Abstract
In Mycobacterium tuberculosis, the stringent response to amino acid starvation is mediated by the M. tuberculosis Rel (RelMtb) enzyme, which transfers a pyrophosphate from ATP to GDP or GTP to synthesize ppGpp and pppGpp, respectively. (p)ppGpp then influences numerous metabolic processes. RelMtb also encodes a second, distinct catalytic domain that hydrolyzes (p)ppGpp into pyrophosphate and GDP or GTP. RelMtb is required for chronic M. tuberculosis infection in mice; however, it is unknown which catalytic activity of RelMtb mediates pathogenesis and whether (p)ppGpp itself is necessary. In order to individually investigate the roles of (p)ppGpp synthesis and hydrolysis during M. tuberculosis pathogenesis, we generated RelMtb point mutants that were either synthetase dead (RelMtbH344Y) or hydrolase dead (RelMtbH80A). M. tuberculosis strains expressing the synthetase-dead RelMtbH344Y mutant did not persist in mice, demonstrating that the RelMtb (p)ppGpp synthetase activity is required for maintaining bacterial titers during chronic infection. Deletion of a second predicted (p)ppGpp synthetase had no effect on pathogenesis, demonstrating that RelMtb was the major contributor to (p)ppGpp production during infection. Interestingly, expression of an allele encoding the hydrolase-dead RelMtb mutant, RelMtbH80A, that is incapable of hydrolyzing (p)ppGpp but still able to synthesize (p)ppGpp decreased the growth rate of M. tuberculosis and changed the colony morphology of the bacteria. In addition, RelMtbH80A expression during acute or chronic M. tuberculosis infection in mice was lethal to the infecting bacteria. These findings highlight a distinct role for RelMtb-mediated (p)ppGpp hydrolysis that is essential for M. tuberculosis pathogenesis.
INTRODUCTION
At least 30% of the world's population is infected with latent Mycobacterium tuberculosis, which will reactivate in some individuals and cause an estimated 1.4 million deaths a year (1). Significant obstacles in controlling the epidemic result from the chronic nature of M. tuberculosis infection, which necessitates prolonged treatment and generates a large reservoir of latently infected people. This health crisis is exacerbated by the alarming emergence of drug-resistant strains. The inadequacies of present tuberculosis therapies demand the discovery of new agents to treat M. tuberculosis infection, which requires insight into the pathways used by the pathogen to survive in the host. During infection, the host restrains mycobacteria from proliferating by imposing an arsenal of defenses, including oxidative stress, hypoxia, acid stress, genotoxic stress, cell surface stress, and starvation (reviewed in reference 2). Despite this onslaught of attacks, M. tuberculosis is able to persist for the lifetime of the host, indicating that this pathogen has substantial molecular mechanisms to resist host-inflicted damage.
One way bacteria resist stress is via the stringent response. The stringent response is a conserved global stress response in bacteria that involves the production of the hyperphosphorylated guanine nucleotides ppGpp and pppGpp [collectively called (p)ppGpp]. (p)ppGpp metabolism is controlled by Rel/SpoT homolog proteins (RSHs), named for their sequence similarity to RelA and SpoT enzymes in Escherichia coli (3). The activity of RelA homologs is best characterized during amino acid starvation when an uncharged tRNA enters the A site of the ribosome and stimulates RelA-mediated (p)ppGpp production (4). (p)ppGpp then regulates a number of cellular processes, including transcription, replication, cell surface morphology, and nucleotide levels (reviewed in references 5 and 6).
In M. tuberculosis, the stringent response is initiated by the M. tuberculosis Rel (RelMtb) enzyme, which transfers the 5′-β,γ-pyrophosphate from ATP to the 3′-OH of GDP or GTP to synthesize ppGpp and pppGpp, respectively (7). The (p)ppGpp synthetase domain is in the middle of the RelMtb protein, and a mutation of a conserved histidine (H344) or glycine (G241) residue in this domain abolishes synthetase activity (8, 9). The C terminus of RelMtb is not required for catalytic activity but is necessary for association with the RelMtb activating complex, which consists of ribosomes, mRNA, and tRNAs and regulates (p)ppGpp synthesis (10, 11). RelMtb also hydrolyzes (p)ppGpp into diphosphate pyrophosphate (PPi) and GDP or GTP via an N-terminal catalytic HD superfamily domain that is distinct from the (p)ppGpp synthetase domain (9). HD superfamily members are phosphohydrolases with conserved histidine and aspartate residues that are involved in the coordination of divalent cations, which is essential for their activity (12). Alanine substitution of the conserved histidine (H80) or aspartate (D81) residue in RelMtb abolishes hydrolase activity in vitro without affecting (p)ppGpp synthesis (9). The crystal structure of the RelMtb homolog in Streptococcus dysgalactiae subsp. equisimilis, RelSeq, indicates that at any given time only one enzymatic function is active, thereby preventing simultaneous synthesis and hydrolysis of (p)ppGpp (13).
M. tuberculosis mutants deleted for the entire relMtb gene (ΔrelMtb) are compromised for long-term survival in culture (7) and are unable to persist in mouse models of infection (14, 15). In Mycobacterium smegmatis, deletion of relMsm causes changes in cellular and colony morphology, decreased long-term survival, and increased sensitivity to nutrient starvation and hypoxia (16). However, it is still unclear which catalytic activities of RelMtb contribute to these diverse phenotypes and what role (p)ppGpp has in these processes. In addition to RelMtb, M. tuberculosis also encodes a homolog to a group of RSHs called small alarmone synthetases (SAS), which encode the catalytic domain to produce (p)ppGpp but not the hydrolase domain (3). The role of the M. tuberculosis SAS, Rv1366, in maintaining (p)ppGpp levels, responding to stress, and virulence has yet to be investigated. In many other bacteria, SAS activity is toxic in the absence of a functional (p)ppGpp hydrolase (17–21). If this were the case in M. tuberculosis, Rv1366-mediated (p)ppGpp synthesis in the ΔrelMtb strain could be the cause for attenuation of this strain. We have addressed these questions regarding the roles of (p)ppGpp metabolism by studying strains of M. tuberculosis that express alleles of RelMtb that are defective in individual catalytic activities and strains deficient in the rv1366 gene.
MATERIALS AND METHODS
M. tuberculosis strains and media.
All M. tuberculosis strains were derived from Erdman and were grown planktonically at 37°C in 7H9 (broth) or 7H10 (agar) (Difco) medium supplemented with 10% oleic acid-albumin-dextrose-catalase (OADC), 0.5% glycerol, and 0.05% Tween 80 (broth). Biofilm cultures were grown in 24-well dishes at 37°C by inoculating 1.5 ml of Sauton's medium with 150 μl of saturated planktonic culture. The 24-well dish was placed in a tightly sealed Tupperware dish for 3 weeks, at which point the Tupperware was opened and incubated for another week before photographing. This is similar to previously described conditions used to form M. tuberculosis biofilms (22).
The ΔrelMtb strain was made by infecting M. tuberculosis Erdman with a specialized transducing phage (phAE87) containing homology to M. tuberculosis nucleotides 2907267 to 2907826 and 2910191 to 2910844, which replaced all but the first two codons and the stop codon of the endogenous relMtb gene with a hygromycin resistance cassette. The Δrv1366 strain was made by infecting wild-type (WT) M. tuberculosis Erdman with phAE87 containing homology to M. tuberculosis nucleotides 1537706 to 1538406 and 1539203 to 1539884, which replaced all but the first 6 codons and the last 2 codons of the endogenous rv1366 gene with a hygromycin resistance cassette flanked by two loxP sites. The deletion of rv1366 in this strain was confirmed by EcoRI digestion of genomic DNA and Southern blot analysis using a radiolabeled fragment containing M. tuberculosis nucleotides 1537706 to 1538406 as the probe. The hygromycin resistance cassette was cured from the Δrv1366 strain by transforming the strain with an unstable episomal plasmid (pmsg381) that expresses the Cre recombinase. Loss of the hygromycin resistance cassette was confirmed by both PCR and plating in the presence or absence of hygromycin. The relMtb gene was deleted from the Δrv1366 strain to make the ΔrelMtb Δrv1366 strain by infecting the Δrv1366 strain, which had been cured of the hygromycin resistance cassette, with the phAE87 phage that replaces the endogenous relMtb gene with a hygromycin resistance cassette. The deletion of the relMtb gene in this strain was confirmed by Southern blot analysis of SmaI-digested genomic DNA using a radiolabeled fragment containing M. tuberculosis nucleotides 2910191 to 2910844 as a probe.
To express the relMtbWT, relMtbH344Y, relMtbH80A, relMtbH344Y-H80A, or rv1366 allele from the attB site of the gene deletion strains, the ΔrelMtb, Δrv1366, or ΔrelMtb Δrv1366 strain was transformed with a pMSG430 plasmid that integrates into the attB site of the genome, confers kanamycin resistance, and expresses the respective allele from a constitutive pmyc1tetO promoter. Control strains were made by transforming each deletion mutant with an empty pMSG430 vector. The relMtb and rv1366 genes from each transformant were sequenced to confirm the presence of the correct sequence. There is only a single copy each of the relMtb and rv1366 genes in all strains of mycobacteria used in this paper.
To achieve conditional expression of relMtbH80A, the ΔrelMtb strain was cotransformed with pMSG430-relMtbH80A and pTES-2MOX (kindly provided by Dirk Schnappinger and Sabine Ehrt, Cornell University, NY), which confers streptomycin resistance and expresses the WT tetracycline repressor (TetR). This generated the Tet-RelMtbH80A strain, wherein the WT TetR allows for induced expression of the relMtbH80A allele in the presence of tetracycline (Tet) analogs. A control strain was constructed by cotransforming WT M. tuberculosis Erdman with empty pMSG430 and pTES-2MOX.
M. smegmatis strains and media.
All M. smegmatis strains were isogenic to strain mc2155 and were grown at 37°C in LB (broth and agar) or 7H10 (agar) supplemented with 0.5% dextrose, 0.5% glycerol, and 0.05% Tween 80 (broth). The ΔrelMsm strain was made by infecting mc2155 with phAE87 containing homology to mc2155 nucleotides 3027905 to 3028545 and 3030884 to 3031534, which deleted all but the first 4 codons and the last 14 codons of the relMsm gene. The M. smegmatis Tet-RelMtbH80A strain was made as described for M. tuberculosis. The M. smegmatis control strain was made by cotransforming the ΔrelMsm strain with pMSG430-relMtbWT and pTES-2MOX.
Antibiotics.
Twenty μg/ml kanamycin, 50 μg/ml hygromycin, 20 μg/ml streptomycin, and 50 ng/ml of anhydrotetracycline (ATc) were used for both mycobacterial species.
Mouse infections.
Before infection, exponentially replicating M. tuberculosis bacteria were washed in phosphate-buffered saline (PBS) plus 0.05% Tween 80 and sonicated to disperse clumps. Eight- to 9-week-old female C57BL/6 mice (Jackson Laboratory) were exposed to 8 × 107 CFU of the appropriate strain in an inhalation exposure system (Glas-Col), which delivers ∼100 bacteria to the lung per animal. Bacterial burden was determined by plating serial dilutions of lung and spleen homogenates onto 7H10 agar plates in the absence or presence of kanamycin and streptomycin. Plates were incubated at 37°C in 5% CO2 for 3 weeks prior to counting colonies. When indicated, the standard diet was replaced with doxycycline-containing mouse chow (2,000 ppm; Research Diets). All procedures involving animals were conducted by following the National Institutes of Health guidelines for housing and care of laboratory animals and were performed in accordance with institutional regulations after protocol review and approval by the Institutional Animal Care and Use Committee of The Washington University in St. Louis School of Medicine (protocol 20100190; Analysis of Mycobacterial Pathogenesis). Washington University is registered as a research facility with the U.S. Department of Agriculture and is fully accredited by the American Association of Accreditation of Laboratory Animal Care. The Animal Welfare Assurance is on file with OPRR-NIH. All animals used in these experiments were subjected to no or minimal discomfort. All mice were euthanized by CO2 asphyxiation, which is approved by the Panel on Euthanasia of the American Veterinary Association.
qRT-PCR.
RNA was extracted from exponentially growing mycobacteria using TRIzol (Invitrogen), cDNA was prepared using Superscript III (Invitrogen), and quantitative reverse transcription-PCR (qRT-PCR) was performed using the Bio-Rad Sso Advanced SYBR green kit. Levels of relMtb and relMsm transcript were normalized to sigA transcript levels as previously described (23).
Assaying nucleotide and metabolite levels using LC-MS/MS.
M. smegmatis Tet-RelMtbH80A and control strains were grown to exponential phase and treated with 50 ng/ml ATc for 6 h, and 5.9 × 109 CFU per sample was collected and washed in PBS. ATP, GTP, ppGpp, and pppGpp were assayed using a liquid chromatography-tandem mass spectrometry (LC-MS/MS) method. For the nucleotide extraction, 10 μl of 0.1 mM internal standard Br-ATP was spiked in each sample. Six hundred μl of cold 50% ethanol was added to each sample, and samples were homogenized with TissueLyser II for 5 min at a frequency of 20 Hz/s. After centrifugation at 16,000 × g for 5 min at 4°C, the supernatants were transferred to new tubes and the pellets were reextracted as previously described. The second supernatant was combined with the first one and dried down. The pellets were dissolved in water for LC-MS/MS analysis.
The LC-MS/MS system used is composed of a Shimadzu LC system with two Shimadzu solvent delivery pumps (model LC10AD), a Shimadzu integrated controller (SCL10Avp), a Valco two-position diverter valve, and a LEAP CTC PAL autosampler with a 50-μl sample loop. This LC system is interfaced with an AB Sciex 4000 QTRAP mass spectrometer equipped with a TurboIonSpray (TIS) electrospray ion source. Source parameters were set as the following: curtain gas, 10 arbitrary units (AU); source gas 1, 50 AU; source gas 2, 50 AU; collision activated dissociation, high; interface heater, on; temperature, 650°C; ionspray voltage, −4,500. Both quadruples (Q1 and Q3) were set to unit resolution. Analyst software (version 1.4.2) was used to control sample acquisition and data analysis. The 4000 QTRAP mass spectrometer was tuned and calibrated according to the manufacturer's recommendations. Each compound was detected using 3 multiple reaction monitoring (MRM) transitions that were previously optimized using standards described in Table 1, with the exception of pppGpp, for which no commercial standard is available. Only 2 MRM transitions for pppGpp were used based on the ppGpp standard.
Table 1.
Mass spectrometry of nucleotide standardsa
| Q1 MS | Q3 MS | Time (ms) | Name | DP (V) | CE (V) |
|---|---|---|---|---|---|
| 506.2 | 408.0 | 50 | ATP1 | −70 | −30 |
| 506.2 | 158.8 | 50 | ATP2 | −70 | −35 |
| 506.2 | 78.8 | 50 | ATP3 | −70 | −80 |
| 522.2 | 423.9 | 50 | GTP1 | −70 | −30 |
| 522.2 | 158.8 | 50 | GTP2 | −70 | −40 |
| 522.2 | 78.8 | 50 | GTP3 | −70 | −80 |
| 602.2 | 504.0 | 50 | ppGpp1 | −70 | −35 |
| 602.2 | 423.9 | 50 | ppGpp2 | −70 | −45 |
| 602.2 | 158.8 | 50 | ppGpp3 | −70 | −60 |
| 682.2 | 584.0 | 50 | pppGpp1 | −70 | −35 |
| 682.2 | 504.0 | 50 | pppGpp2 | −70 | −45 |
| 586.0 | 273.0 | 50 | Br-ATP1 | −55 | −60 |
| 586.0 | 159.0 | 50 | Br-ATP2 | −55 | −50 |
| 586.0 | 79.0 | 50 | Br-ATP3 | −55 | −95 |
DP, declustering potential; CE, collision energy.
For LC separation, a Synergi 2.5-U Hydro-RP 100A column (100 by 2 mm; Phenomenex) was used at a flow rate of 0.11 ml/min. The gradient started at 100% solvent A (10 mM tributylamine in 5% methanol, pH 4.8), was held for 2 min, and then moved to 20% solvent B (100% methanol) in 2 min. The gradient then increased to 50% solvent B in 33 min. It was finally ramped up to 90% solvent B in 1 min and then ramped back to initial conditions (100% solvent A) in 1 min and reequilibrated for an additional 7 min. For quantification, a series of standard samples containing different concentrations of compounds was prepared. Calibration curves for all of the compounds were achieved by integration of the peak area under the curve of the standards and then normalized to the internal standard Br-ATP.
RESULTS
RelMtb-mediated (p)ppGpp synthesis is required for WT rates of growth on solid media and formation of biofilms.
Deletion of M. tuberculosis relMtb results in slightly slower growth, compromised long-term survival in cultures (7), and an inability to persist in mouse models of infection (14, 15). relMtb encodes two different enzymatic activities, (p)ppGpp synthesis and hydrolysis, and it is unknown if the phenotypes of the ΔrelMtb strain result exclusively from the loss of (p)ppGpp production. A RelMtbH344Y point mutant is unable to synthesize (p)ppGpp but retains hydrolase activity in vitro (9). To determine whether RelMtb-mediated (p)ppGpp synthesis is specifically important for mycobacteria, we constructed M. tuberculosis strains deleted for the endogenous relMtb gene and constitutively expressing either the relMtbWT allele or the relMtbH344Y synthetase-dead mutant from the attB site of the genome. As previously reported, the M. tuberculosis ΔrelMtb mutant grew more slowly than a strain complemented with the relMtbWT allele, which is particularly evident on 7H10 solid media (Fig. 1A) (7). Expression of the RelMtbH344Y mutant did not fully complement this growth defect, demonstrating that (p)ppGpp synthesis by RelMtb is necessary for optimal growth on solid media. A lower growth rate was defined by smaller colony size on 7H10 plates, as illustrated in Fig. 1A at the 10−3 and 10−4 dilutions. In addition to the slow growth on solid media, we also observed that loss of RelMtb-mediated (p)ppGpp synthetase activity resulted in delayed formation of biofilms and pellicles in static liquid cultures (Fig. 1B). This illustrates another in vitro condition that requires RelMtb-mediated (p)ppGpp synthesis. Biofilms may serve as a model for persister cell formation, in that they harbor a larger number of drug-tolerant cells than planktonic M. tuberculosis cultures (22). Thus, this defect in biofilm formation by the ΔrelMtb mutant may be due to the interference of the same pathways necessary to persist in the mouse infection model.
Fig 1.
RelMtb-mediated (p)ppGpp synthesis is necessary for optimal M. tuberculosis growth and biofilm formation in vitro. M. tuberculosis ΔrelMtb strains containing a vector that constitutively expresses RelMtbWT (WT) or RelMtbH344Y (H344Y) or containing an empty vector (−) integrated at the attB site in the genome were normalized to the same ODλ600 and either diluted and plated on 7H10 plates (A) or grown in Sauton's medium in static, biofilm-forming conditions in a 24-well dish (B). (A) 7H10 plates were incubated for 3 weeks at 37°C. (B) Biofilms were incubated for 4 weeks at 37°C. The parent strain used is labeled above each photo, and the protein expressed from the attB site is labeled below the photo.
(p)ppGpp synthesis by RelMtb is required for chronic infection in mice.
M. tuberculosis infection in mice begins with an initial phase of unrestricted bacterial growth termed acute infection. The onset of adaptive immunity stops the bacterial titers from increasing at 3 to 4 weeks postinfection, but bacteria are still able to persist for the lifetime of the mouse. M. tuberculosis ΔrelMtb aerosol infection of mice results in normal initial bacterial growth but impaired chronic infection, resulting in less severe gross pathology of the disease (Fig. 2) (14). To determine whether RelMtb-mediated (p)ppGpp synthesis was specifically required for chronic infection, we infected C57BL/6 mice with M. tuberculosis ΔrelMtb expressing relMtbWT, relMtbH344Y, or no relMtb allele from the attB site. M. tuberculosis strains expressing the RelMtbH344Y mutant phenocopied the null mutant by not persisting in the murine lungs and spleens and causing less tissue pathology than control strains, demonstrating that RelMtb (p)ppGpp synthetase activity is required for chronic M. tuberculosis infection (Fig. 2).
Fig 2.
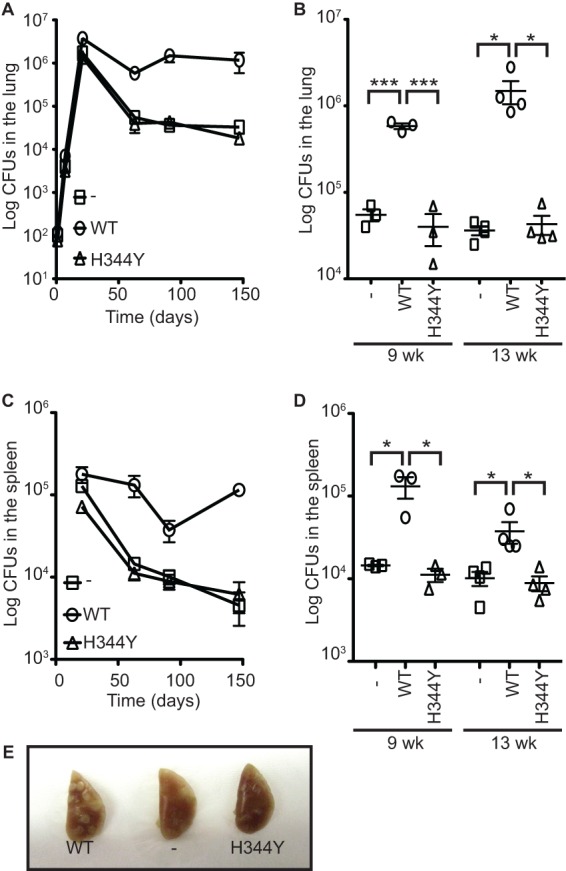
RelMtb-mediated (p)ppGpp synthesis is necessary for chronic M. tuberculosis infection in mice. C57BL/6 mice were infected with M. tuberculosis ΔrelMtb strains containing a vector that constitutively expresses RelMtbWT (WT; open circles) or RelMtbH344Y (H344Y; open triangles) or containing an empty vector (−; open squares) integrated at the attB site in the genome. Shown are bacterial titers from lungs (A and B) and spleens (C and D) at various time points. Each time point is the means ± standard errors of the means (SEM) of data from 3 to 4 mice per strain. (B and D) Significance of differences compared to the RelMtbWT-expressing strain at 9 and 13 weeks postinfection was determined by calculating P values by Student's t tests; one asterisk indicates significance with a P value of <0.05, and three asterisks indicates a significance with a P value of <0.005. (E) Gross pathology of representative left lungs from each strain at 13 weeks postinfection shows more lesions in the RelMtbWT-expressing strain.
Rv1366 activity does not contribute to phenotypes of a ΔrelMtb mutant and is dispensable for pathogenesis in the mouse model of infection.
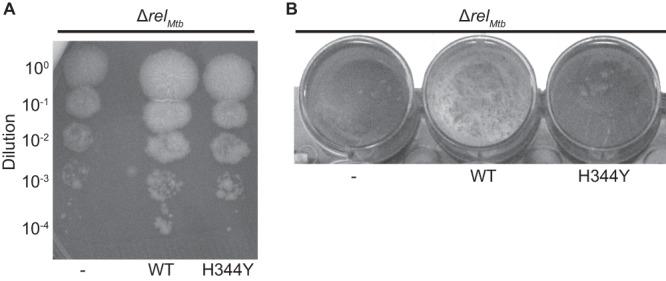
The SAS homolog Rv1366 is a second predicted (p)ppGpp synthetase in M. tuberculosis and could contribute to (p)ppGpp metabolism; however, it has yet to be characterized. To investigate the function of Rv1366, we constructed an M. tuberculosis strain deleted for the predicted (p)ppGpp synthetase rv1366 (Δrv1366 strain) and an M. tuberculosis double mutant of relMtb and rv1366 (ΔrelMtb Δrv1366 strain), which were both confirmed by Southern blot analysis (Fig. 3A and B), and monitored their growth rate and ability to form biofilms. The Δrv1366 mutant did not have a growth defect compared to controls on either solid media or under biofilm conditions (Fig. 3C and D). Additionally, the ΔrelMtb Δrv1366 double mutant phenocopied the ΔrelMtb single mutant on both solid media (data not shown) and under biofilm conditions (Fig. 3D). Expression of RelMtbWT was also able to fully complement the growth and biofilm phenotypes in the ΔrelMtb Δrv1366 double mutant (Fig. 3C and D). These data demonstrate that Rv1366 does not provide a function necessary for growth in nutrient-rich media or under biofilm conditions regardless of the presence of RelMtb-mediated (p)ppGpp synthesis. We then examined whether Rv1366 was necessary during M. tuberculosis infection of mice by infecting C57BL/6 mice with the Δrv1366 mutant and the ΔrelMtb Δrv1366 double mutant. The Δrv1366 single mutant was not attenuated during infection in either the lung or the spleen, and the ΔrelMtb Δrv1366 double mutant phenocopied the ΔrelMtb mutant and was fully complemented by expression of only RelMtbWT (Fig. 4). These data show that Rv1366 is not necessary for M. tuberculosis growth or survival in vitro or in vivo in the models tested.
Fig 3.
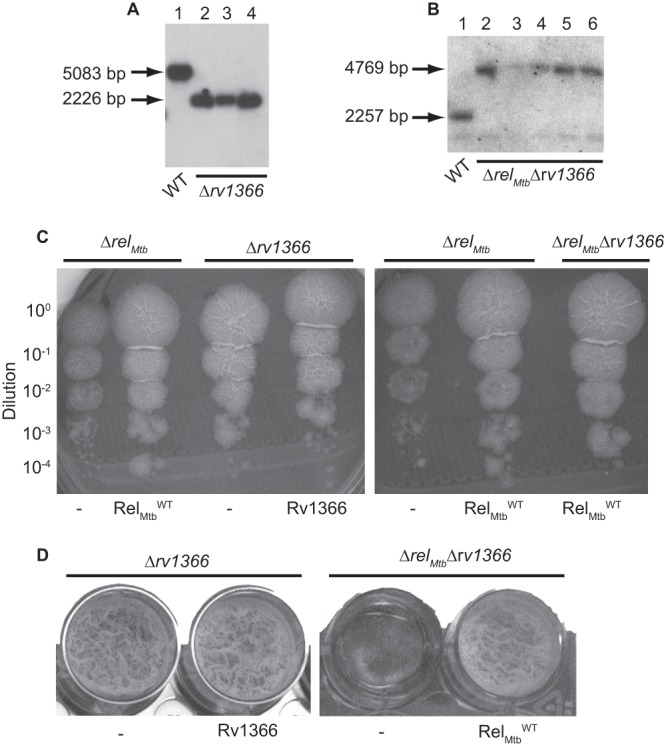
Rv1366 is not necessary for M. tuberculosis growth or biofilm formation in vitro. (A) Southern blot analysis of WT M. tuberculosis Erdman (lane 1) and the Δrv1366 strain (lanes 2 to 4) using EcoRI-digested genomic DNA and a probe that spans nucleotides 1537706 to 1538406 of the M. tuberculosis genome. EcoRI digestion of WT M. tuberculosis yields a band at 5,083 bp. EcoRI digestion of Δrv1366 results in a 2,226-bp band due to the replacement of the rv1366 gene with a hygromycin resistance cassette. (B) Southern blot analysis of WT M. tuberculosis Erdman (lane 1) and the ΔrelMtb Δrv1366 strain (lanes 2 to 6) using SmaI-digested genomic DNA and a probe that spans nucleotides 2910191 to 2910844 of the M. tuberculosis genome. SmaI digestion of WT M. tuberculosis yields a band at 2,257 bp. SmaI digestion of the ΔrelMtb Δrv1366 strain results in a 4,769-bp band due to the replacement of the relMtb gene with a hygromycin resistance cassette. (C and D) M. tuberculosis ΔrelMtb, Δrv1366, and ΔrelMtbΔrv1366 strains containing a vector that constitutively expresses RelMtbWT or Rv1366 or containing an empty vector (−) integrated at the attB site in the genome were normalized to the same ODλ600 and either diluted and plated on 7H10 plates (C) or grown in Sauton's media in static, biofilm-forming conditions in a 24-well dish (D). (C) 7H10 plates were incubated for 3 weeks at 37°C. (D) Biofilms were incubated for 4 weeks at 37°C. The parent strain used is labeled above each photo, and the protein expressed from the attB site is labeled below the photo.
Fig 4.
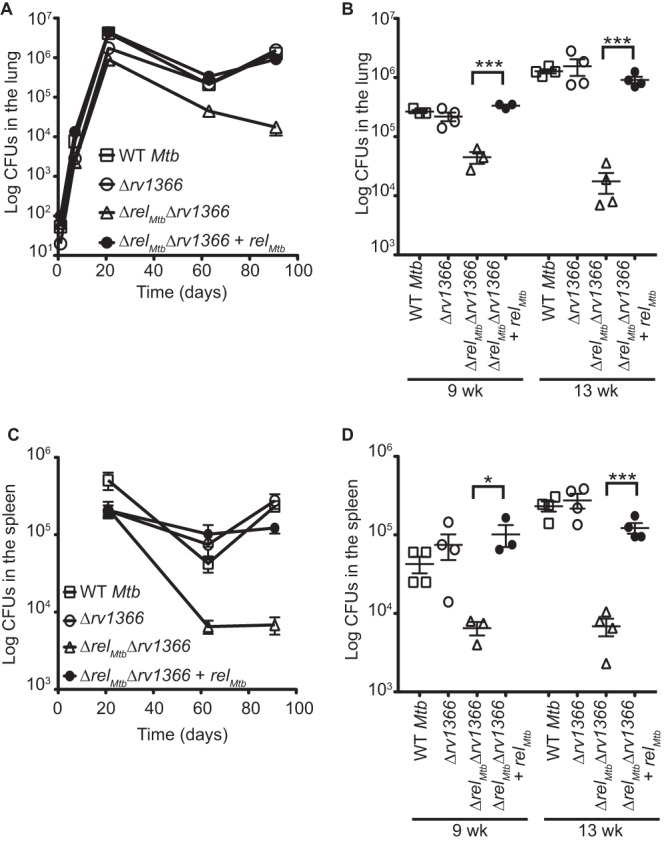
Rv1366 is not necessary for M. tuberculosis pathogenesis in mice. C57BL/6 mice were infected with the WT M. tuberculosis Erdman strain (open squares), the Δrv1366 strain (open circles), the ΔrelMtb Δrv1366 strain containing an empty vector integrated at the attB site in the genome (ΔrelMtb Δrv1366; open triangles), or the ΔrelMtb Δrv1366 strain containing a vector that constitutively expresses RelMtbWT from the attB site in the genome (ΔrelMtb Δrv1366 + relMtb; closed circles). Shown are bacterial titers from lungs (A and B) and spleens (C and D) at various time points. Each time point is the means ± SEM of data from 3 to 4 mice per strain. (B and D) Significance of differences compared to the ΔrelMtb Δrv1366 strain at 9 and 13 weeks postinfection was determined by calculating P values by Student's t tests; one asterisk indicates significance with a P value of <0.05, and three asterisks indicates significance with a P value of <0.005.
In Bacillus subtilis, the phenotypes of the ΔrelA strain are attributed in part to the absence of (p)ppGpp hydrolase activity in the presence of two SAS homologs, such that suppressors of the B. subtilis ΔrelA mutant have inactivating mutations in the SAS genes (17). Therefore, we wanted to confirm that the M. tuberculosis rv1366 gene was intact in the ΔrelMtb strain. We sequenced rv1366 along with 159 nucleotides upstream and 182 nucleotides downstream of the rv1366 gene in WT M. tuberculosis and ΔrelMtb strains. The nucleotide sequences were identical between strains and were the same as the sequence deposited in NCBI for the M. tuberculosis H37Rv strain. Therefore, the role of Rv1366 in M. tuberculosis remains unknown, but its function is not redundant with (p)ppGpp production by RelMtb under the conditions examined.
RelMtb hydrolase activity is critical for mycobacterial physiology when (p)ppGpp synthesis is intact.
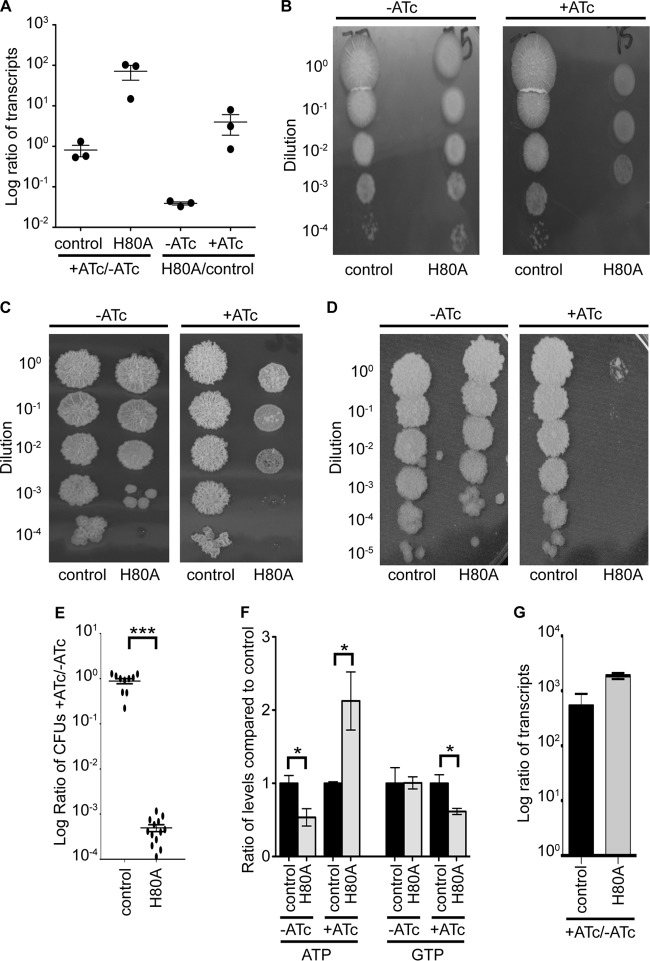
In addition to (p)ppGpp synthesis, RelMtb is the only known enzyme in M. tuberculosis that is capable of hydrolyzing (p)ppGpp for turnover and renewal of GTP and GDP by removal of pyrophosphate. In order to investigate the roles of the RelMtb hydrolase activity during growth and survival, we attempted to generate strains of M. tuberculosis expressing a single copy of RelMtb that was hydrolase dead but retained (p)ppGpp synthetase activity (RelMtbH80A) or was inactive for both synthesis and hydrolysis (RelMtbH80A-H344Y). Although we were able to obtain the M. tuberculosis RelMtbH80A-H344Y-expressing strain, we did not recover clones expressing the relMtbH80A allele. This suggested that the retention of the RelMtb (p)ppGpp synthetase activity is toxic in the absence of a functional hydrolase. To investigate this further, we used a tetracycline-inducible expression system (24) and engineered an M. tuberculosis ΔrelMtb strain to induce expression of the relMtbH80A allele from a cassette at the attB site in the presence of tetracycline analogs, such as anhydrotetracycline (ATc), due to the expression of a tetracycline repressor (TetR) from an episomal plasmid. We named this strain Tet-RelMtbH80A. We also constructed a control strain by inserting an empty vector into the attB site of WT M. tuberculosis and transforming this strain with the same TetR-expressing plasmid as that in Tet-RelMtbH80A. relMtb expression from its endogenous locus in the control strain is not affected by ATc (Fig. 5A). Induction of relMtbH80A transcription in the presence of ATc in the Tet-RelMtbH80A strain was confirmed by quantitative real-time PCR (qRT-PCR) (Fig. 5A).
Fig 5.
RelMtb-mediated (p)ppGpp hydrolysis is required for growth in vitro and maintenance of ATP and GTP levels when (p)ppGpp is being produced. (A and B) An M. tuberculosis ΔrelMtb strain containing an episomal vector expressing the WT TetR that induces expression of relMtbH80A from a cassette integrated at the attB site in the genome in the presence of ATc (Tet-RelMtbH80A strain, designated H80A here) and a WT M. tuberculosis strain containing an empty vector in the attB site and transformed with the same TetR-expressing plasmid (control strain). (A) Transcript levels in exponential-growth-phase cultures in liquid 7H9 media. relMtbH80A transcripts from the cassette at the attB site were detected in the Tet-RelMtbH80A strain, and transcripts from the endogenous relMtb gene were detected in the control strain. The first comparison shows the ratio of transcript levels in the presence compared to the absence of ATc in each strain (+ATc/−ATc), and it illustrates the induction of relMtbH80A in the Tet-RelMtbH80A strain when exposed to ATc and the unresponsiveness of the relMtb gene to ATc in the control strain. The second comparison shows the ratio of transcript levels in the Tet-RelMtbH80A strain compared to the control strain (H80A/control) under each condition to illustrate the low level of transcription of relMtbH80A in the absence of ATc compared to that of induced cultures. (B) M. tuberculosis Tet-RelMtbH80A and control strains were diluted and plated on 7H10 plates in the absence or presence of ATc. (C to G) M. smegmatis ΔrelMsm strains containing an episomal vector expressing the WT TetR that turns on expression of relMtbWT (Tet-RelMtbWT strain; designated the control) or relMtbH80A (Tet-RelMtbH80A strain; designated H80A here) from cassettes integrated at the attB site in the genome in the presence of ATc. (C and D) M. smegmatis strains were diluted and plated on 7H10 (C) and LB (D) plates in the absence or presence of ATc. (E) Graphic representation of the number of M. smegmatis CFU that grew in the presence compared to the absence of ATc when grown on LB, where all Tet-RelMtbH80A strain bacteria that grew in the presence of ATc on LB were suppressors and were no longer responsive to ATc treatment. Data are means ± SEM from 13 replicates. (F) ATP and GTP levels in M. smegmatis strains grown in the absence or presence of ATc for 6 h to an ODλ600 of ∼0.6 in LB liquid media were measured by LC-MS/MS. The ratio of levels in the Tet-RelMtbH80A strain (H80A) to those in the Tet-RelMtbWT-expressing strain (control) under the same conditions is graphed. Data are the means ± SEM from 4 to 6 replicates. (E and F) The significance of differences was determined by calculating P values by Student's t tests; one asterisk indicates significance with a P value of <0.05, and three asterisks indicates a significance with a P value of <0.005. (G) Log ratio of transcript levels in the presence of ATc compared to the absence of ATc, illustrating the induction of relMtbWT and relMtbH80A transcripts within 6 h of ATc treatment in Tet-RelMtbWT and Tet-RelMtbH80A strains, respectively. Data are the means ± SEM from 3 replicates.
We plated the M. tuberculosis control and Tet-RelMtbH80A strains onto 7H10 agar in the absence or presence of ATc and found that although the strains had equivalent survival rates under both conditions, the Tet-RelMtbH80A strain exhibited smaller, rounder, and smoother colonies than control bacteria in the presence of ATc (Fig. 5B). This demonstrates that inducing expression of RelMtbH80A, which will synthesize (p)ppGpp but will not hydrolyze (p)ppGpp, has an impact on M. tuberculosis physiology. Thus, (p)ppGpp turnover is required to maintain normal colony morphology. There was a mild effect on colony morphology in the Tet-RelMtbH80A strain in the absence of ATc (Fig. 5B), suggesting that even low levels of relMtbH80A expression in the absence of inducer due to the inherent leakiness of the TetR system was enough to affect bacterial physiology. This low level of relMtbH80A transcription in the Tet-RelMtbH80A strain in the absence of ATc was detectable by qRT-PCR and was close to 100-fold lower than in induced conditions (Fig. 5A).
We also engineered M. smegmatis ΔrelMsm strains to induce expression of either the WT relMtb allele (Tet-RelMtbWT) or the relMtbH80A allele (Tet-RelMtbH80A) in the presence of ATc. Similar to the results from M. tuberculosis experiments, when we plated M. smegmatis Tet-RelMtbWT and Tet-RelMtbH80A on 7H10 agar in the presence of ATc, the Tet-RelMtbH80A strain exhibited smaller, smoother colonies than the Tet-RelMtbWT strain (Fig. 5C). This demonstrated that the role of (p)ppGpp turnover in colony morphology is conserved in M. tuberculosis and M. smegmatis. Interestingly, when we plated the M. smegmatis Tet-RelMtbWT and Tet-RelMtbH80A strains on LB agar in the absence or presence of ATc, the induced expression of the relMtbH80A mutant allele inhibited growth (Fig. 5D and E) and all of the Tet-RelMtbH80A colonies that grew were no longer responsive to the presence of ATc, as determined by plating and qRT-PCR. Therefore, in order to grow on nutrient-rich LB media, the M. smegmatis Tet-RelMtbH80A strain must mutate the Tet-inducible system and permanently repress the expression of relMtbH80A to survive. This proved that inhibition of the RelMtb (p)ppGpp hydrolase activity in the presence of a functional (p)ppGpp synthetase is toxic to M. smegmatis in nutrient-rich LB media and links nutrient availability with the importance of (p)ppGpp turnover. (p)ppGpp production has been associated with colony morphology in mycobacteria (16, 25), but this is the first time in mycobacteria that the hydrolysis of (p)ppGpp has been shown to be required for maintenance of WT colony morphology and growth in nutrient-rich conditions.
(p)ppGpp levels were measured in the M. smegmatis Tet-RelMtbWT and Tet-RelMtbH80A strains by liquid chromatography-tandem mass spectrometry (LC-MS/MS), but because of the low levels of (p)ppGpp in the samples, the measurements observed were not confidently reproduced. However, the same LC-MS/MS approach was used to monitor ATP and GTP levels in LB liquid cultures of M. smegmatis Tet-RelMtbWT and Tet-RelMtbH80A in the presence and absence of ATc (Fig. 5F). We found that following induction of relMtbH80A expression in M. smegmatis for 6 h, GTP levels were significantly decreased compared to those of strains expressing RelMtbWT. Conversely, ATP levels during induction of relMtbH80A expression were actually higher than those of control strains. The increased transcription of the relMtbH80A gene within 6 h of ATc treatment in the M. smegmatis Tet-RelMtbH80A strain was confirmed by qRT-PCR (Fig. 5G). These data demonstrate that inhibition of (p)ppGpp hydrolysis impacts not only (p)ppGpp levels but also ATP and GTP levels within the bacteria.
Inhibition of RelMtb (p)ppGpp hydrolase activity is lethal for acute and chronic M. tuberculosis infection in mice.
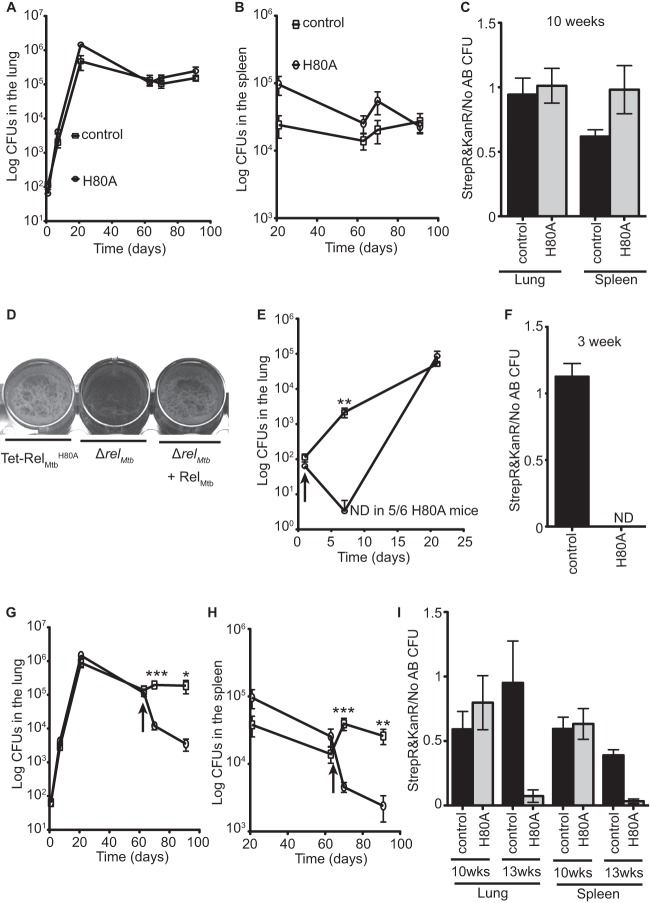
We next investigated whether inhibition of (p)ppGpp hydrolysis, while keeping (p)ppGpp synthesis intact, would compromise M. tuberculosis pathogenesis. We tested this by infecting mice with either the control M. tuberculosis strain that expresses relMtb from its native locus or the Tet-RelMtbH80A strain that allows for conditional expression of the relMtbH80A allele in the presence of doxycycline. We then determined bacterial titers in the lungs and spleens during both acute and chronic infection (Fig. 6). In the absence of doxycycline, control and Tet-RelMtbH80A M. tuberculosis strains displayed similar kinetics and virulence in mice (Fig. 6A and B). Since both strains express TetR from a plasmid that also confers streptomycin resistance and contain a kanamycin resistant cassette integrated into the attB site that, in the case of Tet-RelMtbH80A, carries the relMtbH80A allele, we could monitor the retention of the Tet-inducible system during infection. By plating the lung and spleen homogenates on plates containing streptomycin and kanamycin, we showed that the Tet-inducible system was intact for at least 10 weeks postinfection in both strains in the absence of doxycycline (Fig. 6C). It was surprising that the Tet-RelMtbH80A strain was able to maintain high titers during chronic infection, since the levels of relMtbH80A transcripts were a log lower than those in the control strain (Fig. 5A), and we have shown that the absence of relMtb expression leads to lower titers during chronic infection (Fig. 2). This indicated that the low-level leaky expression of the relMtbH80A allele in the absence of doxycycline was able to result in enough (p)ppGpp synthesis to phenocopy WT M. tuberculosis instead of the ΔrelMtb strain (Fig. 2) but not too much (p)ppGpp synthesis to confer toxicity in the absence of a functional hydrolase. In order to confirm that the low levels of relMtbH80A transcription allowed for synthesis of (p)ppGpp in the Tet-RelMtbH80A strain in the absence of inducer, we analyzed the ability of this strain to form biofilms, which is a phenotype we had previously shown requires (p)ppGpp synthesis (Fig. 1B). In the absence of ATc, when only low levels of relMtbH80A transcription are occurring, the Tet-RelMtbH80A strain was able to form biofilms similar to those observed in WT RelMtb-expressing strains, further supporting that the low level of relMtbH80A transcription in the absence of inducer is enough to allow for adequate (p)ppGpp synthesis to confer normal bacterial physiology (Fig. 6D).
Fig 6.
RelMtb (p)ppGpp hydrolase activity is essential for acute and chronic M. tuberculosis infection of mice. Bacterial titers in the lungs (A, E, and G) and spleens (B and H) of C57BL/6 mice infected with either control or Tet-RelMtbH80A (designated H80A in the figure) M. tuberculosis strains. The parent strain for the control is WT M. tuberculosis Erdman, and the parent strain for Tet-RelMtbH80A is M. tuberculosis ΔrelMtb. Both strains express TetR from a plasmid that confers streptomycin resistance and contain a kanamycin resistance cassette integrated into the attB site that, in the case of Tet-RelMtbH80A, carries the relMtbH80A allele. The strain symbols in panel A are the same for all panels. (A to C) Mice were given normal mouse chow throughout infection. (E and F) Mice were administered doxycycline-containing mouse chow starting at day 1 postinfection (designated by the arrow). (G to I) Mice were administered doxycycline-containing mouse chow starting at day 63 postinfection (designated by the arrow). (C, F, and I) The ratio of CFU from the lungs or spleens grown on 7H10 plates containing streptomycin and kanamycin compared to 7H10 containing no antibiotics (No AB). ND denotes when no colonies were recovered after plating 5% of the lung homogenate (limit of detection, 20 CFU). Data are means ± SEM of 6 Tet-RelMtbH80A strain-infected mice and 3 control strain-infected mice per time point from two replicate experiments. (E, G, and H) The significance of differences were determined by calculating P values by Student's t tests; two asterisks indicate significance with a P value of <0.01, and three asterisks indicate significance with a P value of <0.005. (D) M. tuberculosis Tet-RelMtbH80A, ΔrelMtb, and ΔrelMtb complemented with WT relMtb were normalized to the same ODλ600 and grown in Sauton's media under static, biofilm-forming conditions in a 24-well dish. Biofilms were incubated for 4 weeks at 37°C.
Separate groups of mice were infected with either control or Tet-RelMtbH80A M. tuberculosis strains and administered doxycycline starting on day 1 (acute infection) (Fig. 6E and F) or day 63 (chronic infection) (Fig. 6G to I) postinfection to determine the effect of inhibiting RelMtb (p)ppGpp hydrolase activity during acute and chronic infection in mice. After 1 week of inducing RelMtbH80A expression with doxycycline at day 1 postinfection in Tet-RelMtbH80A, the bacteria were unable to replicate and decreased in titers to the point that 5 out of 6 animals tested had no detectable M. tuberculosis in the lungs, with a limit of detection of 20 CFU. Additionally, none of the surviving Tet-RelMtbH80A cells at day 21 retained the Tet-inducible expression system, as determined by the lack of colony growth from lung homogenates on plates containing streptomycin and kanamycin (Fig. 6F). In contrast, the control strain that expressed the endogenous relMtb allele was not affected by doxycycline treatment at this stage of infection (Fig. 6E) and retained the Tet-inducible expression system (Fig. 6F). These data demonstrate that even though M. tuberculosis was capable of growth on 7H10 media during inhibition of RelMtb (p)ppGpp hydrolase activity (Fig. 5B), this catalytic activity is essential for acute infection in mice.
During chronic infection, expression of RelMtbH80A caused a 2-log decrease of titers in the lungs (Fig. 6G) and a 1-log decrease in the spleens (Fig. 6H) over a 4-week period. In the 4 weeks following induction of RelMtbH80A expression during chronic infection, 93% of the Tet-RelMtbH80A bacteria lost the plasmids encoding the Tet-inducible system and most likely reverted to relMtb null by losing the allele and linked kanamycin resistant cassette at the attB site (Fig. 6I). This would explain why the bacterial titers did not recover after loss of the Tet-inducible system, since an M. tuberculosis ΔrelMtb strain is attenuated for chronic infection (Fig. 2 and 6G and H). These data demonstrate that inhibition of the RelMtb (p)ppGpp hydrolase activity in both acute and chronic infection compromises M. tuberculosis survival and presents the RelMtb hydrolase domain as a promising drug target. In addition, suppression of the toxic effect of inhibiting the RelMtb (p)ppGpp hydrolase activity by losing RelMtb-mediated (p)ppGpp synthetase activity still leaves the bacteria unable to persist in the mouse model of infection.
DISCUSSION
A previous study reported that an M. tuberculosis ΔrelMtb mutant is attenuated for chronic infection in mice (14). In the same study, microarray analyses revealed that more than a quarter of the genome was differentially expressed in the ΔrelMtb strain in both nutrient-rich and depleted conditions, suggesting that there are extensive metabolic alterations in strains lacking relMtb. The transcripts affected included virulence factors, cell wall biosynthetic enzymes, heat shock proteins, and secreted antigens, any of which may contribute to the attenuation in vivo (14). In addition, WT M. tuberculosis, but not the ΔrelMtb mutant, downregulated the cellular translation machinery during nutrient starvation, which is consistent with RelMtb's role in the stringent response and may also be necessary for M. tuberculosis virulence (14).
RelMtb is a large protein encoding two distinct catalytic activities. Despite its importance for pathogenesis, it was still unknown what function(s) of RelMtb contribute to its role in vivo. We have dissected the roles of the two enzymatic activities encoded by RelMtb during both growth in culture and pathogenesis in mice. By studying a point mutant that specifically abolishes (p)ppGpp synthesis by RelMtb, we conclude that (p)ppGpp production by RelMtb is required for efficient rates of growth and biofilm formation in culture, as well as for maintaining titers during chronic infection in the mouse model of infection. It is worth noting that we do not have an antibody specific for RelMtb; therefore, we cannot confirm that the RelMtbH344Y protein is stably expressed and that the phenotypes are not due to decreased RelMtb protein rather than loss of synthetase activity. However, there are multiple lines of evidence that support that it is the loss of synthetase activity, and not protein instability, that results in these phenotypes. First, our group and others are able to purify the mutant protein easily, and it is stable in vitro with no loss in hydrolase activity, suggesting that the protein can fold normally and other domains can function in the presence of the H344Y substitution (9 and data not shown). Second, data presented here indicate that lower levels of RelMtb would not phenocopy complete loss of the relMtb allele. For instance, infection of mice with the Tet-RelMtbH80A strain in the absence of doxycycline, where relMtbH80A transcript levels are a log lower than transcripts from the allele in WT M. tuberculosis (Fig. 5A), indicated that even this low level of synthetase-active RelMtb expression is able to sustain WT titers during chronic infection of mice, unlike the ΔrelMtb- and RelMtbH344Y-expressing strains (Fig. 2 and 6).
(p)ppGpp has been shown to directly impact many processes in other bacteria, including transcription of the translation machinery (26–29), GTP and ATP levels (30–33), DNA replication (34–37), and metabolism (38–40). In general, the many effects of (p)ppGpp are complex and seem to vary greatly among different organisms. Any number of these mechanisms may be conserved in mycobacteria, and determining which functions of (p)ppGpp make its production necessary for chronic M. tuberculosis infection requires further study. Since it has been shown that (p)ppGpp accumulates in mycobacteria during starvation (7, 23, 25), oxidative stress (23), and stationary phase (7), these would be good conditions to begin to investigate a molecular mechanism for (p)ppGpp in M. tuberculosis.
Our experiments also demonstrate that the production of (p)ppGpp is not the only role of RelMtb during pathogenesis. By expressing a relMtb point mutant that retains the ability to synthesize (p)ppGpp but is unable to hydrolyze (p)ppGpp, our experiments have highlighted a critical role for RelMtb-mediated hydrolysis of (p)ppGpp in colony morphology, survival, and virulence. In contrast to (p)ppGpp synthesis, which is necessary for chronic but not acute infection, hydrolysis of (p)ppGpp is required for all stages of M. tuberculosis infection in mice, suggesting a role for this catalytic activity in general homeostasis. We predict that expression of the RelMtbH80A hydrolase mutant leads to increased abundance of (p)ppGpp, which may directly impact cellular pathways in an uncontrolled manner that is detrimental for the bacteria. In addition, we identify a dysregulation of ATP and GTP levels during relMtbH80A expression, which also may contribute to the lethality of expressing this allele. A recently published study on B. subtilis highlighted a role for (p)ppGpp in directly inhibiting GTP biosynthesis to maintain GTP levels within a range that supports viability (33). To investigate whether the phenotypes caused by relMtbH80A expression in M. tuberculosis are due to the low levels of GTP, we attempted to increase GTP production by adding guanosine to the media, as was previously done in B. subtilis (33). Unfortunately, guanosine supplementation did not restore WT GTP levels, colony morphology, or growth rate during RelMtbH80A expression in mycobacteria. However, this may be due to inability of the nucleoside to access the mycobacterial cytosol.
Evidence of the importance of controlling (p)ppGpp levels specifically by hydrolysis activity comes from studies of small alarmone synthetase (SAS) homologs in B. subtilis (17, 18), Streptomyces mutans (19), Enterococcus faecalis (20), and Vibrio cholerae (21). SAS proteins encode the catalytic domain to produce (p)ppGpp but not the domain to hydrolyze it. Expression of a V. cholerae or S. mutans SAS homolog in E. coli is toxic in the absence of SpoT-mediated (p)ppGpp hydrolysis (19, 21), supporting the theory that production of (p)ppGpp in the absence of a functional hydrolase is deleterious. In addition, deletion of the respective SAS homologs largely abolishes the slow-growth phenotypes of ΔrelA strains of S. mutans, E. faecalis, and B. subtilis (17–20), which, like M. tuberculosis ΔrelMtb, lack the only known enzyme capable of hydrolyzing (p)ppGpp. These data suggest that in S. mutans, E. faecalis, and B. subtilis, the slow growth of the ΔrelA mutant is due to the lack of (p)ppGpp hydrolase activity in the presence of (p)ppGpp synthesis.
With these data in mind, we have also reported the first investigations into the M. tuberculosis SAS homolog, rv1366. Interestingly, another important finding from our experiments is that a Δrv1366 strain exhibited WT rates of growth in liquid shaking cultures, on plates, and in biofilms as well as WT levels of virulence in the mouse model of infection. This indicates that under the conditions we tested and during pathogenesis in the mouse model, RelMtb is the main producer of (p)ppGpp. Deletion of rv1366 had no effect on the phenotypes of the ΔrelMtb strain, demonstrating that, unlike in other bacteria, the presence of an intact M. tuberculosis SAS homolog allele was not a source of toxicity in strains lacking RelMtb-mediated (p)ppGpp hydrolysis. Therefore, the question regarding the role of Rv1366 in M. tuberculosis remains unanswered. Murdeshwar et al. recently reported a study of the M. smegmatis SAS homolog, which they term MS_RHII-RSD because, unlike the M. tuberculosis homolog, MS_RHII-RSD encodes an amino-terminal RNase HII domain (41). The authors show that a small amount of (p)ppGpp is produced in an M. smegmatis ΔrelMsm mutant that is dependent on MS_RHII-RSD; however, the importance of this activity for survival has not been explored. Interestingly, the authors were unable to detect (p)ppGpp synthetase activity in vitro using the region of MS_RHII-RSD with homology to Rv1366, raising the possibility that MS_RHII-RSD has evolved functions distinct from those of Rv1366.
In summary, our data point toward a Goldilocks model of (p)ppGpp levels in mycobacteria, where too little (p)ppGpp is harmful for the bacteria in certain situations, like chronic infection, and too much is always deleterious. Recent antibiotic development efforts aim to target RelMtb (p)ppGpp synthetase activity as a therapeutic option (42, 43). We propose that tipping the balance of (p)ppGpp levels in the other direction by inhibiting (p)ppGpp hydrolysis is also an attractive approach to targeting M. tuberculosis. Our data imply that if we were to inhibit (p)ppGpp hydrolysis with an antibiotic, mutants that develop resistance by abolishing (p)ppGpp production would be unable to persist in the host. Our findings would also apply to other bacteria that require a balance of (p)ppGpp production.
ACKNOWLEDGMENTS
C.L.S. is supported by an Interdisciplinary Research Initiative grant from the Children's Discovery Institute of Washington University and St. Louis Children's Hospital.
We thank the other members of the Stallings laboratory for careful reading of the manuscript and Jessica Schneider for assistance with mouse harvests. The LC-MS/MS experiments were performed by Sophie Alvarez and Leslie Hicks at the Proteomics and Mass Spectrometry Facility at the Donald Danforth Plant Sciences Center.
Footnotes
Published ahead of print 11 October 2013
REFERENCES
- 1.WHO 2012. Global tuberculosis report 2012. WHO, Geneva, Switzerland [Google Scholar]
- 2.Stallings CL, Glickman MS. 2010. Is Mycobacterium tuberculosis stressed out? A critical assessment of the genetic evidence. Microbes Infect. 12:1091–1101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Atkinson GC, Tenson T, Hauryliuk V. 2011. The RelA/SpoT homolog (RSH) superfamily: distribution and functional evolution of ppGpp synthetases and hydrolases across the tree of life. PLoS One 6:e23479. 10.1371/journal.pone.0023479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Haseltine WA, Block R. 1973. Synthesis of guanosine tetra- and pentaphosphate requires the presence of a codon-specific, uncharged transfer ribonucleic acid in the acceptor site of ribosomes. Proc. Natl. Acad. Sci. U. S. A. 70:1564–1568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dalebroux ZD, Swanson MS. 2012. ppGpp: magic beyond RNA polymerase. Nat. Rev. Microbiol. 10:203–212 [DOI] [PubMed] [Google Scholar]
- 6.Potrykus K, Cashel M. 2008. (p)ppGpp: still magical? Annu. Rev. Microbiol. 62:35–51 [DOI] [PubMed] [Google Scholar]
- 7.Primm TP, Andersen SJ, Mizrahi V, Avarbock D, Rubin H, Barry CE., III 2000. The stringent response of Mycobacterium tuberculosis is required for long-term survival. J. Bacteriol. 182:4889–4898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gropp M, Strausz Y, Gross M, Glaser G. 2001. Regulation of Escherichia coli RelA requires oligomerization of the C-terminal domain. J. Bacteriol. 183:570–579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Avarbock A, Avarbock D, Teh JS, Buckstein M, Wang ZM, Rubin H. 2005. Functional regulation of the opposing (p) ppGpp synthetase/hydrolase activities of RelMtb from Mycobacterium tuberculosis. Biochemistry 44:9913–9923 [DOI] [PubMed] [Google Scholar]
- 10.Avarbock D, Salem J, Li LS, Wang ZM, Rubin H. 1999. Cloning and characterization of a bifunctional RelA/SpoT homologue from Mycobacterium tuberculosis. Gene 233:261–269 [DOI] [PubMed] [Google Scholar]
- 11.Jain V, Saleem-Batcha R, China A, Chatterji D. 2006. Molecular dissection of the mycobacterial stringent response protein Rel. Protein Sci. 15:1449–1464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aravind L, Koonin EV. 1998. The HD domain defines a new superfamily of metal-dependent phosphohydrolases. Trends Biochem. Sci. 23:469–472 [DOI] [PubMed] [Google Scholar]
- 13.Hogg T, Mechold U, Malke H, Cashel M, Hilgenfeld R. 2004. Conformational antagonism between opposing active sites in a bifunctional RelA/SpoT homolog modulates (p) ppGpp metabolism during the stringent response. Cell 117:57–68 [DOI] [PubMed] [Google Scholar]
- 14.Dahl JL, Kraus CN, Boshoff HI, Doan B, Foley K, Avarbock D, Kaplan G, Mizrahi V, Rubin H, Barry CE., III 2003. The role of RelMtb-mediated adaptation to stationary phase in long-term persistence of Mycobacterium tuberculosis in mice. Proc. Natl. Acad. Sci. U. S. A. 100:10026–10031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Karakousis PC, Yoshimatsu T, Lamichhane G, Woolwine SC, Nuermberger EL, Grosset J, Bishai WR. 2004. Dormancy phenotype displayed by extracellular Mycobacterium tuberculosis within artificial granulomas in mice. J. Exp. Med. 200:647–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dahl JL, Arora K, Boshoff HI, Whiteford DC, Pacheco SA, Walsh OJ, Lau-Bonilla D, Davis WB, Garza AG. 2005. The relA homolog of Mycobacterium smegmatis affects cell appearance, viability, and gene expression. J. Bacteriol. 187:2439–2447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Natori Y, Tagami K, Murakami K, Yoshida S, Tanigawa O, Moh Y, Masuda K, Wada T, Suzuki S, Nanamiya H, Tozawa Y, Kawamura F. 2009. Transcription activity of individual rrn operons in Bacillus subtilis mutants deficient in (p) ppGpp synthetase genes, relA, yjbM, and ywaC. J. Bacteriol. 191:4555–4561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nanamiya H, Kasai K, Nozawa A, Yun CS, Narisawa T, Murakami K, Natori Y, Kawamura F, Tozawa Y. 2008. Identification and functional analysis of novel (p)ppGpp synthetase genes in Bacillus subtilis. Mol. Microbiol. 67:291–304 [DOI] [PubMed] [Google Scholar]
- 19.Lemos JA, Lin VK, Nascimento MM, Abranches J, Burne RA. 2007. Three gene products govern (p) ppGpp production by Streptococcus mutans. Mol. Microbiol. 65:1568–1581 [DOI] [PubMed] [Google Scholar]
- 20.Abranches J, Martinez AR, Kajfasz JK, Chavez V, Garsin DA, Lemos JA. 2009. The molecular alarmone (p)ppGpp mediates stress responses, vancomycin tolerance, and virulence in Enterococcus faecalis. J. Bacteriol. 191:2248–2256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Das B, Pal RR, Bag S, Bhadra RK. 2009. Stringent response in Vibrio cholerae: genetic analysis of spoT gene function and identification of a novel (p) ppGpp synthetase gene. Mol. Microbiol. 72:380–398 [DOI] [PubMed] [Google Scholar]
- 22.Ojha AK, Baughn AD, Sambandan D, Hsu T, Trivelli X, Guerardel Y, Alahari A, Kremer L, Jacobs WR, Jr, Hatfull GF. 2008. Growth of Mycobacterium tuberculosis biofilms containing free mycolic acids and harbouring drug-tolerant bacteria. Mol. Microbiol. 69:164–174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stallings CL, Stephanou NC, Chu L, Hochschild A, Nickels BE, Glickman MS. 2009. CarD is an essential regulator of rRNA transcription required for Mycobacterium tuberculosis persistence. Cell 138:146–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ehrt S, Guo XV, Hickey CM, Ryou M, Monteleone M, Riley LW, Schnappinger D. 2005. Controlling gene expression in mycobacteria with anhydrotetracycline and Tet repressor. Nucleic Acids Res. 33:e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ojha AK, Mukherjee TK, Chatterji D. 2000. High intracellular level of guanosine tetraphosphate in Mycobacterium smegmatis changes the morphology of the bacterium. Infect. Immun. 68:4084–4091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Artsimovitch I, Patlan V, Sekine S, Vassylyeva MN, Hosaka T, Ochi K, Yokoyama S, Vassylyev DG. 2004. Structural basis for transcription regulation by alarmone ppGpp. Cell 117:299–310 [DOI] [PubMed] [Google Scholar]
- 27.Barker MM, Gaal T, Josaitis CA, Gourse RL. 2001. Mechanism of regulation of transcription initiation by ppGpp. I. Effects of ppGpp on transcription initiation in vivo and in vitro. J. Mol. Biol. 305:673–688 [DOI] [PubMed] [Google Scholar]
- 28.Paul BJ, Barker MM, Ross W, Schneider DA, Webb C, Foster JW, Gourse RL. 2004. DksA: a critical component of the transcription initiation machinery that potentiates the regulation of rRNA promoters by ppGpp and the initiating NTP. Cell 118:311–322 [DOI] [PubMed] [Google Scholar]
- 29.Paul BJ, Berkmen MB, Gourse RL. 2005. DksA potentiates direct activation of amino acid promoters by ppGpp. Proc. Natl. Acad. Sci. U. S. A. 102:7823–7828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Krasny L, Tiserova H, Jonak J, Rejman D, Sanderova H. 2008. The identity of the transcription +1 position is crucial for changes in gene expression in response to amino acid starvation in Bacillus subtilis. Mol. Microbiol. 69:42–54 [DOI] [PubMed] [Google Scholar]
- 31.Lopez JM, Dromerick A, Freese E. 1981. Response of guanosine 5′-triphosphate concentration to nutritional changes and its significance for Bacillus subtilis sporulation. J. Bacteriol. 146:605–613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Krasny L, Gourse RL. 2004. An alternative strategy for bacterial ribosome synthesis: Bacillus subtilis rRNA transcription regulation. EMBO J. 23:4473–4483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kriel A, Bittner AN, Kim SH, Liu K, Tehranchi AK, Zou WY, Rendon S, Chen R, Tu BP, Wang JD. 2012. Direct regulation of GTP homeostasis by (p)ppGpp: a critical component of viability and stress resistance. Mol. Cell 48:231–241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang JD, Sanders GM, Grossman AD. 2007. Nutritional control of elongation of DNA replication by (p)ppGpp. Cell 128:865–875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maciag M, Kochanowska M, Lyzen R, Wegrzyn G, Szalewska-Palasz A. 2010. ppGpp inhibits the activity of Escherichia coli DnaG primase. Plasmid 63:61–67 [DOI] [PubMed] [Google Scholar]
- 36.Lesley JA, Shapiro L. 2008. SpoT regulates DnaA stability and initiation of DNA replication in carbon-starved Caulobacter crescentus. J. Bacteriol. 190:6867–6880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Boutte CC, Henry JT, Crosson S. 2012. ppGpp and polyphosphate modulate cell cycle progression in Caulobacter crescentus. J. Bacteriol. 194:28–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Eichel J, Chang YY, Riesenberg D, Cronan JE., Jr 1999. Effect of ppGpp on Escherichia coli cyclopropane fatty acid synthesis is mediated through the RpoS sigma factor (sigmaS). J. Bacteriol. 181:572–576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Heath RJ, Jackowski S, Rock CO. 1994. Guanosine tetraphosphate inhibition of fatty acid and phospholipid synthesis in Escherichia coli is relieved by overexpression of glycerol-3-phosphate acyltransferase (plsB). J. Biol. Chem. 269:26584–26590 [PubMed] [Google Scholar]
- 40.Kanjee U, Gutsche I, Alexopoulos E, Zhao B, El Bakkouri M, Thibault G, Liu K, Ramachandran S, Snider J, Pai EF, Houry WA. 2011. Linkage between the bacterial acid stress and stringent responses: the structure of the inducible lysine decarboxylase. EMBO J. 30:931–944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Murdeshwar MS, Chatterji D. 2012. MS_RHII-RSD: a dual function RNase HII-(p)ppGpp synthetase from Mycobacterium smegmatis. J. Bacteriol. 194:4003–4014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wexselblatt E, Katzhendler J, Saleem-Batcha R, Hansen G, Hilgenfeld R, Glaser G, Vidavski RR. 2010. ppGpp analogues inhibit synthetase activity of Rel proteins from Gram-negative and Gram-positive bacteria. Bioorg. Med. Chem. 18:4485–4497 [DOI] [PubMed] [Google Scholar]
- 43.Wexselblatt E, Oppenheimer-Shaanan Y, Kaspy I, London N, Schueler-Furman O, Yavin E, Glaser G, Katzhendler J, Ben-Yehuda S. 2012. Relacin, a novel antibacterial agent targeting the Stringent Response. PLoS Pathog. 8:e1002925. 10.1371/journal.ppat.1002925 [DOI] [PMC free article] [PubMed] [Google Scholar]