Abstract
Colistin is an old antibiotic which has been used as a therapeutic option for carbapenem- and multidrug-resistant Gram-negative bacteria, like Acinetobacter baumannii. This pathogen produces life-threatening infections, mainly in patients admitted to intensive care units. Rapid detection of resistance to colistin may improve patient outcomes and prevent the spread of resistance. For this purpose, Micromax technology was evaluated in four isogenic A. baumannii strains with known mechanisms of resistance to colistin and in 66 isolates (50 susceptible and 16 resistant). Two parameters were determined, DNA fragmentation and cell wall damage. To assess DNA fragmentation, cells trapped in a microgel were incubated with a lysing solution to remove the cell wall, and the released nucleoids were visualized under fluorescence microscopy. Fragmented DNA was observed as spots that diffuse from the nucleoid. To assess cell wall integrity, cells were incubated with a lysis solution which removes only weakened cell walls, resulting in nucleoid release exclusively in affected cells. A dose-response relationship was demonstrated between colistin concentrations and the percentages of bacteria with DNA fragmentation and cell wall damage, antibiotic effects that were delayed and less frequent in resistant strains. Receiver operating characteristic (ROC) curves demonstrated that both DNA fragmentation and cell wall damage were excellent parameters for identifying resistant strains. Obtaining ≤11% of bacteria with cell wall damage after incubation with 0.5 μg/ml colistin identified resistant strains of A. baumannii with 100% sensitivity and 96% specificity. Results were obtained in 3 h 30 min. This is a simple, rapid, and accurate assay for detecting colistin resistance in A. baumannii, with strong potential value in critical clinical situations.
INTRODUCTION
Acinetobacter baumannii is a Gram-negative coccobacillus that survives in the hospital niche and has the ability to persist for long periods on desiccated surfaces (1, 2). This microorganism predominates in intensive care units (ICUs) and can produce severe mechanical ventilation-associated pneumonia, central venous catheter-associated bloodstream infections, and catheter-associated urinary tract infections as well as infections in the central nervous system, skin, soft tissues, and bone (2–4).
Worldwide rates of multidrug-resistant A. baumannii strains are increasing alarmingly, and even panresistance is emerging (5). For example, 36.8% of A. baumannii strains isolated from patients with ventilator-associated pneumonia in the United States are resistant to carbapenems, and resistance rates are as high as 85% in isolates from intensive care units in Greece. Resistance may also extend to fluoroquinolones, cephalosporins, etc. (6).
Colistin (polymyxin E), an old polymyxin antibiotic discovered in 1949, has undergone a resurgence in use in recent years as a therapeutic option for carbapenem- or multidrug-resistant Gram-negative bacteria (7–12). Polymyxins consist of a hydrophilic polycationic peptide ring and a tripeptide side chain with a lipophilic fatty acid tail (13). They are bactericidal antibiotics that bind to lipopolysaccharides (LPSs) and phospholipids in the outer cell membrane of Gram-negative bacteria. The polycationic peptide ring has a strong affinity for divalent cations and competitively displaces calcium and magnesium from the negatively charged phosphate groups of the lipid A of LPSs (14, 15). It alters the stability of the LPS molecule, leading to disruption of the outer cell membrane and thus altering cellular permeability. More polymyxin molecules are inserted into the outer membrane through the hydrophobic fatty acyl chain domain, producing a detergent-like effect that causes transient cracks (15). This results in leakage of intracellular contents and possibly more uptake of the polymyxin, i.e., a self-promoted uptake pathway, resulting in bacterial death (13).
Resistance to colistin is relatively rare, possibly due to its infrequent use in these last 50 years and perhaps also because of its detergent properties. The majority of the mechanisms of resistance to polymyxins are based on modifications of lipid A of the LPS, which decrease its net negative charge, thus preventing or reducing the initial binding. These changes may be associated with mutations in the PmrA/B two-component system, which increase the level of expression of the lipid A-modifying phosphoethanolamine transferase, PmrC, resulting in the addition of phosphoethanolamine to lipid A (16–19). Resistance may also be a consequence of the replacement of lipid A with aminoarabinose, which requires the products of the ugd and pbg loci (20, 21). Unfortunately, there have recently been several outbreaks of infections caused by colistin-resistant A. baumannii emerging from Asia (22, 23). The emergence of colistin-resistant A. baumannii is very likely to have major public health implications, since no novel antibiotics with activity against Gram-negative bacteria are expected to be available within the coming years (24). As a consequence, there is an urgent need to optimize the clinical use of colistin and reduce the development and spread of resistant strains.
Given the elevated risk of mortality in patients with infections acquired in the intensive care unit (ICU), early adequate antibiotic therapy has been shown to significantly improve patient outcomes and reduce health care costs (8, 25, 26). Standard antibiograms require nearly 24 h or longer after bacterial isolation and identification to provide results. But in these critical clinical situations, the rapid availability of information regarding resistance to carbapenems and colistin may be essential.
In a previously reported study, a simple procedure for the rapid assessment of resistance to carbapenems and ciprofloxacin in A. baumannii using Micromax technology was validated (27). In the case of carbapenems, cells immersed in a microgel on a slide were incubated with a lysing solution that removes the cell wall only in those bacteria in which peptidoglycan synthesis has been affected by the beta-lactam. Thus, only susceptible bacteria release the nucleoid, which is stained and visualized under fluorescence microscopy. Unaffected bacteria retain their standard shape (27, 28). For ciprofloxacin assessment, the lysis procedure removes the cell wall in all bacteria. Those affected by the quinolone, i.e., with double-strand DNA breaks as a consequence of DNA gyrase and topoisomerase IV trapping, show diffusion of DNA fragments emerging from the residual central core, whereas resistant strains reveal only intact nucleoids (29–31).
In addition to carbapenems and quinolones, rapid determination of resistance of A. baumannii to colistin is of remarkable interest. It not only has decisive clinical value for patients, it also could contribute to optimizing the medical use of colistin, thereby helping to prevent the spread of strains resistant to a last-resort antibiotic. In this study we characterized the efficacy of using both DNA damage and cell wall damage assays to rapidly determine the resistance of A. baumannii to colistin.
MATERIALS AND METHODS
Bacterial strains.
The procedures for determining chromosomal DNA fragmentation and cell wall damage in situ were first assayed for control A. baumannii strains harboring known mechanisms of colistin resistance. These were laboratory isogenic strains derived from susceptible strains from the American Type Culture Collection (ATCC; Manassas, VA), ATCC 19606 ColS (MIC, 0.25 μg/ml), ATCC 19606 ColR (MIC, 16 μg/ml), ATCC 19606 ColR lpxA (MIC, 32 to 64 μg/ml), and ATCC 19606 ColR lpxD (MIC, >64 μg/ml) (18, 32, 33).
Afterward, 66 A. baumannii isolates obtained from the University Hospital A Coruña and Virgen del Rocío Hospital, Seville, between 2001 and 2012 were analyzed. Repetitive extragenic palindromic (REP)-PCR was performed in some cases to rule out clonality (17). All of the isolates were clinical isolates except for a group of 4 and another group of 5 laboratory-derived isogenic strains. The MICs were determined by automated microdilution (MicroScan WalkAway; Siemens) repeated a minimum of 6 times. According to the CLSI breakpoint concentration of susceptibility (2 μg/ml), 50 strains were categorized as susceptible (MIC, ≤2 μg/ml), and 16 were categorized as resistant (MIC, ≥4 μg/ml) to colistin (five with an MIC of 4 μg/ml, five with an MIC of 8 μg/ml, two with an MIC of 16 μg/ml, two with an MIC of 32 μg/ml, and two with an MIC of >32 μg/ml). Of these resistant strains, 10 were laboratory derived and 6 naturally occurring clinical isolates.
DNA damage and cell wall damage assays.
Determinations of DNA fragmentation and cell wall integrity were carried out as blind procedures, without knowledge of the MICs, by three independent technicians. Two variants of the Micromax kit for fluorescence microscopy (research use only) (Halotech DNA SL, Madrid, Spain), Micromax Q and Micromax WG, have been employed to evaluate the integrity of the nucleoid and of the cell wall, respectively. Their only difference lies in the composition of the lysing solution (27).
Bacteria were routinely grown on Mueller-Hinton agar at 37°C for 24 h. A colony was selected and incubated in 2 ml Mueller-Hinton broth at 37°C for 90 min. The bacteria were diluted to an optical density at 600 nm (OD600) of 0.1 in Mueller-Hinton broth and incubated at 37°C in 200-μl tubes with colistin for 60 min, in a final volume of 30 μl. In initial experiments, the four control ATCC 19606 strains were incubated with 11 doses of colistin ranging from 0 to 64 μg/ml. Afterward, the other 66 strains were treated with colistin concentrations of 0, 0.4, 0.5, 0.75, and 1 μg/ml. An aliquot of each sample was diluted to a concentration of 5 to 10 million microorganisms/ml in Mueller-Hinton broth. The kit includes 0.5-ml snap-cap microcentrifuge tubes containing gelled aliquots of low-melting-point agarose. The tube was placed in a water bath at 90 to 100°C for about 5 min to melt the agarose completely and then placed in a water bath at 37°C. Thirty microliters of the diluted sample was added to the tube and mixed with the melted agarose. A 10-μl aliquot of the sample-agarose mixture was pipetted onto a precoated slide and covered with an 18- by 18-mm coverslip. The slide was placed on a cold plate in the refrigerator (4°C) for 5 min to allow the agarose to produce a microgel with the trapped intact cells inside. The coverslip was removed gently, and the slide was immediately immersed horizontally in 10 ml of the specific lysing solution for 5 min at room temperature The slide was washed horizontally in a tray with abundant distilled water for 3 min, dehydrated by incubating horizontally in cold (−20°C) ethanol of increasing concentrations (70%, 90%, and 100%) for 2 min each, and air dried in an oven. The dried slide was incubated in a microwave oven at 750 W for 4 min, and then the DNA was stained with 100 μl of the fluorochrome SYBR gold (Molecular Probes, Eugene, OR) diluted 1:200 in Tris-borate-EDTA (TBE) buffer (0.09 M Tris-borate, 0.002 M EDTA [pH 7.5]) for 2 min in the dark, with a glass coverslip. After a brief wash in phosphate buffer (pH 6.88) (Merck, Darmstadt, Germany), a 24- by 60-mm coverslip was added. The slides were visualized under fluorescence microscopy and 200 to 500 bacteria were scored per experimental point. The scoring criterion of DNA fragmentation is the presence of diffused DNA fragments from the nucleoid (27, 30). Otherwise, cell wall damage is evidenced when the nucleoid is released (27, 28). The time required for the assay included 1 h 30 min of incubation of the bacteria in Mueller-Hinton broth, 1 h of incubation with colistin, 40 min of technical processing (10 min of microgel enclosing, 5 min of lysis, 15 min of washing and dehydrating, and 10 min of drying and staining), and 5 to 15 min of scoring under the microscope, for a total of 3 h 15 min to 3 h 30 min.
Statistical analysis.
Data were normally distributed as ascertained by the Kolmogorov-Smirnov test. The Student t test was performed to check homogeneity. Receiver operating characteristic (ROC) curves were constructed for establishing the optimal concentration of colistin to determine possible resistance and calculate cutoff values for DNA fragmentation and cell wall damage in predicting resistance to colistin, using the Youden index (J). Data were analyzed using the IBM SPSS Statistics 21 software package for Windows (IBM). For determination of the Youden index we used the Epidat 3.1 software package (Consellería de Sanidade, Xunta de Galicia, Spain, and Panamerican Health Organization, Washington, DC). Differences were defined as significant at P values of <0.05.
RESULTS AND DISCUSSION
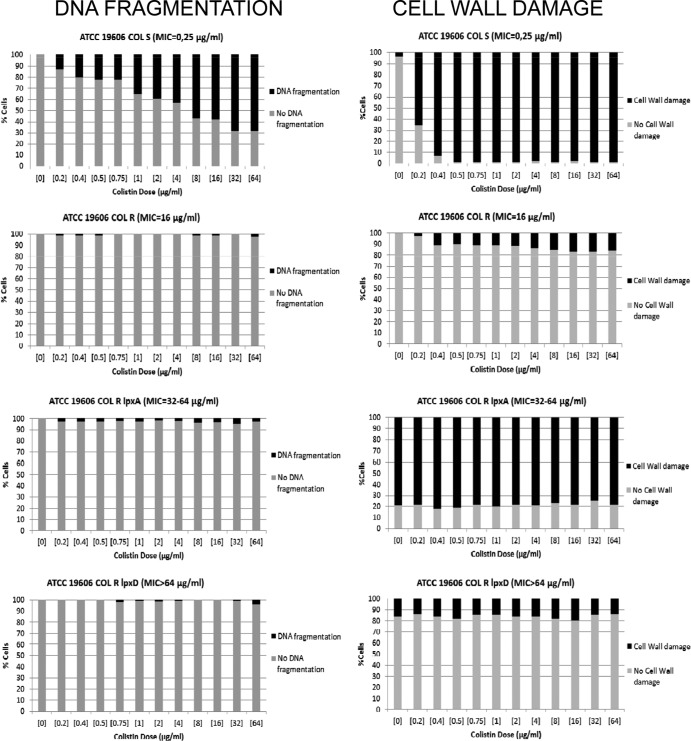
The procedures for the determination of DNA fragmentation and cell wall damage were initially assessed by incubating four isogenic A. baumannii strains with a high range of concentrations of colistin, from 0 to 64 μg/ml. In the susceptible strain ATCC 19606 ColS (MIC, 0.25 μg/ml), after incubation with the lowest concentration (0.2 μg/ml), 13% of cells showed DNA fragmentation whereas 65% appeared affected at the cell wall. Incubation with the next concentration (0.4 μg/ml) resulted in 20.5% of cells with DNA fragmentation and 93% (almost all of them) with cell wall damage (Fig. 1).
Fig 1.
Distributions of the frequencies of cells with DNA fragmentation (left) and levels of cell wall damage (right) after incubation with increasing concentrations of colistin of isogenic A. baumannii strains susceptible (ATCC 19606 ColS) and with defined mechanisms of resistance to colistin (ATCC 19606 ColR, ATCC 19606 ColR lpxA, ATCC 19606 ColR lpxD).
The laboratory-derived colistin-resistant strain ATCC 19606 ColR (MIC, 16 μg/ml) possesses a single nucleotide Ala227Val mutation in pmrB (32) that is adjacent to the conserved histidine at the site of phosphorylation (His228), which is critical for phosphatase activity. The final result is the addition of phosphoethanolamine to hepta-acylated lipid A (18). Although less immunogenic than the wild type, this LPS modification confers colistin resistance. Unlike the parental strain, the mutant strain achieved only 2.5% of cells with fragmented DNA after incubation with the highest concentration of colistin (64 μg/ml) and 17% of cells with affected cell walls after incubation with 16 to 64 μg/ml (Fig. 1). A discrete intercellular background of DNA fragments was visualized in the cultures of the susceptible strains at concentrations of 0.4 μg/ml and higher when the lysing solution was used to determine cell wall integrity, but not when lysis was employed to assess nucleoids. This background was absent in the cultures of the resistant strain.
The isogenic strains ATCC 19606 ColR lpxA (MIC, 32 to 64 μg/ml) and ATCC 19606 ColR lpxD (MIC, >64 μg/ml) were also studied. These laboratory derivatives carry mutations in the genes implicated in lipid A biosynthesis, lpxA and lpxD, respectively, leading to loss of LPS production and increased MICs of colistin (33). After technical processing, even in the absence of the antibiotic, these strains evidenced decreased fitness and showed high intercellular backgrounds of DNA fragments spontaneously released into the culture medium. This background was maintained with the lysing solution used to check cell wall damage but tended to disappear when the stronger lysing solution was used to release the nucleoids. In both strains, no more than 4 to 4.5% of cells showed fragmented DNA, and there were no relationships between the percentages of fragmentation and the doses of colistin (Fig. 1). Interestingly, the lpxA mutant showed 75 to 80% of cells with released nucleoids when incubated with the lysing solution to assess cell wall integrity, without antibiotic and without changes with increasing concentrations (Fig. 1). This suggests a constitutively weaker cell wall. This spontaneous effect on the cell wall was also detected in the lpxD mutant strain, but only in 14 to 20% of cells, also without significant increases with higher concentrations (Fig. 1). The less resistant cell wall in the lpxA mutant is in accordance with the previously described changes in its membrane potential, which were probably due to effects on outer membrane integrity and loss of affinity to colistin (34).
It is doubtful that A. baumannii strains without LPS can survive in nature given their low fitness. Moreover, their possible virulence should be greatly compromised. In any case, these strains were detected by the technique to assess cell wall damage, showing a strong background of DNA fragments and exhibiting weak cell walls, even in the absence of colistin treatment. Colistin-resistant strains in nature should be more closely related to ATCC 19606 ColR, which exhibits a modified LPS in the outer cell membrane, than to strains without LPS. Consequently, taking into account the results with the susceptible ATCC 19606 and the resistant ATCC 19606 ColR strains, the techniques of detection of DNA fragmentation and cell wall damage appear adequate for making rapid distinctions between A. baumannii strains susceptible and resistant to colistin. The cell wall assay seemed especially discriminative. In a quantitative validation attempt, 66 mostly clinical strains, 50 susceptible and 16 resistant according to the CLSI criteria, were studied. MICs were equilibrated and ranged from 0.03 to >32 μg/ml.
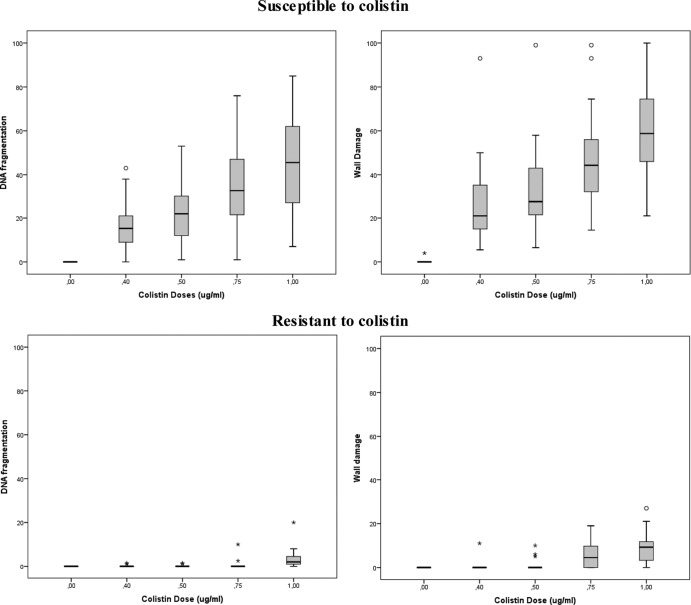
Figure 2 shows the frequencies of damage to the DNA and the cell wall after incubation with increasing concentrations of colistin up to 1 μg/ml for the susceptible and the resistant strains. Representative images are presented in Fig. 3 for DNA damage and in Fig. 4 for cell wall damage for strains with MICs of 0.03 to 0.25, 16, and >32 μg/ml. Unlike the susceptible strains, almost all of the resistant strains exhibited little damage, and this damage was evident in only a few cells after incubation at the highest concentration. In the susceptible strains, more cells appeared affected at the cell wall than at the DNA level for each colistin concentration (Student's t test, P < 0.001). This also occurred in the resistant strains after incubation with colistin concentrations of 0.75 μg/ml (P = 0.002) and 1 μg/ml (P < 0.001).
Fig 2.
Box-and-whisker plots of the dose response of A. baumannii strains susceptible (above) and resistant (below) to colistin, for DNA fragmentation (left) and for cell wall damage (right). The horizontal line in the box represents the median, the lower line of the box the first quartile, and the upper line of the box the third quartile data values, and the whiskers (the end of the vertical lines) represent the maximum and minimum data values. The dots outside the box correspond to abnormally high values.
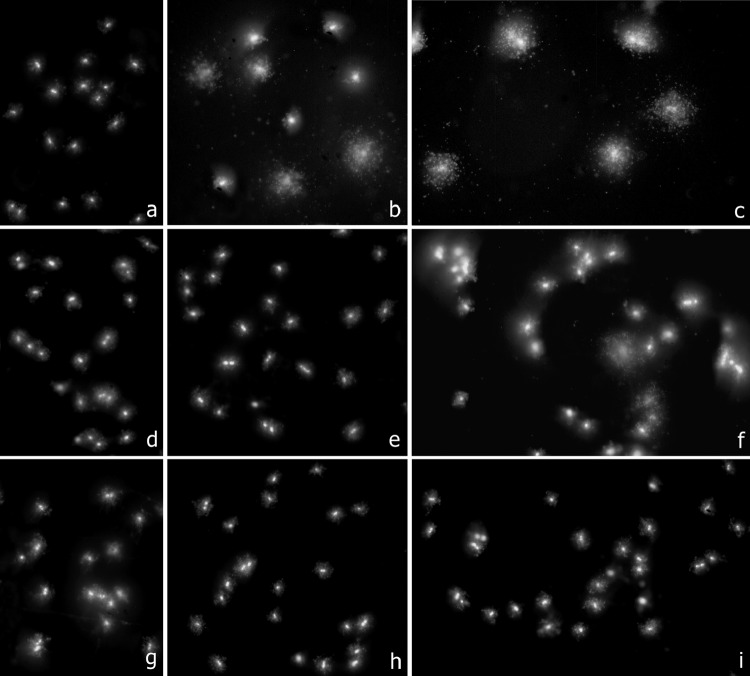
Fig 3.
Three representative clinical strains of A. baumannii, incubated with colistin for 60 min at 0 μg/ml (first column, a, d, and g), 0.5 μg/ml (second column, b, e, and h), and 1 μg/ml (third column, c, f, and i). Bacteria were processed to determine DNA fragmentation evaluated through diffusion of DNA spots from the nucleoids after technical processing. First row (a, b, and c), susceptible strain 1380 (MIC, 0.03 to 0.25 μg/ml); second row (d, e, and f), strain 1384 (MIC, 16 μg/ml); third row (g, h, and i), resistant strain 1382 (MIC, >32 μg/ml). Fragmentation of the nucleoids was visualized in the susceptible 1380 strain after 0.5 μg/ml (40% of affected cells) and 1 μg/ml (80% of affected cells). Only 21% of affected cells were scored after 1 μg/ml in the 1384 strain. Nucleoids practically always appeared intact in the highly resistant 1382 strain (2% of affected cells after 1 μg/ml).
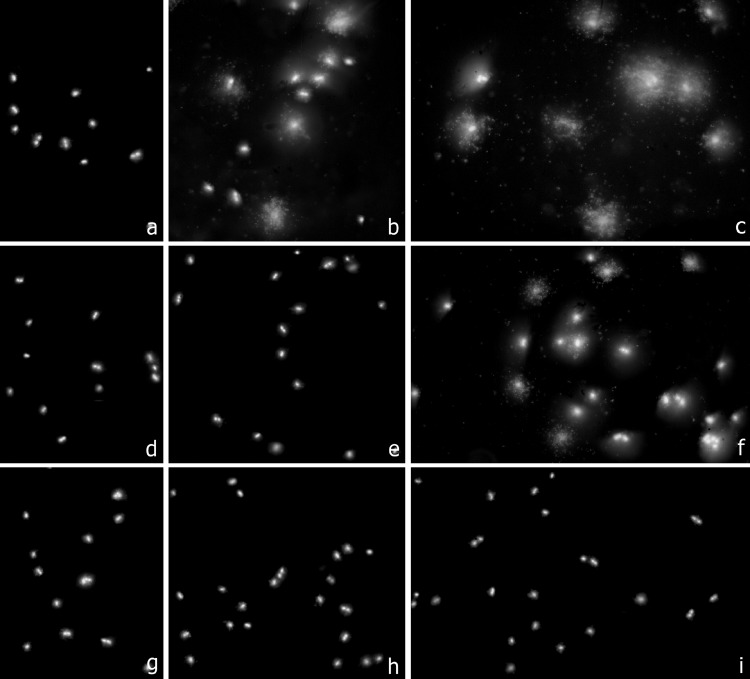
Fig 4.
Three representative clinical strains of A. baumannii, incubated with colistin for 60 min at 0 μg/ml (first column, a, d, and g), 0.5 μg/ml (second column, b, e, and h), and 1 μg/ml (third column, c, f, and i). Bacteria were processed to determine effects on the cell wall evaluated through nucleoid release after technical processing. First row (a, b, and c), susceptible strain 1380 (MIC, 0.03 to 0.25 μg/ml); second row (d, e, and f), strain 1384 (MIC, 16 μg/ml); third row (g, h, and i), resistant strain 1382 (MIC, >32 μg/ml). Release of the nucleoids was obvious in the susceptible 1380 strain after 0.5 μg/ml (41% of affected cells) and 1 μg/ml of colistin (100% of affected cells), but with lower frequency after 1 μg/ml in the 1384 strain (31% of affected cells). A discrete background of extracellular DNA fragments was visualized in the colistin-affected cultures. A. baumannii bacteria of the highly resistant 1382 strain almost always maintained their morphological appearance (2% of affected cells after 1 μg/ml).
Previously reported studies evaluating this technology were focused on antibiotics whose primary target was the DNA, like quinolones, which produce DNA fragmentation (27, 30), or the cell wall, like beta-lactams, which inhibit peptidoglycan synthesis (28). In those cases results were qualitative, affected versus not affected, since practically the whole population appeared damaged or not. Moreover, the data regarding susceptibility had been obtained after incubation of the bacteria with the CLSI breakpoint concentration for susceptibility. In the case of colistin, neither the DNA nor the cell wall is the primary target of the antibiotic. Nevertheless, DNA fragmentation and cell wall damage were evidenced, and these occurred only in a fraction of cells which increased with colistin concentration. This damage should be secondary, after the primary effect on lipid A and disruption of the cell membrane with subsequent alteration of permeability (13–15). One possibility is that the DNA and cell wall damages are collateral to the production of large amounts of reactive oxygen species (ROS) during the cell death process, as reported after treatment with bactericidal antibiotics like beta-lactams or quinolones (35). Given the quantitative nature of this damage in the population, even resistant strains may show a fraction of cells affected at the DNA and/or the cell wall when incubated with concentrations corresponding to the CLSI breakpoints for resistance. Better discrimination between susceptible and resistant strains could be achieved when incubating with lower concentrations, which produce damaged cells at much higher frequencies in the susceptible than in the resistant strains.
Since the foremost goal of the rapid test was to confidently identify resistant strains, the concentrations of colistin and the cell wall damage rates or DNA damage frequencies which optimally distinguish resistant from susceptible strains were investigated. For this purpose, we calculated ROC curves for each dose and analyzed the data with the aim of obtaining a 0% false-negative rate, because the most undesirable situation is to administer the antibiotic to a patient infected with a resistant strain, wrongly identified by the rapid test as susceptible. The ROC curves were excellent for every dose of colistin assayed, but incubation with a concentration of 0.5 μg/ml was found to be the most discriminative, with similar values of areas under curve for DNA fragmentation and for cell wall damage, 0.998 ± 0.003(P = 0.0001) (95% confidence interval [CI] for DNA damage, 0.993 to 1.000; 95% CI for cell wall damage, 0.991 to 1.000). The optimal cutoff values were 8.5% and 11% for DNA fragmentation and for cell wall damage, respectively. This resulted in the desirable 0% false-negative rate in both cases, i.e., 100% sensitivity (95% CI for both DNA damage and cell wall damage, 0.969 to 1.000). Moreover, the false-positive rate, i.e., for a susceptible strain which the test identifies as resistant, was 10% for DNA fragmentation and only 4% for cell wall damage, i.e., 90% and 96% specificity, respectively (95% CI for DNA damage, 0.807 to 0.993; 95% CI for cell wall damage, 0.896 to 1.000). Thus, the Youden index (J) values were 0.90 and 0.96 for DNA fragmentation and for cell wall damage, respectively (95% CI for DNA damage, 0.82 to 0.98; 95% CI for cell wall damage, 0.91 to 1.01).
In conclusion, both DNA fragmentation and cell wall damage are excellent parameters for discriminating susceptible and resistant strains, but the latter assay appears to be more specific. Obtaining ≤11% of bacteria with cell wall damage after incubation with 0.5 μg/ml colistin identifies resistant strains of A. baumannii with 100% sensitivity and 96% specificity according to CLSI standards. The assay requires 1 h of incubation with colistin concentrations of 0 and 0.5 μg/ml, 40 min of technical processing, and 5 to 15 min of scoring of 200 to 500 cells, a total of approximately 2 h. If the bacteria were not exponentially growing, 90 min of previous incubation in Mueller-Hinton broth is desirable in order to achieve exponential growth before the subsequent addition of colistin. This also eliminates the possible background of cells with fragmented DNA and cell wall damage in the control 0 μg/ml dose. Taking this into account, the results could be available in 3 h 30 min. Simultaneous processing of a control susceptible and a control resistant strain is adequate for validation of results. In comparison with the routine automatic microdilution procedure, the present methodology is much faster, 3 h 30 min versus 6 to 8 h, and allows a cell-by-cell assessment, so heterogeneity in damage induction and evolution with time, heteroresistance, or related phenomena could possibly be ascertained. Otherwise, the methodology is manual or semiautomatic and requires an epifluorescence microscope, as well as cell scoring. The latter can be easily automated through a motorized microscope coupled with a digital camera and image analysis software. Finally, the testing is restricted to colistin and a few other antibiotics previously validated (27, 28, 30, 31), unlike the commercial microdilution panels which evaluate 15 to 20 antibiotics. Nevertheless, more antibiotics will be assayed and validated in the future, for incorporation into the Micromax assay.
This is a rapid, simple, and reliable procedure for determining resistance to a relevant antibiotic like colistin in clinical A. baumannii strains. This may help facilitate the administration of early adequate antibiotic therapy in cases of life-threatening infections caused by A. baumannii and promote the responsible use of this therapeutic option, preserving it and avoiding the spread of resistances.
ACKNOWLEDGMENTS
We thank L. Rivas for the gift of the ATCC 19606 ColS and ColR strains.
This work has been supported by MagicBullet, Xunta de Galicia 10CSA916020PR, and by REIPI, the Spanish Network for Research in Infectious Diseases (Instituto de Salud Carlos III, REIPI RD12/0015) and the Fondo de Investigaciones Sanitarias (FIS PI12/00552) to G.B. M.J.M. is supported by the Subprograma Miguel Servet from the Ministerio de Economía y Competitividad of Spain (CP11/00314). MagicBullet is a project funded by the European Union–Directorate General for Research and Innovation through the Seventh Framework Program for Research and Development (grant agreement 278232) and has been running since 1 January 2012 (36 months' duration).
Footnotes
Published ahead of print 28 August 2013
REFERENCES
- 1. Jawad A, Seifert H, Snelling AM, Heritage J, Hawkey PM. 1998. Survival of Acinetobacter baumannii on dry surfaces: comparison of outbreak and sporadic isolates. J. Clin. Microbiol. 36:1938–1941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Peleg AY, Hooper DC. 2010. Hospital-acquired infections due to Gram-negative bacteria. N. Engl. J. Med. 362:1804–1813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Gootz TD, Marra A. 2008. Acinetobacter baumannii: an emerging multidrug-resistant threat. Expert Rev. Anti Infect. Ther. 6:309–325 [DOI] [PubMed] [Google Scholar]
- 4. Peleg AY, Seifert H, Paterson DL. 2008. Acinetobacter baumannii: emergence of a successful pathogen. Clin. Microbiol. Rev. 21:538–582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Taccone FS, Rodriguez-Villalobos H, De Backer D, De Moor V, Deviere J, Vincent JL, Jacobs F. 2006. Successful treatment of septic shock due to pan-resistant Acinetobacter baumannii using combined antimicrobial therapy including tigecycline. Eur. J. Clin. Microbiol. Infect. Dis. 25:257–260 [DOI] [PubMed] [Google Scholar]
- 6. Falagas ME, Bliziotis IA. 2007. Pandrug-resistant Gram-negative bacteria: the dawn of the post-antibiotic era? Int. J. Antimicrob. Agents 29:630–636 [DOI] [PubMed] [Google Scholar]
- 7. Falagas ME, Kasiakou SK. 2005. Colistin: the revival of polymyxins for the management of multidrugresistant Gram-negative bacterial infections. Clin. Infect. Dis. 40:1333–1341 [DOI] [PubMed] [Google Scholar]
- 8. Falagas ME, Kasiakou SK, Rafailidis PI, Zouglakis G, Morfou P. 2006. Comparison of mortality of patients with Acinetobacter baumannii bacteraemia receiving appropriate and inappropriate empirical therapy. J. Antimicrob. Chemother. 57:1251–1254 [DOI] [PubMed] [Google Scholar]
- 9. Landman D, Georgescu C, Martin DA, Quale J. 2008. Polymyxins revisited. Clin. Microbiol. Rev. 21:449–465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Li J, Nation RL, Turnidge JD, Milne RW, Coulthard K, Rayner CR, Paterson DL. 2006. Colistin: the re-emerging antibiotic for multidrug-resistant Gram-negative bacterial infections. Lancet Infect. Dis. 6:589–601 [DOI] [PubMed] [Google Scholar]
- 11. Nation RL, Li J. 2009. Colistin in the 21st Century. Curr. Opin. Infect. Dis. 22:535–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zavascki AP, Goldani LZ, Li J, Nation RL. 2007. Polymyxin B for the treatment of multidrug-resistant pathogens: a critical review. J. Antimicrob. Chemother. 60:1206–1215 [DOI] [PubMed] [Google Scholar]
- 13. Hancock REW. 1997. Peptide antibiotics. Lancet 349:418–422 [DOI] [PubMed] [Google Scholar]
- 14. Evans ME, Feola DJ, Rapp RP. 1999. Polymyxin B sulfate and colistin: old antibiotics for emerging multiresistant Gram-negative bacteria. Ann. Pharmacother. 33:960–977 [DOI] [PubMed] [Google Scholar]
- 15. Hermsen ED, Sullivan CJ, Rotschafer JC. 2003. Polymyxins: pharmacology, pharmacokinetics, pharmacodynamics, and clinical applications. Infect. Dis. Clin. North. Am. 17:545–562 [DOI] [PubMed] [Google Scholar]
- 16. Abraham N, Kwon DH. 2009. A single amino acid substitution in PmrB is associated with polymyxin B resistance in clinical isolate of Pseudomonas aeruginosa. FEMS Microbiol. Lett. 298:249–254 [DOI] [PubMed] [Google Scholar]
- 17. Adams MD, Nickel GC, Bajaksouzian S, Lavender H, Murthy AR, Jacobs MR, Bonomo RA. 2009. Resistance to colistin in Acinetobacter baumannii associated with mutations in the PmrAB two-component system. Antimicrob. Agents Chemother. 53:3628–3634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Beceiro A, Llobet E, Aranda J, Bengoechea JA, Doumith M, Hornsey M, Dhanji H, Chart H, Bou G, Livermore DM, Woodford N. 2011. Phosphoethanolamine modification of lipid A in colistin resistant variants of Acinetobacter baumannii mediated by the pmrAB two-component regulatory system. Antimicrob. Agents Chemother. 55:3370–3379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lee H, Hsu FF, Turk J, Groisman EA. 2004. The PmrA-regulated pmrC gene mediates phosphoethanolamine modification of lipid A and polymyxin resistance in Salmonella enterica. J. Bacteriol. 186:4124–4133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Nummila K, Kilpeläinen I, Zähringer U, Vaara M, Helander IM. 1995. Lipopolysaccharides of polymyxin B-resistant mutants of Escherichia coli are extensively substituted by 2-aminoethyl pyrophosphate and contain aminoarabinose in lipid A. Mol. Microbiol. 16:271–278 [DOI] [PubMed] [Google Scholar]
- 21. Trent MS, Ribeiro AA, Lin S, Cotter RJ, Raetz CR. 2001. An inner membrane enzyme in Salmonella and Escherichia coli that transfers 4-amino-4-deoxy-l-arabinose to lipid A: induction on polymyxin-resistant mutants and role of a novel lipid-linked donor. J. Biol. Chem. 276:43122–43131 [DOI] [PubMed] [Google Scholar]
- 22. Ko KS, Suh JY, Kwon KT, Jung SI, Park KH, Kang CI, Chung DR, Peck KR, Song JH. 2007. High rates of resistance to colistin and polymyxin B in subgroups of Acinetobacter baumannii isolates from Korea. J. Antimicrob. Chemother. 60:1163–1167 [DOI] [PubMed] [Google Scholar]
- 23. Park YK, Jung SI, Park KH, Cheong HS, Peck KR, Song JH, Ko KS. 2009. Independent emergence of colistin-resistant Acinetobacter spp. isolates from Korea. Diagn. Microbiol. Infect. Dis. 64:43–51 [DOI] [PubMed] [Google Scholar]
- 24. Payne DJ, Gwynn MN, Holmes DJ, Pompliano DL. 2007. Drugs for bad bugs: confronting the challenges of antibacterial discovery. Nat. Rev. Drug Discov. 6:29–40 [DOI] [PubMed] [Google Scholar]
- 25. Iregui M, Ward S, Sherman G, Fraser VJ, Kollef MH. 2002. Clinical importance of delays in the initiation of appropriate antibiotic treatment for ventilator-associated pneumonia. Chest 122:262–268 [DOI] [PubMed] [Google Scholar]
- 26. Kollef KE, Schramm GE, Wills AR, Reichley RM, Micek ST, Kollef MH. 2008. Predictors of 30-day mortality and hospital costs in patients with ventilator-associated pneumonia attributed to potentially antibiotic-resistant Gram-negative bacteria. Chest 134:281–287 [DOI] [PubMed] [Google Scholar]
- 27. Bou G, Otero FM, Santiso R, Tamayo M, Fernández MC, Tomás M, Gosálvez J, Fernández JL. 2012. Fast assessment of resistance to carbapenems and ciprofloxacin of clinical strains of Acinetobacter baumannii. J. Clin. Microbiol. 50:3609–3613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Santiso R, Tamayo M, Gosálvez J, Bou G, Fernández MC, Fernández JL. 2011. A rapid in situ procedure for determination of bacterial susceptibility or resistance to antibiotics that inhibit peptidoglycan biosynthesis. BMC Microbiol. 11:191. 10.1186/1471-2180-11-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Fernández JL, Cartelle M, Muriel L, Santiso R, Tamayo M, Goyanes V, Gosálvez J, Bou G. 2008. DNA fragmentation in microorganisms assessed in situ. Appl. Environ. Microbiol. 74:5925–5933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Santiso R, Tamayo M, Fernández JL, Fernández MC, Molina F, Villanueva R, Gosálvez J, Bou G. 2009. Rapid and simple determination of ciprofloxacin resistance in clinical strains of Escherichia coli. J. Clin. Microbiol. 47:2593–2595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Tamayo M, Santiso R, Gosálvez J, Bou G, Fernández JL. 2009. Rapid assessment of ciprofloxacin effect on chromosomal DNA from Escherichia coli with an in situ DNA fragmentation assay. BMC Microbiol. 9:69. 10.1186/1471-2180-9-69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Fernández-Reyes M, Rodríguez-Falcón M, Chiva C, Pachón J, Andreu D, Rivas L. 2009. The cost of resistance to colistin in Acinetobacter baumannii: a proteomic perspective. Proteomics 9:1632–1645 [DOI] [PubMed] [Google Scholar]
- 33. Moffatt JH, Harper M, Harrison P, Hale JD, Vinogradov E, Seemann T, Henry R, Crane B, St Michael F, Cox AD, Adler B, Nation RL, Li J, Boyce JD. 2010. Colistin resistance in Acinetobacter baumannii is mediated by complete loss of lipopolysaccharide. Antimicrob. Agents Chemother. 54:4971–4977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Soon RL, Nation RL, Cockram S, Moffatt JH, Harper M, Adler B, Boyce JD, Larson I, Li J. 2011. Different surface charge of colistin-susceptible and -resistant Acinetobacter baumannii cells measured with zeta potential as a function of growth phase and colistin treatment. J. Antimicrob. Chemother. 66:126–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kohanski MA, Dwyer DJ, Hayete B, Lawrence CA, Collins JJ. 2007. A common mechanism of cellular death induced by bactericidal antibiotics. Cell 130:797–810 [DOI] [PubMed] [Google Scholar]