Abstract
We assessed the performance of a duplex real-time PCR assay for blaKPC and blaNDM performed directly (D-PCR) on perianal and perirectal swabs and stool. Spiked specimens and 126 clinical surveillance swabs (comprising a sensitivity panel of 46 perirectal double swabs previously determined to be culture positive for blaKPC-PCR-positive Enterobacteriaceae and a specificity panel of 80 perianal swabs from patients at risk of carbapenemase-producing Enterobacteriaceae [CPE] colonization) were studied. For the surveillance swabs, D-PCR was compared to PCR after broth enrichment (BE-PCR) and two culture-based methods: the HardyCHROM ESBL agar (HC-A) and the CDC screening (CDC-A) methods. PCR was performed on morphologically distinct colonies that were isolated by culture. All of the initial PCR testing was done without extraction using a simple lysis procedure. The analytical sensitivities of D-PCR for blaKPC were 9 CFU/μl (for swabs) and 90 CFU/μl (for stool), and for blaNDM, it was 1.9 CFU/μl (for both swabs and stool). In the clinical sensitivity panel, D-PCR and BE-PCR were initially positive for blaKPC in 41/46 (89.1%) and 43/46 (93.5%) swabs, respectively. The swabs that were initially negative by D-PCR (n = 5) and BE-PCR (n = 3) were visibly stool soiled; all swabs were blaKPC positive upon repeat testing after lysate extraction. The CDC-A and HC-A yielded blaKPC-positive Enterobacteriaceae from 36/46 (78.3%) and 35/46 (76.1%) swabs, respectively (sensitivities of D-PCR/BE-PCR postextraction of soiled specimens versus HC-A, P = 0.0009, and versus CDC-A, P = 0.0016). All swabs in the specificity panel were negative for CPE by all four methods. D-PCR allows for the timely detection of blaKPC and blaNDM carriage with excellent sensitivity when specimens visibly soiled with stool undergo preparatory extraction.
INTRODUCTION
Active surveillance for carbapenemase-producing Enterobacteriaceae (CPE) is a component of successful outbreak control (1–3). The recently updated Centers for Disease Control and Prevention (CDC) recommendations include screening hospitalized patients who are epidemiologically linked contacts of patients who carry CPE or who have had a history of hospitalization overseas in the last 6 months (4). Klebsiella pneumoniae carbapenemase (blaKPC) and New Delhi metallo-β-lactamases (blaNDM) are the carbapenemases of foremost current concern in the United States.
Various culture-based methods utilizing rectal and perianal swabs or stool have been used for the detection of CPE colonization. These include several commercially available chromogenic media (5–7), none of which are currently cleared by the FDA, and laboratory-developed methods which utilize selective enteric media containing carbapenems (6) or onto which carbapenem disks are placed (6, 8, 9). The CDC-recommended culture method entails overnight selective broth enrichment culture of surveillance swabs, followed by plate culture and testing of suspect colonies using the modified Hodge test (MHT) or formal antimicrobial susceptibility testing (10). Culture methods, while yielding organisms for further characterization, require an incubation period of at least overnight and can take days. Also, while culture methods may yield carbapenem-resistant Gram-negative bacilli (CR-GNB), these may not necessarily be carbapenemase-producing organisms, as carbapenem resistance mediated by other mechanisms (e.g., AmpC β-lactamases or extended-spectrum β-lactamases with porin loss) may result in carbapenem nonsusceptibility, diminishing the specificity of culture-based screening tests for the detection of CPE (7). The MHT itself lacks both sensitivity (especially for the metallo-β-lactamases) and specificity (11).
The importance of accurate and rapid characterization of CPE for infection control was illustrated recently during an outbreak of blaKPC-positive K. pneumoniae at the National Institutes of Health Clinical Center, in which prompt laboratory response and infection control measures, including the implementation of molecular testing directly from swabs, mitigated the outbreak (12, 13).
Several laboratory-developed PCR-based nucleic acid amplification tests (NAATs) have been described for the detection of carbapenemases in surveillance swabs and stool (8, 14–19). Commercial assays, including a nucleic acid sequence-based amplification (NASBA)-based assay, the NucliSENS EasyQ KPC assay (bioMérieux SA, Marcy l'Etoile, France), have also been evaluated on surveillance swabs (20, 21). A multiplex PCR assay (hyplex SuperBug ID and hyplex-MBL ID; Amplex Diagnostics GmbH, Gars-Bahnhof, Germany) has been developed for use directly on clinical material for the detection of carbapenemases (22, 23), although data on its performance on surveillance swabs are lacking. More recently, a multiplex PCR assay targeting Klebsiella pneumoniae carbapenemase (KPC), New Delhi metallo-β-lactamases (NDM), the OXA-48 group, and VIM, the Check-Direct CPE (Check-Points Health B.V., Wageningen, the Netherlands) was developed for performance on surveillance swabs or cultured isolates (24), although there are no published data on this assay at the time of this writing.
Most of the abovementioned assays performed on surveillance swabs have been limited in one or more ways; most are single-plex assays (8, 14, 15, 18–21) targeting only blaKPC; and some entail a broth enrichment step that delays the turnaround time by a day (15, 18, 19) or require a prior extraction step (8, 14, 20, 21, 24).
NAAT testing for the two most commonly encountered carbapenemases in the United States performed directly on surveillance swab specimens with minimal preparatory steps, preferably avoiding upfront nucleic acid extraction, should provide results that translate into timelier implementation of infection control measures. Recently, we described a duplex blaKPC and blaNDM assay for the direct testing of isolated colonies (25). In this study, we applied our assay to spiked surveillance specimens (perianal or perirectal swabs and stool). We also compared the performance of the duplex PCR assay done directly (D-PCR) on a set of clinical surveillance perianal or perirectal swabs after a simple lysis procedure and after a broth enrichment step, alongside two culture-based methods, the CDC-recommended method (CDC-A) (10), and a commercially available chromogenic medium, HardyCHROM ESBL agar (HC-A) (Hardy Diagnostics, Santa Maria, CA).
(This paper was presented in part at the 9th International Symposium of Antimicrobial Agents and Resistance, 13 to 15 March 2013, in Kuala Lumpur, Malaysia.)
MATERIALS AND METHODS
Limit of detection.
The perianal swabs and stool used for the spiking studies were routine specimens submitted to our clinical laboratory for testing for other reasons and were shown to be negative for blaKPC and blaNDM by our assay. Serial 10-fold dilutions (10−1 to 10−8) of two reference isolates, KPC-positive K. pneumoniae ATCC BAA 1705 and NDM-positive K. pneumoniae NCTC 13443, were made in saline with a starting inoculum equivalent to a 0.5 McFarland standard. Quantitative culture was performed on the final dilution. Whole organisms in each dilution series (90,000 to 0.9 CFU/μl for KPC-positive K. pneumoniae ATCC BAA 1705, and 190,000 to 1.9 CFU/μl for NDM-positive K. pneumoniae NCTC 13443) were then heat killed and spiked into perianal swabs (BBL CultureSwab with Stuart's medium; Becton Dickinson, Franklin Lakes, NJ), and stool in 50% Stool Recovery and Transport (STAR) medium (Roche Applied Science, Indianapolis, IN). The stool specimens were originally collected in Cary-Blair transport medium (Para-Pak C&S; Meridian Bioscience, Cincinnati, OH); a cotton swab was used to inoculate stool into 1-ml aliquots of STAR medium. The spiked stool specimens in STAR medium were extracted in triplicate on the MagNA Pure LC 1.0 instrument using the MagNA Pure total nucleic acid protocol/kit (Roche Applied Science). The spiked specimens were tested by the duplex blaKPC and blaNDM PCR assay, a real-time fluorescent resonance energy transfer (FRET) hybridization probe-based real-time PCR assay targeting a 160-bp region of the blaKPC-2 gene and a 156-bp region of the blaNDM-1 gene, performed on the LightCycler 2.0 instrument (Roche Molecular Diagnostics, Indianapolis, IN) (25).
Accuracy studies (clinical surveillance swabs). (i) Sensitivity panel.
We tested a sensitivity panel consisting of 47 previously characterized perirectal double-swab specimens (Becton Dickinson) that were determined at the Rush University Medical Center, Chicago, to be culture positive for blaKPC PCR-positive Enterobacteriaceae using the direct ertapenem disk screening method described by Lolans et al. (9) and a blaKPC PCR assay described by Cole et al. (26), which was modified to include a 16S rRNA gene target as an internal control (18). These perirectal surveillance swabs were collected between September and November 2012 as part of a longitudinal study on CPE colonization in patients in Chicago long-term acute care facilities (27). The perirectal swabs were inserted approximately 1 cm past the anal verge and obtained by study personnel. One swab from each double-swab specimen was plated during this initial screen and then aseptically replaced in the original transport tube containing a sponge reservoir with Stuart's transport medium. The swabs were stored at 4°C and tested for this study within 10 to 41 days (median, 20 days) of collection.
(ii) Specificity panel.
The specificity panel consisted of 80 self- or provider-collected perianal double swabs obtained from consecutively enrolled international patients hospitalized at the Mayo Clinic, Rochester, MN, from February to April 2013 as part of a study assessing CPE carriage in at-risk patients (institutional review board [IRB] no. 12-008283). These swabs were stored at 4°C and tested within 7 days of collection.
Testing of clinical surveillance swabs.
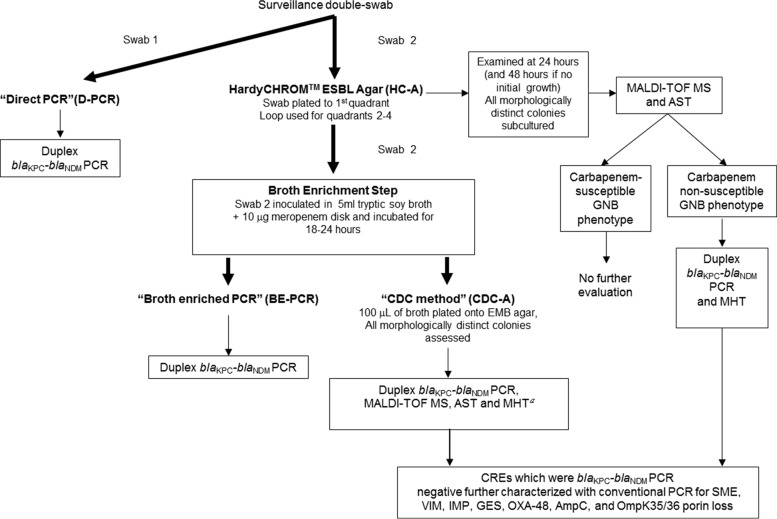
The general protocol for the testing of clinical surveillance swabs is outlined in Fig. 1 and described in detail below.
Fig 1.
Study schema for clinical surveillance swabs. MALDI-TOF MS, matrix-assisted laser desorption–time of flight mass spectrometry; AST, antimicrobial susceptibility testing (by agar dilution); CRE, carbapenem-resistant Enterobacteriaceae (per CLSI 2012 interpretative criteria); EMB agar, eosin methylene blue agar; MHT, modified Hodge test; GNB, Gram-negative bacilli. MHT was performed on isolates from the sensitivity panel (a).
Direct PCR method.
The first swab (swab 1) from each specimen was placed in a LightCycler Advanced lysis tube (Roche Molecular Diagnostics) and subjected to heat lysis on a Thermomixer R (Eppendorf AG, Germany) for 6 min at 99°C and 1,400 rpm, followed by centrifugation at 20,800 × g for 20 s. Five microliters of supernatant was added to 15 μl of PCR mastermix in a 20-μl LightCycler reaction cuvette and tested with the duplex blaKPC-blaNDM assay (25).
HardyCHROM ESBL agar.
The second swab (swab 2) was first plated by rotating it 360° across the first quadrant of a HardyCHROM ESBL plate. The plate was streaked for isolation to four quadrants using a sterile loop. The plates were incubated at 35°C in ambient air for 18 to 24 h and observed for growth. If no growth was observed at 24 h, the plates were reincubated for another 24 h, according to the manufacturer's instructions. All morphologically distinct colonies were identified using Bruker Biotyper matrix-assisted laser desorption–time of flight mass spectrometry (MALDI-TOF MS) (Bruker Daltonics, Billerica, MA) and tested for antimicrobial susceptibility using agar dilution. The blaKPC-blaNDM assay and the MHT were performed on isolates that were carbapenem resistant according to CLSI criteria (28).
Broth-enriched PCR (BE-PCR).
After plating on HC-A, swab 2 was placed into tryptic soy broth (TSB) (BD BBL; Becton Dickinson) with a 10-μg meropenem disk (BD BBL Sensi-Disc; Becton Dickinson), vortexed for 5 s, and incubated for 18 to 24 h at 35°C in ambient air with loosened caps. A 6-μl aliquot of the TSB, after incubation, was heat lysed in the lysis tube, as described above, and tested with the duplex PCR assay.
CDC method for CPE screening.
The CDC-A was performed using the protocol recommended by the CDC (10), with a slight modification in that eosin methylene blue (EMB) agar (Becton Dickinson) was used instead of MacConkey agar. Briefly, a 100-μl aliquot of the TSB incubated overnight with swab 2 was plated on EMB and streaked for isolation. The plate was incubated for 18 to 24 h at 35°C in ambient air. All morphologically distinct colonies were tested with the duplex PCR assay, identified using MALDI-TOF MS, and subjected to agar dilution antibiotic susceptibility testing. In addition, the MHT was performed on isolates from the sensitivity panel.
Categorization of isolates for analysis.
For the purposes of this analysis, the isolates were considered by species and susceptibility according to breakpoints defined by the CLSI (28). Resistance was classified into three categories: (i) carbapenem-resistant GNB (CR-GNB), defined as Enterobacteriaceae that were nonsusceptible to either ertapenem (MIC > 0.5 μg/ml) or meropenem (MIC > 1 μg/ml) and nonfermenting GNB that were nonsusceptible to meropenem (MIC > 2 μg/ml for Pseudomonas aeruginosa and MIC > 4 μg/ml for other nonfermenting GNB); (ii) third- or fourth-generation cephalosporin (oxyimino-cephalosporin)-resistant GNB (CephR-GNB), defined as Enterobacteriaceae that were nonsusceptible to ceftriaxone (MIC > 1 μg/ml), ceftazidime (MIC > 4 μg/ml), or cefepime (MIC > 8 μg/ml) and nonfermenting GNB that were nonsusceptible to for ceftriaxone, ceftazidime, and cefepime (all MIC > 8 μg/ml); only ceftazidime and cefepime were considered for P. aeruginosa isolates; (iii) susceptible GNB (S-GNB), defined as any GNB that was fully susceptible to the tested carbapenems and oxyimino-cephalosporins.
Further molecular characterization of carbapenem-resistant Enterobacteriaceae isolates negative for blaKPC and blaNDM.
If negative for blaKPC and blaNDM, carbapenem-resistant Enterobacteriaceae isolates from HC-A and CDC-A were further evaluated for SME (29), VIM (30), IMP (31), GES (32), OXA-48 (33), AmpC (34), and OmpK35/36 porin loss (35) using conventional PCR. Molecular confirmation of the presence of carbapenemase genes, either by the real-time blaKPC-blaNDM assay or conventional PCR, was considered to be the gold standard for assigning a CPE status.
Accuracy studies (spiked specimens).
In order to supplement the clinical sample data, blinded randomized spiking studies were performed using perianal swabs and Cary-Blair transported stool (predetermined to be negative for blaKPC and blaNDM). Briefly, samples were spiked with heat-killed organisms near the limit of detection of the blaKPC-blaNDM assay. This corresponded to 395 CFU/μl of K. pneumoniae NCTC 13443 (blaNDM) and/or 550 CFU/μl K. pneumoniae ATCC BAA-1705 (blaKPC). The spiked samples were then assayed with the duplex blaKPC-blaNDM assay with swabs processed with the D-PCR method, and with stool specimens that had undergone extraction prior to PCR.
Statistical analysis.
Data analysis was performed using SAS version 9.3 (SAS Institute Inc., Cary, NC). McNemar's test was used for comparisons of paired variables. A P value of <0.05 was considered significant.
RESULTS
Limit of detection.
The limit of detection of the PCR assay was 9 CFU/μl and 90 CFU/μl for blaKPC in perianal swabs and stool samples, respectively, and 1.9 CFU/μl for blaNDM in both perianal swabs and stool.
Accuracy studies (clinical surveillance swabs). (i) Molecular methods (D-PCR and BE-PCR).
For the sensitivity panel, 47 swabs, each collected from an individual patient and previously determined to be culture positive for blaKPC Enterobacteriaceae, were tested. One swab was negative by all four methods (both molecular and culture-based methods; the culture yielded no growth at all), implying the loss of organisms with possible initial low inoculum; this specimen was excluded from the analysis. Of the remaining 46 swabs assessed, D-PCR and BE-PCR were positive for 41/46 (89.1%) and 43/46 swabs (93.5%) without prior extraction. The swabs that were initially negative by D-PCR (n = 5) and BE-PCR (n = 3) were visibly soiled with stool, and all were positive for blaKPC upon repeat testing after the lysates were extracted. Thus, in an approach in which swabs visibly soiled with stool underwent extraction prior to PCR, both the D-PCR and BE-PCR yielded 100% clinical sensitivity (Table 1). All 80 swabs in the specificity panel tested negative by D-PCR and BE-PCR.
Table 1.
Clinical sensitivity of the evaluated methods for detecting blaKPC in rectal surveillance swabs
| Swab testing method | No. of positive swabs/total no. of swabs | Sensitivity (% [95% confidence interval])c |
|---|---|---|
| Direct PCRa | 46/46 | 100 (92.3–100) |
| Broth-enriched PCRa | 46/46 | 100 (92.3–100) |
| CDC methodb | 36/46 | 78.3 (64.4–78.7) |
| HardyCHROM ESBL agarb | 35/46 | 76.1 (62.11–86.1) |
Five swabs with the D-PCR and 3 swabs with the BE-PCR with visible soiling were negative on initial testing, but were positive on retesting after nucleic acid extraction on the MagNA Pure Compact system.
For the CDC method, blaKPC and blaNDM PCR was performed on all morphologically distinct colonies. For the HardyCHROM ESBL agar method, PCR was performed on carbapenem-resistant isolates. A specimen was considered positive if any isolate from that specimen was blaKPC positive by PCR.
McNemar's test D/BE-PCR versus HC-A, P = 0.0009; D/BE-PCR versus CDC-A, P = 0.0016; CDC-A versus HC-A, P = 0.76.
(ii) Culture-based methods (HC-A and CDC-A).
Overall, when considered by species and susceptibility categories, 148 distinct isolates were cultured from the 46 swabs previously determined to be culture positive for blaKPC Enterobacteriaceae, 56 by both HC-A and CDC-A, 44 by CDC-A alone, and 48 by HC-A alone. Of the 148 isolates, 109 were Enterobacteriaceae and 39 were nonfermenting GNB; 74 were CR-GNB, 49 were CephR-GNB, and 25 were S-GNB (Table 2).
Table 2.
Breakdown of cultured Gram-negative bacilli from blaKPC-positive rectal surveillance swabs by culture method, identification, susceptibility phenotype, and blaKPC status
| Culture method(s) detectiona | Organism characteristics |
No. with susceptibility phenotypeb: |
blaKPC PCR-positive (% of CR-GNB)c | ||||
|---|---|---|---|---|---|---|---|
| Nonfermenter or Enterobacteriaceae | Genus/species | CR-GNB | CephR-GNB | S-GNB | Total (all susceptibility phenotypes) | ||
| CDC-A and HC-A | Nonfermenter | Acinetobacter spp. | 2 | 1 | 0 | 3 | 0 (0) |
| Pseudomonas aeruginosa | 10 | 2 | 2 | 14 | 0 (0) | ||
| Enterobacteriaceae | Escherichia coli | 1 | 4 | 0 | 5 | 1 (100) | |
| Klebsiella pneumoniae | 28 | 2 | 0 | 30 | 28 (100) | ||
| Morganella morganii | 0 | 1 | 0 | 1 | |||
| Proteus mirabilis | 0 | 1 | 0 | 1 | |||
| Providencia stuartii | 0 | 2 | 0 | 2 | |||
| CDC-A alone | Nonfermenters | Acinetobacter spp. | 3 | 0 | 0 | 3 | 0 (0) |
| Pseudomonas aeruginosa | 7 | 1 | 6 | 14 | 0 (0) | ||
| Alcaligenes faecalis | 0 | 0 | 1 | 1 | |||
| Enterobacteriaceae | Citrobacter spp. | 1 | 0 | 0 | 1 | 1 (100) | |
| E. coli | 1 | 0 | 5 | 6 | 0 (0) | ||
| K. pneumoniae | 4 | 1 | 1 | 6 | 4 (100) | ||
| M. morganii | 0 | 0 | 1 | 1 | |||
| P. mirabilis | 0 | 2 | 5 | 7 | |||
| P. stuartii | 1 | 1 | 3 | 5 | 1 (100) | ||
| HC-A alone | Nonfermenters | Acinetobacter spp. | 2 | 0 | 0 | 2 | 0 (0) |
| P. aeruginosa | 2 | 0 | 0 | 2 | 0 (0) | ||
| Enterobacteriaceae | Citrobacter spp. | 0 | 3 | 0 | 3 | ||
| Enterobacter aerogenes | 1 | 0 | 0 | 1 | 1 (100) | ||
| Enterobacter cloacae complex | 2 | 1 | 0 | 3 | 0 (0) | ||
| E. coli | 3 | 8 | 0 | 11 | 0 (0) | ||
| K. pneumoniae | 6 | 8 | 0 | 14 | 6 (100) | ||
| M. morganii | 0 | 4 | 1 | 5 | |||
| P. mirabilis | 0 | 4 | 0 | 4 | |||
| P. stuartii | 0 | 2 | 0 | 2 | |||
| Serratia marcescens | 0 | 1 | 0 | 1 | |||
| Total | Nonfermenters | Acinetobacter spp. | 7 | 1 | 0 | 8 | 0 (0) |
| A. faecalis | 0 | 0 | 1 | 1 | |||
| P. aeruginosa | 19 | 3 | 8 | 30 | 0 (0) | ||
| Enterobacteriaceae | Citrobacter spp. | 1 | 3 | 0 | 4 | 1 (100) | |
| E. aerogenes | 1 | 0 | 0 | 1 | 1 (100) | ||
| E. cloacae complex | 2 | 1 | 0 | 3 | 0 (0) | ||
| E. coli | 5 | 12 | 5 | 22 | 1 (20) | ||
| K. pneumoniae | 38 | 11 | 1 | 50 | 38 (100) | ||
| M. morganii | 0 | 5 | 2 | 7 | |||
| P. mirabilis | 0 | 7 | 5 | 12 | |||
| P. stuartii | 1 | 5 | 3 | 9 | 1 (100) | ||
| S. marcescens | 0 | 1 | 0 | 1 | |||
| All genera/species | 74 | 49 | 25 | 148 | 42 (56.8) | ||
CDC-A: CDC-recommended method; HC-A: HardyCHROM Agar ESBL.
CR-GNB, carbapenem-resistant GNB; CephR-GNB, 3rd- or 4th-generation cephalosporin-resistant GNB (oxyimino-cephalosporin, i.e., ceftriaxone, ceftazidime, or cefepime). For P. aeruginosa isolates, only ceftazidime and cefepime were considered. S-GNB, susceptible GNB (susceptible to carbapenems or oxyimino-cephalosporins). Isolates testing “intermediate” in susceptibility were classified together with resistant isolates. See “Categorization of isolates for analysis” in the text for detailed definitions.
blaKPC and blaNDM PCR was performed for all isolates cultured by CDC-A and for carbapenem-nonsusceptible isolates cultured by HC-A for the sensitivity panel. All isolates from the clinical surveillance swabs that were blaKPC positive in this study were CR-GNB.
CDC-A yielded 100 distinct isolates, including 58 CR-GNB and 42 carbapenem-susceptible GNB (18 CephR-GNB and 24 S-GNB). Of the 100 isolates, the MHT was positive for 46 and the blaKPC PCR positive for 35 of 46 (76.1%) (Table 3). HC-A yielded 104 distinct isolates, including 57 CR-GNB and 47 carbapenem-susceptible GNB (44 CephR-GNB and 3 S-GNB). Of the 57 CR-GNBs, 44 (77.2%) were MHT positive, of which 36 (81.8%) were blaKPC PCR positive. The blaKPC-positive isolates had MIC50s of >4 μg/ml to ertapenem and 8 μg/ml to meropenem by both plate methods. The other carbapenem-resistant Enterobacteriaceae in the sensitivity panel that were negative for blaKPC and blaNDM were also negative for the other less commonly encountered carbapenemases assessed (Table 4).
Table 3.
blaKPC and modified Hodge test status by susceptibility phenotype and culture method in Gram-negative bacilli cultured from blaKPC-positive rectal surveillance swabs
| Testing method and results by GNB group | Data by modified Hodge test results: |
||
|---|---|---|---|
| Positive | Negative | Noninterpretable | |
| CDC method | |||
| Carbapenem-resistant Gram-negative bacilli (n = 58) | |||
| blaKPC PCR positive | 35 | 0 | 0 |
| blaKPC PCR negative | 7 | 14 | 2 |
| 3rd-/4th-generation (oxyimino-cephalosporin)-resistant Gram-negative bacilli (n = 18) | |||
| blaKPC PCR positive | 0 | 0 | 0 |
| blaKPC PCR negative | 2 | 15 | 1 |
| Susceptible Gram-negative bacilli (n = 24) | |||
| blaKPC PCR positive | 0 | 0 | 0 |
| blaKPC PCR negative | 2 | 17 | 5 |
| HardyCHROM ESBL | |||
| Carbapenem-resistant Gram-negative bacilli (n = 57) | |||
| blaKPC PCR positive | 36 | 0 | 0 |
| blaKPC PCR negative | 8 | 10 | 3 |
Table 4.
Characteristics of carbapenem-resistant Enterobacteriaceae isolated from clinical surveillance swabs that were negative for blaKPC and blaNDM
| Isolate no. | Panel/method isolated froma: | Species | Ertapenem/meropenem MIC (μg/ml) | Results by: |
Other resistance mechanisms | |
|---|---|---|---|---|---|---|
| Modified Hodge test | blaKPC/blaNDM PCR | |||||
| 1 | Sensitivity panel, CDC-A | Escherichia coli | >4/>8 | Negative | Negative | Unknown mechanism, ESBL phenotype |
| 2 | Sensitivity panel, HC-A | E. coli | >4/≤1 | Negative | Negative | Unknown mechanism, ESBL phenotype |
| 3 | Sensitivity panel, HC-A | E. coli | 1/≤1 | Inhibited (noninterpretable) | Negative | AmpC |
| 4 | Sensitivity panel, HC-A | Enterobacter cloacae complex | 1/≤1 | Positive (ertapenem disk positive, meropenem disk weak positive) | Negative | Unknown mechanism, ESBL phenotype |
| 5 | Sensitivity panel, HC-A | E. coli | >4/8 | Positive (ertapenem and meropenem disks) | Negative | AmpC |
| 6 | Sensitivity panel, HC-A | E. cloacae complex | 1/≤1 | Positive (ertapenem and meropenem disks) | Negative | AmpC |
| 7 | Specificity panel, HC-A | E. coli | >4/2 | Negative | Negative | AmpC |
CDC-A, CDC method; HC-A, HardyCHROM ESBL.
Of 80 swabs in the specificity panel, one carbapenem-resistant Enterobacteriaceae isolate was cultured from one specimen (by both CDC-A and HC-A). This was an Escherichia coli isolate with a meropenem MIC of 2 μg/ml and an ertapenem MIC of >4 μg/ml. The isolate was positive for AmpC and negative by MHT, blaKPC/blaNDM PCR, and the other less commonly encountered carbapenemases tested. This isolate was designated a non-CPE (Table 4). Thus, in the specificity panel, no CPE were detected by any evaluated method (100% agreement).
Overall, the sensitivities of the culture-based methods studied were inferior to that of PCR: the sensitivity of the CDC-A was 78.3%, versus 100% for PCR (P = 0.0016) and the sensitivity of HC-A was 76.1% versus 100% for PCR (P = 0.0009).
Accuracy studies (spiked specimens).
The sensitivities of blaKPC PCR in spiked surveillance swabs and stool were 93.3% and 100%, respectively. The sensitivity of blaNDM PCR in both spiked surveillance swabs and stool was 100% (data not shown).
DISCUSSION
In this study, we demonstrated that our assay (D-PCR) accurately detected blaKPC and blaNDM directly from perirectal and perianal swabs. We believe that NAAT testing performed directly from surveillance specimens for the detection of CPE represents the most sensitive and straightforward methodology. The majority of the clinical surveillance swabs in our study were not visibly soiled with stool and were able to be processed with a simple lysis procedure, without upfront extraction, which saves money and shortens the turnaround time. Additionally, we found that compared to D-PCR, a broth enrichment step (BE-PCR) did not improve sensitivity or completely eliminate inhibition in swabs that were visibly soiled with stool. Our approach using D-PCR without extraction has a turnaround time of 90 min from swab collection to the results and can be performed for the majority of swab specimens. Nucleic acid extraction should be performed on swabs that are soiled with stool or if testing is performed directly on stool. D-PCR is anticipated to allow for accurate and rapid test results and will be especially helpful in outbreak settings where screening volumes are increased and the timely implementation of infection control measures is paramount. When isolates are required for further characterization, cultures can be performed from a second swab (if double swabs are used) or from the sponge from the swab-holding tube.
The D-PCR approach was more sensitive than the two culture methods we assessed, the widely used CDC-recommended method (CDC-A) and HardyCHROM ESBL agar (HC-A) method. The CDC-A was labor-intensive and had a long turnaround time. Despite the selective broth incubation step with meropenem, a substantial amount of breakthrough growth of carbapenem-susceptible isolates was noted with the CDC-A (42% of isolates cultured by the CDC-A). We chose to use an extended-spectrum β-lactamase (ESBL) selective chromogenic medium (HC-A) for our study, as this detects both ESBLs and CPE and is theoretically more sensitive than carbapenem-resistant Enterobacteriaceae selective media, especially for CPE with low carbapenem MICs. However, the performance of the D-PCR/BE-PCR was superior to that of the HC-A. The HC-A is easy to read because colony color allows for the presumptive identification of an organism as E. coli (pink) or Klebsiella/Enterobacter species (blue), but like in the CDC-A, isolated colonies require further testing (e.g., MHT, antimicrobial susceptibility testing) to determine CPE status, and molecular testing is required for confirmation and characterization.
There were several limitations to our study. First, the prevalence of CPE is low at Mayo Clinic, and so part of this study was performed using spiked specimens. Accuracy studies were performed using swabs previously shown to be culture positive for blaKPC-positive Enterobacteriaceae that were collected in a region where blaKPC is endemic; there was a median time interval of 20 days from when these swabs were collected to when this study was done. We believe that there was minimal loss of viability of bacteria on the swabs and a consequent bias against the culture-based methods studied, as the swabs were stored at 4°C, and all except one (which was negative by all four methods and thus was excluded) yielded viable bacteria (although not always CPE). Additionally, all blaKPC-positive swabs that were negative by the HC-A were detected by the CDC-A (and vice versa), except for five swabs. A second limitation is that blaNDM, while reported, is not endemic in the United States, and as such we were unable to assess the clinical accuracy of detection of this carbapenemase in our clinical surveillance swab panel. However, the accuracy as assessed by spiking studies performed near the limit of detection showed excellent (100%) sensitivity for blaNDM.
Emerging and novel carbapenemases will likely make the detection of CPE a challenge and a “moving goal post,” so it is important to keep abreast of and be able to detect the most prevalent CPE mechanisms in one's area of practice. For example, a recent hospital outbreak of blaNDM was reported in Denver (36) where affected patients had not traveled internationally and it was not clear how blaNDM was introduced. It may be increasingly difficult to differentiate blaNDM and blaKPC carriers by their type of exposure, thus justifying the need for rapid and accurate tests that can differentiate between the carbapenemases. While culture-based methods can be considered truly multiplexed in that various CPE may potentially be detected, we did not detect other more rarely occurring carbapenemases in our study (Table 4). Institutions should screen for the carbapenemases that are most commonly encountered in their areas of practice. In the United States, this would include routine assessment for blaNDM and blaKPC at a minimum, as is recommended by the CDC (4). Isolates suspected of being CPE and that are negative for blaNDM and blaKPC should be referred to public health laboratories for further characterization.
In summary, PCR performed directly on perirectal and perianal surveillance swab specimens after a simple lysis procedure (when swabs are not visibly soiled with stool) provides timely and accurate results. This approach can also be applied to stool and swabs soiled with stool after a nucleic acid extraction step. Where laboratory resources allow, we believe that NAAT testing performed directly on surveillance specimens represents the most sensitive and straightforward screening approach for the detection of CPE.
ACKNOWLEDGMENTS
We thank the technologists in the antimicrobial susceptibility section of the bacteriology laboratory, Mayo Clinic, Rochester, Minnesota, for their assistance with antimicrobial susceptibility testing for this project.
Footnotes
Published ahead of print 21 August 2013
REFERENCES
- 1. Kochar S, Sheard T, Sharma R, Hui A, Tolentino E, Allen G, Landman D, Bratu S, Augenbraun M, Quale J. 2009. Success of an infection control program to reduce the spread of carbapenem-resistant Klebsiella pneumoniae. Infect. Control Hosp. Epidemiol. 30:447–452 [DOI] [PubMed] [Google Scholar]
- 2. Munoz-Price LS, Hayden MK, Lolans K, Won S, Calvert K, Lin M, Stemer A, Weinstein RA. 2010. Successful control of an outbreak of Klebsiella pneumoniae carbapenemase-producing K. pneumoniae at a long-term acute care hospital. Infect. Control Hosp. Epidemiol. 31:341–347 [DOI] [PubMed] [Google Scholar]
- 3. Chitnis AS, Caruthers PS, Rao AK, Lamb J, Lurvey R, Beau De Rochars V, Kitchel B, Cancio M, Török TJ, Guh AY, Gould CV, Wise ME. 2012. Outbreak of carbapenem-resistant Enterobacteriaceae at a long-term acute care hospital: sustained reductions in transmission through active surveillance and targeted interventions. Infect. Control Hosp. Epidemiol. 33:984–992 [DOI] [PubMed] [Google Scholar]
- 4.Centers for Disease Control and Prevention Health Alert Network 2013. CDC health advisory: new carbapenem-resistant enterobacteriaceae warrant additional action by healthcare providers. CDC Health Alert Network, Atlanta, GA: http://www.bt.cdc.gov/HAN/han00341asp [Google Scholar]
- 5. Panagea T, Galani I, Souli M, Adamou P, Antoniadou A, Giamarellou H. 2011. Evaluation of CHROMagar KPC for the detection of carbapenemase-producing Enterobacteriaceae in rectal surveillance cultures. Int. J. Antimicrob. Agents 37:124–128 [DOI] [PubMed] [Google Scholar]
- 6. Adler A, Navon-Venezia S, Moran-Gilad J, Marcos E, Schwartz D, Carmeli Y. 2011. Laboratory and clinical evaluation of screening agar plates for detection of carbapenem-resistant Enterobacteriaceae from surveillance rectal swabs. J. Clin. Microbiol. 49:2239–2242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Girlich D, Poirel L, Nordmann P. 2013. Comparison of the SUPERCARBA, CHROMagar KPC, and Brilliance CRE screening media for detection of Enterobacteriaceae with reduced susceptibility to carbapenems. Diagn. Microbiol. Infect. Dis. 75:214–217 [DOI] [PubMed] [Google Scholar]
- 8. Samra Z, Bahar J, Madar-Shapiro L, Aziz N, Israel S, Bishara J. 2008. Evaluation of CHROMagar KPC for rapid detection of carbapenem-resistant Enterobacteriaceae. J. Clin. Microbiol. 46:3110–3111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lolans K, Calvert K, Won S, Clark J, Hayden MK. 2010. Direct ertapenem disk screening method for identification of KPC-producing Klebsiella pneumoniae and Escherichia coli in surveillance swab specimens. J. Clin. Microbiol. 48:836–841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Centers for Disease Control and Prevention 2008. Laboratory protocol for detection of carbapenem-resistant or carbapenemase-producing, Klebsiella spp. and E. coli from rectal swabs. U.S. Department of Health and Human Services, CDC, Atlanta, GA [Google Scholar]
- 11. Girlich D, Poirel L, Nordmann P. 2012. Value of the modified Hodge test for detection of emerging carbapenemases in Enterobacteriaceae. J. Clin. Microbiol. 50:477–479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Snitkin ES, Zelazny AM, Thomas PJ, Stock F, NISC Comparative Sequencing Program Group. Henderson DK, Palmore TN, Segre JA. 2012. Tracking a hospital outbreak of carbapenem-resistant Klebsiella pneumoniae with whole-genome sequencing. Sci. Transl. Med. 4:148ra116. 10.1126/scitranslmed.3004129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lau A, Stock F, Palmore T, Zelazny A. 2012. Laboratory response to a KPC outbreak at the NIH Clinical Center. Abstr. 112th Gen. Meet. Am. Soc. Microbiol., 16–19 June 2012, San Francisco, CA [Google Scholar]
- 14. Hindiyeh M, Smollen G, Grossman Z, Ram D, Davidson Y, Mileguir F, Vax M, Ben David D, Tal I, Rahav G, Shamiss A, Mendelson E, Keller N. 2008. Rapid detection of blaKPC carbapenemase genes by real-time PCR. J. Clin. Microbiol. 46:2879–2883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Schechner V, Straus-Robinson K, Schwartz D, Pfeffer I, Tarabeia J, Moskovich R, Chmelnitsky I, Schwaber MJ, Carmeli Y, Navon-Venezia S. 2009. Evaluation of PCR-based testing for surveillance of KPC-producing carbapenem-resistant members of the Enterobacteriaceae family. J. Clin. Microbiol. 47:3261–3265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Naas T, Cotellon G, Ergani A, Nordmann P. 2013. Real-time PCR for detection of blaOXA-48 genes from stools. J. Antimicrob. Chemother. 68:101–104 [DOI] [PubMed] [Google Scholar]
- 17. Naas T, Ergani A, Carrer A, Nordmann P. 2011. Real-time PCR for detection of NDM-1 carbapenemase genes from spiked stool samples. Antimicrob. Agents Chemother. 55:4038–4043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mangold KA, Santiano K, Broekman R, Krafft CA, Voss B, Wang V, Hacek DM, Usacheva EA, Thomson RB, Jr, Kaul KL, Peterson LR. 2011. Real-time detection of blaKPC in clinical samples and surveillance specimens. J. Clin. Microbiol. 49:3338–3339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Singh K, Mangold KA, Wyant K, Schora DM, Voss B, Kaul KL, Hayden MK, Chundi V, Peterson LR. 2012. Rectal screening for Klebsiella pneumoniae carbapenemases: comparison of real-time PCR and culture using two selective screening agar plates. J. Clin. Microbiol. 50:2596–2600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Richter SN, Frasson I, Biasolo MA, Bartolini A, Cavallaro A, Palù G. 2012. Ultra-rapid detection of blaKPC1/2-12 from perirectal and nasal swabs by use of real-time PCR. J. Clin. Microbiol. 50:1718–1720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. McEwan AS, Derome A, Meunier D, Burns PJ, Woodford N, Dodgson AR. 2013. Evaluation of the NucliSENS EasyQ KPC assay for detection of Klebsiella pneumoniae carbapenemase-producing Enterobacteriaceae. J. Clin. Microbiol. 51:1948–1950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Avlami A, Bekris S, Ganteris G, Kraniotaki E, Malamou-Lada E, Orfanidou M, Paniara O, Pantazatou A, Papagiannitsis CC, Platsouka E, Stefanou I, Tzelepi E, Vagiakou H, Miriagou V. 2010. Detection of metallo-β-lactamase genes in clinical specimens by a commercial multiplex PCR system. J. Microbiol. Methods 83:185–187 [DOI] [PubMed] [Google Scholar]
- 23. Kaase M, Szabados F, Wassill L, Gatermann SG. 2012. Detection of carbapenemases in Enterobacteriaceae by a commercial multiplex PCR. J. Clin. Microbiol. 50:3115–3118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Nijhuis RHT, Savelkoul PHM, van Zwet AA. 2013. A new real-time PCR for rapid detection of VIM, OXA-48, NDM and KPC carbapenemases in Gram negative bacteria directly from rectal swabs. Abstract P-1846 23rd European Congress of Clinical Microbiology and Infectious Diseases, 27–30 April 2013, Berlin, Germany [Google Scholar]
- 25. Cunningham SA, Noorie T, Meunier D, Woodford N, Patel R. 2013. Rapid and simultaneous detection of genes encoding Klebsiella pneumoniae carbapenemase (blaKPC) and New Delhi metallo-β-lactamase (blaNDM) in Gram-negative bacilli. J. Clin. Microbiol. 51:1269–1271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Cole JM, Schuetz AN, Hill CE, Nolte FS. 2009. Development and evaluation of a real-time PCR assay for detection of Klebsiella pneumoniae carbapenemase genes. J. Clin. Microbiol. 47:322–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Thurlow CJ, Prabaker K, Lin MY, Lolans K, Weinstein RA, Hayden MK, Centers for Disease Control and Prevention Epicenters Program 2013. Anatomic sites of patient colonization and environmental contamination with Klebsiella pneumoniae carbapenemase-producing Enterobacteriaceae at long-term acute care hospitals. Infect. Control Hosp. Epidemiol. 34:56–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Clinical and Laboratory Standards Institute 2013. Performance standards for antimicrobial susceptibility testing; 23rd informational supplement. CLSI M100-S23. Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 29. Hong SS, Kim K, Huh JY, Jung B, Kang MS, Hong SG. 2012. Multiplex PCR for rapid detection of genes encoding class A carbapenemases. Ann. Lab. Med. 32:359–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Poirel L, Naas T, Nicolas D, Collet L, Bellais S, Cavallo JD, Nordmann P. 2000. Characterization of VIM-2, a carbapenem-hydrolyzing metallo-beta-lactamase and its plasmid- and integron-borne gene from a Pseudomonas aeruginosa clinical isolate in France. Antimicrob. Agents Chemother. 44:891–897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Senda K, Arakawa Y, Ichiyama S, Nakashima K, Ito H, Ohsuka S, Shimokata K, Kato N, Ohta M. 1998. PCR detection of metallo-β-lactamase gene (blaIMP) in Gram-negative rods resistant to broad-spectrum β-lactams. J. Clin. Microbiol. 34:2909–2913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Poirel L, Le Thomas I, Naas T, Karim A, Nordmann P. 2000. Biochemical sequence analyses of GES-1, a novel class A extended-spectrum beta-lactamase, and the class 1 integron In52 from Klebsiella pneumoniae. Antimicrob. Agents Chemother. 44:622–632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Poirel L, Héritier C, Tolün V, Nordmann P. 2004. Emergence of oxacillinase-mediated resistance to imipenem in Klebsiella pneumoniae. Antimicrob. Agents Chemother. 48:15–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Pérez-Pérez FJ, Hanson ND. 2002. Detection of plasmid-mediated AmpC beta-lactamase genes in clinical isolates by using multiplex PCR. J. Clin. Microbiol. 40:2153–2162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kaczmarek FM, Dib-Hajj F, Shang W, Gootz TD. 2006. High-level carbapenem resistance in a Klebsiella pneumoniae clinical isolate is due to the combination of blaACT-1 beta-lactamase production, porin OmpK35/36 insertional inactivation, and down-regulation of the phosphate transport porin PhoE. Antimicrob. Agents Chemother. 50:3396–3406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Centers for Disease Control and Prevention 2013. Notes from the field: hospital outbreak of carbapenem-resistant Klebsiella pneumoniae producing New Delhi metallo-beta-lactamase—Denver, Colorado, 2012. MMWR Morb. Mortal. Wkly. Rep. 62:108. [PMC free article] [PubMed] [Google Scholar]