Abstract
The global spread of methicillin-resistant Staphylococcus aureus (MRSA) is a serious problem, particularly in mainland China. In order to better understand the national molecular epidemiology and resistance profiles of hospital-associated MRSA (HA-MRSA) in China, a laboratory-based multicenter surveillance study was conducted. Sixty-nine hospitals in 45 large cities in 27 provinces were involved, and a total of 1,141 HA-MRSA isolates were collected during the 6-month study period in 2011. All MRSA isolates were characterized by multilocus sequence typing (MLST), staphylococcal cassette chromosome mec (SCCmec) typing, spa typing, detection of the Panton-Valentine leukocidin (PVL) locus (lukS-PV and lukF-PV), and antibiogram analysis. ST239-III-t030, ST239-III-t037, and ST5-II-t002 were the predominant HA-MRSA clones (overall prevalence rates, 57.1%, 12.9%, and 8.1%, respectively), although the prevalence rates of these major clones varied markedly in different administrative regions. Of note, 6.6% of the HA-MRSA isolates were found to belong to ST59, which had typical community-associated MRSA (CA-MRSA) features, including carriage of SCCmec type IV or V and PVL and less antimicrobial resistance than other major HA-MRSA clones. Moreover, among 36 MLST sequence types (STs) identified, 15 STs, accounting for 3.5% of total isolates, were novel. A novel ST designated ST2590, which is a single-locus variant of ST5-II-t002, was identified in three hospitals in two large cities, with a total of 17 isolates. To further monitor trends in HA-MRSA prevalence, epidemic clonal shifts, clone emergence, and transmission between community and health care settings, longitudinal national MRSA surveillance is required.
INTRODUCTION
Methicillin-resistant Staphylococcus aureus (MRSA) is an infamous multidrug-resistant organism (MDRO) that can cause life-threatening infections such as septic shock, endocarditis, and severe pneumonia. The spread of MRSA, including hospital-associated MRSA (HA-MRSA) and community-associated MRSA (CA-MRSA), has been a major problem worldwide (1–4). The proportion of MRSA isolates among S. aureus isolates from hospitals is very high in many Asian countries (2, 5, 6). In mainland China, the prevalence of MRSA has reached 50 to 70% of total S. aureus isolates, based on previous laboratory-based surveillance data (4, 6).
Analyzing the genotypic characteristics of MRSA clones is valuable for the understanding of MRSA evolution and dissemination (3, 7, 8). Pulsed-field gel electrophoresis (PFGE) patterns (9), carriage of lukS-PV and lukF-PV (the genes encoding Panton-Valentine leukocidin [PVL]) (8, 10), staphylococcal cassette chromosome mec (SCCmec) typing (11, 12), spa typing (13), and multilocus sequence typing (MLST) (14) have proven useful for monitoring pandemic MRSA clones. Previous studies have documented several major MRSA clones in different geographic regions, e.g., the HA-MRSA clone ST239-III prevails in most Asian countries (2, 3, 5) and the CA-MRSA clone US300 (CMRSA-10) is prevalent in North America (5, 15, 16). In addition, the spread of major HA-MRSA clones to communities, as well as the introduction of CA-MRSA clones into hospitals, has been reported in many countries (2, 5, 17).
Several studies have analyzed the epidemiology of MRSA in China using molecular tools and susceptibility testing (2, 7, 18–20). However, considering China's vast territory, the number of hospitals represented in those studies was limited. In addition, the extent to which CA-MRSA clones have spread into Chinese hospitals remains largely unknown. In order to produce a more comprehensive national description of the molecular epidemiology and resistance profiles of HA-MRSA in China, we characterized 1,141 HA-MRSA isolates collected from a number of geographically dispersed Chinese hospitals.
MATERIALS AND METHODS
Study design.
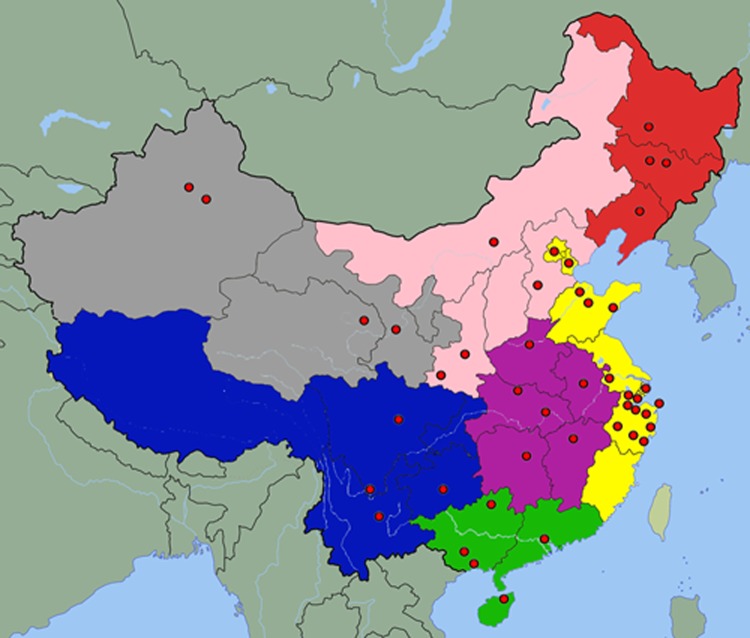
This was a laboratory-based multicenter study of HA-MRSA, involving the voluntary participation of 69 hospitals in 45 large cities located in 27 provinces of mainland China (Fig. 1; see also Table S1 in the supplemental material). The inclusion criteria for these institutions were (i) a general hospital located in a city with a population of over 2 million, (ii) certified as a rank A tertiary hospital by the Ministry of Health of China, and (iii) having >1,000 hospital beds and >0.4 million outpatient visits per year.
Fig 1.
Map of China, its seven administrative regions (i.e., northeast [red], north [pink], east [yellow], middle [purple], south [green], southwest [blue], and northwest [gray]), and the locations of hospitals (cities) participating in the surveillance (red circles).
From January to June 2011, the first 50 nonrepetitive HA-MRSA isolates (or all HA-MRSA isolates if the total number of nonrepetitive S. aureus isolates was less than 50) cultured at each participating hospital were included in the study. HA-MRSA was defined as isolates cultured from patients after 48 hours of admission or from patients who, in the 6 months prior to the culture date, had a history of hospitalization for >24 hours, had undergone dialysis or surgery, or had resided in a long-term health care facility (21).
Isolates and data collection.
All HA-MRSA isolates that met the inclusion criteria were forwarded to the central laboratory (Department of Clinical Laboratory, Peking Union Medical College Hospital) for identification confirmation, susceptibility testing, and genotyping. Isolates were stored at −80°C prior to testing. For each HA-MRSA isolate, clinical data collection was completed by the collecting hospital using a standard electronic report form. The information collected included patient age and gender, ward location of the patient at the time of sample collection, date of sample collection, date of isolation, and body site of the sample.
DNA extraction.
Suspensions of overnight S. aureus cultures on blood agar were incubated with lysostaphin and proteinase K, boiled for 15 min, and then centrifuged (22). The resulting cell lysates were used as the templates for the following PCR methods.
Confirmative identification of MRSA and detection of PVL.
A multiplex PCR was used for simultaneous amplification of 16S rRNA, femA, mecA, and lukS-PV/lukF-PV genes as described previously (10). MRSA reference strain MW2 was used as the positive control (7).
Molecular typing methods.
Three typing methods, i.e., spa typing, MLST, and SCCmec typing, previously proposed for national MRSA surveillance (3, 23) were used in the present study. SCCmec types were determined by a multiplex PCR method that detects types I to V (11, 12); other SCCmec types were listed as nontypeable (NT). MLST and spa typing were performed using standard methods (see http://saureus.mlst.net and http://www.ridom.de/spaserver) (13, 14). Assignment of related sequence types (STs) into clonal complexes (CCs) was conducted using eBURST (24). MRSA clones were named in the format of ST-SCCmec type-spa type-PVL (if detected) (e.g., ST239-III-t030 and ST59-IV-t437-PVL). An international well-characterized panel of MRSA strains (7) was used for quality control.
Antimicrobial susceptibility testing.
Antimicrobial susceptibility profiles were determined in the central laboratory by agar dilution, following CLSI methods presented in document M7-A8 (25). MICs were determined for the following 14 antimicrobial agents: cefoxitin, ceftaroline, ciprofloxacin, levofloxacin, moxifloxacin, erythromycin, clindamycin, trimethoprim-sulfamethoxazole, rifampin, vancomycin, norvancomycin, linezolid, teicoplanin, and tigecycline. S. aureus ATCC 29213 and Enterococcus faecalis ATCC 29212 were used as quality controls. Interpretative criteria were consistent with CLSI document M100-S22 (26) except for norvancomycin, for which vancomycin breakpoints were used, and tigecycline and ceftaroline, for which breakpoints from the U.S. Food and Drug Administration were applied (27, 28).
Submission of novel STs.
Novel alleles in each novel ST were confirmed twice by sequencing in both directions. Fifteen novel STs identified in the present study have been deposited in the MLST database, with assigned numbers of ST2579 to ST2593.
RESULTS
HA-MRSA isolates.
A total of 1,141 HA-MRSA isolates were identified during the study period. The numbers of participating hospitals from the seven administrative regions of China (Fig. 1) (6) and isolates collected in each region were as follows: east, 18 hospitals and 298 isolates; middle, 11 hospitals and 171 isolates; northwest, 10 hospitals and 219 isolates; north, 9 hospitals and 132 isolates; south, 9 hospitals and 101 isolates; northeast, 6 hospitals and 116 isolates; southwest, 6 hospitals and 104 isolates (Table 1; Fig. 1).
Table 1.
Epidemiology of 1,141 MRSA isolates in seven regions in China, including distributions of SCCmec, spa, and MLST types
| Parameter | Value |
|||||||
|---|---|---|---|---|---|---|---|---|
| Total | East | Middle | North | Northeast | Northwest | South | Southwest | |
| No. of hospitals | 69 | 18 | 11 | 9 | 6 | 10 | 9 | 6 |
| No. of MRSA isolates | 1,141 | 298 | 171 | 132 | 116 | 219 | 101 | 104 |
| No. of clones identified | 86 | 43 | 20 | 12 | 11 | 12 | 20 | 11 |
| ST-SCCmec type (%)a | ||||||||
| ST239-III | 74.6 | 55.7 | 79.5 | 84.8 | 62.1 | 92.7 | 68.3 | 89.4 |
| ST5-II | 9.3 | 16.1 | 5.3 | 4.5 | 32.8 | 0.5 | 2.0 | 1.9 |
| ST59-IV | 5.1 | 5.7 | 6.4 | 7.6 | 0.9 | 5.9 | 1.0 | 4.8 |
| Other | 11.0 | 22.5 | 8.8 | 3.0 | 4.3 | 0.9 | 28.7 | 3.8 |
| spa type (%a) | ||||||||
| t030 | 57.5 | 38.9 | 60.8 | 64.4 | 52.6 | 82.6 | 35.6 | 70.2 |
| t037 | 13.1 | 12.4 | 10.5 | 10.6 | 9.5 | 8.7 | 32.7 | 16.3 |
| t002 | 10.1 | 19.5 | 6.4 | 4.5 | 31.0 | 0.5 | 2.0 | 1.0 |
| t437 | 5.5 | 6.0 | 7.6 | 9.1 | 0.9 | 4.1 | 4.0 | 5.8 |
| Other | 13.8 | 23.2 | 14.6 | 11.4 | 6.0 | 4.1 | 25.7 | 6.7 |
| lukS-PV/lukF-PV positive (%) | 5.5 | 5.4 | 7.6 | 9.8 | 0 | 2.7 | 8.9 | 5.8 |
Percentage of each molecular type in the administrative region.
SCCmec types.
Five SCCmec types (types I to V) were identified in 1,114 isolates, and an additional 27 isolates were nontypeable by the SCCmec typing method used. SCCmec type III was the predominant type in all administrative regions, accounting for 75.4% (n = 860) of isolates. This type was less prevalent in east, northeast, and south China (prevalence rates of 57 to 69%) than in other regions (rates of >80%). SCCmec type II was the second most common type, accounting for 12.0% (n = 137) of isolates, and was particularly prevalent in northeast and east China (rates of 32.8% and 25.5%, respectively) but was rare in other regions (rates of 0.5 to 6.4%). SCCmec types IV and V accounted for 8.2% and 1.9% of MRSA isolates, respectively. A single isolate was found to carry SCCmec type I.
MLST.
Overall, 36 STs were identified, representing seven CCs and nine singletons (Table 2). The most common ST identified was ST239. This ST accounted for more than one-half of the isolates in all seven administrative regions, with prevalence rates ranging from 55.7% in east China to 92.7% in northwest China (overall prevalence rate of 74.6%). ST5 was the second most common ST nationwide and was found with greatest frequency in northeast and east China (32.8% and 16.1%, respectively), with frequencies of <10% in other regions. The prevalence of ST59 was 6.6% overall but varied from 0.9% to 9.9% in different administrative regions. By eBURST analysis, CC8 (composed of ST239 and 7 closely related STs) was the most common (75.4%), followed by CC5 (12.6%) and CC59 (7.3%).
Table 2.
Multilocus sequence types identified among 1,141 MRSA isolates
| CC type (n)a | ST | No. of isolates | Allelic profile |
||||||
|---|---|---|---|---|---|---|---|---|---|
| arcC | aroE | glpF | gmk | pta | tpi | yqiL | |||
| CC8 (860) | ST239 | 851 | 2 | 3 | 1 | 1 | 4 | 4 | 3 |
| ST630 | 2 | 12 | 3 | 1 | 1 | 4 | 4 | 3 | |
| ST2589 | 2 | 2 | 3 | 1 | 1 | 4 | 4 | 292 | |
| ST1296 | 1 | 2 | 3 | 1 | 111 | 4 | 4 | 3 | |
| ST2582 | 1 | 2 | 3 | 319 | 1 | 4 | 4 | 3 | |
| ST2586 | 1 | 2 | 3 | 1 | 1 | 282 | 4 | 3 | |
| ST2587 | 1 | 2 | 3 | 1 | 1 | 283 | 4 | 3 | |
| ST2588 | 1 | 2 | 3 | 1 | 1 | 4 | 281 | 3 | |
| CC5 (144) | ST5 | 106 | 1 | 4 | 1 | 4 | 12 | 1 | 10 |
| ST2590 | 17 | 1 | 4 | 1 | 4 | 12 | 1 | 293 | |
| ST764 | 11 | 1 | 136 | 1 | 4 | 12 | 1 | 10 | |
| ST965 | 5 | 1 | 4 | 1 | 4 | 119 | 1 | 10 | |
| ST258 | 3 | 1 | 372 | 1 | 4 | 12 | 1 | 10 | |
| ST2585 | 2 | 1 | 4 | 1 | 4 | 281 | 1 | 10 | |
| CC59 (83) | ST59 | 75 | 19 | 23 | 15 | 2 | 19 | 20 | 15 |
| ST338 | 6 | 19 | 23 | 15 | 48 | 19 | 20 | 15 | |
| ST2579 | 1 | 292 | 23 | 15 | 2 | 19 | 20 | 15 | |
| ST2591 | 1 | 19 | 23 | 15 | 2 | 19 | 20 | 294 | |
| CC1 (11) | ST1 | 8 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| ST2583 | 2 | 1 | 1 | 1 | 197 | 1 | 1 | 1 | |
| ST2592 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 295 | |
| CC9 (11) | ST9 | 10 | 3 | 3 | 1 | 1 | 1 | 1 | 10 |
| ST2593 | 1 | 3 | 3 | 1 | 1 | 1 | 1 | 296 | |
| CC88 (5) | ST88 | 4 | 22 | 1 | 14 | 23 | 12 | 4 | 31 |
| ST2584 | 1 | 22 | 1 | 14 | 23 | 280 | 4 | 31 | |
| CC30 (4) | ST1777 | 3 | 139 | 2 | 2 | 2 | 6 | 3 | 2 |
| ST30 | 1 | 2 | 2 | 2 | 2 | 6 | 3 | 2 | |
| Singletons | ST45 | 7 | 10 | 14 | 8 | 6 | 10 | 3 | 2 |
| ST2580 | 5 | 293 | 2 | 2 | 2 | 6 | 3 | 291 | |
| ST7 | 3 | 5 | 4 | 1 | 4 | 4 | 6 | 3 | |
| ST72 | 3 | 1 | 4 | 1 | 8 | 4 | 4 | 3 | |
| ST6 | 1 | 12 | 4 | 1 | 4 | 12 | 1 | 3 | |
| ST22 | 1 | 7 | 6 | 1 | 5 | 8 | 8 | 6 | |
| ST93 | 1 | 6 | 64 | 44 | 2 | 43 | 55 | 51 | |
| ST398 | 1 | 3 | 35 | 19 | 2 | 20 | 26 | 39 | |
| ST1774 | 1 | 10 | 1 | 1 | 8 | 1 | 1 | 2 | |
CC, clonal complex; ST, sequence type.
Fifteen STs identified in this study were not found in the MLST database (Tables 2 and 3). Fourteen of these novel STs (subsequently designated ST2579 to ST2589 and ST2591 to ST2593) presented rarely, with less than five isolates identified, and each was detected in only a single hospital. ST2590 was an exception, however, with 17 isolates identified in two hospitals in east China and one in middle China. ST2590 differed from ST5 by only one point mutation in the yqiL gene (Table 2); all ST2590 MRSA isolates identified were spa type t002 and SCCmec type II (Table 3).
Table 3.
Characterization of novel STs
| ST (n)a | CC | SCCmec-spa-PVLb | Distribution |
|---|---|---|---|
| ST2579 (1) | CC59 | IV-t437-PVL | North China |
| ST2580 (5) | Singleton | IV-t3351-PVL | South China |
| ST2581 (3) | CC5 | II-t002 | East China |
| ST2582 (1) | CC8 | III-t030 | Southwest China |
| ST2583 (2) | CC1 | IV-t1381-PVL | South China |
| ST2584 (1) | CC88 | NT-t2310 | South China |
| ST2585 (2) | CC5 | NT-t062 | South China |
| ST2586 (1) | CC8 | III-t233 | Middle China |
| ST2587 (1) | CC8 | III-t037 | East China |
| ST2588 (1) | CC8 | III-t030 | Middle China |
| ST2589 (2) | CC8 | III-t030 | Middle China |
| ST2590 (17) | CC5 | II-t002 | East and Middle China |
| ST2591 (1) | CC59 | IV-t163 | East China |
| ST2592 (1) | CC1 | NT-t1381 | South China |
| ST2593 (1) | CC9 | IV-t899 | Middle China |
ST, sequence type; CC, clonal complex; NT, nontypeable.
PVL indicates that the genes encoding Panton-Valentine leukocidin were detected.
spa types.
Typing of 1,141 MRSA isolates yielded 54 spa types in all. Substantial correlations were observed among spa types, SCCmec types, and STs (Table 4). t030 and t037 were the predominant spa types, constituting 57.5% and 13.1% of all isolates, respectively (Table 1). All isolates of these two types carried SCCmec type III. In addition, 99.4% of t030 isolates and 98.7% of t037 isolates belonged to CC8-ST239 (Table 4). Isolates of the third most common spa type, t002, accounted for 10.1% of isolates (Table 1), belonged to CC5, including both ST5 (80.0%) and ST2590 (14.8%), and carried SCCmec type II (100%) (Table 4). Isolates of spa type t437, which carried SCCmec type IV, type V, or NT, made up 5.5% of the collection, 89.9% of which belonged to ST59 (Table 4). Notably, although 15 novel STs were identified, no novel spa types were found among the 1,141 MRSA isolates tested.
Table 4.
Characterization of common HA-MRSA molecular types and their antimicrobial resistance profiles
| spa type (n)a | Predominant ST-CC (%)b | Predominant SCCmec type (%) | PVL (%) | Predominant antimicrobial resistance profile (%)c | % of isolates with resistance to: |
% of isolates with no resistance to C, R, or S | |||
|---|---|---|---|---|---|---|---|---|---|
| CR | CS | C | Other | ||||||
| t030 (656) | ST239-CC8 (99.4) | III (100) | 0 | FCLM(E)(D)R (86.1) | 98.6 | 0.3 | 0.2 | 0.6 | 0.3 |
| t037 (149) | ST239-CC8 (98.7) | III (100) | 0 | FCL(M)EDS (61.7) | 8.1 | 62.4 | 24.2 | 3.4 | 2 |
| t002 (115) | ST5-CC5 (80.0), ST12-CC5 (14.8) | II (100) | 0 | FCL(M)ED (84.3) | 6.1 | 0 | 93.9 | 0 | 0 |
| t437 (63) | ST59-CC59 (88.9) | IV (73.0), V (17.5), NT (9.5) | 71.4 | FED (87.3) | 1.6 | 0 | 6.3 | 4.8 | 87.3 |
| t601 (18) | ST5-CC5 (55.6), ST764-CC5 (44.4) | II (100) | 0 | FCLMED (83.3) | 5.6 | 0 | 94.4 | 0 | 0 |
| t459 (16) | ST239-CC8 (93.8) | III (100) | 0 | FCLMEDR (93.8) | 93.8 | 0 | 0 | 0 | 6.3 |
spa types with a >1% prevalence (or >12 of 1,141 isolates) in the present study are listed.
Percentages indicate the percentage of MRSA isolates belonging to the given type. ST, sequence type; CC, clonal complex; F, cefoxitin; C, ciprofloxacin; L, levofloxacin; M, moxifloxacin; E, erythromycin; D, clindamycin; S, trimethoprim-sulfamethoxazole; R, rifampin.
Antimicrobial agents in parentheses were ignored in calculations of the percentages of isolates with the indicated resistance profile.
Prevalence of PVL.
Overall, PVL was detected in 63 MRSA isolates (5.5%) distributed in six of seven administrative regions in China (all except northeast China). The majority (77.8%) of PVL-positive isolates belonged to CC59 (49 of 63 isolates), including 29 isolates of ST59-IV-t437-PVL and seven of ST59-V-t437-PVL. SCCmec type IV, type V, and NT were found in 65.1%, 23.8%, and 11.1% of PVL-positive isolates, respectively, and no PVL-positive isolate carried SCCmec type I, type II, or type III.
Antimicrobial resistance.
No resistance to linezolid, vancomycin, norvancomycin, or teicoplanin was detected in any of the isolates. Additionally, high rates of susceptibility to tigecycline and trimethoprim-sulfamethoxazole were found (94.7% and 89.3%, respectively). In contrast, the novel cephalosporin ceftaroline had poor activity against MRSA (susceptibility rate, 55.8%). All other antibiotics tested had susceptibility rates of less than 37%.
There was a strong correlation between molecular typing and antimicrobial resistance profiles (Table 4). In particular, resistance to ciprofloxacin, rifampin, or trimethoprim-sulfamethoxazole was characteristic of particular MRSA clones. For instance, the majority of the ST239-III-t030 isolates were cross-resistant to ciprofloxacin and rifampin but not to trimethoprim-sulfamethoxazole (98.6%) (Table 4), while only 8.2% of the ST239-III-t037 isolates exhibited this resistance profile; 62.6% of the isolates were cross-resistant to ciprofloxacin and trimethoprim-sulfamethoxazole but not to rifampin, and 23.8% were resistant to ciprofloxacin only (Table 4). In comparison, ST5-II-t002 isolates were more likely to be resistant to ciprofloxacin (93.5%) (Table 4). The majority (90.9%) of ST59-IV-t437 isolates were susceptible to ciprofloxacin, rifampin, and trimethoprim-sulfamethoxazole (Table 4).
DISCUSSION
Antimicrobial resistance is a global public health crisis, leading to increasing rates of morbidity and death from bacterial infections and posing a serious threat to patient safety in hospitals (5, 6, 29). In China, the crisis of antimicrobial resistance is particularly severe, by virtue of previous unregulated overuse of antimicrobials (6). Surveillance for bacterial resistance is considered essential for understanding the magnitude of the problem of MDROs and for controlling their spread (6, 29). In an effort to control the emergence and spread of MDROs in China, the Ministry of Health of China in 2012 issued an order on the management of antimicrobial use in clinical practice (http://www.gov.cn/flfg/2012-05/08/content_2132174.htm, in Chinese). This order included the establishment of a nationwide antimicrobial resistance surveillance program.
MRSA is considered the most significant MDRO causing infections in mainland China, given its high prevalence rates (2, 4–6), yet the surveillance of MRSA utilizing molecular typing methods was previously limited to a few large hospitals (2, 7, 8, 22). To obtain a more representative picture of the epidemiology of HA-MRSA in China, this laboratory-based surveillance study was initiated in 2011.
We found that ST239-III-t030, ST239-III-t037, and ST5-II-t002 were the most common clones, although the prevalence of these clones varied significantly across administrative regions. ST239-III (also known as the Brazilian or Hungarian clone) was reported previously to be endemic in most Asian countries (2, 3, 7, 30) but is seen less commonly in the United States (31, 32), while ST5-II (the New York/Japan clone) has been documented as a major clone in South Korea and Japan (2, 33, 34). Interestingly, clonal replacement of ST239-III-t037 with ST239-III-t030 was observed in one Chinese hospital between 2000 and 2002, indicating a possible survival advantage of the latter clone (22). Such phenomena emphasize the need for ongoing molecular surveillance.
Importantly, 75 ST59-IV/V/NT (Taiwanese clone) isolates were identified in our HA-MRSA collection, of which over one-half were PVL positive. In addition, the ST59 isolates had less antibiotic resistance than ST239-III and ST5-II isolates. Clones harboring SCCmec type IV or type V and carrying PVL are usually reported in the context of community-onset infections, and ST59 is known to be the most common CA-MRSA clone among Chinese children (19, 35). In Hong Kong and Taiwan, CA-MRSA clones including ST59 have spread into hospital settings (2). Our findings further suggest that hospitals in mainland China are facing the same situation.
Another important finding from this study is that isolates belonging to novel STs contributed a small but significant proportion of HA-MRSA cases, indicating that HA-MRSA epidemiology in China continues to evolve. Moreover, one of these novel types (ST2590) has disseminated in three hospitals in two administrative regions. This ST has not been previously documented in Chinese studies of MRSA, including a recent study in Shanghai (36). It is likely that ST2590 evolved from ST5 by a single point mutation in yqiL, as these two sequence types share the same genetic background (spa type t002, SCCmec type II, PVL negative).
Nationwide MRSA epidemiological surveillance utilizing multiple typing methods (MLST, spa typing, SCCmec typing, and detection of PVL) is expensive, time-consuming, and labor-intensive. However, incorporation of these four typing methods in our study proved complementary. For instance, none of the MRSA isolates with novel STs in the present study carried novel spa types, although spa typing is generally reported to have higher discriminatory power and lower cost than MLST (3, 9, 23). To best track the changing epidemiology of MRSA clones across China's vast territory, ongoing surveillance with a combination of typing methods, in addition to antimicrobial resistance profiling, will be necessary.
In summary, our study observed that the major HA-MRSA clones in China are ST239-III-t030, ST239-III-t037, and ST5-II-t002, with varying distributions among administrative regions. In addition, the previously described CA-MRSA clone ST59-IV/V/NT(±PVL) has spread in hospitals in China. Fifteen novel STs accounted for a significant proportion of HA-MRSA isolates, including one (ST2590) found to be circulating in three hospitals in two large cities. We recommend ongoing monitoring to demonstrate trends in HA-MRSA prevalence, epidemic clonal shifts, new clone emergence, and transmission of MRSA clones between community and health care settings, using longitudinal nationwide MRSA surveillance.
Supplementary Material
ACKNOWLEDGMENTS
We thank Timothy Kudinha (Charles Sturt University, Orange, Australia) for his critical reading of the paper. We also thank the staff at all of the hospitals that participated in the study.
The present study is supported by the Research Special Fund for Public Welfare Industry of Health, Research and Application for the Prevention and Control of Nosocomial Infections Caused by Multi-Drug Resistant Bacteria (grant 201002021), and National Science and Technology Specific Projects, Clinical Specimen Resource Bank, a Sub-subject of Essential Drug Research and Development (grant 2011ZX09307-001).
We have no conflicts of interest to declare.
Footnotes
Published ahead of print 28 August 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JCM.01375-13.
REFERENCES
- 1. Klein EY, Sun L, Smith DL, Laxminarayan R. 2013. The changing epidemiology of methicillin-resistant Staphylococcus aureus in the United States: a national observational study. Am. J. Epidemiol. 177:666–674 [DOI] [PubMed] [Google Scholar]
- 2. Song JH, Hsueh PR, Chung DR, Ko KS, Kang CI, Peck KR, Yeom JS, Kim SW, Chang HH, Kim YS, Jung SI, Son JS, So TM, Lalitha MK, Yang Y, Huang SG, Wang H, Lu Q, Carlos CC, Perera JA, Chiu CH, Liu JW, Chongthaleong A, Thamlikitkul V, Van PH. 2011. Spread of methicillin-resistant Staphylococcus aureus between the community and the hospitals in Asian countries: an ANSORP study. J. Antimicrob. Chemother. 66:1061–1069 [DOI] [PubMed] [Google Scholar]
- 3. Stefani S, Chung DR, Lindsay JA, Friedrich AW, Kearns AM, Westh H, Mackenzie FM. 2012. Meticillin-resistant Staphylococcus aureus (MRSA): global epidemiology and harmonisation of typing methods. Int. J. Antimicrob. Agents 39:273–282 [DOI] [PubMed] [Google Scholar]
- 4. Zhao C, Sun H, Wang H, Liu Y, Hu B, Yu Y, Sun Z, Chu Y, Cao B, Liao K, Lei J, Hu Z, Zhang L, Zhang X, Xu Y, Wang Z, Chen M. 2012. Antimicrobial resistance trends among 5608 clinical Gram-positive isolates in China: results from the Gram-Positive Cocci Resistance Surveillance program (2005-2010). Diagn. Microbiol. Infect. Dis. 73:174–181 [DOI] [PubMed] [Google Scholar]
- 5. Molton JS, Tambyah PA, Ang BS, Ling ML, Fisher DA. 2013. The global spread of healthcare-associated multidrug-resistant bacteria: a perspective from Asia. Clin. Infect. Dis. 56:1310–1318 [DOI] [PubMed] [Google Scholar]
- 6. Xiao YH, Giske CG, Wei ZQ, Shen P, Heddini A, Li LJ. 2011. Epidemiology and characteristics of antimicrobial resistance in China. Drug Resist. Updat. 14:236–250 [DOI] [PubMed] [Google Scholar]
- 7. Liu Y, Wang H, Du N, Shen E, Chen H, Niu J, Ye H, Chen M. 2009. Molecular evidence for spread of two major methicillin-resistant Staphylococcus aureus clones with a unique geographic distribution in Chinese hospitals. Antimicrob. Agents Chemother. 53:512–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Yu F, Li T, Huang X, Xie J, Xu Y, Tu J, Qin Z, Parsons C, Wang J, Hu L, Wang L. 2012. Virulence gene profiling and molecular characterization of hospital-acquired Staphylococcus aureus isolates associated with bloodstream infection. Diagn. Microbiol. Infect. Dis. 74:363–368 [DOI] [PubMed] [Google Scholar]
- 9. Hallin M, Deplano A, Denis O, De Mendonca R, De Ryck R, Struelens MJ. 2007. Validation of pulsed-field gel electrophoresis and spa typing for long-term, nationwide epidemiological surveillance studies of Staphylococcus aureus infections. J. Clin. Microbiol. 45:127–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Al-Talib H, Yean CY, Al-Khateeb A, Hassan H, Singh KK, Al-Jashamy K, Ravichandran M. 2009. A pentaplex PCR assay for the rapid detection of methicillin-resistant Staphylococcus aureus and Panton-Valentine leucocidin. BMC Microbiol. 9:113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Boye K, Bartels MD, Andersen IS, Moller JA, Westh H. 2007. A new multiplex PCR for easy screening of methicillin-resistant Staphylococcus aureus SCCmec types I–V. Clin. Microbiol. Infect. 13:725–727 [DOI] [PubMed] [Google Scholar]
- 12. Zhang K, McClure JA, Elsayed S, Louie T, Conly JM. 2005. Novel multiplex PCR assay for characterization and concomitant subtyping of staphylococcal cassette chromosome mec types I to V in methicillin-resistant Staphylococcus aureus. J. Clin. Microbiol. 43:5026–5033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Koreen L, Ramaswamy SV, Graviss EA, Naidich S, Musser JM, Kreiswirth BN. 2004. spa typing method for discriminating among Staphylococcus aureus isolates: implications for use of a single marker to detect genetic micro- and macrovariation. J. Clin. Microbiol. 42:792–799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Enright MC, Day NP, Davies CE, Peacock SJ, Spratt BG. 2000. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J. Clin. Microbiol. 38:1008–1015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Simor AE, Gilbert NL, Gravel D, Mulvey MR, Bryce E, Loeb M, Matlow A, McGeer A, Louie L, Campbell J. 2010. Methicillin-resistant Staphylococcus aureus colonization or infection in Canada: national surveillance and changing epidemiology, 1995-2007. Infect. Control Hosp. Epidemiol. 31:348–356 [DOI] [PubMed] [Google Scholar]
- 16. Tenover FC, Goering RV. 2009. Methicillin-resistant Staphylococcus aureus strain USA300: origin and epidemiology. J. Antimicrob. Chemother. 64:441–446 [DOI] [PubMed] [Google Scholar]
- 17. Hetem DJ, Westh H, Boye K, Jarlov JO, Bonten MJ, Bootsma MC. 2012. Nosocomial transmission of community-associated methicillin-resistant Staphylococcus aureus in Danish hospitals. J. Antimicrob. Chemother. 67:1775–1780 [DOI] [PubMed] [Google Scholar]
- 18. Chu H, Zhao L, Zhang Z, Gui T, Han L, Ni Y. 2013. Antibiotic resistance and molecular epidemiology of methicillin-resistant Staphylococcus aureus from lower respiratory tract: multi-resistance and high prevalence of SCCmec III type. Cell Biochem. Biophys. [Epub ahead of print.] 10.1007/s12013-013-9542-7 [DOI] [PubMed] [Google Scholar]
- 19. Geng W, Yang Y, Wu D, Zhang W, Wang C, Shang Y, Zheng Y, Deng L, Fu Z, Li X, Yu S, Shen X. 2010. Community-acquired, methicillin-resistant Staphylococcus aureus isolated from children with community-onset pneumonia in China. Pediatr. Pulmonol. 45:387–394 [DOI] [PubMed] [Google Scholar]
- 20. Zhao C, Liu Y, Zhao M, Yu Y, Chen H, Sun Q, Jiang W, Han S, Xu Y, Chen M, Cao B, Wang H. 2012. Characterization of community acquired Staphylococcus aureus associated with skin and soft tissue infection in Beijing: high prevalence of PVL+ ST398. PLoS One 7:e38577. 10.1371/journal.pone.0038577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wang SH, Sun ZL, Guo YJ, Yang BQ, Yuan Y, Wei Q, Ye KP. 2010. Meticillin-resistant Staphylococcus aureus isolated from foot ulcers in diabetic patients in a Chinese care hospital: risk factors for infection and prevalence. J. Med. Microbiol. 59:1219–1224 [DOI] [PubMed] [Google Scholar]
- 22. Chen H, Liu Y, Jiang X, Chen M, Wang H. 2010. Rapid change of methicillin-resistant Staphylococcus aureus clones in a Chinese tertiary care hospital over a 15-year period. Antimicrob. Agents Chemother. 54:1842–1847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. David MZ, Taylor A, Lynfield R, Boxrud DJ, Short G, Zychowski D, Boyle-Vavra S, Daum RS. 2013. Comparing pulsed-field gel electrophoresis with multilocus sequence typing, spa typing, staphylococcal cassette chromosome mec (SCCmec) typing, and PCR for Panton-Valentine leukocidin, arcA, and opp3 in methicillin-resistant Staphylococcus aureus isolates at a U.S. medical center. J. Clin. Microbiol. 51:814–819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Feil EJ, Li BC, Aanensen DM, Hanage WP, Spratt BG. 2004. eBURST: inferring patterns of evolutionary descent among clusters of related bacterial genotypes from multilocus sequence typing data. J. Bacteriol. 186:1518–1530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Clinical and Laboratory Standards Institute 2009. Antimicrobial susceptibility test for bacteria that grow aerobically—eighth edition. Approved standard M7-A8. Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 26.Clinical and Laboratory Standards Institute 2012. Performance standards for antimicrobial susceptibility testing. Twenty-second informational supplement M100-S22. Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 27. Kronvall G, Karlsson I, Walder M, Sorberg M, Nilsson LE. 2006. Epidemiological MIC cut-off values for tigecycline calculated from Etest MIC values using normalized resistance interpretation. J. Antimicrob. Chemother. 57:498–505 [DOI] [PubMed] [Google Scholar]
- 28. Zhang H, Xiao M, Yang QW, Wang Y, Wang H, Zhao Y, Brown M, Zhao HR, Kong F, Xu YC. 2013. High ceftaroline non-susceptibility in Staphylococcus aureus isolated from acute skin infections in 15 tertiary hospitals in China. J. Med. Microbiol. 62:496–497 [DOI] [PubMed] [Google Scholar]
- 29.WHO 2001. WHO global strategy for containment of antimicrobial resistance. WHO, Geneva, Switzerland [Google Scholar]
- 30. Harris SR, Feil EJ, Holden MT, Quail MA, Nickerson EK, Chantratita N, Gardete S, Tavares A, Day N, Lindsay JA, Edgeworth JD, de Lencastre H, Parkhill J, Peacock SJ, Bentley SD. 2010. Evolution of MRSA during hospital transmission and intercontinental spread. Science 327:469–474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Tenover FC, Tickler IA, Goering RV, Kreiswirth BN, Mediavilla JR, Persing DH. 2012. Characterization of nasal and blood culture isolates of methicillin-resistant Staphylococcus aureus from patients in United States hospitals. Antimicrob. Agents Chemother. 56:1324–1330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wang SH, Khan Y, Hines L, Mediavilla JR, Zhang L, Chen L, Hoet A, Bannerman T, Pancholi P, Robinson DA, Kreiswirth BN, Stevenson KB. 2012. Methicillin-resistant Staphylococcus aureus sequence type 239-III, Ohio, USA, 2007-2009. Emerg. Infect. Dis. 18:1557–1565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Chongtrakool P, Ito T, Ma XX, Kondo Y, Trakulsomboon S, Tiensasitorn C, Jamklang M, Chavalit T, Song JH, Hiramatsu K. 2006. Staphylococcal cassette chromosome mec (SCCmec) typing of methicillin-resistant Staphylococcus aureus strains isolated in 11 Asian countries: a proposal for a new nomenclature for SCCmec elements. Antimicrob. Agents Chemother. 50:1001–1012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kwon JC, Kim SH, Park SH, Choi SM, Lee DG, Choi JH, Park C, Shin NY, Yoo JH. 2011. Molecular epidemiologic analysis of methicillin-resistant Staphylococcus aureus isolates from bacteremia and nasal colonization at 10 intensive care units: multicenter prospective study in Korea. J. Korean Med. Sci. 26:604–611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wang L, Liu Y, Yang Y, Huang G, Wang C, Deng L, Zheng Y, Fu Z, Li C, Shang Y, Zhao C, Sun M, Li X, Yu S, Yao K, Shen X. 2012. Multidrug-resistant clones of community-associated meticillin-resistant Staphylococcus aureus isolated from Chinese children and the resistance genes to clindamycin and mupirocin. J. Med. Microbiol. 61:1240–1247 [DOI] [PubMed] [Google Scholar]
- 36. Liu Q, Han L, Li B, Sun J, Ni Y. 2012. Virulence characteristic and MLST-agr genetic background of high-level mupirocin-resistant, MRSA isolates from Shanghai and Wenzhou, China. PLoS One 7:e37005. 10.1371/journal.pone.0037005 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.