Abstract
We modified and automated a highly sensitive chemiluminescent enzyme immunoassay (CLEIA) for surface antigen (HBsAg) detection using a combination of monoclonal antibodies, each for a specific epitope of HBsAg, and by improving an earlier conjugation technique. Of 471 hepatitis B virus (HBV) carriers seen in our hospital between 2009 and 2012, 26 were HBsAg seronegative as determined by the Abbott Architect assay. The Lumipulse HBsAg-HQ assay was used to recheck those 26 patients who demonstrated seroclearance by the Abbott Architect assay. The performance of the Lumipulse HBsAg-HQ assay was compared with that of a quantitative HBsAg detection system (Abbott Architect) and the Roche Cobas TaqMan HBV DNA assay (CTM) (lower limit of detection, 2.1 log copies/ml) using blood serum samples from patients who were determined to be HBsAg seronegative by the Abbott Architect assay. Ten patients had spontaneous HBsAg loss. Of 8 patients treated with nucleotide analogues (NAs), two were HBsAg seronegative after stopping lamivudine therapy and 6 were HBsAg seronegative during entecavir therapy. Eight acute hepatitis B (AH) patients became HBsAg seronegative. Of the 26 patients, 16 were HBsAg positive by the Lumipulse HBsAg-HQ assay but negative by the Abbott Architect assay. The differences between the two assays in terms of detectable HBsAg persisted over the long term in the spontaneous loss group (median, 10 months), the NA-treated group (2.5 months), and the AH group (0.5 months). In 9 patients, the Lumipulse HBsAg-HQ assay detected HBsAg when HBV DNA was negative by the CTM assay. HBsAg was also detected by the Lumipulse HBsAg-HQ assay in 4 patients with an anti-HBs concentration of >10 mIU/ml, 3 of whom had no HBsAg escape mutations. The automatic, highly sensitive HBsAg CLEIA Lumipulse HBsAg-HQ is a convenient and precise assay for HBV monitoring.
INTRODUCTION
Today, >400 million people worldwide are hepatitis B virus (HBV) carriers (1). We have monitored HBV markers, such as HBV DNA, hepatitis B surface antigen (HBsAg), hepatitis B e antigen (HBeAg), and HB core-related antigen (HBcrAg), in chronic hepatitis B patients. The measurement of HBV DNA levels by a PCR-based method is the state-of-the-art technique for monitoring HBV replication in clinical practice (2). However, it is suboptimal for chronic hepatitis B patients who are medicated with nucleotide analogues (NAs), as those, in many cases, can decrease HBV DNA to below the limit of detection.
HBsAg is a secreted envelope protein that is continuously shed into the blood as long as HBV infection persists, irrespective of viral replication. Recent advances in HBsAg quantification (qHBsAg) have opened up new perspectives in the study of HBV; qHBsAg levels are correlated with intrahepatic covalently closed circular (ccc) DNA, which is used as a template for viral transcription and maintains the chronic HBV infection state (3–5). Additionally, a correlation between qHBsAg and HBV DNA has been suggested, with the possibility of a role for qHBsAg as a surrogate marker for viral replication put forward, which might identify chronic hepatitis B patients who are likely to be cured with pegylated alpha interferon (6–9).
In Japan, two HBsAg quantification assays are available: the Architect HBsAg-QT (Abbott Japan) (detection range, 50 to 250,000 mIU/ml) and the HISCL HBsAg (Sysmex) (detection range, 30 to 2,500,000 mIU/ml). These two methods have a good correlation and are sensitive over a wide detection range. Recently, Matsubara et al. (10) reported a novel highly sensitive chemiluminescent enzyme immunoassay (CLEIA) that was developed for quantitative HBsAg detection by combining monoclonal antibodies, each specific for a different epitope of the antigen, and employing an improved conjugation technique. It is as sensitive as nucleic acid testing for detecting early HBV infection. We further modified and improved the high-sensitivity assay reagent described above for adaptation to both ferrite microparticles as the solid phase and the automated analyzer system by modification of the optimum combination of monoclonal antibodies. As was recently reported (11), this assay (Lumipulse HBsAg-HQ) had good accuracy, reproducibility, specificity, and sensitivity, and the results correlate well with those of the Abbott Architect. The coefficient of variation in the Lumipulse HBsAg-HQ is <5.9% for samples with a low concentration of HBsAg (11), and the assay was approved by the Japanese government in 2013.
The sensitivity of this assay (5 mIU/ml) was approximately 10-fold higher than that of the Abbott Architect assay (50 mIU/ml). Here, we adapted this assay to monitor chronic hepatitis B patients with apparent HBsAg seroclearance as determined by the Abbott Architect assay.
MATERIALS AND METHODS
Samples.
Four hundred seventy-one patients with chronic HBV infection visited our hospital from 2009 to 2012. One hundred eighty-one patients were asymptomatic carriers, 232 had chronic hepatitis B (CHB), and 58 had liver cirrhosis. Of these, 13 patients took lamivudine, one adefovir, 19 lamivudine plus adefovir, 140 entecavir, 8 entecavir plus adefovir, and 9 tenofovir. Thirty patients with acute HB (AH) infection (8 of whom developed chronic hepatitis) visited our hospital from January 2009 to 2012. We determined HBsAg seroclearance according to the Abbott Architect assay in 26 HBV-infected patients during the observation period. Of these, 10 were not treated with nucleotide analogues (spontaneous HBsAg loss group) and 8 were treated (NA-treated group). Of the 8 NA-treated patients, 2 on lamivudine therapy were HBsAg seronegative after stopping therapy, and the other 6 were HBsAg seronegative during entecavir therapy. Eight AH patients became HBsAg seronegative.
The study protocol conformed to the 1975 Declaration of Helsinki and was approved by the ethics committees of our institutions, and informed consent was obtained from each carrier. We rechecked HBsAg status of the patients by the Lumipulse HBsAg-HQ assay in their serial blood serum samples and compared the results with those of the Architect HBsAg-QT assay.
Methods. (i) Measurement of HBsAg by Lumipulse HBsAg-HQ assay.
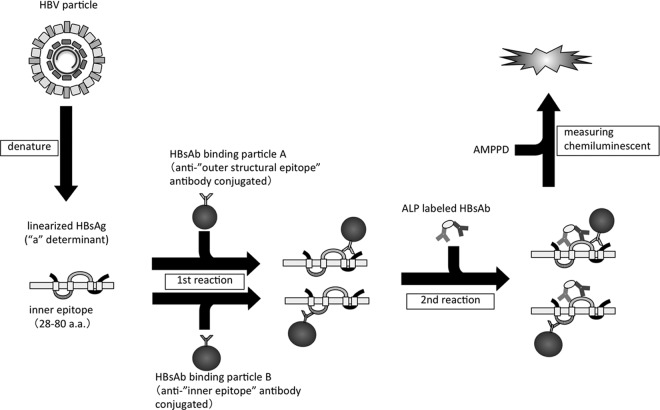
HBsAg was measured on the two-step sandwich assay principle with a fully automated chemiluminescent enzyme immunoassay system (Lumipulse G1200; Fujirebio, Inc.). The assay principle for this new reagent was based on that previously reported by Matsubara et al. (10). Briefly, samples were pretreated with a solution, including surfactant to disrupt HBV particles, to dissociate HBsAg from HBsAg–anti-HBs complexes and to denature epitopes to a linear form. Linearized HBsAg were then detected using two monoclonal antibodies against external structural regions as determinant “a” and the internal epitope as a capture reagent, with two monoclonal antibodies coupled to alkaline phosphatase as the detector. For the assay procedures, 100 μl blood serum and/or plasma samples together with 20 μl pretreatment solution were incubated with the monoclonal antibodies binding ferrite microparticles at 37°C for 10 min. After automatic washing, 250 μl of the alkaline phosphatase-labeled antibodies were added and further incubated at 37°C for 10 min. After the washing step, 200 μl substrate solution (AMPPD [3-(2′-spiroadamantane)-4-methoxy-4-(3″-phosphoryloxy)phenyl-1,2-dioxetane disodium salt]) (Applied Biosystems, Bedford, MA) was added and incubated at 37°C for 5 min. The relative intensity of chemiluminescence was measured and the HBsAg concentration was calculated by comparison with a standard curve. The range of HBsAg concentrations assayed was 5 to 150,000 mIU/ml, and retesting was accepted with a 200-fold dilution of samples that exceeded this range. In the present study, the cutoff value of HBsAg concentration was set at 5 mIU/ml. HBsAg in blood serum was also quantified at the same intervals using the Abbott Architect HBsAg-QT assay (cutoff value, 50 mIU/ml) (Fig. 1).
Fig 1.
The principle of Lumipulse HBsAg-HQ.
(ii) Quantification of HBV DNA.
Serum HBV DNA was measured using the TaqMan PCR assay (Cobas TaqMan; Roche Molecular Systems [lower limit of detection, 2.1 log copies/ml]).
(iii) Quantification of HBcrAg.
Serum HBcrAg was measured using CLEIA, as described previously (12, 13). Briefly, sodium dodecyl sulfate pretreated serum was incubated with monoclonal antibodies against denatured HBcAg and HBeAg. After washing and incubation with alkaline phosphatase-labeled secondary antibodies, the relative chemiluminescence intensity was measured, and the HBcAg concentration was calculated by comparison with a standard curve generated using a known concentration of recombinant HBeAg-containing peptide. The cutoff value of HBcrAg was 3 log U/ml.
(iv) Quantification of anti-HBs.
Serum anti-HBs was measured using the Architect system's anti-HBs. A specimen was considered positive for anti-HBs when the concentration was ≥10.0 mIU/ml.
RESULTS
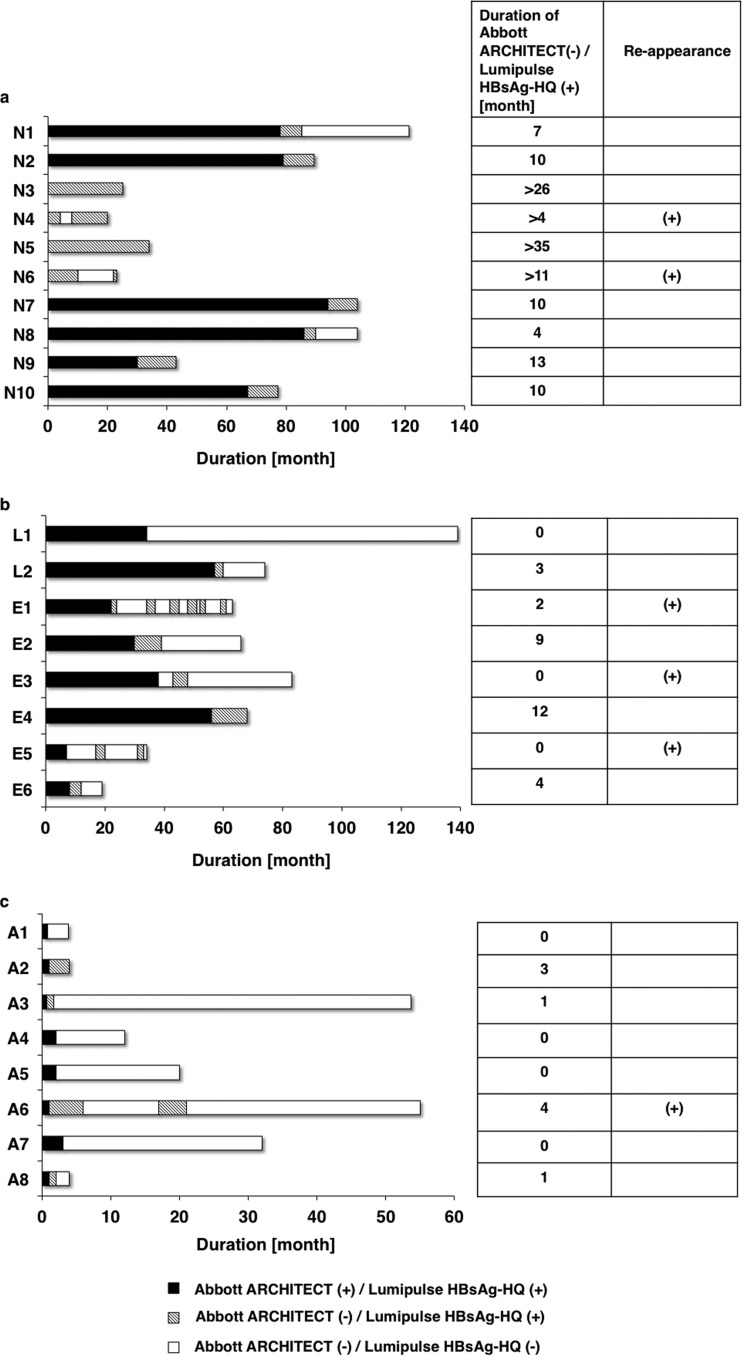
Table 1 shows clinical data at baseline for the three groups with HBsAg seroclearance according to data from the Abbott Architect assay. In four of 10 spontaneous HBsAg loss cases, HBsAg had already been <50 mIU/ml as measured by the Abbott Architect assay at the first visit. Table 1 shows the characteristics of all 26 patients in these 3 groups. The HBV DNA and HBcrAg levels at baseline were significantly higher in the NA-treated and AH groups than in the spontaneous HBsAg loss group. The HBsAg levels at baseline were also significantly higher in the AH group and the NA-treated group than in the spontaneous HBsAg loss group. However, HBsAg became undetectable by the Abbott Architect assay immediately in the AH group (median, 1 month), compared with the NA-treated group (32 months) and the spontaneous HBsAg loss group (78.5 months [excluding 4 patients with HBsAg of ≤50 mIU/ml by the Abbott Architect assay at the first visit]). In 19 of the 26 cases, the HBsAg levels were still detectable by the Lumipulse HBsAg-HQ assay at the time point when they were undetectable by the Abbott Architect assay. At the last time point with detectable HBsAg by Lumipulse HBsAg-HQ assay, the Abbott Architect assay could not detect HBsAg in all 10 spontaneous HBsAg loss patients, but the Abbott Architect assay was also able to detect at the last time point in three (case no. L1, E3, and E5) of eight NA-treated group patients and four (case no. A1, A4, A5, and A7) of eight AH patients. In the spontaneous HBsAg loss group, the decline in HBsAg was slower than in the NA-treated and AH groups (Fig. 2a to 2c). Differences in the median duration between the Abbott Architect and Lumipulse HBsAg-HQ assays were seen at 10 months (excluding 4 patients with HBsAg of <50 mIU/ml by the Abbott Architect assay at the first visit), 2.5 months, and 0.5 months in the spontaneous HBsAg loss group, NA-treated group, and AH group, respectively. We observed the reappearance of HBsAg measured by Lumipulse HBsAg-HQ assay in 2 patients (case no. N4 and N6) in the spontaneous HBsAg loss group, 3 (case no. E1, E3, and E5) in the NA-treated group, and one (case no. A6) in the AH group (Fig. 2a to 2c). At the last time point with detectable HBsAg by the Lumipulse HBsAg-HQ assay, HBV DNA was undetectable by the Cobas TaqMan assay in 4 of 10 spontaneous HBsAg loss patients (40%), 4 of 8 NA-treated patients (50%), and one of 8 AH patients (12.5%). At the last time of detection by the Lumipulse HBsAg-HQ assay, HBcrAg was <3 log U/ml in 8 of 10 spontaneous HBsAg loss patients (80%), 2 of 8 NA-treated patients (25%), and none of the 10 AH patients (0%). At the last time point of detection by the Lumipulse HBsAg-HQ assay, anti-HBs was positive in one of 10 spontaneous HBsAg loss patients (10%), none of the 8 NA-treated patients (0%), and 2 of 10 AH patients (20%) (Tables 2 to 4). In case no. A1 and A7, HBsAg was relatively high at the last time point at which HBsAg was detectable by the Lumipulse HBsAg-HQ assay (Table 4). In case no. A1, however, HBsAg was undetectable by the Abbott Architect and Lumipulse HBsAg-HQ assays after 1 month. In case no. A7, HBsAg was undetectable by the Abbott Architect and Lumipulse HBsAg-HQ assays after 3 months.
Table 1.
Clinical data at baseline of 3 groups with HBsAg seroclearance as determined by the Abbott Architect assay
| Patient characteristic | Data for group (n): |
||
|---|---|---|---|
| Spontaneous HBsAg loss (10) | NA treated (8)a | Acute hepatitis (8) | |
| Age at first visit or medication (yr) | 60.6 ± 12.6 | 46.8 ± 12.2 | 50.5 ± 10.8 |
| Sex (no. of males/no. of females) | 10/0 | 7/1 | 8/0 |
| Route of infection (no. of vertical/no. of horizontal) | 10/0 | 4/4 | 0/8 |
| No. with genotype Aa/Ae/Ba/Bj/C | 0/0/0/2/8 | 1/1/1/1/4 | 1/4/1/0/2 |
| Clinical data | |||
| ALT (median [range]) (IU/liter) | 23.5 (8–51) | 76 (11–220) | 1,682 (455–3,622) |
| HBeAg (no. positive/no. negative) | 0/10 | 5/3 | 8/0 |
| HBV DNA (median [range]) (log copies/ml) | 2.3 (<2.1 to 3.4) | 7.4 (4.1 to >9.1) | 6.5 (3.8–8.5) |
| HBcrAg (median [range]) (log IU/ml) | <3 (<3 to 3.3) | 6.8 (4.2–8.6) | 7.1 (6.6–8) |
| Abbott Architect HBsAg-QT detection (median [range]) (mIU/ml) | 1,300 (<50 to 10,880) | 2,676,800 (9,680–89,679,600) | 362,500 (91,200–40,000,000) |
| NA therapy (no. with none/no. with LVD/no. with ETV)b | 10/0/0 | 0/2/6 | 5/0/3 |
NA, nucleotide analogue.
LVD, lamivudine; ETV, entecavir.
Fig 2.
HBsAg dynamics by the Abbott Architect and Lumipulse HBsAg-HQ assays in the spontaneous HBsAg loss group (a), the NA-treated group (b), and the AH group (c).
Table 2.
Clinical data of spontaneous HBsAg loss patients at the last time point at which HBsAg was detectable by the Lumipulse HBsAg-HQ assay
| Clinical data | Values for patient no.: |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| N1 | N2b | N3a,b | N4a,b | N5a,b | N6a,b | N7b | N8 | N9b | N10b | |
| Nucleotide analogue therapy | None | None | None | None | None | None | None | None | None | None |
| Age (yr) | 61 | 54 | 91 | 50 | 76 | 63 | 71 | 62 | 62 | 65 |
| HBeAg (+/−) | − | − | − | − | − | − | − | − | − | − |
| Abbott Architect HBsAg-QT detection (mIU/ml) | <50 | <50 | <50 | <50 | <50 | <50 | <50 | <50 | <50 | <50 |
| Lumipulse HBsAg-HQ detection (mIU/ml) | 8.0 | 51.0 | 12.0 | 8.9 | 10.4 | 5 | 5.8 | 20.4 | 11.7 | 30.3 |
| HBV DNA (log copies/ml) | Not detected | Not detected | <2.1 | <2.1 | 2.9 | 2.6 | <2.1 | Not detected | 2.7 | Not detected |
| HBcrAg (log IU/ml) | <3 | 3 | <3 | <3 | 3.2 | <3 | <3 | <3 | <3 | <3 |
| Anti-HBs (mIU/ml) | <10 | 973.8 | <10 | <10 | <10 | <10 | <10 | <10 | <10 | <10 |
Abbott Architect HBsAg-QT assay (IU/ml) was already negative at first visit.
Lumipulse HBsAg-HQ assay was still able to detect HBsAg at the last observation time.
Table 4.
Clinical data of AH patients at the last time point at which HBsAg was detectable by Lumipulse HBsAg-HQassay
| Clinical data | Values for patient no.: |
|||||||
|---|---|---|---|---|---|---|---|---|
| A1 | A2 | A3 | A4 | A5 | A6 | A7 | A8 | |
| Nucleotide analogue therapy | None | None | None | None | None | ETV | ETV | ETV |
| Age (yr) | 62 | 34 | 53 | 50 | 39 | 39 | 53 | 54 |
| HBeAg (+/−) | − | − | − | − | − | − | + | + |
| Abbott Architect HBsAg-QT detection (mIU/ml) | 91,200a | <50 | <50 | 240a | 680a | <50 | 11,500a | <50 |
| Lumipulse HBsAg-HQ detection (mIU/ml) | 112,289.3 | 5.6 | 13.6 | 180.4 | 771.9 | 7.6 | 12,358.4 | 34.3 |
| HBV DNA (copies/ml) | 3.8 | Not detected | 2.3 | 2.2 | 3 | <2.1 | <2.1 | <2.1 |
| HBcrAg (log IU/ml) | 6.8 | 4.0 | 5.4 | 4.9 | 3.2 | 3.1 | 3.7 | 4.3 |
| Anti-HBs (mIU/ml) | <10 | 24.41 | <10 | <10 | <10 | 23.18 | <10 | <10 |
HBsAg was detectable by both assays at this point, but HBsAg became undetectable at the next point.
Table 3.
Clinical data of NA-treated patients at the last time point at which HBsAg was detectable by the Lumipulse HBsAg-HQ assay
| Clinical data | Values for patient no.: |
|||||||
|---|---|---|---|---|---|---|---|---|
| L1 | L2 | E1 | E2 | E3 | E4a | E5 | E6 | |
| Nucleotide analogue therapy | LVD | LVD | ETV | ETV | ETV | ETV | ETV | ETV |
| Age (yr) | 62 | 49 | 53 | 40 | 44 | 44 | 67 | 39 |
| HBeAg (+/−) | − | − | − | − | − | − | − | − |
| Abbott Architect HBsAg-QT detection (mIU/ml) | 80b | <50 | <50 | <50 | 90b | <50 | 90b | <50 |
| Lumipulse HBsAg-HQ detection (mIU/ml) | 77.3 | 5 | 14.7 | 8 | 44.6 | 6.5 | 42.5 | 89 |
| HBV DNA (log copies/ml) | <2.1 | Not detected | Not detected | Not detected | 3.3 | 2.2 | <2.1 | Not detected |
| HBcrAg (log IU/ml) | <3 | 3.3 | 4.3 | 4.1 | 3.2 | <3 | 3.8 | 4.3 |
| Anti-HBs (mIU/ml) | <10 | <10 | <10 | <10 | <10 | <10 | <10 | <10 |
The Lumipulse HBsAg-HQ assay was still able to detect HBsAg at the last observation time.
HBsAg was detectable by both assays at this point, but HBsAg became undetectable at the next point.
To elucidate possible HBs escape mutants, we examined the S gene sequences of all 26 patients at the first visit. Patient N2 had an amino acid G145S mutation, L1 had an amino acid S143T mutation, and L2 had amino acid I126N and F134Y mutations. None had an amino acid G145R mutation. At the last time point that HBsAg was detected by the Abbott Architect assay, anti-HBs was positive in patient N2 (from the spontaneous HBsAg loss group) with an amino acid G145S mutation. We performed an inhibition assay for samples N1 and N2 at the time of Abbott Architect undetectability but Lumipulse HBsAg-HQ detectability to confirm whether the identification of HBsAg by the Lumipulse HBsAg-HQ assay was specific. HBsAg detection of these samples was inhibited, indicating that the Lumipulse HBsAg-HQ assay was indeed specific. The following are three representative cases.
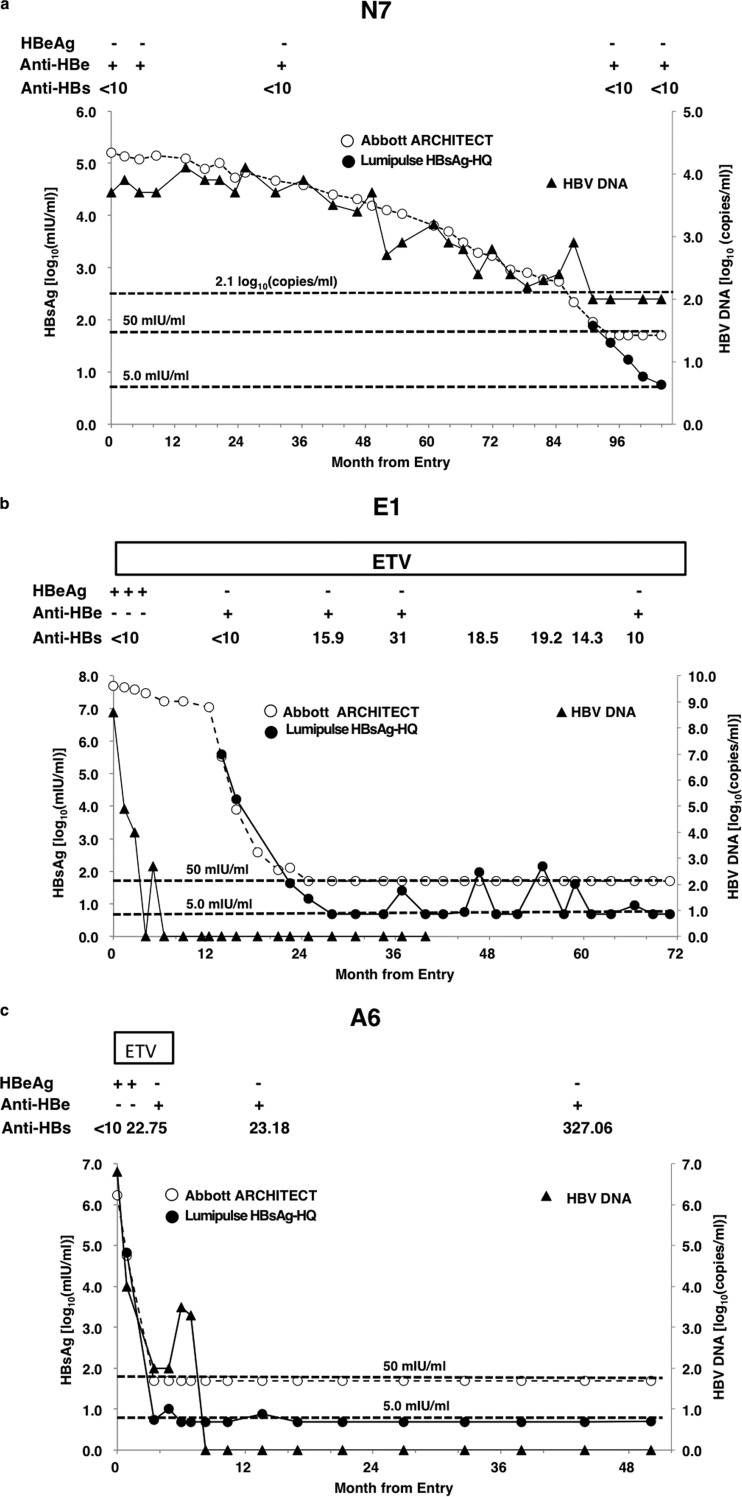
(i) Case no. N7 was a 71-year-old male. His alanine transaminase (ALT) was 19 IU/liter, HBV DNA was 3.7 log copies/ml at his first visit, the HBV genotype was C, HBeAg was negative, and anti-HBe was positive. The HBsAg level as measured by the Abbott Architect assay was 162,000 mIU/ml. The patient was followed as an inactive HB carrier. The last time at which HBsAg was detectable by the Abbott Architect assay was 87 months after the first visit, and it became undetectable in 3 months. However, it was still detectable by the Lumipulse HBsAg-HQ assay (78 mIU/ml). HBV DNA by Cobas TaqMan assay decreased to <2.1 log copies/ml. The Lumipulse HBsAg-HQ assay was still positive even 10 months after the Abbott Architect assay became negative. The HBsAg level measured by the Lumipulse HBsAg-HQ assay was 5.8 mIU/ml at this time (Fig. 3a).
Fig 3.
(a) HBsAg and HBV DNA dynamics of case no. N7. The Lumipulse HBsAg-HQ was still positive even 10 months after Abbott Architect results became negative. (b) HBsAg and HBV DNA dynamics of case no. E1. The HBsAg level as measured by the Lumipulse HBsAg-HQ assay was detectable for 3 months after HBsAg became negative by the Abbott Architect assay. After 1 year, HBsAg became detectable by the Lumipulse HBsAg-HQ assay, although HBV DNA was undetectable by the Cobas TaqMan and HBsAg was undetectable by the Abbott Architect assay. At 5 points, HBsAg was detectable by the Lumipulse HBsAg-HQ assay, and the anti-HBs concentration was >10 mIU/ml. (c) HBsAg and HBV DNA dynamics of case no. A6. HBsAg was detectable by the Lumipulse HBsAg-HQ assay for 3 months after HBsAg became negative by the Abbott Architect assay.
(ii) Case no. E1 was a 51-year-old male who had been infected with HBV by transfusion in adulthood and had developed chronic hepatitis B. His ALT was 57 IU/liter, HBV DNA was 8.6 copies/ml by the Cobas TaqMan assay, the HBV DNA genotype was Ba, HBeAg was positive, and anti-HBe was negative. The HBsAg level as measured by the Abbott Architect assay was 4,983,730 mIU/ml. The patient was treated with entecavir. After 24 months, HBsAg became undetectable by the Abbott Architect assay, and from this point to the last observation point, the Abbott Architect assay was continuously unable to detect HBsAg. The HBsAg level as measured by the Lumipulse HBsAg-HQ assay was 14.7 mIU/ml at the first point that was undetectable by the Abbott Architect assay, and it had been detectable for 3 months. After 3 months, HBsAg became undetectable by the Lumipulse HBsAg-HQ assay and anti-HBs reached >10 mIU/ml. From this point, anti-HBs was continually >10 mIU/ml. Interestingly, after 1 year, HBsAg measured by Lumipulse HBsAg-HQ assay became detectable again (25.2 mIU/ml), although HBV DNA by the Cobas TaqMan and HBsAg by the Abbott Architect assays remained undetectable. At some time points, HBsAg as determined by the Lumipulse HBsAg-HQ assay was detectable, and at the same time, anti-HBs was >10 mIU/ml (Fig. 3b).
(iii) Case no. A6 was a 38-year-old male diagnosed as having acute hepatitis B. After 1 month, HBeAg became seronegative and anti-HBe became seropositive. Three months after the first visit, HBV DNA was <2.1 log copies/ml, HBsAg became undetectable by the Abbott Architect assay, anti-HBs was 22.75 IU/ml, and the Lumipulse HBsAg-HQ assay detected HBsAg. After this time, anti-HBs was continually >10 mIU/ml. Thirteen months after the first visit, the Lumipulse HBsAg-HQ assay detected the reappearance of HBsAg (7.6 mIU/ml), although anti-HBs was still positive at 23.18 IU/ml (Fig. 3c).
DISCUSSION
The Lumipulse HBsAg-HQ assay showed improved sensitivity after disrupting HBV particles, dissociating HBsAg from HBsAg/anti-HBs complexes, and denaturing epitopes into linear forms. A major difference between the Abbott Architect and the Lumipulse HBsAg-HQ assays is that the latter detects HBsAg-anti-HBs complexes as well as small S proteins, which are present 10,000 to 1,000,000 times more frequently than Dane particles. The detection limit of the Lumipulse HBsAg-HQ assay (5 mIU/ml) was 10 times lower than that of the Abbott Architect assay, but there was otherwise a good correlation between the two. In clinical practice, more precise and broader HBsAg dynamics might therefore be followed by using the Lumipulse HBsAg-HQ assay. Differences between the two assays in detectable HBsAg persisted for a long time in the spontaneous HBsAg loss group (median, 10 months), followed by the NA-treated group (2.5 months) and the AH group (0.5 months).
In addition to the significant decrease or loss of all HBV replication in the blood serum, the long-term outcome after HBsAg seroclearance is good if there is no preexisting cirrhosis or viral superinfection. This view is supported by studies showing increased survival, a lower rate of hepatic decompensation, and a reduced frequency of hepatocellular carcinoma (HCC) in patients who have cleared HBsAg (14, 15). In carriers without cirrhosis and with no evidence of viral superinfection (hepatitis C virus [HCV] and/or hepatitis D virus [HDV]) at HBsAg seroclearance, liver function can improve or remain stable and hepatic decompensation rarely occurs; however, the incidence of HCC varies significantly, as was previously reported (16, 17). These discrepancies might depend on concurrent hepatitis, the severity of liver disease, age, and other factors. Yuen et al. (17) reported that HBsAg seroclearance of patients aged ≥50 years was associated with a higher risk of developing HCC than in patients of age <50 years, suggesting that we have to consider the age at which HBsAg becomes undetectable.
In most patients in our study (9 of 10 in the spontaneous HBsAg loss group and 7 of 8 in each of the NA-treated and AH groups), HBV DNA or HBcrAg was still detectable by the Abbott Architect assay at the time of HBsAg seroclearance (data not shown). Suzuki et al. (18) reported that HBcrAg correlates with intrahepatic covalently closed circular DNA in chronic hepatitis B patients. Hence, as the current CLEIA HBsAg quantification methods are inadequate for following some cases of HBV infection, the use of the Lumipulse HBsAg-HQ assay together with HBcrAg and HBV DNA testing might be valuable for evaluating patient response to treatment with interferon and NAs. Additionally, we reported that the measurement of HBcrAg is useful for predicting relapse after the cessation of lamivudine therapy for chronic hepatitis B; an HBcrAg level of <3.4 log U/ml at this time was the only independent predictive factor for the absence of posttreatment relapse (19). Thus, the combination of highly sensitive HBsAg detection by the Lumipulse HBsAg-HQ assay and HBcrAg might improve the accuracy of predicting response to treatment and relapse. Highly sensitive HBsAg detection by the Lumipulse HBsAg-HQ assay might be useful for several clinical applications. First, the Lumipulse HBsAg-HQ assay might replace HBV DNA monitoring by a PCR-based method for blood screening. As shown in Tables 2 to 4, at the last time point that HBsAg was detectable by the Lumipulse HBsAg-HQ assay, HBV DNA was undetectable in 9 of 26 patients (34%) by the Cobas TaqMan assay. This suggests that the sensitivity of the Lumipulse HBsAg-HQ assay for HBV detection was at least as high as that for the Cobas TaqMan assay at some time points. The Lumipulse HBsAg-HQ assay is simpler, more convenient, and less expensive than HBV DNA quantification by real-time PCR. At present in Japan, nucleic acid testing is used for detecting HBV in blood donors, but the Lumipulse HBsAg-HQ assay might substitute for nucleic acid testing for screening HBV if the sensitivity could be improved.
Second, the Lumipulse HBsAg-HQ assay may be useful for detecting occult HBV infection as well as HBV reactivation. Occult HBV infection is defined as infection with detectable HBV DNA but undetectable HBsAg with or without antibodies to HBV core antigen (anti-HBc) and/or anti-HBs (20–22). Recent interest in occult HBV infection has focused on the potential of donors with such infections to transmit the virus to susceptible recipients (23, 24). In this study, we detected HBsAg by the Lumipulse HBsAg-HQ assay in occult hepatitis B virus infection (OBI) patients, including those with HBsAg clearance as determined by the Architect assay (case no. N1, N3, N4, N5, N6, N7, N10, E3, E4, E5, E6, A3, A6, A8, and A9). In case no. N5, even >35 months after HBsAg became undetectable by the Abbott Architect assay, HBsAg was still detectable by the Lumipulse HBsAg-HQ assay. The Lumipulse HBsAg-HQ assay may change the diagnosis of patients defined as having current occult HBV infection. In case no. E1, HBsAg was detectable by the Lumipulse HBsAg-HQ assay at some time points, although HBV DNA by the Cobas TaqMan assay and HBsAg by Abbott Architect assay remained undetectable. In many cases (cases N1, N2, N4, N6, N8, N10, L2, E1, E2, E3, E5, E6, A2, A4, and A6), the HBV DNA and Lumipulse HBsAg-HQ results did not correlate. Interestingly, the original highly sensitive HBsAg assay reported by Matsubara et al. (10) had a similar sensitivity with HBV DNA detection during the acute phase of HBV infection. If the sensitivity of the Lumipulse HBsAg-HQ assay is improved, it would be sensitive enough to monitor HBV reactivation instead of needing to rely on HBV DNA monitoring. More importantly, there have been cases of HBV reactivation in patients with resolved infection (HBsAg-negative, anti-HBc, and/or anti-HBs positive) during the course of chemotherapy and/or immunotherapy (especially therapy with rituximab plus steroids), sometimes proving fatal (25–29). The Lumipulse HBsAg-HQ assay might be more convenient for such screening than TaqMan PCR.
Third, previous CLEIA HBsAg quantification methods, including the Abbott Architect assay, apply monoclonal/polyclonal antibodies against external structural regions within the determinant “a” loop. HBsAg escape mutations, such as G130D, T131N, M133T, and G145R, were found in patients who were positive for anti-HBs but negative for HBsAg (9, 30). Oon et al. (32) reported that HBV carriers, including HCC patients who were negative for HBsAg but positive for anti-HBc and anti-HBs, had the T126S, Q129D, M133L, T140I, and G145R mutations within the S region. Wu et al. (31) reported that amino acid residues at positions 122 and 145 of HBsAg had a major effect on antigenicity and immunogenicity. HBsAg mutants can escape current detection and persist in HBV-infected individuals after the loss of HBsAg (32). In the present study, we therefore determined the HBs amino acid sequences of all cases (with detectable HBV DNA), some of which had amino acid I126N, F134Y, S143T, and G145S (not G145R) mutations. It is possible that these HBsAg mutants escape detection by current HBsAg assays and the sensitivity becomes low (33). Based on the pretreatment, however, the Lumipulse HBsAg-HQ assay was able to detect HBsAg mutants because it uses two monoclonal antibodies against the external structural region as determinant “a” and the internal epitope as the capture target. Additionally, the Lumipulse HBsAg-HQ assay can detect HBsAg from samples with anti-HBs.
In conclusion, the automatic, highly sensitive HBsAg CLEIA Lumipulse HBsAg-HQ assay is a very convenient and precise assay for HBV monitoring in clinical practice.
ACKNOWLEDGMENTS
This study was supported by a grant-in-aid from the Ministry of Education, Culture, Sports, Science, and Technology and a grant-in-aid from the Ministry of Health, Labor, and Welfare of Japan.
The authors declare no conflicts of interest.
Footnotes
Published ahead of print 14 August 2013
REFERENCES
- 1.Pan CQ, Zhang JX. 2005. Natural history and clinical consequences of hepatitis B virus infection. Int. J. Med. Sci. 2:36–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kohmoto M, Enomoto M, Tamori A, Habu D, Takeda T, Kawada N, Sakaguchi H, Seki S, Shiomi S, Nishiguchi S. 2005. Quantitative detection of hepatitis B surface antigen by chemiluminescent microparticle immunoassay during lamivudine treatment of chronic hepatitis B virus carriers. J. Med. Virol. 75:235–239 [DOI] [PubMed] [Google Scholar]
- 3.Chan HL, Wong VW, Tse AM, Tse CH, Chim AM, Chan HY, Wong GL, Sung JJ. 2007. Serum hepatitis B surface antigen quantitation can reflect hepatitis B virus in the liver and predict treatment response. Clin. Gastroenterol. Hepatol. 5:1462–1468 [DOI] [PubMed] [Google Scholar]
- 4.Werle B, Cinquin K, Marcellin P, Pol S, Maynard M, Trépo C, Zoulim F. 2004. Evolution of hepatitis B viral load and viral genome sequence during adefovir dipivoxil therapy. J. Viral Hepat. 11:74–83 [DOI] [PubMed] [Google Scholar]
- 5.Wursthorn K, Lutgehetmann M, Dandri M, Volz T, Buggisch P, Zollner B, Longerich T, Schirmacher P, Metzler F, Zankel M, Fischer C, Currie G, Brosgart C, Petersen J. 2006. Peginterferon alpha-2b plus adefovir induce strong cccDNA decline and HBsAg reduction in patients with chronic hepatitis B. Hepatology 44:675–684 [DOI] [PubMed] [Google Scholar]
- 6.Brunetto MR, Moriconi F, Bonino F, Lau GK, Farci P, Yurdaydin C, Piratvisuth T, Luo K, Wang Y, Hadziyannis S, Wolf E, McCloud P, Batrla R, Marcellin P. 2009. Hepatitis B virus surface antigen levels: a guide to sustained response to peginterferon alfa-2a in HBeAg-negative chronic hepatitis B. Hepatology 49:1141–1150 [DOI] [PubMed] [Google Scholar]
- 7.Chen CH, Lee CM, Wang JH, Tung HD, Hung CH, Lu SN. 2004. Correlation of quantitative assay of hepatitis B surface antigen and HBV DNA levels in asymptomatic hepatitis B virus carriers. Eur. J. Gastroenterol. Hepatol. 16:1213–1218 [DOI] [PubMed] [Google Scholar]
- 8.Deguchi M, Yamashita N, Kagita M, Asari S, Iwatani Y, Tsuchida T, Iinuma K, Mushahwar IK. 2004. Quantitation of hepatitis B surface antigen by an automated chemiluminescent microparticle immunoassay. J. Virol. Methods 115:217–222 [DOI] [PubMed] [Google Scholar]
- 9.Martinot-Peignoux M, Maylin S, Moucari R, Ripault MP, Boyer N, Cardoso AC, Giuily N, Castelnau C, Pouteau M, Stern C, Aupérin A, Bedossa P, Asselah T, Marcellin P. 2009. Virological response at 4 weeks to predict outcome of hepatitis C treatment with pegylated interferon and ribavirin. Antivir. Ther. 14:501–511 [PubMed] [Google Scholar]
- 10.Matsubara N, Kusano O, Sugamata Y, Itoh T, Mizuii M, Tanaka J, Yoshizawa H. 2009. A novel hepatitis B virus surface antigen immunoassay as sensitive as hepatitis B virus nucleic acid testing in detecting early infection. Transfusion 49:585–595 [DOI] [PubMed] [Google Scholar]
- 11.Shinkai N, Tanaka Y, Matsuura K, Kani S, Naganuma H, Mizokami M. 2010. Evaluation and application of a newly developed highly sensitive HBsAg chemiluminescent enzyme immunoassay for chronic hepatitis B patients. Rinsho Byori 58:1078–1084 (Article in Japanese.) [PubMed] [Google Scholar]
- 12.Kimura T, Rokuhara A, Sakamoto Y, Yagi S, Tanaka E, Kiyosawa K, Maki N. 2002. Sensitive enzyme immunoassay for hepatitis B virus core-related antigens and their correlation to virus load. J. Clin. Microbiol. 40:439–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wong DK, Tanaka Y, Lai CL, Mizokami M, Fung J, Yuen MF. 2007. Hepatitis B virus core-related antigens as markers for monitoring chronic hepatitis B infection. J. Clin. Microbiol. 45:3942–3947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moucari R, Korevaar A, Lada O, Martinot-Peignoux M, Boyer N, Mackiewicz V, Dauvergne A, Cardoso AC, Asselah T, Nicolas-Chanoine MH, Vidaud M, Valla D, Bedossa P, Marcellin P. 2009. High rates of HBsAg seroconversion in HBeAg-positive chronic hepatitis B patients responding to interferon: a long-term follow-up study. J. Hepatol. 50:1084–1092 [DOI] [PubMed] [Google Scholar]
- 15.van Zonneveld M, Honkoop P, Hansen BE, Niesters HG, Darwish Murad S, de Man RA, Schalm SW, Janssen HL. 2004. Long-term follow-up of alpha-interferon treatment of patients with chronic hepatitis B. Hepatology 39:804–810 [DOI] [PubMed] [Google Scholar]
- 16.Simonetti J, Bulkow L, McMahon BJ, Homan C, Snowball M, Negus S, Williams J, Livingston SE. 2010. Clearance of hepatitis B surface antigen and risk of hepatocellular carcinoma in a cohort chronically infected with hepatitis B virus. Hepatology 51:1531–1537 [DOI] [PubMed] [Google Scholar]
- 17.Yuen MF, Wong DK, Fung J, Ip P, But D, Hung I, Lau K, Yuen JC, Lai CL. 2008. HBsAg seroclearance in chronic hepatitis B in Asian patients: replicative level and risk of hepatocellular carcinoma. Gastroenterology 135:1192–1199 [DOI] [PubMed] [Google Scholar]
- 18.Suzuki F, Miyakoshi H, Kobayashi M, Kumada H. 2009. Correlation between serum hepatitis B virus core-related antigen and intrahepatic covalently closed circular DNA in chronic hepatitis B patients. J. Med. Virol. 81:27–33 [DOI] [PubMed] [Google Scholar]
- 19.Shinkai N, Tanaka Y, Orito E, Ito K, Ohno T, Hirashima N, Hasegawa I, Sugauchi F, Ueda R, Mizokami M. 2006. Measurement of hepatitis B virus core-related antigen as predicting factor for relapse after cessation of lamivudine therapy for chronic hepatitis B virus infection. Hepatol Res. 36:272–276 [DOI] [PubMed] [Google Scholar]
- 20.Hollinger FB. 2008. Hepatitis B virus infection and transfusion medicine: science and the occult. Transfusion 48:1001–1026 [DOI] [PubMed] [Google Scholar]
- 21.Raimondo G, Pollicino T, Cacciola I, Squadrito G. 2007. Occult hepatitis B virus infection. J. Hepatol. 46:160–170 [DOI] [PubMed] [Google Scholar]
- 22.van Hemert FJ, Zaaijer HL, Berkhout B, Lukashov VV. 2008. Occult hepatitis B infection: an evolutionary scenario. Virol. J. 5:146. 10.1186/1743-422X-5-146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dettori S, Candido A, Kondili LA, Chionne P, Taffon S, Genovese D, Iudicone P, Miceli M, Rapicetta M. 2009. Identification of low HBV-DNA levels by nucleic acid amplification test (NAT) in blood donors. J. Infect. 59:128–133 [DOI] [PubMed] [Google Scholar]
- 24.Satake M, Taira R, Yugi H, Hino S, Kanemitsu K, Ikeda H, Tadokoro K. 2007. Infectivity of blood components with low hepatitis B virus DNA levels identified in a lookback program. Transfusion 47:1197–1205 [DOI] [PubMed] [Google Scholar]
- 25.Fukushima N, Mizuta T, Tanaka M, Yokoo M, Ide M, Hisatomi T, Kuwahara N, Tomimasu R, Tsuneyoshi N, Funai N, Sueoka E. 2009. Retrospective and prospective studies of hepatitis B virus reactivation in malignant lymphoma with occult HBV carrier. Ann. Oncol. 20:2013–2017 [DOI] [PubMed] [Google Scholar]
- 26.Law JK, Ho JK, Hoskins PJ, Erb SR, Steinbrecher UP, Yoshida EM. 2005. Fatal reactivation of hepatitis B post-chemotherapy for lymphoma in a hepatitis B surface antigen-negative, hepatitis B core antibody-positive patient: potential implications for future prophylaxis recommendations. Leuk. Lymphoma 46:1085–1089 [DOI] [PubMed] [Google Scholar]
- 27.Pei SN, Chen CH, Lee CM, Wang MC, Ma MC, Hu TH, Kuo CY. 2010. Reactivation of hepatitis B virus following rituximab-based regimens: a serious complication in both HBsAg-positive and HBsAg-negative patients. Ann. Hematol. 89:255–262 [DOI] [PubMed] [Google Scholar]
- 28.Wu JM, Huang YH, Lee PC, Lin HC, Lee SD. 2009. Fatal reactivation of hepatitis B virus in a patient who was hepatitis B surface antigen negative and core antibody positive before receiving chemotherapy for non-Hodgkin lymphoma. J. Clin. Gastroenterol. 43:496–498 [DOI] [PubMed] [Google Scholar]
- 29.Yeo W, Chan TC, Leung NW, Lam WY, Mo FK, Chu MT, Chan HL, Hui EP, Lei KI, Mok TS, Chan PK. 2009. Hepatitis B virus reactivation in lymphoma patients with prior resolved hepatitis B undergoing anticancer therapy with or without rituximab. J. Clin. Oncol. 27:605–611 [DOI] [PubMed] [Google Scholar]
- 30.Chen WN, Oon CJ. 2000. Hepatitis B virus surface antigen (HBsAg) mutants in Singapore adults and vaccinated children with high anti-hepatitis B virus antibody levels but negative for HBsAg. J. Clin. Microbiol. 38:2793–2794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wu C, Deng W, Deng L, Cao L, Qin B, Li S, Wang Y, Pei R, Yang D, Lu M, Chen X. 2012. Amino acid substitutions at positions 122 and 145 of hepatitis B virus surface antigen (HBsAg) determine the antigenicity and immunogenicity of HBsAg and influence in vivo HBsAg clearance. J. Virol. 86:4658–4669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Oon CJ, Chen WN, Goh KT, Mesenas S, Ng HS, Chiang G, Tan C, Koh S, Teng SW, Toh I, Moh MC, Goo KS, Tan K, Leong AL, Tan GS. 2002. Molecular characterization of hepatitis B virus surface antigen mutants in Singapore patients with hepatocellular carcinoma and hepatitis B virus carriers negative for HBsAg but positive for anti-HBs and anti-HBc. J. Gastroenterol. Hepatol. 17(Suppl):S491–S496 [DOI] [PubMed] [Google Scholar]
- 33.Coleman PF. 2006. Detecting hepatitis B surface antigen mutants. Emerg. Infect. Dis. 12:198–203 [DOI] [PMC free article] [PubMed] [Google Scholar]