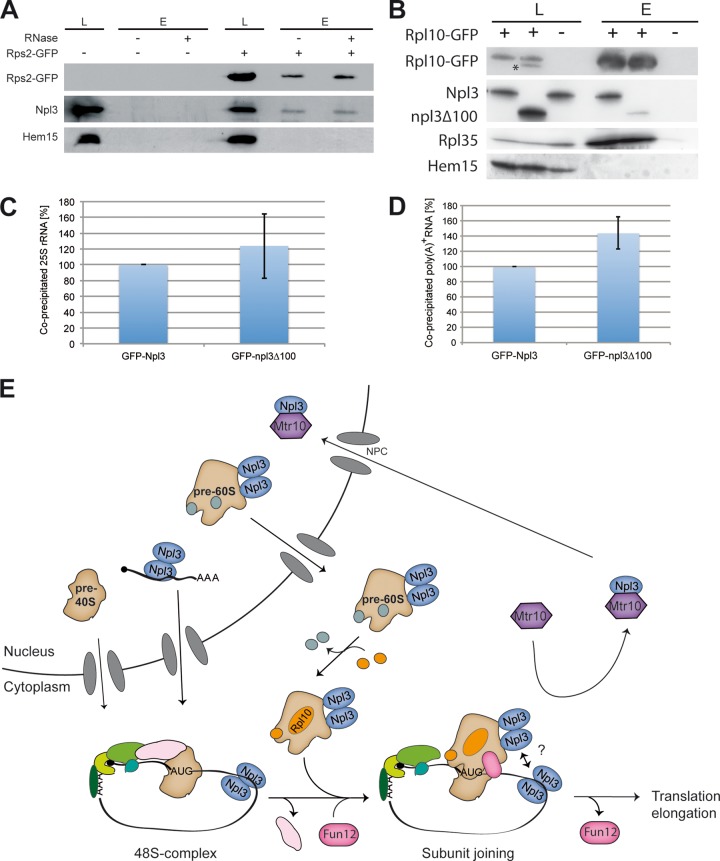
All of the genetic evidence points to a function of Npl3 in ribosomal subunit joining and, although Npl3 is not involved in the export of the small ribosomal subunit (6), it should be possible to detect a physical interaction of Npl3 with the 40S particle that occurs during monosome formation. Indeed, coimmunoprecipitations of Npl3 with Rps2-GFP show a clear interaction (Fig. 6A). This interaction is not disrupted by RNase A treatment, which would occur if Npl3 was only bound to the mRNA, due to its function in mRNA export. To specifically support the genetic evidence for a collaborative function of Npl3 and Rpl10 in subunit joining, we investigated a potential physical interaction of both proteins and found them to be in a complex that is not disrupted by the addition of RNase (Fig. 6B). Strikingly, the interaction of npl3 and Rpl10 is significantly reduced in npl3Δ100 cells, in contrast to the interaction of Rpl10 with Rpl35 (Fig. 6B). This indicates that the incorporation of Rpl10 into the 60S particle does not depend on Npl3 and, in particular, its C terminus, but that the Npl3-Rpl10 interaction does. Only full-length Npl3 seems to be properly located to the Rpl10 complex, where it in association with Rpl10 might support monosome formation.
Fig 6.
Full-length Npl3 forms a complex with Rpl10. (A) Npl3 physically interacts with the small ribosomal subunit. Coimmunoprecipitations were performed with Rps2-GFP and Npl3. The results are shown in a Western blot. Hem15 served as a negative control. (B) The binding of npl3Δ100 to Rpl10 is disturbed. RNase was added to all samples. Western blots of coimmunoprecipitations of Rpl10-GFP with Npl3, npl3Δ100, Rpl35, and the negative control protein Hem15 were performed. The asterisk marks the npl3Δ100 protein from a previous detection. L, lysate; E, eluate. (C) The dimerization domain of Npl3 is not essential for 25S rRNA binding. RNA coimmunoprecipitations were performed with GFP-tagged Npl3 or npl3Δ100. The coprecipitated 25S rRNA was detected by quantitative reverse transcription-PCR. Binding of npl3Δ100 to the rRNA was quantified in three different experiments relative to the wild-type protein. (D) The dimerization domain of Npl3 is not essential for mRNA binding. RNA coimmunoprecipitations were performed with GFP-tagged Npl3 or npl3Δ100. The coprecipitated poly(A)+ RNA was detected by dot blot hybridization with a 32P-labeled oligo(dT) probe and set into relation to the immunoprecipitated protein amount. The mRNA binding of GFP-npl3Δ100 was compared in three different experiments to the wild type. (E) Model for the novel function of Npl3 in translation initiation. Npl3 mediates the nuclear export of mRNA. In the cytoplasm the initiation complex assembles on the mRNA and, upon scanning and AUG recognition, the 40S ribosomal subunit waits for the joining of the mature 60S ribosomal subunit. In addition, Npl3 mediates the export of pre-60S particles and stays bound to the subunit during cytoplasmic maturation. The mature Rpl10-containing 60S subunit joins the 48S initiation complex, which is supported by Fun12 and dimerized Npl3, possibly by its multimerization with the mRNA-bound counterparts. eIF5B/Fun12 dissociates from the complex and allows translation elongation. Finally, the import receptor Mtr10 dissociates Npl3 and recycles it back to the nucleus for other rounds of transport.
Strikingly, although the interaction of the dimerization-defective mutant of Npl3 with Rpl10 is reduced (Fig. 6B), it is still able to bind to the 60S particle via the 25S rRNA (Fig. 6C), which was shown to be a contact point of Npl3 to the pre-60S (6). Similarly, the binding of npl3Δ100 to the poly(A)+ RNA is not reduced (Fig. 6D). However, since the dimerization-defective mutant is not visibly entering the nucleus any longer (Fig. 1E), due to the fact that it also lacks the Mtr10 import receptor domain, it might be possible that the nuclear functions of Npl3 either are not essential or do not require large amounts of the protein. Rather, the cytoplasmic function of Npl3 requires (i) its dimerization domain and (ii) the interaction with the Rpl10 complex. Here, Npl3 supports monosome formation, possibly by establishing the proper joining face of the 60S particle, perhaps by positioning itself or other proteins properly. This might also be important for quality control reasons since Npl3 might, upon cytoplasmic maturation and proper Rpl10 incorporation, signal that subunit joining should occur. It is tempting to speculate that at that point it might also recognize the 40S-bound mRNA by oligomerization with other Npl3 molecules associated with the mRNA, which could stimulate monosome formation on translatable mRNAs (Fig. 6E).
The dissection of the individual functions of this SR protein not only revealed a novel essential role but also helped to allocate domains to different responsibilities. In addition, these data now allow us to assign the first active cytoplasmic function to the shuttling SR domain of Npl3, which is required for dimerization, interaction with Rpl10, and bringing together its two transport cargoes in docking the large ribosomal subunit to the mRNA-bound small ribosomal subunit to generate a functional monosome required for translation initiation.
DISCUSSION
Shuttling SR proteins are required for mRNA export (4). Recently, the SR protein Npl3 has also been shown to export the large ribosomal subunit (6) and to be involved in the recruitment of the early spliceosome (21). Few descriptions of the functions in mRNA translation exist; however, no general active role in this process has yet been described. We have identified a mutant with a truncated C terminus (npl3Δ100) that shows growth defects at 25°C and is lethal at 37°C due to defects in translation (Fig. 1D and Fig. 2A and B). Interestingly, and in contrast to a former described repressive function in translation (8, 9), we show here that npl3Δ100 is defective in interacting with the Rpl10 complex (Fig. 6B) and that its absence causes defects in ribosomal subunit joining (Fig. 3A and B).
A stimulatory role on the translation of certain mRNAs has also been described for the human shuttling SR protein SFRS1 (ASF/SF2) upon tethering to a reporter RNA or upon its overexpression (22). However, our findings demonstrate for the first time a general stimulating impact on translation for an SR protein in vivo.
In contrast to our in vivo data, Rajyaguru et al. suggested an inhibitory function of Npl3 on translation (9). This is due to their finding that Npl3 binds to eIF4G (which is both recombinantly expressed and purified from bacteria), and this interaction inhibited the translation of a reporter transcript in vitro. However, we could not detect a protein-protein binding of Npl3 to eIF4G in vivo (data not shown), suggesting that Npl3 does not contact eIF4G in vivo. Thus, the situation in vivo might be different because important interacting partners, modifications of the proteins, and the multitude of mRNAs are missing. The other repressive function of Npl3 was determined with a mutant of Npl3 (npl3-27) that shows dissociation defects from mRNAs engaged in translation (8). However, this protein might simply inhibit ribosomes from moving forward on the mRNA.
Interestingly, overexpression of the dimerization domain of Npl3 inhibits dimer formation and induces defects in the formation of monosomes (Fig. 4A and B). This observation suggests that the disturbed dimerization domain of Npl3 either prevents the right localization of another protein to the 60S surface or dimerization of Npl3 itself is necessary for the formation of the monosome. Although we show that dimer formation of Npl3 occurs in both compartments, the nucleus and the cytoplasm, it is likely that Npl3 dimers are primarily formed in the nucleus, because guiding the dimerization domain to the cytoplasm by the addition of an NES eliminates the toxic effect on monosome formation (Fig. 4B and E).
The C-terminal region of Npl3 is methylated and phosphorylated in vivo, and both modifications influence its nucleocytoplasmic shuttling (23–25). In fact, several residues of the C-terminal region of Npl3 are methylated (26). However, this seems not to be crucial for translation, because deletion of the methyltransferase neither leads to a hypersensitivity to the translational inhibitor cycloheximide, nor does it show genetic interaction with fun12Δ (data not shown). The phosphorylation site at position S411 of Npl3 is well characterized (23). However, since this site is already missing in the npl3Δ50 mutant, which shows no defects in translation (Fig. 2A), it seems unlikely that the missing phosphorylation is the cause of the subunit joining defects. However, it is still possible that other modifications of the dimerization domain in Npl3 influence translation.
Due to the fact that the npl3Δ100 mutant has major defects in the global protein synthesis (Fig. 2A), a process must be inhibited that impacts a huge variety of translated mRNAs. These might be defects in transcription, mRNA export, or transport of ribosomal subunits. None of these processes are visibly altered (Fig. 1H and I and data not shown). The halfmer phenotype seen in the polysome profiles of the dimerization-defective mutant and upon overexpression of the dimerization domain instead suggests malfunctions in monosome formation. This could be caused by defects in the cytoplasmic maturation of the 60S particle or subsequent processes of translation initiation such as scanning, AUG recognition or in the joining of the 60S ribosomal subunit with the AUG-bound 40S subunit. Since we show here that npl3Δ100 is absent from the Rpl10 surface of the 60S ribosomal subunit (Fig. 6B) and we present genetic and high-copy-number suppression data linking Npl3 to Rpl10 and eIF5B/Fun12 (Fig. 5), we suggest that Npl3 rather functions in ribosomal subunit joining. Rpl10 is the last component that is incorporated into the mature 60S particle, a prerequisite for subunit joining. eIF5B/Fun12 associates with the 48S initiation complex prior to the 60S association to support and finally approve proper monosome formation by its dissociation. This subsequently leads to the entry into translation elongation (11, 13, 14). Thus, both Rpl10 and eIF5B/Fun12 are important late in translation initiation for the formation of the functional ribosome. Interestingly, although we found a strong physical interaction of Npl3 and Rpl10, an interaction with eIF5B/Fun12 could not be detected (Fig. 6B and data not shown). This might be due to a potentially rather transient interaction.
The incorporation of Rpl10 into the 60S ribosomal subunit and the consequent dissociation of Nmd3 constitute the last step of the cytoplasmic maturation of the 60S ribosomal subunit and lead to full translation competence (12). The release of Nmd3 has been suggested to lead to a relaxation of the surface of the 60S particle, which brings Rpl10 into its final position (27). Since it seems very unlikely that Npl3 upon transport dissociates and reassociates with the 60S particle, its interaction with Rpl10 suggests that Npl3 remains associated during transport and the complete cytoplasmic maturation procedure.
The cytoplasmic maturation process follows an ordered progression of rearrangements (28). trans-Acting and transport factors that were recruited in the nucleus dissociate from the pre-60S particle in the cytoplasm to enable joining competence. These rearrangements require several cytoplasmic factors (29). Npl3 could possibly also be one of these stimulatory proteins that support the transfiguration into the mature 60S particle; however, mutations in these maturation factors often show a halfmer phenotype in ribosomal profiles in combination with pre-60S export defects and the degradation of 60S particles (30, 31). Interestingly, and in contrast to the maturation factors, the npl3Δ100 mutant possesses a halfmer phenotype without having defects in the export of pre-60S particles or reduced amounts of 60S (Fig. 1I and 3C). Moreover, the proper incorporation of Rpl10 (Fig. 3D), as well as the genetic interaction with fun12Δ (Fig. 5A) and the suppression effects of high-copy-number NPL3 in fun12Δ (Fig. 5B), instead suggests a function of Npl3 in monosome formation.
We further show that high-copy-number NPL3 but not NMD3 can suppress the growth defect and the halfmer formation visible in ribosomal gradients of rpl10(G161D) cells (Fig. 5B and C). In contrast, the nuclear accumulation of the pre-60S ribosomal subunit is not suppressed by the overexpression of NPL3 (Fig. 5D). Additionally, we found that rpl10(G161D) is present in the 60S particles in similar amounts as its wild-type counterpart (data not shown). Thus, dissociation of rpl10(G161D) or degradation of the mutated protein cannot be the cause of the phenotype, since protein levels are not visibly different. The mutated protein might instead change the surface of the 60S particle and therefore diminish monosome formation efficiency. Additional copies of Npl3 might support monosome formation in this situation more directly.
The opposite experiment, in which we investigated whether high-copy-number NMD3, RPL10, or FUN12 would suppress npl3Δ100, showed suppression only for high-copy-number FUN12 (Fig. 5E). This indicates that (i) a low transport level of pre-60S particles seems not to be the cause of the phenotype, since NMD3 did not act as a suppressor, (ii) elevating the level of Rpl10, a structural component of the 60S particle, might not replace the subunit joining stimulatory function of Npl3, and (iii) both Fun12 and Npl3 might support each other in their monosome formation activity.
Since Npl3 could not be detected in a complex with initiation factors (data not shown), it seems unlikely that Npl3 represents a canonical translation initiation factor. Possibly, Npl3 acts as a factor that supports monosome formation more directly at the joining surface by stabilizing the interaction of the subunits. An attractive, though purely speculative possibility is that multiple copies of Npl3 oligomerize to connect the 40S-bound mRNA to the mature 60S particle.
Taken together, data indicate that the SR protein Npl3 not only is involved in the transport of mRNAs and large ribosomal subunits but, as we show here, is required for the subunit joining step in translation initiation (Fig. 6E). Formation of the monosome is a crucial step in translation and might involve several control steps. Since Npl3 participates in both pathways—the mRNA export and the pre-60S export pathways—and is a shuttling protein, it has the potential to control successful monosome formation.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to M. Ashe, G. H. Braus, E. Hurt, R. Lill, U. G. Maier, M. Seedorf, P. A. Silver, and F. Winston for providing strains and antibodies. We also thank S. Khoshnevis for technical assistance and H. Bastians, E. Hurt, and W. Kramer for helpful discussions.
This study was supported by a grant of the Deutsche Forschungsgemeinschaft, SFB860 to H.K.
Footnotes
Published ahead of print 7 October 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/MCB.00873-13.
REFERENCES
- 1. Dimaano C, Ullman KS. 2004. Nucleocytoplasmic transport: integrating mRNA production and turnover with export through the nuclear pore. Mol. Cell. Biol. 24:3069–3076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Gilbert W, Guthrie C. 2004. The glc7p nuclear phosphatase promotes mRNA export by facilitating association of mex67p with mRNA. Mol. Cell 13:201–212 [DOI] [PubMed] [Google Scholar]
- 3. Huang Y, Gattoni R, Stevenin J, Steitz JA. 2003. SR splicing factors serve as adapter proteins for TAP-dependent mRNA export. Mol. Cell 11:837–843 [DOI] [PubMed] [Google Scholar]
- 4. Huang Y, Steitz JA. 2005. SRprises along a messenger's journey. Mol. Cell 17:613–615 [DOI] [PubMed] [Google Scholar]
- 5. Hacker S, Krebber H. 2004. Differential export requirements for shuttling serine/arginine-type mRNA-binding proteins. J. Biol. Chem. 279:5049–5052 [DOI] [PubMed] [Google Scholar]
- 6. Hackmann A, Gross T, Baierlein C, Krebber H. 2011. The mRNA export factor Npl3 mediates the nuclear export of large ribosomal subunits. EMBO Rep. 12:1024–1031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Senger B, Simos G, Bischoff FR, Podtelejnikov A, Mann M, Hurt E. 1998. Mtr10p functions as a nuclear import receptor for the mRNA-binding protein Npl3p. EMBO J. 17:2196–2207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Windgassen M, Sturm D, Cajigas IJ, Gonzalez CI, Seedorf M, Bastians H, Krebber H. 2004. Yeast shuttling SR proteins Npl3p, Gbp2p, and Hrb1p are part of the translating mRNPs, and Npl3p can function as a translational repressor. Mol. Cell. Biol. 24:10479–10491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Rajyaguru P, She M, Parker R. 2012. Scd6 targets eIF4G to repress translation: RGG motif proteins as a class of eIF4G-binding proteins. Mol. Cell 45:244–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Michlewski G, Sanford JR, Caceres JF. 2008. The splicing factor SF2/ASF regulates translation initiation by enhancing phosphorylation of 4E-BP1. Mol. Cell 30:179–189 [DOI] [PubMed] [Google Scholar]
- 11. Hinnebusch AG, Lorsch JR. 2012. The mechanism of eukaryotic translation initiation: new insights and challenges. Cold Spring Harbor Perspect. Biol. 4:1–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lo KY, Li Z, Bussiere C, Bresson S, Marcotte EM, Johnson AW. 2010. Defining the pathway of cytoplasmic maturation of the 60S ribosomal subunit. Mol. Cell 39:196–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hedges J, West M, Johnson AW. 2005. Release of the export adapter, Nmd3p, from the 60S ribosomal subunit requires Rpl10p and the cytoplasmic GTPase Lsg1p. EMBO J. 24:567–579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Shin BS, Maag D, Roll-Mecak A, Arefin MS, Burley SK, Lorsch JR, Dever TE. 2002. Uncoupling of initiation factor eIF5B/IF2 GTPase and translational activities by mutations that lower ribosome affinity. Cell 111:1015–1025 [DOI] [PubMed] [Google Scholar]
- 15. Gross T, Siepmann A, Sturm D, Windgassen M, Scarcelli JJ, Seedorf M, Cole CN, Krebber H. 2007. The DEAD-box RNA helicase Dbp5 functions in translation termination. Science 315:646–649 [DOI] [PubMed] [Google Scholar]
- 16. Yu MC, Bachand F, McBride AE, Komili S, Casolari JM, Silver PA. 2004. Arginine methyltransferase affects interactions and recruitment of mRNA processing and export factors. Genes Dev. 18:2024–2035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Eisinger DP, Dick FA, Trumpower BL. 1997. Qsr1p, a 60S ribosomal subunit protein, is required for joining of 40S and 60S subunits. Mol. Cell. Biol. 17:5136–5145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kemmler S, Occhipinti L, Veisu M, Panse VG. 2009. Yvh1 is required for a late maturation step in the 60S biogenesis pathway. J. Cell Biol. 186:863–880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kerppola TK. 2006. Visualization of molecular interactions by fluorescence complementation. Nat. Rev. 7:449–456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gadal O, Strauss D, Kessl J, Trumpower B, Tollervey D, Hurt E. 2001. Nuclear export of 60s ribosomal subunits depends on Xpo1p and requires a nuclear export sequence-containing factor, Nmd3p, that associates with the large subunit protein Rpl10p. Mol. Cell. Biol. 21:3405–3415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kress TL, Krogan NJ, Guthrie C. 2008. A single SR-like protein, Npl3, promotes pre-mRNA splicing in budding yeast. Mol. Cell 32:727–734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sanford JR, Gray NK, Beckmann K, Caceres JF. 2004. A novel role for shuttling SR proteins in mRNA translation. Genes Dev. 18:755–768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gilbert W, Siebel CW, Guthrie C. 2001. Phosphorylation by Sky1p promotes Npl3p shuttling and mRNA dissociation. RNA 7:302–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Shen EC, Henry MF, Weiss VH, Valentini SR, Silver PA, Lee MS. 1998. Arginine methylation facilitates the nuclear export of hnRNP proteins. Genes Dev. 12:679–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Xu C, Henry MF. 2004. Nuclear export of hnRNP Hrp1p and nuclear export of hnRNP Npl3p are linked and influenced by the methylation state of Npl3p. Mol. Cell. Biol. 24:10742–10756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. McBride AE, Cook JT, Stemmler EA, Rutledge KL, McGrath KA, Rubens JA. 2005. Arginine methylation of yeast mRNA-binding protein Npl3 directly affects its function, nuclear export, and intranuclear protein interactions. J. Biol. Chem. 280:30888–30898 [DOI] [PubMed] [Google Scholar]
- 27. Sengupta J, Bussiere C, Pallesen J, West M, Johnson AW, Frank J. 2010. Characterization of the nuclear export adaptor protein Nmd3 in association with the 60S ribosomal subunit. J. Cell Biol. 189:1079–1086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Panse VG. 2011. Getting ready to translate: cytoplasmic maturation of eukaryotic ribosomes. Chimia 65:765–769 [DOI] [PubMed] [Google Scholar]
- 29. Panse VG, Johnson AW. 2010. Maturation of eukaryotic ribosomes: acquisition of functionality. Trends Biochem. Sci. 35:260–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Karbstein K. 2013. Quality control mechanisms during ribosome maturation. Trends Cell Biol. 23:242–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Strunk BS, Karbstein K. 2009. Powering through ribosome assembly. RNA 15:2083–2104 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.