Abstract
CTnDOT is a 65-kb conjugative transposon (CTn) in Bacteroides spp. that confers resistance to the antibiotics erythromycin and tetracycline (Tc). Conjugative transfer of CTnDOT is regulated upon exposure of cells to Tc. In the absence of Tc, no transfer is detectable; however, a cascade of regulatory events results in the conjugative transfer of CTnDOT upon Tc induction. Previous studies addressing regulation of CTnDOT conjugative transfer focused primarily on the 13-kb transfer (tra) operon, which encodes the proteins required for assembly of the mating apparatus. We report here that the mob operon that encodes the relaxase and coupling proteins required for mobilization of CTnDOT are regulated at the transcriptional level upon Tc induction. The Xis2d and Exc excision proteins are required for the upregulation of mob transcription upon Tc induction, and yet a deletion of xis2c has no effect. We also show preliminary evidence suggesting that the integrase, IntDOT, may play a regulatory role, as pLYL72 transfer is not detectable when intDOT is provided in trans.
INTRODUCTION
Bacteroides spp. are Gram-negative obligate anaerobes that are primarily found in the human colon. Bacteroides are abundant members of the gut microbiota and play an important role by aiding in processes such as nutrient acquisition and pathogen exclusion (1–3). However, Bacteroides spp. can be detrimental to the host if they escape the colon, causing opportunistic infections that may be difficult to treat. These infections may not respond well to antibiotic therapy due to the increasing prevalence of antibiotic resistance within Bacteroides, which is often linked to mobile genetic elements (3–5). One such element is CTnDOT, a 65-kb conjugative transposon that confers resistance to the antibiotics erythromycin (ermF) and tetracycline (tetQ).
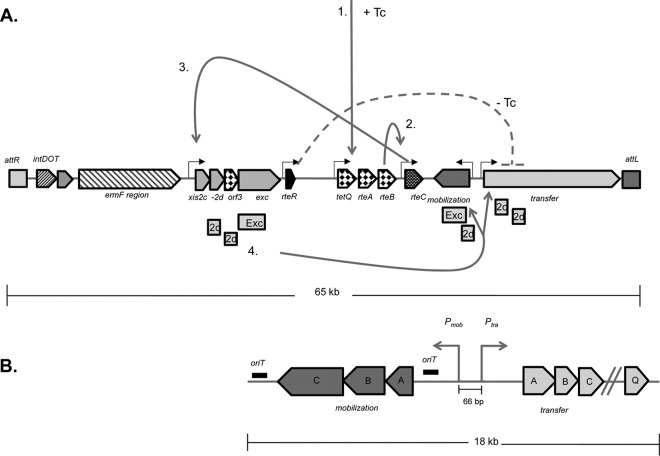
An interesting feature of CTnDOT is that excision and conjugative transfer of CTnDOT are dependent upon tetracycline exposure (6–8). In the absence of tetracycline (Tc), there is no detectable transfer of CTnDOT (9). Upon exposure to low (<1 μg/ml) levels of Tc, a regulatory cascade (Fig. 1A) initiates through the tetQ-rteA-rteB operon, which is regulated by a translational attenuation mechanism. RteB activates the transcription of rteC, and RteC in turn activates transcription of the excision operon that contains xis2c, xis2d, orf3, and exc (8, 10). Not only do Xis2c, −2d, and Exc function to excise CTnDOT from the chromosome, but Xis2c and −2d also activate transcription of the tra operon, which encodes the mating apparatus (11, 12). We have yet to identify any regulatory or structural role for Orf3. An overview of the steps involved in the regulation of CTnDOT conjugative transfer is summarized in Fig. 1.
Fig 1.
Tetracycline-dependent regulation of CTnDOT transfer. (A) Upon tetracycline (Tc) induction (solid gray lines), translation of TetQ, RteA, and RteB resumes, as this region is regulated by a translational attenuation mechanism (step 1). In step 2, RteB activates the transcription of rteC, which activates (step 3) transcription of the excision operon. In step 4, the proteins encoded in the excision operon then activate transcription of the transfer and mobilization operons, thereby inducing conjugative transfer of CTnDOT. In the absence of Tc (dashed gray line), no transfer is detectable due to RteR-mediated premature transcription termination in the tra operon. (B) The 18-kb region of CTnDOT containing the mobilization and transfer regions is present on the pLYL72 self-transmissible plasmid. The mob and tra promoters are 66 bp apart.
Many studies have described transcriptional regulation of both excision and transfer of CTnDOT (10, 11, 13, 14), but thus far we have yet to characterize a very important set of genes involved in CTnDOT conjugative transfer: the mobilization region. The CTnDOT mob genes encode proteins that are involved in formation of the relaxosome that is required to nick the oriT and for coupling the CTnDOT circular intermediate to the mating bridge (15).
We report here that the mob genes of CTnDOT are part of a 4-kb operon that is divergently transcribed from the transfer (tra) operon (Fig. 1B). Like the tra operon, the mob genes are also regulated at the transcriptional level by Tc induction. CTnDOT mob mRNA is detectable from the element in the chromosome only when cells have been exposed to Tc. However, when detected from the self-transmissible plasmid pLYL72 (containing the 18-kb region of CTnDOT shown in Fig. 1B), a transcript was seen whether or not Tc was present, demonstrating that the mob operon is actually transcribed constitutively with respect to Tc exposure. This observation suggests that CTnDOT encodes transcriptional activators that are enhancing the level of transcription upon Tc induction and that a repressor of mob transcription is also present on CTnDOT that prevents mob transcription in the absence of Tc. Xis2d and Exc are required for enhancement of mob transcription upon Tc induction. Xis2c, which is required for activation of the neighboring tra genes, does not appear to play a role in mob activation as a deletion of xis2c shows no detectable defect in mob transcription. This observation was surprising to us given the close proximity between the mob and tra promoters (Fig. 1B). Nonetheless, this further demonstrates the complex nature of coordinating excision and transfer of CTnDOT.
Not only did we characterize the transcriptional regulation of the mob operon, but we also wanted to confirm the location of the oriT, which is the DNA site nicked by the relaxases to initiate mobilization of CTnDOT. We had previously identified an oriT that was downstream of mobC; however, the oriT on a CTnDOT-like element (CTn341) is in fact upstream of mobA (15). The upstream mobA regions of CTnDOT and CTn341 are approximately 94% identical, which prompted us to evaluate whether this region on CTnDOT also contained an oriT. We report that the upstream mobA region from CTnDOT does contain an oriT, similar to CTn341. However, the CTnDOT-like element CTnERL, which is remarkably similar to CTnDOT, does not recognize the upstream mobA oriT region (16, 17). We have confirmed that CTnDOT does possess a second oriT that is located in a 900-bp region downstream of mobC and that CTnERL was able to recognize this oriT region. It is unusual that CTnDOT appears to harbor two oriT regions, as most elements have a single oriT.
Last, we report that the enhancement of mob transcription upon Tc induction does not appear to directly correspond with the frequency of conjugative transfer. A deletion of exc, which is required for mob transcriptional enhancement but has no role in tra regulation, shows no substantial defect in the transfer frequency of pLYL72. Further, we show that the mob genes are transcribed at a higher level on CTnDOT than on CTnERL and yet these two CTns possess the same frequency of conjugative transfer.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
All bacterial strains and plasmids used in this study are summarized in Table 1. Unless otherwise noted, all Bacteroides strains were initially cultured in chopped meat broth (Remel) and then transferred to anaerobic Trypticase-yeast extract-glucose (TYG) medium (18) for overnight growth with antibiotics when appropriate. Subculturing of Bacteroides strains was also done in TYG liquid medium, and all Bacteroides culturing was performed anaerobically at 37°C. Escherichia coli was grown aerobically in Luria-Bertani (LB) broth at 37°C with antibiotics when appropriate. Unless otherwise noted, antibiotic concentrations used for culturing were as follows: ampicillin at 100 μg/ml; cefoxitin at 10 μg/ml; chloramphenicol at 10 μg/ml; erythromycin at 10 μg/ml; gentamicin at 200 μg/ml; kanamycin at 100 μg/ml; rifampin at 2 μg/ml; streptomycin at 100 μg/ml; and tetracycline at 1 μg/ml.
Table 1.
Bacterial strains and plasmids
| Strain or plasmid | Relative phenotype(s)a | Description and/or reference |
|---|---|---|
| Strains | ||
| B. thetaiotaomicron BT4001 | (Rifr) | Spontaneous B. thetaiotaomicron Rifr mutant |
| B. thetaiotaomicron BT4001 ΩQABC | (Thy−, Tpr, Tcr) | BT4100 with a site-specific insertion of the regulatory region tetQ-rteA-rteB-rteC (9) |
| B. thetaiotaomicron BT4001 ΩQABC Ω2.6pGERM | (Rifr, Tcr) | BT4001 ΩQABC containing an insertion of pDJE2.3, which contains intDOT, integrated into the chromosome (26) |
| B. thetaiotaomicron BT4004 | (Rifr, Emr) | BT4001 containing CTnERL integrated into the chromosome |
| B. thetaiotaomicron BT4007 | (Rifr, Tcr, Emr) | BT4001 containing CTnDOT integrated into the chromsome |
| B. thetaiotaomicron BT4104 | (Thy−, Tpr, Tcr) | BT4100 containing a chromosomal copy of CTnERL (27) |
| E. coli DH5aMCR | RecA | Gibco BRL |
| E. coli HB101 | RecA, Strr | E. coli strain used as a conjugative transfer recipient (28) |
| E. coli HB101 RP1 | RecA, Strr | HB101 containing the IncPα plasmid RP1 (18) |
| Plasmids | ||
| pAFD1 | Apr (Emr) | E. coli-Bacteroides shuttle vector containing ermF (A. M. Stevens, unpublished data) |
| pGRW53Δ2C | Apr (Emr) | pGW46 insert containing a 114-aa in-frame deletion of xis2c, ligated into pAFD1 (13) |
| pGRW53Δ2D | Apr (Emr) | pGW46 insert containing an in-frame deletion of xis2d from aa 3 through the end of orf3, ligated into pAFD1 (13) |
| pGRW53ΔExc | Apr (Emr) | pGW46 insert containing a 670-aa in-frame deletion of the C-terminal end of Exc, ligated into pAFD1 (13) |
| pGRW54 | Apr (Cfr) | A 900-bp fragment containing the region between rteC and mobC from CTnDOT, cloned into pLYL7oriT(RK2) (this study) |
| pGRW55 | Apr (Cfr) | A 200-bp fragment containing the putative CTnDOT oriT region upstream of mobA, cloned into pLYL7oriT(RK2) (this study) |
| pGW45 | Apr (Cfr) | A 1.6-kb fragment containing orf2b, xis2c, xis2d, exc, orf2e, and rteR cloned into pLYL05 (9) |
| pGW46 | Apr (Cfr) | A 1.3-kb fragment containing xis2c, xis2d, exc, orf2e, and rteR cloned into pLYL05 (9) |
| pHopp1 | Apr (Emr) | pAFD1 containing xis2c, xis2d, orf3, exc, and rteR (13) |
| pKS04 | Apr (Cfr) | A 4.9-kb fragment containing orf2a, orf2b, orf3, and a truncation of exc where a 840-bp sequence was deleted from the 3′ end (26) |
| pLYL05 | Apr (Cfr) | E. coli-Bacteroides shuttle vector (L. Y. Li, unpublished data) |
| pLYL72 | Knr, Cmr (Cmr) | Self-transmissible plasmid containing mob genes, tra operon, and OriT from CTnDOT (19) |
| pLYL7oriTRK2 | Apr (Cfr) | E. coli-Bacteroides shuttle vector that is mobilizable in E. coli by the IncP plasmid RP4 but is not mobilizable in Bacteroides (29) |
| pYS41 | Apr (Cfr) | A 1.5-kb fragment ligated into pLYL05 containing orf2a, orf2b, exc, and rteR (Y. Sutanto, unpublished data) |
Bacteroides phenotypes are shown in parentheses, whereas E. coli phenotypes are shown without parentheses. Abbreviations: aa, amino acid; Ap, ampicillin; Cf, cefoxitin; Cm, chloramphenicol; Em, erythromycin; Kn, kanamycin; Rif, rifampin; Str, streptomycin; Tc, tetracycline; Thy, thymidine auxotroph; Tp, trimethoprim.
Bacterial mating assays.
E. coli strain HB101 containing the RPI IncPα plasmid was used to mobilize plasmids from an E. coli donor strain to Bacteroides recipients, as described previously (18).
Mating-out assays measuring the frequency of transfer of pLYL72 from a Bacteroides donor to recipient E. coli (HB101) were performed as described previously (19). The transfer frequency of pLYL72 is represented as the ratio of the number of transconjugants per recipient.
RNA isolation.
Bacteroides strains were grown in anaerobic TYG medium to an optical density at 650 nm (OD650) of 0.3 to 0.5. RNA was then isolated using a QIAgen RNeasy kit. Removal of contaminating genomic DNA was performed using Ambion Turbo DNA-free reagents, and RNA samples were subsequently tested to confirm that no DNA was remaining by performing PCR. RNA concentrations were measured using a NanoDrop 2000c spectrophotometer (Thermo Scientific).
Qualitative RT-PCR.
Reverse transcription-PCR (RT-PCR) was performed using an Access RT-PCR system (Promega). Reactions that omitted reverse transcriptase were tested to ensure that all genomic DNA had been adequately removed. Samples were electrophoresed on a 1.2% to 1.5% agarose gel for analysis. Amplification of rpoD was used as a control to ensure that equal concentrations of RNA were present in all samples tested. Sequences of the primers used for qualitative RT-PCR are shown in Table 2.
Table 2.
Primers used in this study
| Primer function and name | 5′ to 3′ sequence |
|---|---|
| Cloning | |
| GRW54 fwd | GATGACTGCAGGGAATCACAAT |
| GRW54 rev | TCTTGATAAGAGCTCTAGAAGTCATT |
| GRW55 fwd | TGAAAGGTATCTGCAGTCCGT |
| GRW55 rev | GGATCGAGCTCGGGATTTCT |
| RT-PCR | |
| mobA fwd | TCAAGGTGCTGAAGGTGGACAAGA |
| mobA rev | TCTGACGGCTCAGTTTCACGAGTT |
| mobB fwd | TTTCCGTGCTGCAAGGCACTTATG |
| mobB rev | ATGAAAGCCTCTTCCTCGGCATCT |
| mobC fwd | TGAACCGCCGCTACGAGGATATTT |
| mobC rev | TCGGGATTGTTGATGTCCAGCGTA |
| mobA-B fwd | TGGACTACTACACCAAGCTATCGG |
| mobA-B rev | TTTCCAGATACTCGCGGGC |
| mobB-C fwd | ACAACCGCCGATACCGTTATCCAT |
| mobB-C rev | CCCTTTCGTACCCAGACAGGACA |
| RpoD 67 fwd | TTTAATCTGACGAACGCGCTCACG |
| RpoD 548 rev | ACGCTGTATGGTGGATTCGTCAGT |
| RT-qPCR | |
| mobA fwd | ACAACCGTTTCCTCGCCATGTTTG |
| mobA rev | TCTTGTCCACCTTCAGCACCTTGA |
| mobA-B fwd | AACTCGTGAAACTGAGCCGTCAGA |
| mobA-B rev | TGTAGATGCGGTTGGTGGTGAGAA |
| mobB fwd | ACAACCGCCGATACCGTTATCCAT |
| mobB rev | ATGAAAGCCTCTTCCTCGGCATCT |
| RpoD 467 fwd | ACCTGATTCAACGGGAGACGACA |
| RpoD 548 rev | ACGCTGTATGGTGGATTCGTCAGT |
RT-quantitative PCR (RT-qPCR).
Synthesis of cDNA was performed using a Bio-Rad iScript kit per the manufacturer's protocol, with the addition of 200 ng random pentadecamers (Integrated DNA Technologies) per reaction. Approximately 1 μg of DNase-treated RNA was used for each reaction. Quantitative PCR was then performed using a Realplex2 Mastercycler (Eppendorf), with Bio-Rad Ssofast EvaGreen as a signal reporter. Amplification of mobB from various strains was used as a marker to measure the effect on transcription of various CTnDOT regions in trans to the mob operon. Amplification of rpoD was used as a reference marker to normalize for variations in the quantity of RNA template, as rpoD is a single-copy gene and the expression of rpoD is not altered by Tc induction (14). The sequences of the primers used are listed in Table 2. The final levels of reaction components were as follows: 1 μM primers, 5 μl cDNA, and 10 μl Ssofast EvaGreen Supermix (Bio-Rad). The reaction conditions were as follows: initial denaturation for 2 min at 98.0°C and amplification and quantification at 98.0°C for 5 s and 55°C for 10 s for 40 cycles. A melting curve was performed at 95.0°C for 15 s, followed by 55.0°C for 15 s, with subsequent heating to 95.0°C over the course of 20 min with continuous fluorescence measurement, followed by a final incubation at 95.0°C for 15 s. Each measurement was performed in triplicate, and relative quantification determinations were performed using the Pfaffl equation N = 2−ΔΔCT, where N is the relative fold difference. ΔCT represents the CT (cycle threshold) value difference within a sample between the target gene (mob) and the reference marker (rpoD). ΔΔCT then represents the difference in ΔCT values between tetracycline-induced and noninduced samples (20).
Localization of the oriT regions recognized by CTnDOT and CTnERL.
A 900-bp region containing the intergenic region between rteC and mobC was PCR amplified using primers GRW54 fwd and rev (Table 2) from BT4007 and cloned into pLYL7oriT(RK2), which is mobilizable by the IncP plasmid RP4 but cannot be mobilized by Bacteroides. The resulting plasmid, pGRW54, was then introduced into both BT4007 and BT4004. Mating assays were performed to evaluate mobilization of pGRW54.
Similarly, a 200-bp region containing the region upstream of mobA was PCR amplified from BT4007 using primers GRW55 fwd and rev (Table 2). The resulting product was then subcloned into pLYL7oriT(RK2), and the resulting pGRW55 plasmid was introduced into BT4007 and BT4004 via a triparental mating, as described above. Mating assays were then performed to determine whether CTnERL (BT4004) and/or CTnDOT (BT4007) could mobilize pGRW55. Tetracycline (1 μg/ml) was added to the donor growth medium, as it is required for conjugative transfer from these strains.
RESULTS
The CTnDOT mob genes are regulated by tetracycline induction and are assembled in an operon.
A recent study of a CTnDOT-like element, CTn341, revealed that the CTn341 mob genes comprise an operon that is induced at the transcriptional level by tetracycline (15). To determine if the CTnDOT mob genes are regulated in response to tetracycline (Tc) exposure, we performed qualitative RT-PCR detecting mobA, -B, and -C from BT4007, a B. thetaiotaomicron strain that contains a single copy of CTnDOT integrated into the chromosome (Table 1). BT4007 cultures were grown in the absence of Tc or with low levels of Tc (1 μg/ml). As shown in Fig. 2, mobA, -B, and -C transcripts are detectable only when cells are exposed to Tc, demonstrating that the CTnDOT mob genes are in fact directly or indirectly regulated by Tc exposure.
Fig 2.
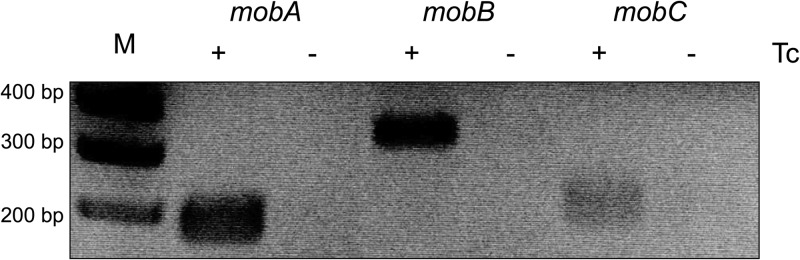
The mob genes are detected from CTnDOT only upon Tc induction. To establish whether the transcription of the mob genes is regulated by Tc induction, RT-PCR was performed, detecting mobA, mobB, and mobC from BT4007, which contains a copy of CTnDOT integrated into the chromosome. RT-PCR was performed on BT4007 RNA samples from cells that were noninduced or induced with (1 μg/ml) tetracycline (Tc). An amplicon pertaining to each of the mob genes was detected only upon Tc induction, thus confirming that the transcription of the mob genes is dependent upon exposure of cells to Tc.
To establish whether the mob genes are assembled in an operon, we tested for transcriptional linking, where the intergenic regions of mobA and mobB (mobA-B) and mobB-C were amplified using qualitative RT-PCR. If these genes are transcriptionally linked, and thus organized in an operon, we would expect to detect these intergenic transcripts. Conversely, if each mob gene was transcribed from an independent promoter, no transcript pertaining to the intergenic region would be detected. An amplification product was detected for both the mobA-B and mobB-C transcripts, which suggests that the mob genes are in an operon (Fig. 3). Further supporting these findings is the recent demonstration by Peed and colleagues that the mob genes of a CTnDOT-like element, CTn341, are also in an operon that is regulated by Tc induction (15).
Fig 3.
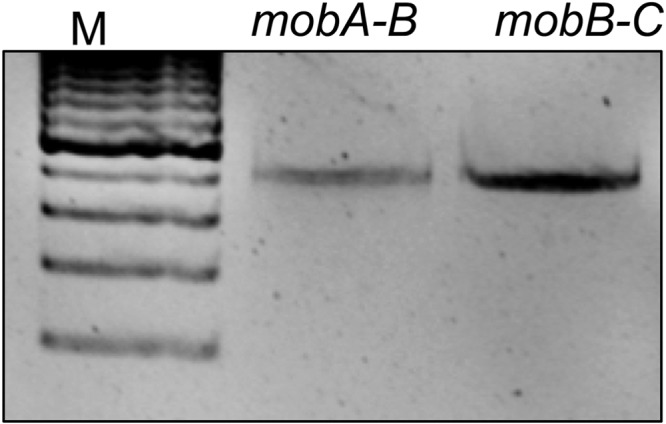
The CTnDOT mob genes are transcriptionally linked. To address whether the mob genes form a polycistronic transcript, RT-PCR was performed using primers that amplified the regions of the mobA-B and mobB-C intergenic junctions from BT4001 pLYL72 pGW45 induced with tetracycline (1 μg/ml). Amplicons for both of these primer sets were observed, thus indicating that the mob genes are transcriptionally linked and organized as an operon.
CTnDOT appears to harbor two OriT regions that are located upstream of mobA and downstream of mobC.
Prior studies of CTnDOT suggested that the oriT of CTnDOT was within the approximately 1-kb region that is located between rteC and mobC. However, a recent study characterizing the Mob proteins on the CTn341 CTnDOT-related element suggested that the oriT of CTn341 was just upstream of mobA. The previous studies that localized the CTnDOT oriT used subcloned regions in trans to another element, CTnERL, which is also considered to be virtually identical to CTnDOT with the exception that CTnERL lacks the 13-kb ermF region (17, 21). Collectively, these observations prompted us to re-evaluate the location of the oriT regions on CTnDOT.
A 900-bp region located between rteC and mobC, previously reported to contain the CTnDOT oriT, was cloned onto a vector that cannot be mobilized from Bacteroides. The resulting plasmid, pGRW54, was then introduced into a Bacteroides strain that contained either CTnDOT (BT4007) or CTnERL (BT4004) integrated into the chromosome. Mating assays were performed using both of these Bacteroides strains to see whether pGRW54 was able to transfer to an E. coli recipient (HB101). If transconjugants were detected, this would suggest that an oriT region was present on pGRW54 and could be mobilized by the conjugative transposon present in the donor Bacteroides strain. Plasmid pGRW54 was able to transfer from both BT4004 and BT4007, demonstrating that an oriT downstream of mobC is mobilized by both CTnERL and CTnDOT (Table 3).
Table 3.
Mobilization of putative oriT regions
| Donor strain | Transfer frequencya |
|---|---|
| BT4004 pGW54 | 1.3 × 10−6 |
| BT4007 pGRW54 | 6.3 × 10−4 |
| BT4004 pGRW55 | <10−9 |
| BT4007pGRW55 | 4.2 × 10−3 |
The transfer frequency is represented as the average ratio of the number of transconjugants per recipient cell.
A 200-bp region upstream of mobA that is almost identical to the CTn341 oriT region was cloned into a vector that is not mobilizable in Bacteroides. Transfer of the resulting pGRW55 plasmid was detected only from BT4007 and not from BT4004, thus suggesting that an oriT was present upstream of mobA that is mobilizable by CTnDOT and yet not by CTnERL (Table 3). While these data strongly indicate that CTnDOT contains two oriT regions as opposed to the single oriT region on CTnERL, future studies performing mutational analysis on these regions would further support the notion that they are true origins of transfer.
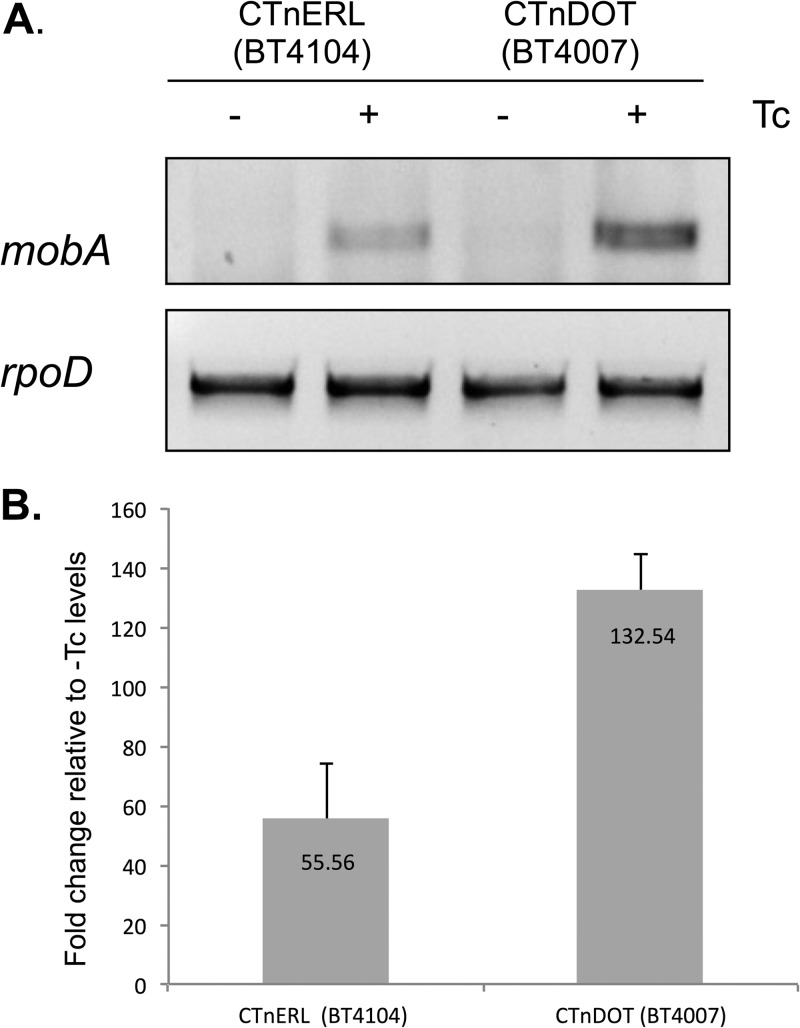
Due to this observation that the oriT recognition patterns may differ between CTnDOT and CTnERL, we wanted to investigate whether the CTnERL mob genes are also regulated at the transcriptional level in response to Tc induction. We performed both qualitative and quantitative RT-PCR to detect the mob genes from CTnERL (BT4014) and CTnDOT (BT4007). The mob genes from both CTnERL and CTnDOT were regulated by exposure to Tc (Fig. 4A). However, much more transcript was observed from CTnDOT than from CTnERL upon Tc induction. CTnERL showed an approximately 55-fold induction of detectable mob transcript upon Tc induction, whereas CTnDOT had an over 130-fold increase in transcript (Fig. 4B). Despite the difference in mob transcription results, the two elements are reported to transfer at similar rates, suggesting that the level of mob expression does not directly affect the frequency of conjugative transfer (21, 22).
Fig 4.
More mob transcript is detectable from CTnDOT than from CTnERL. (A) Qualitative RT-PCR was performed using primers to detect mobA from a strain containing CTnERL (BT4104) and CTnDOT (BT4007) both with and without Tc induction. No transcript is detectable in the absence of Tc, and yet, upon exposure to Tc, a mob transcript is detected. However, more mobA transcript is seen in cells containing CTnDOT than in cells containing CTnERL. rpoD is a reference marker used to ensure equivalent RNA concentrations in samples. (B) RT-qPCR was performed using primers to detect mobB. RNA content was normalized to the reference marker rpoD, and the ΔΔCT was calculated comparing induced (+) to noninduced (− Tc) levels.
Providing the excision operon in trans to the mobilization region is sufficient for enhancement of mob transcription upon Tc induction.
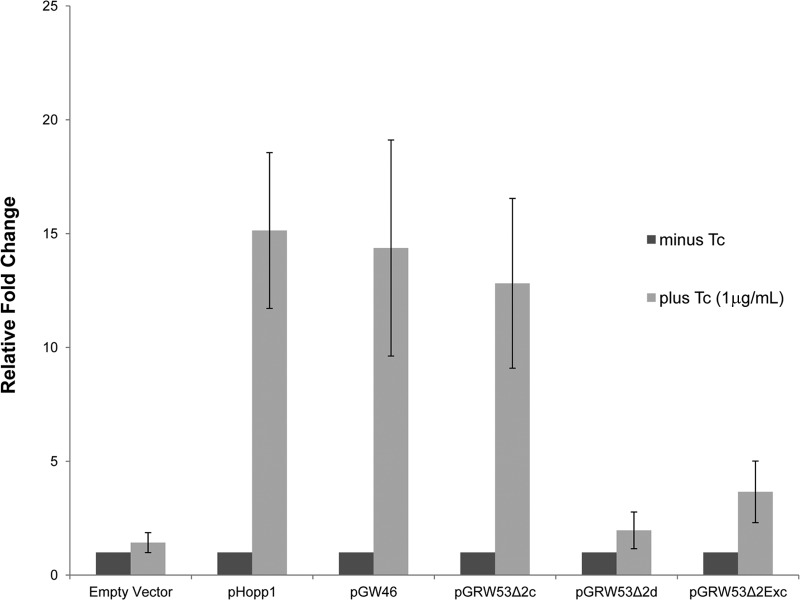
After observing that the mob genes from CTnDOT are detected only upon exposure to Tc, we predicted that proteins encoded within the excision operon are acting as regulators of the mob operon. We have previously reported that when the excision operon is provided in trans with the self-transmissible plasmid pLYL72, there is transcriptional activation of the tra genes as well as enhanced pLYL72 transfer (9, 11, 13). Because of the close proximity of the tra promoter and the putative mob promoter, we predicted that the interactions that regulate the tra operon could also be mediating mob transcriptional activation. Using both qualitative and quantitative RT-PCR, we first amplified the mob genes from BT4001 ΩQABC pLYL72 pHopp1 both with Tc (1 μg/ml) and without Tc. As shown in Fig. 5, an increase in mob transcript was observed upon Tc induction, which suggests that proteins encoded within the excision operon can act as transcriptional activators of the mob operon.
Fig 5.
The mob genes are transcribed constitutively from pLYL72, and the excision proteins Xis2d and Exc are required for sufficient enhancement of transcription upon Tc induction. RT-qPCR was performed to localize potential regulators of mob transcription; the relative fold changes in transcription measuring mobB relative to the levels in the absence of tetracycline (−) are shown. Fold changes were calculated using the Pfaffl ΔΔCT equation, and samples were normalized using the reference marker rpoD (20). The observations described above were also confirmed using qualitative RT-PCR (not shown). The mob genes were transcribed constitutively from pLYL72, and the excision operon in trans was sufficient for transcriptional activation. A deletion of xis2C appears to have no deleterious effect on mob transcription, whereas a deletion in either xis2d or exc drops transcription to the levels determined for constitutive empty vector.
We then wanted to see if we could determine which excision proteins were necessary for enhancement of mob transcription. Various subclones of the excision region were provided in trans to pLYL72 to detect the regulation of mob. As shown in Fig. 5, a plasmid containing xis2c xis2d exc rteR (pGW45) still resulted in mob activation, demonstrating that orf3 is not required. However, a deletion of either xis2d or exc decreased the detectable mob transcript to constitutive levels in the absence of Tc, suggesting that Xis2d and Exc are required for the upregulation of mob transcription. A deletion of xis2c had no noticeable effect on the mob transcriptional activation. This was unexpected, as we predicted that a deletion of xis2c would greatly diminish mob activation based on the observation that xis2c is required for tra operon activation (13). Further, a deletion of exc had no effect on the tra operon and yet brought mob activation down to the constitutive levels in the absence of Tc. These observations suggest that the patterns of transcriptional regulation of the mob and tra operons are not as similar as we had predicted based on their close proximity.
A deletion of xis2d abolishes conjugative transfer of pLYL72.
Mating assays were performed to detect the frequency of transfer of pLYL72 from BT4001 ΩQABC containing various excision operon deletions on a second plasmid provided in trans to pLYL72. When the complete excision operon (pGW45) or a plasmid with a deletion of orf 3 (pGW46) was present, both enhancement and repression of pLYL72 transfer were observed with and without the addition of Tc, respectively. When we investigated the ability of pGRW53ΔExc to regulate pLYL72 transfer, the transfer frequency was similar to that seen with a strain containing the fully intact excision operon, where both enhancement and repression were observed in response to Tc induction. This observation suggests that Exc is not required for enhanced transfer of pLYL72.
A deletion of xis2c (pGRW53Δ2c) resulted in a loss of enhancement, and pLYL72 transferred at a rate similar to that seen with the empty vector control upon Tc induction, where transconjugants were detected at a frequency of approximately 10−5 to 10−6 transconjugants per recipient. However, a deletion of xis2d (pGRW53Δ2d) resulted in no pLYL72 transfer even with the induction of Tc. Similarly, a deletion of both xis2c and xis2d (pYS41) resulted in no detectable pLYL72 transfer, likely due to the requirement for Xis2d. For each of the plasmids described above, no transconjugants were detectable without Tc added to the growth medium due to RteR-mediated negative regulation.
We also tested the effect of the CTnDOT integrase, IntDOT, on the transfer of pLYL72. We hypothesized that IntDOT could be a candidate for the negative regulator of mob transcription in the absence of Tc since the integrase of Tn916 is thought to bind the oriT in an effort to prevent premature conjugative transfer (23). Our results demonstrated that the mob genes are transcribed constitutively from pLYL72 when intDOT is in trans (Fig. 6). RT-qPCR confirmed that the mob genes are transcribed constitutively from this strain (data not shown). However, no transfer of pLYL72 was detected with or without Tc induction with intDOT present. This suggests that IntDOT may play a role in preventing premature conjugative transfer, although IntDOT does not appear to do so by preventing transcription of the mob operon.
Fig 6.
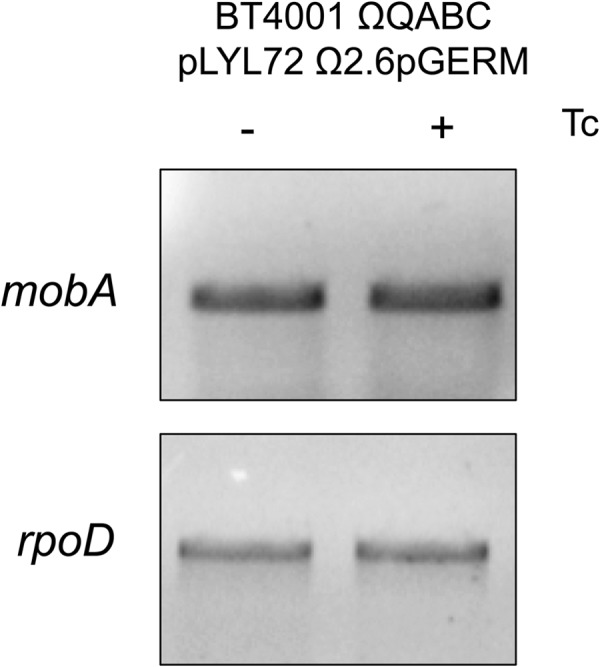
The mob genes are transcribed when intDOT is present. Qualitative RT-PCR was performed detecting mobA from strain BT4001 ΩQABC pLYL72 Ω2.6pGERM, which contains a copy of intDOT integrated into the chromosome. Mob transcript was detectable whether or not Tc was present, which suggests that IntDOT was not preventing transcription of the mob operon.
DISCUSSION
Coordinating the events of CTnDOT excision, mobilization, transfer, and integration results in a mobile element whose transfer is very tightly regulated. Such coordination is crucial, as it is important to delay the initiation of mobilization and transfer functions until CTnDOT has properly excised from the chromosome. If transfer were to happen prematurely, an Hfr-like transfer would be the consequence, which could be rather costly to the donor cell. Many studies have detailed the coordinated regulation of excision and transfer of CTnDOT (10, 13, 14, 24), while this report is the first to characterize the transcriptional regulation of CTnDOT mobilization functions.
We have shown that the mob genes of CTnDOT are part of an operon that is transcribed upon induction with low levels of the antibiotic tetracycline (Tc), while no transcript is detectable in the absence of Tc. This tetracycline-dependent mob regulation is similar to observations made with respect to a related conjugative transposon, CTn341. Peed et al. reported that the rteA and rteB genes were required in trans for transcriptional activation of the mobCTn341 operon, a fact that prompted us to further investigate the regulation of the CTnDOT mob operon (15).
From what we know about the CTnDOT regulatory cascade initiated upon Tc induction (Fig. 1), RteA and RteB are required for the activation of a regulatory protein, RteC, which acts as a transcriptional activator of the excision operon (8, 10, 14, 25). Because RteA and RteB are not known to directly regulate any regions of CTnDOT other than the rteC regulatory protein-encoding gene, we predicted that the genes located in the excision region could be acting as transcriptional activators of the mob genes. We report here that the Xis2d and Exc excision proteins are required for the activation of mob transcription upon Tc induction (Fig. 5). Although Exc is required for enhancement of mob expression, a deletion of exc has no obvious effect on pLYL72 transfer (Table 4). This suggests that there is no direct correlation between the level of mob expression and the frequency of conjugative transfer. This idea is further supported by the observation that although there is a large difference between CTnERL and CTnDOT in mob expression levels, they are reported to transfer at similar rates (Fig. 5 and Table 4).
Table 4.
Effect of excision protein deletions on the transfer frequency of pLYL72a
| Plasmid used with donor strain BT4001 ΩQABC pLYL72 | Transfer frequency of pLYL72 |
Regulation of transferb | Source or reference | |
|---|---|---|---|---|
| +Tc | −Tc | |||
| pLYL05 | 10−5–10−6 | 10−5–10−6 | E−, R− | 5 |
| pKS04 | 10−5–10−6 | 10−5–10−6 | E−, R− | 5 |
| pGW46 | 5.5 × 10−3 | <10−9 | E+, R+ | This study |
| pGRW53Δ2c | 1.8 × 10−4 | <10−9 | E−, R+ | This study |
| pGRW53Δ2d | <10−9 | <10−9 | R+, R+ | This study |
| pGRW53ΔExc | 1.2 × 10−3 | <10−9 | E+, R+ | This study |
| pYS41 | <10−9 | <10−9 | R+, R+ | This study |
| Ω2.6pGERM | <10−9 | <10−9 | R+, R+ | This study |
The transfer frequency of pLYL72 is expressed as the average ratio of the number of transconjugants per recipient cell. +, present; −, absent.
E, enhancer; R, repressor.
A deletion of xis2c has no detectable effect on mob expression but decreases the transfer of pLYL72 to a rate of 1.8 × 10−4 transconjugants per recipient. A deletion of xis2d, on the other hand, results in no detectable transfer of pLYL72 upon Tc induction. Previous studies have demonstrated that a deletion of xis2c or xis2d abolishes the enhancement of tra transcription upon Tc induction, and yet the different effects these deletions have on pLYL72 conjugative transfer suggest that a deletion of xis2d has greater consequences than deletion of xis2c. This intimates that there may be a greater requirement of Xis2d for transcriptional regulation and that Xis2c is possibly acting more as an accessory protein that is facilitating Xis2d-mediated regulation.
Our findings have also demonstrated that a negative regulator prevents transcription of the mob operon when no Tc is present. The mob genes are transcribed constitutively from the self-transmissible plasmid pLYL72, and yet a transcript is detected from CTnDOT only upon Tc induction. We investigated the possibility that the CTnDOT integrase (IntDOT) was a potential repressor, as the integrase from Tn916 can bind the oriT of Tn916 and is thought to prevent premature conjugative transfer (23). RT-PCR demonstrated that the mob genes were transcribed constitutively when intDOT was provided in trans; however, no pLYL72 transfer was detectable (Fig. 6 and Table 4). Although IntDOT is not directly acting as the negative regulator of the mob operon, this finding suggests that IntDOT plays a regulatory role in preventing conjugative transfer. Nevertheless, this observation is preliminary and warrants further investigation.
Concluding remarks.
In summary, our findings have added another layer of complexity to understanding the already intricate regulation of CTnDOT. These observations further support the idea of the importance of highly coordinated regulation of excision and conjugative transfer. It would be advantageous to initiate these steps as soon as possible so that antibiotic resistance determinants could readily disseminate throughout the population but not prematurely to the disadvantage of the donor cell.
ACKNOWLEDGMENT
This work was supported by grant AI 22383 from the National Institutes of Health.
Footnotes
Published ahead of print 27 September 2013
REFERENCES
- 1.Hooper LV, Midtvedt T, Gordon JI. 2002. How host-microbial interactions shape the nutrient environment of the mammalian intestine. Annu. Rev. Nutr. 22:283–307 [DOI] [PubMed] [Google Scholar]
- 2.Round JL, Mazmanian SK. 2009. The gut microbiota shapes intestinal immune responses during health and disease. Nat. Rev. Immunol. 9:313–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wexler HM. 2007. Bacteroides: the good, the bad, and the nitty-gritty. Clin. Microbiol. Rev. 20:593–621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nguyen M, Vedantam G. 2011. Mobile genetic elements in the genus Bacteroides, and their mechanism(s) of dissemination. Mob. Genet. Elements 1:187–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Whittle G, Shoemaker NB, Salyers AA. 2002. The role of Bacteroides conjugative transposons in the dissemination of antibiotic resistance genes. Cell. Mol. Life Sci. 59:2044–2054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Salyers AA, Shoemaker NB. 1997. Conjugative transposons. Genet. Eng. (N Y) 19:89–100 [DOI] [PubMed] [Google Scholar]
- 7.Shoemaker NB, Wang GR, Stevens AM, Salyers AA. 1993. Excision, transfer, and integration of NBU1, a mobilizable site-selective insertion element. J. Bacteriol. 175:6578–6587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stevens AM, Shoemaker NB, Li LY, Salyers AA. 1993. Tetracycline regulation of genes on Bacteroides conjugative transposons. J. Bacteriol. 175:6134–6141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Whittle G, Shoemaker NB, Salyers AA. 2002. Characterization of genes involved in modulation of conjugal transfer of the Bacteroides conjugative transposon CTnDOT. J. Bacteriol. 184:3839–3847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Park J, Salyers AA. 2011. Characterization of the Bacteroides CTnDOT regulatory protein RteC. J. Bacteriol. 193:91–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jeters RT, Wang G-R, Moon K, Shoemaker NB, Salyers AA. 2009. Tetracycline-associated transcriptional regulation of transfer genes of the Bacteroides conjugative transposon CTnDOT. J. Bacteriol. 191:6374–6382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Keeton CM, Gardner JF. 2012. Roles of Exc protein and DNA homology in the CTnDOT excision reaction. J. Bacteriol. 194:3368–3376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Keeton CM, Park J, Wang G-R, Hopp CM, Shoemaker NB, Gardner JF, Salyers AA. 2013. The excision proteins of CTnDOT positively regulate the transfer operon. Plasmid 69:172–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moon K, Shoemaker NB, Gardner JF, Salyers AA. 2005. Regulation of excision genes of the Bacteroides conjugative transposon CTnDOT. J. Bacteriol. 187:5732–5741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Peed L, Parker AC, Smith CJ. 2010. Genetic and functional analyses of the mob operon on conjugative transposon CTn341 from Bacteroides spp. J. Bacteriol. 192:4643–4650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bonheyo G, Graham D, Shoemaker NB, Salyers AA. 2001. Transfer region of a bacteroides conjugative transposon, CTnDOT. Plasmid 45:41–51 [DOI] [PubMed] [Google Scholar]
- 17.Whittle G, Hund BD, Shoemaker NB, Salyers AA. 2001. Characterization of the 13-kilobase ermF region of the Bacteroides conjugative transposon CTnDOT. Appl. Environ. Microbiol. 67:3488–3495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shoemaker NB, Getty C, Guthrie EP, Salyers AA. 1986. Regions in Bacteroides plasmids pBFTM10 and pB8-51 that allow Escherichia coli-Bacteroides shuttle vectors to be mobilized by IncP plasmids and by a conjugative Bacteroides tetracycline resistance element. J. Bacteriol. 166:959–965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li LY, Shoemaker NB, Salyers AA. 1995. Location and characteristics of the transfer region of a Bacteroides conjugative transposon and regulation of transfer genes. J. Bacteriol. 177:4992–4999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pfaffl MW. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29:e45. 10.1093/nar/29.9.e45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Whittle G, Hamburger N, Shoemaker NB, Salyers AA. 2006. A bacteroides conjugative transposon, CTnERL, can transfer a portion of itself by conjugation without excising from the chromosome. J. Bacteriol. 188:1169–1174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Franco AA, Cheng RK, Chung G-T, Wu S, Oh H-B, Sears CL. 1999. Molecular evolution of the pathogenicity island of enterotoxigenic Bacteroides fragilis strains. J. Bacteriol. 181:6623–6633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hinerfeld D, Churchward G. 2001. Specific binding of integrase to the origin of transfer (oriT) of the conjugative transposon Tn916. J. Bacteriol. 183:2947–2951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Waters JL, Salyers AA. 2012. The small RNA RteR inhibits transfer of the Bacteroides conjugative transposon CTnDOT. J. Bacteriol. 194:5228–5236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang Y, Shoemaker NB, Salyers AA. 2004. Regulation of a Bacteroides operon that controls excision and transfer of the conjugative transposon CTnDOT. J. Bacteriol. 186:2548–2557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cheng Q, Sutanto Y, Shoemaker NB, Gardner JF, Salyers AA. 2001. Identification of genes required for excision of CTnDOT, a Bacteroides conjugative transposon. Mol. Microbiol. 41:625–632 [DOI] [PubMed] [Google Scholar]
- 27.Shoemaker NB, Salyers AA. 1987. Facilitated transfer of IncP beta R751 derivatives from the chromosome of Bacteroides uniformis to Escherichia coli recipients by a conjugative Bacteroides tetracycline resistance element. J. Bacteriol. 169:3160–3167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Boyer HW, Roulland-Dussoix D. 1969. A complementation analysis of the restriction and modification of DNA in Escherichia coli. J. Mol. Biol. 41:459–472 [DOI] [PubMed] [Google Scholar]
- 29.Li LY, Shoemaker NB, Salyers AA. 1993. Characterization of the mobilization region of a Bacteroides insertion element (NBU1) that is excised and transferred by Bacteroides conjugative transposons. J. Bacteriol. 175:6588–6598 [DOI] [PMC free article] [PubMed] [Google Scholar]