Abstract
Conventional blood culturing using automated instrumentation with phenotypic identification requires a significant amount of time to generate results. This study investigated the speed and accuracy of results generated using PCR and pyrosequencing compared to the time required to obtain Gram stain results and final culture identification for cases of culture-confirmed bloodstream infections. Research and physician-ordered blood cultures were drawn concurrently. Aliquots of the incubating research blood culture fluid were removed hourly between 5 and 8 h, at 24 h, and again at 5 days. DNA was extracted from these 6 time point aliquots and analyzed by PCR and pyrosequencing for bacterial rRNA gene targets. These results were then compared to those of the physician-ordered blood culture. PCR and pyrosequencing accurately identified 92% of all culture-confirmed cases after a mean enrichment time of 5.8 ± 2.9 h. When the time needed to complete sample processing was included for PCR and pyrosequencing protocols, the molecular approach yielded results in 11.8 ± 2.9 h compared to means of 27.9 ± 13.6 h to obtain the Gram stain results and 81.6 ± 24.0 h to generate the final culture-based identification. The molecular approach enabled accurate detection of most bacteria present in incubating blood culture bottles on average about 16 h sooner than Gram stain results became available and approximately 3 days sooner than the phenotypic identification was entered in the Laboratory Information System. If implemented, this more rapid molecular approach could minimize the number of doses of unnecessary or ineffective antibiotics administered to patients.
INTRODUCTION
Bloodstream infections (BSIs) are the third most common cause of hospital mortality and are the 7th and 11th leading cause of all U.S. deaths for infants and adults, respectively (1, 2). In a 7-year cohort study of 49 U.S. hospitals reported in 2004, mortality rates directly attributable to BSIs were estimated to be between 16% and 40%. According to surveillance data from U.S. hospitals, 87% of BSIs are caused by a single infecting microorganism, with Gram-positive bacteria accounting for 65% of BSIs, Gram-negative bacteria for 25%, and fungi for 9.5% (3).
The current gold standard for diagnosing bloodstream infections utilizes automated blood culturing, usually performed on at least two separate blood draws taken at different times (4, 5). Studies have found that the sensitivity of blood culturing is between 65% and 96%, depending on the number of cultures taken and the volume of blood obtained for each culture. Within 5 days of growth, 99% of BSIs can be detected using Bactec blood culturing bottles (6). Automated blood culture systems can detect as few as 1 CFU/ml, while molecular methods have demonstrated limits of detection between 10 and 100 CFU/ml (1, 7).
While these characteristics are certainly benefits of conventional blood culture, a major weakness is that it requires an extended amount of time for the organism(s) to grow to detectable levels and then for isolation and identification (ID) of the organism(s). One study found that the culturing time required for bacteria to grow to levels detectable by an automated instrument averaged 12 to 24 h for Gram-negative organisms and 24 to 48 h for Gram-positive organisms (7). A separate study of neonates at the Magee Women's Hospital in Pittsburgh, PA, determined that the mean time to detect bacterial growth using an automated blood culture instrument was 18 h (range, 11 to 28 h), while the mean time to complete the organism's final identification was 49 h (range, 23 to 73 h) (8).
The relatively long time required to diagnose a BSI using current methods presents a significant challenge given the small window in which effective antibiotic treatment can be successfully administered and the public health risk associated with administering ineffective and/or unnecessary antibiotics. A number of potential molecular methods of diagnosis, including the use of PCR assays and nucleotide sequencing, have been investigated as a way to address this issue.
Significant success has been seen using PCR protocols in neonates, who typically have higher bacterial loads (9) and fewer bacterial species causing sepsis than adult populations (8). Studies using real-time PCR to amplify universal sequences of the 16S rRNA gene were shown to have high sensitivity and specificity, as well as a very high negative predictive value (99.2%) when used in neonatal populations (7, 10). The high negative predictive value of the PCR assay means that a negative result can be obtained within hours, not days, thus leading to a significant reduction in the duration of unnecessary antibiotic therapy (11).
Pyrosequencing is a valuable tool used in conjunction with PCR to diagnose BSIs and can eliminate the 1 to 2 additional days needed to isolate the organism for phenotypic identification. Previous studies combining real-time PCR and pyrosequencing have been successful in identifying bacteria in blood culture bottles from both infants and adults that demonstrate detectable growth. One such study found an overall agreement of 97.8% between molecular and culture-based methods of diagnosis, with a slightly better agreement (98.8%) when infections were monomicrobial (12).
In this study, we assessed whether screening blood culture fluids in the first few hours of their incubation would permit more rapid detection of culture-confirmed bacterial BSIs than conventional automated blood culturing. To that end, we investigated the accuracy and speed of our molecular approach in identifying the microorganism(s) in cases of culture-confirmed BSIs and compared the times required to the times required to obtain the Gram stain result and final culture identification.
(M. S. Moore completed this work for her senior thesis in partial fulfillment of the requirements for the degree of bachelor of science at Georgetown University, Washington, DC.)
MATERIALS AND METHODS
Enrollment.
This study was designed as a prospective, observational study in an urban academic emergency department (ED) and intensive care unit (ICU). Participants were eligible if they had a physician-ordered blood culture and a complete blood count (CBC) sample drawn. A total of 1,144 eligible study participants were provided information about the study by clinical staff and consented to participate. Potential participants were excluded from the study if they declined to participate or if a blood sample could not be obtained. This study describes the results from the subset of participants with culture-confirmed BSIs. The Institutional Review Board of The George Washington University (GWU) and Medical Center, Office of Human Research approved this study.
Automated blood culturing and phenotypic identification.
Blood (8 to 10 ml) for conventional culture was collected by either a venous or arterial draw and inoculated directly into one each of a BD Bactec Plus Aerobic/F medium blood culture bottle (catalog no. 442192; Becton, Dickinson [BD], Sparks, MD) and a standard Anaerobic/F medium blood culture bottle (catalog no. 442191; BD). Physician-ordered blood cultures were drawn and sent to the hospital clinical microbiology lab for routine blood culture performed using a Bactec 9240 bottle (BD). Each bottle was loaded into the Bactec 9240 automated blood culture instrument within 1 h from the time of receipt in the clinical laboratory in accordance with the manufacturer's recommendation.
Bottles flagged by the instrument for detectable levels of CO2 production had fluid removed for Gram staining and subculturing onto appropriate agar-based culture plates. Purified bacterial colonies were analyzed either by an automated identification system (Vitek 2; bioMérieux, Durham, NC) or with the appropriate biochemical reagents for manual phenotypic identification.
Research blood sample incubation.
The entire 8 to 10 ml of the research blood sample, drawn at the same time as the routine clinical blood sample using the same needle stick, was collected in a tube containing sodium polyanethole sulfonate-sodium chloride (catalog no. 364960; BD) was inoculated in a Bactec Plus Aerobic/F blood culture bottle (BD catalog no. 442192) and then immediately placed in a shaking incubator at 37°C and 150 rpm (Excella E24 incubator shaker series; New Brunswick Scientific, Enfield, CT). The time of inoculation was noted, and beginning at 5 h after inoculation and for every subsequent hour through 8 h, again at 24 h, and after 5 days, 1.5-ml aliquots of blood were removed aseptically for DNA extraction. At the terminal 5-day time point, a blood smear for Gram staining was also prepared, air dried, heat fixed, and stained. In all, aliquots at 6 time points were removed from each incubating research blood culture bottle for DNA extraction, PCR, and pyrosequencing. Blood samples were rejected if the blood was grossly hemolyzed and/or clotted or if the volume of blood was inadequate.
DNA extraction of blood culture fluids obtained from time point aliquots.
DNA was extracted from each of the six time point aliquots removed from the incubating blood culture bottle. Five hundred microliters of blood was centrifuged for 5 min at 5,000 × g at room temperature (RT), and the resulting supernatants were discarded, after which 100 μl of 5 mM guanidinium-HCl in 100 mM Tris-HCl (pH 8.0) was added to resuspend each pellet. The resuspended samples were added to 0.24 g 0.1-mm zirconium silica beads (catalog no. 11079101z; Biospec Products, Bartlesville, OK) and vortexed for 5 min. To each sample was added 400 μl distilled H2O (dH2O) and 800 μl 99% benzyl alcohol, followed by centrifugation for 5 min at 5,000 × g. The aqueous supernatant was retained, and this extraction step was repeated. Next, a 1/10 volume of 3 M sodium acetate (NaOAC) and an equal volume of 99% isopropyl alcohol and 1 μl glycogen were added to each sample, and the samples were centrifuged for 15 min at 4°C and 13,000 × g. The pellets were washed with 100 μl 70% ethanol (EtOH) and centrifuged for 5 min at 4°C and 13,000 × g. The supernatant was removed, and the pellets were allowed to air dry. Once they were dry, 50 μl of 1× TE (10 mM Tris-HCl [pH 8.0], 1 mM EDTA) was added to each pellet.
Real-time PCR.
DNA extracts generated from the 6 time point aliquots were screened by PCR using slightly modified sequences from previously published primers (Table 1) (10, 12), including a 16S rRNA gene universal target and, when applicable, either a Staphylococcus 16S rRNA gene, Streptococcus 23S rRNA gene, or enteric Gram-negative rod (GNR) 23S rRNA gene target(s). PCR mixtures of 25 μl were set up, with 0.5 μl each of 10 μM concentrations of the forward and reverse primers (one primer of each pair was biotinylated), 12.5 μl of 2× SYBR Premix Ex Taq polymerase master mix (catalog no. RR420A; TaKaRa Biotechnology, Dalian Corp., Ltd.), and 5 μl of each DNA extract. PCR cycling conditions for the universal, Staphylococcus, and Streptococcus targets consisted of 95°C for 60 s, then 40 cycles of 95°C for 20 s, 60°C for 60 s, and 72°C for 15 s. Cycling conditions for the enteric Gram-negative rod target consisted of 95°C for 120 s, then 40 cycles of 95°C for 20 s and 62°C for 30 s. All PCR testing was performed using Smart Cycler instruments (Cepheid Inc., Sunnyvale, CA). An appropriate positive control for each group was included: 1 μl DNA extracts from purified isolates of either Staphylococcus aureus or coagulase-negative Staphylococcus species (CoNS) (Staphylococcus target), Streptococcus pneumoniae or an Enterococcus sp. (Streptococcus target), or Escherichia coli or Klebsiella pneumoniae (enteric Gram-negative rod target). Any of the above bacterial DNA extracts served as a positive control for the universal target. Two no-template negative controls, one at the beginning and one at the end, were also included in each PCR run.
Table 1.
Primer sequences used for analysis of bacterial rRNA genes
| Organism group | rRNA gene target | Real-time PCR primer seta (5′–3′) | Pyrosequencing primer(s)b (5′–3′) |
|---|---|---|---|
| Staphylococcus | 16S | FB, TGCCTAATACATGCAAGTCGAGCG; R, GTTGCCTTGGTAAGCCGTTACCTT | GTGTTACTCACCCGTCCGCCGCTA |
| Streptococcus | 23S | FB, GCCTTTTGTAGAATGAACCGGCGA; R, CGTTTGGAATTTCTCCGCTACCCA | TCACATGGTTTCGGGTCTA |
| Enteric | 23S | F1, CTAAGGCGAGGCCGAAAG; F2, CTAAGGCGAGGCTGAAAAG; RB, CTACCTGACCACCTGTGTCG | S1, GGTTGTCCCGGTTTA; S2, GGTCGTCCCGGTTCA |
| Universal | 16S | FB, AACTGGAGGAAGGTGGGGAT; R, AGGAGGTGATCCAACCGCA | TACAAGGCCCGGGAACGTATTCA |
FB, biotinylated forward primer; R, reverse primer; RB, biotinylated reverse primer; F1 and F2, degenerate forward primers used in combination.
S1 and S2, degenerate pyrosequencing primers used in combination.
Pyrosequencing.
The resulting 25 μl of biotin-labeled PCR product was immobilized onto 5 μl of streptavidin-Sepharose high-performance beads (catalog no. 17-5113-01; GE Healthcare, Little Chalfont, United Kingdom) suspended in 50 μl of binding buffer (catalog no. 979006; Qiagen, Germantown, MD) in a deep-well plate (catalog no. NC9650319; Phenix Research Products, Candler, NC) that was placed on a Thermomixer (catalog no. 5350 24744; Eppendorf AG, Hamburg, Germany) and shaken at 1,400 rpm for 10 min at RT. Using the pyrosequencing vacuum prep tool (catalog no. 9001516; Qiagen), the beads were immersed in 70% EtOH, followed first by 0.2 M NaOH and then by wash buffer (catalog no. 979008; Qiagen). The resulting single-stranded biotinylated PCR product was bound to beads and transferred into another deep-well plate containing 2 μl of 10 μM sequencing primer (Table 1) along with 38 μl annealing buffer (catalog no. 979009; Qiagen) in each well. The deep-well plate was placed in a heated shaker (1,400 rpm) at 90°C for 1 min, followed by RT for 5 min to allow complementary strands to anneal. The prep tool carried the beads containing the single-stranded biotinylated PCR product-primer hybrid through 70% EtOH followed by wash buffer (catalog no. 979008; Qiagen) before being transferred into a low-well plate (catalog no. 979002; Qiagen) containing 40 μl of annealing buffer (catalog no. 979009; Qiagen) in each well. The plate containing the prepared sample was analyzed using the PyroMark ID pyrosequencer (Qiagen) with PyroMark Gold Q96 reagents (catalog no. 972804; Qiagen) and a dispensation program of either an 11(ACTG) deoxynucleoside triphosphate (dNTP) for universal, Staphylococcus, and Streptococcus rRNA gene targets or an AGC-12(CTGA) dNTP for the enteric rod rRNA gene target. Using PyroMark Identifire v1.0.5.0 software, the resulting sequences were compared to our reference library (8, 10, 12), which included validated sequences from previously identified clinical isolates, ATCC reference sequences, and sequences found in the NCBI GenBank using the BLAST algorithm (8, 10, 12). One hundred percent identity was required to make a genus or species level identification.
Calculating the time required to obtain results using the molecular approach and time saved compared to conventional automated blood culturing.
For each set of DNA extracts generated from the time point series, we recorded the first time point aliquot of the six tested that was positive by real-time PCR and pyrosequencing. This information was used to determine the mean enrichment time needed to obtain a positive result. A generous estimate of 6 h was used to reflect the additional time needed to complete sample processing, PCR, and pyrosequencing, which reflected the time required to process the six time point aliquots from one participant. The sum of the mean enrichment time plus 6 h was used to calculate the total time needed for this molecular approach. The time required for a clinical diagnosis of BSI using conventional automated blood culturing was based on electronic medical record data and included the dates and times of the physician-ordered blood draw for culture and of the telephone call made to the unit with the Gram-stain result and lastly the date when the final identification was entered into the laboratory information system (LIS); the software used by the hospital records only the date, and not time, when the final identification is entered into the LIS. When comparing the times required, a two-tailed t test was utilized to assess statistical significance.
RESULTS
Description of bacteria present in culture-confirmed cases of bloodstream infections.
One hundred bacterial isolates were cultured from 97 blood culture bottles obtained from 95 enrolled participants. There were 3 polymicrobial infections (2 from the ED and 1 from the ICU). Culture-confirmed cases containing strict anaerobes or yeast were not included in this evaluation. The 100 blood culture isolates included 51 Staphylococcus sp. isolates (19 S. aureus isolates and 32 CoNS sp. isolates), 28 Streptococcus sp. isolates (8 Enterococcus sp. isolates, 5 S. pneumoniae isolates, 5 viridans group streptococcal isolates, 4 group G Streptococcus [GGS] isolates, 2 group A Streptococcus [GAS]isolates, 2 group B Streptococcus [GBS]isolates, and 2 group C Streptococcus [GCS] isolates), 15 enteric Gram-negative rod isolates (8 E. coli isolates and 1 isolate each of K. pneumoniae, Serratia marcescens, Providencia rettgeri, Enterobacter aerogenes, Citrobacter freundii, Proteus mirabilis, and a Salmonella sp.), 5 nonfermentative Gram-negative rod isolates (4 Pseudomonas aeruginosa isolates and 1 Stenotrophomonas maltophilia isolate), and 1 Bacillus sp. isolate (not Bacillus anthracis).
Of the 97 positive blood culture bottles, 47 (48.5%) originated from ED patients, while 50 (51.5%) were from the ICU. One hundred percent (10/10) of the blood cultures containing S. pneumoniae or viridans group streptococci and 80% (12/15) of blood cultures containing an enteric Gram-negative rod originated from participants enrolled in the ED, while 87.5% (7/8) of all Enterococcus spp. and 75% (3/4) of P. aeruginosa isolates came from blood cultures originating from participants in the ICU.
The molecular approach demonstrated a high rate of agreement with conventional automated blood culturing for detecting and classifying bacteria.
There was 92% (92/100) agreement between the molecular approach and phenotypically based culture ID (Table 2), with 98% agreement (50/51) for Staphylococcus spp., 96.4% (27/28) for Streptococcus spp., 73.3% (11/15) for enteric Gram-negative rods (GNRs), 80% (4/5) for nonfermenting GNRs, and 100% (1/1) for the Bacillus sp.
Table 2.
Number and percent of results in agreement, disagreement, or undetected using the molecular method compared to the corresponding physician-ordered automated blood culture resulta
| Primer grouping | Total nb | Agreement |
Disagreement |
Not detected |
|||
|---|---|---|---|---|---|---|---|
| n | % | n | % | n | % | ||
| Staphylococcus spp.c | 51 | 50 | 98.0 | 1d | 0.0 | 0 | 2.0 |
| Streptococcus spp.e | 28 | 27 | 96.4 | 1f | 3.6 | 0 | 0.0 |
| Enteric GNRsg,h | 15 | 11 | 73.3 | 0 | 0.0 | 4i | 26.7 |
| Universal onlyj | 6 | 4 | 66.7 | 0 | 0.0 | 2 | 33.3 |
| All | 100 | 92 | 92 | 2 | 2.0 | 6 | 6.0 |
Numbers and percentages of the 100 isolates detected by PCR/pyrosequencing using a target-specific and/or universal primer set(s). Purified isolates were available for all 8 discordant samples; DNA extracts were analyzed using PCR/pyrosequencing, and all isolates were correctly identified using both the universal and target-specific primer sets (data not shown).
n, number of isolates.
Results for correct identification of Staphylococcus spp. are as follows: S. aureus (including both methicillin-sensitive S. aureus [MSSA] and methicillin-resistant S. aureus [MRSA]), 18/19 isolates (94.7%); CoNS, 32/32 isolates (100%).
While the universal primer set correctly classified the organism as a Staphylococcus sp., the DNA sequences generated using the Staphylococcus-specific primers failed to match any known sequence in our library or the GenBank database.
Results for correct identification of Streptococcus and Enterococcus spp. (n = 28) are as follows: GAS, 2/2 isolates (100%); GBS, 2/2 isolates (100%); GCS, 2/2 isolates (100%); GGS, 3/4 isolates (75%); Enterococcus spp., 8/8 isolates (100%); S. pneumoniae, 5/5 isolates (100%); viridans group streptococci, 5/5 isolates (100%).
Culture-based identification was GGS. While the universal primer set correctly classified the organism as a Streptococcus sp., the Streptococcus-specific primer set identified the organism as GCS. The difference in classification between GGS and GCS was a single base in the 233-bp region of the Streptococcus-specific 23S rRNA gene target being amplified.
GNRs, Gram-negative rods.
Results for correct identification of enteric GNRs (n = 15) are as follows: E. coli, 6/8 isolates (75%); Enterobacter/Citrobacter spp., 2/2 isolates (100%); S. marcescens, 1/1 isolate (100%); Salmonella sp., 1/1 isolate (100%); P. rettgeri, 1/1 isolate (100%); P. mirabilis, 0/1 isolate (0%); K. pneumoniae, 0/1 isolate (0%).
PCR/pyrosequencing reactions from both the universal primers and the enteric GNR primers failed to detect 2 E. coli isolates and 1 isolate each of P. mirabilis and K. pneumoniae.
Results for correct identification using the universal target only (n = 6) are as follows: P. aeruginosa, 3/4 isolates (75%); Bacillus sp., 1/1 isolate (100%); S. maltophilia: 0/1 isolate (0%).
Of the 8 discordant results, 6 were from blood cultures containing GNRs, including 4 enteric GNR and 2 nonfermentative GNR species not detected by the molecular approach; the former 4 samples were not detected by either the 16S rRNA gene universal primer set or the 23S rRNA gene enteric GNR-specific primer set, while the latter 2 samples were not detected by the universal primer set. Three of these 6 (50%) discordant results were from blood cultures containing polymicrobial growth. Presence of PCR inhibitors was ruled out by performing PCR on these time point aliquot DNA extracts spiked with 1,000 CFU/ml of purified bacterial DNA using the 16S rRNA gene universal primer set (data not shown).
PCR/pyrosequencing was also performed on 300 DNA extracts generated from sets of 6 time point aliquots of 50 culture-negative specimens using the universal 16S rRNA gene primer set. These blood cultures were obtained from the same patient population (ED or ICU) as the other samples tested in this study. All 300 DNA extracts tested were negative for bacterial DNA (data not shown).
Most bacteria within the research blood culture bottles from culture-confirmed BSIs were first detected at the 5-h time point.
Figure 1 illustrates the percent distribution of the first time point aliquot in each set that was found to be positive for bacteria using the molecular approach. The results clearly show that the majority (72.8%, 67/92) of bacteria from the 92 concordant culture-confirmed cases of BSI were first detected and correctly classified in DNA extracts made after 5 h of enrichment. After 8 h, 93.5% (86/92) of culture-confirmed cases were detected and accurately classified, with only 6.5% (6/92) requiring more than 8 h for detection. Because it was not feasible to collect hourly time points between 8 h and 24 h, the specific incubation time required for this subset of samples is unknown. Three isolates were first detected from the 24-hour-time-point aliquot (one each of S. aureus, S. marcescens, and P. rettgeri), while 3 were detected only from the 5-day-time-point aliquot (one each of an E. coli, CoNS, and Enterococcus faecium).
Fig 1.
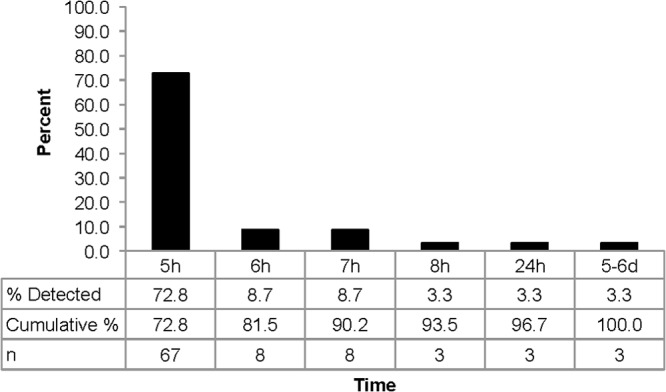
Bar graph depicting the percentages, cumulative percentages, and numbers of isolates detected at the indicated time points by the molecular method from culture-confirmed blood culture samples.
The molecular approach provided bacterial identification from culture-confirmed bloodstream infections sooner than did the Gram stain or phenotypic culture.
The overall mean enrichment time needed to detect bacteria using the molecular approach was 5.8 ± 2.9 h and ranged from 5.0 ± 0.2 h to first detect Streptococcus spp. to 7.1 ± 5.7 h to first detect enteric GNRs (Table 3).
Table 3.
Mean times required to obtain first positive molecular result compared to mean times required to obtain Gram stain result or final identification from corresponding physician-ordered blood culture
| Bacterial grouping | Result (mean h ± SD) for: |
P by t test | |||
|---|---|---|---|---|---|
| PCR/pyrosequencing |
Physician-ordered blood culture |
||||
| First positive time pointd (n = 89) | TATb for MDxc IDd (n = 89) | TAT for Gram staine (n = 76) | TAT for phenotypic culture ID (n = 92) | ||
| Staphylococcus spp. | 5.9 ± 2.8 | 11.9 ± 2.8 | 31.8 ± 12.9 | 81.6 ± 24.0 | <0.001 |
| Streptococcus spp. | 5.0 ± 0.2 | 11.0 ± 0.2 | 26.3 ± 16.6 | 79.2 ± 24.0 | <0.001 |
| Enteric GNRsa | 7.1 ± 5.7 | 13.1 ± 5.7 | 20.3 ± 7.5 | 81.6 ± 24.0 | <0.001 |
| All bacteria | 5.8 ± 2.9 | 11.8 ± 2.9 | 27.9 ± 13.6 | 81.6 ± 24.0 | <0.001 |
GNRs, Gram-negative rods.
TAT, turnaround time.
MDx, molecular.
For the purposes of these calculations, 3 outliers were excluded from this analysis, including 1 each of a Staphylococcus sp., a Streptococcus sp., and an enteric GNR species, which were detected and accurately identified by PCR/pyrosequencing only from the final 5-day-time-point aliquot.
Electronic medical records from 11 of the physician-ordered, culture-confirmed positive blood culture bottles lacked documentation for the specific time that the Gram stain result was called to the unit and were therefore excluded from this analysis. For the purposes of these calculations, 5 outliers were excluded from this analysis, including 3 Staphylococcus isolates, with Gram stain times of 61.2, 123.3, and 121.9 h, and 2 Streptococcus isolates, with Gram stain times of 76.2 and 66.6 h. Removal of these 5 outliers was considered acceptable because the adjusted mean times to Gram stain were not appreciably different from the calculated medians.
Calculating total turnaround times (TAT) to detect and classify bacteria using molecular testing included a generous estimate of 6 h in order to complete DNA extraction, PCR, and pyrosequencing protocols. This resulted in an overall mean total TAT of 11.8 ± 2.9 h; mean total TAT ranged from 11.0 ± 0.2 h for Streptococcus spp. to 13.1 ± 5.7 h for enteric GNRs. In contrast, Gram stain results took an average of 27.9 ± 13.6 h to be called to the hospital unit, ranging from 20.3 ± 7.5 h for the enteric GNRs to 31.8 ± 12.9 h for Staphylococcus spp. Phenotypic identification took an average of 3.4 days to be entered into the LIS. The molecular approach allowed for a more specific bacterial classification ∼16 h sooner than the less informative Gram stain result and ∼3 days sooner than the conventional phenotypic culture-based ID. Based on a two-tailed t test, there was a statistically significant decrease in the time required for the molecular approach to generate an identification compared to the Gram stain result for all organisms (P < 0.001) as well as for each specific grouping of bacteria (Table 3).
Gram stain results generated from the clinical bottles were compared to those obtained from the terminal 5-day-time-point aliquot of the research samples. There was perfect correlation between the clinical and research Gram stain results from the 92 concordant cases (data not shown). Between the two discordant samples (Table 2) in which DNA sequences either generated a different ID (GGS versus GCS using the Streptococcus-specific target) or did not match any known sequences in the library (S. aureus using the Staphylococcus-specific target), there was concordance between the clinical and research Gram stain results—Gram-positive cocci either in chains or clusters were observed (data not shown). In contrast, in the 6 research samples where the molecular approach did not detect any organisms, no bacteria were seen in the Gram stains of the blood smears from the 5-day-time-point aliquots (data not shown).
DISCUSSION
This study, which focused primarily on a subset of participants with culture-proven BSIs, found that the molecular approach was significantly faster at detecting BSIs than a conventional culture-based approach, while still maintaining a high degree of accuracy. A 2006 study by Kumar et al. found that in a critically ill ICU population, every hour of delay in administering antibiotic therapy resulted in a 7.6% decrease in survival rate (15). A study by Ibrahim et al. in a similar population determined that administration of inadequate or inappropriate antimicrobial treatment was also associated with increased hospital mortality (13). Rapid detection of BSI has the potential to not only improve patient outcomes due to quicker administration of appropriate antibiotics but also improve antibiotic stewardship by reducing patient exposures to ineffective or unnecessary broad-spectrum antibiotics (11, 14). Additionally, the fact that 72.8% of all research samples from this study tested positive by the molecular approach after just 5 h of enrichment suggests that a large percentage may actually have been detected using this molecular approach even sooner.
The biggest limitation of this study was the failure to detect six culture-confirmed cases (all GNRs) by both the universal and, if applicable, the enteric-bacterium-specific target. It was determined that the universal and/or target-specific PCR primer sets used here did recognize DNA made from the purified bacterial isolates (data not shown). Additionally, the presence of PCR inhibitors was ruled out by performing spiking experiments with the DNA extracts made from these time point aliquots (data not shown). It is therefore possible that bacteria in the research blood culture bottles may have been present below the limit of detection. It is important to note that no evidence of bacteria was found in the Gram stains made from the 5-day-time-point aliquots of these six research blood culture bottles (data not shown). Therefore, it is also possible that, while the bottles were drawn consecutively, bacteria may have been present in the clinical bottle and not in the research sample. Alternatively, the research blood culture bottles could have been incubated longer than 5 days to see if the bacteria could have been detected. However, this is the standard time used in the clinical labs, and we wanted to be consistent with their practice.
Additionally, the groups of primers used here were not designed to determine the species of every etiologic agent known to cause BSI. For example, the pyrograms generated using the enteric primers are identical for those for Citrobacter spp. and Enterobacter spp. (12). However, when making decisions about antibiotic choices, physicians rely heavily on hospital antibiogram data, which often group several related species together to generate the antimicrobial susceptibility testing data. Nevertheless, the information provided by our molecular approach was much more specific than the results from the Gram stain, which currently is the first piece of information provided to physicians that is used to reassess antibiotic decisions.
This study was also limited in its ability to detect bacterial growth between 8 and 24 h. Samples that were first detected at 24 h may have been positive sooner but, due to issues of feasibility, could not have been tested before the 24-hour time point. Similarly, we did not sample daily after 24 h, so the small number that were not picked up until the 5-day-time-point aliquot may also have detected sooner. It is therefore possible that the calculated mean TAT for the molecular approach may in reality be lower than was calculated (though the 5-day time points were excluded from the mean TAT, the 3 samples first positive at 24 h were included).
For streamlining purposes, it is possible to envision developing a testing algorithm that consisted of batch testing of all newly inoculated bottles from the previous shift or previous day rather than processing and testing individual samples one at a time. Automation of some or all of the steps would also help improve efficiency and reduce hands-on time.
Further research efforts in molecular diagnosis of BSIs should be directed toward continuing to reduce TATs, detecting bacteria directly from whole-blood samples, and addressing the issue of antibiotic resistance. Although a few molecular methods have been successfully developed to detect the more common antibiotic-resistant organisms seen in the clinical setting (methicillin-resistant S. aureus and vancomycin-resistant Enterococcus spp.), there is still a large array of genes conferring antibiotic resistance that are found in many BSI-causing bacteria other than S. aureus and Enterococcus spp. for which molecular detection methods would still need to be developed (9).
For the near future, molecular testing won't be a substitute for conventional culture. If molecular methods are implemented, routine culturing will still be needed to enhance sensitivity and perform susceptibility testing. In summary, the results of this study clearly show that it was possible to quickly and accurately diagnose BSIs using a molecular approach. The ability to detect and classify 72.8% of the cases after only ∼11 to 13 h can mean a dramatic reduction in the number of inappropriate antibiotic doses given to patients over the course of their treatment. By allowing physicians to tailor their antibiotic treatment of BSI hours or even days sooner, this molecular approach has the potential to improve patient outcome and lessen the selective pressures that promote antibiotic resistance.
ACKNOWLEDGMENT
This research was funded by grant R01 AI0 73342 from NIH NIAID (to J.A.J., principal investigator).
Footnotes
Published ahead of print 28 August 2013
REFERENCES
- 1. Klouche M. 2008. Rapid diagnostic tests to detect pathogenic microorganisms. Clin. Chem. Lab. Med. 46:885–887 [DOI] [PubMed] [Google Scholar]
- 2. Kochanek KD, Xu J, Murphy SL, Miniño AM, Kung HC. 2011. Deaths: final data for 2009. Natl. Vital Stat. Rep. 60(3):1–116 [PubMed] [Google Scholar]
- 3. Wisplinghoff H, Bischoff T, Tallent SM, Seifert H, Wenzel RP, Edmond MB. 2004. Nosocomial bloodstream infections in US hospitals: analysis of 24,179 cases from a prospective nationwide surveillance study. Clin. Infect. Dis. 39:309–317 [DOI] [PubMed] [Google Scholar]
- 4. O'Grady NP, Barie PS, Bartlett JG, Bleck T, Carroll K, Kalil AC, Linden P, Maki DG, Nierman D, Pasculle W, Masur H. 2008. Guidelines for evaluation of new fever in critically ill adult patients: 2008 update from the American College of Critical Care Medicine and the Infectious Diseases Society of America. Crit. Care Med. 36:1330–1349 [DOI] [PubMed] [Google Scholar]
- 5. Ntusi N, Aubin L, Oliver S, Whitelaw A, Mendelson M. 2010. Guideline for the optimal use of blood cultures. S. Afr. Med. J. 100:839–843 [DOI] [PubMed] [Google Scholar]
- 6. Cockerill FR, III, Wilson JW, Vetter EA, Goodman KM, Torgerson CA, Harmsen WS, Schleck CD, Ilstrup DM, Washington JA, II, Wilson WR. 2004. Optimal testing parameters for blood cultures. Clin. Infect. Dis. 38:1724–1730 [DOI] [PubMed] [Google Scholar]
- 7. Jordan JA, Durso MB. 2005. Real-time polymerase chain reaction for detecting bacterial DNA directly from blood of neonates being evaluated for sepsis. J. Mol. Diagn. 7:575–581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Jordan JA, Butchko AR, Durso MB. 2005. Use of pyrosequencing of 16S rRNA fragments to differentiate between bacteria responsible for neonatal sepsis. J. Mol. Diagn. 7:105–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Klouche M, Schroder U. 2008. Rapid methods for diagnosis of bloodstream infections. Clin. Chem. Lab. Med. 46:888–908 [DOI] [PubMed] [Google Scholar]
- 10. Jordan JA, Durso MB, Butchko AR, Jones JG, Brozanski BS. 2006. Evaluating the near term infant for early onset sepsis: progress and challenges to consider with 16S rDNA polymerase chain reaction testing. J. Mol. Diagn. 8:357–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Brozanski BS, Jones JG, Krohn MJ, Jordan JA. 2006. Use of polymerase chain reaction as a diagnostic tool for neonatal sepsis can result in a decrease in use of antibiotics and total neonatal intensive care unit length of stay. J. Perinatol. 26:688–692 [DOI] [PubMed] [Google Scholar]
- 12. Jordan JA, Jones-Laughner J, Durso MB. 2009. Utility of pyrosequencing in identifying bacteria directly from positive blood culture bottles. J. Clin. Microbiol. 47:368–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ibrahim EH, Sherman G, Ward S, Fraser VJ, Kollef MH. 2000. The influence of inadequate antimicrobial treatment of bloodstream infections on patient outcomes in the ICU setting. Chest 118:146–155 [DOI] [PubMed] [Google Scholar]
- 14. Garnacho-Montero J, Garcia-Garmendia JL, Barrero-Almodovar A, Jimenez-Jimenez FJ, Perez-Paredes C, Ortiz-Leyba C. 2003. Impact of adequate empirical antibiotic therapy on the outcome of patients admitted to the intensive care unit with sepsis. Crit. Care Med. 31:2742–2751 [DOI] [PubMed] [Google Scholar]
- 15. Kumar A, Roberts D, Wood KE, Light B, Parrillo JE, Sharma S, Suppes R, Feinstein D, Zanotti S, Tailberg L, Gurka D, Kumar A, Cheang M. 2006. Duration of hypotension before initiation of effective antimicrobial therapy is the critical determinant of survival in human septic shock. Crit. Care Med. 34:1589–1596 [DOI] [PubMed] [Google Scholar]


