Abstract
Mycobacterium bovis populations in countries with persistent bovine tuberculosis usually show a prevalent spoligotype with a wide geographical distribution. This study applied mycobacterial interspersed repetitive-unit–variable-number tandem-repeat (MIRU-VNTR) typing to a random panel of 115 M. bovis isolates that are representative of the most frequent spoligotype in the Iberian Peninsula, SB0121. VNTR typing targeted nine loci: ETR-A (alias VNTR2165), ETR-B (VNTR2461), ETR-D (MIRU4, VNTR580), ETR-E (MIRU31, VNTR3192), MIRU26 (VNTR2996), QUB11a (VNTR2163a), QUB11b (VNTR2163b), QUB26 (VNTR4052), and QUB3232 (VNTR3232). We found a high degree of diversity among the studied isolates (discriminatory index [D] = 0.9856), which were split into 65 different MIRU-VNTR types. An alternative short-format MIRU-VNTR typing targeting only the four loci with the highest variability values was found to offer an equivalent discriminatory index. Minimum spanning trees using the MIRU-VNTR data showed the hypothetical evolution of an apparent clonal group. MIRU-VNTR analysis was also applied to the isolates of 176 animals from 15 farms infected by M. bovis SB0121; in 10 farms, the analysis revealed the coexistence of two to five different MIRU types differing in one to six loci, which highlights the frequency of undetected heterogeneity.
INTRODUCTION
Bovine tuberculosis (Mycobacterium bovis) remains a concern in Spain; although herd prevalence has been considerably reduced since the systematic implementation of the national eradication program in the 1990s, infection still affected 1.33% of herds in 2011 (per the Ministry of the Environment and Rural and Marine Affairs). In consideration of the economic and public health implications of this zoonosis, tracing the source of infection by genotyping has become a key element within the program. The most widely used technique to type M. bovis isolates is direct variable repeat (DVR) spoligotyping (1). However, in some geographical settings, this cost-effective high-throughput technique does not offer adequate discrimination (2, 3, 4) and is thus complemented with mycobacterial interspersed repetitive-unit–variable-number tandem-repeat (MIRU-VNTR) typing (5, 6). The MIRU-VNTR technique has been evaluated in many countries using different sets of makers (2, 4, 7–15).
In recent surveys in the Iberian Peninsula, SB0121 was found to be the most frequent spoligotype of M. bovis (26.3% in Portugal [16] and 27.9% in Spain [17]). These studies showed that high levels of discrimination could be achieved by spoligotyping (discriminatory index [D] = 0.89 and 0.87 in Portugal and Spain, respectively), although subsequent studies from Portugal found that discrimination actually increased with MIRU-VNTR typing (12, 13).
In this study, we used nine MIRU-VNTR markers to type a panel of randomly selected Spanish M. bovis isolates with the spoligotype SB0121. The allelic diversity and overall discriminatory power of the technique were evaluated, using several different formats. Additionally, MIRU-VNTR was applied to all the infected animals from farms infected with M. bovis SB0121 to evaluate if heterogeneity was underdetected.
MATERIALS AND METHODS
Mycobacterial strains.
The study was carried out in M. bovis isolates with spoligotype SB0121 that were sampled from across Spain between 1997 and 2010. To evaluate the discriminatory power of MIRU-VNTR, we used a panel (n = 115) (see Table S1 in the supplemental material) comprising 111 isolates selected randomly from 3,863 M. bovis SB0121 isolates from the Spanish strain collection at the VISAVET Health Surveillance Centre affecting animals plus four additionally selected M. bovis isolates from less frequently affected species. The isolates originated from cattle (Bos taurus) (n = 98), goats (Capra aegagrus hircus) (n = 2), wild boars (Sus scrofa) (n = 7), red deer (Cervus elaphus) (n = 2), fallow deer (Dama dama) (n = 2), badgers (Meles meles) (n = 2 [18]), and domestic pigs (Sus scrofa domestica) (n = 2).
To evaluate potential heterogeneity within the farms infected by M. bovis SB0121, we randomly selected 15 farms from 1,342 cattle farms from which isolates were included in the strain collection to design a second panel that comprised all the SB0121 isolates obtained from infected animals from these farms (total n = 176; number of isolates per farm, 2 to 31).
Genotyping methods. (i) Spoligotyping.
Spoligotyping was performed following the protocol described by Kamerbeek et al. (1), and authoritative names (written as the prefix SB followed by four digits) were assigned according to the Mycobacterium bovis Spoligotype Database (see http://www.Mbovis.org) (19).
(ii) Variable-number tandem-repeat typing: analysis of discriminatory power.
For the first panel, VNTR typing was carried out targeting nine VNTR loci: ETR-A (alias VNTR2165), ETR-B (VNTR2461), ETR-D (MIRU4, VNTR580), ETR-E (MIRU31, VNTR3192), MIRU26 (VNTR2996), QUB11a (VNTR2163a), QUB11b (VNTR2163b), QUB26 (VNTR4052), and QUB3232 (VNTR3232) (5, 8, 20–22). Six of these markers (ETR-A, ETR-B, ETR-D, QUB11a, QUB11b, and QUB3232) were recommended for typing M. bovis by the Veterinary Network of Laboratories Researching into Improved Diagnosis and Epidemiology of Mycobacterial Diseases (VENoMYC) Consortium (EU Coordination Action SSPE-CT-2004-501903 [23]), and the other three loci were previously used for typing Spanish strains (ETR-E [24, 25], MIRU26 [24], and QUB26 [9]). PCR was performed using the HotStarTaq DNA polymerase kit (Qiagen) in a Bio-Rad MyCycler thermal cycler. The reaction mixtures without mycobacterial DNA were used as a negative control and Mycobacterium bovis BCG strain Danish (CCUG 27863) (Culture Collection, University of Gothenburg, Sweden) as a positive control. The positive control was sequenced at each of the nine loci using the Applied Biosystems ABI Prism 3730 DNA sequencer (Secugen S.L. and CIB sequencing facilities, Madrid, Spain), and a correlating allele calling table was built according to previous publications (5, 6, 21, 26). The number of tandem repeats (alleles) was estimated after electrophoresis on 2.5% agarose gels at 45 V during 5 h with a 100-bp ladder (Biotools B&M Labs, Madrid, Spain) according to the allele calling table.
(iii) Variable-number tandem-repeat typing: analysis of farms with heterogenous infections.
For the second panel, we applied a two-step analysis scheme to first screen for and then confirm heterogeneity within each farm. First, we compared the mobility patterns in agarose electrophoresis (2% MS-8; Pronadisa, Madrid, Spain) at 45 V for 18.5 h of the amplification products obtained with two triplex-PCRs, including the most discriminatory loci (mix 1, ETR-A, ETR-B, and QUB11a, and mix 2, MIRU26, QUB26, and QUB3232). The final reaction mixture (50 μl) included 25 μl of PCR master mix (Qiagen multiplex PCR kit), 5 μl or 7 μl (mixes including QUB3232) of Q solution (Qiagen multiplex PCR kit), and 0.25 μM each oligonucleotide. The thermocycling conditions applied were 15 min at 95°C followed by 35 cycles of denaturation at 95°C for 50 s, annealing at 57°C for 60 s, extension at 72°C for 1.50 min, and a final extension for 7 min at 72°C.
Subsequently, isolates from the same farm showing different mobility patterns in the screening step were reanalyzed to assign precise allelic values in order to obtain the complete nine-loci MIRU-VNTR type. Next, multiplex PCR was performed, including the use of unlabeled and labeled primers, and the PCR products were analyzed by capillary electrophoresis using an ABI Prism 3100 genetic analyzer (Applied Biosystems, NL Lab Centraal B.V., Haarlem, The Netherlands). In certain cases, some loci that did not amplify in the multiplex PCR were assigned by simplex PCR. When a unique genotype was found in only one infected animal in a farm, the PCR and allele calling were repeated to confirm the result.
Data analysis.
A MIRU-VNTR (MV) type number was assigned for isolates with a complete typing profile, while profiles with nonamplifiable loci or loci presenting double bands were excluded. Isolates were named based on the results obtained by the MIRU-VNTR profile so that MV type “n” with the four-loci approach could be further discriminated into MV types “nA,” “nB,” etc., when applying the six- and nine-loci approaches (see Table S1 in the supplemental material). The index of discrimination (D) (27, 28) was calculated to determine the overall discriminatory power of the MIRU-VNTR typing technique, as well as the individual allelic diversity of the nine loci. We used the in silico website of the University of the Basque Country (see http://insilico.ehu.es/), filling in the number of unrelated isolates with each MIRU-VNTR type or allele, respectively. The MIRU-VNTRplus website (http://www.miru-vntrplus.org) was used for the construction of the minimum spanning trees (29, 30); the loci QUB11a and QUB3232 are not included in the standardized data sets of MIRU-VNTRplus and were therefore arbitrarily assigned to database fields of standard loci. Confidence intervals (CI) were calculated using the NCSS8 statistical software (NCSS, LLC, Kaysville, UT, USA).
RESULTS
Discriminatory power of MIRU-VNTR typing.
MIRU-VNTR typing using nine loci (ETR-A, ETR-B, ETR-D, ETR-E, MIRU26, QUB11a, QUB11b, QUB26, and QUB3232) split the 115 M. bovis isolates with spoligotype SB0121 into 65 different MV types (see Table S1 in the supplemental material). Twelve isolates were not assigned an MV type because a complete pattern could not be obtained due to either a lack of amplification at one or more loci and/or by the presence of double alleles at one or two loci. The loci that most often presented amplification problems were ETR-E and QUB-11a (Table 1).
Table 1.
Allelic diversity of the individual variable-number tandem-repeat loci and their different combinations in the panel of 115 Mycobacterium bovis isolates
| MIRU-VNTR typing analysis | No. of isolates with MIRU-VNTR allele no.: |
Dd | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | Nb | DBc | ||
| Individual MIRU-VNTR locus (alias) | ||||||||||||||||||
| QUB3232 (3232) | 2 | 6 | 17 | 32a | 30 | 7 | 11 | 5 | 1 | 1 | 2 | 1 | 0 | 0 | 0.8242 | |||
| ETR-A (2165) | 1 | 3 | 10 | 49 | 46a | 1 | 1 | 1 | 1 | 2 | 0.6438 | |||||||
| ETR-B (2461) | 9 | 17 | 76a | 11 | 1 | 0 | 1 | 0.5336 | ||||||||||
| QUB11a (2163a) | 1 | 2 | 1 | 10 | 1 | 6 | 83a | 1 | 3 | 1 | 1 | 5 | 0 | 0.4424 | ||||
| QUB26 (4052) | 2 | 6 | 11 | 89a | 2 | 1 | 2 | 1 | 0.3471 | |||||||||
| MIRU26 (2996) | 6 | 6 | 92a | 8 | 3 | 0 | 0.3450 | |||||||||||
| ETR-D (580) | 6 | 104a | 5 | 0 | 0 | 0.2038 | ||||||||||||
| ETR-E (3192) | 4 | 106a | 5 | 0 | 0.0807 | |||||||||||||
| QUB11b (2163b) | 3 | 109a | 1 | 2 | 0 | 0.0790 | ||||||||||||
| Combined MIRU-VNTR approaches | ||||||||||||||||||
| Four locie | 6 | 0.9689 | ||||||||||||||||
| Six locif | 8 | 0.9826 | ||||||||||||||||
| Nine locig | 12 | 0.9856 | ||||||||||||||||
Number of isolates at the allele that correspond to the most frequent MIRU-VNTR type.
N, number of isolates not amplifiable and therefore of undetermined MIRU-VNTR type.
DB, number of isolates with double bands.
D, discriminatory index.
Combination of QUB3232, ETR-A, ETR-B, and QUB11a.
Four-loci analysis plus QUB26 and MIRU26.
Six-loci analysis plus ETR-D, ETR-E, and QUB11b.
The largest cluster was MV type 1A (9 isolates), followed by MV type 3 (6 isolates), MV type 2A (5 isolates), MV types 5A and 7A (four isolates each), and MV types 4A and 6A (3 isolates each); 11 MV types clustered two isolates, and 47 types were unique (see Table S1 in the supplemental material).
We observed the following allelic diversities for the different loci (in order of decreasing D value; see Table 1): QUB3232 (D = 0.82), ETR-A (D = 0.64), ETR-B (D = 0.53), QUB11a (D = 0.44), QUB26 (D = 0.35), MIRU26 (D = 0.35), ETR-D (D = 0.20), ETR-E (D = 0.08), and QUB11b (D = 0.08). The overall discriminatory index of the MIRU-VNTR typing technique using these nine loci was 0.9856.
To evaluate whether a more cost-effective typing format would work with similar discriminatory power, we analyzed the effect of reducing the number of markers for the analysis. First, we selected the six most discriminatory loci (QUB3232, ETR-A, ETR-B, QUB11a, QUB26, and MIRU26) that resulted in 65 MV types and a discriminatory index of 0.9826. Compared with the nine-loci approach, this panel did not distinguish between MV types 5A and 5B, 8A and 8B (both presenting a change at ETR-D), and 1D and 1E (due to variation at QUB11b); however, three MV types could be assigned that were undetermined using the nine-loci approach because of the repeated failure to amplify at ETR-E or QUB11b in that analysis. Second, using only the four most discriminatory loci (QUB3232, ETR-A, ETR-B, and QUB11a), the isolates clustered in 52 MV types, achieving a discrimination index of 0.9689. In this case, the largest cluster included MV type 1 (16 isolates), followed by MV type 2 (8 isolates), MV types 3 and 4 (6 isolates each), MV types 5, 6, and 7 (five isolates each), and MV type 8 (3 isolates); 11 MV types had two isolates, 33 types were unique, and six isolates could not be assigned to an MV type using this approach.
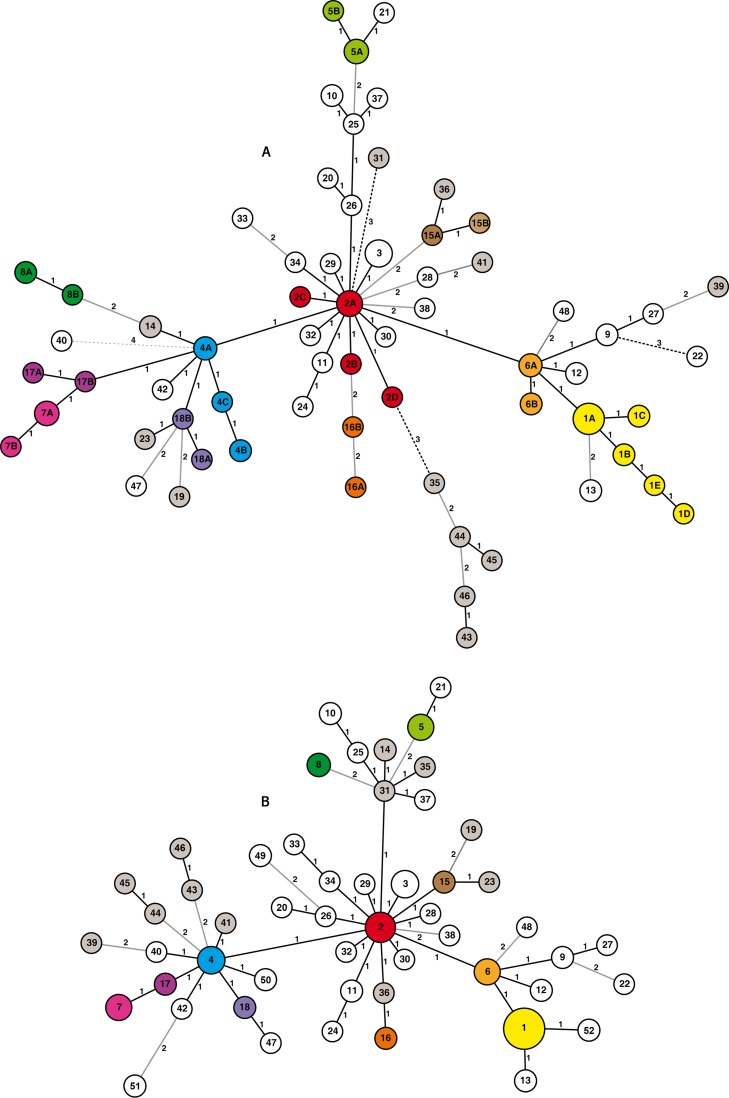
The panel also included a small group of isolates from animal species other than cattle. These isolates clustered either in the largest groups of the MV types or were related to MV types found in cattle. Even with the nine-loci approach, the isolates from cattle and other species seem to be identical (wild boar and cattle MV type 3, goat and cattle isolates MV type 4B, red deer and fallow deer MV type 9) or closely related by variation in a single locus (for example, wild boar MV type 1E and cattle MV types 1B and 1D, wild boar MV type 18B and cattle MV types 4A, 18A, and 23) (Fig. 1).
Fig 1.
Minimum spanning trees (MSTs) of the 115 Mycobacterium bovis isolates with the 65 MIRU-VNTR types obtained with the nine-loci approach (A) and the 51 MIRU-VNTR types obtained with the four-loci approach (B). The MSTs were created using the online application MIRU-VNTRplus (30). The colored nodes show MIRU-VNTR types that were subdivided when the number of markers was increased; MIRU-VNTR types that clustered at different central nodes when reducing the set of markers are shown in gray.
Minimum spanning trees.
The construction of minimum spanning trees for the three different combinations of loci (nine loci [Fig. 1], six loci [not shown], and four loci [Fig. 1]) revealed that most of the MIRU-VNTR types are closely related. Despite the high diversity, most of the genotypes (57 MV types [87.69%]), including 95 (92.23%) of the typeable isolates (95% confidence interval [CI], 0.8527 to 0.9659) belonged to the same clonal complex when applying the default setting where the maximum difference allowed within a group was two loci (double locus variant [DLV]). The eight genotypes excluded were MV types 22 and 31, the cluster of MV type 35 and its relatives MV types 43, 44, 45, and 46 (differences at three loci), and MV type 40 (differences at four loci); these genotypes included one isolate each. Using a less stringent criterion, i.e., allowing triple locus variants within a clonal complex, all MV types (except from MV type 40, the singleton with variations at four loci) including 102 (99.01%) of the typeable isolates (95% CI, 0.9471 to 0.9998) cluster in a single clonal group. The minimum spanning tree for the 6-loci approach when choosing the default setting also clustered the typeable isolates in two clonal complexes, with the bigger one clustering 60 MV types, including 102 (95.33%) of the isolates (95% CI, 0.8943 to 0.9847); no singletons were found. The analysis of the panel with only four loci considering DLVs as members of the same clonal complex grouped all isolates in a single group. Furthermore, most of the genotypes (41 MV types, 78.84%), clustering 91 (83.49%) of the typeable isolates (95% CI, 0.7516 to 0.8991), showed a single locus variation, while only 11 MV types that included 18 isolates (95% CI, 0.1009 to 0.2483) were DLVs.
Study of heterogeneity within farms.
The high discriminatory power observed by the MIRU-VNTR analysis of the SB0121 isolates encouraged us to evaluate whether the systematic application of MIRU-VNTR could reveal infections caused by more than one strain in farms that previously had been considered to be homogeneously infected by SB0121. We therefore evaluated a panel consisting of multiple isolates from the same farm (15 premises, n = 176). Only five out of the 15 farms were confirmed to be truly homogenous (Table 2); these farms comprised six to 31 isolates each, obtained over a time span of up to 5 years. In the remaining 10 farms, with four to 30 isolates each, the MIRU-VNTR typing scheme revealed a marked heterogeneity, identifying two to five different MV types per farm.
Table 2.
Systematic MIRU-VNTR typing of all Mycobacterium bovis SB0121 isolates (n = 176) from 15 farmsa
| Farm no. | No. of alleles at MIRU-VNTR locus (VNTR alias)b |
No. of isolates | Nature of outbreak (no. of types) | Other spoligotype(s) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| QUB3232 short (3232) | ETR-A (2165) | ETR-B (2461) | QUB11a (2163a) | QUB26 (4052) | MIRU26 (2996) | ETR-D (580) | ETR-E (3192) | QUB11b (2163b) | ||||
| 1 | 7 | 6 | 4 | 10 | 3 | 5 | 3 | 3 | 2 | 9 | Homogeneous | |
| 2 | 6 | 5 | 3 | 9 | 5 | 5 | 3 | 3 | 2 | 17 | Homogeneous | |
| 3 | 7 | 6 | 4 | 10 | 5 | 5 | 3 | 3 | 2 | 6 | Homogeneous | |
| 4 | 10 | 6 | 5 | 10 | 5 | 5 | 3 | 3 | 2 | 31 | Homogeneous | SB0339, SB0818, SB0140 |
| 5 | 4 | 5 | 4 | 10 | 5 | 5 | 3 | 3 | 2 | 7 | Homogeneous | |
| 6 | 7 | 6 | 4 | 10 | 6 | 5 | 3 | 3 | 2 | 5 | Heterogeneous (2) | |
| 7 | 6 | 4 | 10 | 5 | 2 | 3 | 3 | 2 | 4 | |||
| 7 | 7 | 6 | 4 | 10 | 5 | 5 | 3 | 3 | 2 | 11 | Heterogeneous (2) | SB0339, SB1142 |
| 7 | 5 | 3 | 7 | 5 | 5 | 3 | 3 | 2 | 1 | |||
| 8 | 7 | 6 | 3 | 10 | 4 | 5 | 3 | 3 | 2 | 29 | Heterogeneous (2) | SB0339 |
| 10 | 5 | 3 | 10 | 5 | 5 | 4 | 3 | 2 | 1 | |||
| 9 | 8 | 5 | 4 | 10 | 5 | 5 | 3 | 3 | 2 | 2 | Heterogeneous (3) | |
| 8 | 5 | 4 | 9 | 5 | 5 | 3 | 3 | 2 | 1 | |||
| 8 | 6 | 4 | 8 | 5 | 5 | 3 | 3 | 2 | 1 | |||
| 10 | 9 | 5 | 4 | 10 | 5 | 5 | 4 | 3 | 2 | 3 | Heterogeneous (3) | SB0295 |
| 10 | 5 | 3 | 10 | 5 | 5 | 3 | 3 | 2 | 1 | |||
| 7 | 6 | 4 | 10 | 2 | 2 | 3 | 3 | 2 | 1 | |||
| 11 | 9 | 3 | 3 | 10 | 5 | 5 | 3 | 3 | 2 | 9 | Heterogeneous (2) | |
| 9 | 5 | 3 | 10 | 5 | 5 | 3 | 3 | 2 | 6 | |||
| 12 | 15 | 5 | 4 | 9 | 5 | 5 | 3 | 3 | 1 | 6 | Heterogeneous (3) | SB0265, SB0807 |
| 11 | 5 | 4 | 9 | 5 | 5 | 3 | 3 | 1 | 1 | |||
| 8 | 12 | 5 | Nc | 4 | 4 | 3 | 3 | 2 | 1 | |||
| 13 | 10 | 5 | 4 | 10 | 5 | 5 | 3 | 3 | 2 | 6 | Heterogeneous (4) | SB0295, SB0416 |
| 6 | 5 | 4 | 12 | 4 | 6 | 3 | 3 | 2 | 1 | |||
| 6 | 6 | 4 | 10 | 5 | 5 | 3 | 3 | 2 | 1 | |||
| 7 | 5 | 4 | 10 | 5 | 6 | 3 | 3 | 2 | 1 | |||
| 14 | 6 | 5 | 2 | 7 | 5 | 5 | 3 | 3 | 2 | 3 | Heterogeneous (5) | SB0152, SB1320 |
| 8 | 6 | 4 | 10 | 5 | 5 | 3 | 3 | 2 | 1 | |||
| 6 | 6 | 4 | 10 | 5 | 5 | 3 | 3 | 2 | 1 | |||
| 7 | 5 | 4 | 10 | 4 | 5 | 3 | 3 | 2 | 1 | |||
| 7 | 6 | 4 | 10 | 5 | 5 | 3 | 3 | 2 | 1 | |||
| 15 | 7 | 6 | 4 | 10 | 6 | 5 | 3 | 3 | 2 | 2 | Heterogeneous (5) | SB0265, SB0875 |
| 7 | 5 | 5 | 10 | 5 | 5 | 3 | 2 | 2 | 2 | |||
| 14 | 6 | 4 | 13 | 4 | 6 | 3 | 3 | 2 | 1 | |||
| 8 | 5 | 5 | 10 | 5 | 5 | 3 | 2 | 2 | 1 | |||
| 7 | 6 | 3 | 7 | 5 | 5 | 3 | 3 | 2 | 1 | |||
This analysis revealed heterogeneity in 10 out of the 15 farms.
MIRU-VNTR loci with corresponding VNTR alias (markers have been ordered according to the D value).
N, not amplifiable.
Among the 10 farms with heterogeneous isolates, we observed (i) one farm with all of its isolates showing MV types differing in only one locus, (ii) five farms with all their MV types differing between two and six loci, and (iii) four farms in which MV types differing in only one locus coexisted with other types varying at more than one locus. In farms 7 and 8, some MV types were markedly underrepresented (only one isolate out of 11 and 29, respectively).
In all except one farm, heterogeneity was already revealed by the four-loci combination; most of the isolates were distinguished by allelic differences in two to four markers. The ETR-A locus was the main contributor (9 farms), followed by QUB3232 (6 farms, but with higher allelic diversity) and ETR-B and QUB11a (5 farms each), while only in five cases, the isolates differed in a single locus (ETR-A in one case, QUB-11a in one case, and QUB-3232 in three cases). In one farm, diversity was detected only by changes in MIRU26 and QUB26; this was the only case that needed the six-loci approach for resolution. The changes in these loci in other farms provided only redundant information.
DISCUSSION
Spoligotyping has been extensively used to characterize M. bovis in Spain because of the high degree of strain diversity in this pathogen (17) and the convenience of being able to record the results in a database at the national level (31). A common feature of M. bovis populations is that each geographical area presents a clearly more frequent spoligotype that can be subtyped by MIRU-VNTR analysis to some extent. Strains with the spoligotype SB0121 are the most common representatives of the European 2 clonal complex and its putative recent common ancestor (32). This spoligotype is also present at a high frequency in Portugal (16), is less prevalent in France (33) and Italy (10), and is almost absent in the British Isles (3, 4). The present study applied MIRU-VNTR typing with different purposes: first to determine the degree of diversity within strains with spoligotype SB0121 and then suggest the hypothetical evolution of this clonal complex, second, to evaluate the most suitable format of MIRU-VNTR for a cost-efficient typing of SB0121 isolates, and finally, to assess the reliability or limitations of spoligotyping for epidemiological investigation when M. bovis SB0121 is involved.
A random selection of isolates was chosen to represent the Spanish M. bovis population with spoligotype SB0121. Areas where M. bovis is highly endemic or areas and farms that were intensively sampled might therefore be overrepresented; nonetheless, we consider this subpopulation to be a representative snapshot of the national situation. The MIRU-VNTR characterization of this random panel of M. bovis SB0121 isolates revealed a high degree of diversity. This is consistent with previous reports from Portugal (12, 13) and Italy (10) where the MIRU-VNTR type diversity of the most prevalent spoligotypes (SB0121 and SB0120, respectively) is high compared to that of the European 1 clonal complex in the United Kingdom and the Republic of Ireland (34). The underlying reasons for this high diversity in the Iberian M. bovis population remain unknown. It might result from a higher mutation rate, or it may be due to the earlier introduction of the pathogen in mainland Europe, which gave more opportunity for evolution.
In order to screen for large polymorphisms that might hint at an explanation of the expansion of certain genotypes, we analyzed three isolates with spoligotype SB0121 and different MV types using DNA microarray according to Garcia-Pelayo et al. (35). The three analyzed strains, MI05/00611, MI06/00001, and MI05/00050 (see Table S1 in the supplemental material), were a cattle isolate with MV type 1A, the largest cluster found in the study, a cattle isolate with MV type 2D, a unique type, and a wild boar isolate with MV type 44, also a unique type, respectively. No large polymorphisms common to these clonal subgroups were identified (see Table S2 in the supplemental material).
The hypothetical relationships among the isolates are shown in the minimum spanning tree created using the data obtained from the nine-loci analysis (Fig. 1). The putative founder of the cluster (central node) is MV type 2A, although the pattern is not the most frequent one in this panel; a similar situation was previously described for strains with spoligotype SB0140 in the United Kingdom (36). Loci with a higher D value are the main contributors to the expansion in the diversity of the clonal group. We have observed that differences between strains in this panel were mainly caused by single or double allele variations. Moreover, in this snapshot, we did not observe clustering of distinct MIRU-VNTR types in specific geographic areas or animal species with any of the loci combinations, in contrast to the report from the United Kingdom (36). The inability to obtain and type each and every isolate when there is high prevalence of bovine tuberculosis together with high genetic diversity suggests a reasonable adaptation of the current criteria for interpretation of MIRU-VNTR analysis. We suggest adopting a less stringent interpretation (i.e., considering SLVs to be related sources) for disease tracing in case a perfect MIRU-VNTR type match to source cannot be identified (37).
The marked subdivision of SB0121 by applying MIRU-VNTR leads us to consider spoligotyping as being limited in its ability to precisely track transmissions, and it is probably responsible for the misinterpretation of supposedly related SB0121 isolates that might actually differ by MIRU-VNTR analysis. Before extending the genotyping effort from using spoligotyping to MIRU-VNTR for the analysis of this prevalent spoligotype, it was convenient to apply cost-effective techniques with an optimized level of discrimination. Obviously, the more markers included in the analysis, the better the resolution of genotypes and accuracy of results (8, 10). However, in practical terms (both economic and technical), this is not feasible when handling a large number of isolates. This also might be the situation in other countries with high herd prevalences. For this reason, in this study, we compared the results obtained with different combinations of loci. Our results show that the use of a minimum set of the four loci ETR-A, ETR-B, QUB11a, and QUB3232 is satisfactory for routine application with M. bovis isolates with spoligotype SB0121 from a high-diversity setting. The analysis of the panel of isolates with the four-loci approach still maintains a good discrimination value and does not change the main structure of the minimum spanning tree, as shown in Fig. 1. These four loci were among the loci proposed by the VENoMYC Consortium for the MIRU-VNTR typing of M. bovis strains (EU coordination action SSPE-CT-2004-501903 [23]); therefore, this selection maintains the advantage of being able to exchange data between laboratories.
The optimal minimum set of markers may depend on the spoligotype and/or epidemiological setting; therefore, in the absence of specific data, we recommend extending the set of markers to include MIRU26 and QUB26. This six-loci approach includes markers previously reported to be useful for MIRU-VNTR typing of diverse M. bovis spoligotypes in Ireland (26), Belgium (8), Italy (10), and Portugal (11); thus, it might be suitable for application as a general scheme for M. bovis isolates. For a more specific use, the markers ETR-D, ETR-E, and QUB11b can be added to complete the nine-loci approach, though their performances are variable. ETR-D and ETR-E did not achieve a high level of discrimination in either this study or in the above-mentioned publications. Remarkably, the discrimination power of MIRU-VNTR locus QUB11b varies with different spoligotypes and strain collections, e.g., in Doñana National Park (9) or bullfighting cattle (38).
In order to evaluate whether a MIRU-VNTR-based strategy could change the epidemiological interpretation of those farms infected by M. bovis SB0121, we analyzed multiple isolates from 15 farms. While five out of the 15 farms showed homogeneous SB0121 strains, 10 showed a variable degree of heterogeneity, which was detected in almost all cases with the four-loci scheme. The cases that differed in a single locus could be considered related, as they might reflect a microevolutionary event in the farm. However, differences in several markers between isolates in the other cases would exclude any close phylogenetic relationship of these strains isolated from one farm. Among these heterogeneous farms, we found farms (i) with SLVs present, (ii) that were infected simultaneously by two to five different strains, and (iii) in which both phenomena coexisted. In farms with a coexistence of clonal variants, these variants are likely to be related and might have appeared by way of microevolutionary events. The impacts that these events might have on the precise definition of “epidemiologically linked” must be addressed in further studies. Interestingly, the presence of different strains and/or clonal variants would have remained undetected on some farms if an exhaustive analysis of all positive animals had not been performed.
The finding of such diversity within the spoligotype SB0121 is relevant to the evaluation of the eradication program and the interpretation of persistent infection on a farm. The persistence of infection in the farms may not, in some cases, be due to the failure of diagnostic tests but rather to the existence of a continuous risk of reinfection, such as that which occurs in the trade of infected cattle, with contact with infected wildlife, or with inadequate biosafety measures. This analysis is the basis of future approaches to tracking M. bovis infection within a defined area. The presence of multiple sources of infection is also strongly suggested by the fact that M. bovis with spoligotypes other than SB0121 were identified in eight of the selected 15 farms (Table 2).
The occurrence of double bands in the MIRU-VNTR analysis, as observed in three isolates in this study, might be due to mixed infection (9, 39) or to microevolutionary events (40). Although we could not exclude or confirm any of these possibilities, we think that these are likely due to mixed infections, because the presence of double bands affected two or three markers, except for in one isolate. This effect has been previously described in M. bovis isolates from high-prevalence settings (9).
In summary, MIRU-VNTR analysis revealed genetic variability among Spanish M. bovis isolates with spoligotype SB0121 and delineated the cluster as an expanding clonal group in which isolates are mainly linked by single or double locus variants. In practical terms, this also implies the need to subtype M. bovis isolates using MIRU-VNTR when this common spoligotype is identified, at least using a minimum combination of four loci QUB3232, ETR-A, ETR-B, and QUB11a. This lack of variability within spoligotype SB0121 strains has hindered, until now, the identification of farms that are simultaneously infected by several strains and the existence of microevolved clonal variants; these variants may well interfere with our standard concepts for the interpretation of the source of infection for these herds.
Supplementary Material
ACKNOWLEDGMENTS
This research was funded by the Spanish Ministry of Environment and Rural and Marine Affairs (MARM), EU project TB-STEP (KBBE-2007-1-3-04, no. 212414) and Fondo de Investigaciones Sanitarias (FIS) (S09/02205, PI12/02080). S.R.C. was financed by a PhD studentship (AP2006-01630) of the Spanish Ministry of Education. Research by Y.N. was partly supported by a PICATA predoctoral fellowship (BE55/11) from the Moncloa Campus of International Excellence (UCM-UPM, Instituto de Investigación Sanitaria Gregorio Marañón). N.H.S., P.G., and G.R.H. were funded by the Department of Environment, Food and Rural Affairs, United Kingdom (project SB4020).
We thank the National Animal Health authorities, especially J. L. Sáez (MARM), for their continuous encouragement. We are grateful to the Regional Laboratories for remittance of samples, especially A. Balseiro, M. F. Copano, E. Fernández, and I. Merediz (Asturias), C. Fernández, F. M. Fernández, M. G. Gradillas, M. Gutiérrez, and E. Sola (Cantabria), C. Calvo, D. Fernández, M. López, J. E. Mourelo, and M. Muñoz (Galicia), J. A. Anguiano, I. Burón, J. Cermeño, C. Domínguez, F. Fernández, A. Grau, S. Marques, O. Martín, C. Martínez, O. Mínguez, F. Moreno, and I. Romero (Castile and León), and C. Fornell, J. M. Gómez, A. Jiménez, I. Muñoz, J. A. Téllez, and E. J. Villalba (Andalusia).
Footnotes
Published ahead of print 28 August 2013
For this multi-institutional research network, see http://www.ciberes.org/.
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JCM.01271-13.
REFERENCES
- 1. Kamerbeek J, Schouls L, Kolk A, van Agterveld M, van Soolingen D, Kuijper S, Bunschoten A, Molhuizen H, Shaw R, Goyal M, van Embden J. 1997. Simultaneous detection and strain differentiation of Mycobacterium tuberculosis for diagnosis and epidemiology. J. Clin. Microbiol. 35:907–914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Skuce RA, McDowell SW, Mallon TR, Luke B, Breadon EL, Lagan PL, McCormick CM, McBride SH, Pollock JM. 2005. Discrimination of isolates of Mycobacterium bovis in Northern Ireland on the basis of variable numbers of tandem repeats (VNTRs). Vet. Rec. 157:501–504 [DOI] [PubMed] [Google Scholar]
- 3. Hewinson RG, Vordermeier HM, Smith NH, Gordon SV. 2006. Recent advances in our knowledge of Mycobacterium bovis: a feeling for the organism. Vet. Microbiol. 112:127–139 [DOI] [PubMed] [Google Scholar]
- 4. Smith NH, Gordon SV, de la Rua-Domenech R, Clifton-Hadley RS, Hewinson RG. 2006. Bottlenecks and broomsticks: the molecular evolution of Mycobacterium bovis. Nat. Rev. Microbiol. 4:670–681 [DOI] [PubMed] [Google Scholar]
- 5. Frothingham R, Meeker-O'Connell WA. 1998. Genetic diversity in the Mycobacterium tuberculosis complex based on variable numbers of tandem DNA repeats. Microbiology 144(Pt 5):1189–1196 [DOI] [PubMed] [Google Scholar]
- 6. Supply P, Mazars E, Lesjean S, Vincent V, Gicquel B, Locht C. 2000. Variable human minisatellite-like regions in the Mycobacterium tuberculosis genome. Mol. Microbiol. 36:762–771 [DOI] [PubMed] [Google Scholar]
- 7. Hilty M, Diguimbaye C, Schelling E, Baggi F, Tanner M, Zinsstag J. 2005. Evaluation of the discriminatory power of variable number tandem repeat (VNTR) typing of Mycobacterium bovis strains. Vet. Microbiol. 109:217–222 [DOI] [PubMed] [Google Scholar]
- 8. Allix C, Walravens K, Saegerman C, Godfroid J, Supply P, Fauville-Dufaux M. 2006. Evaluation of the epidemiological relevance of variable-number tandem-repeat genotyping of Mycobacterium bovis and comparison of the method with IS6110 restriction fragment length polymorphism analysis and spoligotyping. J. Clin. Microbiol. 44:1951–1962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Romero B, Aranaz A, Sandoval A, Alvarez J, de Juan L, Bezos J, Sánchez C, Galka M, Fernández P, Mateos A, Domínguez L. 2008. Persistence and molecular evolution of Mycobacterium bovis population from cattle and wildlife in Doñana National Park revealed by genotype variation. Vet. Microbiol. 132:87–95 [DOI] [PubMed] [Google Scholar]
- 10. Boniotti MB, Goria M, Loda D, Garrone A, Benedetto A, Mondo A, Tisato E, Zanoni M, Zoppi S, Dondo A, Tagliabue S, Bonora S, Zanardi G, Pacciarini ML. 2009. Molecular typing of Mycobacterium bovis strains isolated in Italy from 2000 to 2006 and evaluation of variable-number tandem repeats for a geographically optimized genotyping. J. Clin. Microbiol. 47:636–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Duarte EL, Domingos M, Amado A, Cunha MV, Botelho A. 2010. MIRU-VNTR typing adds discriminatory value to groups of Mycobacterium bovis and Mycobacterium caprae strains defined by spoligotyping. Vet. Microbiol. 143:299–306 [DOI] [PubMed] [Google Scholar]
- 12. Skuce RA, Mallon TR, McCormick CM, McBride SH, Clarke G, Thompson A, Couzens C, Gordon AW, McDowell SW. 2010. Mycobacterium bovis genotypes in Northern Ireland: herd level surveillance (2003 to 2008). Vet. Rec. 167:684–689 [DOI] [PubMed] [Google Scholar]
- 13. Cunha MV, Matos F, Canto A, Albuquerque T, Alberto JR, Aranha JM, Vieira-Pinto M, Botelho A. 2011. Implications and challenges of tuberculosis in wildlife ungulates in Portugal: a molecular epidemiology perspective. Res. Vet. Sci. 92:225–235 [DOI] [PubMed] [Google Scholar]
- 14. McLernon J, Costello E, Flynn O, Madigan G, Ryan F. 2010. Evaluation of mycobacterial interspersed repetitive-unit-variable-number tandem-repeat analysis and spoligotyping for genotyping of Mycobacterium bovis isolates and a comparison with restriction fragment length polymorphism typing. J. Clin. Microbiol. 48:4541–4545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lari N, Bimbi N, Rindi L, Tortoli E, Garzelli C. 2011. Genetic diversity of human isolates of Mycobacterium bovis assessed by spoligotyping and variable number tandem repeat genotyping. Infect. Genet. Evol. 11:175–180 [DOI] [PubMed] [Google Scholar]
- 16. Duarte EL, Domingos M, Amado A, Botelho A. 2008. Spoligotype diversity of Mycobacterium bovis and Mycobacterium caprae animal isolates. Vet. Microbiol. 130:415–421 [DOI] [PubMed] [Google Scholar]
- 17. Rodríguez S, Romero B, Bezos J, de Juan L, Alvarez J, Castellanos E, Moya N, Lozano F, González S, Sáez-Llorente JL, Mateos A, Domínguez L, Aranaz A, Spanish Network on Surveillance and Monitoring of Animal Tuberculosis 2010. High spoligotype diversity within a Mycobacterium bovis population: clues to understanding the demography of the pathogen in Europe. Vet. Microbiol. 141:89–95 [DOI] [PubMed] [Google Scholar]
- 18. Balseiro A, Rodríguez O, González-Quiros P, Merediz I, Sevilla IA, Davé D, Dalley DJ, Lesellier S, Chambers MA, Bezos J, Muñoz M, Delahay RJ, Gortázar C, Prieto JM. 2011. Infection of Eurasian badgers (Meles meles) with Mycobacterium bovis and Mycobacterium avium complex in Spain. Vet. J. 190:e21–e25. 10.1016/j.tvjl.2011.04.012 [DOI] [PubMed] [Google Scholar]
- 19. Smith NH, Upton P. 2011. Naming spoligotype patterns for the RD9-deleted lineage of the Mycobacterium tuberculosis complex: www.Mbovis.org. Infect. Genet. Evol. 12:873–876 [DOI] [PubMed] [Google Scholar]
- 20. Supply P, Lesjean S, Savine E, Kremer K, van Soolingen D, Locht C. 2001. Automated high-throughput genotyping for study of global epidemiology of Mycobacterium tuberculosis based on mycobacterial interspersed repetitive units. J. Clin. Microbiol. 39:3563–3571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Skuce RA, McCorry TP, McCarroll JF, Roring SM, Scott AN, Brittain D, Hughes SL, Hewinson RG, Neill SD. 2002. Discrimination of Mycobacterium tuberculosis complex bacteria using novel VNTR-PCR targets. Microbiology 148:519–528 [DOI] [PubMed] [Google Scholar]
- 22. Supply P, Allix C, Lesjean S, Cardoso-Oelemann M, Rüsch-Gerdes S, Willery E, Savine E, de Haas P, van Deutekom H, Roring S, Bifani P, Kurepina N, Kreiswirth B, Sola C, Rastogi N, Vatin V, Gutierrez MC, Fauville M, Niemann S, Skuce R, Kremer K, Locht C, van Soolingen D. 2006. Proposal for standardization of optimized mycobacterial interspersed repetitive unit-variable-number tandem repeat typing of Mycobacterium tuberculosis. J. Clin. Microbiol. 44:4498–4510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Supply P. 2006. Protocol and guidelines for multilocus variable number tandem repeat genotyping of M. bovis, p 15–16 VENoMYC (Veterinary Network of Laboratories Researching into Improved Diagnosis and Epidemiology of Mycobacterial Diseases) WP7 workshop, Toledo, Spain, 19 to 22 October 2006. WP7 Workshop VENoMYC Coordination Action EU SSPE-CT-2004-501903
- 24. Gortázar C, Vicente J, Samper S, Garrido JM, Fernández-De-Mera IG, Gavín P, Juste RA, Martín C, Acevedo P, De La Puente M, Höfle U. 2005. Molecular characterization of Mycobacterium tuberculosis complex isolates from wild ungulates in south-central Spain. Vet. Res. 36:43–52 [DOI] [PubMed] [Google Scholar]
- 25. Parra A, Larrasa J, García A, Alonso JM, de Mendoza JH. 2005. Molecular epidemiology of bovine tuberculosis in wild animals in Spain: a first approach to risk factor analysis. Vet. Microbiol. 110:293–300 [DOI] [PubMed] [Google Scholar]
- 26. Roring S, Scott A, Brittain D, Walker I, Hewinson G, Neill S, Skuce R. 2002. Development of variable-number tandem repeat typing of Mycobacterium bovis: comparison of results with those obtained by using existing exact tandem repeats and spoligotyping. J. Clin. Microbiol. 40:2126–2133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hunter PR. 1990. Reproducibility and indices of discriminatory power of microbial typing methods. J. Clin. Microbiol. 28:1903–1905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hunter PR, Gaston MA. 1988. Numerical index of the discriminatory ability of typing systems: an application of Simpson's index of diversity. J. Clin. Microbiol. 26:2465–2466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Allix-Béguec C, Harmsen D, Weniger T, Supply P, Niemann S. 2008. Evaluation and strategy for use of MIRU-VNTRplus, a multifunctional database for online analysis of genotyping data and phylogenetic identification of Mycobacterium tuberculosis complex isolates. J. Clin. Microbiol. 46:2692–2699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Weniger T, Krawczyk J, Supply P, Niemann S, Harmsen D. 2010. MIRU-VNTRplus: a web tool for polyphasic genotyping of Mycobacterium tuberculosis complex bacteria. Nucleic Acids Res. 38:W326–W331. 10.1093/nar/gkq351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Rodriguez-Campos S, González S, de Juan L, Romero B, Bezos J, Casal C, Álvarez J, Fernández-de-Mera IG, Castellanos E, Mateos A, Sáez-Llorente JL, Domínguez L, Aranaz A, Spanish Network on Surveillance Monitoring of Animal Tuberculosis 2012. A database for animal tuberculosis (mycoDB.es) within the context of the Spanish national programme for eradication of bovine tuberculosis. Infect. Genet. Evol. 12:877–882 [DOI] [PubMed] [Google Scholar]
- 32. Rodriguez-Campos S, Schürch AC, Dale J, Lohan AJ, Cunha MV, Botelho A, De Cruz K, Boschiroli ML, Boniotti MB, Pacciarini M, Garcia-Pelayo MC, Romero B, de Juan L, Domínguez L, Gordon SV, van Soolingen D, Loftus B, Berg S, Hewinson RG, Aranaz A, Smith NH. 2011. European 2–a clonal complex of Mycobacterium bovis dominant in the Iberian Peninsula. Infect. Genet. Evol. 12:866–872 [DOI] [PubMed] [Google Scholar]
- 33. Haddad N, Ostyn A, Karoui C, Masselot M, Thorel MF, Hughes SL, Inwald J, Hewinson RG, Durand B. 2001. Spoligotype diversity of Mycobacterium bovis strains isolated in France from 1979 to 2000. J. Clin. Microbiol. 39:3623–3632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Smith NH, Berg S, Dale J, Allen A, Rodriguez S, Romero B, Matos F, Ghebremichael S, Karoui C, Donati C, Machado Ada C, Mucavele C, Kazwala RR, Hilty M, Cadmus S, Ngandolo BN, Habtamu M, Oloya J, Müller A, Milian-Suazo F, Andrievskaia O, Projahn M, Barandiaran S, Macías A, Müller B, Zanini MS, Ikuta CY, Rodriguez CA, Pinheiro SR, Figueroa A, Cho SN, Mosavari N, Chuang PC, Jou R, Zinsstag J, van Soolingen D, Costello E, Aseffa A, Proaño-Perez F, Portaels F, Rigouts L, Cataldi AA, Collins DM, Boschiroli ML, Hewinson RG, Ferreira Neto JS, Surujballi O, Tadyon K, Botelho A, Zárraga AM, et al. 2011. European 1: a globally important clonal complex of Mycobacterium bovis. Infect. Genet. Evol. 11:1340–1351 [DOI] [PubMed] [Google Scholar]
- 35. Garcia-Pelayo MC, Caimi KC, Inwald JK, Hinds J, Bigi F, Romano MI, van Soolingen D, Hewinson RG, Cataldi A, Gordon SV. 2004. Microarray analysis of Mycobacterium microti reveals deletion of genes encoding PE-PPE proteins and ESAT-6 family antigens. Tuberculosis (Edinb.) 84:159–166 [DOI] [PubMed] [Google Scholar]
- 36. Smith NH, Dale J, Inwald J, Palmer S, Gordon SV, Hewinson RG, Smith JM. 2003. The population structure of Mycobacterium bovis in Great Britain: clonal expansion. Proc. Natl. Acad. Sci. U. S. A. 100:5271–15275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Rodriguez-Campos S, Aranaz A, de Juan L, Sáez-Llorente JL, Romero B, Bezos J, Jiménez A, Mateos A, Domínguez L. 2011. Limitations of spoligotyping and variable-number tandem-repeat typing for molecular tracing of Mycobacterium bovis in a high diversity setting. J. Clin. Microbiol. 49:3361–3364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Rodriguez-Campos S. 2012. Molecular epidemiology of Mycobacterium bovis and Mycobacterium caprae in Spain. PhD thesis Universidad Complutense de Madrid, Madrid, Spain [Google Scholar]
- 39. García de Viedma D, Marín M, Ruiz-Serrano MJ, Alcalá L, Bouza E. 2003. Polyclonal and compartmentalized infection by Mycobacterium tuberculosis in patients with both respiratory and extrarespiratory involvement. J. Infect. Dis. 187:695–699 [DOI] [PubMed] [Google Scholar]
- 40. Al-Hajoj SA, Akkerman O, Parwati I, al-Gamdi S, Rahim Z, van Soolingen D, van Ingen J, Supply P, van der Zanden AG. 2010. Microevolution of Mycobacterium tuberculosis in a tuberculosis patient. J. Clin. Microbiol. 48:3813–3816 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.