Abstract
Human microbiome-derived strains of Lactobacillus reuteri potently suppress proinflammatory cytokines like human tumor necrosis factor (TNF) by converting the amino acid l-histidine to the biogenic amine histamine. Histamine suppresses mitogen-activated protein (MAP) kinase activation and cytokine production by signaling via histamine receptor type 2 (H2) on myeloid cells. Investigations of the gene expression profiles of immunomodulatory L. reuteri ATCC PTA 6475 highlighted numerous genes that were highly expressed during the stationary phase of growth, when TNF suppression is most potent. One such gene was found to be a regulator of genes involved in histidine-histamine metabolism by this probiotic species. During the course of these studies, this gene was renamed the Lactobacillus reuteri-specific immunoregulatory (rsiR) gene. The rsiR gene is essential for human TNF suppression by L. reuteri and expression of the histidine decarboxylase (hdc) gene cluster on the L. reuteri chromosome. Inactivation of rsiR resulted in diminished TNF suppression in vitro and reduced anti-inflammatory effects in vivo in a trinitrobenzene sulfonic acid (TNBS)-induced mouse model of acute colitis. A L. reuteri strain lacking an intact rsiR gene was unable to suppress colitis and resulted in greater concentrations of serum amyloid A (SAA) in the bloodstream of affected animals. The PhdcAB promoter region targeted by rsiR was defined by reporter gene experiments. These studies support the presence of a regulatory gene, rsiR, which modulates the expression of a gene cluster known to mediate immunoregulation by probiotics at the transcriptional level. These findings may point the way toward new strategies for controlling gene expression in probiotics by dietary interventions or microbiome manipulation.
INTRODUCTION
Probiotics are defined as “living microorganisms, which when administered in adequate amounts confer a health benefit on the host” (1, 2). In 1907, Metchnikoff and Mitchell introduced the concept of beneficial microbes to the scientific community through their seminal discovery of the positive effects of fermented milk product consumption on the health and longevity of people in Eastern Europe (3). Since that time, probiotics have become increasingly popular as dietary supplements or functional foods. Investigations into the beneficial effects of probiotics and mechanisms of probiosis have demonstrated that several probiotic species produce metabolites that modulate the host's mucosal immune system. For example, l-lactic acid production by Lactobacillus casei strain Shirota may work via Toll-like receptor 4 (TLR4) signaling to suppress indomethacin-induced myeloperoxidase activity and tumor necrosis factor (TNF) production by human myeloid (THP-1) cells in a rat model of small intestine injury (4). Bifidobacterium breve strain BbC50 and Streptococcus thermophilus strain St065 also secrete small, digestive-enzyme-resistant metabolites that were found to be able to inhibit TNF production from lipopolysaccharide (LPS)-activated THP-1 cells (5). Several probiotic species convert dietary components into bioactive molecules that affect the host's physiological functions. Many probiotics produce short-chain fatty acids (SCFAs) as a product of dietary fiber catabolism (6). SCFAs have anti-inflammatory effects on human immune cells and the gut through binding with G-protein-coupled receptor 43 (GPR43), and this interaction plays a key role in the resolution of several inflammatory conditions, such as arthritis, colitis, and asthma (7). Finally, a recent study demonstrated increased longevity in mice treated with Bifidobacterium animalis subsp. lactis LKM12 compared to control mice, possibly due to the anti-inflammatory effects of polyamines produced by the bacteria (8).
Amino acid decarboxylation and biogenic amine synthesis in bacteria (for example, the conversion of histidine to histamine) are proposed to have at least two major functions: maintaining intracellular pH homeostasis, especially in an acidic environment, and providing energy via proton motive force (9, 10). Histamine biosynthesis through decarboxylation of l-histidine has been extensively studied in both Gram-negative and Gram-positive bacteria. Two different families of histidine decarboxylase (HDC) enzymes have been identified and characterized: pyridoxal phosphate-dependent HDC and pyruvoyl-dependent HDC are present in Gram-negative bacteria and Gram-positive bacteria, respectively. The first HDC identified in lactobacilli was purified from Lactobacillus saerimneri ATCC 33222 (formerly known as Lactobacillus sp. strain 30a), an isolate from a horse's stomach (11). Subsequently, several other Lactobacillus species were found to contain a functional hdc gene cluster, which consists of the histidine decarboxylase pyruvoyl type (hdcA), a putative helper protein (hdcB), and a histidine/histamine antiporter (hdcP) (12). The hdcA and hdcB genes are cotranscribed as a single bicistronic mRNA, and hdcA and hdcB expression is coregulated under the PhdcAB promoter, which lies directly upstream of hdcA (13, 14). Expression of hdcP is regulated by a different promoter. Factors affecting PhdcAB promoter activity and the expression of genes in the hdc cluster have been identified in several Gram-positive bacteria, like Staphylococcus capitis IFIJ12 (13), Lactobacillus saerimneri ATCC 33222, Lactobacillus sp. strain w53 (15), and L. hilgardii 464 (16, 17). These include acidic pH, supplemental l-histidine, histamine, and other molecules, like glucose, fructose, malic acid, and citric acid, in the growth medium. The exact regulatory mechanism of hdc gene cluster expression is still not well characterized.
The model probiotic organism L. reuteri ATCC PTA 6475 (L. reuteri 6475) also produces histamine (18). L. reuteri-derived histamine suppressed TNF production by TLR2-activated THP-1 cells via activation of the histamine receptor type 2 (H2) and inhibition of MEK/extracellular signal-regulated kinase mitogen-activated protein (MAP) kinase signaling. Supplementation of l-histidine in L. reuteri 6475 growth medium increased expression of the hdc gene cluster and production of TNF-inhibitory histamine (18).
In this study, we investigated the role of the Lactobacillus reuteri-specific immunoregulatory (rsiR) gene, a novel regulator of genes involved in histidine-histamine metabolism, in L. reuteri-mediated immunomodulation and histamine production. We characterized the immunomodulatory phenotype of L. reuteri 6475 mutants deficient in RsiR compared to that of the wild type and investigated the regulatory role of RsiR in the expression of the hdc gene cluster and L. reuteri-derived histamine production. We found that RsiR was necessary for L. reuteri-mediated immunomodulation in vitro and in vivo. Inactivation of RsiR resulted in decreased expression of the hdc gene cluster and L. reuteri-derived histamine production compared to the levels for the wild type. Moreover, an RsiR-deficient mutant demonstrated defective upregulation of hdc gene expression and histamine production in the presence of supplemental l-histidine. On the basis of the evidence presented in this report, RsiR regulates the expression of hdcA and hdcB genes at the transcriptional level.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
All bacterial strains used in this study are described in Table S1 in the supplemental material. L. reuteri strains were cultured under anaerobic conditions for 16 to 18 h in deMan, Rogosa, Sharpe (MRS) medium (Difco, Franklin Lakes, NJ) and inoculated into a semidefined medium, LDMIII (the optical density at 600 nm [OD600] was adjusted to 0.1), as previously described (18). Each LDMIII culture was incubated for 24 h at 37°C in an anaerobic workstation (MACS MG-500; Microbiology International, Frederick, MD) supplied with a mixture of 10% CO2, 10% H2, and 80% N2. At mid-exponential phase (6 to 8 h) or stationary phase (24 h), the cells were collected by centrifugation (4,000 × g, 10 min). Cell pellets and bacterial cell-free supernatants were further processed for TNF inhibition, histamine enzyme-linked immunosorbent assay (ELISA), RNA isolation, and GusA reporter assays.
Cell line and reagents.
In vitro experiments were performed with THP-1 cells (human monocytoid cell line, ATCC number TIB-202; ATCC, Manassas, VA) maintained in RPMI (ATCC) and heat-inactivated fetal bovine serum (Invitrogen, Carlsbad, CA) at 37°C in 5% CO2. All other reagents were obtained from Sigma (St. Louis, MO), unless otherwise stated.
Analysis of cDNA microarray data.
We analyzed microarray data from data sets previously deposited under NCBI Gene Expression Omnibus (GEO) series accession number GSE24415. Data from three biological replicates of spotted two-color cDNA microarray experiments comparing the gene expression profiles of L. reuteri 6475 between mid-exponential phase (8 h) and stationary phase (24 h) (samples GSM601520, GSM601521, GSM601522, GSM601523, GSM601524, and GSM601525) were analyzed using the R- and Bioconductor-based web interface for microarray data analysis CARMAweb (19). Data preprocessing included the removal of flagged spots, background correction by use of the normexp algorithm, within-array normalization using the loess method, and between-array normalization with quantile normalization. Analysis of moderated t statistics followed by P-value correction was conducted using the Benjamini and Hochberg method provided by the limma package to identify differentially expressed genes.
Inactivation of rsiR in L. reuteri by targeted insertional mutagenesis (6475::rsiR).
All plasmids, primers, and oligonucleotides are described in Table S1 in the supplemental material. The Lactobacillus reuteri-specific immunoregulatory gene (rsiR, HMPREF0536_10683) was identified in the genome of L. reuteri 6475 (GenBank accession numbers NZ_ACGX02000001 through NZ_ACGX02000007). Inactivation of the rsiR gene was targeted by site-specific integration of plasmid pORI28 (20) into the chromosome, as previously described (21, 22). Briefly, internal rsiR gene fragments were PCR amplified (see Table S1 in the supplemental material) and cloned into pORI28. The resulting construct was integrated into rsiR by site-specific homologous recombination. The targeted insertion was confirmed by sequencing.
Inactivation of rsiR in L. reuteri by ssDNA recombineering (6475rsiR-Stop and 6475 ΔrsiR).
The rsiR gene was inactivated by single-stranded DNA (ssDNA) recombineering as previously described (23, 24). Briefly, L. reuteri 6475/pJP042 carrying recombinase recT was cultured in MRS medium with 10 μg/ml erythromycin to mid-exponential phase (OD600, 0.6) at 37°C. Expression of recT was induced by incubation with pSip peptide (125 μg/ml) at 37°C for 20 min. Recovered cells were plated for single colonies on 10 μg/ml chloramphenicol and incubated anaerobically at 37°C for 16 to 20 h. Colonies were passaged in MRS medium twice and screened by PCR using primers homologous to the stop codon mutations and the 3′ end of rsiR (KJP33 and KJP16; see Table S1 in the supplemental material) or to the sequences flanking the deletion site (KJP15 and KJP16; see Table S1 in the supplemental material). Plasmids utilized in the recombineering reaction were cured by continuous passaging until susceptibility to 10 μg/ml erythromycin and 10 μg/ml chloramphenicol was observed. Mutations were verified by sequencing.
Complementation of 6475::rsiR mutant (6475::rsiR/pJKS104).
The Escherichia coli-L. reuteri shuttle vector (pJKS100) was utilized for rsiR complementation in the 6475::rsiR mutant strain (25). The rsiR gene with its putative promoter was PCR amplified from purified wild-type strain 6475 genomic DNA (primers rsiR-F and rsiR-R; see Table S1 in the supplemental material) and cloned into pCR2.1-TOPO using a TOPO-TA cloning kit (Invitrogen, Carlsbad, CA) to create pCR2.1-rsiR. Full-length rsiR was subcloned from pCR2.1-rsiR into pJKS100 using BstXI restriction sites, resulting in pJKS104 (see Fig. S2 in the supplemental material). The construct was introduced into the 6475::rsiR insertion mutant via electroporation and confirmed by PCR amplification of the cat gene (see Table S1 in the supplemental material).
Global transcriptomic analysis of L. reuteri.
Wild-type strain L. reuteri 6475 and the 6475rsiR-Stop mutant strain were cultured in LDMIII as described above and harvested at 16 h postinoculation. Total RNA was isolated and used as the template to synthesize cDNA libraries using an Ovation prokaryotic RNA-seq system (Nugen Technologies, San Carlos, CA) for high-throughput sequencing on an Illumina HiSeq platform (Illumina, San Diego, CA). Coverage of sequence data was evaluated using the Artemis tool (Wellcome Trust Sanger Institute, Hinxton, United Kingdom). Comparative transcriptomic analyses were performed using the Bowtie (version 0.12.9) tool (26) for sequence mapping and alignment, followed by DNASTAR ArrayStar/QSeq 5 software (DNASTAR, Madison, WI) and the GFOLD algorithm (27), with L. reuteri 6475 draft genome contigs (GenBank accession numbers NZ_ACGX02000001 through NZ_ACGX02000007) used as a reference for transcript mapping. Data were normalized across all experiments by assigned reads per kilobase of feature per million mapped reads (RPKM), and the relative gene expression level was calculated for each open reading frame. Lists of genes whose expression was most affected by genetic inactivation of RsiR (>1.5-fold compared to wild type) were analyzed using subsequent functional clustering analysis with the DAVID bioinformatics tool (28, 29) to identify genetic enrichments of pathways that may be affected by RsiR mutation.
Gene expression studies of the L. reuteri histidine decarboxylase cluster.
Wild-type L. reuteri 6475 and the 6475 ΔrsiR mutant strain were grown in LDMIII as described above in the presence or absence of 4 g/liter supplemental l-histidine and harvested at 16 h postinoculation. Isolation of RNA from collected cell pellets and cDNA synthesis from total RNA were performed as previously described (18). Expression of the hdcA, hdcB, and hdcP genes was analyzed using quantitative reverse transcription-PCR (RT-qPCR). All primers used in this study were designed using the Universal ProbeLibrary Assay Design Center (Roche Applied Science, Indianapolis, IN) and are described in Table S1 in the supplemental material. The RNA polymerase β-subunit (rpoB) gene, which was unaffected by inactivation of rsiR (RNA-seq analysis; data not shown), was used as a reference gene. RT-qPCR mixtures included 2× FastStart Universal Probe Master (Rox; Roche Applied Science) and prepared cDNA as the template, along with the corresponding probes and primers at final concentrations of 100 nM and 200 nM, respectively. Serially diluted genomic DNA of wild-type L. reuteri 6475 was also included to create the standard curve used in the analysis. PCRs were performed using a ViiA (version 7) real-time PCR system (Life Technologies, Grand Island, NY) with the cycling parameters described previously (18). The relative standard curve method (ViiA [version 7] data analysis software) was used to calculate relative changes in gene expression.
Quantification of histamine by ELISA.
The production of histamine by L. reuteri strains was measured by quantitative histamine ELISA, as previously described (18). Briefly, wild-type L. reuteri 6475 and the 6475 ΔrsiR mutant strain were grown in LDMIII in the presence or absence of 4 g/liter supplemental l-histidine. Cultures were harvested at 24 h, centrifuged, and filter sterilized with 0.22-μm-pore-size polyvinylidene difluoride filters (EMD Millipore, Billerica, MA). Histamine concentrations were determined using a histamine ELISA kit (Neogen, Lexington, KY) as per the manufacturer's instructions. The absorbance was measured with a Bio-Rad Spectramax 340PC spectrophotometer. Data were normalized to those for the LDMIII culture at an OD of 1.0 and corrected with the values obtained from the background control.
β-Glucuronidase (GusA) promoter assay.
The activity of the hdcAB promoter was tested in a β-glucuronidase (GusA) promoter assay. The putative promoter of the hdcAB genes (PhdcAB) from L. reuteri 6475 was predicted using the Neural Network Promoter Prediction tool (30). The promoter DNA sequence was amplified by primers PhdcAB-F and PhdcAB-R (described in Table S1 in the supplemental material) and cloned into an expression vector, pJKS100, using KpnI and EcoRI restriction sites, replacing the original P23 promoter. A hyperactive β-glucuronidase reporter gene, gusA3, from pGK12 (31) was cloned directly downstream from PhdcAB using an EcoRI site to create pPH-R1 (see Fig. S3 in the supplemental material). Wild-type L. reuteri 6475 or the 6475 ΔrsiR mutant strain carrying pPH-R1 was grown anaerobically in LDMIII as described above in the presence or absence of 4 g/liter supplemental l-histidine to mid-exponential or stationary phase. Cell pellets were collected and assayed for GusA activity using a protocol from Axelsson et al. (32), with some modifications. Briefly, each pellet was resuspended in 200 μl of 50 mM NaH2PO4, pH 7.0. Fifty microliters of cell suspension was added to 450 μl of GUS buffer (50 mM NaH2PO4, pH 7.0, 10 mM β-mercaptoethanol, 1 mM EDTA, 0.1% Triton X-100). To this mixture, 12.5 μl of 0.1% sodium dodecyl sulfate and 25 μl of chloroform were added. After 5 min incubation at 37°C, 12.5 μl of 4 mg/ml of p-nitrophenyl-β-d-glucoronide was added. Each reaction mixture was incubated at 37°C for 3 min, and the reaction was stopped by adding 250 μl of 1 M Na2CO3. After centrifugation at 8,000 × g for 5 min, supernatants were transferred into cuvettes and optical densities at 420 nm were measured using a SmartSpec Plus spectrophotometer (Bio-Rad Laboratories, Hercules, CA). Data were converted to Miller units and analyzed using two-way analysis of variance (ANOVA) on GraphPad Prism (version 5) software (GraphPad Inc., La Jolla, CA).
TNF inhibition bioassay.
A TNF inhibition bioassay and a TNF ELISA were performed as previously described (18). Briefly, supernatants from L. reuteri LDMIII cultures were filter sterilized and size fractionated to select for factors smaller than 3 kDa in size. The filtrate was speed vacuum dried, resuspended in RPMI medium, and normalized by volume to an OD600 of 1.5. Supernatants were tested for their ability to modulate TNF production in monocytoid cells. THP-1 cells (5 × 104 cells) were treated with L. reuteri supernatant (5%, vol/vol) and subsequently activated by 100 ng/ml Pam3Cys-SKKKK × 3 HCl (EMC Microcollections, Tübingen, Germany), as previously described (33). Cells were incubated at 37°C in 5% CO2 for 3.5 h and then pelleted (3,000 × g, 5 min, 4°C). Quantitative ELISAs were used to determine the concentration of TNF in THP-1 cell supernatants according to the manufacturer's instructions (R&D Systems, Minneapolis, MN). Data were analyzed using an unpaired t test on GraphPad Prism (version 5) software.
Mice.
Female BALB/c mice (45 days old) were received from Harlan Laboratories (Houston, TX) and maintained under specific-pathogen-free (SPF) conditions. Mice were kept in filter-top cages (5 mice per cage) and had free access to distilled water and Harlan rodent chow 2918. After 10 days of acclimatization, mice were randomly divided into several experimental groups. All mouse experiments were performed in an SPF animal facility according to an Institutional Animal Care and Use Committee (IACUC)-approved mouse protocol at the Baylor College of Medicine, Houston, TX.
Preparation of supernatant from L. reuteri strains and administration to mice.
Bacterial supernatants from LDMIII cultures of wild-type L. reuteri and the 6475::rsiR mutant were prepared as described above. All supernatants were filter sterilized, size fractionated, and concentrated to 20× with speed vacuum drying before administration to mice. Each mouse received two intraperitoneal (i.p.) injections of bacterial supernatant or medium control (0.1 ml each time), with the first dose given 18 h before a trinitrobenzene sulfonic acid (TNBS) rectal enema (described below) and the second dose given 2 min before the TNBS enema.
Induction of acute colitis by intrarectal instillation of TNBS.
Two minutes after i.p. injection of the second dose of bacterial supernatant or medium control, mice were anesthetized by constant isoflurane inhalation. A 5% (vol/vol) TNBS (Sigma-Aldrich, St. Louis, MO) solution in water was diluted with an equal volume of absolute ethanol and administered intrarectally via a catheter (Braintree Scientific, Braintree, MA) at a dose of 100 mg/kg of body weight 4 cm distal to the anus. Mice were kept head down in a vertical position for 2 min after the enema to ensure complete retention of the enema in the colon. Procedure control mice received 50% ethanol in phosphate-buffered saline (PBS) as an enema and two i.p. injections of the medium control. Colitis-positive mice received a TNBS enema and two i.p. injections of the medium control, while test mice received a TNBS enema and two i.p. injections of the prepared bacterial supernatant.
Macroscopic assessment of TNBS-induced colitis.
The colons were collected 48 h after colitis induction and opened longitudinally, and images were recorded with a digital camera. Colonic inflammation and damage in the distal colon were determined according to the Wallace criteria (34). In brief, the grading scale was as follows: score 0, normal/healthy appearance; score 1, focal hyperemia, slight thickening, and no ulcers; score 2, hyperemia, prominent thickening, and no ulcers; score 3, ulceration with inflammation at one site; score 4, ulceration with inflammation at two or more sites; score 5, major sites of damage extending >1 cm; scores 6 to 10, when the area of damage extended >2 cm, an increase of 1 score unit for each additional 1 cm of tissue involvement. Each colon was scored blindly by one individual.
Plasma measurements of mouse SAA.
Blood samples were collected from mice via cardiac puncture, stored with anticoagulant, and centrifuged (10 min, 17,000 × g) to isolate plasma. Serum amyloid A protein (SAA) concentrations in plasma were measured using an SAA ELISA kit (Alpco, Salem, NH) according to the manufacturer's instructions.
RESULTS
Discovery of rsiR gene and structural predictions.
Based on prior data showing robust TNF suppression from stationary-phase bacterial supernatants (35, 36), we hypothesized that genes upregulated in the stationary phase of growth would likely be involved in L. reuteri-mediated immunomodulation. We compared the gene expression profiles of L. reuteri 6475 between exponential phase (8 h) and stationary phase (24 h) from previously deposited sets of data for cDNA microarray experiments from our L. reuteri metabolic modeling study (35). The 20 most highly upregulated genes from this comparison are listed in Table 1. Of particular interest was a 354-bp open reading frame (GenBank gene locus tag HMPREF0536_10683) predicted to encode a 118-amino-acid hypothetical protein with a molecular mass of 12.9 kDa and a calculated isoelectric point of 8.76. The open reading frame is located on the minus strand of the chromosome, positioned between a protein disulfide isomerase (frnE) gene and several tRNA genes (see Fig. S1 in the supplemental material). Nucleotide BLAST searches against the GenBank microbial genome and Whole Genome Shotgun (WGS) databases identified full-length hits with between 97 and 100% identity in 10 L. reuteri genomes. Of these 10 genomes, 3 were those of L. reuteri strains that possessed TNF-suppressive activity (strains ATCC PTA 4659, JCM 1112, DSM 20016) (36) (J. K. Spinler, unpublished data).
Table 1.
The 20 most highly upregulated genes in L. reuteri 6475 in stationary phase compared to gene expression in mid-exponential phasea
| Microarray identifier | Locus tag | Gene name | Corrected P value | Fold upregulated |
|---|---|---|---|---|
| NT01LR0789 | HMPREF0536_11158 | Conserved hypothetical protein | 4.83E−04 | 34.60 |
| NT01LR0977 | HMPREF0536_10683 | Conserved hypothetical protein (rsiR) | 3.54E−05 | 32.20 |
| NT01LR1511 | HMPREF0536_10555 | Dihydrofolate:folylpolyglutamate synthetase | 3.61E−04 | 26.96 |
| NT01LR0625 | HMPREF0536_10297 | Conserved hypothetical protein | 3.15E−04 | 26.30 |
| NT01LR1631 | HMPREF0536_11291 | Aldo/keto reductase family oxidoreductase | 1.53E−04 | 25.27 |
| NT01LR1311 | HMPREF0536_11270 | Hypothetical protein | 3.29E−04 | 24.36 |
| NT01LR1510 | HMPREF0536_10556 | DNA repair protein (radC) | 9.78E−05 | 23.53 |
| NT01LR1386 | HMPREF0536_10299 | Conserved hypothetical protein | 1.85E−03 | 22.62 |
| xth_1 | HMPREF0536_11294 | Exodeoxy-RNase III | 1.36E−04 | 20.93 |
| cbiD | HMPREF0536_11708 | Cobalamin biosynthesis protein (cbiD) | 4.26E−04 | 19.02 |
| cobD | HMPREF0536_11710 | Cobalamin biosynthesis protein (cobD) | 5.33E−04 | 18.09 |
| NT01LR1337 | HMPREF0536_11674 | Amino acid permease | 9.78E−05 | 16.65 |
| NT01LR1312 | HMPREF0536_11269 | LacI family sugar-binding transcriptional regulator | 9.78E−05 | 16.41 |
| NT01LR0974 | HMPREF0536_10863 | Phage antirepressor protein | 3.71E−04 | 16.21 |
| NT01LR1675 | HMPREF0536_10858 | Hypothetical protein | 1.48E−04 | 16.21 |
| NT01LR1937 | HMPREF0536_10005 | Competence CoiA family protein | 1.05E−04 | 14.94 |
| NT01LR1936 | HMPREF0536_10006 | Dipeptidase A | 9.78E−05 | 14.68 |
| NT01LR1346 | HMPREF0536_11683 | Conserved hypothetical protein | 1.13E−03 | 14.54 |
| NT01LR1336 | HMPREF0536_11673 | Alpha/beta fold family hydrolase | 9.78E−05 | 14.42 |
| NT01LR1345 | HMPREF0536_11682 | Hypothetical protein | 1.01E−04 | 14.11 |
These data represent microarray comparisons of a previously published study (35).
Functional domain prediction by InterProScan analysis (37) did not reveal any conserved functional domains but predicted the presence of a putative signal peptide at the N terminus (amino acids 1 to 26) and three additional transmembrane domains (amino acids 44 to 64, 69 to 89, and 93 to 113), indicating that RsiR may be a transmembrane protein. Prediction of the function of RsiR by use of the Protein Function Prediction tool (38) suggested involvement in folic acid transport (GO: 0015884), indole derivative metabolism (GO: 0042434), or cofactor transport and metabolism (GO: 0051181 and 0051186). Ab initio three-dimensional structure prediction using Phyre2 software (39) suggested the presence of three transmembrane helices lying in parallel to each other. These observations suggest that rsiR may encode a transmembrane protein that plays a role in the transport or metabolism of cofactors that are involved in L. reuteri-mediated immunomodulation.
The rsiR gene product regulates genes involved in histidine transport and metabolism.
To study the role of rsiR in L. reuteri-mediated immunomodulation, we created RsiR-deficient L. reuteri 6475 mutants by two methods: (i) targeted insertional mutagenesis and (ii) ssDNA recombineering. The rsiR insertional mutant 6475::rsiR was generated by homologous recombination of plasmid pORI28 into the 5′ end of rsiR (21). Genetic recombineering (23) was used to create (i) an rsiR mutant containing two premature translational stop codons near the 5′ end of the gene (6475rsiR-Stop mutant) and (ii) a mutant with a complete deletion of rsiR (the 6475 ΔrsiR mutant). No significant differences in growth under standard culture conditions (see Materials and Methods) were found between wild-type L. reuteri 6475 and the three RsiR-deficient mutants (data not shown).
To elucidate the role of rsiR in L. reuteri-mediated immunomodulation and in other bacterial metabolic pathways, we performed a global comparative analysis of stationary-phase transcriptomes from L. reuteri 6475 and 6475rsiR-Stop. Paired-end sequencing resulted in 162,737,873 and 195,032,924 100-bp reads from wild-type L. reuteri 6475 and 6475rsiR-Stop, respectively. The sequencing data demonstrated complete coverage of the entire genome, with all predicted transcripts appearing at least once. All transcripts were mapped to L. reuteri 6475 draft genome contigs. After filtering out low-count transcripts (<200 transcripts per open reading frame), the gene expression profiles of the two strains were compared. We identified 195 genes (9.3% of the genome) that were downregulated more than 1.5-fold in 6475rsiR-Stop compared to their level of expression in wild-type L. reuteri 6475 (see Table S2 in the supplemental material). Of note, two highly downregulated genes included the histidine decarboxylase (hdcA) and histidine-tRNA ligase (hisS2) genes (which were downregulated 3.76- and 3.70-fold, respectively) (see Table S2 in the supplemental material). In addition, the putative amino acid-polyamine-organocation (APC) family lysine transporter gene (lysP2) was downregulated, and a homolog of this transporter in Lactococcus lactis subsp. cremoris NZ9000 appears to function as a secondary l-histidine transporter (40). We also identified 143 genes that were highly upregulated (>1.5-fold) in 6475rsiR-Stop, including hdcP, while most genes were involved in cell redox homeostasis pathways, N-acetyltransferase activity, and hypothetical proteins of unknown function (see Table S3 in the supplemental material). DAVID functional clustering analysis of genes whose expression was most affected by RsiR inactivation highlighted several metabolic pathways with altered expression (see Table S4 in the supplemental material). In the rsiR mutant, genes involved in cysteine/methionine metabolism, fatty acid biosynthesis, and glycerolipid metabolism were most downregulated, while genes involved in cell redox homeostasis, glycerophospholipid metabolism, and cellular homeostasis were most upregulated. These results suggested that RsiR may regulate several essential metabolic functions in L. reuteri and may explain why it is conserved in some non-human-derived L. reuteri strains that lack the hdc gene cluster. Since hdcA was the most downregulated gene according to the RNA-seq analysis and histidine decarboxylation has been shown to be involved in L. reuteri-mediated immunomodulation (18), we decided to focus on the expression of hdc cluster genes in the rsiR mutant.
To validate the results of RNA sequence analysis and confirm the decreased expression of hdc cluster genes in the rsiR mutant, stationary-phase expression of the hdcA, hdcB, and hdcP genes in wild-type L. reuteri 6475 was compared to that in the 6475 ΔrsiR mutant using RT-qPCR. The expression of hdcA and hdcB in the 6475 ΔrsiR mutant was dramatically decreased compared to that in L. reuteri 6475, while the expression of hdcP was unaffected (Fig. 1A). In the presence of 4 mg/ml l-histidine, all 3 genes (hdcA, hdcB, and hdcP) in wild-type L. reuteri 6475 were upregulated compared to the level of expression in unsupplemented medium. However, the hdcA, hdcB, and hdcP genes were not affected in the 6475 ΔrsiR mutants, when cells were grown in the presence of l-histidine (Fig. 1B). Taken together, these data suggest that rsiR may play a role in the transcriptional regulation of genes involved in histamine biosynthesis.
Fig 1.
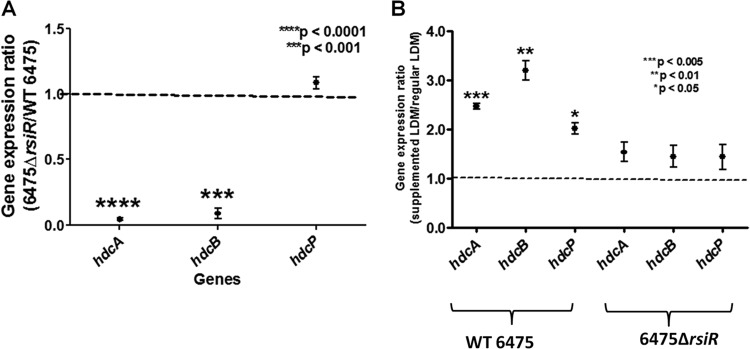
Effects of rsiR and l-histidine on hdc gene expression in L. reuteri. Gene expression in the histidine decarboxylase (hdc) cluster was suppressed in the rsiR mutant compared to its expression in wild-type (WT) L. reuteri. (A) RT-qPCR demonstrating decreased expression of the hdcA and hdcB genes in the RsiR-deficient mutant compared to wild-type strain 6475. Expression ratios of each gene (rsiR mutant versus wild type) were calculated, and results represent the mean ± SD (n = 3). P values were determined using a one-sample t test and are in comparison to the theoretical mean of 1. (B) RT-qPCR demonstrating expression of all genes in the hdc cluster when L. reuteri was grown in the presence or absence of supplemental l-histidine. The expression ratios of each gene (histidine-supplemented versus unsupplemented) were calculated. Results represent the mean ± SD (n = 3). P values are in comparison to the theoretical mean of 1.0. All RT-qPCR data were normalized to those for a reference gene, rpoB.
Inactivation of rsiR diminished the expression of histamine biosynthesis genes (Fig. 1; see Table S2 in the supplemental material) and, coincidentally, histamine production (Fig. 2). Histamine concentrations in culture supernatants from L. reuteri 6475 grown to stationary phase were approximately 40 μg/ml and increased more than 2-fold in medium supplemented with 4 mg/ml l-histidine. In contrast, histamine production from the 6475 ΔrsiR mutant was significantly reduced, and supplementing the medium with l-histidine did not significantly increase the histamine concentration in the supernatants (Fig. 2).
Fig 2.
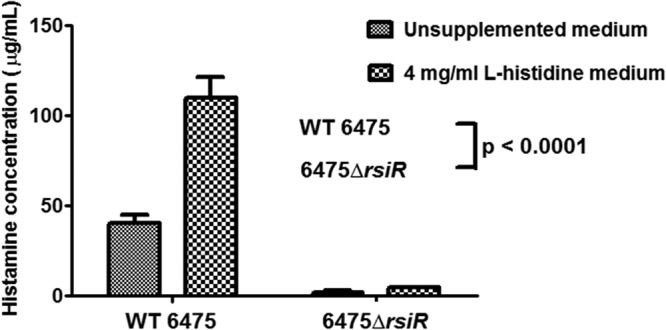
Effects of rsiR and l-histidine on histamine production by L. reuteri. Histamine production was diminished in the rsiR mutant compared to that in wild-type L. reuteri even in the presence of supplemental l-histidine. The histamine produced by L. reuteri was quantified using ELISA. Data were analyzed by two-way ANOVA. Results represent the mean ± SD (n = 3). P was <0.0001 compared to wild-type 6475.
Defining the hdcAB promoter region by reporter gene studies.
We used the Neural Network Promoter Prediction tool (30) to identify a 117-bp region directly upstream of hdcAB (PhdcAB) in wild-type L. reuteri 6475. The predicted sequence had 57.1% homology to the PhdcAB promoter sequence previously identified in Staphylococcus capitis IFIJ12 (13). The activity of this putative PhdcAB promoter sequence was tested in both wild-type and rsiR mutant strains of L. reuteri using a reporter plasmid, pPH-R1, containing the hyperactive β-glucuronidase reporter gene, gusA3 (31) (see Fig. S3 in the supplemental material). During stationary phase in the absence of supplemental histidine, wild-type L. reuteri containing pPH-R1 averaged 458.7 Miller units of GusA activity, while 6475 ΔrsiR produced very little GusA activity (Fig. 3). In the presence of 4 mg/ml supplemental l-histidine, wild-type L. reuteri 6475 yielded increased GusA reporter activity (P < 0.0001), while the GusA activity with the 6475 ΔrsiR mutant was unchanged.
Fig 3.
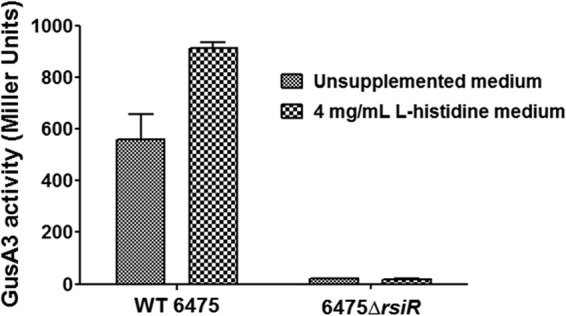
Reporter assays defining the promoter region affected by rsiR. A mutation in rsiR resulted in decreased expression of the gusA3 reporter gene driven by the putative hdcAB promoter. Wild-type strain 6475 or the 6475 ΔrsiR mutant expressing reporter gene gusA3 was grown in the presence or absence of supplemental l-histidine. GusA3 activity was measured in Miller units. Data were analyzed using two-way ANOVA (P < 0.0001) and the Bonferroni posttest to compare the GusA3 activity between the wild type and mutant at each concentration of supplemental l-histidine (P < 0.0001 for both 0 and 4 mg/ml l-histidine).
The rsiR gene is essential for suppression of human TNF production by L. reuteri.
We characterized the immunomodulatory role of rsiR by studying the effects of rsiR inactivation on suppression of TNF production by activated human myeloid (THP-1) cells. Bacterial culture supernatants from wild-type L. reuteri 6475 and the 6475 ΔrsiR mutant were evaluated for the ability to inhibit human TNF production by TLR2-activated THP-1 cells. Wild-type L. reuteri 6475 suppressed human production (P < 0.0001), while the mutant 6475 ΔrsiR strain yielded a diminished ability to inhibit TNF production (Fig. 4). Supplementation of growth medium with 4 mg/ml l-histidine was able to partially restore the TNF-inhibitory phenotype of the rsiR mutant. Other rsiR mutants (the 6475rsiR-Stop and 6475::rsiR mutants) also yielded a reduced ability to suppress TNF production (data not shown).
Fig 4.
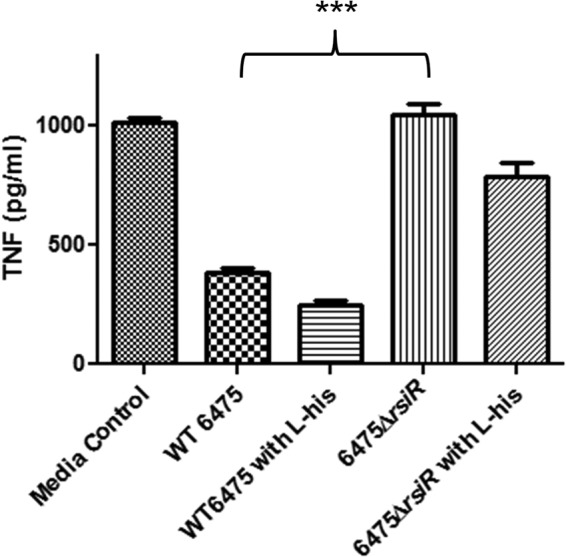
Importance of rsiR in human TNF suppression by L. reuteri. The rsiR mutant could not suppress TNF production by activated THP-1 cells. Genetic inactivation of rsiR resulted in a complete loss of TNF suppression in THP-1 cells, which was not fully complemented in the presence of supplemental histidine. ***, P = 0.0001 using an unpaired t test comparing wild-type strain 6475 and the 6475 ΔrsiR mutant.
The rsiR gene is essential for L. reuteri to suppress intestinal inflammation in a mouse model.
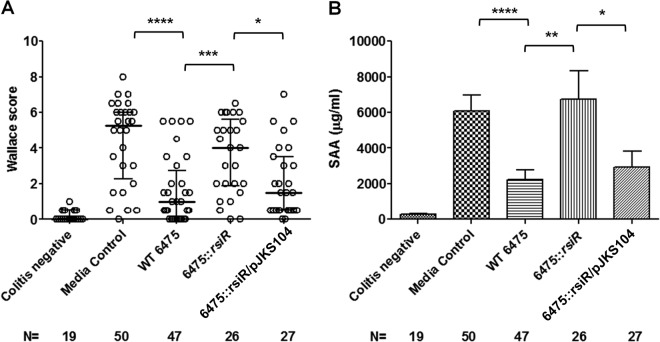
The in vivo effect of rsiR inactivation on intestinal and systemic inflammation was evaluated in a TNBS-induced colitis mouse model. Eight-week-old, female BALB/c mice received L. reuteri culture supernatants by intraperitoneal injection. Bacteria tested included wild-type L. reuteri 6475 and the 6475::rsiR mutant. TNBS was instilled intrarectally to induce acute intestinal inflammation. Colitis was evaluated by macroscopic examination of colons and quantified using the Wallace grade scoring system (34). Mice pretreated with control medium (LDMIII) followed by PBS rectal instillation (colitis-negative control) did not develop acute colitis, while mice instilled with TNBS (colitis-positive control) and lacking bacterial immunomodulatory factors developed significant colitis. Pretreatment with culture supernatants from wild-type L. reuteri 6475 significantly reduced the severity of acute colitis compared to that in colitis-positive control mice. However, protective effects were abolished in mice pretreated with culture supernatants from rsiR-deficient L. reuteri 6475. The alleviation of colitis was partially restored in mice receiving culture supernatants of L. reuteri with an intact rsiR gene on a plasmid (6475::rsiR/pJKS104) (Fig. 5A).
Fig 5.
Role of rsiR in suppression of intestinal inflammation in vivo by L. reuteri. The L. reuteri 6475::rsiR mutant conferred a diminished protective effect in a TNBS-induced colitis mouse model compared to that conferred by wild-type strain 6475. Complementation of the L. reuteri 6475::rsiR mutant (6475::rsiR/pJKS104) restored the anti-inflammatory and protective effects. (A) Macroscopic evaluation of colitis was performed using Wallace scoring criteria. Data are presented using scatter dot plots. Statistical significance among all groups was determined at a P value of <0.0001 by the nonparametric Kruskal-Wallis test. Statistical significance between two groups was assessed by the Mann-Whitney U test followed by a Bonferroni adjustment. (B) Plasma concentrations of SAA were measured by ELISA. Statistical significance among all groups was determined as described for the Wallace score for panel A. Differences between experimental groups are shown as the mean concentration ± SEM. ****, P ≤ 0.0001; ***, P < 0.001; **, P < 0.005; *, P < 0.05.
The plasma concentrations of acute-phase reactant protein SAA were measured (Fig. 5B). SAA is a biomarker of colonic mucosal inflammation, and its concentration correlates with the severity of pathology in the TNBS-induced colitis mouse model (41, 42). Elevated SAA concentrations were observed in colitis-positive control mice, while mice pretreated with L. reuteri 6475 culture supernatants yielded significantly reduced SAA concentrations (P < 0.005). The SAA concentrations in mice treated with culture supernatants from rsiR-deficient bacteria were not significantly different from the SAA concentrations in the colitis-positive control mice. Mice that received the culture supernatants from the complemented strain (6475::rsiR/pJKS104) demonstrated significant reductions in SAA concentrations compared to those in mice that received the same treatment with the rsiR mutant strain. These observations suggest that the bacterial rsiR gene may play an important role in L. reuteri-mediated immunoregulation and amelioration of systemic and intestinal inflammation in vivo.
DISCUSSION
In this study, we characterized a hypothetical protein, RsiR, and investigated its role in immunomodulation, histamine production, and regulation of histidine decarboxylase gene cluster expression. Comparative transcriptomic analysis revealed downregulation of genes involved in histamine biosynthesis in RsiR-deficient mutants compared to their expression in the wild-type strain, and the finding was confirmed by quantitative reverse transcription-PCR. We found that l-histidine supplementation did not affect hdc cluster gene expression or histamine production in mutant L. reuteri deficient in RsiR. Promoter studies of PhdcAB suggested that RsiR may regulate the expression of the hdc gene cluster at the transcriptional level. Inactivation of RsiR resulted in the relative inability to induce hdc gene expression, produce histamine, suppress TNF production by human myeloid cells, and protect animals in a TNBS colitis mouse model.
RsiR was predicted to be a transmembrane protein, and characterization of rsiR-deficient L. reuteri mutants demonstrated the regulatory role of rsiR in hdc gene cluster expression. In silico structural analysis of RsiR did not predict the presence of a DNA binding domain. We identified putative promoter regions for genes whose expression was most affected by rsiR inactivation (the 5 most downregulated genes and the 5 most upregulated genes) but failed to demonstrate significant homology among these sequences (data not shown). Transcriptional factors without a DNA binding domain have been previously characterized in many prokaryotes, for example, Spx found in Bacillus subtilis and other low-GC-content Gram-positive bacteria (43–45) and TsrA found in Vibrio cholerae (46). These proteins were global transcriptional regulators controlling expression of genes involved in the toxic stress response and virulence. Instead of binding to promoter DNA sequence, these proteins may interact with the α C-terminal domain (α-CTD) of RNA polymerases and suppress or induce gene expression (44, 47). Similarly, RsiR may indirectly regulate gene expression via interaction with other transcriptional factors. Protein-protein interaction studies, such as bacterium two-hybrid analysis or bimolecular fluorescence complementation (BiFC), would help identify the interactions between RsiR and other proteins and elucidate the mechanism of RsiR-dependent gene regulation. Results from protein function prediction indicated a role for RsiR in the metabolism and transport of folic acid and indole derivatives, which are physiologically relevant bacterial metabolites in the human gastrointestinal tract (48, 49). With the presence of transmembrane domains and a predicted role in transporting indole derivatives, it is possible that RsiR could form multimers and function as a histidine transporter. This hypothesis is unlikely to be true, since RsiR does not contain any known amino acid transporter domain with key residues that would be able to bind and transport amino acids. RsiR may instead play a role in the metabolism of folate or indole compounds that may be present in the intestinal milieu. Orally consumed nutrients, such as vitamins, amino acids, or other cofactors, may be metabolized by members of the intestinal microbiome and converted in the intestinal lumen to biologically active molecules (50), including short-chain fatty acids (SCFAs), biogenic amines (such as histamine), or other amino acid-derived metabolites, like serotonin, tryptophan, or gamma-aminobutyric acid (GABA). These small bacterial metabolites may be able to affect the physiological functions of the host, such as the immune system or the cardiovascular or central nervous system (51, 52).
The possible involvement of RsiR in amino acid metabolism and transport in L. reuteri is of particular interest, since these processes have been suggested to play a role in intestinal physiology and immunology. Amino acid metabolism is an essential functional process that plays an important role in the biochemical pathways of prokaryotic cells, such as energy harvest and acid stress survival. In a recent DNA microarray transcriptomic study of L. casei, Zhang and colleagues identified 162 genes that were upregulated during soymilk fermentation (53). Approximately 38.4% of these genes are involved in amino acid metabolism and transport, especially histidine and lysine. Metagenomic analysis of the human intestinal microbiome has revealed the presence of genetic elements involved in amino acid biosynthesis and metabolism encoded in the genomes of bacteria that comprise the gut microbiota (54). A recent study demonstrated an association between amino acid malnutrition and susceptibility to chemical colitis in mice deficient in angiotensin I-converting enzyme 2 (Ace2), which facilitates amino acid transport (33). Ace2-deficient mice had an altered intestinal microbiota compared to that of wild-type mice. Transplantation of these altered microbial communities into germfree mice resulted in transmission of the susceptibility to developing severe colitis, which suggested the role of the gut microbiome in the regulation of intestinal inflammation. A recent metagenomic study in humans revealed a depletion of genes involved in the metabolism and biosynthesis of amino acids, especially histidine and lysine, in patients suffering from inflammatory bowel disease (28). This evidence suggests that bioactive molecules produced by intestinal microbes may be able to affect the integrity of the gut barrier and the proliferation of intestinal epithelial cells and to modulate the host immune response. RsiR may have a regulatory role in L. reuteri-mediated luminal conversion of certain cofactors that are involved in immunomodulation, as suggested by our in vitro and in vivo assays.
In order to characterize bacterial pathways that were affected by genetic inactivation of RsiR, we performed a comparative transcriptomic analysis using RNA-seq. To our knowledge, this was the first attempt at implementing RNA-seq in an L. reuteri transcriptomic study. Compared with DNA microarray analysis, whole-transcriptome sequencing has a more extensive detection range with no background and allows the absolute quantification of gene expression (55). Moreover, data obtained from different RNA-seq experiments can also be easily normalized and compared (56). Our analysis revealed several metabolic and regulatory pathways with altered expression in the rsiR mutant, which suggested that rsiR may possess a global regulatory function across many physiological pathways. A global regulatory role may underlie the conservation of rsiR across the L. reuteri strains sequenced to date. From our RNA-seq analysis, we identified hdcA and hisS2 as highly downregulated genes in the rsiR mutant, along with a 2-fold downregulation of putative secondary l-histidine transporter lysP2. A homologue of lysP2 in L. lactis subsp. cremoris NZ9000 was proposed to play a role in histidine transport and was essential for growth in low concentrations of free l-histidine (40). We also validated the results of the analysis using GFOLD software (27), which gives more biologically meaningful results when no biological replicate is available. The results from the GFOLD analysis were similar to those from an analysis with Qseq software (data not shown). The association between histidine metabolism and L. reuteri-mediated immunomodulation is supported by a recent study showing histamine to be one immunomodulatory factor produced by L. reuteri 6475 (18). Purified bacterium-derived histamine inhibited TNF production at the level of transcription. The loss of TNF inhibition in the rsiR mutant is most likely a result of impaired histamine production.
Quantitative histamine ELISA data demonstrated significantly diminished histamine production by the rsiR mutant even in the presence of supplemental histidine, the major substrate to the histidine decarboxylation pathway. Increased histamine production and increased expression of the hdc gene cluster in the presence of histidine have been described in other histaminogenic Gram-positive bacteria, such as Lactobacillus hilgardii ISE 5211 (57), L. hilgardii 464, Pediococcus parvulus P270, Oenococcus oeni 4042 (16), and Streptococcus thermophilus PRI60 (12). However, the regulatory mechanisms affecting hdc gene expression in Gram-positive bacteria are currently unknown. The inability of RsiR-deficient mutants to increase production of histamine when supplemented with l-histidine suggests that RsiR may have a modulatory role on histidine production, most likely via regulation of hdc gene expression. This finding was affirmed by the RT-qPCR results, which demonstrated significant downregulation of hdcA and hdcB, but not hdcP. In the presence of supplemental l-histidine, hdc genes were not upregulated in the rsiR mutant compared to the level of expression in wild-type L. reuteri. It has previously been shown in histaminogenic lactobacilli, including L. reuteri 6475 (data not shown), that hdcA and hdcB are coregulated by a single common promoter (PhdcAB) and transcribed as a bicistronic mRNA, while hdcP is regulated by a different promoter (14, 58).
To our knowledge, our PhdcAB promoter study is the first report of a GusA promoter assay in L. reuteri. The low basal level of GusA activity, along with the lack of responsiveness to supplemental l-histidine in the rsiR mutant, suggested a regulatory role of RsiR for genes under the control of the PhdcAB promoter. However, RsiR could act as a global transcriptional activator and regulate other key metabolic pathways in L. reuteri; RNA-seq analysis along with the subsequent DAVID functional clustering analysis (28, 29) revealed downregulation of genes involved in other biological processes. Pathways highly downregulated in the RsiR mutant included cysteine and methionine metabolism, fatty acid biosynthesis, and glycerolipid metabolism (see Tables S2 and S3 in the supplemental material). Moreover, the loss of TNF suppression in human myeloid cells by the RsiR-deficient mutant strain was greater than what was seen in cells treated with culture supernatants from hdcA, hdcB, and hdcP mutants (18). These results suggest that genetic activation of RsiR may globally affect pathways involved in the L. reuteri-mediated production of other immunomodulatory factors besides histamine, ultimately resulting in a lack of TNF inhibition through multiple mechanisms. Further promoter analysis or a DNA-protein interaction study, such as by an in vivo chromatin immunoprecipitation (ChIP) assay, is needed in order to characterize the putative global regulatory role of RsiR in L. reuteri.
In addition to the hypothesis that the gene product of rsiR may function as a global transcriptional activator, it is possible that transcripts of rsiR may also function as small RNA (sRNA) regulators. Noncoding regulatory RNAs in bacteria, which can range from 50 to 500 nucleotides in size, have been extensively characterized and shown to regulate translation or the stability of target mRNAs (20, 59). Instead of increasing the degradation and inhibiting the translation of target genes, some sRNAs that contain partial complementarity (at least 6 to 8 contiguous base pairs) with target mRNAs enhance mRNA stability and prevent the formation of inhibitory secondary RNA structures, allowing more efficient translation (60). A BLAST search of rsiR against the L. reuteri 6475 genome revealed seeding sequence complementarity of 10 bp or more between rsiR and genes that were differentially expressed in the 6474rsiR-Stop mutant compared to the wild type (see Table S5 in the supplemental material). rsiR transcripts may function as global trans-encoded base-pairing sRNAs and alter the stability of mRNAs involved in different metabolic pathways. Mutations in rsiR transcripts (like those in our 6475rsiR-Stop mutant) may affect its secondary structure and regulatory function, resulting in a decreased or increased mRNA stability of its target genes, with downstream effects on various biological processes.
As discussed above, the production of bioactive microbial metabolites that are products of amino acid conversion by human intestinal microbes may play an important role in interkingdom interactions and promote the health of the host. The current study has characterized the effects of supplemental l-histidine on the production of immunomodulatory histamine and has demonstrated a possible regulatory mechanism of histidine decarboxylation in L. reuteri, which is a member of the human intestinal microbiome. Studies of RsiR and similar transcriptional/translational regulators of the human microbiome may shed light on the connections between diet, the microbiome, and innate immunity. These potential connections are highlighted by a recent study in mice fed a Westernized fast-food diet, where the authors demonstrated a significant shift in the proinflammatory immune response in the host, which included a reduction in focal inflammation in abdominal fat and weight gain in animals treated with L. reuteri 6475, the same strain studied here (61). According to the authors, these effects were interleukin-10 dependent and may have been a result of Foxp3+ regulatory T cell activation by the probiotic bacteria. By understanding how dietary components such as amino acids (e.g., l-histidine) regulate gene expression through specific regulators (e.g., RsiR), the gut microbiome and its effects on immunity could be modulated or manipulated by a combination of nutritional and probiotic interventions in the future.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported in part by research support from the National Institutes of Health (NIH; R01 AT004326, R01 DK065075, UH3 DK083990). We also acknowledge the support of the NIH (NIDDK)-funded (DK56338) Texas Medical Center Digestive Diseases Center.
We thank Eamonn Connolly (BioGaia AB, Stockholm, Sweden) for providing the L. reuteri strains, Lisa White and Rebecca Thornton for assistance with RNA-seq, and Ashley Grimm French and Yue (Moon) Shang for assistance with RT-qPCR.
Footnotes
Published ahead of print 11 October 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.00261-13.
REFERENCES
- 1.Food and Agriculture Organization of the United Nations and World Health Organization 2001. Health and nutritional properties of probiotics in food including powder milk with live lactic acid bacteria. Report of a Joint FAO/WHO Expert Consultation on Evaluation of Health and Nutritional Properties of Probiotics in Food Including Powder Milk with Live Lactic Acid Bacteria. Food and Agriculture Organization of the United Nations and World Health Organization, Rome, Italy [Google Scholar]
- 2.Food and Agriculture Organization of the United Nations and World Health Organization 2002. Guidelines for the evaluation of probiotics in food. Report of a Joint FAO/WHO Working Group on Drafting Guidelines for the Evaluation of Probiotics in Food. Food and Agriculture Organization of the United Nations and World Health Organization, Rome, Italy [Google Scholar]
- 3.Metchnikoff E, Mitchell PC. 1907. The prolongation of life: optimistic studies. G. P. Putnam's Sons, London, United Kingdom [Google Scholar]
- 4.Watanabe T, Nishio H, Tanigawa T, Yamagami H, Okazaki H, Watanabe K, Tominaga K, Fujiwara Y, Oshitani N, Asahara T, Nomoto K, Higuchi K, Takeuchi K, Arakawa T. 2009. Probiotic Lactobacillus casei strain Shirota prevents indomethacin-induced small intestinal injury: involvement of lactic acid. Am. J. Physiol. Gastrointest. Liver Physiol. 297:G506–G513 [DOI] [PubMed] [Google Scholar]
- 5.Menard S, Candalh C, Bambou JC, Terpend K, Cerf-Bensussan N, Heyman M. 2004. Lactic acid bacteria secrete metabolites retaining anti-inflammatory properties after intestinal transport. Gut 53:821–828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.van Zanten GC, Knudsen A, Roytio H, Forssten S, Lawther M, Blennow A, Lahtinen SJ, Jakobsen M, Svensson B, Jespersen L. 2012. The effect of selected synbiotics on microbial composition and short-chain fatty acid production in a model system of the human colon. PLoS One 7:e47212. 10.1371/journal.pone.0047212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Maslowski KM, Vieira AT, Ng A, Kranich J, Sierro F, Yu D, Schilter HC, Rolph MS, Mackay F, Artis D, Xavier RJ, Teixeira MM, Mackay CR. 2009. Regulation of inflammatory responses by gut microbiota and chemoattractant receptor GPR43. Nature 461:1282–1286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Matsumoto M, Kurihara S, Kibe R, Ashida H, Benno Y. 2011. Longevity in mice is promoted by probiotic-induced suppression of colonic senescence dependent on upregulation of gut bacterial polyamine production. PLoS One 6:e23652. 10.1371/journal.pone.0023652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tabanelli G, Torriani S, Rossi F, Rizzotti L, Gardini F. 2012. Effect of chemico-physical parameters on the histidine decarboxylase (HdcA) enzymatic activity in Streptococcus thermophilus PRI60. J. Food Sci. 77:M231–M237 [DOI] [PubMed] [Google Scholar]
- 10.Molenaar D, Bosscher JS, ten Brink B, Driessen AJ, Konings WN. 1993. Generation of a proton motive force by histidine decarboxylation and electrogenic histidine/histamine antiport in Lactobacillus buchneri. J. Bacteriol. 175:2864–2870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rodwell AW. 1953. The histidine decarboxylase of a species of Lactobacillus; apparent dispensability of pyridoxal phosphate as coenzyme. J. Gen. Microbiol. 8:233–237 [DOI] [PubMed] [Google Scholar]
- 12.Rossi F, Gardini F, Rizzotti L, La Gioia F, Tabanelli G, Torriani S. 2011. Quantitative analysis of histidine decarboxylase gene (hdcA) transcription and histamine production by Streptococcus thermophilus PRI60 under conditions relevant to cheese making. Appl. Environ. Microbiol. 77:2817–2822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de las Rivas B, Rodriguez H, Carrascosa AV, Munoz R. 2008. Molecular cloning and functional characterization of a histidine decarboxylase from Staphylococcus capitis. J. Appl. Microbiol. 104:194–203 [DOI] [PubMed] [Google Scholar]
- 14.Copeland WC, Domena JD, Robertus JD. 1989. The molecular cloning, sequence and expression of the hdcB gene from Lactobacillus 30A. Gene 85:259–265 [DOI] [PubMed] [Google Scholar]
- 15.Pessione E, Mazzoli R, Giuffrida MG, Lamberti C, Garcia-Moruno E, Barello C, Conti A, Giunta C. 2005. A proteomic approach to studying biogenic amine producing lactic acid bacteria. Proteomics 5:687–698 [DOI] [PubMed] [Google Scholar]
- 16.Landete JM, Pardo I, Ferrer S. 2006. Histamine, histidine, and growth-phase mediated regulation of the histidine decarboxylase gene in lactic acid bacteria isolated from wine. FEMS Microbiol. Lett. 260:84–90 [DOI] [PubMed] [Google Scholar]
- 17.Landete JM, Pardo I, Ferrer S. 2008. Regulation of hdc expression and HDC activity by enological factors in lactic acid bacteria. J. Appl. Microbiol. 105:1544–1551 [DOI] [PubMed] [Google Scholar]
- 18.Thomas CM, Hong T, van Pijkeren JP, Hemarajata P, Trinh DV, Hu W, Britton RA, Kalkum M, Versalovic J. 2012. Histamine derived from probiotic Lactobacillus reuteri suppresses TNF via modulation of PKA and ERK signaling. PLoS One 7:e31951. 10.1371/journal.pone.0031951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rainer J, Sanchez-Cabo F, Stocker G, Sturn A, Trajanoski Z. 2006. CARMAweb: comprehensive R- and Bioconductor-based web service for microarray data analysis. Nucleic Acids Res. 34:W498–W503. 10.1093/nar/gkl038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Waters LS, Storz G. 2009. Regulatory RNAs in bacteria. Cell 136:615–628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Walter J, Chagnaud P, Tannock GW, Loach DM, Dal Bello F, Jenkinson HF, Hammes WP, Hertel C. 2005. A high-molecular-mass surface protein (Lsp) and methionine sulfoxide reductase B (MsrB) contribute to the ecological performance of Lactobacillus reuteri in the murine gut. Appl. Environ. Microbiol. 71:979–986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maguin E, Duwat P, Hege T, Ehrlich D, Gruss A. 1992. New thermosensitive plasmid for gram-positive bacteria. J. Bacteriol. 174:5633–5638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van Pijkeren JP, Britton RA. 2012. High efficiency recombineering in lactic acid bacteria. Nucleic Acids Res. 40:e76. 10.1093/nar/gks147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Van Pijkeren JP, Neoh KM, Sirias D, Findley AS, Britton RA. 2012. Exploring optimization parameters to increase ssDNA recombineering in Lactococcus lactis and Lactobacillus reuteri. Bioengineered 3:209–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Santos F, Spinler JK, Saulnier DM, Molenaar D, Teusink B, de Vos WM, Versalovic J, Hugenholtz J. 2011. Functional identification in Lactobacillus reuteri of a PocR-like transcription factor regulating glycerol utilization and vitamin B12 synthesis. Microb. Cell Fact. 10:55. 10.1186/1475-2859-10-55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Langmead B, Trapnell C, Pop M, Salzberg SL. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10:R25. 10.1186/gb-2009-10-3-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Feng J, Meyer CA, Wang Q, Liu JS, Shirley Liu X, Zhang Y. 2012. GFOLD: a generalized fold change for ranking differentially expressed genes from RNA-seq data. Bioinformatics 28:2782–2788 [DOI] [PubMed] [Google Scholar]
- 28.Morgan XC, Tickle TL, Sokol H, Gevers D, Devaney KL, Ward DV, Reyes JA, Shah SA, LeLeiko N, Snapper SB, Bousvaros A, Korzenik J, Sands BE, Xavier RJ, Huttenhower C. 2012. Dysfunction of the intestinal microbiome in inflammatory bowel disease and treatment. Genome Biol. 13:R79. 10.1186/gb-2012-13-9-r79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gossringer M, Hartmann RK. 2012. 3′-UTRs as a source of regulatory RNAs in bacteria. EMBO J. 31:3958–3960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Romby P, Charpentier E. 2010. An overview of RNAs with regulatory functions in gram-positive bacteria. Cell Mol. Life Sci. 67:217–237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Callanan MJ, Russell WM, Klaenhammer TR. 2007. Modification of Lactobacillus beta-glucuronidase activity by random mutagenesis. Gene 389:122–127 [DOI] [PubMed] [Google Scholar]
- 32.Axelsson L, Lindstad G, Naterstad K. 2003. Development of an inducible gene expression system for Lactobacillus sakei. Lett. Appl. Microbiol. 37:115–120 [DOI] [PubMed] [Google Scholar]
- 33.Hashimoto T, Perlot T, Rehman A, Trichereau J, Ishiguro H, Paolino M, Sigl V, Hanada T, Hanada R, Lipinski S, Wild B, Camargo SM, Singer D, Richter A, Kuba K, Fukamizu A, Schreiber S, Clevers H, Verrey F, Rosenstiel P, Penninger JM. 2012. ACE2 links amino acid malnutrition to microbial ecology and intestinal inflammation. Nature 487:477–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wallace JL, MacNaughton WK, Morris GP, Beck PL. 1989. Inhibition of leukotriene synthesis markedly accelerates healing in a rat model of inflammatory bowel disease. Gastroenterology 96:29–36 [DOI] [PubMed] [Google Scholar]
- 35.Saulnier DM, Santos F, Roos S, Mistretta TA, Spinler JK, Molenaar D, Teusink B, Versalovic J. 2011. Exploring metabolic pathway reconstruction and genome-wide expression profiling in Lactobacillus reuteri to define functional probiotic features. PLoS One 6:e18783. 10.1371/journal.pone.0018783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lin YP, Thibodeaux CH, Pena JA, Ferry GD, Versalovic J. 2008. Probiotic Lactobacillus reuteri suppress proinflammatory cytokines via c-Jun. Inflamm. Bowel Dis. 14:1068–1083 [DOI] [PubMed] [Google Scholar]
- 37.Zdobnov EM, Apweiler R. 2001. InterProScan—an integration platform for the signature-recognition methods in InterPro. Bioinformatics 17:847–848 [DOI] [PubMed] [Google Scholar]
- 38.Hawkins T, Luban S, Kihara D. 2006. Enhanced automated function prediction using distantly related sequences and contextual association by PFP. Protein Sci. 15:1550–1556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kelley LA, Sternberg MJ. 2009. Protein structure prediction on the Web: a case study using the Phyre server. Nat. Protoc. 4:363–371 [DOI] [PubMed] [Google Scholar]
- 40.Trip H, Mulder NL, Lolkema JS. 2013. Cloning, expression, and functional characterization of secondary amino acid transporters of Lactococcus lactis. J. Bacteriol. 195:340–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.de Villiers WJ, Varilek GW, de Beer FC, Guo JT, Kindy MS. 2000. Increased serum amyloid A levels reflect colitis severity and precede amyloid formation in IL-2 knockout mice. Cytokine 12:1337–1347 [DOI] [PubMed] [Google Scholar]
- 42.Uhlar CM, Whitehead AS. 1999. Serum amyloid A, the major vertebrate acute-phase reactant. Eur. J. Biochem. 265:501–523 [DOI] [PubMed] [Google Scholar]
- 43.Nakano S, Nakano MM, Zhang Y, Leelakriangsak M, Zuber P. 2003. A regulatory protein that interferes with activator-stimulated transcription in bacteria. Proc. Natl. Acad. Sci. U. S. A. 100:4233–4238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kajfasz JK, Mendoza JE, Gaca AO, Miller JH, Koselny KA, Giambiagi-Demarval M, Wellington M, Abranches J, Lemos JA. 2012. The Spx regulator modulates stress responses and virulence in Enterococcus faecalis. Infect. Immun. 80:2265–2275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zuber P. 2004. Spx-RNA polymerase interaction and global transcriptional control during oxidative stress. J. Bacteriol. 186:1911–1918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zheng J, Shin OS, Cameron DE, Mekalanos JJ. 2010. Quorum sensing and a global regulator TsrA control expression of type VI secretion and virulence in Vibrio cholerae. Proc. Natl. Acad. Sci. U. S. A. 107:21128–21133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nakano S, Kuster-Schock E, Grossman AD, Zuber P. 2003. Spx-dependent global transcriptional control is induced by thiol-specific oxidative stress in Bacillus subtilis. Proc. Natl. Acad. Sci. U. S. A. 100:13603–13608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rossi M, Amaretti A, Raimondi S. 2011. Folate production by probiotic bacteria. Nutrients 3:118–134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bansal T, Alaniz RC, Wood TK, Jayaraman A. 2010. The bacterial signal indole increases epithelial-cell tight-junction resistance and attenuates indicators of inflammation. Proc. Natl. Acad. Sci. U. S. A. 107:228–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hemarajata P, Versalovic J. 2013. Effects of probiotics on gut microbiota: mechanisms of intestinal immunomodulation and neuromodulation. Ther. Adv. Gastroenterol. 6:39–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Marques TM, Wall R, Ross RP, Fitzgerald GF, Ryan CA, Stanton C. 2010. Programming infant gut microbiota: influence of dietary and environmental factors. Curr. Opin. Biotechnol. 21:149–156 [DOI] [PubMed] [Google Scholar]
- 52.Desbonnet L, Garrett L, Clarke G, Bienenstock J, Dinan TG. 2008. The probiotic Bifidobacteria infantis: an assessment of potential antidepressant properties in the rat. J. Psychiatr. Res. 43:164–174 [DOI] [PubMed] [Google Scholar]
- 53.Wang JC, Zhang WY, Zhong Z, Wei AB, Bao QH, Zhang Y, Sun TS, Postnikoff A, Meng H, Zhang HP. Transcriptome analysis of probiotic Lactobacillus casei Zhang during fermentation in soymilk. J. Ind. Microbiol. Biotechnol. 39:191–206 [DOI] [PubMed] [Google Scholar]
- 54.Human Microbiome Project Consortium 2012. Structure, function and diversity of the healthy human microbiome. Nature 486:207–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mutz KO, Heilkenbrinker A, Lonne M, Walter JG, Stahl F. 2013. Transcriptome analysis using next-generation sequencing. Curr. Opin. Biotechnol. 24:22–30 [DOI] [PubMed] [Google Scholar]
- 56.Febrer M, McLay K, Caccamo M, Twomey KB, Ryan RP. 2011. Advances in bacterial transcriptome and transposon insertion-site profiling using second-generation sequencing. Trends Biotechnol. 29:586–594 [DOI] [PubMed] [Google Scholar]
- 57.Mazzoli R, Lamberti C, Coisson JD, Purrotti M, Arlorio M, Giuffrida MG, Giunta C, Pessione E. 2009. Influence of ethanol, malate and arginine on histamine production of Lactobacillus hilgardii isolated from an Italian red wine. Amino Acids 36:81–89 [DOI] [PubMed] [Google Scholar]
- 58.Martin MC, Fernandez M, Linares DM, Alvarez MA. 2005. Sequencing, characterization and transcriptional analysis of the histidine decarboxylase operon of Lactobacillus buchneri. Microbiology 151:1219–1228 [DOI] [PubMed] [Google Scholar]
- 59.Gottesman S, Storz G. 2011. Bacterial small RNA regulators: versatile roles and rapidly evolving variations. Cold Spring Harb. Perspect. Biol. 3:pii=a003798. 10.1101/cshperspect.a003798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gottesman S. 2005. Micros for microbes: non-coding regulatory RNAs in bacteria. Trends Genet. 21:399–404 [DOI] [PubMed] [Google Scholar]
- 61.Poutahidis T, Kleinewietfeld M, Smillie C, Levkovich T, Perrotta A, Bhela S, Varian BJ, Ibrahim YM, Lakritz JR, Kearney SM, Chatzigiagkos A, Hafler DA, Alm EJ, Erdman SE. 2013. Microbial reprogramming inhibits Western diet-associated obesity. PLoS One 8:e68596. 10.1371/journal.pone.0068596 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.