Abstract
Streptococcus mutans, a dental pathogen, secretes different kinds of lantibiotic and nonlantibiotic bacteriocins. For self-protection, a bacteriocin producer strain must possess one or more cognate immunity mechanisms. We report here the identification of one such immunity complex in S. mutans strain GS-5 that confers protection against Smb, a two-component lantibiotic. The immunity complex that we identified is an ABC transporter composed of two proteins: SmbF (the ATPase component) and SmbT (the permease component). Both of the protein-encoding genes are located within the smb locus. We show that GS-5 becomes sensitized to Smb upon deletion of smbT, which makes the ABC transporter nonfunctional. To establish the role SmbFT in providing immunity, we heterologously expressed this ABC transporter complex in four different sensitive streptococcal species and demonstrated that it can confer resistance against Smb. To explore the specificity of SmbFT in conferring resistance, we tested mutacin IV (a nonlantibiotic), nisin (a single peptide lantibiotics), and three peptide antibiotics (bacitracin, polymyxin B, and vancomycin). We found that SmbFT does not recognize these structurally different peptides. We then tested whether SmbFT can confer protection against haloduracin, another two-component lantibiotic that is structurally similar to Smb; SmbFT indeed conferred protection against haloduracin. SmbFT can also confer protection against an uncharacterized but structurally similar lantibiotic produced by Streptococcus gallolyticus. Our data suggest that SmbFT truly displays immunity function and confer protection against Smb and structurally similar lantibiotics.
INTRODUCTION
Oral biofilm is complex and diverse in nature. As many as 700 different bacterial species colonizes in our mouth; unfortunately, the identity of many of them is still unknown (1, 2). The complexity of biofilm greatly depends on the interspecies interaction during both early and maturing phases of biofilm formation. The participation of these organisms is not restricted to dental disease only, since they can cause bacteremia and in some cases infective endocarditis (3–5). Endocarditis is generally caused by viridans group of streptococci. Some of these viridians streptococci are commensal in oral cavity, and the others are involved in disease formation (5, 6). Some of the commensal streptococci have a protective role against oral disease development (7). The exact in vivo mechanisms for this interspecies interference are currently not well understood; however, several studies strongly suggest an important role for hydrogen peroxide and bacteriocin in this process (8–10).
In general, bacteriocins are ribosomally synthesized small peptides with bactericidal or bacteriostatic activity on other species. Bacteriocins are broadly categorized into two groups: nonlantibiotic and lantibiotic. The nonlantibiotic group contains peptides that do not require any modifications for their biological activity (11). In contrast, the lantibiotic groups contain peptides that require posttranslational modification for their antimicrobial activity (12, 13). In lantibiotics, most of the serine and threonine residues are dehydrated to dehydroalanine (Dha) and dehydrobutyrine (Dhb), respectively, by modifying enzymes encoded within the lantibiotic synthesis operon. With neighboring cysteine residues, Dha and Dhb can form thioether-linked lanthionine and 3-methyl lanthionine bridges, respectively. Sometimes Dha, Dhb, and other modified residues can be present as unlinked residues (for reviews, see references 13, 14, 15, 16, and 17). Based on the nature of the enzymes responsible for the modifications, lantibiotics are divided into three classes (13). Class I lantibiotics (such as nisin and subtilin) are modified by two enzymes generally referred to as LanBC system. Class II lantibiotics (such as lacticin 481 and mersacidin) are modified by a single enzyme often referred to as LanM-type enzyme. Class II lantibiotics also include two-component lantibiotics (such as lacticin 3147 and haloduracin), and the antimicrobial activity requires synergistic interaction of both peptides (18, 19). Lantibiotics that belong to class III are lanthionine peptides that have no antimicrobial activity (20, 21). The advantage of bearing modified residues, as well as cyclization, is to make lantibiotics more stable against heat, pH, and protease degradation compared to nonlantibiotics (22). This stable nature of lantibiotics makes them a major candidate for commercial application; for example, nisin is extensively used as a preservative in the food industry (23).
Lantibiotics are generally produced by Gram-positive bacteria and usually have a variable spectrum of inhibition (15, 22–25). The producer organisms usually encodes specific immunity proteins that protect themselves from the deleterious effect of their own lantibiotics, and the immunity protein encoding genes are often present within the same lantibiotic biosynthesis operon. Currently, two types of immunity protein have been identified. The first type includes dedicated ABC transporters that presumably pumps lantibiotics out of the membrane and thus prevent accumulation to inhibitory levels (for reviews, see references 14 and 26). The second type includes small proteins that are weakly associated with the membrane and often sequester specific lantibiotics at the cell surface before they could cause cell damage (27–29).
Streptococcus mutans, an oral streptococci and a major etiological agent of human dental caries, often produces several kinds of bacteriocins, collectively known as mutacins. A majority of the mutacins characterized to date belong to lantibiotics, such as the mutacins I, II, III, K8, B-Ny266, Smb, and 1140 (30–37). Based on the primary amino acid sequences, mutacins belonging to lantibiotics are further subdivided into two subclasses: AI and AII (38–40). Subclass AI contains the most well-characterized peptides such as mutacins I and III (mutacin 1140), which are similar to nisin and subtilin. Detailed structural information of these mutacins is currently lacking. Although the lantibiotic mutacins are widely present in S. mutans (37, 41), the first sequenced reference strain UA159, surprisingly, does not encode any lantibiotic but only encodes nonlantibiotics (42). In contrast, another recently sequenced S. mutans strain GS-5 encodes both Smb and nonlantibiotics (35, 43). The majority of the mutacins encoding genes are acquired by the S. mutans by horizontal gene transfer mechanism. For example, the strains that produce mutacin K8 contains the K8-encoding muk locus that is inserted in between SMU.1811 and SMU.1812 genes of the corresponding UA159 genome (37). Similarly, the smb locus, which contains all of the genes necessary for Smb biosynthesis, is integrated in between SMU.1942 and syl locus. Smb is the only two-component lantibiotic identified thus far in S. mutans and appears to be encoded only by a few strains (37).
The smb locus contains seven open reading frames in the following order: smbM1, smbF, smbT, smbM2, smbG, smbA, and smbB. Putative transposase encoding genes are also present near the smb locus (35). Two genes, smbA and smbB, are the structural genes for the Smb prepeptides and two putative modification enzymes encoded by smbM1 and smbM2 are also present in the smb locus. A recent report indicated that SmbG appears to play a role in Smb immunity and sensitivity to antimicrobial agents (44). Two other genes, smbF and smbT, show homology with the ATP binding cassette transporter and were previously predicted to be involved in processing and secretion of Smb prepeptides (35). We noticed that this SmbFT ABC transporter complex does not have a signal peptidase domain and hence might not be involved in the processing and secretion of the Smb prepeptide. We predicted that SmbFT might provide immunity to Smb-producing S. mutans strains. In the present study, we provide evidence that SmbFT indeed confers resistance to Smb in the producer strain and show that deletion of smbT gene in GS-5 leads to sensitivity toward Smb. We also demonstrate that SmbFT can confer resistance to sensitive streptococci belonging to nonmutans group. Furthermore, we show that SmbFT has a narrow specificity, since it only recognizes structurally similar two-peptide lantibiotics.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Escherichia coli was routinely grown in Luria-Bertani (LB) medium and, when necessary, supplemented with 100 μg of kanamycin/ml. S. mutans and other Gram-positive bacteria were normally grown at 37°C in Todd-Hewitt medium (BBL, BD) supplemented with 0.2% yeast extract (THY) under microaerophilic conditions. When necessary, 5 μg of erythromycin/ml or 500 μg of kanamycin/ml was included in the THY medium. All of the streptococcal strains, except for S. pyogenes, were transformed by means of natural transformation according to a standard protocol with the addition of competence stimulating peptides (45). For S. pyogenes, electrotransformation was carried out as previously described (46).
Inactivation of smbAB and smbT genes in GS-5.
A previously described fusion PCR method was used for the construction of ΔsmbAB and ΔsmbT mutant strains (47). In short, 500-bp upstream (up) and downstream (dn) flanking regions of the respective genes were separately PCR amplified using GS-5 genomic DNA as a template (see Table 1 for the primers). A third fragment (middle) containing an erythromycin resistance cassette was amplified using pIBM01 (I. Biswas, unpublished data), a plasmid containing an ermB gene, as a template. This plasmid was created by amplifying the ermB gene from pGhost9 plasmid (48) and inserted into the pGEM-T Easy vector by TA cloning. The 5′ end of this middle PCR fragment has a 20-bp complementary sequence to the 3′ end of the up fragments, whereas the 3′ end of this middle fragment has a 20-bp complementary region to the dn fragment. To generate fusion PCR products, equal molar ratios of the three fragments were mixed and used as a template for next round of PCR amplification with the flanking primer pairs of the respective genes. The final PCR products were verified by gel electrophoresis for correct sizes and transformed into S. mutans GS-5 using standard protocol. Transformants were selected on THY agar plate containing erythromycin.
Table 1.
Oligonucleotides used in this study
| Oligonucleotide | Sequence (5′-3′) | Purposea |
|---|---|---|
| fsnsmbAupF | GTTGGCGAAAGTGGTTCTGGTAAG | Inactivation of smbAB (up) |
| fsnsmbAupR | GCCGCCATGGCGGCCGGGAGTAATAAATTACTTTTCATTTTA | Inactivation of smbAB (up) |
| fsnsmbBdnF | CGCGGCCGCCTGCAGGTCGACCGAATGCATGAGAAATTGTAG | Inactivation of smbAB (dn) |
| fsnsmbAdnR | GACCGCTTTCATATTGTTCAGCAC | Inactivation of smbAB (dn) |
| locussmbAF | GGAAGGAATATAGGGTGAAAAG | smbAB locus amplification |
| locussmbdnR | CGTATTACTTACTACAATTCTCATGC | smbAB locus amplification |
| NcoI-Kan-D7-F | CTCCCGGCCGCCATGGCGGCCGC | Antibiotic cassette (middle) |
| PstI-Kan-D7-R | GGTCGACCTGCAGGCGGCCGCG | Antibiotic cassette (middle) |
| fsnSmbTupF | CCAGCCACATCTTACAAAATTTGGAGC | Deletion of smbT (up) |
| fsncrtSmbTupR | GCGGCCGCCATGGCGGCCGGGAGGGTCCTTTTAAATTCTCAATAG | Deletion of smbT (up) |
| fsnSmbTdnF | CGCGGCCGCCTGCAGGTCGACGCTGATTTGACTAAATATATTCC | Deletion of smbT (dn) |
| fsnSmbTdnR | CCTTTGAATATAATTACAAATAACAAC | Deletion of smbT (dn) |
| SMBFT-XHO-R | GCCCTCGAGCCACTATGCATAACCCCATTTACGATAAATC | Complementation of smbFT |
| SMBFT-BGLII-RBSGTF-F | CGCAGATCTTGGAGGTTCCTAATGAGAATTTTAGACATTCAAAATTTAAA | Complementation of smbFT |
up, upstream; dn, downstream.
Construction of plasmids for complementation.
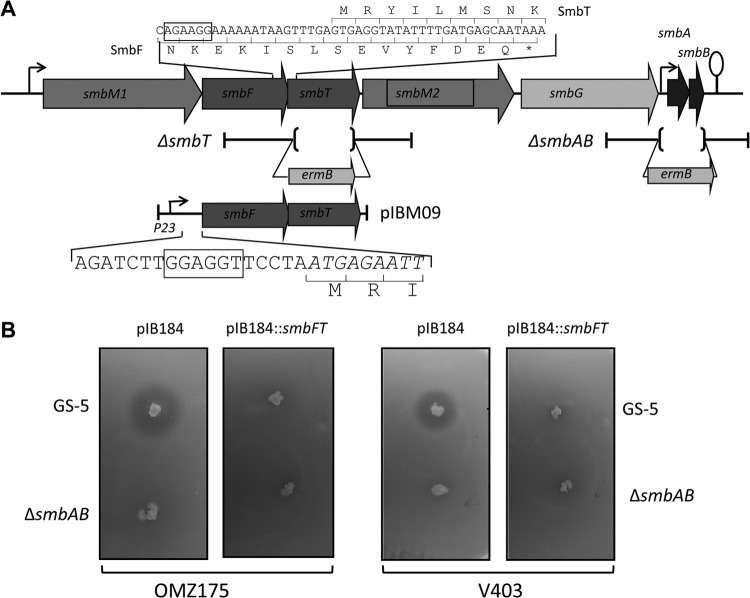
Plasmid pIB184-Km was used as vector for cloning (47). This plasmid contains P23 promoter from lactococcal phage (pOri23). To clone the smbFT genes, a fragment containing the coding regions was amplified from GS-5 genomic DNA. The upstream primer was designed in such a way that it also includes the ribosome-binding site (see Fig. 2). The amplified fragment was digested with BglII and XhoI and cloned into BamHI-XhoI-digested pIB184-Km to generate pIBM09. This plasmid and the vector were introduced into various streptococci.
Fig 2.
Smb and its immunity protein. (A) Genomic organization of smb locus in GS-5. The regions used for fusion PCR for deletion constructions are shown at the bottom. Brackets indicate the regions that were deleted from the genome, and an erythromycin resistance gene (ermB) was inserted. A plasmid used for the heterologous expression of smbFT (pIBM09) is shown. This plasmid contains a ribosome-binding site from the gftB gene and a P23 promoter used for smbFT expression. Relevant sequences near the smbF start codon and the overlapping region between smbF and smbT are shown. Bent arrows and lollipops indicate promoters and transcription termination sites. Block arrows indicate gene orientation. (B). Immunity activity of SmbFT in S. mutans strains. GS-5 and an ΔsmbAB strain were stabbed into THY agar, followed by incubation overnight at 37°C under microaerophilic conditions. The plates were overlaid with soft agar containing the indicator strains. The ZOIs of the indicator strains were evaluated after overnight incubation. The indicator strains are OMZ175 (left panel) and V403 (right panel) containing the vector (pIB184) or the vector with SmbFT. These plates are representative of three independent assays.
Bacteriocin assay (zone of inhibition).
GS-5 and its mutant derivatives were stabbed on THY-agar plates and incubated overnight under microaerophilic condition at 37°C (49). The following day, the plates were overlaid with freshly grown indicator strain cultures by mixing with soft agar. When the indicator strains contain plasmids, kanamycin was also included in the soft agar. The overlaid plates were incubated again overnight under the same condition as described above. The diameter of the clearing zone was measured afterward. Assays were repeated at least twice with four replicates.
Antibiotic sensitivity assay.
Disk diffusion assays were performed for vancomycin, bacitracin, and nisin against different strains, including wild-type (WT) and ΔsmbT strains. Briefly, disks (BRL) were placed on THY agar plates and overlaid with soft agar containing overnight grown S. mutans or other strains of choice. The plates were incubated at 37°C under microaerophilic condition. Zones of inhibition were measured after 24 h.
RESULTS
Smb-sensitive strains do not encode smb locus.
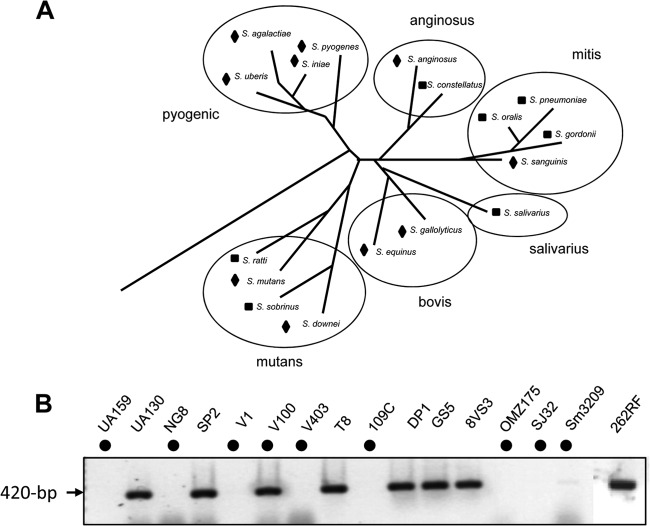
Smb is the only lantibiotic that is secreted by S. mutans GS-5 (43), and a previous study suggested that Smb is active against many oral streptococci (50). Since most of the lantibiotics, including nisin, have wider spectra than the nonlantibiotics (11, 22, 39), we wanted to determine whether Smb could inhibit various streptococci belonging to different phylogenetic groups (51). We first constructed an smbAB deletion mutant strain (ΔsmbAB) to include in our inhibition assays to make sure that the activity that we observed was due to lantibiotic Smb and not to other bacteriocins. We then used GS-5 and ΔsmbAB strains as testers against 18 streptococci and 16 S. mutans isolates (Fig. 1A). We observed that all of the streptococci strains, except some S. mutans isolates (see below), were inhibited by Smb. We divided the activity spectra in to two classes, a zone of inhibition (ZOI) smaller than 10 mm and larger than 10 mm (Fig. 1A). Two groups of streptococci, the pyogenic and the bovis groups, consistently produced larger ZOI than the other groups, suggesting that the streptococci belonging to these groups are more susceptible to Smb than the other groups. S. mutans strains generally do not inhibit the growth of other S. mutans strains. However, we found that 9 of 16 S. mutans also produced large ZOI (Table 2). Furthermore, two S. ratti strains also produced some ZOI, while the FA strain produced small (∼8 mm) but clear ZOI, and the BHT strain produced diffused ZOI (∼10 mm). Among the mitis group, most of the isolates produced smaller ZOI except S. sanguinis SK36, which produced a large ZOI (∼20 mm).
Fig 1.
Smb activity spectrum and distribution of smb locus in various S. mutans strains. (A) Member species of different phylogenetic group of streptococci were tested against mutacin Smb produced by GS-5. GS-5 and a ΔsmbAB mutant strain were stabbed onto THY-agar plate and incubated overnight in microaerophilic condition at 37°C. The next day, an overnight-grown culture of indicator strain was mixed with soft agar and overlaid onto the plate. The overlaid plates were incubated again overnight under the same condition. The following day, the diameter of the zone of inhibition (ZOI) was measured. The observation is based on two separate experiments with four replicates. Diamonds and squares indicate ZOIs of >10 mm or <10 mm in diameter, respectively. (B) Presence of the smb locus in 16 different S. mutans strains. Analysis of PCR products of smbAB genes by agarose gel electrophoresis. Solid circles indicate the strains that are susceptible to Smb inhibition (see Table S1 in the supplemental material). The observation is based on at least three independent experiments.
Table 2.
Susceptibility of various S. mutans strains to Smb
| S. mutans straina | Mean zone of inhibition diam (mm) ± SD |
|
|---|---|---|
| GS-5 strain | ΔsmbAB mutant | |
| UA159* | 11.0 ± 1.0 | 2.0 ± 2.0 |
| NG-8* | 11.0 ± 1.0 | 0.0 ± 0.0 |
| GS-5 | 0.0 ± 0.0 | 0.0 ± 0.0 |
| SP-2 | 0.0 ± 0.0 | 0.0 ± 0.0 |
| Sm3209 | 9.5 ± 1.5 | 2.5 ± 1.5 |
| T8 | 0.0 ± 0.0 | 0.0 ± 0.0 |
| SJ32 | 13.5 ± 1.5 | 0.0 ± 0.0 |
| 8VS3 | 4.1 ± 1.0 | 0.0 ± 0.0 |
| 109C* | 16.5 ± 0.5 | 0.0 ± 0.0 |
| OMZ175* | 15.5 ± 0.5 | 0.0 ± 0.0 |
| V100 | 11.5 ± 1.5 | 0.0 ± 0.0 |
| 262RF | 0.0 ± 0.0 | 0.0 ± 0.0 |
| V403* | 18.0 ± 2.0 | 0.0 ± 0.0 |
| UA130 | 0.0 ± 2.0 | 0.0 ± 0.0 |
| DP1 | 0.0 ± 1.0 | 0.0 ± 0.0 |
| V1 | 10.0 ± 1.0 | 0.0 ± 0.0 |
*, strains used for heterologous expression of SmbFT.
It was surprising to observe that many of the S. mutans isolates were insensitive to Smb. Since the lantibiotics have the potential to attack the producer strains, the producer strains must contain some self-protection mechanisms to protect against their own lantibiotics. Therefore, it is possible that the S. mutans strains that were insensitive may actually encode the smb locus. To examine the presence of smb locus, we PCR amplified the smbAB structural genes from all of the 16 S. mutans isolates. Eleven of these isolates belong to serotype c, including common laboratory strains (UA159, NG8, and GS5), and one each from serotype e (V100) and serotype f (OMZ175); the rest were taken from unknown serotypes. We found the presence of an expected 420-bp size band in eight of the isolates. As expected, these strains were also immune to Smb, indicating the presence of the same immune mechanism(s) in these strains as in the producer strain. The only exception was V100 strain, which encodes the smb locus but was sensitive to Smb-mediated inhibition. The eight strains that did not produce a PCR product all produced clear ZOIs against Smb. These data support the notion that strains that encode the smb locus are all immune to Smb-mediated killing, except for strain V100. V100 is unique among the S. mutans strains since does not develop natural competence (data not shown). We speculate that it does not express the smb locus.
SmbFT, a putative ABC transporter complex, provides immunity against Smb.
Since the Smb-encoding operon also encodes smbFT that shares high homology to a putative ABC transporter complex, and since ABC transporters often function as immunity proteins, we wanted to verify whether SmbFT can protect S. mutans from Smb-mediated killing. SmbFT transporter is composed of two polypeptides. SmbT encodes permease component and is 238 residues long with six transmembrane domains. SmbF, on the other hand, encodes a 274 residues long protein with ATPase signature sequences (52). The promoter for smb locus lies ∼3.0 kb upstream of the smbFT genes. Sequence analysis suggests the presence of a weak ribosome-binding site located five nucleotides upstream of smbF start codon and an overlap of nine codons between the smbF and smbT coding sequences (Fig. 2A). Because the promoter lies far from the smbF and since smbFT are linked together, we decided to express both smbFT from a low-copy-number plasmid under a heterologous promoter (P23) as a single transcript. We selected a stable theta-replicating plasmid, pIB184Km (46), to clone and express smbFT. We also replaced the native weak ribosome-binding sequence with a strong sequence from the gftB gene. To examine the role in immunity, we heterologously expressed SmbFT in five S. mutans strains that are susceptible to Smb. These strains are UA159, NG-8, V403, 109c, and OMZ175; all of these strains produced a clear ZOI in our susceptibility assay (Fig. 1). As a control, we also transformed the vector plasmid into the S. mutans strains. These strains were then used as indicators in an agar plate assay. Heterologous expression of SmbFT in all of the five S. mutans strains provided complete protection against Smb compared to the empty-vector carrying strains (Fig. 2A, data not shown). Thus, our findings support the prediction that SmbFT indeed functions as an immunity protein against Smb.
Inactivation of smbT makes the strain susceptible to Smb.
Since SmbFT can confer protection against Smb lantibiotic, we wanted to determine whether SmbFT has a redundant function in providing immunity against Smb in GS-5. This is because a previous study suggests that two other factors, Bip (SMU.1914) and SmbG (encoded by smb operon), also provide protection against Smb (44). We first constructed an smbT deletion mutant (ΔsmbT) in GS-5 by replacing the smbT coding sequence with a nonpolar erythromycin resistance cassette. We used the ΔsmbT mutant as an indicator strain in previously described plate assays against Smb producer. As shown in Fig. 3A, we observed that ΔsmbT is more susceptible to Smb compared to the GS-5 strain. The ZOI produced by the ΔsmbT strain was very clear and as large as the ZOI produced by the S. salivarius strain. This result supports that SmbFT functions primarily as an immunity protein in GS-5, and its absence makes the strain more susceptible to the lantibiotic.
Fig 3.
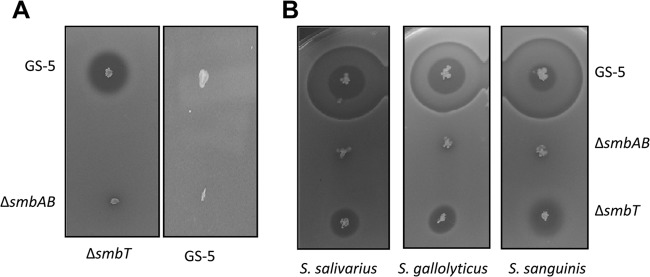
Sensitivity of the ΔsmbT strain to Smb and the secretion of Smb by the ΔsmbT strain. (A) Agar diffusion assays were carried out with GS-5 and ΔsmbAB strains as tester strains with GS-5 and ΔsmbT strains as indicator strains. (B) The production of Smb by the ΔsmbT strain was verified by agar diffusion assay using S. salivarius BAA491, S. gallolyticus BAA2069, and S. sanguinis SK36 as indicator strains. The GS-5 and ΔsmbAB strains were used as positive and negative controls, respectively. The plates are representative of three independent replicates.
To investigate the influence of SmbFT in Smb production, we performed a plate assay to measure ZOI formation by ΔsmbT strain against S. salivarius indicator strain with GS-5 and ΔsmbAB strains as controls. Although GS-5 produced a larger and clear ZOI, including a secondary halo, the ZOI produced by the ΔsmbT mutant was clear and small. On the other hand, the ΔsmbAB mutant did not produce a visible ZOI. The larger secondary ZOI that was seen with GS-5 is not always visible, and it depends on the age of the indicator culture (Fig. 3B, data not shown). To confirm that the ΔsmbT mutant could produce a clear ZOI, we used two other indicator strains, S. gallolyticus and S. sanguinis. As shown in Fig. 3B (right panels), the ΔsmbT mutant produced a clear ZOI in both strains compared to the ΔsmbAB control strain, which did not produce a halo. As expected, the GS-5 strain produced a larger ZOI with a secondary halo against both indicator strains. Taken together, our data suggest that Smb is secreted at low levels even in the absence of smbT.
SmbFT can confer immunity in heterologous streptococcal hosts.
Oral cavity harbors over 700 different bacterial species and streptococci constitute the majority (53). Many streptococci contain ABC transporters that are similar to SmbFT, and yet they are sensitive to Smb. To verify whether SmbFT can confer resistance in other oral streptococci, we selected a mitis group of streptococci, S. sanguinis, which plays a beneficial role in the oral cavity and is known to compete with S. mutans for subsequent colonization (8, 10). Another reason for selecting S. sanguinis was that the organism is naturally competent and therefore easy to manipulate (54). When we overexpressed SmbFT from the plasmid in S. sanguinis SK36, we observed a complete protection against Smb-mediated killing (Fig. 4A). In fact, there was very little or no ZOI in strain overexpressing SmbFT, whereas ZOI of ∼20 mm in diameter was observed in strain containing the vector only (Fig. 4A).
Fig 4.
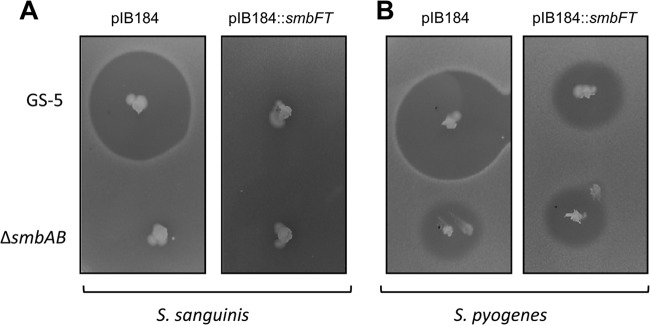
SmbFT confers protection in heterologous hosts agar diffusion assays were carried out with the GS-5 and ΔsmbAB strains as tester strains. The indicator strains are S. sanguinis SK36 (left panel) and S. pyogenes JRS4 (right panel) containing the vector (pIB184) or the vector with SmbFT. These plates are representative of three independent assays.
Since SmbFT conferred protection against Smb in streptococcus belonging to mitis group, we wanted to test whether it can confer protection in other group. For this we selected the pyogenic group, since all of the four tested species produced larger and clearer ZOI (Fig. 1A). We introduced the plasmid overexpressing SmbFT and the vector control in S. pyogenes JRS4 strain, an M6 serotype strain. As shown in Fig. 4B, the overexpression of SmbFT in S. pyogenes also provided protection against Smb in the S. pyogenes strain. However, the level of protection was less drastic, and the ΔsmbAB strain, which was included as a control, also produced a clear ZOI in both vector- and SmbFT-overexpressing strains. Nevertheless, SmbFT conferred protection against this lantibiotic in S. pyogenes. Taken together, our results suggest that SmbFT can function as a universal immunity protein for Smb in streptococci.
The immunity provided by SmbFT is specific toward Smb and related lantibiotics.
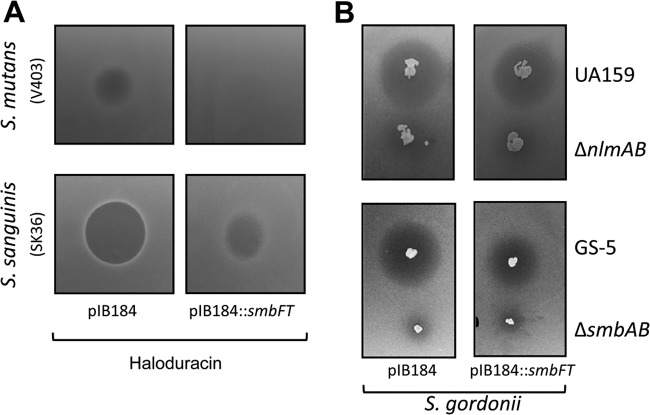
Cross-immunity is a rare occurrence among lantibiotic producer strains and has only been found in strains that produces very closely related lantibiotics, such as nukacin ISK-1 and lacticin 481 (55). Since SmbFT was able to confer protection in various streptococci, we wondered whether SmbFT could provide cross-protection against other bacteriocins. Toward this end, we first tested against haloduracin, a two-component lantibiotic produced by Bacillus halodurans and shares structural similarity with Smb (18). S. sanguinis and S. mutans V403 strains containing either smbFT expression plasmid or the vector alone were used as indicator strains in an agar diffusion assay with 10 μl of 100 μM purified haloduracin (kindly provided by W. A. van der Donk) spotted onto a plate, and the results are shown in Fig. 5A. Haloduracin produced a clearer and larger ZOI in S. sanguinis containing the vector, whereas S. sanguinis expressing SmbFT produced a much smaller and cloudier ZOI. The ZOI in S. mutans V403 with the vector was much smaller and cloudier, suggesting that haloduracin is not very effective against S. mutans. However, no ZOI was observed in V403 expressing SmbFT, indicating that, as in S. sanguinis, SmbFT provided protection against haloduracin in S. mutans.
Fig 5.
Substrate specificity of SmbFT. (A) Purified haloduracin (1:1 mixture of Halα and Halβ) was spotted on THY plate and then overlaid with indicator strains containing the vector or the vector with SmbFT. (B) UA159 and ΔnlmAB strains were stabbed into THY agar, and a deferred antagonism assay was carried out with S. gordonii DL-1 strain containing the vector or the vector with SmbFT. GS-5 and ΔsmbAB strains were used as control. Assays were repeated at least three times, and a representative plate is shown.
S. gallolyticus BAA-2069 possess a unique 23-kb genetic island in the genome, coding for bacteriocin-associated genes (SGGBAA2069_c00810-c00960). These genes encode for a lantibiotic that share high degree of similarity with Smb and haloduracin. Hinse et al. (56) have shown that the BAA-2069 strain indeed produces a lantibiotic that is active. Therefore, we tested S. mutans V403, S. sanguinis SK36, and S. pyogenes JRS4 strains overexpressing SmbFT or containing the vector. All of these streptococci produced much smaller ZOIs when SmbFT was present compared to vector-only strains (data not shown). This finding indicates that SmbFT is also active against the lantibiotic produced by BAA-2069.
We then tested whether SmbFT could provide protection against other bacteriocins. For this, we chose S. mutans UA159 that primarily produces mutacin IV (NlmAB, a two-component nonlantibiotic). S. gordonii is generally used as an indicator strain for mutacin IV. As shown in Fig. 5B, S. gordonii with or without SmbFT produced similar size ZOI, indicating that SmbFT could not confer protection (the control ΔnlmAB strain did not produce any ZOI in either cases). Next, we checked the effect of SmbFT on protection against purified nisin, a well-studied monopeptide lantibiotic that is structurally different from Smb. As expected, SmbFT did not provide protection against nisin (data not shown). ABC transporters are often associated with tolerance to various peptide antibiotics, such as vancomycin and bacitracin (57). Therefore, we also checked sensitivity to different cell wall-specific peptide antibiotics, such as bacitracin, polymyxin B, and vancomycin, by disc diffusion assay. We did not observe any noticeable differences between the strain containing SmbFT or the vector control (data not shown). Taken together, our results suggest that SmbFT is specific in conferring protection against Smb or closely related lantibiotics.
DISCUSSION
Lantibiotic producer strains possess multiple mechanisms to protect themselves against their own inhibitory activity. One of the most effective mechanisms that provide the maximum protection is through various dedicated ABC transporters. ABC transporters that are involved in self-immunity or protection against other lantibiotics are generally classified into multiple categories (58). ABC transporters such as NisT and SunT family members are encoded by a single polypeptide, where the ATPase domain is fused with the permease domain. Some members of SunT also encode an N-terminal peptidase C39 domain. SmbG, which was shown to confer protection against Smb (44), belongs to the SunT family. The other ABC transporter families all contain two separate polypeptides, one with permease activity and the other with ATPase function. SmbFT, the ABC transporter that we studied here, belong to the BcrAB family that contains two separate polypeptides. ABC transporters belonging to BcrAB family are not extensively characterized, except for two transporters from bacilli (59, 60). A recent evolutionary relationship study indicates that BcrAB family proteins are closely related to LanFEG family transporters. The latter family proteins, which contain two separate permeases (LanE and LanG) and one ATPase (LanF), are all involved in self-immunity of lantibiotic-producing bacteria. On the other hand, BcrAB family transporters are involved in resistance against bacitracin (59, 61). Since the genes are encoded within the biosynthesis locus for bacitracin, BcrAB family proteins appear to confer self-immunity (61). The SmbFT transporter that we studied here is the first example of a BcrAB family protein that is involved in lantibiotic resistance.
As already mentioned above, the SmbFT transporter is composed of two polypeptides: SmbT, which encodes the permease component, and SmbF, which encodes the ATPase where two-thirds of the N-terminal region comprises of P-loop containing nucleoside triphosphate hydrolase signature sequences (62). Although S. mutans encodes numerous permease proteins (42), a sequence similarity search with SmbT against the genome returned no results, suggesting that SmbT encodes a unique sequence. In contrast, a search using SmbF as the query against the genome returned several ABC transporter-related ATPases. Four ATPases—SMU.238, SMU.654, SMU.1035, and SMU.1811—showed maximum similarity (E-value ≥30) with SmbF (data not shown). At present, we do not know whether any of these ATPases can form a productive complex with SmbT permease to counteract Smb.
We expressed SmbFT in at least four different streptococcal strains other than S. mutans. In each of these streptococcal strains, the heterologous expression of SmbFT conferred full protection against the Smb lantibiotic. This means that SmbFT does not need any other accessory proteins for its activity. Alternatively, the accessory protein could be a highly conserved protein present in all of the streptococcal strains tested here. As previously mentioned, lantibiotic producer strains often encode two types of transporters for full protection. For example, a Lactococcus lactis strain that produces lacticin 3147, a two-component lantibiotic similar to Smb, possesses both LtnI, a lipid-anchored membrane protein, and LtnFE systems (63). Draper et al. (63) have demonstrated that, in a heterologous system, LtnI and LntFE act synergistically to confer complete protection. The smb locus does not encode any LntI homolog, but it does encode SmbG, a SunT homolog, containing the ABC transporter motif and a C39 peptidase motif. It has been proposed that SmbG functions as an immunity protein against Smb (44); however, an smbG mutant strain is not susceptible to Smb. An immunity-like protein encoded by SMU.1913 (referred to as Bip in reference 44) has also been proposed to function as an Smb immunity protein. Surprisingly, a bip single-mutant strain is also not susceptible to Smb, indicating other proteins can confer resistance. In contrast, our data suggest that SmbFT alone can confer complete protection against Smb in a variety of streptococcal strains; none of these strains contain either SmbG or Bip. Therefore, unlike other lantibiotic immunity systems, we speculate that SmbG and SmbFT provide different functions in S. mutans. We believe that the primary role of SmbG is to secrete Smb peptides from the cytoplasm to the milieu and, when SmbG is absent, the strain does not secrete active Smb. Indeed, we found that an smbG mutant is unable to produce Smb and functionally behave like the ΔsmbAB mutant. We also believe that the primary function of SmbFT is to confer self-protection and not secretion. This is because we found that the ΔsmbT mutant produced a clear ZOI against multiple strains (Fig. 3), indicating Smb production; however, the ZOI was much smaller than that observed with the GS-5 strain. There are several reasons that can explain the smaller ZOI produced by the ΔsmbT strain. Since the strain was constructed by insertion of an ermB gene (erythromycin resistance), it might have interfered with the transcription of the downstream genes in the smb operon and so, as a result, less smbAB was produced. Often lantibiotic production is subject to feedback inhibition and is positively influenced by the immunity proteins (64, 65). Therefore, it is also possible that SmbFT positively influences smb production. Whether SmbFT has a positive role in Smb production remains to be tested.
It appears that SmbFT could confer protection not only against Smb but also against haloduracin and an uncharacterized lantibiotic produced by BAA-2069 (we named this lantibiotic gallolacticin). All three lantibiotics share high degree of sequence similarity (see Fig. S1 in the supplemental material) and probably structurally similar as well. All three lantibiotics are composed of two modified peptides, α and β. Although no information is available for Smb or gallolacticin, a detailed structure has been proposed for haloduracin that is based on mass spectrometry data and modeled on nuclear magnetic resonance structure of lacticin 3147 (66). The structural information, together with the sequence similarity, suggests that all α-peptides have the same topology with three C-terminal rings (13) (see Fig. S1 in the supplemental material). The β-peptides appear to contain three or four rings; however, the B ring is present only in haloduracin and is absent in all of the others. Since SmbFT recognized all three lantibiotics, we speculate that the transporter complex predominantly recognizes the conserved topology and structurally similar lantibiotics. Based on this observation, we also speculate that SmbFT will confer protection against two-component lantibiotics that share this overall topology. Interestingly, the β-peptides of two lantibiotics, lacticin 3147 and staphylococcus strain C55 lack the first ring (ring A) but contain three other rings (rings B to D). It would be of interest to test whether SmbFT could recognize lacticin 3147 and staphylococcin C55.
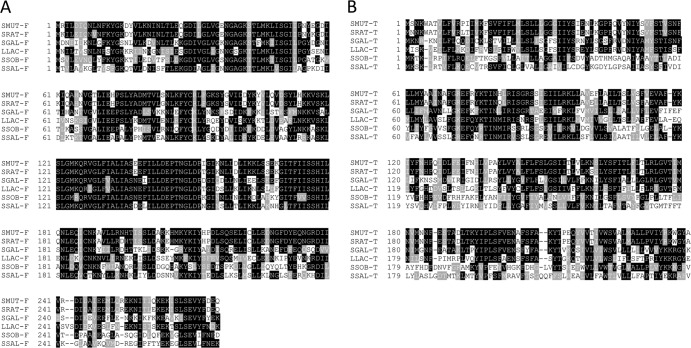
Thus far, all of the ABC transporters that belong to BcrAB family have been shown to act specifically against cyclic peptide bacitracin (59, 61). Although sequence similarity indicates that SmbFT belongs BcrAB family, we found that SmbFT does not recognize bacitracin; it also recognizes neither vancomycin nor polymyxin B, two other cyclic peptide antibiotics. SmbFT also did not recognize nisin, a heavily modified single-peptide lantibiotic. A BLASP search using SmbF or SmbT peptides as a query sequences against the GenBank database returned sequences mostly from streptococci. The top hit sequences include S. ratti (strains BHT and FA-1), S. gallolyticus BAA-2069, S. sobrinus DSM20742, S. salivarius CCHSS3, and Lactococcus lactis (Fig. 6). We propose that SmbFT and its homolog constitute an ABC transporter family distinct from BcrAB and specifically recognize two-component lantibiotics. It should be noted that although S. gallolyticus BAA2069 produces gallolacticin and encodes an SmbFT homolog, which we named GalFT; the strain was sensitive to Smb (data not shown). Therefore, we speculate that whereas SmbFT confers protection against gallolacticin, GalFT from BAA2069 cannot confer protection against Smb. A sequence alignment between SmbF and SmbT with the corresponding GalF and GalT sequences identified presence of several unique amino acid residues in both GalF and GalT. A hybrid analytical approach is under way wherein we are combining SmbF with GalT and vice versa to identify the subunit necessary for substrate recognition and/or discrimination.
Fig 6.
Multiple sequence alignment of SmbF (A) and SmbT (B) with their closest homologs. The alignment was performed using CLUSTAL W. Degree of shading was done using BoxShade where black and gray blocks indicate identical and similar amino acids, respectively. Sequences were obtained from GenBank (accession numbers are in parentheses). The following strains were tested: S. ratti (SRAT-F, WP_003086779.1; SRAT-T, WP_003086777.1), S. gallolyticus (SGAL-F, YP_004287007.1; SGAL-T, YP_004287008.1), L. lactis (LLAC-F, YP_004761484.1; LLAC-T, YP_004761485.1), S. sobrinus (SSOB-F, WP_019770819.1; SSOB-T, WP_019776383.1), and S. salivarius (SSAL-F, YP_004728647.1 SSAL-T, YP_004728646.1).
Although the mechanisms related to protection against lantibiotics have been extensively studied, the molecular mechanism of lantibiotic transport is still poorly understood. Several recent studies suggest that, unlike general ABC transporters that transport substrates across the membranes, the ABC transporters related to immunity function transport specific lantibiotics from the membrane to the extracellular space (28, 29, 55). A recent study identified the presence of a conserved motif, termed the E-loop, in the ABC transporters belonging to LanFEG and BcrAB families (52). This E-loop structure plays an important role in the function of these transporters and perhaps induces structural changes in the transmembrane domains. Whether the E-loop is involved in substrate recognition and/or binding remains to be evaluated. Additional structure-function studies are necessary in order to understand the molecular mechanism of lantibiotic transport by immunity-related ABC transporter systems.
Supplementary Material
ACKNOWLEDGMENTS
We thank Wilfred A. van der Donk, University of Illinois at Urbana—Champaign, for kindly providing purified haloduracin. We also thank Pierce O'Neil for some technical help.
This study was supported in part by an NIH-NIDCR grant (DE021664) to I.B.
Footnotes
Published ahead of print 11 October 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.01060-13.
REFERENCES
- 1.Aas JA, Paster BJ, Stokes LN, Olsen I, Dewhirst FE. 2005. Defining the normal bacterial flora of the oral cavity. J. Clin. Microbiol. 43:5721–5732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Norder Grusell E, Dahlen G, Ruth M, Ny L, Quiding-Jarbrink M, Bergquist H, Bove M. 2013. Bacterial flora of the human oral cavity, and the upper and lower esophagus. Dis. Esophagus 26:84–90 [DOI] [PubMed] [Google Scholar]
- 3.Que YA, Moreillon P. 2011. Infective endocarditis. Nat. Rev. Cardiology 8:322–336 [DOI] [PubMed] [Google Scholar]
- 4.Nakano K, Ooshima T. 2009. Serotype classification of Streptococcus mutans and its detection outside the oral cavity. Future Microbiol. 4:891–902 [DOI] [PubMed] [Google Scholar]
- 5.Mitchell J. 2011. Streptococcus mitis: walking the line between commensalism and pathogenesis. Mol. Oral Microbiol. 26:89–98 [DOI] [PubMed] [Google Scholar]
- 6.Doern CD, Burnham CA. 2010. It's not easy being green: the viridans group streptococci, with a focus on pediatric clinical manifestations. J. Clin. Microbiol. 48:3829–3835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burton JP, Wescombe PA, Cadieux PA, Tagg JR. 2011. Beneficial microbes for the oral cavity: time to harness the oral streptococci? Beneficial Microbes 2:93–101 [DOI] [PubMed] [Google Scholar]
- 8.Kreth J, Merritt J, Shi W, Qi F. 2005. Competition and coexistence between Streptococcus mutans and Streptococcus sanguinis in the dental biofilm. J. Bacteriol. 187:7193–7203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Qi F, Kreth J. 2010. Characterization of anti-competitor activities produced by oral bacteria. Methods Mol. Biol. 666:151–166 [DOI] [PubMed] [Google Scholar]
- 10.Kreth J, Zhang Y, Herzberg MC. 2008. Streptococcal antagonism in oral biofilms: Streptococcus sanguinis and Streptococcus gordonii interference with Streptococcus mutans. J. Bacteriol. 190:4632–4640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van Belkum MJ, Stiles ME. 2000. Nonlantibiotic antibacterial peptides from lactic acid bacteria. Natural Product Rep. 17:323–335 [DOI] [PubMed] [Google Scholar]
- 12.Islam MR, Nishie M, Nagao J, Zendo T, Keller S, Nakayama J, Kohda D, Sahl HG, Sonomoto K. 2012. Ring A of nukacin ISK-1: a lipid II-binding motif for type-A(II) lantibiotic. J. Am. Chem. Soc. 134:3687–3690 [DOI] [PubMed] [Google Scholar]
- 13.Willey JM, van der Donk WA. 2007. Lantibiotics: peptides of diverse structure and function. Annu. Rev. Microbiol. 61:477–501 [DOI] [PubMed] [Google Scholar]
- 14.Chatterjee C, Paul M, Xie L, van der Donk WA. 2005. Biosynthesis and mode of action of lantibiotics. Chemical Rev. 105:633–684 [DOI] [PubMed] [Google Scholar]
- 15.Diep DB, Nes IF. 2002. Ribosomally synthesized antibacterial peptides in Gram positive bacteria. Curr. Drug Targets 3:107–122 [DOI] [PubMed] [Google Scholar]
- 16.Woodyer RD, Li G, Zhao H, van der Donk WA. 2007. New insight into the mechanism of methyl transfer during the biosynthesis of fosfomycin. Chem. Commun. 4:359–361 [DOI] [PubMed] [Google Scholar]
- 17.Knerr PJ, van der Donk WA. 2012. Discovery, biosynthesis, and engineering of lantipeptides. Annu. Rev. Biochem. 81:479–505 [DOI] [PubMed] [Google Scholar]
- 18.McClerren AL, Cooper LE, Quan C, Thomas PM, Kelleher NL, van der Donk WA. 2006. Discovery and in vitro biosynthesis of haloduracin, a two-component lantibiotic. Proc. Natl. Acad. Sci. U. S. A. 103:17243–17248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McAuliffe O, Hill C, Ross RP. 2000. Each peptide of the two-component lantibiotic lacticin 3147 requires a separate modification enzyme for activity. Microbiology 146(Pt 9):2147–2154 [DOI] [PubMed] [Google Scholar]
- 20.Kodani S, Lodato MA, Durrant MC, Picart F, Willey JM. 2005. SapT, a lanthionine-containing peptide involved in aerial hyphae formation in the streptomycetes. Mol. Microbiol. 58:1368–1380 [DOI] [PubMed] [Google Scholar]
- 21.Tillotson RD, Wosten HA, Richter M, Willey JM. 1998. A surface active protein involved in aerial hyphae formation in the filamentous fungus Schizophyllum commune restores the capacity of a bald mutant of the filamentous bacterium Streptomyces coelicolor to erect aerial structures. Mol. Microbiol. 30:595–602 [DOI] [PubMed] [Google Scholar]
- 22.Bierbaum G, Sahl HG. 2009. Lantibiotics: mode of action, biosynthesis, and bioengineering. Curr. Pharm. Biotechnol. 10:2–18 [DOI] [PubMed] [Google Scholar]
- 23.Cotter PD, Hill C, Ross RP. 2005. Bacteriocins: developing innate immunity for food. Nat. Rev. Microbiol. 3:777–788 [DOI] [PubMed] [Google Scholar]
- 24.Cotter PD, Hill C, Ross RP. 2005. Bacterial lantibiotics: strategies to improve therapeutic potential. Curr. Protein Peptide Sci. 6:61–75 [DOI] [PubMed] [Google Scholar]
- 25.Heng CK WP, Burton JP, Jack RW, Tagg JR. 2007. The diversity of bacteriocins in Gram-positive bacteria, p 45–92 In Riley MA. (ed), Bacteriocins: ecology and evolution. Springer-Verlag, Berlin, Germany [Google Scholar]
- 26.Draper LA, Ross RP, Hill C, Cotter PD. 2008. Lantibiotic immunity. Curr. Protein Peptide Sci. 9:39–49 [DOI] [PubMed] [Google Scholar]
- 27.Hoffmann A, Schneider T, Pag U, Sahl HG. 2004. Localization and functional analysis of PepI, the immunity peptide of Pep5-producing Staphylococcus epidermidis strain 5. Appl. Environ. Microbiol. 70:3263–3271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stein T, Heinzmann S, Dusterhus S, Borchert S, Entian KD. 2005. Expression and functional analysis of the subtilin immunity genes spaIFEG in the subtilin-sensitive host Bacillus subtilis MO1099. J. Bacteriol. 187:822–828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stein T, Heinzmann S, Solovieva I, Entian KD. 2003. Function of Lactococcus lactis nisin immunity genes nisI and nisFEG after coordinated expression in the surrogate host Bacillus subtilis. J. Biol. Chem. 278:89–94 [DOI] [PubMed] [Google Scholar]
- 30.Hillman JD, Novak J, Sagura E, Gutierrez JA, Brooks TA, Crowley PJ, Hess M, Azizi A, Leung K, Cvitkovitch D, Bleiweis AS. 1998. Genetic and biochemical analysis of mutacin 1140, a lantibiotic from Streptococcus mutans. Infect. Immun. 66:2743–2749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Qi F, Chen P, Caufield PW. 1999. Purification of mutacin III from group III Streptococcus mutans UA787 and genetic analyses of mutacin III biosynthesis genes. Appl. Environ. Microbiol. 65:3880–3887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Qi F, Chen P, Caufield PW. 1999. Functional analyses of the promoters in the lantibiotic mutacin II biosynthetic locus in Streptococcus mutans. Appl. Environ. Microbiol. 65:652–658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Qi F, Chen P, Caufield PW. 2000. Purification and biochemical characterization of mutacin I from the group I strain of Streptococcus mutans, CH43, and genetic analysis of mutacin I biosynthesis genes. Appl. Environ. Microbiol. 66:3221–3229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tsang P, Merritt J, Nguyen T, Shi W, Qi F. 2005. Identification of genes associated with mutacin I production in Streptococcus mutans using random insertional mutagenesis. Microbiology 151:3947–3955 [DOI] [PubMed] [Google Scholar]
- 35.Yonezawa H, Kuramitsu HK. 2005. Genetic analysis of a unique bacteriocin, Smb, produced by Streptococcus mutans GS5. Antimicrob. Agents Chemother. 49:541–548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mota-Meira M, Lacroix C, LaPointe G, Lavoie MC. 1997. Purification and structure of mutacin B-Ny266: a new lantibiotic produced by Streptococcus mutans. FEBS Lett. 410:275–279 [DOI] [PubMed] [Google Scholar]
- 37.Robson CL, Wescombe PA, Klesse NA, Tagg JR. 2007. Isolation and partial characterization of the Streptococcus mutans type AII lantibiotic mutacin K8. Microbiology 153:1631–1641 [DOI] [PubMed] [Google Scholar]
- 38.de Vos WM, Kuipers OP, van der Meer JR, Siezen RJ. 1995. Maturation pathway of nisin and other lantibiotics: posttranslationally modified antimicrobial peptides exported by gram-positive bacteria. Mol. Microbiol. 17:427–437 [DOI] [PubMed] [Google Scholar]
- 39.Sahl HG, Bierbaum G. 1998. Lantibiotics: biosynthesis and biological activities of uniquely modified peptides from gram-positive bacteria. Annu. Rev. Microbiol. 52:41–79 [DOI] [PubMed] [Google Scholar]
- 40.Nicolas GG, Lavoie MC, LaPointe G. 2007. Molecular genetics, genomics and biochemistry of mutacins, vol 1 Global Science Books, London, United Kingdom [Google Scholar]
- 41.Kamiya RU, Hofling JF, Goncalves RB. 2008. Frequency and expression of mutacin biosynthesis genes in isolates of Streptococcus mutans with different mutacin-producing phenotypes. J. Med. Microbiol. 57:626–635 [DOI] [PubMed] [Google Scholar]
- 42.Ajdic D, McShan WM, McLaughlin RE, Savic G, Chang J, Carson MB, Primeaux C, Tian R, Kenton S, Jia H, Lin S, Qian Y, Li S, Zhu H, Najar F, Lai H, White J, Roe BA, Ferretti JJ. 2002. Genome sequence of Streptococcus mutans UA159, a cariogenic dental pathogen. Proc. Natl. Acad. Sci. U. S. A. 99:14434–14439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Biswas S, Biswas I. 2012. Complete genome sequence of Streptococcus mutans GS-5, a serotype c strain. J. Bacteriol. 194:4787–4788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Matsumoto-Nakano M, Kuramitsu HK. 2006. Role of bacteriocin immunity proteins in the antimicrobial sensitivity of Streptococcus mutans. J. Bacteriol. 188:8095–8102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Biswas I, Drake L, Johnson S, Thielen D. 2007. Unmarked gene modification in Streptococcus mutans by a cotransformation strategy with a thermosensitive plasmid. Biotechniques 42:487–490 [DOI] [PubMed] [Google Scholar]
- 46.Biswas I, Jha JK, Fromm N. 2008. Shuttle expression plasmids for genetic studies in Streptococcus mutans. Microbiology 154:2275–2282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hossain MS, Biswas I. 2012. An extracellular protease, SepM, generates functional competence-stimulating peptide in Streptococcus mutans UA159. J. Bacteriol. 194:5886–5896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Maguin E, Prevost H, Ehrlich SD, Gruss A. 1996. Efficient insertional mutagenesis in lactococci and other gram-positive bacteria. J. Bacteriol. 178:931–935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Banas JA, Biswas S, Zhu M. 2011. Effects of DNA methylation on expression of virulence genes in Streptococcus mutans. Appl. Environ. Microbiol. 77:7236–7242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Petersen FC, Fimland G, Scheie AA. 2006. Purification and functional studies of a potent modified quorum-sensing peptide and a two-peptide bacteriocin in Streptococcus mutans. Mol. Microbiol. 61:1322–1334 [DOI] [PubMed] [Google Scholar]
- 51.Facklam R. 2002. What happened to the streptococci: overview of taxonomic and nomenclature changes. Clin. Microbiol. Rev. 15:613–630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Okuda K, Yanagihara S, Sugayama T, Zendo T, Nakayama J, Sonomoto K. 2010. Functional significance of the E loop, a novel motif conserved in the lantibiotic immunity ATP-binding cassette transport systems. J. Bacteriol. 192:2801–2808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Aas JA, Griffen AL, Dardis SR, Lee AM, Olsen I, Dewhirst FE, Leys EJ, Paster BJ. 2008. Bacteria of dental caries in primary and permanent teeth in children and young adults. J. Clin. Microbiol. 46:1407–1417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xu P, Ge X, Chen L, Wang X, Dou Y, Xu JZ, Patel JR, Stone V, Trinh M, Evans K, Kitten T, Bonchev D, Buck GA. 2011. Genome-wide essential gene identification in Streptococcus sanguinis. Sci. Rep. 1:125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Aso Y, Okuda K, Nagao J, Kanemasa Y, Thi Bich Phuong N, Koga H, Shioya K, Sashihara T, Nakayama J, Sonomoto K. 2005. A novel type of immunity protein, NukH, for the lantibiotic nukacin ISK-1 produced by Staphylococcus warneri ISK-1. Biosci. Biotechnol. Biochem. 69:1403–1410 [DOI] [PubMed] [Google Scholar]
- 56.Hinse D, Vollmer T, Ruckert C, Blom J, Kalinowski J, Knabbe C, Dreier J. 2011. Complete genome and comparative analysis of Streptococcus gallolyticus subsp. gallolyticus, an emerging pathogen of infective endocarditis. BMC Genomics 12:400. 10.1186/1471-2164-12-400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dintner S, Staron A, Berchtold E, Petri T, Mascher T, Gebhard S. 2011. Coevolution of ABC transporters and two-component regulatory systems as resistance modules against antimicrobial peptides in Firmicutes Bacteria. J. Bacteriol. 193:3851–3862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gebhard S. 2012. ABC transporters of antimicrobial peptides in Firmicutes bacteria: phylogeny, function, and regulation. Mol. Microbiol. 86:1295–1317 [DOI] [PubMed] [Google Scholar]
- 59.Podlesek Z, Comino A, Herzog-Velikonja B, Zgur-Bertok D, Komel R, Grabnar M. 1995. Bacillus licheniformis bacitracin-resistance ABC transporter: relationship to mammalian multidrug resistance. Mol. Microbiol. 16:969–976 [DOI] [PubMed] [Google Scholar]
- 60.Butcher BG, Lin YP, Helmann JD. 2007. The yydFGHIJ operon of Bacillus subtilis encodes a peptide that induces the LiaRS two-component system. J. Bacteriol. 189:8616–8625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Neumuller AM, Konz D, Marahiel MA. 2001. The two-component regulatory system BacRS is associated with bacitracin ‘self-resistance' of Bacillus licheniformis ATCC 10716. Eur. J. Biochem. 268:3180–3189 [DOI] [PubMed] [Google Scholar]
- 62.Davidson AL, Chen J. 2004. ATP-binding cassette transporters in bacteria. Annu. Rev. Biochem. 73:241–268 [DOI] [PubMed] [Google Scholar]
- 63.Draper LA, Grainger K, Deegan LH, Cotter PD, Hill C, Ross RP. 2009. Cross-immunity and immune mimicry as mechanisms of resistance to the lantibiotic lacticin 3147. Mol. Microbiol. 71:1043–1054 [DOI] [PubMed] [Google Scholar]
- 64.Valsesia G, Medaglia G, Held M, Minas W, Panke S. 2007. Circumventing the effect of product toxicity: development of a novel two-stage production process for the lantibiotic gallidermin. Appl. Environ. Microbiol. 73:1635–1645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Puramattathu TV, Islam MR, Nishie M, Yanagihara S, Nagao J, Okuda K, Zendo T, Nakayama J, Sonomoto K. 2012. Enhanced production of nukacin D13E in Lactococcus lactis NZ9000 by the additional expression of immunity genes. Appl. Microbiol. Biotechnol. 93:671–678 [DOI] [PubMed] [Google Scholar]
- 66.Cooper LE, McClerren AL, Chary A, van der Donk WA. 2008. Structure-activity relationship studies of the two-component lantibiotic haloduracin. Chem. Biol. 15:1035–1045 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.