Abstract
Riboflavin (vitamin B2) is the precursor of flavin mononucleotide and flavin adenine dinucleotide, which are cofactors essential for a host of intracellular redox reactions. Microorganisms synthesize flavins de novo to fulfill nutritional requirements, but it is becoming increasingly clear that flavins play a wider role in cellular physiology than was previously appreciated. Flavins mediate diverse processes beyond the cytoplasmic membrane, including iron acquisition, extracellular respiration, and interspecies interactions. While investigating the regulation of flavin electron shuttle biosynthesis in the Gram-negative gammaproteobacterium Shewanella oneidensis, we discovered that a riboflavin biosynthetic gene (ribBA) annotated as encoding a bifunctional 3,4-dihydroxy-2-butanone 4-phosphate (DHBP) synthase/GTP cyclohydrolase II does not possess both functions. The novel gene, renamed ribBX here, encodes an amino-terminal DHBP synthase domain. The carboxy-terminal end of RibBX not only lacks GTP cyclohydrolase II activity but also has evolved a different function altogether in S. oneidensis, regulating the activity of the DHBP synthase domain. Phylogenetic analysis revealed that the misannotation of ribBX as ribBA is rampant throughout the phylum Proteobacteria (40% of 2,173 annotated ribBA genes) and that ribBX emerged early in the evolution of this group of microorganisms. We examined the functionality of representative ribBX genes from Beta-, Gamma-, and Epsilonproteobacteria and found that, consistent with sequence-based predictions, the encoded GTP cyclohydrolase II domains lack catalytic activity. The persistence of ribBX in the genomes of so many phylogenetically divergent bacterial species lends weight to the argument that ribBX has evolved a function which lends a selective advantage to the host.
INTRODUCTION
Riboflavin (vitamin B2), the precursor molecule for flavin mononucleotide (FMN) and flavin adenine dinucleotide (FAD) (here referred to collectively as flavins), is synthesized de novo by plants and microorganisms (1). Traditionally thought of only as redox-active cofactors of cellular proteins, flavins have been studied extensively for essential roles played in oxidative metabolism and other intracellular processes. More recently, a wider role for flavins in the physiology of microorganisms is coming to light, as a number of bacteria have been found to use free, extracytoplasmic flavins to carry out vital processes beyond the borders of the cell. Flavins are important for assimilatory iron reduction in Campylobacter jejuni, Helicobacter pylori, and three species of methanotrophic bacteria (2–4). Shewanella oneidensis and Geothrix fermentans use secreted flavin electron shuttles to accelerate respiration of insoluble minerals and electrodes (5–8). Secretion of riboflavin by symbiotic nodule-forming Sinorhizobium meliloti enhances root respiration in alfalfa (9, 10). Finally, flavins secreted by the alga Chlamydomonas reinhardtii have even been shown to mimic the bacterial quorum sensing signals of Pseudomonas aeruginosa, manipulating quorum sensing-controlled gene expression in a competing organism (11).
Regulation of flavin biosynthesis has been characterized in a number of organisms, usually occurring at the level of synthesis or activity of GTP cyclohydrolase II or 3,4-dihydroxy-2-butanone 4-phosphate (DHBP) synthase (1), the enzymes that catalyze the rate-limiting steps in the pathway (12). In bacteria, riboflavin biosynthesis is regulated by the RFN element, a highly conserved riboswitch present in the 5′-untranslated region of mRNAs that encode riboflavin biosynthesis enzymes. Intracellular FMN binds to the RFN element, inhibiting transcription and/or translation of the downstream gene (1, 13). The diverse functional roles occupied by flavins hint at a larger range of regulatory mechanisms controlling flavin biosynthesis than is currently appreciated. Secretion of flavins into the surrounding environment presents a problem for the above-mentioned regulatory schemes, which rely on the inhibition of gene expression via the accumulation of flavins in the cytoplasm. While our appreciation of the diverse roles played by flavins continues to grow, additional regulatory mechanisms of flavin biosynthesis remain to be discovered.
S. oneidensis strain MR-1 is a Gram-negative gammaproteobacterium that employs flavin electron shuttles to enhance electron transfer to insoluble extracellular metals and carbon electrodes during anaerobic respiration (14, 15). Given the importance of secreted flavins in the anaerobic respiratory strategy of MR-1, we wanted to examine the regulation of riboflavin biosynthesis, with the goal of increasing extracellular electron transfer through genetic manipulation. MR-1, like the majority of microorganisms, is able to synthesize flavins de novo to satisfy nutritional requirements for the redox cofactor. MR-1 also secretes significant quantities of flavins into the surrounding medium under laboratory conditions (5–7). Genetic tractability combined with a simple fluorescence-based assay for flavin detection makes MR-1 an ideal model system for studying the production/regulation of flavins intended for extracytoplasmic function. Here we report the discovery of a novel regulatory mechanism which controls riboflavin biosynthesis in MR-1 and show that, in doing so, we also uncovered widespread misannotation of the ribBA gene. The canonical ribBA gene encodes a bifunctional 3,4-dihydroxy-2-butanone 4-phosphate (DHBP) synthase/GTP cyclohydrolase II. We have determined that 40% (871 of 2,173 genes) of annotated ribBA genes encode a widespread variant that we have termed ribBX, which encodes a protein with an amino-terminal DHBP synthase domain and a carboxy-terminal domain that appears to regulate the DHBP synthase activity of the enzyme in MR-1. A homology search revealed that ribBX is present in the genomes of a highly diverse group of medically and environmentally important bacterial taxa. Characterization of ribBX from Pseudomonas putida, Vibrio parahaemolyticus, and Burkholderia cenocepacia confirms the lack of GTP cyclohydrolase II activity and lends further weight to the assertion that ribBX is widespread in the phylum Proteobacteria.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
Strains and plasmids used in this study are listed in Table S1 in the supplemental material. Escherichia coli strains were maintained on lysogeny broth (LB) agar plates supplemented with the following as necessary: 50 μg/ml kanamycin, 10 μg/ml gentamicin, 200 μM riboflavin, and/or 250 μM 2,6-diaminopimelic acid. E. coli flavin auxotrophs (16) were obtained from the Coli Genetic Stock Center (http://cgsc.biology.yale.edu). During routine manipulation and strain construction, MR-1 was maintained on LB agar containing 50 μg/ml kanamycin as necessary. For growth assays, MR-1 was grown in or on Shewanella basal medium (SBM) containing 5 ml/liter vitamin mix, 5 ml/liter mineral mix (5), 0.01% Casamino Acids, 20 mM sodium dl-lactate, and 40 mM sodium fumarate and supplemented with 50 μg/ml kanamycin when required. MR-1 flavin determination was performed as follows. Strains stored in glycerol at −80°C were freshly streaked onto LB agar plates and incubated at 30°C for ∼16 h, after which single colonies were inoculated into LB medium and shaken at 30°C for 6 to 8 h. LB cultures were subcultured in SBM and shaken at 30°C for ∼16 h, after which cultures were pelleted by centrifugation, washed twice with SBM, and used to inoculate fresh SBM to a final optical density at 600 nm (OD600) of 0.025 to 0.05. Anaerobic cultures were stoppered with butyl rubber and flushed with nitrogen gas for 15 min following inoculation (17). After ∼6 to 8 h of growth, cell-free supernatants were harvested for determination of flavin content.
Flavin fluorescence measurements.
Measurements of secreted flavins present in culture supernatants were taken as previously described (18), with minor modifications. Briefly, samples were extracted from MR-1 cultures and centrifuged to pellet cells, and 200 μl was then transferred to a clear 96-well plate (for bulk fluorescence measurements). Bulk fluorescence measurements were taken using a Molecular Devices SpectraMax M2 plate reader with a 440-nm excitation and a 525-nm emission cutoff.
Deletion and complementation.
Primers used to amplify portions of the MR-1 chromosome for cloning deletion and/or complementation constructs are listed in Table S2 in the supplemental material. PCR products were cloned using standard laboratory molecular biology protocols. Regions flanking deletion targets were amplified using PCR and cloned into the pSMV3 suicide vector. In-frame gene deletions in MR-1 were generated using homologous recombination as previously described (5). For complementation constructs, gene coding regions and 30 bp upstream were amplified by PCR and cloned into pBBR1MCS-2 or pBBR1MCS-5. Complementation constructs were moved into E. coli as previously described (19) or into MR-1 by conjugal transfer from an E. coli donor. All plasmid constructs and gene deletions were verified by sequencing.
Bacterial mono- and two-hybrid assays.
A detailed protocol outlining the LexA bacterial mono- and two-hybrid system is available (20). LexA fusion protein expression vectors (pSR658 and pSR659) and E. coli strains carrying a LexA-repressible lacZ reporter (SU101 and SU202) were used for all interaction studies (20). Briefly, reporter strains were transformed with plasmids expressing various LexA-Rib fusion proteins and selected for on LB agar with the appropriate antibiotic(s). Single colonies were picked from LB agar and streaked onto fresh MacConkey agar plates for colorimetric determination of β-galactosidase activity.
Sequence analysis.
Amino acid sequences sharing similarity with RibA (GTP cyclohydrolase II), RibB (3,4-dihydroxy-2-butanone 4-phosphate synthase), and RibBA were identified in the protein database at the NCBI website (http://www.ncbi.nlm.nih.gov/protein), using a Web-based gapped-BLAST algorithm (http://blast.ncbi.nlm.nih.gov) (21). Mutant sequences and short peptide fragments (<150 residues) were discarded. Sequences were aligned using Muscle (22) as implemented in SeaView (23), with minor adjustments done manually. Partial sequences were removed unless the missing sites comprised less than 5% of the sequence (commonly seen at peptide amino and carboxy termini). Only a single wild-type representative in clusters of sequences sharing more than 98% identity was retained. Sequence sections homologous to RibB were used to build trees. Bootstrapped maximum likelihood and neighbor-joining trees were constructed with RAXML (24), using a JTT (25) substitution matrix and SeaView (23) with a Poisson correction.
RESULTS
A diverse group of Proteobacteria has multiple copies of riboflavin biosynthetic genes enabling regulatory plasticity.
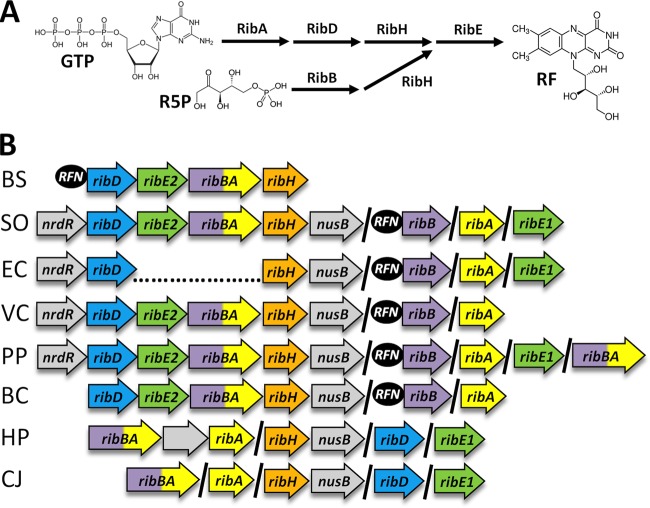
To search for novel regulatory mechanisms controlling flavin biosynthesis in MR-1, we first examined the riboflavin biosynthetic genes annotated in the genomes of organisms belonging to the phylum Proteobacteria. The biochemical pathway for flavin biosynthesis has been characterized extensively, and a comprehensive review of the field is available (1). Nomenclature for riboflavin biosynthesis genes from different organisms can be confusing, and accordingly, a single nomenclature system is used throughout this article, with gene products indicated. In Escherichia spp., one molecule of GTP and two molecules of ribulose-5-phosphate are converted into one molecule of riboflavin in a stepwise manner by the proteins encoded by the ribA (GTP cyclohydrolase II), ribB (3,4-dihydroxy-2-butanone 4-phosphate synthase), ribD (pyrimidine deaminase/reductase), ribH (lumazine synthase), and ribE (riboflavin synthase) genes (Fig. 1A). Riboflavin is then converted first into FMN, and then into FAD, by the protein encoded by the ribF gene (riboflavin kinase/FAD synthase). In Bacillus spp., the same pathway is used; however, the ribA and ribB genes are fused into a bifunctional ribBA gene. Despite significant differences in the organization of riboflavin biosynthetic genes between Bacillus and Escherichia, only one gene encoding an enzyme with each biochemical function is found in the genome (Fig. 1B). In contrast to the examples given above, a phylogenetically diverse set of Proteobacteria appears to have multiple copies of ribA, ribB, ribBA, ribH, and/or ribE (26). MR-1, for example, has two genes annotated to encode the functions of RibA, RibB, and RibE, and the genetic organization is reminiscent of those of both Bacillus and Escherichia (Fig. 1B). The superficial appearance of functional duplication may indicate that individual flavin biosynthetic genes are differentially regulated, possibly in response to the need for flavins that perform functions distinct from nutritional requirements.
Fig 1.
Riboflavin biosynthetic pathway and genetic basis in representative bacterial species. (A) Riboflavin (RF) is synthesized from one molecule of GTP and two molecules of ribulose-5-phosphate (R5P). Note that the first dedicated reactions for GTP and R5P are catalyzed by RibA and RibB, respectively. (B) Genomic context of riboflavin biosynthesis genes in representative bacterial species. Gene color is based on function. Gray genes are not involved in riboflavin biosynthesis but are included to show their shared genomic context. A forward slash indicates that genes are not adjacent on the chromosome. RFN elements, denoted by black circles, were predicted by Vitreschak and coworkers (26), with the exception of the B. subtilis RFN element, which was demonstrated experimentally (13). Genome abbreviations: BS, Bacillus subtilis subsp. subtilis 168; SO, Shewanella oneidensis MR-1; EC, Escherichia coli MG1655; VP, Vibrio parahaemolyticus RIMD 2210633; PP, Pseudomonas putida KT2440; BC, Burkholderia cenocepacia PC184; HP, Helicobacter pylori 83; CJ, Campylobacter jejuni NCTC11168.
The ribBA gene from MR-1 does not encode an enzyme with GTP cyclohydrolase II activity.
To verify the functionality of annotated riboflavin biosynthesis genes from MR-1, we utilized a set of E. coli riboflavin auxotrophs with transposon insertions in ribA, ribB, or ribE (16). E. coli mutants were transformed with plasmids expressing the ribA, ribB, ribBA, ribE1, and ribE2 genes from MR-1, and the resulting strains were cultivated on agar plates that did not contain exogenously added riboflavin to assess functional complementation of riboflavin biosynthesis. With the exception of the MR-1 ribBA gene, each gene complemented the corresponding E. coli mutant for growth in the absence of exogenous riboflavin (Table 1). We know that the ribB domain of ribBA can be active (see below), so the inability to complement the E. coli ribB auxotroph suggests that the amount of activity of the full-length protein is insufficient. To ensure that the failure of the MR-1 ribBA gene to complement either a ribA or ribB mutation in E. coli was not simply due to the gene fusion (ribA and ribB are carried separately in E. coli), we demonstrated functional complementation of both mutants with a plasmid expressing ribBA from Bacillus subtilis (ribBABS) (Table 1).
Table 1.
Functional complementation of E. coli riboflavin auxotrophs
| Plasmid | Complementation of E. coli auxotropha |
||
|---|---|---|---|
| ribA::Tn5 | ribB::Tn5 | ribE::Tn5 | |
| pBBR1MCS-2 vector | − | − | − |
| ribAEC | +++ | − | ND |
| ribBEC | − | +++ | ND |
| ribA | +++ | − | ND |
| ribB | − | +++ | ND |
| ribBA | − | − | ND |
| ribBX-NTD | − | +++ | ND |
| ribBX-CTD | − | − | ND |
| ribBABS | +++ | +++ | ND |
| ribBABS-NTD | − | +++ | ND |
| ribBABS-CTD | +++ | − | ND |
| ribEEC | ND | ND | +++ |
| ribE1 | ND | ND | +++ |
| ribE2 | ND | ND | +++ |
| ribBXVP_0681 | − | +++ | ND |
| ribBXPP_0516 | − | +++ | ND |
| ribBXPP_3813 | − | +++ | ND |
| ribBXBcenP_01000846 | − | +++ | ND |
−, no complementation; +++, complementation by the plasmid; ND, not determined.
We next examined the functions of ribA, ribB, ribBA, ribE1, and ribE2 in MR-1 by attempting to construct an in-frame deletion of each gene and measuring the accumulation of flavins in culture medium as an indicator of riboflavin production/secretion in the resulting strains. Deletion of the ribE1, ribE2, and ribB genes in MR-1 had very little effect on the accumulation of extracellular flavins (Fig. 2A). This result was interesting because it indicated that, at least under the conditions tested, the ribBA gene product could provide nearly wild-type levels of flavin electron shuttles in a ribB mutant background, despite the inability of ribBA to complement E. coli riboflavin auxotrophs (Table 1). Deletion of ribBA, on the other hand, resulted in a 2-fold decrease in the level of secreted flavins (Fig. 2A). Despite repeated efforts, we were unable to delete ribA, and saturating transposon-sequencing (Tn-seq) experiments failed to detect insertions in the ribA coding sequence (27). In a similar situation, another group was unable to delete the gene encoding RibA in C. jejuni (3), despite the presence of a gene annotated ribBA (Fig. 1B). Given these results, we conclude that the ribA gene product is essential for the production of riboflavin in MR-1.
Fig 2.
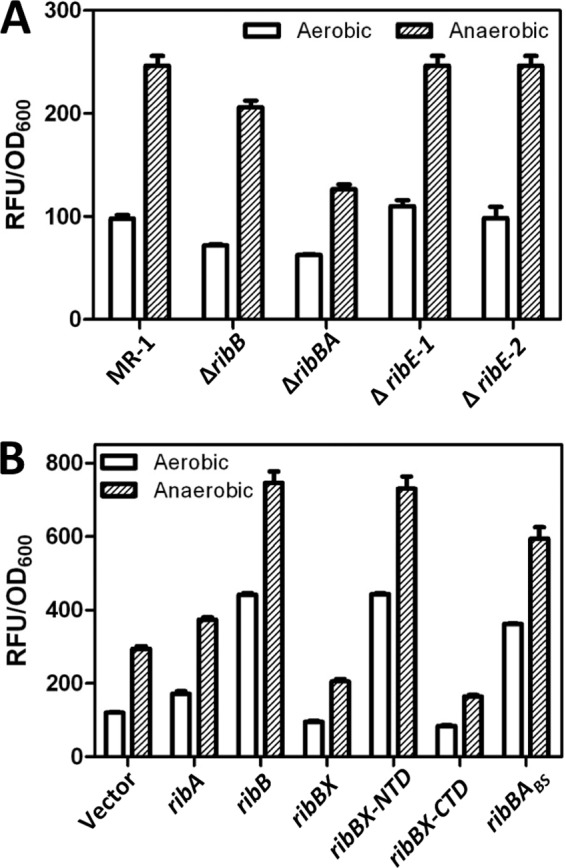
Effect of deletion or overexpression of riboflavin biosynthetic genes on the accumulation of extracellular flavins in S. oneidensis culture supernatants. S. oneidensis strains carrying deletions in riboflavin biosynthetic genes (A) or plasmids expressing riboflavin biosynthetic genes in multiple copies (B) were grown aerobically and anaerobically in SBM. Culture supernatants were harvested after 7 h, and the fluorescence intensity (in RFU) was measured and normalized to the OD600. Reported values for both panels are the averages for three independent experiments, and error bars represent the standard errors of the means (SEM).
It is possible that the inability of the MR-1 ribBA gene to complement riboflavin auxotrophy in E. coli is simply because the ribBA gene product is not expressed/active in this heterologous host. We therefore transformed MR-1 with a vector expressing the ribA, ribB, or ribBA gene from MR-1 and measured the accumulation of extracellular flavins. Expression of the ribA gene resulted in a modest but reproducible increase in the accumulation of flavins (294.5 ± 6.9 versus 374.0 ± 6.0 relative fluorescence units [RFU]/OD600 unit; P < 0.001) (Fig. 2B). Expression of the ribB gene, on the other hand, resulted in a 2.5-fold increase in the concentration of secreted flavins (Fig. 2B). In addition to overexpression of a rate-limiting enzyme in riboflavin biosynthesis, the large increase upon overexpression of ribB was likely due to the fact that the ribB coding sequence was cloned without the upstream RFN element, freeing the plasmid-encoded ribB transcript from feedback inhibition. Surprisingly, expression of ribBA resulted in a reproducible, albeit small, decrease (∼33%) in the concentration of secreted flavins (294.5 ± 6.9 versus 205.4 ± 6.3 RFU/OD600 unit; P < 0.001) (Fig. 2B). Expression of ribBA from B. subtilis (ribBABS) in MR-1 resulted in a significant increase in the secretion of flavins (Fig. 2B). These results are in agreement with the failure of ribBA from MR-1 to complement E. coli riboflavin auxotrophs and warranted further investigation.
Sequence analysis and homology modeling of RibBA from MR-1.
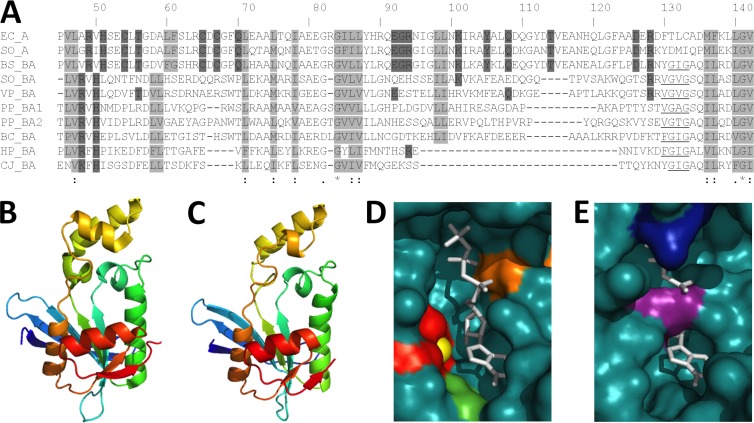
The ribBA gene of MR-1 is annotated as coding for an enzyme with an amino-terminal DHBP synthase domain (ribBA-NTD) and a carboxy-terminal GTP cyclohydrolase II domain (ribBA-CTD), which are encoded separately, by the ribB and ribA genes, respectively, in E. coli. Due to the failure of the ribBA gene from MR-1 to complement a ribA or ribB mutation in E. coli (Table 1) or to increase flavin secretion when overexpressed in MR-1 (Fig. 2B), we examined the predicted amino acid sequence for insight into the apparent lack of function. The three-dimensional structures of the DHBP synthase (RibB) and GTP cyclohydrolase II (RibA) enzymes from E. coli have been solved, and the amino acids that participate in the catalytic mechanism are known (28–30). The active site residues of RibB from E. coli are completely conserved in the ribB gene product of MR-1 and the ribBA gene products from MR-1 and B. subtilis (see Fig. S1 in the supplemental material). While the active site residues of RibA from E. coli are completely conserved in the ribA gene product of MR-1 and the ribBA gene product from B. subtilis, the ribBA gene product from MR-1 lacks most of the amino acids critical for GTP cyclohydrolase II activity (Fig. 3A). To further examine the active site of the RibBA-CTD from MR-1, we generated structural models based on the crystal structure of RibA from E. coli (28). The RibBA-CTD (residues 188 to 367) from MR-1 was threaded onto the crystal structure of RibA bound to a GTP analog (doi:10.2210/pdb2bz0/pdb) by using PHYRE 2.0 (31), and the predicted structure was visualized using PyMOL display software (www.pymol.org). Superficially, the overall tertiary structures are highly similar between the two proteins (Fig. 3B and C), and amino acids comprising the hydrophobic core are especially well conserved (Fig. 3A, light gray boxes). The substrate-binding pocket of RibA from E. coli is clearly visible, as is the zinc ion coordinated by Cys54, Cys65, and Cys67, which is required to activate a water molecule for nucleophilic attack on the guanine C-8 atom (Fig. 3D). In the RibBA-CTD from MR-1, Arg269 and Arg350 protrude into the substrate-binding pocket and the coordinated zinc ion is no longer present, as Cys54, Cys65, and Cys67 are absent in this protein (Fig. 3E). Two other amino acids which directly participate in the catalytic reaction, Arg94 and Tyr105, are also absent. These data led us to predict that the RibBA-CTD from MR-1 does not possess GTP cyclohydrolase II activity and has evolved a different function altogether. We propose renaming the ribBA gene from S. oneidensis ribBX, to differentiate it from ribBA genes in other organisms which encode a bifunctional DHBP synthase/GTP cyclohydrolase II (e.g., B. subtilis). Here we refer to the domain formerly annotated as a GTP cyclohydrolase II as the RibBX-CTD.
Fig 3.
Alignment and structural modeling of GTP cyclohydrolase II domains from Proteobacteria. (A) ClustalW alignment of the E. coli and MR-1 RibA sequences and the RibBA-CTD sequences of select Proteobacteria. Based on the published crystal structure of E. coli RibA (28), nonpolar residues that form the hydrophobic core are boxed in light gray, and residues that directly participate in the catalytic mechanism are boxed in dark gray. The underlined stretch of nonpolar amino acids is conserved only in the RibBA sequences. Numbering is based on the E. coli RibA sequence. (B to E) Tertiary structure prediction of the RibBA-CTD from MR-1 (C), based upon the crystal structure of RibA from E. coli (B). The RibBA-CTD (residues 188 to 367) was threaded onto the crystal structure of RibA by using PHYRE2 (www.sbg.bio.ic.ac.uk/phyre2/), and the resulting model was visualized using PyMOL (www.pymol.org). The active sites of RibA (D) and the RibBA-CTD (E) are displayed using space-filling models. The GTP analog phosphomethylphosphonic acid guanylate ester (GMPCPP), cocrystallized with RibA from E. coli, is shown in white and superimposed onto the RibBA-CTD for comparison. Active site residues Cys54, Cys65, and Cys67 (red), a coordinated zinc ion (yellow), Tyr105 (green), and Arg128 (orange) are also indicated. The active site residues and coordinated zinc ion are not highlighted in panel E because they are not present in the RibBA-CTD. Arg269 (purple) and Arg350 (blue), the residues extending into the binding pocket of the RibBA-CTD, are indicated instead.
The ribBX gene from MR-1 encodes a protein with two functional domains.
The prototrophic nature of the ribB mutant for riboflavin demonstrates that the RibBX-NTD from MR-1 possesses DHBP synthase activity, at least in the absence of the ribB gene (Fig. 2A). We cloned the regions of ribBX from MR-1 encoding the NTD (amino acids 1 to 207) and CTD (amino acids 188 to 367) into an expression vector, transformed the E. coli ribA and ribB mutants with each plasmid, and cultivated the resulting strains on agar plates without added riboflavin. While ribBX-CTD was unable to complement a ribA mutation in E. coli, ribBX-NTD was now able to complement a ribB mutation for growth in the absence of exogenous riboflavin (Table 1). To ensure that the location where we chose to divide the ribBX coding sequence did not inactivate the RibBX-CTD, we cloned the analogous regions from the B. subtilis ribBABS gene into an expression vector, and expression of B. subtilis ribBABS-NTD and ribBABS-CTD complemented the E. coli ribB and ribA mutants, respectively (Table 1). To further investigate the functionality of ribBX-NTD and ribBX-CTD, we introduced the expression plasmids into MR-1 and measured the accumulation of flavins in the culture supernatant. Expression of ribBX-NTD resulted in a significant increase in secreted flavins compared to the level in vector-only controls, now mirroring that of MR-1 carrying the same vector overexpressing ribB (Fig. 2B). Expression of ribBX-CTD resulted in a small (∼44%) but reproducible decrease in the concentration of secreted riboflavin (294.5 ± 6.9 versus 164.8 ± 3.3 RFU/OD600 unit; P < 0.001), similar to that observed when ribBX was overexpressed in MR-1 (Fig. 2B). We conclude that ribBX-NTD encodes a functional DHBP synthase that is inhibited by the presence of ribBX-CTD.
To elucidate a possible mechanism for the function of the RibBX-CTD, we used bacterial LexA mono- and two-hybrid reporter systems to look for protein-protein interactions with other riboflavin biosynthetic genes. The reporter systems rely on the DNA-binding domain of LexA, a transcriptional repressor that must dimerize in order to bind DNA and repress transcription of a β-galactosidase reporter (20). The self-association of RibA and RibB from E. coli and RibBA from B. subtilis has been described previously (32–34). We wondered if the RibBX-CTD would multimerize, since it has not retained GTP cyclohydrolase II function, or possibly interact with the RibBX-NTD or another riboflavin biosynthetic protein to achieve the observed regulatory effect (Fig. 2B). In the monohybrid assay, compared to LexA lacking a dimerization domain, expression of a LexA-RibA, -RibB, -RibBX, -RibBX-NTD, -RibBX-CTD, or -RibBABS fusion protein resulted in strong repression of the β-galactosidase reporter (Table 2). A LexA-chloramphenicol acetyltransferase (CAT) fusion, which is known to multimerize, was used as a positive control (Table 2) (20). We next employed the LexA two-hybrid system to look for interactions between the RibBX-CTD and other riboflavin biosynthetic genes. Coexpression of LexA-RibBX-CTD with LexA-RibBX-CTD or LexA-RibBX resulted in repression of the β-galactosidase reporter. Significant interaction was not detected between LexA-RibBX-CTD and any other riboflavin biosynthesis protein, including the RibBX-NTD. LexA fusions to Jun and Fos, protein domains known to interact, were used as a positive control (20). While the mono- and two-hybrid assays demonstrated that the RibBX-CTD retains the ability to self-associate, if protein-protein interaction with other riboflavin biosynthetic proteins is the method of regulation, these interactions are too weak to be detected in these assays.
Table 2.
Results of LexA mono- and two-hybrid assays
| Assay and plasmid | Resulta |
|---|---|
| SU101 monohybrid assay | |
| Vector | − |
| CAT | ++ |
| ribA | +++ |
| ribB | +++ |
| ribBX | +++ |
| ribBX-NTD | +++ |
| ribBX-CTD | +++ |
| ribBABS | +++ |
| SU202 two-hybrid assay with vector expressing ribBX-CTD and indicated second vector | |
| Fosb | +++ |
| Vector | − |
| ribA | − |
| ribB | − |
| ribBX | ++ |
| ribBX-NTD | − |
| ribBX-CTD | ++ |
−, high activity; ++, moderate activity; +++, low activity.
Positive control with Jun.
The ribBX gene is widespread in the Proteobacteria.
Having demonstrated that ribBX from MR-1 does not encode an enzyme with GTP cyclohydrolase II activity, we wanted to know if ribBX was confined to the genus Shewanella or if the misannotation of ribBA was more widespread. A simple search of genomes in the phylum Proteobacteria that have both ribBA and ribA genes revealed numerous genes similar to ribBX, in that catalytic amino acids are absent in the encoded RibBX-CTD (representative examples are shown in Fig. 3A). We chose genes annotated ribBA from Vibrio parahaemolyticus (VP_0681), Pseudomonas putida (PP_0516 and PP_3813), and Burkholderia cenocepacia (BcenP_01000846) (Fig. 1B and 3A) and expressed them from a plasmid in the E. coli ribA and ribB mutants. Each of the four genes was able to complement the ribB mutant for growth without exogenous riboflavin, but none of the genes complemented the ribA mutant (Table 1). These results are consistent with a report demonstrating the same pattern of complementation with the gene annotated ribBA from Helicobacter pylori (2). ribBX appears to be present beyond the genus Shewanella, lending weight to the argument that ribBX-CTD encodes a protein domain with a function that confers a selective advantage for certain bacteria.
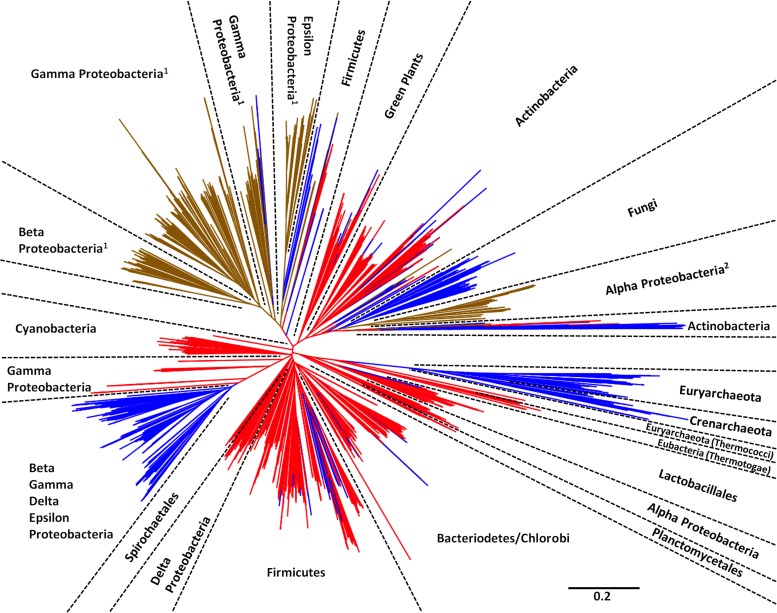
We next expanded our analysis to include all amino acid sequences sharing similarity with RibA, RibB, RibBA, and RibBX in the protein database maintained by NCBI (http://www.ncbi.nlm.nih.gov/protein), using a gapped-BLAST algorithm. A phylogenetic analysis of the resulting sequences was performed (see Materials and Methods), and the results for sequences containing a RibB-like domain (i.e., RibB, RibBA, and RibBX) are illustrated in Fig. 4. Interestingly, the phylogenetic evidence overwhelmingly supports a scenario in which RibBA is the ancestral state that gave rise to RibB and RibBX. RibA and RibB probably originated independently of each other, as the folds can be expressed separately (Table 1) but became fused in the lineage leading to the last common ancestor of all organisms for which sequence data are available. Since then, there is no evidence for the generation of RibBA or RibBX via gene fusion, because in no case was a RibBA or RibBX protein derived from a RibB protein (Fig. 4). Loss of the RibA domain from a RibBA protein or the RibBX-CTD from a RibBX protein has given rise to RibB multiple times across the different phyla, except the Cyanobacteria. This is true even when ignoring single RibB lineages that might be products of errors in annotation. RibBX lineages have arisen independently on at least two occasions in the Proteobacteria (Fig. 4, superscripts 1 and 2). The fact that the same groups appear in multiple positions in the tree (e.g., the Alphaproteobacteria and the eukaryotic green plants and fungi) is readily explained by postulating either early gene duplication with subsequent different copies surviving in different lineages or horizontal gene transfer.
Fig 4.
Phylogenetic analysis of RibB, RibBA, and RibBX. The diagram shows the unrooted maximum likelihood phylogeny of RibB, RibBA, and RibBX protein sequences from the NCBI database. Named phyla are well supported (>80% bootstrap support, most with >90% support), while short branches render little support for any branching order deep in the phylogeny. RibBX (brown) and RibB (blue) were derived from the ancestral RibBA protein (red). RibBX evolved independently at least twice in the Proteobacteria, as denoted by superscript numbers. Deletion of RibA or the RibBX-CTD to yield RibB occurred multiple times (red lines changing to blue), most notably in the archaeal, fungal, actinobacterial, and proteobacterial lineages.
DISCUSSION
Flavins have been studied intensely because of their vital role as redox-active cofactors of enzymes involved in cellular metabolic reactions (1). In the last decade, numerous examples have been described where microorganisms secrete flavins into the extracellular environment to perform functions beyond the cell wall (2–7, 9–11). Regulation of production and secretion of extracellular flavins in nonindustrial strains have not been studied, despite the growing body of literature concerning their use. The majority of studies examining flavin biosynthetic enzymes and/or regulation of flavin biosynthesis in bacteria have been performed in B. subtilis and E. coli, two organisms that are not known to utilize flavins to perform extracellular functions (1). Accordingly, regulation of flavin biosynthesis in both these organisms is dependent on intracellular pools of riboflavin, and one would expect regulation to be different in organisms that actively secrete flavins. The genomes of B. subtilis and E. coli contain only a single gene encoding each enzyme in the riboflavin biosynthetic pathway, while many bacteria have multiple copies of ribA, ribB, ribBA, ribH, ribE, and/or ribBX (Fig. 1B) (26). We initially speculated that multiple copies of flavin biosynthetic genes would allow for differential regulation of individual genes in response to the need for extracellular flavins. In this study, we investigated the functions of riboflavin biosynthetic genes in MR-1, discovered the misannotation of ribBA (now called ribBX), and identified ribBX genes in the genomes of a number of important pathogenic and environmental species of bacteria.
Flavins are essential for life, and if the RibBX-CTD does not appear to be an enzyme with GTP cyclohydrolase II activity, then another gene encoding this enzyme is required. Accordingly, despite an extensive search of the genomes maintained by the DOE Joint Genomics Institute (JGI), we have been unable to identify a free-living bacterium which has a ribBX gene but not a ribA gene or ribBA gene, giving further evidence for our assertion that the RibBX-CTD is not a GTP cyclohydrolase II. RibBX may have evolved when the selective pressure to maintain the GTP cyclohydrolase II activity of the ancestral RibBA protein was lost with the acquisition of another gene encoding a GTP cyclohydrolase II. While the presence of ribBX is by no means an assurance of a species utilization of extracellular flavins, a more careful examination of the role that flavins play in the physiology of microorganisms that carry the gene is warranted. In this study, we examined ribBX, but multiple copies of ribB, ribH, and/or ribE are also present in a number of bacterial genomes, raising questions as to the functionality of these genes as well.
In MR-1, the RibBX-CTD appears to regulate the activity of the RibBX-NTD, inhibiting DHBP synthase activity (Fig. 2B and Table 1). Interestingly, repression of DHBP activity of RibBX was alleviated in a ribB deletion strain of S. oneidensis, as evidenced by the ability of this strain to grow and secrete flavins at nearly wild-type levels (Fig. 2A). If the RibBX-CTD of RibBX modulates activity of the RibBX-NTD by sensing cytoplasmic flavin biosynthetic intermediates, then these pools are likely altered in a ribB mutant background, allowing for DHBP activity from RibBX. Differences in biosynthetic intermediates and/or other protein factors may also explain why RibBX is active in the S. oneidensis ribB mutant background (Fig. 2) but not in the E. coli ribB mutant background (Table 1). The mechanism of regulation remains unknown and is likely to be different in other systems. Regulation was not observed with RibBX enzymes from other Proteobacteria, at least under the conditions used in this study (Table 1). In all 4 cases tested, and in a published report using H. pylori (2), ribBX complemented an E. coli ribB mutant for growth in medium without riboflavin supplementation. The function of the RibBX-CTD remains unknown; however, it is not surprising that the RibBX enzymes from other organisms are not regulated in the same way as that in MR-1. The conditions under which extracellular flavins are advantageous are vastly different depending on the organism. For example, flavin secretion in S. meliloti is linked to the root nodule environment (9, 10), whereas flavin secretion in H. pylori is tied to iron limitation (2). MR-1 secretes large amounts of flavins under all conditions tested (5, 6), but a significant increase is observed under anaerobic conditions (see Fig. S2 in the supplemental material). The RibBX-CTD from other Proteobacteria could regulate the activity of the RibBX-NTD in specialized circumstances, as is the case for MR-1, or have a different function altogether.
In this study, we were unable to determine regulatory modulators of the RibBX-CTD in MR-1, but it is highly unlikely that GTP plays a role. GTP, the original substrate for GTP cyclohydrolase II, cannot fit into the modeled binding pocket of RibBX (Fig. 3E). Additionally, our data show that the RibBX-CTD effects regulatory change on its own, as expression of the RibBX-CTD decreased the secretion of flavins by MR-1. Despite a lack of experimental data regarding the mechanism of the RibBX-CTD-mediated regulation in MR-1 and other organisms, ribBX is widespread in the phylum Proteobacteria, and an alignment of the predicted amino acid sequences of ribBX gene products used in this study reveals a high level of conservation between the C-terminal domains, which is indicative of purifying selection (Fig. 3A). Most compelling is the nearly complete conservation of the nonpolar amino acids which comprise the hydrophobic core of the protein (Fig. 3A, light gray boxes), likely the reason that RibBX is always annotated RibBA by automated pipelines. Selective pressure has maintained ribBX-CTD in the genomes of many distantly related bacteria, indicating an important yet undiscovered role in cellular physiology.
Supplementary Material
ACKNOWLEDGMENTS
We thank Joshua M. Boster for technical assistance and our laboratory colleagues for helpful discussions.
This work was supported by the Office of Naval Research (grant N000141210309) and the University of Minnesota Initiative for Renewable Energy and Environment (grant RC-0003-12 awarded to J.A.G.).
Footnotes
Published ahead of print 4 October 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.00651-13.
REFERENCES
- 1.Abbas CA, Sibirny AA. 2011. Genetic control of biosynthesis and transport of riboflavin and flavin nucleotides and construction of robust biotechnological producers. Microbiol. Mol. Biol. Rev. 75:321–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fassbinder F, Kist M, Bereswill S. 2000. Structural and functional analysis of the riboflavin synthesis genes encoding GTP cyclohydrolase II (ribA), DHBP synthase (ribBA), riboflavin synthase (ribC), and riboflavin deaminase/reductase (ribD) from Helicobacter pylori strain P1. FEMS Microbiol. Lett. 191:191–197 [DOI] [PubMed] [Google Scholar]
- 3.Crossley RA, Gaskin DJ, Holmes K, Mulholland F, Wells JM, Kelly DJ, van Vliet AH, Walton NJ. 2007. Riboflavin biosynthesis is associated with assimilatory ferric reduction and iron acquisition by Campylobacter jejuni. Appl. Environ. Microbiol. 73:7819–7825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Balasubramanian R, Levinson BT, Rosenzweig AC. 2010. Secretion of flavins by three species of methanotrophic bacteria. Appl. Environ. Microbiol. 76:7356–7358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Coursolle D, Baron DB, Bond DR, Gralnick JA. 2010. The Mtr respiratory pathway is essential for reducing flavins and electrodes in Shewanella oneidensis. J. Bacteriol. 192:467–474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.von Canstein H, Ogawa J, Shimizu S, Lloyd JR. 2008. Secretion of flavins by Shewanella species and their role in extracellular electron transfer. Appl. Environ. Microbiol. 74:615–623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Marsili E, Baron DB, Shikhare ID, Coursolle D, Gralnick JA, Bond DR. 2008. Shewanella secretes flavins that mediate extracellular electron transfer. Proc. Natl. Acad. Sci. U. S. A. 105:3968–3973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mehta-Kolte MG, Bond DR. 2012. Geothrix fermentans secretes two different redox-active compounds to utilize electron acceptors across a wide range of redox potentials. Appl. Environ. Microbiol. 78:6987–6995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Phillips DA, Joseph CM, Yang GP, Martinez-Romero E, Sanborn JR, Volpin H. 1999. Identification of lumichrome as a sinorhizobium enhancer of alfalfa root respiration and shoot growth. Proc. Natl. Acad. Sci. U. S. A. 96:12275–12280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang G, Bhuvaneswari TV, Joseph CM, King MD, Phillips DA. 2002. Roles for riboflavin in the Sinorhizobium-alfalfa association. Mol. Plant-Microbe Interact. 15:456–462 [DOI] [PubMed] [Google Scholar]
- 11.Rajamani S, Bauer WD, Robinson JB, Farrow JM, 3rd, Pesci EC, Teplitski M, Gao M, Sayre RT, Phillips DA. 2008. The vitamin riboflavin and its derivative lumichrome activate the LasR bacterial quorum-sensing receptor. Mol. Plant-Microbe Interact. 21:1184–1192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hümbelin M, Griesser V, Keller T, Schurter W, Haiker M, Hohmann HP, Ritz H, Richter G, Bacher A, van Loon APGM. 1999. GTP cyclohydrolase II and 3,4-dihydroxy-2-butanone 4-phosphate synthase are rate-limiting enzymes in riboflavin synthesis of an industrial Bacillus subtilis strain used for riboflavin production. J. Ind. Microbiol. Biotechnol. 22:1–7 [Google Scholar]
- 13.Winkler WC, Cohen-Chalamish S, Breaker RR. 2002. An mRNA structure that controls gene expression by binding FMN. Proc. Natl. Acad. Sci. U. S. A. 99:15908–15913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nealson KH, Scott J. 2006. Ecophysiology of the genus Shewanella, p 1133–1151 In Prokaryotes, vol 6 Springer, New York, NY [Google Scholar]
- 15.Hau HH, Gralnick JA. 2007. Ecology and biotechnology of the genus Shewanella. Annu. Rev. Microbiol. 61:237–258 [DOI] [PubMed] [Google Scholar]
- 16.Bandrin SV, Rabinovich PM, Stepanov AI. 1983. 3 linkage groups of the genes of riboflavin biosynthesis in Escherichia coli. Genetika 19:1419–1425 [PubMed] [Google Scholar]
- 17.Balch WE, Wolfe RS. 1976. New approach to the cultivation of methanogenic bacteria: 2-mercaptoethanesulfonic acid (HS-CoM)-dependent growth of Methanobacterium ruminantium in a pressurized atmosphere. Appl. Environ. Microbiol. 32:781–791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Covington ED, Gelbmann CB, Kotloski NJ, Gralnick JA. 2010. An essential role for UshA in processing of extracellular flavin electron shuttles by Shewanella oneidensis. Mol. Microbiol. 78:519–532 [DOI] [PubMed] [Google Scholar]
- 19.Chung CT, Niemela SL, Miller RH. 1989. One-step preparation of competent Escherichia coli: transformation and storage of bacterial cells in the same solution. Proc. Natl. Acad. Sci. U. S. A. 86:2172–2175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Daines DA, Granger-Schnarr M, Dimitrova M, Silver RP. 2002. Use of LexA-based system to identify protein-protein interactions in vivo. Methods Enzymol. 358:153–161 [DOI] [PubMed] [Google Scholar]
- 21.Johnson M, Zaretskaya I, Raytselis Y, Merezhuk Y, McGinnis S, Madden TL. 2008. NCBI BLAST: a better web interface. Nucleic Acids Res. 36:W5–W9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32:1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gouy M, Guindon S, Gascuel O. 2010. SeaView version 4: a multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol. Biol. Evol. 27:221–224 [DOI] [PubMed] [Google Scholar]
- 24.Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22:2688–2690 [DOI] [PubMed] [Google Scholar]
- 25.Jones DT, Taylor WR, Thornton JM. 1992. The rapid generation of mutation data matrices from protein sequences. Comput. Appl. Biosci. 8:275–282 [DOI] [PubMed] [Google Scholar]
- 26.Vitreschak AG, Rodionov DA, Mironov AA, Gelfand MS. 2002. Regulation of riboflavin biosynthesis and transport genes in bacteria by transcriptional and translational attenuation. Nucleic Acids Res. 30:3141–3151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brutinel ED, Gralnick JA. 2012. Anomalies of the anaerobic tricarboxylic acid cycle in Shewanella oneidensis revealed by Tn-seq. Mol. Microbiol. 86:273–283 [DOI] [PubMed] [Google Scholar]
- 28.Ren J, Kotaka M, Lockyer M, Lamb HK, Hawkins AR, Stammers DK. 2005. GTP cyclohydrolase II structure and mechanism. J. Biol. Chem. 280:36912–36919 [DOI] [PubMed] [Google Scholar]
- 29.Liao DI, Calabrese JC, Wawrzak Z, Viitanen PV, Jordan DB. 2001. Crystal structure of 3,4-dihydroxy-2-butanone 4-phosphate synthase of riboflavin biosynthesis. Structure 9:11–18 [DOI] [PubMed] [Google Scholar]
- 30.Kelly MJ, Ball LJ, Krieger C, Yu Y, Fischer M, Schiffmann S, Schmieder P, Kuhne R, Bermel W, Bacher A, Richter G, Oschkinat H. 2001. The NMR structure of the 47-kDa dimeric enzyme 3,4-dihydroxy-2-butanone-4-phosphate synthase and ligand binding studies reveal the location of the active site. Proc. Natl. Acad. Sci. U. S. A. 98:13025–13030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kelley LA, Sternberg MJ. 2009. Protein structure prediction on the Web: a case study using the Phyre server. Nat. Protoc. 4:363–371 [DOI] [PubMed] [Google Scholar]
- 32.Schramek N, Bracher A, Bacher A. 2001. Biosynthesis of riboflavin. Single turnover kinetic analysis of GTP cyclohydrolase II. J. Biol. Chem. 276:44157–44162 [DOI] [PubMed] [Google Scholar]
- 33.Richter G, Volk R, Krieger C, Lahm HW, Rothlisberger U, Bacher A. 1992. Biosynthesis of riboflavin: cloning, sequencing, and expression of the gene coding for 3,4-dihydroxy-2-butanone 4-phosphate synthase of Escherichia coli. J. Bacteriol. 174:4050–4056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Boretskii Iu R, Skoblov Iu S, Khodova OM, Rabinovich PM. 1992. Purification and properties of GTP-cyclohydrolase from Bacillus subtilis. Biokhimiia 57:1021–1030 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.