Abstract
Neurons have their own systems for regulating RNA. Several multigene families encode RNA binding proteins (RNABPs) that are uniquely expressed in neurons, including the well-known neuron-specific markers ELAV and NeuN, and the disease antigen NOVA. New technologies have emerged in recent years to assess the function of these proteins in vivo, and the answers are yielding insights into how and why neurons may regulate RNA in special ways—to increase cellular complexity, to spatially localize mRNA, and to regulate their expression in response to synaptic stimuli. The functions of such restricted neuronal proteins is likely to be complimented by more widely expressed RNABPs that may themselves have developed specialized functions in neurons, including Argonaute/miRNAs. Here we review what is known about such RNABPs, and explore the potential biologic and neurologic significance of neuronal RNA regulatory systems.
Keywords: HITS-CLIP, Nova, Elavl, FMRP, microRNA, neuron-specific splicing factors
I. Introduction
The idea that neurons have developed unique systems for regulating RNA metabolism—processing, localization, and expression—has several interesting roots. First are observations on memory. A large body of evidence suggests that memory, and physiologic correlates such as long-term potentiation (LTP), require protein synthesis during a brief window after a memorable experience (Lynch, 2004). Physical evidence—the presence of detectable mRNA and ribosomes, and the ability to detect local translation (Aakalu et al., 2001; Schuman et al., 2006; Steward and Schuman, 2001)—and physiologic evidence—the protein synthesis requirement for axon guidance (Holt and Bullock, 2009; Ming et al., 2002; Zhang and Poo, 2002) or for long term potentiation after Hebbian stimuli (Lynch, 2004) — support the hypothesis that synaptic plasticity requires the regulation of some mRNA transcripts within the synapse itself.
Second, early characterization of neuronal transcripts suggested that pre-mRNAs are differentially processed in the brain relative to other tissues. This idea was raised by the work of Amara and Evans, who discovered that the transcript encoding calcitonin, normally made in the thyroid gland, had two isoforms in thyroid tumors. The alternate isoform was a poly(A)/splice variant normally found only in neurons, where it encoded an entirely different protein, the neuropeptide transmitter calcitonin-gene related peptide, CGRP (Amara et al., 1982). This observation, and many follow-up studies, underlies the idea that neurons leverage RNA processing to generate orders of magnitude increases in complexity from a limited size protein-coding genome (Licatalosi and Darnell, 2010).
A third line of evidence came from the study of neuronal markers. Neurobiologists have long sought out genes that are uniquely expressed in neurons, to demarcate neurons histologically and to develop genetic tools. While many markers have been identified that are specific to neurons within the nervous system (versus glia, for example), most are robustly expressed in other tissues at some point in development. However, a few were identified that are extremely specific to neurons throughout life (with one common exception being germ cells). The first such marker was the Drosophila ELAV (embryonic lethal abnormal visual) protein. ELAV was identified through a screen for behavioral defects in flies (Homyk et al., 1980) that were found to have abnormal electroretinograms and abnormal axonal tracts. Ultimately, RNA and protein expression studies revealed that ELAV was expressed in all neurons, but not in neuroblasts, glia, or any other tissue type (Campos et al., 1987; Bier et al., 1988; Yao et al., 1993). Subsequently, ELAV became widely used as a specific neuronal marker, and as a neuron-specific driver for genetic studies.
In mammalian systems, early putative neuronal markers such as neuron-specific enolase were found to label glia (Magavi and Macklis, 2008), prompting empiric searches for additional markers. These led to the mammalian homologue of ELAV (original termed the Hu antigen, discussed below, now termed ELAV-like (ELAVL)), and NeuN (for “neuronal nuclei”). NeuN, identified with a monoclonal antibody generated against brain nuclei and originally thought to be a transcription factor, was found to reliably identify mature neurons in the adult brain (Wolf et al., 1996).
Amazingly, each of these classical neuron-specific markers, Elav/Elavl and NeuN (see below), were found to encode RNABPs. These discoveries have since been augmented with an expanding list of additional neuron-specific RNABPs, and with the recognition that at least some more generally expressed RNABPs may act in concert with the neuronal RNABPs (Table 1).
Table 1.
Neuronal RNA binding proteins
| Neuron-specific RNA binding proteins | ||||||
|---|---|---|---|---|---|---|
| RNABP | Comments | Expression | Other Paralogs | Position-dependent splicing map/other function | Expression References | Knockout Mouse |
| Nova1-2 | Nova1 - first neuronal RNABP for which a genetic-null mouse was engineered | CNS-specific Nova2: Trace expression in lung (unknown cell type) |
none | yes | Nova1 (Jensen et al., 2000b) Nova2 (Huang et al., 2005) |
|
| Elavl2-4 | Elavl4: Frequent marker neuron-specific marker | Elavl2: Most weakly expressed Elavl3: most unique expression patterns Extra-CNS expression in autonomic neurons (e.g. intestinal expression in myenteric plexus neurons) |
Elavl1 | yes | (Okano and Darnell, 1997) | Elavl3 (Akamatsu et al., 2005; Ince-Dunn et al., 2012) Elavl4 (Akamatsu et al., 2005) |
| Rbfox3 (NeuN) | Frequent neuron-specific marker; Rbfox1 implicated in autism | Intestinal expression restricted to myenteric plexus neurons | Rbfox1/2 | yes | Refs for Rbfox3 Refs Geshwind |
Rbfox1 (Gehman et al., 2011) |
| Ptbp2 | Expression in rare subsets of glial cells and in mitotic neuronal progenitors | Ptbp1 | yes | (Licatalosi et al., 2012) | (Licatalosi et al., 2012) | |
| nSR100 | Highly neural enriched by non-quantitative RT-PCR | ? | (Calarco et al., 2009) | |||
| RNABPs not unique to neurons, but of biologic interest | ||||||
| CELF | Family includes CELF1 (CUGBP1), a splicing factor implicated in myotonic dystrophy; literature unclear re: neuronal specificity of other family members | Celf4: Weak RNA expression by Northern blot outside nervous system, but question of cross reactivity among paralogs unclear | Celf1, 2, 4-6 | ? | (Meins et al., 2002; Cooper et al., 2009) | (Wagnon et al., 2011) |
| FMRP | Neuron-specific in brain; widely expressed in other tissues | FXR1/2 (wide expression) | Inhibits translation by arresting ribosomal elongation | (Darnell et al., 2011) | Fmr1 null (Bakker et al., 1994) Fmr1 inactive (Zang et al., 2009) |
|
| Mbnl1-3 | Mbnl2: Neuron-specific in brain; robust expression in lung | yes | Mbnl1 (Kanadia et al., 2003) Mbnl2 (Charizanis et al., 2012) |
|||
| Msi1 | Expressed in mitotic neuronal progenitors in embryonic and adult brain; also expressed in some astrocytes, some neurons (e.g. deep cerebellar nuclei, stellate neurons, as well as ovaries and small intestine (unknown cell type) | Msi2 | ? | Msi1 (Sakakibara et al., 2002) | ||
| ZBP | ZBP1 particularly well-defined functions in mRNA localization and translation | Widely expressed in brain and other tissues | ZBP2 | ? | (Hansen et al., 2004) | |
A fourth and equally unexpected line of evidence was the discovery of neuron-specific RNA binding proteins in humans that came from the work on a set of cancer-related autoimmune neurologic disorders, termed the paraneoplastic neurologic disorders (Darnell and Posner, 2011). Autoimmune antisera from these patients identified neuron-specific proteins that were ectopically expressed in tumor cells, triggering an anti-tumor immune response that ultimately crossed into the brain, leading to the neurologic disease. In the 1980's, the Darnell laboratory established that these antisera could be used to clone cDNAs encoding these antigens using λgt11 expression vectors, and two different multigene families of genes turned out to encode the neuron-specific RNA binding proteins Nova and Elavl (Darnell, 1996; Musunuru and Darnell, 2001; Darnell and Posner, 2011).
The significance of the discovery that the brain expresses its own neuronal RNABPs relates to our attempt to understand what underlies complexity in the brain function. To a first approximation, the genomic protein-coding capacity of the human and the worm are very similar (in number and types of protein coding genes). This observation has shifted interest in understanding complexity as a consequence of the ways in which these genes are deconvoluted into the RNA world...into how pre-mRNA gene copies are alternatively spliced and polyadenylated, edited, localized throughout the neuron and translationally regulated. This much more robust complexity, relative to the control of DNA transcription, is likely to play a key role in the evolution of complex cellular function, neuronal plasticity and brain function (Licatalosi and Darnell, 2010). This review will describe the approaches used to identify the functions of neuronal RNABP's, what is known about each in brain function and disease, followed by a discussion of future directions.
II. Approaches to studying neuronal RNA binding protein function
To appreciate the work done by many laboratories in establishing the roles of neuronal RNABPs, it is essential to appreciate the methods used to establish their functions. Three major approaches have been established, which, when used in combination with modern bioinformatics, combine to form a powerful means of defining in vivo functions for RNABPs. A prior review detailed the combined use of these approaches (Licatalosi and Darnell, 2010), which are outlined below.
Traditional biochemical approaches
The importance of knowing whether a neuronal protein is an RNABP is underscored by the original reports that described NeuN was a transcription factor. The traditional means for defining a protein as an RNABP came from the lab of Gideon Dreyfuss, who characterized a large number of hnRNP proteins as RNABPs. The fundamental assay, still valid as a screen, was to bind purified proteins to ribohomopolymer columns, and measure their retention under increasingly stringent salt washes. In this way, for example, after the gene encoding the NOVA1 protein was cloned, NOVA1 was found to bind ribohomopolymers in up to 1.0 M salt, evidence of robust RNA affinity (Buckanovich et al., 1996), while the Fragile-X mental retardation protein, FMRP, also bound to ribohomopolymers, but with much less affinity (Siomi et al., 1993). These approaches allowed Dreyfuss and colleagues to classify RNABPs according to the presence of several canonical motifs (Burd and Dreyfuss, 1994). This in turn accelerated the classification of many newly discovered proteins as RNABPs, although it should be noted that new high affinity RNA binding motifs continue to be described.
A second level of analysis was to identify preferred RNA binding motifs in vitro. This strategy relied on the development of methods to use recombinant proteins to affinity purify RNA sequences from random RNA libraries in an iterative manner, termed in vitro RNA selection (developed using affinity chromatography (Ellington and Szostak, 1990; Green et al., 1991)) or RNA SELEX (developed using filter binding strategies (Tuerk and Gold, 1990)). Early validation of these methods included their use to identify RNAs bound to the HIV-1 Rev protein (Ellington and Szostak, 1990), to T4 DNA polymerase (Tuerk and Gold, 1990), and to confirm binding of U1 snRNP-A to sequences in U1 RNA (Tsai et al., 1991). These approaches have been used to identify in vitro RNA ligands for many of the mammalian neuronal RNABPs discussed in this review (Table 2), and these have proved to be extremely valuable in cross-checking binding motifs identified by complimentary methods described below.
Table 2.
Neuron-specific RNABPs–RNA binding sites
| Neuron-specific RNABP | Methods used in identification | Binding motif | Comment | Reference |
|---|---|---|---|---|
| NOVA1 NOVA2 |
In vitro RNA selection CLIP HITS-CLIP Bayesian network |
Stem, loop sequence: UCAU × 3 YCAY clusters |
Stem sequence variable; may play structural role to keep YCAY binding site available in sequence single loop | (Buckanovich and Darnell, 1997; Yang et al., 1998; Jensen et al., 2000a) |
| nELAVL2 (Hel-N1) | In vitro RNA selection | UUUAUUU | (Gao et al., 1994) | |
| nELAVL2/3/4 |
In vitro RNA selection HITS-CLIP |
U-rich element with single purine | G slightly preferred over A as purine | (Ince-Dunn et al., 2012) |
| RBFOX1 |
In vitro RNA selection HITS-CLIP |
GCAUG UGCAUG |
(Jin et al., 2003; Yeo et al., 2009) | |
| PTBP2 | HITS-CLIP | CU-repeat and UCUY-rich elements | (Licatalosi et al., 2012) |
Mammalian genetics
Crucially, for many neuronal RNABPs, in vitro biochemistry has been complimented by validation of predicted functions in the brains of RNABP-knock-out mice. This is important, since neither cell quality (particularly the specialized neuronal cell types in the brain), cell biology (particularly the complex synaptic interactions among many cell types), nor the stoichiometry of RNA-protein interactions can be faithfully reproduced in tissue culture cells or primary neurons.
The first neuronal RNABP for which a genetic-null mouse was engineered was the neuron-specific RNABP Nova1 (Jensen et al., 2000b). Subsequently null mice have been generated for most of the RNABPs discussed in this review (see Table 1). In addition, both null mice (Bakker et al., 1994) and mice harboring an inactivating point mutant (Zang et al., 2009) have been generated for Fmr1, the gene encoding the Fragile X protein FMRP, providing useful overlapping means of validating biochemical predictions of in vivo biology (Darnell et al., 2011). For each of these RNABPs, as detailed below, the most robust data regarding their function in neurons relates predictions from biochemical experiments to examination of RNA variants in wild-type compared with knock-out mice.
Global RNA analyses
One significant limitation of traditional biochemical approaches is that they study one RNA-protein interaction at a time, such that making generalizations from such data is difficult. The past decade or so saw an emergence and maturation of methods to analyze RNA complexity on a global scale. By complexity, we refer to the unique species of RNA present in a given biological sample. The initial enumeration of RNA complexity came from analysis of microarrays. As previously discussed (Blencowe et al., 2009; Mortazavi et al., 2008), the difficulties with such arrays was their inherent signal:noise problem—different probesets had varying sensitivity and specificity in detecting a range of transcript levels, leading to burdensome and sometimes inaccurate normalization requirements. Nonetheless, such arrays were important in providing genome-wide means for approximating transcript levels in different tissues.
A subsequent generation of exon arrays allowed analysis of alternative splice variants, using probesets that spanned exon-exon junctions. These exon junction microarrays were especially informative in the analysis of neuronal RNA complexity. They initially were used to enumerate alternative splice variants present in brain relative to other tissues (Johnson et al., 2003; Pan et al., 2004), and then more specifically to analyze splicing alterations altered when specific factors were missing. Such analyses included genome-wide descriptions of splice variants in Nova null mice (Ule et al., 2005b) and, in Drosophila, in flies in which various splicing factors were knocked down (Blanchette et al., 2005). Subsequent studies expanded our understanding of splicing variation in different tissues (Sugnet et al., 2006), and of splicing variants related to a number of different neuronal RNABPs. These include studies of splicing dependent on PTBP2 in N2A neuroblastoma cells (Boutz et al., 2007), the neural-specific SR protein nSR100 in tissue culture and zebrafish (Calarco et al., 2009), and, in knockout mouse brain, PTBP2 (Licatalosi et al., 2012)), MBNL1 (Du et al., 2010), RBFOX1 (Gehman et al., 2011), and ELAVL3 (Ince-Dunn et al., 2012).
A more refined and accurate picture of transcript variants in brain (and other tissues) has now been afforded by the application of next-generation sequencing—RNA-seq—originally developed to quantify transcriptomes in yeast (Nagalakshmi et al., 2008) and mammalian tissues (Mortazavi et al., 2008; Wang et al., 2008), including brain. Even with early generation high throughput sequencing, read depth was sufficient to begin delineating alternative splicing patterns unique to brain (Mortazavi et al., 2008; Wang et al., 2008). Such studies, combined with pair-end sequencing methods and decreasing costs of sequencing, promise to become the most sensitive and specific means of enumerating RNA variants. Recent applications include analysis of RNA variants generated by alternative splicing (Calarco et al., 2011), alternative polyadenylation (Weill et al., 2012) and RNA editing (Silberberg and Ohman, 2011).
HITS-CLIP
A critical view of data enumerating RNA variants underscores its correlative nature (Licatalosi and Darnell, 2010). For example, alternative exons whose levels vary in proportion to expression of a neuronal splicing factor may be directly regulated by that factor, or indirectly regulated, by, for example an action on a transcript encoding an intermediate RNABP. Moreover, to understand RNABP function, it is important to identify both the target RNAs they directly act upon, as well as the actual sites of action. Although traditional biochemical experiments may demarcate potential binding interactions, even with high level bioinformatic analysis they have not been able to fully and accurately predict the range and variability of interactions seen in living cells. Resolving these issues is crucial to understanding the mechanisms of RNABP actions in the brain.
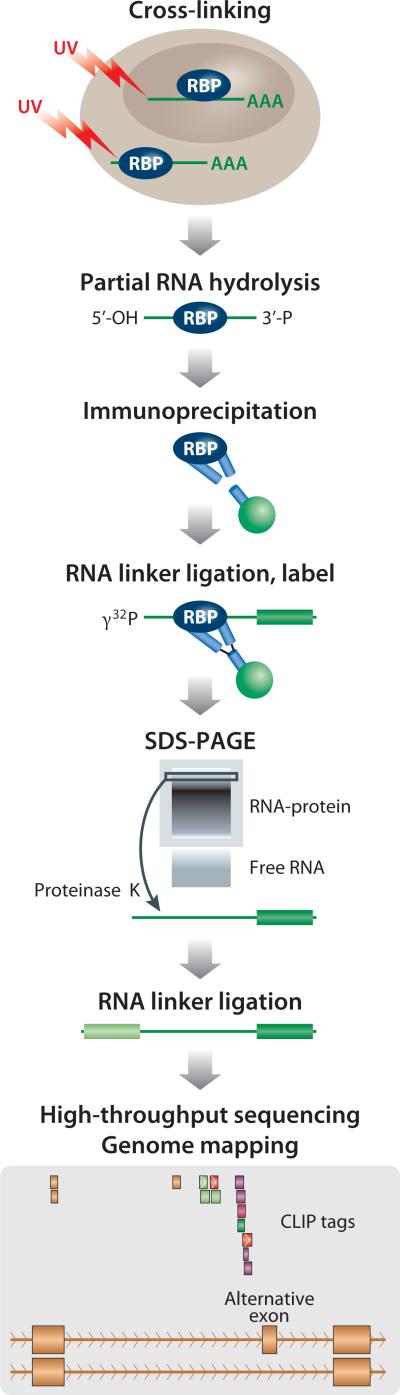
Among several approaches taken, those involving crosslinking of RNA-protein complexes have become the gold standard for identifying sites of functional interaction in vivo, including in the brain (reviewed in (Licatalosi and Darnell, 2010; Darnell, 2010; Calarco et al., 2011; Konig et al., 2012). These methods originated with the development of crosslinking immunoprecipitation (CLIP) (Figure 1) for identifying Nova-RNA interactions in the brain (Ule et al., 2003). CLIP was initially adopted slowly, although it proved versatile for a host of species and RNABPs (see (Darnell, 2010)). However, the application of high throughput sequencing methods to CLIP (HITS-CLIP; (Licatalosi et al., 2008); see Sidebar), provided a breakthrough to the field and is now in wide use.
Figure 1. Schematic of the HITS-CLIP Method.
Schematic of major steps in HITS-CLIP (modified from (Licatalosi and Darnell, 2010); see SIDEBAR for details about CLIP). In brief, tissues or cells to be analyzed are irradiated with UV-light, inducing crosslinking between RNA-protein complexes within the cell, in situ. RNA is partially hydrolyzed, using nucleases or chemical hydrolysis, to a desired modal size (e.g. 30-50 nt), RNA-protein complexes are purified (typically by immunoprecipitation), RNA is labeled and run on autoradiograms to separate crosslinked RNA-protein complexes by size and transferred to nitrocellulose (through which any free RNA passes). The RNABP is the removed by Proteinase K treatment, RNA linker ligation is completed, and RNA PCR amplified and sequenced.
III. Neuron-specific RNA binding proteins
Elavl
ELAV, the Drosophila homologue of the mammalian neuronal ELAVL proteins, was first discovered in a Drosophila screen for behavioral defects (Homyk et al., 1980), and named for the phenotype “embryonic lethal abnormal visual.” Work on ELAV function was spearheaded by Kalpana White and colleagues, who helped document the neuron-specific nature of its expression (see Introduction), and went on to recognize that it contained three RRM-type RNA binding motifs (Robinow et al., 1988). This led to a series of studies using candidate gene approaches to identify RNAs whose isoform usage correlated with ELAV levels in mutant flies, including neuroglian (Koushika et al., 1996), encoding a Drosophila protein with homology to neural adhesion molecule L1, the erect wing gene, encoding a transcription factor, and armadillo, whose vertebrate homologue is β-catenin (Koushika et al., 2000; Soller and White, 2003). More recent evidence suggested that ELAV bound an intronic AU-rich element could mediate effects on ewg splicing (Soller and White, 2005).
Studies of paraneoplastic neurologic disorders led to the discovery of three mammalian homologues of Elav, each with expression that is entirely restricted to neurons, termed Elavl2-4 (originally HuB, HuC and HuD; there is also one non-neuronal paralogue, Elavl1, or HuA/HuR). These neuronal ELAVL (termed nELAVL) proteins were discovered using autoantibodies to screen for the target antigen in the paraneoplastic subacute sensory neuropathy/encephalomyelopathy syndrome, or the Hu syndrome neurologic disorders (see SIDEBAR). A careful description of their expression revealed that the three nELAVL proteins were indeed restricted to central and peripheral neurons, in overlapping but unique patterns in all postmitotic neurons, with expression persisting throughout adulthood (Okano and Darnell, 1997).
RNA selection experiments were first performed with nELAVL2/HuB, which revealed a consensus sequence for U-rich elements interspersed with purines, interpreted as a preference for binding AU-rich elements (AREs) (Gao et al., 1994; Levine et al., 1993). Moreover, in elegant biochemical studies, nELAVL1/HuA co-purified with the ARE in the c-fos gene (Myer et al., 1997). ARE elements mediate rapid decay of mRNAs, through shortening of the poly(A) tail, followed by mRNA degradation (Chen and Shyu, 2011) (see (Meisner and Filipowicz, 2011) for review of nELAVL1 function). A number of studies demonstrated that overexpression of mammalian ELAVL proteins could stabilize ARE-containing messages (including studies of ELAVL1 (Dean et al., 2001; Fan et al., 1997; Fan and Steitz, 1998; Peng et al., 1998), nELAVL2 (Antic et al., 1999; Jain et al., 1997) and nELAVL4 (Bolognani and Perrone-Bizzozero, 2008)). In vitro and tissue culture experiments indicated that ELAVL could affect stability and/or translation of a large number of proteins, including tau, GAP-43, p21 (for review, see (Hinman and Lou, 2008)) and even the neuronal splicing factor Nova1 (Ratti et al., 2008). Following the studies from White and colleagues in flies, a number of studies in mammalian cells also contributed to evidence that the ELAVL proteins could act as splicing factors, including in vitro RNA-protein binding assays, minigene overexpression studies and analyses of conserved AU-rich elements adjacent to alternative exons (Zhu et al., 2006; Wang et al., 2010a; Zhu et al., 2008; Wang et al., 2010a) (reviewed in (Hinman and Lou, 2008)).
Together, these reports provided interesting suggestive data, but were restricted by uncertainty regarding which targets were direct and physiologically relevant, given the concerns raised above in Section II. Until recently, a list of actual neuronal target RNAs and the processes they regulated remained unclear, with the most complete effort coming from a list RNAs coprecipitating with nELAVL4 in transgenic mice overexpressing nELAVL4 (Bolognani and Perrone-Bizzozero, 2008). Nonetheless, this body of work stimulated great interest in the nELAVL proteins, and was further fueled by functional studies, including the observations that the nELAVL3 and nELAVL4 proteins were necessary for neuronal maturation in mice (Akamatsu et al., 1999; Akamatsu et al., 2005). This interest laid the groundwork for the generation of nElavl3 (Ince-Dunn et al., 2012) and nElavl4 (Akamatsu et al., 2005) null mice, which have proven critical in demarcating nELAVL function in vivo.
nElavl4 null mice were recently used to provide support for a non-cannonical role for nELAVL4 in pancreatic beta cells (Lee et al., 2012). This interesting data needs confirmation (RNA could not be detected in beta cells, although protein could be, raising concern about antibody specificity). Notably, the origin of pancreatic beta cells remains unclear; progenitor cells in the adult mouse pancreas are able to give rise to neurons and pancreatic cells, and beta cells express many neuronal proteins and pathways (Dor et al., 2004; Smukler et al., 2011), including GABA and synaptic like microvesicles (Reetz et al., 1991)). Examination of nElavl4 null mice demonstrated abnormally high levels of insulin, and this was correlated with the ability of nELAVL4 to bind to the transcript encoding insulin in vitro (Lee et al., 2012). Patients with paraneoplastic autoimmunity to the Hu antigen do not develop diabetes (Darnell and Posner, 2011) even though beta cells are a common autoimmune target, so aspects of these observations remain puzzling.
A more precise in vivo picture of nELAVL-RNA interactions in neurons within the mouse brain has recently been afforded by a combination of experiments using nElavl null mice, HITS-CLIP and bioinformatic analysis (Ince-Dunn et al., 2012). In mouse brain, nELAVL proteins were found to bind directly to 3’ UTR elements, and, to a lesser degree, intronic elements. An interesting relationship with the original in vitro RNA selection studies was revealed, in which the in vivo binding sites were U-rich elements harboring a purine, more commonly a G than an A residue (so rather than binding AU-rich elements, the nELAVL proteins bind purine/U-rich elements). These binding sites were confirmed by analysis of crosslink-induced mutations (Zhang and Darnell, 2011), and were found to be functional. In particular, several intronic binding sites were able to mediate alternative splicing regulation, as evidenced by their ability to predict splicing changes in nELAVL-null mouse brain. In addition, nELAVL 3’ UTR binding sites predicted changes in mRNA steady-state levels.
A major goal of identifying RNABP-RNA interaction sites is to gain insight into the relevant biology. Gene ontology (GO) analysis demonstrated prominent roles for nELAVL-splicing regulation of transcripts involved in synaptic cytoskeletal dynamics, and nELAVL-steady-state regulation of transcripts involved in amino acid biosynthesis (Ince-Dunn et al., 2012). One outstanding target in both GO analyses was the pathway involved in glutamine synthesis, in particular the gene encoding glutaminase, which catalyzes formation of glutamate, the major excitatory neurotransmitter in the brain. Indeed, glutamate levels were reduced in nELAV3 null mouse brain, and both these mice and haploinsufficient mice had spontaneous epilepsy. These studies illustrate how traditional and modern (HITS-CLIP) biochemistry, combined together with mouse genetics and bioinformatics, provides a potent means of discovering the role of neuronal RNABP function in the brain.
Nova
This triad of biochemistry, mouse genetics and bioinformatics to define neuronal RNABP function was first established in studies of the RNABP Nova (as reviewed in (Licatalosi and Darnell, 2010)). As with the nElavl, the genes encoding the NOVA1 and NOVA2 proteins were identified using autoimmune sera from patients with paraneoplastic opsoclonus-myoclonus ataxia (Buckanovich et al., 1993; Yang et al., 1998). In this neurologic disorder, patients with lung or gynecologic tumors develop excessive motor movements, attributable neurologically (Darnell and Posner, 2011) to a failure of motor inhibition, which sometimes progresses to encephalopathy (Luque et al., 1991; Hormigo et al., 1994). Characterization of Nova1 and Nova2 expression revealed expression that is extremely restricted to post-mitotic neurons (Yano et al., 2010) in the central, but not peripheral, nervous system, with Nova1 expressed primarily in the hindbrain and ventral spinal cord, and Nova2 primarily in the neocortex (Buckanovich et al., 1993; Yang et al., 1998; Racca et al., 2010).
Traditional biochemical studies established that Nova proteins bind to RNA in a sequence-specific manner and are able to regulate alternative splicing in vitro. Nova proteins function as RNABPs (Buckanovich and Darnell, 1997; Buckanovich et al., 1996) and harbor three KH-type RNA binding motifs. RNA selection defined their in vitro sequence targets as clusters of YCAY motifs (where Y is a pyrimidine) (Jensen et al., 2000a; Yang et al., 1998; Buckanovich and Darnell, 1997). The mechanism of KH-domain sequence-specificity was first revealed by X-ray crystallographic studies of NOVA-RNA complexes (Lewis et al., 2000; Jensen et al., 2000a; Lewis et al., 1999; Teplova et al., 2011), which showed that the NOVA KH domains fold into a highly conserved structure in which side chain amino acids are exactly positioned to provide Watson-Crick hydrogen bond donor and acceptor groups to specific precisely the CA dinucleotide, and, to a lesser degree, restrict binding of the surrounding nucleotides to pyrimidines.
These data were used to identify three Nova-regulated pre-mRNAs harboring YCAY clusters, targets that were confirmed with cellular and in vitro splicing assays. Interestingly, these targets included two transcripts encoding inhibitory neurotransmitter receptors encoding the α2 subunit of the glycine (GlyRα2) receptor (Buckanovich and Darnell, 1997; Jensen et al., 2000b; Racca et al., 2010) and the γ2 subunit of the GABAA (GABAA γ2) receptor (Jensen et al., 2000b; Dredge and Darnell, 2003). In addition, in vitro splicing assays demonstrated that NOVA1 inhibited splicing of its own pre-mRNA by binding YCAY elements to block U1A binding (Dredge et al., 2005; Ule et al., 2006).
However, critical analysis suggested two outstanding unresolved issues. First, biochemical assays lacked physiologic stoichiometry (overexpression assays in vitro or in vivo), leaving uncertainty whether the splicing changes observed were relevant in vivo. This led our lab to generate Nova-null mice (Jensen et al., 2000b; Yang et al., 1998) to re-explore splicing targets in vivo. Second, identification of the GlyRα2 and GABAA γ2 as Nova targets was intriguing in light of the fact that Nova was targeted in PND patients with a failure of motor inhibition, yet our approach to target identification had clearly not been systematic. This prompted the use of genome-wide methods to assess Nova targets, culminating in the development of CLIP.
Initial genome-wide approaches to assess RNA splice variation in Nova-null mice used exon-junction splicing microarrays (Ule et al., 2005b). Analysis of these results, using a bioinformatic algorithm (ASPIRE) designed to look for reciprocal splicing changes in WT and null mice, led to the identification of a robust set of ~50 target RNAs whose splicing was Nova-dependent. GO analysis of this list led to the first compelling evidence that RNABPs could regulate a biologically coherent subset of transcripts in vivo—nearly all 50 transcripts encoded synaptic proteins (Ule and Darnell, 2006; Ule et al., 2005b; Calarco et al., 2011).
Moreover, identifying Nova target RNAs led to prediction and identification of functions for Nova in synaptic function, including roles in mediating inhibitory responses to long-term potentiation (Huang et al., 2005), in formation of the neuromuscular junction and motor neuron function (Ruggiu et al., 2009), and in Reelin signaling and neuronal migration (Yano et al., 2010; Park and Curran, 2010) (see below).
An important adjunct to the biochemical and genetic analysis of Nova targets was a focused set of bioinformatic studies. Initially, these assessed whether the first 50 Nova targets harbored YCAY binding motifs, and led to the unexpected discovery of a position-dependent map governing splicing regulation in neurons (Ule et al., 2006). Specifically, the position of binding within the primary transcript was a key determinant of the outcome of splicing regulation, such that NOVA binding in upstream/within alternative exons mediated their exclusion, while downstream intronic binding enhanced alternative exon inclusion (Licatalosi and Darnell, 2010; Licatalosi et al., 2008). Such positional effects are now recognized (Chen and Manley, 2009; Llorian et al., 2010; Corrionero and Valcarcel, 2009; Blencowe et al., 2009; Witten and Ule, 2011) as a general feature of splicing control applicable to over a dozen RNABPs (Llorian et al., 2010; Konig et al., 2010; Chen and Manley, 2009; Yeo et al., 2009; Xue et al., 2009; Zhang et al., 2008; Kalsotra et al., 2008; Yuan et al., 2007; Licatalosi and Darnell, 2006; Tollervey et al., 2011; Ince-Dunn et al., 2012; Licatalosi et al., 2012; Wang et al., 2012; Charizanis et al., 2012).
The effort to develop unbiased, genome-wide assessments target transcripts directly regulated by NOVA led to the development of CLIP (Ule et al., 2003). These studies, subsequently combined with HITS-CLIP (Licatalosi et al., 2008), and more sophisticated computational methods (e.g. Bayesian network analysis of NOVA datasets; (Zhang et al., 2010)) led to the identification of a robust set of ~700 alternative exons and a smaller number of alternative 3’ UTRs regulated by Nova in the mouse brain (as recently reviewed (Darnell, 2010; Darnell, 2006; Licatalosi and Darnell, 2010)). This data confirmed the biologic coherence of NOVA targets, indicating links between NOVA and a broader set of neurologic disorders, including autism, an observation strengthened by the prediction, experimentally demonstrated, that Nova regulates alternative splicing in a combinatorial manner with RBFOX1 (Zhang et al., 2010), itself a splicing factor implicated in autism (Voineagu et al., 2011).
Moreover, these studies laid the groundwork for applying genome-wide HITS-CLIP to help solve discrete biologic problems. The observation that NOVA null mice have neuronal migration defects similar to those seen with defects in the Reelin pathway led to a focused HITS-CLIP study on developing neocortex (Yano et al., 2010; Park and Curran, 2010). Focusing on transcripts encoding proteins in this pathway, HITS-CLIP was able to identify one NOVA binding target—intronic YCAY elements within the transcript encoding Dab1, a key signaling protein in the Reelin pathway—that regulated an alternative exon encoding a new protein domain in Dab1. Functional studies, including in utero electroporation, demonstrated a specific role for NOVA regulation of this exon in neuronal migration.
The finding that NOVA regulated GlyRα2 splicing (Buckanovich and Darnell, 1997; Jensen et al., 2000b) was linked to GlyRα2 mRNA localization in the dendrite (Racca et al., 1997) by analysis of nuclear and cytoplasmic HITS-CLIP. In this study, HITS-CLIP provided evidence that intronic binding to regulate splicing could be coupled in cis with 3’ UTR binding to regulate mRNA localization and, presumably, dendritic protein translation (Racca et al., 2010). Most recently, NOVA HITS-CLIP was used to study transcripts whose steady-state mRNA levels were altered in Nova null mice. This led to the unexpected finding that large NOVA-dependent changes in the production of proteins were regulated by intronic binding of NOVA to regulate the inclusion of cryptic exons that triggered nonsense mediated mRNA decay (Eom et al., 2013). These included massive changes in the expression of a number of synaptic proteins, including several implicated in epilepsy, and indeed Nova heterozygous mice were found to have spontaneous epilepsy. Taken together, these studies illustrate the potential of HITS-CLIP to provide sets of functionally relevant RNA-protein interactions that are able to predict the role of RNA regulation in neuronal biology, and suggest links between neuronal RNA regulation of splicing and protein production to neuronal excitatory/inhibitory homeostasis.
NeuN/RbFox3
It was only recently recognized that the neuronal marker NeuN is an RNABP in the RBFOX family. This was discovered by using the NeuN monoclonal antibody for immunoprecipitation-mass spectrometry analysis (Kim et al., 2009) and, independently, through expression cDNA screening (Dredge and Jensen, 2011). The RBFOX protein family has three paralogs, RBFOX1-3. Each protein has a single RRM-type RNA binding domain, and has overlapping expression in neurons: RBFOX1 is expressed in neurons, heart and muscle, RBFOX2 (also known as Rbm9) in neurons and other cell types, including hematopoietic and stem cells, while RBFOX3 is neuron-specific (Kuroyanagi, 2009).
To date, the best studied paralogue among these proteins is RBFOX1. RBFOX1 was originally identified in a yeast 2-hybrid screen (Shibata et al., 2000) as a protein that interacts with the spinocerebellar ataxia type 2 gene product SCA2 (hence the original name for RBFOX1 was ataxin-2 binding protein 1, A2BP1); subsequently these interactions were confirmed for other family members (Lim et al., 2006). In vitro RNA selection with RBFOX1 identified a strong consensus binding sequence as (U)GCAUG (Jin et al., 2003).
HITS-CLIP analyses have been accomplished with RBFOX1 and RBFOX2. In ES cells, RBFOX2 bound to GCAUG clusters (Yeo et al., 2009), consistent with the traditional biochemical studies. The position of these clusters around alternative exons conformed to the position-dependent splicing map defined for Nova, consistent with earlier bioinformatics predictions made in analyzing UGCAUG binding sites (Castle et al., 2008), and supporting a role for RBFOX2 as a splicing factor in neurons that was predicted from biochemical analyses (Underwood et al., 2005). A number of binding clusters were also identified that did not harbor this element, and whether this relates to additional biologic roles for RBFOX2 or signal:noise problems in these experiments remains unclear. Interestingly, many of the predicted RBFOX2 alternatively spliced target transcripts themselves encoded splicing factors (Yeo et al., 2009), suggesting the possibility of a higher order regulatory network.
Biologic roles for RBFOX proteins in neurons have been studied in detail by Doug Black and colleagues. Initial studies in tissue culture cells identified position-dependent UGCAUG elements that regulated splicing of two Cav1.1 calcium channel exons, and regulation was abrogated by mutating the elements to UGCGUG (Tang et al., 2009). Studies in P19 cells implicated a role for activity-dependent regulation of alternative splicing for several transcripts (Lee et al., 2009), findings supported by observation of spontaneous seizures and widespread changes in alternative splicing in mice in which RbFox1 had been specifically deleted in neurons with a nestin-Cre driver (Gehman et al., 2011). Interestingly, these splicing changes may be mediated in part by changes in RbFox1 alternative splicing that leads to a shift of the protein from the cytoplasm to the nucleus (Lee et al., 2009). In neurons, the net result of these splicing changes is to impact a biologically coherent set of transcripts encoding proteins mediating synaptic transmission and membrane excitability (Gehman et al., 2011). Since Geschwind and colleagues have implicated RBFOX1-mediated splicing defects as contributory to autism (Voineagu et al., 2011), it will be of interest to assess whether the activity-regulated splicing targets identified in mice overlay those associated with human disease.
Ptbp2
PTBP2 (also termed nPTB or brPTB) is a neuronal RNA binding protein harboring four RRM-type RNA binding motifs. PTBP2 was independently identified in a yeast 2-hybrid screen for NOVA interacting proteins (Polydorides et al., 2000), and in a biochemical purification scheme to identify factors that contribute to alternative splicing of c-src in neurons (Markovtsov et al., 2000). The name derives from the high homology (73% identity) with the polypyrimidine tract binding protein (PTB, now termed PTBP1), a well-studied protein that regulates alternative splicing among other aspects of RNA metabolism (Spellman et al., 2005). While PTBP1 is expressed in most tissues, it is either not expressed or minimally expressed in the brain. Instead, the brain expresses PTBP2, the product of a distinct gene.
Much about PTBP2 function can be inferred from a wealth of studies on PTBP1. In vitro selection and biochemical studies identified UCUU elements in the context of a pyrimidine-rich region (Singh et al., 1995; Perez et al., 1997), as well as CUCUCU (CU-repeats) present in c-src pre-mRNA (Chan and Black, 1997), as high affinity binding sites. While these studies suggested that PTBP1 functioned as a repressor of alternative splicing, splicing microarrays (Boutz et al., 2007; Llorian et al., 2010) and PTBP1 HITS-CLIP studies (Xue et al., 2009) revealed a smaller but significant role for PTBP1 as a splicing enhancer. Careful analysis of the latter studies (Llorian et al., 2010), along with biochemical validation, confirmed that PTBP1 generally acts according to the general rules of the position-dependent splicing map identified for NOVA and other proteins.
Identification of these binding elements in a Nova-regulated transcript revealed that PTBP2 interacts with Nova functionally, antagonizing its ability to enhance GlyRα2 exon 3a inclusion, at least in co-transfection assays (Polydorides et al., 2000). However, these studies, and a series of careful in vitro studies assessing PTBP1/2 regulation of c-src splicing (Markovtsov et al., 2000), including the observation that PTBP1 can inhibit c-src splicing through binding to U1 snRNP (Sharma et al., 2011), do not address the role of PTBP2 in neurons.
Such analyses were first begun by addressing the role of PTBP1/2 in neuronal differentiation. Interestingly, in different cell culture models, undifferentiated cells express PTBP1, which is proposed to bind PTBP2 pre-mRNA and trigger alternative splicing of an isoform harboring a nonsense-mediated decay isoform, thereby suppressing PTBP2 expression (Boutz et al., 2007). During neuronal differentiation, the switch to PTBP2 expression is proposed to be mediated by miR-124 induction in neural cells, since miR-124 is able to suppress PTBP1 expression (Makeyev et al., 2007).
This elegant model for neuronal differentiation was updated following analysis of PTBP2 RNA targets and insight into PTBP2 function in the brain that came from HITS-CLIP studies (Licatalosi et al., 2012). Generation of a PTBP2 binding map confirmed many similarities with PTBP1, including binding to UCU-rich target sites, and use of the same position-dependent map governing the regulation of alternative splicing inhibition and enhancement. In some cases, where transcripts such as Actn1 were expressed in both brain and non-neuronal cells, PTBP2 crosslinked precisely to sites previously mapped as sites governing PTBP1 inhibition of alternative exon inclusion (Matlin et al., 2007). In addition, PTBP2 HITS-CLIP was able to define new sites in the c-src primary transcript associated with alternative splicing of the c-src N1 exon and a second developmentally regulated alternate “N2” exon (Licatalosi et al., 2012).
Early in development, PTBP2 was found to be expressed in both mitotic neuronal progenitors and early post-mitotic neurons (Licatalosi et al., 2012). Moreover, in PTBP2 null mice, ectopic nests of neuronal progenitors were identified, suggesting that PTBP2 acts earlier than thought in the neuronal differentiation switch. Transcripts identified by HITS-CLIP as bound and regulated by PTBP2 in E18.5 embryonic brain included RNAs encoding determinants of the cell division, polarity, and cell fate, including Prkci, Numb, Brat, Trim3, Prox1, Erbb and others (Licatalosi et al., 2012). Taken together, these suggest that PTBP2 may play a role in neuronal progenitors to inhibit cell division, maintain cell polarity, and mediate the temporal control of neurogenesis.
nSR100
Blencowe and colleagues set out to identify SR proteins expressed in a tissue-specific manner, using microarray profiling to assess expression of genes encoding RS domain proteins in 50 mouse cell lines and tissues (Calarco et al., 2009). One of these, termed nSR100, was found to have a highly neural-restricted pattern, by analysis of RT-PCR and Western blot analysis of cell lines, and by RT-PCR analysis of different mouse tissues, although such non-quantitative analysis leaves open the possibility of scattered and low level expression in some other tissues, as evidenced by the authors’ analysis of mRNA expression by microarray.
Interestingly, nSR100 has been previously reported to be a tumor antigen, reminiscent of the identification of the paraneoplastic antigens Nova and Hu/Elavl (Behrends et al., 2003). However, in this instance, nSR100 was identified a screen with low titer (1:100 dilution) antisera taken from pediatric patients with medulloblastoma, and was identified as an immunoreactive protein in 2/5 medulloblastoma patients and 2/40 healthy volunteers, such that the significance of this finding is uncertain. Nonetheless, this observation suggested a possible connection with neural differentiation, and indeed, nSR100 knockdown impaired neuritic extension in differentiating N2A cells, inhibited ESC differentiation into neurospheres, and disrupted neural differentiation in developing zebrafish (Calarco et al., 2009).
These biologic actions correlate with target transcripts whose splicing was regulated by nSR100. In particular, splicing microarrays were used to identify regulated transcripts and a pyrimidine-rich binding motif through which nSR100 can act, suggesting possible interactions with other splicing factors that bind such motifs. Indeed, nSR100 was found to promote the inclusion of the key Ptbp2 exon 10, previously implicated in promoting neural differentiation (Calarco et al., 2009). These studies nicely illustrate the potential interconnections among neuronal-specific RNABPs to promote neuron-specific pathways of development.
IV. RNA binding proteins with special roles in neurons
This category of proteins is not entirely fair to demarcate, since only a few from among a very large number of RNABPs are discussed, their relationship to neuron-specific biology is in some cases unclear, and space constraints severely limits their discussion. Nonetheless, several RNABPs are of particular interest here because their characterization in neurons supports the possibility that neurons may regulate RNA in unique ways. Several excellent reviews discuss RNABPs more generally (Calarco et al., 2011; Cooper et al., 2009; Poulos et al., 2011; Richard, 2010), including those focused on generally expressed RNABPs associated with motor neuron disorders that may have undefined neuronal actions (Da Cruz and Cleveland, 2011; Battle et al., 2006). Additional aspects of RNA complexity that are of great interest in neurons, including alternative polyadenylation and RNA editing, but that go beyond the current discussion have been recently reviewed (Weill et al., 2012; Silberberg and Ohman, 2011).
CELF
CELF proteins are a family of RRM containing proteins with homology to ELAV. The proteins bind to pyrimidine-rich stretches and can activate splicing by competing with PTB family of proteins (Spellman et al., 2005; Chen and Manley, 2009). CELF1 (CUGBP1) is perhaps the best studied member of this family, as it was originally linked to binding of CUG triplet repeats in the pathogenesis of myotonic dystrophy ((Cooper et al., 2009); although see MBNL, below). Most CELF proteins are widely expressed and have been implicated in diverse functions, ranging from splicing regulation to mRNA localization, translation, stability and processing (Barreau et al., 2006). As a group, CELF proteins may also have important actions in the brain; for example, CELF1 specifically has been reported to regulate alternative splicing of the tau exon 10 in the brain (Dhaenens et al., 2011). CELF4 is reported as being neuron-specific in adult brain (Meins et al., 2002), although it is clearly expressed more widely early in development and demarcation of its expression has been complicated by cross reactivity with other family members. Nonetheless, CELF4 haploinsufficient and null mice develop spontaneous seizures; interestingly, this phenotype is only evident when CELF4 is deleted early in development, as deletion in adult mice using a floxed allele does not result in epilepsy (Wagnon et al., 2011).
CPEB
CPEB1 is the best characterized of a family of paralogous proteins that play important roles in translational regulation. CPEB proteins harbor 2 RRM-type RNA binding domains, and act by binding defined 3’ UTR elements to regulate poly(A) tail length and thereby mRNA translation (Richter, 2007). CPEB1 has been found to play important roles in neuronal biology. The protein is present in neuronal dendrites, as well as the cell body, and it induces polyadenylation and translation of several brain transcripts (Richter, 2010). Moreover, CPEB1-null mice have memory deficits and reduced LTP (Alarcon et al., 2004; Richter, 2010). Recently, CPEB1 has also been linked to cell senescence, mediated in part through the fine titration of CPEB1 levels by miR-122, and consequent effects on p53 translation (Burns et al., 2011). Identification of the set of directly bound CPEB1 transcripts will be of great interest in further understanding the action of the RNABP in neuronal plasticity, and, perhaps to neuronal aging.
FMRP
The Fragile X syndrome was the first human neurologic disease clearly linked to dysfunction of an RNABP (O'Donnell and Warren, 2002; Bhakar et al., 2012), following the discoveries that triplet repeat expansions cause loss of expression of the Fmr1 gene, and that the protein product, FMRP, binds to ribohomopolymers (Siomi et al., 1993) and poly(A)+ mRNA (Ashley et al., 1993). Identification of FMRP RNA ligands using in vitro selection identified binding preferences (G-quadruplex RNA that bind the RGG domain (Darnell et al., 2001), and kissing complex RNAs that bind the KH domains (Darnell et al., 2005)), complex structures that have been difficult to use to predict target transcripts. Recently, FMRP HITS-CLIP identified a robust set of target RNAs bound by FMRP in mouse brain, where its expression is largely restricted to neurons (Christie et al., 2009). In contrast to other HITS-CLIP analyses of RNABPs, FMRP bound along the coding sequence of a discrete set of mRNAs (Darnell et al., 2011). This in turn suggested a direct role for FMRP in translational regulation, consistent with its previously postulated function (Bear et al., 2004; Kelleher and Bear, 2008). Development of an in vitro translation system, in which polyribosomes from WT or FMRP-mutant brain were used to assay FMRP-dependent translation on target transcripts, revealed that FMRP does indeed function to inhibit translation in the brain by causing stalling of ribosome elongation (Darnell et al., 2011). How precisely this translational inhibition is itself regulated in neurons remains uncertain, but may involve FMRP dephosphorylation/phosphorylation (Narayanan et al., 2007; Lee et al., 2011), and/or interaction with miRNA-mediated controls (Edbauer et al., 2010; Muddashetty et al., 2011; Jin et al., 2004).
Analysis of the set of FMRP-regulated brain transcripts revealed a robust role in regulating expression of proteins in the pre and post-synaptic proteome. Moreover, consistent with the observation that ~30% of children with the Fragile-X syndrome also have autism spectrum disorders (Wang et al., 2010b), a high degree of overlap with autism candidate genes was found (Darnell et al., 2011). More recently, overlaying FMRP target transcripts with a dataset of de novo mutations identified in autistic children revealed that 1 in 5 autism candidate mutations were also regulated by FMRP (Iossifov et al., 2012), a remarkable overlap that tightens our view of the pathways that may lead to autism. Together, these studies illustrate successful use of the paradigm leading from basic biochemistry to in vivo function, and a means by which careful analysis of RNABP-RNA interactions can lead to new disease insight.
Musashi
Musashi was discovered in a Drosophila screen for genes involved in sensory organ development—mutants failed to develop neurons from sensory organ precursor cells (Nakamura et al., 1994). The protein harbors an RRM domain, and subsequent careful analysis of its expression in Drosophila revealed it to be highly restricted to the nervous system. Interestingly, within the mouse brain expression is largely confined to periventricular neuronal progenitors, with very little immunohistochemical reactivity evident in post-mitotic neurons (Sakakibara et al., 1996; Sakakibara and Okano, 1997). These observations have suggested that Musashi may play a key role in mediating an RNA switch from neuronal progenitors to neurons (Okano et al., 2005), for example through the regulation of expression of key target RNAs such those encoding members of the Notch/Numb signaling pathway (Imai et al., 2001; Okabe et al., 2001).
Mbnl
The MBNL family of proteins have been widely studied in the context of the muscular disorder myotonic dystrophy (DM), where their sequestration by CUG repeats plays a key role in disease pathogenesis. MBNL proteins contain four CCCH zinc finger motifs, and bind to YGCY motifs in pre-mRNA to regulate alternative splicing (Poulos et al., 2011). There has been significant progress in understanding the muscle disease, most notably the finding that mis-splicing of the chloride channel Clcn1, induced by sequestration of MBNL1, is a major contributor to they symptoms of myotonia in DM (Kanadia et al., 2003; Wheeler et al., 2007) and is a target for therapeutic intervention (Wheeler et al., 2009). However, only very recently have studies have begun to consider the central nervous system features of DM. These include sleep disturbance, the most common CNS complaint, as well as sporadic instances of mental retardation, autism, and other cognitive difficulties (Charizanis et al., 2012).
To address the role of MBNL proteins in neurons, two groups recently completed analysis of MBNL-dependent splicing changes together with bioinformatic and HITS-CLIP studies to identify sets of directly-regulated MBNL-regulated transcripts in the brain (Charizanis et al., 2012; Wang et al., 2012). Swanson and colleagues specifically MBNL2-null mice, revealing sleep abnormalities, memory loss, impaired NMDA receptor dependent transmission and hippocampal synaptic plasticity. These defects were correlated with aberrant splicing regulation of MBNL2 target transcripts, defined by exon junction microarray, HITS-CLIP, and validated as targets in both MBNL2-null mice and DM human tissue. These results led to a splicing map showing position-dependent splicing changes similar to that described for Nova, and identified target RNAs that encoding proteins involved in neuronal differentiation, development, axon guidance, and synaptic function (Charizanis et al., 2012). Burge and colleagues evaluated MBNL1 function in a similar set of studies using MBNL1 mutant mice, RNA-Seq and HITS-CLIP (which they term CLIP-Seq) to identify transcripts whose splicing was directly regulated by MBNL1 and a splicing map again consistent with prior position-dependent maps. Interestingly, the authors also did crude subcellular purifications to identify transcripts bound by MBNL1 in 3’ UTRs (identified by CLIP) whose cellular localization was MBNL1-dependent.
ZBP
The ZBP proteins are a family of RNABPs harboring four KH-type RNA binding motifs that have been well studied for their ability to regulate dendritic localization of mRNAs within neurons. This was originally demonstrated in studies by Singer and colleagues, who demonstrated β-actin mRNA localization in neuronal dendrites (Gu et al., 2002), and subsequently linked it to β-actin translation (Huttelmaier et al., 2005). Characterization of a consensus binding motif for ZBP1, a bipartite “zip-code” with a defined spacer, has allowed prediction of a subset of candidate target transcripts (Patel et al., 2012). Extension of these studies more systematically into neurons is ongoing, as are physiologic correlates, such as observations of decreased axonal length and outgrowth in response to injury in ZBP1+/− mice (Donnelly et al., 2011). Analyzing the role of ZBP proteins in neurons provides one of the most robust systems developed for understanding links between mRNA localization in a translationally silent state, and its switch to active translation following cell signaling, for example through protein phosphorylation (Huttelmaier et al., 2005).
Ago-miRNAs in neurons
A burgeoning topic in neurobiology relates to the degree and mechanism by which miRNAs may play special roles in mediating neuronal biology. This topic is worthy of an entire review in and of itself, and fortunately several timely and interesting such pieces are available (O'Carroll and Schaefer, 2012; Siegel et al., 2011; Mendell and Olson, 2012).
Several points are worth mentioning here regarding miRNA regulation specifically in neurons. First, from a methodologic point of view, technologies are now in place to analyze miRNA regulation using Ago HITS-CLIP in the brain (Chi et al., 2009), and it seems likely that Ago HITS-CLIP will be able to be applied to specific populations of neurons (He et al., 2012).
Second, the biology of miRNA action in neurons is rapidly evolving, with several main themes emerging. Studies cataloging miRNA abundance has revealed that several are expressed specifically in the brain (Fiore et al., 2008), and some show enrichment in neuronal dendrites (Siegel et al., 2011). Analysis of one such localized miRNA, miR-134, initially raised the possibility that miRNAs may act within dendrites to modulate local mRNA translation (Schratt et al., 2006). This action on translation was evident after BDNF signaling, an interesting connection given that BDNF is well documented to trigger local protein synthesis in dendrites (Aakalu et al., 2001; Sutton et al., 2004). Another line of studies has indicated that miRNAs can rapidly turn over in neurons in response to stimuli, as first illustrated by Filipowicz and colleagues in the retina (Krol et al., 2010). These observations have been recently connected with the finding that BDNF rapidly induces expression of some miRNAs, as well as neuronal Dicer and Lin28a (an RNABP that can block miRNA production (Heo et al., 2009; Hagan et al., 2009)), and that these coordinated effects may help mediate the specificity of translational response to BDNF (Huang et al., 2012).
Finally, an emerging story from studies in Drosophila suggests that miRNAs may play unexpected roles in chronically suppressing expression of developmental genes in the adult brain, and that such actions may be important in preventing brain aging and neurodegeneration (Liu et al., 2012). Such a role turns the traditional view of miRNAs in brain function—as mediators of developmental changes (miR-124, see discussion of Ptbp2) or of rapid signaling responses—on its head, portraying them as long term guardians against inappropriate gene expression.
V. Future Directions
Technologies are now emerging to analyze RNA regulation within specific neuronal populations. This is crucial, since the brain is composed of many thousands of different cell types, and combining data from each obscures signal:noise.
Indeed this point has been underscored by the wide appreciation of projects such as the Gensat and Allen Brain Atlas projects that enumerated neuron-specific transcripts, and their subsequent use to drive Cre recombinase in specific neuronal cell types. Such technologies have been combined with epitope tagging of RNABPs, a method that works for the purification of cell-specific Ago-miRNA complexes (He et al., 2012), and for ribosomal pull downs, to assess cell-specific translation. For example, in bacTRAP translational profiling, cell-specific Cre drivers derived from GenSat to purify ribosomes from individual neuronal cell types (Doyle et al., 2008). Combined with drivers that discriminate D1 and D2 dopaminergic neurons, Heintz and colleagues were able to identify transcripts specifically impacted by cocaine administration within D1 neurons that are likely to lead to changes in D1-specific synaptic responses (Heiman et al., 2008). In a second example, profiling of cortical layer Va known to be responsive to antidepressant treatment led to the identification of a neuron-specific protein, p11, necessary for physiologic actions on a specific serotonin receptor (Htr4) and behavioral responses to chronic administration of serotonin-specific reuptake inhibitors (Schmidt et al., 2012).
These observations suggest additional approaches that may be applied to HITS-CLIP profiling (Darnell, 2010; Konig et al., 2012) or ribosomal footprinting (Ingolia et al., 2009) to develop detailed maps of functional RNA-protein interactions within individual neuronal cell types. Given the ability to map Ago-miRNA binding sites with HITS-CLIP, they also suggest possible means to map neuron cell-specific miRNA controls. Finally, as high throughput sequencing becomes more potent, perhaps through the use of new direct RNA sequencing methods, these profiles may become applicable to subcellular compartments within neurons.
Supplementary Material
Acknowledgments
The work presented here incorporates the efforts of many tireless colleagues and hours of labor, for which the Author is deeply indebted. Apologies to the many RNABPs that function in neurons but were not discussed here given space issues. This work was supported by NIH R01 NS34389, an NIH Transformative Research Award (NS081706), the Rockefeller University Hospital CTSA (UL1 RR024143), the Starr Cancer Consortium, the ALS Therapy Alliance, a Simons Foundation Research Award, and an Emerald Foundation Distinguished Investigator Award. RBD is an Investigator of the Howard Hughes Medical Institute.
Acronyms
- nELAVL
neuronal embryonic abnormal lethal-like; refers to the 3 neuron-specific paralogs of the mammalian ELAVL proteins (originally termed Hu proteins), each of which show strong homology to the ELAV (embryonic abnormal lethal) protein, a Drosophila a neuron-specific RNABP
- NeuN
“Neuronal nuclei”; a neuron-specific RNA binding protein
- NOVA
Onconeural ventral antigen; a neuron-specific RNA binding protein
- HITS-CLIP
High throughput sequencing, crosslinking immunoprecipitation. Methods for covalently crosslinking RNA-protein complexes and sequencing the RNA binding sites, yielding genome-wide footprints of in vivo interactions. See SIDEBAR.
- PND
Paraneoplastic neurologic disorder; see SIDEBAR.
SIDEBARS
SIDEBAR: HITS-CLIP
HITS-CLIP describes the overlay of genome wide data gathering and bioinformatic strategies with biochemical crosslinking strategies to analyze RNA-protein interactions in living tissues. Originally, the CLIP method was developed to analyze Nova-RNA interactions in the mouse brain. Fundamentally, the method uses UV-irradation to penetrate live cells and crosslink, in situ, RNA-protein complexes that are in direct contact (~1 Å distances). Once crosslinked, RNA can be hydrolyzed to short fragments and protein complexes purified, typically by immunoprecipitation. Epitope-tagged proteins may be used, but attention needs to be paid to expression levels to avoid overexpression artifacts.
Once purified, RNA-protein complexes are treated with proteinase K and linker ligation completed to allow RT-PCR amplification and sequencing. CLIP established that reverse transcriptase (RT) can read through sites of residual amino-acid-RNA crosslinks; in fact, errors at these sites can be leveraged to map crosslink sites with single nucleotide resolution (Zhang and Darnell, 2011). Alternatively, in iCLIP, RT arrest at these sites can be used to map crosslink sites (Wang et al., 2010c). Sequencing the RNA tags yield HITS-CLIP libraries representing genome-wide footprints of RNABP binding. Additional details on HITS-CLIP methods (Ule et al., 2005a; Jensen and Darnell, 2008; Konig et al., 2011) and comparison with other comparable strategies such as PAR-CLIP (Hafner et al., 2010; Kishore et al., 2011) are described in several excellent papers and reviews (Blencowe et al., 2009; Kishore et al., 2010; Darnell, 2010; Calarco et al., 2011; Konig et al., 2012; Sugimoto et al., 2012).
SIDEBAR: Discovery of mammalian neuron-specific RNABPs through study of the Paraneoplastic Neurologic Disorders (PNDs)
In studies analogous to the discovery of snRNPs by Joan Steitz and colleagues made by studies of autoimmune lupus antisera (Lerner et al., 1981), two multigene families of neuron-specific RNA binding proteins were discovered through studies of autoimmune PND antisera. In the PNDs (as discussed in (Darnell and Posner, 2011)), common types of cancers (small cell lung, breast, ovarian, lymphoma, germ cell cancers, etc.) may induce expression of proteins that are normally restricted to the neurons. This triggers an anti-tumor immune response that can be clinically potent (Darnell and Posner, 2003a; Darnell and DeAngelis, 1993; Darnell and Posner, 2003b); in a subset of these patients, this immune response crosses into the nervous system triggering neurologic symptoms and bringing patients to clinical attention. After the original demonstration that PND antisera could be used to screen expression cDNA libraries and identify the genes encoding the target antigens (Darnell et al., 1989; McKeever and Darnell, 1992; Newman et al., 1995), this method was applied to identify the neuron-specific nElavl4 cDNA (Szabo et al., 1991), Nova1 (Buckanovich et al., 1993) and Nova2 (Yang et al., 1998) RNABPs.
Literature Cited
- Aakalu G, Smith WB, Nguyen N, Jiang C, Schuman EM. Dynamic visualization of local protein synthesis in hippocampal neurons. Neuron. 2001;30:489–502. doi: 10.1016/s0896-6273(01)00295-1. [DOI] [PubMed] [Google Scholar]
- Akamatsu W, Fujihara H, Mitsuhashi T, Yano M, Shibata S, Hayakawa Y, Okano HJ, Sakakibara S, Takano H, Takano T, Takahashi T, Noda T, Okano H. The RNA-binding protein HuD regulates neuronal cell identity and maturation. Proc Natl Acad Sci U S A. 2005;102:4625–4630. doi: 10.1073/pnas.0407523102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akamatsu W, Okano HJ, Osumi N, Inoue T, Nakamura S, Sakakibara SI, Miura M, Matsuo N, Darnell RB, Okano H. Mammalian ELAV-like neuronal RNA-binding proteins HuB and HuC promote neuronal development in both the central and the peripheral nervous systems. Proc Natl Acad Sci U S A. 1999;96:9885–9890. doi: 10.1073/pnas.96.17.9885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alarcon JM, Hodgman R, Theis M, Huang YS, Kandel ER, Richter JD. Selective modulation of some forms of schaffer collateral-CA1 synaptic plasticity in mice with a disruption of the CPEB-1 gene. Learn Mem. 2004;11:318–327. doi: 10.1101/lm.72704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amara SG, Jonas V, Rosenfeld MG, Ong ES, Evans R. Alternative RNA processing in calcitonin gene expression generates mRNAs encoding different polypeptide products. Nature. 1982;298:240–244. doi: 10.1038/298240a0. [DOI] [PubMed] [Google Scholar]
- Antic D, Lu N, Keene JD. ELAV tumor antigen, Hel-N1, increases translation of neurofilament M mRNA and induces formation of neurites in human teratocarcinoma cells. Genes Dev. 1999;13:449–461. doi: 10.1101/gad.13.4.449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashley CTJ, Wilkinson KD, Reines D, Warren ST. FMR1 protein: conserved RNP family domains and selective RNA binding. Science. 1993;262:563–566. doi: 10.1126/science.7692601. [DOI] [PubMed] [Google Scholar]
- Bakker CE, Verheij C, Willemsen R, van der Helm R, Oerlemans F, Vermey M, Bygrave V, Hoogeveen AT, Oostra BA. Fmr1 knockout mice: a model to study fragile X mental retardation. The Dutch-Belgian Fragile X Consortium. Cell. 1994;78:23–33. [PubMed] [Google Scholar]
- Barreau C, Paillard L, Mereau A, Osborne HB. Mammalian CELF/Bruno-like RNA-binding proteins: molecular characteristics and biological functions. Biochimie. 2006;88:515–525. doi: 10.1016/j.biochi.2005.10.011. [DOI] [PubMed] [Google Scholar]
- Battle DJ, Kasim M, Yong J, Lotti F, Lau CK, Mouaikel J, Zhang Z, Han K, Wan L, Dreyfuss G. The SMN complex: an assembly machine for RNPs. Cold Spring Harb Symp Quant Biol. 2006;71:313–320. doi: 10.1101/sqb.2006.71.001. [DOI] [PubMed] [Google Scholar]
- Bear MF, Huber KM, Warren ST. The mGluR theory of fragile X mental retardation. Trends Neurosci. 2004;27:370–377. doi: 10.1016/j.tins.2004.04.009. [DOI] [PubMed] [Google Scholar]
- Behrends U, Schneider I, Rossler S, Frauenknecht H, Golbeck A, Lechner B, Eigenstetter G, Zobywalski C, Muller-Weihrich S, Graubner U, Schmid I, Sackerer D, Spath M, Goetz C, Prantl F, Asmuss HP, Bise K, Mautner J. Novel tumor antigens identified by autologous antibody screening of childhood medulloblastoma cDNA libraries. Int J Cancer. 2003;106:244–251. doi: 10.1002/ijc.11208. [DOI] [PubMed] [Google Scholar]
- Bhakar AL, Dolen G, Bear MF. The pathophysiology of fragile X (and what it teaches us about synapses). Annu Rev Neurosci. 2012;35:417–443. doi: 10.1146/annurev-neuro-060909-153138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bier E, Ackerman L, Barbel S, Jan L, Jan YN. Identification and characterization of a neuron-specific nuclear antigen in Drosophila. Science. 1988;240:913–916. doi: 10.1126/science.3129785. [DOI] [PubMed] [Google Scholar]
- Blanchette M, Green RE, Brenner SE, Rio DC. Global analysis of positive and negative pre-mRNA splicing regulators in Drosophila. Genes Dev. 2005;19:1306–1314. doi: 10.1101/gad.1314205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blencowe BJ, Ahmad S, Lee LJ. Current-generation high-throughput sequencing: deepening insights into mammalian transcriptomes. Genes Dev. 2009;23:1379–1386. doi: 10.1101/gad.1788009. [DOI] [PubMed] [Google Scholar]
- Bolognani F, Perrone-Bizzozero NI. RNA-protein interactions and control of mRNA stability in neurons. J Neurosci Res. 2008;86:481–489. doi: 10.1002/jnr.21473. [DOI] [PubMed] [Google Scholar]
- Boutz PL, Stoilov P, Li Q, Lin CH, Chawla G, Ostrow K, Shiue L, Ares MJ, Black DL. A post-transcriptional regulatory switch in polypyrimidine tract-binding proteins reprograms alternative splicing in developing neurons. Genes Dev. 2007;21:1636–1652. doi: 10.1101/gad.1558107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckanovich RJ, Darnell RB. The neuronal RNA binding protein Nova-1 recognizes specific RNA targets in vitro and in vivo. Mol Cell Biol. 1997;17:3194–3201. doi: 10.1128/mcb.17.6.3194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckanovich RJ, Yang YY, Darnell RB. The onconeural antigen Nova-1 is a neuron-specific RNA-binding protein, the activity of which is inhibited by paraneoplastic antibodies. J Neurosci. 1996;16:1114–1122. doi: 10.1523/JNEUROSCI.16-03-01114.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckanovich RJ, Posner JB, Darnell RB. Nova, the paraneoplastic Ri antigen, is homologous to an RNA-binding protein and is specifically expressed in the developing motor system. Neuron. 1993;11:657–672. doi: 10.1016/0896-6273(93)90077-5. [DOI] [PubMed] [Google Scholar]
- Burd CG, Dreyfuss G. Conserved structures and diversity of functions of RNA-binding proteins. Science. 1994;265:615–621. doi: 10.1126/science.8036511. [DOI] [PubMed] [Google Scholar]
- Burns DM, D'Ambrogio A, Nottrott S, Richter JD. CPEB and two poly(A) polymerases control miR-122 stability and p53 mRNA translation. Nature. 2011;473:105–108. doi: 10.1038/nature09908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calarco JA, Superina S, O'Hanlon D, Gabut M, Raj B, Pan Q, Skalska U, Clarke L, Gelinas D, van der Kooy D, Zhen M, Ciruna B, Blencowe BJ. Regulation of vertebrate nervous system alternative splicing and development by an SR-related protein. Cell. 2009;138:898–910. doi: 10.1016/j.cell.2009.06.012. [DOI] [PubMed] [Google Scholar]
- Calarco JA, Zhen M, Blencowe BJ. Networking in a global world: establishing functional connections between neural splicing regulators and their target transcripts. RNA. 2011;17:775–791. doi: 10.1261/rna.2603911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos AR, Rosen DR, Robinow SN, White K. Molecular analysis of the locus elav in Drosophila melanogaster: a gene whose embryonic expression is neural specific. EMBO J. 1987;6:425–431. doi: 10.1002/j.1460-2075.1987.tb04772.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castle JC, Zhang C, Shah JK, Kulkarni AV, Kalsotra A, Cooper TA, Johnson JM. Expression of 24,426 human alternative splicing events and predicted cis regulation in 48 tissues and cell lines. Nat Genet. 2008;40:1416–1425. doi: 10.1038/ng.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan RC, Black DL. The polypyrimidine tract binding protein binds upstream of neural cell- specific c-src exon N1 to repress the splicing of the intron downstream. Mol Cell Biol. 1997;17:4667–4676. doi: 10.1128/mcb.17.8.4667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charizanis K, Lee KY, Batra R, Goodwin M, Zhang C, Yuan Y, Shiue L, Cline M, Scotti MM, Xia G, Kumar A, Ashizawa T, Clark HB, Kimura T, Takahashi MP, Fujimura H, Jinnai K, Yoshikawa H, Gomes-Pereira M, Gourdon G, Sakai N, Nishino S, Foster TC, Ares MJ, Darnell RB, Swanson MS. Muscleblind-like 2-Mediated Alternative Splicing in the Developing Brain and Dysregulation in Myotonic Dystrophy. Neuron. 2012;75:437–450. doi: 10.1016/j.neuron.2012.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CY, Shyu AB. Mechanisms of deadenylation-dependent decay. Wiley Interdiscip Rev RNA. 2011;2:167–183. doi: 10.1002/wrna.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Manley JL. Mechanisms of alternative splicing regulation: insights from molecular and genomics approaches. Nat Rev Mol Cell Biol. 2009 doi: 10.1038/nrm2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi SW, Zang JB, Mele A, Darnell RB. Argonaute HITS-CLIP decodes microRNA-mRNA interaction maps. Nature. 2009;460:479–486. doi: 10.1038/nature08170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christie SB, Akins MR, Schwob JE, Fallon JR. The FXG: a presynaptic fragile X granule expressed in a subset of developing brain circuits. J Neurosci. 2009;29:1514–1524. doi: 10.1523/JNEUROSCI.3937-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper TA, Wan L, Dreyfuss G. RNA and disease. Cell. 2009;136:777–793. doi: 10.1016/j.cell.2009.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corrionero A, Valcarcel J. RNA processing: Redrawing the map of charted territory. Mol Cell. 2009;36:918–919. doi: 10.1016/j.molcel.2009.12.004. [DOI] [PubMed] [Google Scholar]
- Da Cruz S, Cleveland DW. Understanding the role of TDP-43 and FUS/TLS in ALS and beyond. Curr Opin Neurobiol. 2011;21:904–919. doi: 10.1016/j.conb.2011.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell JC, Fraser CE, Mostovetsky O, Stefani G, Jones TA, Eddy SR, Darnell RB. Kissing complex RNAs mediate interaction between the Fragile-X mental retardation protein KH2 domain and brain polyribosomes. Genes Dev. 2005;19:903–918. doi: 10.1101/gad.1276805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell JC, Jensen KB, Jin P, Brown V, Warren ST, Darnell RB. Fragile X mental retardation protein targets G Quartet mRNAs important for neuronal function. Cell. 2001;107:489–99. doi: 10.1016/s0092-8674(01)00566-9. [DOI] [PubMed] [Google Scholar]
- Darnell JC, Van Driesche SJ, Zhang C, Hung KY, Mele A, Fraser CE, Stone EF, Chen C, Fak JJ, Chi SW, Licatalosi DD, Richter JD, Darnell RB. FMRP Stalls Ribosomal Translocation on mRNAs Linked to Synaptic Function and Autism. Cell. 2011;146:247–261. doi: 10.1016/j.cell.2011.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell RB. Developing global insight into RNA regulation. Cold Spring Harb Symp Quant Biol. 2006;71:321–327. doi: 10.1101/sqb.2006.71.002. [DOI] [PubMed] [Google Scholar]
- Darnell RB, DeAngelis LM. Regression of small-cell lung carcinoma in patients with paraneoplastic neuronal antibodies. Lancet. 1993;341:21–22. doi: 10.1016/0140-6736(93)92485-c. [DOI] [PubMed] [Google Scholar]
- Darnell RB, Furneaux H, Posner JB. Characterization of antigens bound by CSF and serum of a patient with cerebellar degeneration: co-expression in Purkinje cells and tumor lines of neuroectodermal origin. Neurology. 1989;39:385. [Google Scholar]
- Darnell RB, Posner JB. Observing the invisible: successful tumor immunity in humans. Nat Immunol. 2003a;4:201. doi: 10.1038/ni0303-201. [DOI] [PubMed] [Google Scholar]
- Darnell RB. HITS-CLIP: panoramic views of protein-RNA regulation in living cells. Wiley Interdiscip Rev RNA. 2010;1:266–286. doi: 10.1002/wrna.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell RB, Posner JB. Paraneoplastic syndromes involving the nervous system. N Engl J Med. 2003b;349:1543–1554. doi: 10.1056/NEJMra023009. [DOI] [PubMed] [Google Scholar]
- Darnell RB, Posner JB. Paraneoplastic Syndromes (Contemporary Neurology Series) 2011 [Google Scholar]
- Dean JL, Wait R, Mahtani KR, Sully G, Clark AR, Saklatvala J. The 3' untranslated region of tumor necrosis factor alpha mRNA is a target of the mRNA-stabilizing factor HuR. Mol Cell Biol. 2001;21:721–730. doi: 10.1128/MCB.21.3.721-730.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhaenens CM, Tran H, Frandemiche ML, Carpentier C, Schraen-Maschke S, Sistiaga A, Goicoechea M, Eddarkaoui S, Van Brussels E, Obriot H, Labudeck A, Gevaert MH, Fernandez-Gomez F, Charlet-Berguerand N, Deramecourt V, Maurage CA, Buee L, de Munain AL, Sablonniere B, Caillet-Boudin ML, Sergeant N. Mis-splicing of Tau exon 10 in myotonic dystrophy type 1 is reproduced by overexpression of CELF2 but not by MBNL1 silencing. Biochim Biophys Acta. 2011;1812:732–742. doi: 10.1016/j.bbadis.2011.03.010. [DOI] [PubMed] [Google Scholar]
- Donnelly CJ, Willis DE, Xu M, Tep C, Jiang C, Yoo S, Schanen NC, Kirn-Safran CB, van Minnen J, English A, Yoon SO, Bassell GJ, Twiss JL. Limited availability of ZBP1 restricts axonal mRNA localization and nerve regeneration capacity. EMBO J. 2011;30:4665–4677. doi: 10.1038/emboj.2011.347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dor Y, Brown J, Martinez OI, Melton DA. Adult pancreatic beta-cells are formed by self-duplication rather than stem-cell differentiation. Nature. 2004;429:41–46. doi: 10.1038/nature02520. [DOI] [PubMed] [Google Scholar]
- Doyle JP, Dougherty JD, Heiman M, Schmidt EF, Stevens TR, Ma G, Bupp S, Shrestha P, Shah RD, Doughty ML, Gong S, Greengard P, Heintz N. Application of a translational profiling approach for the comparative analysis of CNS cell types. Cell. 2008;135:749–762. doi: 10.1016/j.cell.2008.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dredge BK, Darnell RB. Nova regulates GABA(A) receptor gamma2 alternative splicing via a distal downstream UCAU-rich intronic splicing enhancer. Molecular & Cellular Biology. 2003;23:4687–4700. doi: 10.1128/MCB.23.13.4687-4700.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dredge BK, Stefani G, Engelhard CC, Darnell RB. Nova autoregulation reveals dual functions in neuronal splicing. EMBO J. 2005;24:1608–1620. doi: 10.1038/sj.emboj.7600630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dredge BK, Jensen KB. NeuN/Rbfox3 Nuclear and Cytoplasmic Isoforms Differentially Regulate Alternative Splicing and Nonsense-Mediated Decay of Rbfox2. PLoS One. 2011;6:e21585. doi: 10.1371/journal.pone.0021585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du H, Cline MS, Osborne RJ, Tuttle DL, Clark TA, Donohue JP, Hall MP, Shiue L, Swanson MS, Thornton CA, Ares MJ. Aberrant alternative splicing and extracellular matrix gene expression in mouse models of myotonic dystrophy. Nat Struct Mol Biol. 2010;17:187–193. doi: 10.1038/nsmb.1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edbauer D, Neilson JR, Foster KA, Wang CF, Seeburg DP, Batterton MN, Tada T, Dolan BM, Sharp PA, Sheng M. Regulation of synaptic structure and function by FMRP-associated microRNAs miR-125b and miR-132. Neuron. 2010;65:373–384. doi: 10.1016/j.neuron.2010.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellington AD, Szostak JW. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990;346:818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- Fan XC, Myer VE, Steitz JA. AU-rich elements target small nuclear RNAs as well as mRNAs for rapid degradation. Genes Dev. 1997;11:2557–2568. doi: 10.1101/gad.11.19.2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan XC, Steitz JA. Overexpression of HuR, a nuclear-cytoplasmic shuttling protein, increases the in vivo stability of ARE-containing mRNAs. EMBO J. 1998;17:3448–3460. doi: 10.1093/emboj/17.12.3448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiore R, Siegel G, Schratt G. MicroRNA function in neuronal development, plasticity and disease. Biochim Biophys Acta. 2008;1779:471–478. doi: 10.1016/j.bbagrm.2007.12.006. [DOI] [PubMed] [Google Scholar]
- Gao FB, Carson CC, Levine T, Keene JD. Selection of a subset of mRNAs from combinatorial 3' untranslated region libraries using neuronal RNA-binding protein Hel-N1. Proc Natl Acad Sci USA. 1994;91:11207–11211. doi: 10.1073/pnas.91.23.11207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehman LT, Stoilov P, Maguire J, Damianov A, Lin CH, Shiue L, Ares MJ, Mody I, Black DL. The splicing regulator Rbfox1 (A2BP1) controls neuronal excitation in the mammalian brain. Nat Genet. 2011;43:706–711. doi: 10.1038/ng.841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green R, Ellington AD, Bartel DP, Szostak JW. In vitro genetic analysis: selection and amplification of rare functional nucleic acids. Methods Compan Methods Enzymol. 1991;2:75–86. [Google Scholar]
- Gu W, Pan F, Zhang H, Bassell GJ, Singer RH. A predominantly nuclear protein affecting cytoplasmic localization of beta-actin mRNA in fibroblasts and neurons. J Cell Biol. 2002;156:41–51. doi: 10.1083/jcb.200105133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafner M, Landthaler M, Burger L, Khorshid M, Hausser J, Berninger P, Rothballer A, Ascano MJ, Jungkamp AC, Munschauer M, Ulrich A, Wardle GS, Dewell S, Zavolan M, Tuschl T. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell. 2010;141:129–141. doi: 10.1016/j.cell.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagan JP, Piskounova E, Gregory RI. Lin28 recruits the TUTase Zcchc11 to inhibit let-7 maturation in mouse embryonic stem cells. Nat Struct Mol Biol. 2009;16:1021–1025. doi: 10.1038/nsmb.1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen TV, Hammer NA, Nielsen J, Madsen M, Dalbaeck C, Wewer UM, Christiansen J, Nielsen FC. Dwarfism and impaired gut development in insulin-like growth factor II mRNA-binding protein 1-deficient mice. Mol Cell Biol. 2004;24:4448–4464. doi: 10.1128/MCB.24.10.4448-4464.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He M, Liu Y, Wang X, Zhang MQ, Hannon GJ, Huang ZJ. Cell-type-based analysis of microRNA profiles in the mouse brain. Neuron. 2012;73:35–48. doi: 10.1016/j.neuron.2011.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heiman M, Schaefer A, Gong S, Peterson JD, Day M, Ramsey KE, Suarez-Farinas M, Schwarz C, Stephan DA, Surmeier DJ, Greengard P, Heintz N. A translational profiling approach for the molecular characterization of CNS cell types. Cell. 2008;135:738–748. doi: 10.1016/j.cell.2008.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heo I, Joo C, Kim YK, Ha M, Yoon MJ, Cho J, Yeom KH, Han J, Kim VN. TUT4 in concert with Lin28 suppresses microRNA biogenesis through pre-microRNA uridylation. Cell. 2009;138:696–708. doi: 10.1016/j.cell.2009.08.002. [DOI] [PubMed] [Google Scholar]
- Hinman MN, Lou H. Diverse molecular functions of Hu proteins. Cell Mol Life Sci. 2008;65:3168–3181. doi: 10.1007/s00018-008-8252-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt CE, Bullock SL. Subcellular mRNA localization in animal cells and why it matters. Science. 2009;326:1212–1216. doi: 10.1126/science.1176488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homyk TJ, Szidonya J, Suzuki DT. Behavioral mutants of Drosophila melanogaster. III. Isolation and mapping of mutations by direct visual observations of behavioral phenotypes. Mol Gen Genet. 1980;177:553–565. doi: 10.1007/BF00272663. [DOI] [PubMed] [Google Scholar]
- Hormigo A, Dalmau J, Rosenblum MK, River ME, Posner JB. Immunological and pathological study of anti-Ri-associated encephalopathy. Ann Neurol. 1994;36:896–902. doi: 10.1002/ana.410360615. [DOI] [PubMed] [Google Scholar]
- Huang CS, Shi SH, Ule J, Ruggiu M, Barker LA, Darnell RB, Jan YN, Jan LY. Common molecular pathways mediate long-term potentiation of synaptic excitation and slow synaptic inhibition. Cell. 2005;123:105–118. doi: 10.1016/j.cell.2005.07.033. [DOI] [PubMed] [Google Scholar]
- Huang YW, Ruiz CR, Eyler EC, Lin K, Meffert MK. Dual regulation of miRNA biogenesis generates target specificity in neurotrophin-induced protein synthesis. Cell. 2012;148:933–946. doi: 10.1016/j.cell.2012.01.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huttelmaier S, Zenklusen D, Lederer M, Dictenberg J, Lorenz M, Meng X, Bassell GJ, Condeelis J, Singer RH. Spatial regulation of beta-actin translation by Src-dependent phosphorylation of ZBP1. Nature. 2005;438:512–515. doi: 10.1038/nature04115. [DOI] [PubMed] [Google Scholar]
- Imai T, Tokunaga A, Yoshida T, Hashimoto M, Mikoshiba K, Weinmaster G, Nakafuku M, Okano H. The neural RNA-binding protein Musashi1 translationally regulates mammalian numb gene expression by interacting with its mRNA. Mol Cell Biol. 2001;21:3888–3900. doi: 10.1128/MCB.21.12.3888-3900.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ince-Dunn G, Okano HJ, Jensen KB, Park WY, Zhong R, Ule J, Mele A, Fak JJ, Yang CW, Zhang C, Yoo J, Herre M, Okano H, Noebels JL, Darnell RB. Neuronal Elav-like (Hu) proteins regulate RNA splicing and abundance to control glutamate levels and neuronal excitability. Neuron. 2012 doi: 10.1016/j.neuron.2012.07.009. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia NT, Ghaemmaghami S, Newman JR, Weissman JS. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science. 2009;324:218–223. doi: 10.1126/science.1168978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iossifov I, Ronemus M, Levy D, Wang Z, Hakker I, Rosenbaum J, Yamrom B, Lee YH, Narzisi G, Leotta A, Kendall J, Grabowska E, Ma B, Marks S, Rodgers L, Stepansky A, Troge J, Andrews P, Bekritsky M, Pradhan K, Ghiban E, Kramer M, Parla J, Demeter R, Fulton LL, Fulton RS, Magrini VJ, Ye K, Darnell JC, Darnell RB, Mardis ER, Wilson RK, Schatz MC, McCombie WR, Wigler M. De novo gene disruptions in children on the autistic spectrum. Neuron. 2012;74:285–299. doi: 10.1016/j.neuron.2012.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain RG, Andrews LG, Mcgowan KM, Pekala PH, Keene JD. Ectopic expression of Hel-N1, an RNA-binding protein, increases glucose transporter (GluT1) expression in 3T3-L1 adipocytes. Mol Cell Biol. 1997;17:954–962. doi: 10.1128/mcb.17.2.954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen KB, Musunuru K, Lewis HA, Burley SK, Darnell RB. The tetranucleotide UCAY directs the specific recognition of RNA by the Nova K-homology 3 domain. Proceedings of the National Academy of Sciences of the United States of America. 2000a;97:5740–5745. doi: 10.1073/pnas.090553997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen KB, Darnell RB. CLIP: crosslinking and immunoprecipitation of in vivo RNA targets of RNA-binding proteins. Methods Mol Biol. 2008;488:85–98. doi: 10.1007/978-1-60327-475-3_6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen KB, Dredge BK, Stefani G, Zhong R, Buckanovich RJ, Okano HJ, Yang YY, Darnell RB. Nova-1 regulates neuron-specific alternative splicing and is essential for neuronal viability. Neuron. 2000b;25:359–371. doi: 10.1016/s0896-6273(00)80900-9. [DOI] [PubMed] [Google Scholar]
- Jin P, Alisch RS, Warren ST. RNA and microRNAs in fragile X mental retardation. Nat Cell Biol. 2004;6:1048–1053. doi: 10.1038/ncb1104-1048. [DOI] [PubMed] [Google Scholar]
- Jin Y, Suzuki H, Maegawa S, Endo H, Sugano S, Hashimoto K, Yasuda K, Inoue K. A vertebrate RNA-binding protein Fox-1 regulates tissue-specific splicing via the pentanucleotide GCAUG. EMBO J. 2003;22:905–912. doi: 10.1093/emboj/cdg089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson JM, Castle J, Garrett-Engele P, Kan Z, Loerch PM, Armour CD, Santos R, Schadt EE, Stoughton R, Shoemaker DD. Genome-wide survey of human alternative pre-mRNA splicing with exon junction microarrays. Science. 2003;302:2141–2144. doi: 10.1126/science.1090100. [DOI] [PubMed] [Google Scholar]
- Kalsotra A, Xiao X, Ward AJ, Castle JC, Johnson JM, Burge CB, Cooper TA. A postnatal switch of CELF and MBNL proteins reprograms alternative splicing in the developing heart. Proc Natl Acad Sci U S A. 2008;105:20333–20338. doi: 10.1073/pnas.0809045105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanadia RN, Urbinati CR, Crusselle VJ, Luo D, Lee YJ, Harrison JK, Oh SP, Swanson MS. Developmental expression of mouse muscleblind genes Mbnl1, Mbnl2 and Mbnl3. Gene Expr Patterns. 2003;3:459–462. doi: 10.1016/s1567-133x(03)00064-4. [DOI] [PubMed] [Google Scholar]
- Kelleher R. J. r., Bear MF. The autistic neuron: troubled translation? Cell. 2008;135:401–406. doi: 10.1016/j.cell.2008.10.017. [DOI] [PubMed] [Google Scholar]
- Kim KK, Adelstein RS, Kawamoto S. Identification of neuronal nuclei (NeuN) as Fox-3, a new member of the Fox-1 gene family of splicing factors. J Biol Chem. 2009;284:31052–31061. doi: 10.1074/jbc.M109.052969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishore S, Jaskiewicz L, Burger L, Hausser J, Khorshid M, Zavolan M. A quantitative analysis of CLIP methods for identifying binding sites of RNA-binding proteins. Nat Methods. 2011;8:559–564. doi: 10.1038/nmeth.1608. [DOI] [PubMed] [Google Scholar]
- Kishore S, Luber S, Zavolan M. Deciphering the role of RNA-binding proteins in the post-transcriptional control of gene expression. Brief Funct Genomics. 2010;9:391–404. doi: 10.1093/bfgp/elq028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konig J, Zarnack K, Luscombe NM, Ule J. Protein-RNA interactions: new genomic technologies and perspectives. Nat Rev Genet. 2012;13:77–83. doi: 10.1038/nrg3141. [DOI] [PubMed] [Google Scholar]
- Konig J, Zarnack K, Rot G, Curk T, Kayikci M, Zupan B, Turner DJ, Luscombe NM, Ule J. iCLIP reveals the function of hnRNP particles in splicing at individual nucleotide resolution. Nat Struct Mol Biol. 2010;17:909–915. doi: 10.1038/nsmb.1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konig J, Zarnack K, Rot G, Curk T, Kayikci M, Zupan B, Turner DJ, Luscombe NM, Ule J. iCLIP--transcriptome-wide mapping of protein-RNA interactions with individual nucleotide resolution. J Vis Exp. 2011 doi: 10.3791/2638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koushika SP, Lisbin MJ, White K. Elav, A Drosophila neuron-specific protein, mediates the generation of an alternatively spliced neural protein isoform. Curr Biol. 1996;6:1634–1641. doi: 10.1016/s0960-9822(02)70787-2. [DOI] [PubMed] [Google Scholar]
- Koushika SP, Soller M, White K. The neuron-enriched splicing pattern of Drosophila erect wing is dependent on the presence of ELAV protein. Mol Cell Biol. 2000;20:1836–1845. doi: 10.1128/mcb.20.5.1836-1845.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krol J, Busskamp V, Markiewicz I, Stadler MB, Ribi S, Richter J, Duebel J, Bicker S, Fehling HJ, Schubeler D, Oertner TG, Schratt G, Bibel M, Roska B, Filipowicz W. Characterizing light-regulated retinal microRNAs reveals rapid turnover as a common property of neuronal microRNAs. Cell. 2010;141:618–631. doi: 10.1016/j.cell.2010.03.039. [DOI] [PubMed] [Google Scholar]
- Kuroyanagi H. Fox-1 family of RNA-binding proteins. Cell Mol Life Sci. 2009;66:3895–3907. doi: 10.1007/s00018-009-0120-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee EK, Kim W, Tominaga K, Martindale JL, Yang X, Subaran SS, Carlson OD, Mercken EM, Kulkarni RN, Akamatsu W, Okano H, Perrone-Bizzozero NI, de Cabo R, Egan JM, Gorospe M. RNA-binding protein HuD controls insulin translation. Mol Cell. 2012;45:826–835. doi: 10.1016/j.molcel.2012.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HY, Ge WP, Huang W, He Y, Wang GX, Rowson-Baldwin A, Smith SJ, Jan YN, Jan LY. Bidirectional regulation of dendritic voltage-gated potassium channels by the fragile X mental retardation protein. Neuron. 2011;72:630–642. doi: 10.1016/j.neuron.2011.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JA, Tang ZZ, Black DL. An inducible change in Fox-1/A2BP1 splicing modulates the alternative splicing of downstream neuronal target exons. Genes Dev. 2009;23:2284–2293. doi: 10.1101/gad.1837009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerner MR, Boyle JA, Hardin JA, Steitz JA. Two novel classes of small ribonucleoproteins detected by antibodies associated with lupus erythematosus. Science. 1981;211:400–402. doi: 10.1126/science.6164096. [DOI] [PubMed] [Google Scholar]
- Levine TD, Gao F, King PH, Andrews LG, Keene JD. Hel-N1: an autoimmune RNA-binding protein with specificity for 3' uridylate-rich untranslated regions of growth factor mRNA's. Mol Cell Biol. 1993;13:3494–3504. doi: 10.1128/mcb.13.6.3494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis HA, Chen H, Edo C, Buckanovich RJ, Yang YY, Musunuru K, Zhong R, Darnell RB, Burley SK. Crystal structures of Nova-1 and Nova-2 K-homology RNA-binding domains. Structure. 1999;7:191–203. doi: 10.1016/S0969-2126(99)80025-2. [DOI] [PubMed] [Google Scholar]
- Lewis HA, Musunuru K, Jensen KB, Edo C, Chen H, Darnell RB, Burley SK. Sequence-specific RNA binding by a Nova KH domain: implications for paraneoplastic disease and the fragile X syndrome. Cell. 2000;100:323–332. doi: 10.1016/s0092-8674(00)80668-6. [DOI] [PubMed] [Google Scholar]
- Licatalosi DD, Darnell RB. Splicing regulation in neurologic disease. Neuron. 2006;52:93–101. doi: 10.1016/j.neuron.2006.09.017. [DOI] [PubMed] [Google Scholar]
- Licatalosi DD, Darnell RB. RNA processing and its regulation: global insights into biological networks. Nat Rev Genet. 2010;11:75–87. doi: 10.1038/nrg2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Licatalosi DD, Mele A, Fak JJ, Ule J, Kayikci M, Chi SW, Clark TA, Schweitzer AC, Blume JE, Wang X, Darnell JC, Darnell RB. HITS-CLIP yields genome-wide insights into brain alternative RNA processing. Nature. 2008;456:464–469. doi: 10.1038/nature07488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Licatalosi DD, Yano M, Fak JJ, Mele A, Grabinski SE, Zhang C, Darnell RB. Ptbp2 represses adult-specific splicing to regulate the generation of neuronal precursors in the embryonic brain. Genes Dev. 2012;26:1626–1642. doi: 10.1101/gad.191338.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim J, Hao T, Shaw C, Patel AJ, Szabo G, Rual JF, Fisk CJ, Li N, Smolyar A, Hill DE, Barabasi AL, Vidal M, Zoghbi HY. A protein-protein interaction network for human inherited ataxias and disorders of Purkinje cell degeneration. Cell. 2006;125:801–814. doi: 10.1016/j.cell.2006.03.032. [DOI] [PubMed] [Google Scholar]
- Liu N, Landreh M, Cao K, Abe M, Hendriks GJ, Kennerdell JR, Zhu Y, Wang LS, Bonini NM. The microRNA miR-34 modulates ageing and neurodegeneration in Drosophila. Nature. 2012;482:519–523. doi: 10.1038/nature10810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llorian M, Schwartz S, Clark TA, Hollander D, Tan LY, Spellman R, Gordon A, Schweitzer AC, de la Grange P, Ast G, Smith CW. Position-dependent alternative splicing activity revealed by global profiling of alternative splicing events regulated by PTB. Nat Struct Mol Biol. 2010;17:1114–1123. doi: 10.1038/nsmb.1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luque FA, Furneaux HM, Ferziger R, Rosenblum MK, Wray SH, Schold SC, Glantz MJ, Jaeckle KA, Biran H, Lesser M, Paulsen WA, River ME, Posner JB. Anti-Ri: an antibody associated with paraneoplastic opsoclonus and breast cancer. Ann Neurol. 1991;29:241–251. doi: 10.1002/ana.410290303. [DOI] [PubMed] [Google Scholar]
- Lynch MA. Long-term potentiation and memory. Physiol Rev. 2004;84:87–136. doi: 10.1152/physrev.00014.2003. [DOI] [PubMed] [Google Scholar]
- Magavi SS, Macklis JD. Immunocytochemical analysis of neuronal differentiation. Methods Mol Biol. 2008;438:345–352. doi: 10.1007/978-1-59745-133-8_26. [DOI] [PubMed] [Google Scholar]
- Makeyev EV, Zhang J, Carrasco MA, Maniatis T. The MicroRNA miR-124 promotes neuronal differentiation by triggering brain-specific alternative pre-mRNA splicing. Mol Cell. 2007;27:435–448. doi: 10.1016/j.molcel.2007.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markovtsov V, Nikolic JM, Goldman JA, Turck CW, Chou MY, Black DL. Cooperative assembly of an hnRNP complex induced by a tissue-specific homolog of polypyrimidine tract binding protein. Mol Cell Biol. 2000;20:7463–7479. doi: 10.1128/mcb.20.20.7463-7479.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matlin AJ, Southby J, Gooding C, Smith CW. Repression of alpha-actinin SM exon splicing by assisted binding of PTB to the polypyrimidine tract. RNA. 2007;13:1214–1223. doi: 10.1261/rna.219607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKeever MO, Darnell RB. NAP, a human cerebellar degeneration antigen, is a novel, neuron specific, adaptin family member. Soc Neurosci Abstr. 1992;18:1092. [Google Scholar]
- Meins M, Schlickum S, Wilhelm C, Missbach J, Yadav S, Glaser B, Grzmil M, Burfeind P, Laccone F. Identification and characterization of murine Brunol4, a new member of the elav/bruno family. Cytogenet Genome Res. 2002;97:254–260. doi: 10.1159/000066619. [DOI] [PubMed] [Google Scholar]
- Meisner NC, Filipowicz W. Properties of the Regulatory RNA-Binding Protein HuR and its Role in Controlling miRNA Repression. Adv Exp Med Biol. 2011;700:106–123. doi: 10.1007/978-1-4419-7823-3_10. [DOI] [PubMed] [Google Scholar]
- Mendell JT, Olson EN. MicroRNAs in stress signaling and human disease. Cell. 2012;148:1172–1187. doi: 10.1016/j.cell.2012.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ming GL, Wong ST, Henley J, Yuan XB, Song HJ, Spitzer NC, Poo MM. Adaptation in the chemotactic guidance of nerve growth cones. Nature. 2002;417:411–418. doi: 10.1038/nature745. [DOI] [PubMed] [Google Scholar]
- Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- Muddashetty RS, Nalavadi VC, Gross C, Yao X, Xing L, Laur O, Warren ST, Bassell GJ. Reversible inhibition of PSD-95 mRNA translation by miR-125a, FMRP phosphorylation, and mGluR signaling. Mol Cell. 2011;42:673–688. doi: 10.1016/j.molcel.2011.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myer VE, Fan XC, Steitz JA. Identification of HuR as a protein implicated in AUUUA-mediated mRNA decay. EMBO J. 1997;16:2130–2139. doi: 10.1093/emboj/16.8.2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagalakshmi U, Wang Z, Waern K, Shou C, Raha D, Gerstein M, Snyder M. The transcriptional landscape of the yeast genome defined by RNA sequencing. Science. 2008;320:1344–1349. doi: 10.1126/science.1158441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura M, Okano H, Blendy JA, Montell C. Musashi, a neural RNA-binding protein required for Drosophila adult external sensory organ development. Neuron. 1994;13:67–81. doi: 10.1016/0896-6273(94)90460-x. [DOI] [PubMed] [Google Scholar]
- Narayanan U, Nalavadi V, Nakamoto M, Pallas DC, Ceman S, Bassell GJ, Warren ST. FMRP phosphorylation reveals an immediate-early signaling pathway triggered by group I mGluR and mediated by PP2A. J Neurosci. 2007;27:14349–14357. doi: 10.1523/JNEUROSCI.2969-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman LS, McKeever MO, Okano HJ, Darnell RB. ®-NAP, a cerebellar degeneration antigen, is a neuron-specific vesicle coat protein. Cell. 1995;82:773–783. doi: 10.1016/0092-8674(95)90474-3. [DOI] [PubMed] [Google Scholar]
- O'Carroll D, Schaefer A. General Principals of miRNA Biogenesis and Regulation in the Brain. Neuropsychopharmacology. 2012 doi: 10.1038/npp.2012.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Donnell WT, Warren ST. A decade of molecular studies of fragile X syndrome. Annu Rev Neurosci. 2002;25:315–338. doi: 10.1146/annurev.neuro.25.112701.142909. [DOI] [PubMed] [Google Scholar]
- Okabe M, Imai T, Kurusu M, Hiromi Y, Okano H. Translational repression determines a neuronal potential in Drosophila asymmetric cell division. Nature. 2001;411:94–98. doi: 10.1038/35075094. [DOI] [PubMed] [Google Scholar]
- Okano H, Kawahara H, Toriya M, Nakao K, Shibata S, Imai T. Function of RNA-binding protein Musashi-1 in stem cells. Exp Cell Res. 2005;306:349–356. doi: 10.1016/j.yexcr.2005.02.021. [DOI] [PubMed] [Google Scholar]
- Okano HJ, Darnell RB. A hierarchy of Hu RNA binding proteins in developing and adult neurons. J Neurosci. 1997;17:3024–3037. doi: 10.1523/JNEUROSCI.17-09-03024.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Q, Shai O, Misquitta C, Zhang W, Saltzman AL, Mohammad N, Babak T, Siu H, Hughes TR, Morris QD, Frey BJ, Blencowe BJ. Revealing global regulatory features of mammalian alternative splicing using a quantitative microarray platform. Mol Cell. 2004;16:929–941. doi: 10.1016/j.molcel.2004.12.004. [DOI] [PubMed] [Google Scholar]
- Park TJ, Curran T. Alternative splicing disabled by Nova2. Neuron. 2010;66:811–813. doi: 10.1016/j.neuron.2010.06.012. [DOI] [PubMed] [Google Scholar]
- Patel VL, Mitra S, Harris R, Buxbaum AR, Lionnet T, Brenowitz M, Girvin M, Levy M, Almo SC, Singer RH, Chao JA. Spatial arrangement of an RNA zipcode identifies mRNAs under post-transcriptional control. Genes Dev. 2012;26:43–53. doi: 10.1101/gad.177428.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng SS, Chen CY, Xu N, Shyu AB. RNA stabilization by the AU-rich element binding protein, HuR, an ELAV protein. EMBO J. 1998;17:3461–3470. doi: 10.1093/emboj/17.12.3461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez I, Lin CH, McAfee JG, Patton JG. Mutation of PTB binding sites causes misregulation of alternative 3' splice site selection in vivo. RNA. 1997;3:764–778. [PMC free article] [PubMed] [Google Scholar]
- Polydorides AD, Okano HJ, Yang YY, Stefani G, Darnell RB. A brain-enriched polypyrimidine tract-binding protein antagonizes the ability of Nova to regulate neuron-specific alternative splicing. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:6350–6355. doi: 10.1073/pnas.110128397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulos MG, Batra R, Charizanis K, Swanson MS. Developments in RNA splicing and disease. Cold Spring Harb Perspect Biol. 2011;3 doi: 10.1101/cshperspect.a000778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Racca C, Gardiol A, Eom T, Ule J, Triller A, Darnell RB. The neuronal splicing factor Nova co-localizes with target RNAs in the dendrite. Front Neural Circuits. 2010;4:5. doi: 10.3389/neuro.04.005.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Racca C, Gardiol A, Triller A. Dendritic and postsynaptic localizations of glycine receptor alpha subunit mRNAs. J Neurosci. 1997;17:1691–1700. doi: 10.1523/JNEUROSCI.17-05-01691.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratti A, Fallini C, Colombrita C, Pascale A, Laforenza U, Quattrone A, Silani V. Post-transcriptional regulation of neuro-oncological ventral antigen 1 by the neuronal RNA-binding proteins ELAV. J Biol Chem. 2008;283:7531–7541. doi: 10.1074/jbc.M706082200. [DOI] [PubMed] [Google Scholar]
- Reetz A, Solimena M, Matteoli M, Folli F, Takei K, De Camilli P. GABA and pancreatic beta-cells: colocalization of glutamic acid decarboxylase (GAD) and GABA with synaptic-like microvesicles suggests their role in GABA storage and secretion. EMBO J. 1991;10:1275–1284. doi: 10.1002/j.1460-2075.1991.tb08069.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard S. Reaching for the stars: Linking RNA binding proteins to diseases. Adv Exp Med Biol. 2010;693:142–157. [PubMed] [Google Scholar]
- Richter JD. CPEB: a life in translation. Trends Biochem Sci. 2007;32:279–285. doi: 10.1016/j.tibs.2007.04.004. [DOI] [PubMed] [Google Scholar]
- Richter JD. Translational control of synaptic plasticity. Biochem Soc Trans. 2010;38:1527–1530. doi: 10.1042/BST0381527. [DOI] [PubMed] [Google Scholar]
- Robinow S, Campos A, Yao K, White K. The elav gene product of Drosophila, required in neurons, has three RNP consensus motifs. Science. 1988;242:1570–1572. doi: 10.1126/science.3144044. [DOI] [PubMed] [Google Scholar]
- Ruggiu M, Herbst R, Kim N, Jevsek M, Fak JJ, Mann MA, Fischbach G, Burden SJ, Darnell RB. Rescuing Z+ agrin splicing in Nova null mice restores synapse formation and unmasks a physiologic defect in motor neuron firing. Proc Natl Acad Sci U S A. 2009;106:3513–3518. doi: 10.1073/pnas.0813112106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakakibara S, Nakamura Y, Yoshida T, Shibata S, Koike M, Takano H, Ueda S, Uchiyama Y, Noda T, Okano H. RNA-binding protein Musashi family: roles for CNS stem cells and a subpopulation of ependymal cells revealed by targeted disruption and antisense ablation. Proc Natl Acad Sci U S A. 2002;99:15194–15199. doi: 10.1073/pnas.232087499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakakibara S, Imai T, Hamaguchi K, Okabe M, Aruga J, Nakajima K, Yasutomi D, Nagata T, Kurihara Y, Uesugi S, Miyata T, Ogawa M, Mikoshiba K, Okano H. Mouse-musashi-1, a neural RNA binding protein highly enriched in the mammalian CNS stem cell. Devel Biol. 1996;176:230–242. doi: 10.1006/dbio.1996.0130. [DOI] [PubMed] [Google Scholar]
- Sakakibara S, Okano H. Expression of neural RNA-binding proteins in the postnatal CNS: implications of their roles in neuronal and glial cell development. J Neurosci. 1997;17:8300–8312. doi: 10.1523/JNEUROSCI.17-21-08300.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt EF, Warner-Schmidt JL, Otopalik BG, Pickett SB, Greengard P, Heintz N. Identification of the cortical neurons that mediate antidepressant responses. Cell. 2012;149:1152–1163. doi: 10.1016/j.cell.2012.03.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schratt GM, Tuebing F, Nigh EA, Kane CG, Sabatini ME, Kiebler M, Greenberg ME. A brain-specific microRNA regulates dendritic spine development. Nature. 2006;439:283–289. doi: 10.1038/nature04367. [DOI] [PubMed] [Google Scholar]
- Schuman EM, Dynes JL, Steward O. Synaptic regulation of translation of dendritic mRNAs. J Neurosci. 2006;26:7143–7146. doi: 10.1523/JNEUROSCI.1796-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S, Maris C, Allain FH, Black DL. U1 snRNA directly interacts with polypyrimidine tract-binding protein during splicing repression. Mol Cell. 2011;41:579–588. doi: 10.1016/j.molcel.2011.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata H, Huynh DP, Pulst SM. A novel protein with RNA-binding motifs interacts with ataxin-2. Hum Mol Genet. 2000;9:1303–1313. doi: 10.1093/hmg/9.9.1303. [DOI] [PubMed] [Google Scholar]
- Siegel G, Saba R, Schratt G. microRNAs in neurons: manifold regulatory roles at the synapse. Curr Opin Genet Dev. 2011;21:491–497. doi: 10.1016/j.gde.2011.04.008. [DOI] [PubMed] [Google Scholar]
- Silberberg G, Ohman M. The edited transcriptome: novel high throughput approaches to detect nucleotide deamination. Curr Opin Genet Dev. 2011;21:401–406. doi: 10.1016/j.gde.2011.04.009. [DOI] [PubMed] [Google Scholar]
- Singh R, Valcarcel J, Green MR. Distinct binding specificities and functions of higher eukaryotic polypyrimidine tract-binding proteins. Science. 1995;268:1173–116. doi: 10.1126/science.7761834. [DOI] [PubMed] [Google Scholar]
- Siomi H, Siomi MC, Nussbaum RL, Dreyfuss G. The protein product of the fragile X gene, FMR1, has characteristics of an RNA-binding protein. Cell. 1993;74:291–298. doi: 10.1016/0092-8674(93)90420-u. [DOI] [PubMed] [Google Scholar]
- Smukler SR, Arntfield ME, Razavi R, Bikopoulos G, Karpowicz P, Seaberg R, Dai F, Lee S, Ahrens R, Fraser PE, Wheeler MB, van der Kooy D. The adult mouse and human pancreas contain rare multipotent stem cells that express insulin. Cell Stem Cell. 2011;8:281–293. doi: 10.1016/j.stem.2011.01.015. [DOI] [PubMed] [Google Scholar]
- Soller M, White K. ELAV inhibits 3'-end processing to promote neural splicing of ewg pre-mRNA. Genes Dev. 2003;17:2526–2538. doi: 10.1101/gad.1106703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soller M, White K. ELAV multimerizes on conserved AU4-6 motifs important for ewg splicing regulation. Mol Cell Biol. 2005;25:7580–7591. doi: 10.1128/MCB.25.17.7580-7591.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spellman R, Rideau A, Matlin A, Gooding C, Robinson F, McGlincy N, Grellscheid SN, Southby J, Wollerton M, Smith CW. Regulation of alternative splicing by PTB and associated factors. Biochem Soc Trans. 2005;33:457–460. doi: 10.1042/BST0330457. [DOI] [PubMed] [Google Scholar]
- Steward O, Schuman EM. Protein synthesis at synaptic sites on dendrites. Annu Rev Neurosci. 2001;24:299–325. doi: 10.1146/annurev.neuro.24.1.299. [DOI] [PubMed] [Google Scholar]
- Sugimoto Y, Konig J, Hussain S, Zupan B, Curk T, Frye M, Ule J. Analysis of CLIP and iCLIP methods for nucleotide-resolution studies of protein-RNA interactions. Genome Biol. 2012;13:R67. doi: 10.1186/gb-2012-13-8-r67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugnet CW, Srinivasan K, Clark TA, O'Brien G, Cline MS, Wang H, Williams A, Kulp D, Blume JE, Haussler D, Ares MJ. Unusual intron conservation near tissue-regulated exons found by splicing microarrays. PLoS Comput Biol. 2006;2:e4. doi: 10.1371/journal.pcbi.0020004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutton MA, Wall NR, Aakalu GN, Schuman EM. Regulation of dendritic protein synthesis by miniature synaptic events. Science. 2004;304:1979–1983. doi: 10.1126/science.1096202. [DOI] [PubMed] [Google Scholar]
- Szabo A, Dalmau J, Manley G, Rosenfeld M, Wong E, Henson J, Posner JB, Furneaux HM. HuD, a paraneoplastic encephalomyelitis antigen contains RNA-binding domains and is homologous to Elav and sex lethal. Cell. 1991;67:325–333. doi: 10.1016/0092-8674(91)90184-z. [DOI] [PubMed] [Google Scholar]
- Tang ZZ, Zheng S, Nikolic J, Black DL. Developmental control of CaV1.2 L-type calcium channel splicing by Fox proteins. Mol Cell Biol. 2009;29:4757–4765. doi: 10.1128/MCB.00608-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teplova M, Malinina L, Darnell JC, Song J, Lu M, Abagyan R, Musunuru K, Teplov A, Burley SK, Darnell RB, Patel DJ. Protein-RNA and Protein-Protein Recognition by Dual KH1/2 Domains of the Neuronal Splicing Factor Nova-1. Structure. 2011;19:930–944. doi: 10.1016/j.str.2011.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tollervey JR, Curk T, Rogelj B, Briese M, Cereda M, Kayikci M, Konig J, Hortobagyi T, Nishimura AL, Zupunski V, Patani R, Chandran S, Rot G, Zupan B, Shaw CE, Ule J. Characterizing the RNA targets and position-dependent splicing regulation by TDP-43. Nat Neurosci. 2011 doi: 10.1038/nn.2778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai DE, Harper DS, Keene JD. U1-snRNP-A protein selects a ten nucleotide consensus sequence from a degenerate RNA pool presented in various structural contexts. Nucleic Acids Res. 1991;19:4931–4936. doi: 10.1093/nar/19.18.4931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuerk C, Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriphage T4 DNA polymerase. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- Ule J, Darnell RB. RNA binding proteins and the regulation of neuronal synaptic plasticity. Curr Opin Neurobiol. 2006;16:102–110. doi: 10.1016/j.conb.2006.01.003. [DOI] [PubMed] [Google Scholar]
- Ule J, Jensen K, Mele A, Darnell RB. CLIP: a method for identifying protein-RNA interaction sites in living cells. Methods. 2005a;37:376–386. doi: 10.1016/j.ymeth.2005.07.018. [DOI] [PubMed] [Google Scholar]
- Ule J, Jensen KB, Ruggiu M, Mele A, Ule A, Darnell RB. CLIP identifies Nova-regulated RNA networks in the brain. Science. 2003;302:1212–1215. doi: 10.1126/science.1090095. [DOI] [PubMed] [Google Scholar]
- Ule J, Stefani G, Mele A, Ruggiu M, Wang X, Taneri B, Gaasterland T, Blencowe BJ, Darnell RB. An RNA map predicting Nova-dependent splicing regulation. Nature. 2006;444:580–586. doi: 10.1038/nature05304. [DOI] [PubMed] [Google Scholar]
- Ule J, Ule A, Spencer J, Williams A, Hu JS, Cline M, Wang H, Clark T, Fraser C, Ruggiu M, Zeeberg BR, Kane D, Weinstein JN, Blume J, Darnell RB. Nova regulates brain-specific splicing to shape the synapse. Nat Genet. 2005b;37:844–852. doi: 10.1038/ng1610. [DOI] [PubMed] [Google Scholar]
- Underwood JG, Boutz PL, Dougherty JD, Stoilov P, Black DL. Homologues of the Caenorhabditis elegans Fox-1 protein are neuronal splicing regulators in mammals. Mol Cell Biol. 2005;25:10005–10016. doi: 10.1128/MCB.25.22.10005-10016.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voineagu I, Wang X, Johnston P, Lowe JK, Tian Y, Horvath S, Mill J, Cantor RM, Blencowe BJ, Geschwind DH. Transcriptomic analysis of autistic brain reveals convergent molecular pathology. Nature. 2011;474:380–384. doi: 10.1038/nature10110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagnon JL, Mahaffey CL, Sun W, Yang Y, Chao HT, Frankel WN. Etiology of a genetically complex seizure disorder in Celf4 mutant mice. Genes Brain Behav. 2011;10:765–777. doi: 10.1111/j.1601-183X.2011.00717.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ET, Cody NA, Jog S, Biancolella M, Wang TT, Treacy DJ, Luo S, Schroth GP, Housman DE, Reddy S, Lecuyer E, Burge CB. Transcriptome-wide Regulation of Pre-mRNA Splicing and mRNA Localization by Muscleblind Proteins. Cell. 2012;150:710–724. doi: 10.1016/j.cell.2012.06.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Molfenter J, Zhu H, Lou H. Promotion of exon 6 inclusion in HuD premRNA by Hu protein family members. Nucleic Acids Res. 2010a;38:3760–3770. doi: 10.1093/nar/gkq028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang LW, Berry-Kravis E, Hagerman RJ. Fragile X: leading the way for targeted treatments in autism. Neurotherapeutics. 2010b;7:264–274. doi: 10.1016/j.nurt.2010.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Kayikci M, Briese M, Zarnack K, Luscombe NM, Rot G, Zupan B, Curk T, Ule J. iCLIP predicts the dual splicing effects of TIA-RNA interactions. PLoS Biol. 2010c;8:1–16. doi: 10.1371/journal.pbio.1000530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weill L, Belloc E, Bava FA, Mendez R. Translational control by changes in poly(A) tail length: recycling mRNAs. Nat Struct Mol Biol. 2012;19:577–585. doi: 10.1038/nsmb.2311. [DOI] [PubMed] [Google Scholar]
- Wheeler TM, Lueck JD, Swanson MS, Dirksen RT, Thornton CA. Correction of ClC-1 splicing eliminates chloride channelopathy and myotonia in mouse models of myotonic dystrophy. J Clin Invest. 2007;117:3952–3957. doi: 10.1172/JCI33355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler TM, Sobczak K, Lueck JD, Osborne RJ, Lin X, Dirksen RT, Thornton CA. Reversal of RNA dominance by displacement of protein sequestered on triplet repeat RNA. Science. 2009;325:336–339. doi: 10.1126/science.1173110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witten JT, Ule J. Understanding splicing regulation through RNA splicing maps. Trends Genet. 2011;27:89–97. doi: 10.1016/j.tig.2010.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf HK, Buslei R, Schmidt-Kastner R, Schmidt-Kastner PK, Pietsch T, Wiestler OD, Blumcke I. NeuN: a useful neuronal marker for diagnostic histopathology. J Histochem Cytochem. 1996;44:1167–1171. doi: 10.1177/44.10.8813082. [DOI] [PubMed] [Google Scholar]
- Xue Y, Zhou Y, Wu T, Zhu T, Ji X, Kwon YS, Zhang C, Yeo G, Black DL, Sun H, Fu XD, Zhang Y. Genome-wide analysis of PTB-RNA interactions reveals a strategy used by the general splicing repressor to modulate exon inclusion or skipping. Mol Cell. 2009;36:996–1006. doi: 10.1016/j.molcel.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang YY, Yin GL, Darnell RB. The neuronal RNA-binding protein Nova-2 is implicated as the autoantigen targeted in POMA patients with dementia. Proc Natl Acad Sci U S A. 1998;95:13254–13259. doi: 10.1073/pnas.95.22.13254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano M, Hayakawa-Yano Y, Mele A, Darnell RB. Nova2 regulates neuronal migration through an RNA switch in disabled-1 signaling. Neuron. 2010;66:848–858. doi: 10.1016/j.neuron.2010.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao KM, Samson ML, Reeves R, White K. Gene elav of Drosophila melanogaster: a prototype for neuronal-specific RNA binding protein gene family that is conserved in flies and humans. J Neurobiol. 1993;24:723–739. doi: 10.1002/neu.480240604. [DOI] [PubMed] [Google Scholar]
- Yeo GW, Coufal NG, Liang TY, Peng GE, Fu XD, Gage FH. An RNA code for the FOX2 splicing regulator revealed by mapping RNA-protein interactions in stem cells. Nat Struct Mol Biol. 2009;16:130–137. doi: 10.1038/nsmb.1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Y, Compton SA, Sobczak K, Stenberg MG, Thornton CA, Griffith JD, Swanson MS. Muscleblind-like 1 interacts with RNA hairpins in splicing target and pathogenic RNAs. Nucleic Acids Res. 2007;35:5474–5486. doi: 10.1093/nar/gkm601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zang JB, Nosyreva ED, Spencer CM, Volk LJ, Musunuru K, Zhong R, Stone EF, Yuva-Paylor LA, Huber KM, Paylor R, Darnell JC, Darnell RB. A mouse model of the human Fragile X syndrome I304N mutation. PLoS Genet. 2009;5:e1000758. doi: 10.1371/journal.pgen.1000758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Darnell RB. Mapping in vivo protein-RNA interactions at single-nucleotide resolution from HITS-CLIP data. Nat Biotechnol. 2011;29:607–614. doi: 10.1038/nbt.1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Frias MA, Mele A, Ruggiu M, Eom T, Marney CB, Wang H, Licatalosi DD, Fak JJ, Darnell RB. Integrative Modeling Defines the Nova Splicing-Regulatory Network and Its Combinatorial Controls. Science. 2010;329:439–443. doi: 10.1126/science.1191150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Zhang Z, Castle J, Sun S, Johnson J, Krainer AR, Zhang MQ. Defining the regulatory network of the tissue-specific splicing factors Fox-1 and Fox-2. Genes Dev. 2008;22:2550–2563. doi: 10.1101/gad.1703108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Poo MM. Localized synaptic potentiation by BDNF requires local protein synthesis in the developing axon. Neuron. 2002;36:675–688. doi: 10.1016/s0896-6273(02)01023-1. [DOI] [PubMed] [Google Scholar]
- Zhu H, Hinman MN, Hasman RA, Mehta P, Lou H. Regulation of neuron-specific alternative splicing of neurofibromatosis type 1 pre-mRNA. Mol Cell Biol. 2008;28:1240–1251. doi: 10.1128/MCB.01509-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H, Hasman RA, Barron VA, Luo G, Lou H. A nuclear function of Hu proteins as neuron-specific alternative RNA processing regulators. Mol Biol Cell. 2006;17:5105–5114. doi: 10.1091/mbc.E06-02-0099. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.