Summary
Background
The cyanobacterial circadian program exerts genome-wide control of gene expression. KaiC undergoes rhythms of phosphorylation that are regulated by interactions with KaiA and KaiB. The phosphorylation status of KaiC is thought to mediate global transcription via output factors SasA, CikA, LabA, RpaA, and RpaB. Overexpression of kaiC has been reported to globally repress gene expression.
Results
Here we show that the positive circadian component KaiA upregulates “subjective dusk” genes and its overexpression de-activates rhythmic gene expression without significantly affecting growth rates in constant light. We analyze the global patterns of expression that are regulated by KaiA versus KaiC and find in contrast to the previous report of KaiC repression that there is a “Yin-Yang” regulation of gene expression whereby kaiA overexpression activates “dusk genes” and represses “dawn genes,” whereas kaiC overexpression complementarily activates “dawn genes” and represses “dusk genes.” Moreover, continuous induction of kaiA latched KaiABC-regulated gene expression to provide constitutively increased transcript levels of diverse endogenous and heterologous genes that are expressed in the predominant “subjective dusk” phase. In addition to analyzing KaiA regulation of endogenous gene expression, we apply these insights to the expression of heterologous proteins whose products are of potential value, namely human proinsulin, foreign luciferase, and exogenous hydrogenase.
Conclusions
Both KaiC and KaiA complementarily contribute to the regulation of circadian gene expression via Yin-Yang switching. Circadian patterns can be reprogrammed by overexpression of kaiA or kaiC to constitutively enhance gene expression, and this reprogramming can improve 24/7 production of heterologous proteins that are useful as pharmaceuticals or biofuels.
Keywords: Circadian, KaiA, KaiC, Gene Expression, Biotechnology
Introduction
Circadian rhythms are circa-24 h oscillations in biological processes that are controlled by an endogenous biochemical pacemaker to provide a temporal program that allows organisms to optimally adapt to the daily transformation of environmental conditions. This “biological clock” rhythmically orchestrates intracellular activities that range from gene expression, metabolism, cell division, to development and behavior [1]. In eukaryotes, approximately 10% of the genome is regulated by the daily clock in any given tissue [2–5]. In the photoautotrophic cyanobacterium Synechococcus elongatus PCC 7942, virtually all gene expression is controlled at the level of promoter activity by the circadian clock [6,7], and 35–70% of steady-state transcript abundances oscillate [8,9]. The entire chromosome even undergoes daily cycles of topological change and compaction that are related to this genome-wide transcriptional control [9–11].
The KaiA, KaiB, and KaiC proteins form the central clockwork in S. elongatus, and the status of KaiC phosphorylation plays a key role in the central clock mechanism as well as global regulation of output gene expression [12–16]. KaiC is both an autokinase and an autophosphatase, but KaiA promotes phosphorylation of KaiC [17] while KaiB combines with KaiA and KaiC to form a complex that promotes KaiC dephosphorylation [18,19]. There are multiple pathways that link the central KaiABC post-translational oscillator (PTO)[20,21] to its transcriptional outputs that include the proteins SasA, CikA, LabA, RpaA, and RpaB [12,14,16,22]. In particular, RpaA appears to be a major output node that is regulated by KaiABC through independent SasA/CikA/LabA pathways [14]. The global gene expression patterns in S. elongatus are primarily organized into two groups that are phased 180° apart [6]. The “Class I” or “subjective dusk” genes activate at dawn and rise throughout the day to a peak expression at dusk, while the “Class II” or “subjective dawn” genes turn on in the subjective night and peak at dawn [6]. The Class I (subjective dusk) genes are the predominant group. It has been reported that overexpression of kaiC represses the rhythmic components of all genes in the genome [23]. However, a more recent microarray analysis concluded that kaiC overexpression represses the predominant Class I (dusk) genes, while up-regulating Class II (dawn) genes [8].
Insights into the regulation of gene expression in cyanobacteria are important for understanding the basic biology of circadian control, but also have potential applications. Because they derive their energy from the sun and are genetically malleable, photoautotrophic cyanobacteria are attractive bioreactors for synthesizing biofuels and other bioproducts [24–27]. In particular, S. elongatus has become one of the preferred platforms for development of this biotechnology [25–27]. However, despite the appeal of directing photosynthetic carbon fixation towards the production of useful molecules in cyanobacterial hosts, the efficiency of heterologous expression achieved by cyanobacteria is currently too low for industrial application. Furthermore, few tools are available to reprogram metabolic flux in photosynthetic microbes along pathways towards the synthesis of useful bioproducts or their precursors [26,28].
Due to the pervasive circadian control of promoter activities in cyanobacteria, experimental modulation of clock genes could be exploited to tune gene expression to operate maximally under constant-light conditions or to resonate in harmony with periodic light-dark cycling. In this study, we examine the phased expression patterns and find that kaiA- vs. kaiC-overexpression (kaiA-OX vs. kaiC-OX) exhibit opposing actions over promoters such that the genome-wide patterns of both dusk (Class I) and dawn (Class II) genes can be explained. We refer to the opposing actions of kaiA-OX vs. kaiC-OX as a “Yin-Yang” interdependency, based on the Taoist concept of balancing forces that complementarily interact to promote harmony. We then use this basic information to reprogram circadian clock-controlled circuits to switch from cycling to constitutive gene expression as well as manipulating the expression of the kaiA gene to enhance expression of endogenous and foreign genes. As proof of principle, we have applied this strategy towards optimizing the expression of endogenous and foreign [NiFe] hydrogenases for biohydrogen production and expression of foreign genes such as luciferase and human proinsulin (a test case for production of pharmaceuticals in cyanobacteria).
Results
KaiA-OX enhances expression of subjective dusk genes: microarray analyses
In vivo overexpression of kaiC has been claimed to globally repress gene expression in S. elongatus [23], but the converse manipulation of pervasively enhancing gene expression by manipulation of the clock has not been studied. Since the KaiABC-based oscillator globally regulates gene expression in cyanobacteria [6,8,9] and kaiA-OX enhances the expression of the kaiBC promoter [29], we reasoned that KaiA could be enlisted to act as a positive regulator to enhance expression on a genomic scale. Using a luciferase reporter of the expression of the Class I photosynthetic gene psbAI (psbAIp::luxAB), we found that the response of the psbAI promoter to overexpression of kaiA (kaiA-OX) is both acute and sensitive (Figure S1A–C). When kaiA expression was stimulated with the inducer isopropyl β-D-1-thiogalactopyranoside (IPTG), psbAIp::luxAB expression quickly increased to a high level that was essentially arhythmic (Figure S1B), and this response to kaiA-OX was dependent on IPTG dose; concentrations of IPTG as low as 15–20 μM eliminated the clock-controlled luminescence rhythm (Figure S1C). The addition of IPTG to cells that do not harbor an IPTG-derepressible promoter (i.e., trcp) does not elicit any changes in gene expression ([29] and unpublished observations). Moreover, overexpression of kaiA had no marked effect on the growth rates among different reporter strains in constant light, aka LL (Figure S1D).
To evaluate KaiA's genome-wide regulation, we performed microarray assays in the kaiA-overexpressing strains with or without IPTG induction (Figure 1A,Table S1). The expression profile of each clock-controlled gene in kaiA-OX cells with IPTG induction in constant light (LL) from 30 to 48 h was compared with that in the absence of IPTG (LL30–48) (Figure 1A, Table S1). In response to kaiA-OX, about 20% of the genes were up-regulated and about 12% were down-regulated, with the remaining ~68% of genes not clearly affected by kaiA-OX (Table S1). In comparison with the genes that are repressed vs. enhanced by overexpression of kaiC (kaiC-OX) [8], there is a clear trend that kaiA-OX and kaiC-OX have opposite effects for most genes (Figure 1B; Figure S2A & S2B). Among 800 cycling genes revealed by microarrays, kaiA-OX mostly up-regulates “subjective dusk” genes (expression mostly in the daytime, with peak expression at Circadian Time 12 {CT12} ≈ 36 h in LL, aka Class I genes [6]) and down-regulates “subjective dawn” genes (peak expression at CT0 ≈ 24 & 48 h in LL, aka Class II genes [6], Figure 1C–1E), whereas kaiC-OX was shown to repress dusk genes and activate dawn genes [8].
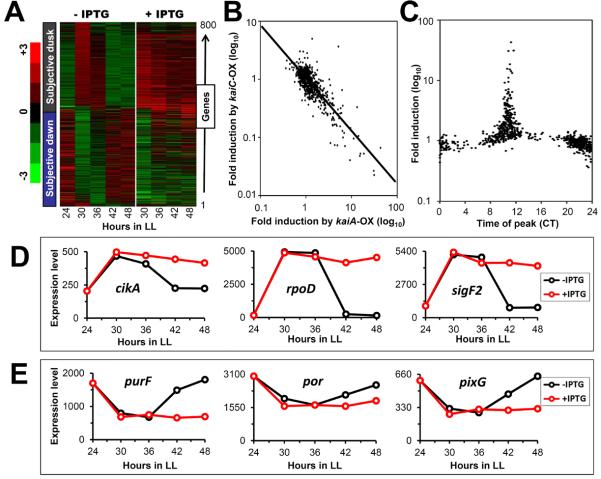
Figure 1.
Microarray profiles of cycling genes in the kaiA-overexpressing (kaiA-OX) strain
(A) Expression profiles of 800 cycling genes in the kaiA-OX strain in LL with or without 1 mM IPTG. These genes were sorted by peak time expressed by wild-type strains [8]. The colors represent normalized data arranged in descending order from red, black, to green representing expression levels from high to low. The average and S.D. over one cycle is 0.0 and 1.0, respectively.
(B) Correlation of the level of induction for ~800 circadian cycling genes. The expression level of each clock-controlled gene in Ptrc∷kaiA cells in the presence of IPTG at LL48 was compared with that in the absence of IPTG at LL48. The abscissa indicates the fold induction by kaiA overexpression at LL48. The ordinate indicates the fold induction by kaiC overexpression at LL33 for the same genes [8] (regression line is R2 = 0.683).
(C) Induction of KaiA up-regulates and down-regulates subjective dusk and dawn genes, respectively. The expression level of each clock-controlled gene in trcp∷kaiA cells in the presence of IPTG at LL48 was compared with that in the absence of IPTG at LL48. The ordinate shows the amount of induction of each gene by kaiA-OX, while the abscissa indicates the peak time of each of the 800 cycling genes in Circadian Time (based on the patterns reported in [8]), (CT0/24 = dawn, CT12 = dusk).
(D) and (E) Temporal expression profiles of representative kaiA-enhanced subjective dusk genes (D) and kaiA-repressed subjective down genes (E) from microarray analysis. Expression profiles of genes in the kaiA-OX strain with (red) or without 1 mM IPTG (black). The number in the ordinate indicates relative expression level.
While there may be some positional effects of these opposing regulatory patterns based on rhythmic changes in chromosomal topology [9–11], there is not an obvious clustering pattern along the chromosome of the genes that are up-regulated vs. down-regulated by kaiA-OX (Figure 2, Figure S2C). On the other hand, there is an obvious correlation of genes along the chromosome that are up-regulated by kaiA-OX with those that are inversely down-regulated by kaiC-OX, and vice-versa (Figure 2), as confirmed by the statistically significant regression shown in Figure 1B (R2 = 0.683). Therefore, kaiA-OX vs. kaiC-OX complementarily regulate circadian expression patterns. As will be shown below, continuous overexpression of kaiA locks the expression of these output genes at constitutively high or low levels and arrests rhythmic expression by the constitutive hyperphosphorylation of KaiC [17,20,30].
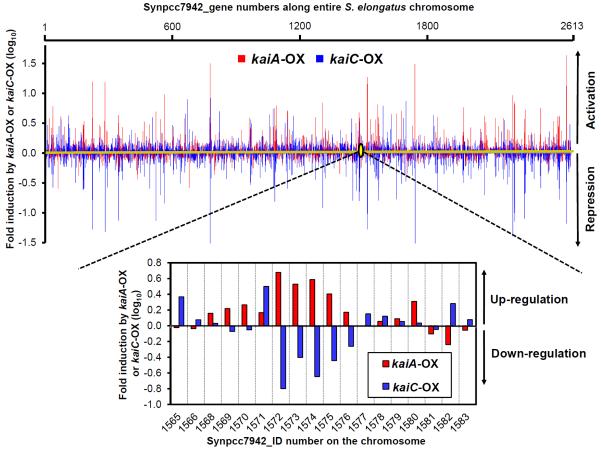
Figure 2.
Spatial patterns of gene expression along the entire S. elongatus chromosome elicited by kaiA/kaiC overexpression.
The S. elongatus chromosome is circular, but it is here shown in linear format with the expression levels of each gene in response to kaiA-OX (red, this study) or kaiC-OX (blue, [8]). Changes of gene expression are shown as the ratio of the transcript abundance in the presence of IPTG to that in the absence of IPTG in LL. Each ratio was arranged in ascending order of Synpcc7942 gene number (i.e., “1200” means gene “Synpcc7942_1200”). The lower panel magnifies the region encompassing Synpcc7942_1565 to Synpcc7942_1583 where changes expression levels regulated by kaiA-OX and kaiC-OX are denoted in red and blue, respectively. Increased gene expression in response to the indicated overexpression is classified as up-regulation (activation), whereas decreased transcript levels are considered down-regulation (repression).
Effect of kaiA-OX on gene expression at “neutral sites” using luminescence reporters
To monitor in real-time the effect of KaiA on promoter activities, we examined luminescence reporters driven by the promoters of diverse S. elongatus genes, including the central clock genes (kaiA and kaiBC), the photosynthesis gene psbAI, the purine biosynthesis pathway gene purF, and the cell division gene ftsZ (Table S2). These genes exemplify both expression patterns: kaiAp, kaiBCp, psbAIp, and ftsZp are Class I promoters, while purFp is a Class II promoter. We also examined heterologous E. coli promoters that are recognized by the transcriptional apparatus of S. elongatus, such as those from the fis factor for site-specific DNA inversion (a marker of local DNA topology [31]), an IPTG-derepressible heterologous promoter trc [32], and the σ70 binding site gene conII [33], all of which are expressed in the Class I (dusk) phase in S. elongatus (Figure S3B). Although all of these reporters were expressed in both E. coli and S. elongatus, their expression levels were quite different between these two bacteria. While reporters driven by cyanobacterial promoters express at much lower levels in E. coli than those from E. coli (a phenomenon that is particularly noticeable in the case of the psbAI promoter; Figure S3A), in S. elongatus the E. coli reporters exhibited both the strongest (e.g. conIIp∷luxAB) and the weakest expression (e.g. fisp∷luxAB; Figure S3B). Nevertheless, all of the reporters–independent of the source of the promoter–were rhythmic in cyanobacteria (Figure S3B), a phenomenon that is likely due to circadian control over chromosomal topology in S. elongatus that modulates promoter activity globally [11]. We then integrated an IPTG-inducible expression cassette of wild-type kaiA with a 5'-untranslated sequence (trcp∷kaiA) into either neutral site I or neutral site II of these reporter strains to examine the impact of kaiA-OX on the activity of these various promoters (Table S2). Overexpression of kaiA constantly enhanced the promoter activities of the central clock genes (kaiAp and kaiBCp) when IPTG was applied to cells at either the beginning of LL treatment (LL0) or 48 h later (LL48), whereas kaiBC-OX repressed kaiBCp activity (Figures 3A, S4A and S4B, also [17,29,32,34]).
Figure 3.
KaiA enhances expression of the central clock genes and E. coli promoters.
(A) Constant kaiA-OX activates expression of the central clock genes and attenuates rhythmicity. Luminescence profiles were measured in a kaiA reporter (kaiAp∷luxAB in NS I, i.e. kaiAp∷lux) or kaiBC reporter (kaiBCp∷luxAB in NS I, i.e. kaiBCp∷lux) co-expressing trcp∷kaiA with or without IPTG. A final concentration of 1mM IPTG was applied either at LL0 (hour 0) or LL48 (hour 48) of constant light (LL) exposure. As a comparison with KaiC-induced repression, the kaiBC reporter strain (kaiBCp∷luxAB, i.e. kaiBCp∷lux) co-expressing trcp∷kaiBC was performed in parallel for IPTG application at LL48.
(B) Northern blot assays for mRNA expression of kaiA, kaiBC, and luxAB in the kaiA-overexpressing reporter strain with or without IPTG in LL (see also Figure S4C & S4D).
(C) KaiA-OX enhances the abundance of KaiB and KaiC proteins and hyper-phosphorylates KaiC. Cultures were collected at Circadian Time 04 (CT04) in LL in presence of different concentrations of IPTG. Ratios of hyper-P KaiC to hypo-P KaiC are shown numerically below the KaiC blots (see also Figure S4E & S4F for the case of {ATG}kaiA-OX). The bottom row shows equivalent loading by Coomassie Brilliant Blue (CBB) staining.
(D) kaiA-OX promotes constant high expression of the luxAB luminescence driven by the E. coli conII promoter in the cyanobacterial reporter strain (conIIp∷luxAB) co-expressing trcp∷kaiA in the absence or presence of IPTG (i.e. H2O [No IPTG] or 1 mM IPTG at LL0 or LL48).
(E) Overall effect of kaiA overexpression on conII promoter activity. Total luminescence units in LL for 7 days were collected from cultures of uniform cell density (30 μl at OD750 = 0.3) placed on agar medium from the conIIp∷luxAB strain co-expressing trcp∷kaiA in the absence (H2O) or presence of 1mM inducer (IPTG). (mean +/− S.D. for triplicates)
(F) KaiA expression potentiates induction by the IPTG-derepressible heterologous trc promoter. Luminescence expression profiles were compared between a strain harboring the trcp∷luxAB reporter alone and a strain co-expressing both the trcp∷luxAB reporter and trcp∷kaiA in the absence or presence of 1 mM IPTG added at LL48.
(G) Co-induction of the reporter activities by the IPTG and KaiA in the trcp∷luxAB reporter strain co-expressing trcp∷kaiA in the absence (No IPTG) or in the presence of 1mM IPTG started at LL0 or LL48. Note the log10 scale for the ordinate.
Moreover, kaiA-OX increased the levels of kaiBC and luxAB transcripts when IPTG was added at LL24 (Figure 3B, Figure S4C & S4D). Increased KaiA also enhanced the abundance of the KaiB and KaiC proteins, and promoted the hyperphosphorylation of KaiC (Figure 3C), which is consistent with KaiA's ability to stimulate KaiC phosphorylation in vitro and in vivo [17,20,30,35,36], and inhibit dephosphorylation [32]. To obtain even stronger production of KaiA, in some of our experiments we used an IPTG-derepressible trcp∷kaiA fusion gene with an ATG start codon, i.e. trcp∷{ATG}kaiA (rather than the aforementioned construct with a GTG start codon) (Figure S4E & S4F). In contrast to the consequences of kaiA-OX, kaiC-OX floods the system with newly synthesized KaiC, which reduces the overall phosphorylation status of the KaiC pool [32,34]. Therefore, kaiA-OX and kaiC-OX have complementary effects on the status of KaiC phosphorylation and therefore mimic opposite points of the endogenous oscillation of KaiC phosphorylation that are 180° out of phase (see Figure 6A later).
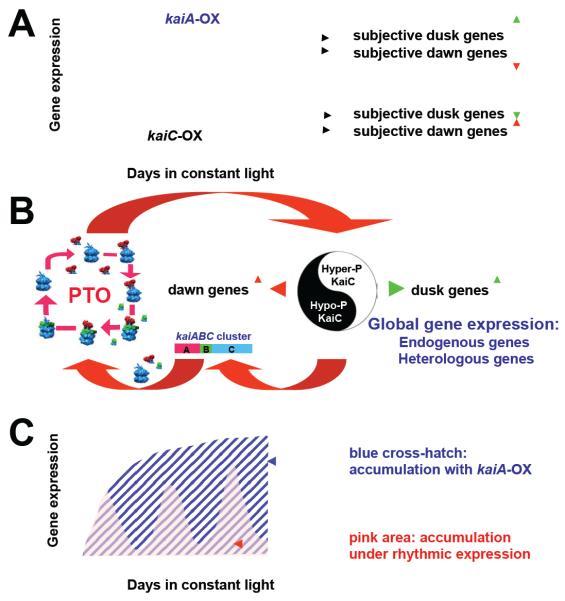
Figure 6.
Complementary regulation by KaiA and KaiC and its manipulation to globally reprogram gene expression.
(A) By constantly overexpressing kaiA (kaiA-OX), subjective dusk (Class I) genes are up-regulated (Class II subjective dawn genes are down-regulated) and rhythmicity of gene expression is lost. This gene expression pattern (red curve) mimics that of subjective dusk genes (peak expression of the kaiBC gene) during the normal rhythm. Constant overexpression of kaiC (kaiC-OX) has the opposite effects on Class I vs. II gene expression and represses activity of subjective dusk genes [23,29].
(B) These complementary gene expression patterns are mediated by the central circadian clock that is composed of a post-translational oscillator (PTO) that regulates the oscillation of KaiC phosphorylation status, leading to a Yin-Yang output of global gene expression (including kaiABC expression) where hyperphosphorylated KaiC up-regulates dusk (Class I) genes and hypophosphorylated KaiC up-regulates dawn (Class II) genes. Overexpression of kaiA vs. kaiC can lock the Yin-Yang patterns into either dusk phase (kaiA-OX) or dawn phase (kaiC-OX). The KaiABC PTO cycles the phosphorylation status of KaiC (blue hexamers) as regulated by interactions with KaiA (red dimers) and KaiB (green monomers).
(C) Latching the Yin-Yang pattern into dusk phase by kaiA-OX enhances the expression of dusk genes–including heterologous genes inserted into NSI or NSII–and leads to a greater accumulation of gene products in constant light (LL) as shown by the blue cross-hatched area than would be possible if the same gene were expressed under control of the native rhythmic system (depicted by the pink area).
An indicator of the global, non-discriminatory control of the S. elongatus genome by the KaiABC-based clock is whether heterologous promoters/genes can be controlled in a similar fashion to endogenous promoters/genes. To determine if kaiA-OX can enhance expression of foreign gene promoters, we tested strains expressing luxAB under the control of conIIp–a strong promoter that is “constitutively” expressed in E. coli [33]. Luminescence activities controlled by conIIp exhibited high levels of rhythmic expression in S. elongatus, and kaiA-OX further enhanced this strong promoter's activity up to ~ 3.5 fold higher than that of controls without kaiA induction (Figure 3D & 3E). This kaiA-OX mediated enhancement of luminescence activity under the control of conIIp appears to occur by boosting the trough levels of the conIIp∷luxAB expression profile to be at or above the peak levels (Figure 3D). KaiA also positively regulates a usefully inducible, non-cyanobacterial promoter, trcp. KaiA-mediated enhancement of luminescence expression is particularly dramatic in a S. elongatus strain co-expressing both IPTG-inducible constructs (trcp∷luxAB reporter and trcp∷kaiA) (Figure S5). In the absence of IPTG induction, the artificial promoter trc could drive circadian luminescence oscillations at low levels in both the reporter strain (trcp∷luxAB) and the reporter/kaiA-coexpressing strain (trcp∷luxAB + trcp∷kaiA; Figure 3F & Figure S5B). In the presence of IPTG, overall promoter activity was increased in the trcp∷luxAB reporter strain but the pattern remained rhythmic (Figure 3F), whereas in the strain co-expressing the trcp∷luxAB reporter and trcp∷kaiA, kaiA overexpression further stimulated the activity of the strong trc promoter but the rhythmic pattern was lost (Figure 3F & 3G). Therefore, kaiA-OX not only enhances the expression of diverse cyanobacterial “subjective dusk” Class I genes in situ (Figure 1), it also stimulates the activity of cyanobacterial kai promoters and of the strong E. coli promoters conIIp and the IPTG-depressible promoter trcp when placed into neutral sites NSI or NSII (Figure 3).
Enhancing expression of endogenous hydrogenase genes by kaiA-OX
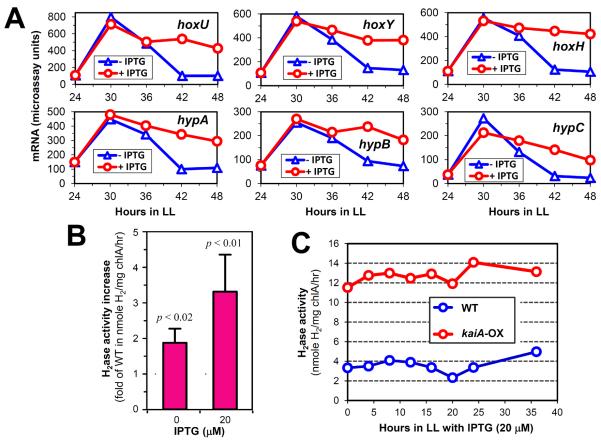
The quest to understand the role of KaiA in regulating KaiC phosphorylation status and therefore gene activity led to the recognition that expression of subjective dusk genes (the predominant Class I genes [6]) could be constitutively enhanced by this rewiring of the circadian circuitry. Thereby, enhanced production of useful bioproducts, such as biofuel compounds encoded by foreign and/or endogenous genes, could be accomplished by kaiA-OX. Hydrogen (H2) is an attractive carbon-free energy storage molecule, and production of H2 using photosynthetic cyanobacteria could provide an alternative to fossil fuels by using solar energy to convert H2O into hydrogen [37]. Hydrogenases catalyze the reversible reduction of protons to H2 and can be divided into three phylogenetically-distinct categories that correlate with the metal composition of the active site: [FeFe], [NiFe], and [Fe]-cluster-free hydrogenases [38,39]. S. elongatus has one native [NiFe] hydrogenase. Interestingly, our microarray analysis revealed that kaiA-OX promoted mRNA profiles of all NAD-reducing hydrogenase subunits (Figure 4A, Table S1). Therefore, we first examined the impact of kaiA-OX (trcp∷{ATG}kaiA) upon the expression of endogenous [NiFe] hydrogenase in S. elongatus. As shown in Figure 4B, endogenous [NiFe] hydrogenase activities were enhanced in the kaiA-OX strain in constant light even without IPTG induction due to “leaky” expression of KaiA in the trcp∷{ATG}kaiA strain. Mild induction of KaiA with 20 μM of IPTG further boosted hydrogenase activity. Overexpression of KaiA also caused consistently high levels of hydrogenase activity in a LL time course experiment in the kaiA-OX strain (+ 20 μM IPTG) relative to wild-type strain (Figure 4C).
Figure 4.
KaiA promotes expression and activity of endogenous Hox hydrogenase (H2ase).
(A) kaiA-enhanced mRNA microarray profiles of all H2ase subunits and H2ase maturation genes (as denoted) in the kaiA-overexpressing strain with (red) or without IPTG (blue).
(B) Hydrogenase activity increases in the {ATG}kaiA-OX strain relative to the wild-type (WT) strain in the absence or presence of IPTG induction. Data are represented as mean +/− SD from three independent experiments and analyzed with t-test. Note that the {ATG}kaiA overexpression construct is slightly “leaky”, and even at 0 μM IPTG, there is some extra expression of KaiA above that in WT.
(C) KaiA causes constantly high levels of hydrogenase activities under continuous light conditions. Following a 12h of dark pulse, the endogenous hydrogenase activities were measured from samples of the WT and {ATG}kaiA-OX strains at the indicated times in LL.
kaiA-OX-enhanced production of foreign proteins in LD, LL, and DD
The kaiA-OX strategy can also be used to enhance the expression of heterologous genes, resulting in the accumulation of foreign proteins. Bacterial luciferase is an example of a foreign protein that is expressed well in S. elongatus as a reporter of promoter activity (e.g., Figure 3). Moreover, our kaiA-OX strategy up-regulates the accumulation of LuxAB protein very significantly (nearly 7-fold in the experiment depicted in Figure 5A & 5B). As indicated above, hydrogenase is a protein of biotechnological interest, but because photosynthesis generates oxygen, production-scale generation of H2 by photosynthetic microbes will ultimately require exogenous hydrogenases that are more tolerant of oxygen [40]. While [FeFe] and [Fe]-hydrogenases are rapidly inactivated by oxygen, [NiFe]-hydrogenases are more active in the presence of photosynthetically produced oxygen [41]. Recently, heterologous expression of a [NiFe]-hydrogenase from Alteromonas macleodii Deep Ecotype with tolerance to partial oxygen was demonstrated in S. elongatus, generating a strain called RC41 [42]. RC41 features a knockout of the endogenous hoxYH genes encoding the bidirectional [NiFe] hydrogenase (HoxYH), and the trcp-driven expression cluster encoding the A. macleodii [NiFe] hydrogenase HynSL and other 11 surrounding accessory proteins were expressed from the NS I site under the control of trcp as shown in Figure S6A. Generally, two subunits (ca. 60 kDa and 30 kDa) are involved in the catalytic core of [NiFe] hydrogenases, and the larger subunit contains the [NiFe] catalytic site that requires an extensive set of accessory proteins to assemble an active catalytic site [43]. While the RC41 strain achieves some hydrogenase activity, low expression of the multiple hydrogenase & accessory protein genes was problematic [43].
Figure 5.
KaiA enhances expression and accumulation of foreign genes and proteins.
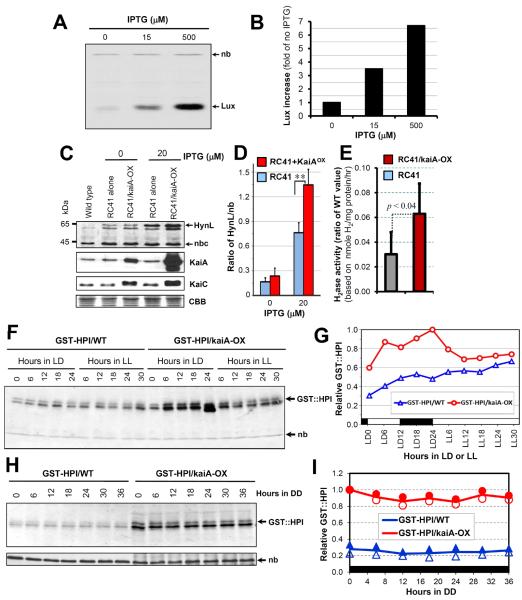
(A) Accumulation of Vibrio harveyi luciferase (Lux) was enhanced by induction of kaiA with or without IPTG (0, 15, 500 μM IPTG) in the strain co-expressing psbAIp∷luxAB and trcp∷{ATG}kaiA. A constitutive nonspecific band is marked “nb.”
(B) Densitometry of the V. harveyi Lux abundance from the immunoblot in panel A, which was calculated from the ratio of Lux:nb abundance (1 = Lux:nb at 0 μM IPTG).
(C) KaiA enhances expression of heterologous hydrogenases. Immunoblot assays for expression of HynL, KaiA, and KaiC in strains of the wild type, RC41, and RC41 co-expressing trcp∷{ATG}kaiA (i.e RC41/kaiA-OX) with (20 μM) or without IPTG (0 μM) for 24 hours. The top row shows both KaiA-enhanced and IPTG-induced expression of the large subunit (HynL, ~69 kDa) of the A. macleodii hydrogenase. “nb” denotes a nonspecific band recognized by the antisera raised against Thiocapsa roseopersicina HynL, which was used as an internal control for quantitative analyses of HynL expression levels. The 2nd and 3rd rows confirm the KaiA overexpression and its enhancement of the hyperphosphorylated KaiC expression in the RC41/kaiA-OX strain. The bottom row shows equivalent loading by CBB staining. Note that the kaiA overexpression construct with an ATG start codon is slightly “leaky” with a higher expression level as compared with WT even without IPTG induction.
(D) Densitometry of expression levels of the A. macleodii hydrogenase subunit HynL from the top panel of (C). Ratios of the HynL/nb signals are averages with standard deviations from three experiments. ** p < 0.001 in a paired t-test.
(E) Activity of the foreign A. macleodii hydrogenase in S. elongatus. After 24 h induction by 20 μM IPTG in light, the hydrogenase activities were determined. Data are the averages with S.D. from four independent experiments, and the hydrogenase activities in the RC41 and RC41/kaiA-OX strains were shown as ratios of the values as compared with wild type.
(F) KaiA enhances accumulation of human proinsulin protein (GST∷HPI fusion protein) in LD and LL (time 0 = beginning of light; LD = 12 h light/12 h dark cycle). Cells expressing conIIp∷GST-HPI (GST-HPI/WT) or co-expressing conIIp∷GST-HPI and trcp∷{ATG}KaiA (GST-HPI/KaiA) were grown in the presence of 1 mM IPTG and collected at indicated LD and LL time points. The immunoblot assay for the fusion protein GST∷HPI was performed using a monoclonal antibody against GST. “nb” denotes a nonspecific band recognized by the GST antibody.
(G) Densitometry of the GST-HPI expression levels from the data of panel (F). Abscissa: black bar indicates the dark portion of a light/dark (LD) cycle, and the white bar indicates illumination in LD or LL.
(H) Constant enhancement of the GST∷HPI production by kaiA overexpression in DD. The LL-grown cells from the strains GST-HPI/WT or GST-HPI/KaiA were given a 12 h dark treatment, then 1 mM IPTG was applied at lights-on. After an additional 12 h growth in light, the cultures were transferred to constant darkness in a shaking water bath with bubbling and cells were collected every 6 h. “nb” denotes a nonspecific band recognized by the GST antibody.
(I) Densitometry of the GST-HPI expression levels from two experiments in DD.
To test if KaiA can stimulate the expression of the foreign [NiFe] hydrogenase from A. macleodii in S. elongatus, we introduced the trcp::{ATG}kaiA construct into the NS II site of the RC41 strain and examined the abundance of the large subunit, HynL, as a marker for expression of the foreign A. macleodii hydrogenase cassette. We found that neither deletion of endogenous hoxYH genes nor overexpression of A. macleodii hydrogenase cluster genes affected the period or phase of the clock in S. elongatus (Figure S6B). When kaiA was additionally expressed in the RC41 strain (+ 20 μM of IPTG), the abundance of the foreign A. macleodii hydrogenase large subunit, HynL, significantly increased relative to a control strain without the trcp::{ATG}kaiA expression cassette (Figure 5C & 5D). Immunoblot assays confirmed that kaiA-OX also enhanced KaiC protein levels in the hoxYH-null mutant strain co-expressing A. macleodii hydrogenase cluster genes and trcp::{ATG}kaiA (Figure 5C & Figure S6C). Compared to the native hydrogenase activity in wild-type S. elongatus, the activity of the foreign A. macleodii hydrogenase in the RC41 strain is lower (Figure 5E), and therefore methods to further enhance activity would be necessary before this strategy could be useful industrially. We conjecture that part of the difficulty could be that this hydrogenase operon is so large (about 13 kb) that not all of the genes are expressed well. Additionally, there may be post-transcriptional constraints to overcome so as to achieve higher hydrogenase activity in vivo in S. elongatus. Nevertheless, overexpression of kaiA increased approximately twofold the activity of H2 production from the foreign [NiFe] hydrogenase as well as HynL abundance in the RC41 strain (Figure 5C–5E).
As an another example illustrating how manipulation of kaiA expression can enhance production of foreign proteins in cyanobacteria, we generated a GST::HPI/KaiA strain, in which a fusion protein between the foreign gene encoding human proinsulin (HPI) and the glutathione S-transferase (GST) tag was expressed under the control of the non-cyanobacterial promoter conIIp in NS I, and the expression cassette trcp::{ATG}kaiA was cloned into NS II (Figure S6D). Under both light:dark (LD) and LL conditions, kaiA-OX increased production of the foreign GST::HPI fusion protein (Figure 5F & 5G, Figure S6E & S6F). We noticed that the accumulation of GST::HPI was particularly high in the dark portion of LD (Figure 5F & 5G), so we tested the expression under constant darkness (DD) and found that kaiA-OX significantly enhanced the accumulation of GST::HPI in extended darkness (Figure 5H & 5I), which was unexpected given that S. elongatus is an obligate photoautotroph.
Discussion
The circadian rhythm of KaiC phosphorylation regulates the global patterns of gene expression in S. elongatus [12–15]. The peak and trough levels of the KaiC phosphorylation rhythm can be mimicked by overexpression of KaiA or KaiC, respectively (Figure 6A). Therefore, the opposing actions of kaiA-OX vs. kaiC-OX form a “Yin-Yang” action, by analogy to the Taoist concept of inverse forces that complementarily interact to form a greater whole (Figure 6A & 6B). Increased KaiA levels stimulate KaiC phosphorylation [17] and inhibits KaiC dephosphorylation [32], thereby promoting KaiC hyperphosphorylation and expression of dusk (Class I) genes (Figure 6B). In the usual post-translational oscillator (PTO) cycle, hyperphosphorylated KaiC interacts with KaiB to form a KaiA/KaiB/KaiC complex that allows KaiC to dephosphorylate [18,19], and this process can be induced by kaiC-OX, which disturbs the normal stoichiometry of Kai A:B:C proteins. The phosphorylation status of the PTO then regulates transcriptional endpoints by output pathways that include SasA, CikA, LabA, RpaA, and RpaB [12,14,16,22]. Therefore, kaiA-OX vs. kaiC-OX inversely switch the KaiC phosphorylation status and gene expression patterns between dusk (kaiA-OX) and dawn (kaiCOX) phases (Figure 6A & 6B). In addition, constant induction of KaiA or KaiC both lead to arhythmic expression patterns [29] (Figure 6A).
Our experimental observations led us to re-evaluate the claim that the “negative element” KaiC is a global repressor of gene expression [23]. In fact, microarray analyses of the impact of kaiC-OX on gene expression in S. elongatus [8] in conjunction with our examination of kaiA-OX herein reveal that BOTH KaiA and KaiC can repress AND enhance transcript abundances, and that they appear to have opposite effects on the expression of many genes (Figures 1 & 2; Figure S2A & S2B). Why then did it appear that KaiC is a global repressor of promoter activities [23]? That conclusion was based upon the insertion of many different randomly chosen promoters into neutral site I (NSI). That particular site in the S. elongatus genome (position 2578661) is downregulated by kaiC-OX [8] and up-regulated by kaiA-OX (Figure S2C). We suggest that the neutral site chosen for the random promoter analysis is topologically regulated by the circadian system [11] so that any promoter placed in that site is repressed by KaiC and enhanced by KaiA as a Class I gene independently of how the promoter is regulated in situ.
We exploit these insights into the fundamental regulation of gene expression in S. elongatus to propose a strategy for maximizing the expression of genes that encode industrially useful products, where non-rhythmically “latching” production at the peak level would be optimal. In this investigation, we report that overexpression of kaiA up-regulates many endogenous genes in situ as well as foreign genes expressed from NS I and NS II. Moreover, kaiA-OX attenuates the circadian rhythm, so that latching of expression at the circadian peak level for many genes is achievable. Surprisingly, this reprogramming of circadian expression patterns does not appear to have significant impact upon growth rates of S. elongatus in constant light. Therefore, enhanced accumulation of a useful product would be expected with kaiA-OX (compare the blue cross-hatched area with the pink area in Figure 6C).
In addition to the impact of kaiA-OX on expression levels of foreign genes expressed from NS I or NS II, the suppression of the circadian rhythm by kaiA-OX can also be advantageous if production of bioindustrial molecules or other gene products is conducted over a grueling 24 h/day, 7 days/week schedule (i.e., “24/7”) under constant illumination. Consistent expression over 24 h with cells maintained in LL opens the possibility of creating bioproduct in both the day phase and the night phase (S. elongatus cells are normally quiescent in the night phase), thereby boosting yield (Figure 6C). The data of Figure 5F–5I also suggest that the expression of some foreign proteins may be stronger in the dark in combination with kaiA-OX. Because the transcription & translation of most endogenous genes is shut down during the dark in S. elongatus [44], this may allow the new synthesis that occurs in darkness to be preferentially weighted to that of foreign genes of industrial interest. We show here the application of stimulating the production of biofuel- and pharmaceutical-related proteins, but this tactic can be potentially used to increase expression of any protein or pathway of industrial importance. Moreover, the overall principle of inactivating the circadian system so that it latches at the peak expression is not restricted to cyanobacteria, but may be useful for 24/7 industrial applications with any organism that has a circadian clock, including eukaryotic organisms where the circadian system regulates 10–20% of the genome (e.g., transgenic expression in plants as “bioreactors”).
Experimental Procedures
Design of endogenous and foreign gene expression constructs, generation of clock-manipulated strains, conditions of cell growth, measurement of luminescence rhythms, performance of microarray assays, and procedures for northern blotting, immunoblotting, quantitative real-time PCR, and hydrogenase activity (as well as statistical analyses) are described in the Supplemental Information.
Supplementary Material
Highlights
KaiA overexpression hyperphosphorylates KaiC and activates “subjective dusk” gene expression
KaiC overexpression complementarily regulates phased gene expression (“dusk” genes down, “dawn” genes up)
KaiA overexpression increased expression of diverse promoters/genes
Circadian reprogramming can improve 24/7 production of useful heterologous proteins in cyanobacteria
Acknowledgements
We are grateful to our colleagues at Vanderbilt for valuable discussions, especially Drs. Jamey Young, Tetsuya Mori, and Brian Robertson, and to Walter A. Vargas for technical assistance. We thank Dr. Susan Golden for the gifts of the neutral site vectors and luminescence reporter plasmids psbAIp∷luxAB and fisp∷luxAB, Dr. Shinsuke Kutsuna for the kaiA-overexpression constructs in NS II, Dr. Peter Greenberg for the antibody to bacterial luciferase, and Dr. Tetsuya Mori for the ftsZp∷luxAB DNA. This research was supported by grants from National Institute of General Medical Sciences (NIGMS R01 GM067152 and R01 GM088595 to CHJ), the Hydrogen, Fuel Cells, and Infrastructure Technology Program of the U.S. Department of Energy (DE-FG36-05GO15027, PW & QX), the Japanese Society for the Promotion of Science (23657138 and 23687002) and the Asahi Glass Foundation to HI, and the Yoshida Scholarship Foundation to MU.
Abbreviations
- LL
Constant light
- DD
Constant darkness
- LD
Light/dark cycle
- CT
Circadian Time (CT0 = subjective dawn, CT12= subjective dusk)
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The authors have declared that no financial conflict of interest exists.
References
- 1.Dunlap JC, Loros JJ, DeCoursey PJ. Chronobiology: biological timekeeping. Sinauer Associates, Inc. Publishers; Massachusetts: 2004. [Google Scholar]
- 2.Dong W, Tang X, Yu Y, Nilsen R, Kim R, Griffith J, Arnold J, Schüttler HB. Systems Biology of the Clock in Neurospora crassa. PLoS ONE. 2008;3:e3105. doi: 10.1371/journal.pone.0003105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ueda HR, Matsumoto A, Kawamura M, Iino M, Tanimura T, Hashimoto S. Genome-wide transcriptional orchestration of circadian rhythms in Drosophila. J. Biol. Chem. 2002;2771:4048–4052. doi: 10.1074/jbc.C100765200. [DOI] [PubMed] [Google Scholar]
- 4.Storch KF, Lipan O, Leykin I, Viswanathan N, Davis FC, Wong WH, Weitz CJ. Extensive and divergent circadian gene expression in liver and heart. Nature. 2002;417:78–83. doi: 10.1038/nature744. [DOI] [PubMed] [Google Scholar]
- 5.Harmer SL, Hogenesch JB, Staume M, Chang HS, Han B, Zhu T, Wang X, Kreps JA, Kay SA. Orchestrated transcription of key pathways in Arabidopsis by the circadian clock. Science. 2000;290:2110–2113. doi: 10.1126/science.290.5499.2110. [DOI] [PubMed] [Google Scholar]
- 6.Liu Y, Tsinoremas NF, Johnson CH, Lebedeva NV, Golden SS, Ishiura M, Kondo T. Circadian orchestration of gene expression in cyanobacteria. Genes Dev. 1995;9:1469–1478. doi: 10.1101/gad.9.12.1469. [DOI] [PubMed] [Google Scholar]
- 7.Nair U, Ditty JL, Min H, Golden SS. Roles for sigma factors in global circadian regulation of the cyanobacterial genome. J. Bacteriol. 2002;184:3530–3538. doi: 10.1128/JB.184.13.3530-3538.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ito H, Mutsuda M, Murayama Y, Tomita J, Hosokawa N, Terauchi K, Sugita C, Sugita M, Kondo T, Iwasaki H. Cyanobacterial daily life with Kai-based circadian and diurnal genome-wide transcriptional control in Synechococcus elongatus. Proc. Natl. Acad. Sci. USA. 2009;106:14168–14173. doi: 10.1073/pnas.0902587106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vijayan V, Zuzow R, O'Shea EK. Oscillations in supercoiling drive circadian gene expression in cyanobacteria. Proc. Natl. Acad. Sci. USA. 2009;106:22564–22568. doi: 10.1073/pnas.0912673106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Smith RM, Williams SB. Circadian rhythms in gene transcription imparted by chromosome compaction in the cyanobacterium Synechococcus elongatus. Proc. Natl. Acad. Sci. USA. 2006;103:8564–856. doi: 10.1073/pnas.0508696103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Woelfle MA, Xu Y, Qin X, Johnson CH. Circadian rhythms of superhelical status of DNA in cyanobacteria. Proc. Natl. Acad. Sci. USA. 2007;104:18819–18824. doi: 10.1073/pnas.0706069104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Takai N, Nakajima M, Oyama T, Kito R, Sugita C, Sugita M, Kondo T, Iwasaki H. A KaiC-associating SasA-RpaA two-component regulatory system as a major circadian timing mediator in cyanobacteria. Proc. Natl. Acad. Sci. USA. 2006;103:12109–12114. doi: 10.1073/pnas.0602955103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Johnson CH, Mori T, Xu Y. A Cyanobacterial Circadian Clockwork. Curr. Biol. 2008;18:R816–R825. doi: 10.1016/j.cub.2008.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Taniguchi Y, Takai N, Katayama M, Kondo T, Oyama T. Three major output pathways from the KaiABC-based oscillator cooperate to generate robust circadian kaiBC expression in cyanobacteria. Proc. Natl. Acad. Sci. USA. 2010;107:3263–3268. doi: 10.1073/pnas.0909924107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Johnson CH, Stewart PL, Egli M. The cyanobacterial circadian system: from biophysics to bioevolution. Annu. Rev. Biophys. 2011;40:143–167. doi: 10.1146/annurev-biophys-042910-155317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gutu A, O'Shea EK. Two antagonistic clock-regulated histidine kinases time the activation of circadian gene expression. Mol. Cell. 2013;50:288–294. doi: 10.1016/j.molcel.2013.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Iwasaki H, Nishiwaki T, Kitayama Y, Nakajima M, Kondo T. KaiA-stimulated KaiC phosphorylation in circadian timing loops in cyanobacteria. Proc. Natl. Acad. Sci. USA. 2002;99:15788–15793. doi: 10.1073/pnas.222467299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rust MJ, Markson JS, Lane WS, Fisher DS, O'Shea EK. Ordered phosphorylation governs oscillation of a three-protein circadian clock. Science. 2007;318:809–812. doi: 10.1126/science.1148596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Qin X, Byrne M, Mori T, Zou P, Williams DR, McHaourab H, Johnson CH. Intermolecular associations determine the dynamics of the circadian KaiABC oscillator. Proc. Natl. Acad. Sci. USA. 2010;107:14805–14810. doi: 10.1073/pnas.1002119107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Qin X, Byrne M, Xu Y, Mori T, Johnson CH. Coupling of a core post-translational pacemaker to a slave transcription/translation feedback loop in a circadian system. PLoS Biol. 2010;8:e1000394. doi: 10.1371/journal.pbio.1000394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nakajima M, Imai K, Ito H, Nishiwaki T, Murayama Y, Iwasaki H, Oyama T, Kondo T. Reconstitution of circadian oscillation of cyanobacterial KaiC phosphorylation in vitro. Science. 2005;308:414–415. doi: 10.1126/science.1108451. [DOI] [PubMed] [Google Scholar]
- 22.Hanaoka M, Takai N, Hosokawa N, Fujiwara M, Akimoto Y, Kobori N, Iwasaki H, Kondo T, Tanaka K. RpaB, another response regulator operating circadian clock-dependent transcriptional regulation in Synechococcus elongatus PCC 7942. J. Biol. Chem. 2012;287:26321–26327. doi: 10.1074/jbc.M111.338251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nakahira Y, Katayama M, Miyashita H, Kutsuna S, Iwasaki H, Oyama T, Kondo T. Global gene repression by KaiC as a master process of prokaryotic circadian system. Proc. Natl. Acad. Sci. USA. 2004;101:881–885. doi: 10.1073/pnas.0307411100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang HH, Camsund D, Lindblad P, Heidorn T. Design and characterization of molecular tools for a Synthetic Biology approach towards developing cyanobacterial biotechnology. Nucleic Acids Res. 2010;38:2577–2593. doi: 10.1093/nar/gkq164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ducat DC, Way JC, Silver PA. Engineering cyanobacteria to generate high-value products. Trends in Biotech. 2011;29:95–103. doi: 10.1016/j.tibtech.2010.12.003. [DOI] [PubMed] [Google Scholar]
- 26.Wang B, Wang J, Zhang W, Meldrum DR. Application of synthetic biology in cyanobacteria and algae. Front Microbiol. 2012;3:344. doi: 10.3389/fmicb.2012.00344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rosgaard L, de Porcellinis AJ, Jacobsen JH, Frigaard NU, Sakuragi Y. Bioengineering of carbon fixation, biofuels, and biochemicals in cyanobacteria and plants. J. Biotechnol. 2012;162:134–47. doi: 10.1016/j.jbiotec.2012.05.006. [DOI] [PubMed] [Google Scholar]
- 28.Sheehan J. Engineering direct conversion of CO2 to biofuel. Nat. Biotechnol. 2009;27:1128–1129. doi: 10.1038/nbt1209-1128. [DOI] [PubMed] [Google Scholar]
- 29.Ishiura M, Kutsuna S, Aoki S, Iwasaki H, Andersson CR, Tanabe A, Golden SS, Johnson CH, Kondo T. Expression of a gene cluster kaiABC as a circadian feedback process in cyanobacteria. Science. 1998;281:1519–1523. doi: 10.1126/science.281.5382.1519. [DOI] [PubMed] [Google Scholar]
- 30.Kitayama Y, Nishiwaki T, Terauchi K, Kondo T. Dual KaiC-based oscillations constitute the circadian system of cyanobacteria. Genes Dev. 2008;22:1513–1521. doi: 10.1101/gad.1661808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gille H, Egan JB, Roth A, Messer W. The fis protein binds and bends the origin of chromosomal DNA replication, oriC, of Escherichia coli. Nucleic Acids Res. 1991;19:4167–4172. doi: 10.1093/nar/19.15.4167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xu Y, Mori T, Johnson CH. The cyanobacterial circadian clockwork: roles of KaiA, KaiB, and the kaiBC promoter in KaiC expression, phosphorylation, and degradation. EMBO J. 2003;22:2117–2126. doi: 10.1093/emboj/cdg168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Elledge SJ, Sugiono P, Guarente L, Davis RW. Genetic selection for genes encoding sequence-specific DNA-binding proteins. Proc. Natl. Acad. Sci. USA. 1989;86:3689–3693. doi: 10.1073/pnas.86.10.3689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xu Y, Mori T, Johnson CH. Circadian clock-protein expression in cyanobacteria: rhythms and phase-setting. EMBO J. 2000;19:3349–3357. doi: 10.1093/emboj/19.13.3349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Williams SB, Vakonakis I, Golden SS, LiWang AC. Structure and function from the circadian clock protein KaiA of Synechococcus elongatus reveals a clock input mechanism. Proc. Natl. Acad. Sci. USA. 2002;99:15357–15362. doi: 10.1073/pnas.232517099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kitayama Y, Iwasaki H, Nishiwaki T, Kondo T. KaiB functions as an attenuator of KaiC phosphorylation in the cyanobacterial circadian clock system. EMBO J. 2003;22:2127–2134. doi: 10.1093/emboj/cdg212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ghirardi ML, Posewitz MC, Maness PC, Dubini A, Yu J, Seibert M. Hydrogenases and hydrogen photoproduction in oxygenic photosynthetic organisms. Ann. Rev. Plant Biol. 2007;58:71–91. doi: 10.1146/annurev.arplant.58.032806.103848. [DOI] [PubMed] [Google Scholar]
- 38.Fontecilla-Camps JC, Volbeda A, Cavazza C, Nicolet Y. Structure/function relationships of [NiFe]- and [FeFe]-hydrogenases. Chem. Rev. 2007;107:4273–4303. doi: 10.1021/cr050195z. [DOI] [PubMed] [Google Scholar]
- 39.Carrieri D, Wawrousek K, Eckert C, Yu J, Maness PC. The role of the bidirectional hydrogenase in cyanobacteria. Bioresour. Technol. 2011;102:8368–8377. doi: 10.1016/j.biortech.2011.03.103. [DOI] [PubMed] [Google Scholar]
- 40.Armstrong FA. Hydrogenases: active site puzzles and progress. Curr. Opin. Chem. Biol. 2004;8:133–140. doi: 10.1016/j.cbpa.2004.02.004. [DOI] [PubMed] [Google Scholar]
- 41.Fritsch J, Lenz O, Friedrich B. Structure, function and biosynthesis of O2-tolerant hydrogenases. Nature Rev. Microbiol. 2013;11:106–114. doi: 10.1038/nrmicro2940. [DOI] [PubMed] [Google Scholar]
- 42.Weyman PD, Vargas WA, Tong Y, Yu J, Maness PC, Smith HO, Xu Q. Heterologous Expression of Alteromonas macleodii and Thiocapsa roseopersicina [NiFe] Hydrogenases in Synechococcus elongatus. PLoS ONE. 2011;6:e20126. doi: 10.1371/journal.pone.0020126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bock A, King PW, Blokesch M, Posewitz MC. Maturation of hydrogenases. Adv. Microb. Physiol. 2006;51:1–71. doi: 10.1016/s0065-2911(06)51001-x. [DOI] [PubMed] [Google Scholar]
- 44.Tomita J, Nakajima M, Kondo T, Iwasaki H. No transcription-translation feedback in circadian rhythm of KaiC phosphorylation. Science. 2005;307:251–254. doi: 10.1126/science.1102540. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.