Abstract
Acinetobacter baumannii pigmented strains are not common in clinical settings. Here, we report an outbreak caused by indigo-pigmented A. baumannii strains isolated in an acute care hospital in Argentina from March to September 2012. Pan-PCR assays exposed a unique pattern belonging to the recently described regional CC113B/CC79P clonal complex that confirms the relevant relationships among the indigo-pigmented A. baumannii strains. All of them were extensively drug resistant and harbored different genetic elements associated with horizontal genetic transfer, such as the transposon Tn2006, class 2 integrons, AbaR-type islands, IS125, IS26, strA, strB, florR, and the small recombinase ISCR2 associated with the sul2 gene preceded by ISAba1.
INTRODUCTION
Acinetobacter baumannii is a well-known significant nosocomial pathogen that causes a variety of diseases (1–3). The ability of this bacterium to survive for long periods on inanimate surfaces and its extensive drug resistance make A. baumannii a successful microorganism that is able to cause outbreaks (4, 5). Many outbreaks due to A. baumannii have been documented in the literature (6–10). However, to date, no outbreaks due to indigo-pigmented A. baumannii strains have been documented.
The production of indigo pigment in the genus Acinetobacter was previously reported only in the environmental Acinetobacter sp. strain ST-550 and in the A. baumannii ATCC 19606 strain in the presence of indole as a carbon source (11–13). This production may be attributed to the activity of a monooxygenase or dioxygenase enzyme (11, 12). Here, we report the molecular characterization of an outbreak of indigo-pigmented A. baumannii strains that began in the traumatology service of an acute hospital in Argentina.
MATERIALS AND METHODS
Bacterial strains.
A total of 13 pigmented A. baumannii strains were isolated in the traumatology service (n = 7), coronary care unit (n = 2), plastic surgery unit (n = 2), and intensive care unit (n = 2) of an acute care hospital in Argentina from March to September 2012 (Table 1). The strains were identified at the species level by using several criteria: (i) analysis on a Vitek 2 Compact (bioMérieux), (ii) amplified ribosomal DNA restriction analysis (ARDRA) using the primers 5′-TGGCTCAGATTGAACGCTGGCGGC and 5′-TACCTTGTTACGACTTCACCCCA with cycling conditions of initial denaturation at 95°C for 5 min, 35 cycles of denaturation at 95°C for 30 s, annealing at 50°C for 45 s, and extension at 72°C for 1 min (14), (iii) amplification and sequencing of the 16S rRNA with the primers fD2 (5′-AGAGTTTGATCATGGCTCAG) and rP2 (5′-ACGGCTACCTTGTTACGACTT) (described by Weisburg et al. [15]) using 35 cycles of denaturation at 95°C for 30 s, annealing at 52°C for 30 s, and extension at 72°C for 1 min as the cycling conditions, and (iv) amplification and sequencing of the ropB gene using the primers Vic4 [5′-GGCGAAATGGC(AGT)GA(AG)AACCA] and Vic6 [GA(AG)TC(CT)TCGAAGTTGTAACC] and the same cycling conditions as described in iii above (see http://www.pasteur.fr/recherche/genopole/PF8/mlst/Abaumannii.html). Antibiotic susceptibility tests were performed with the Vitek 2 system that uses the panel AST-082 (Gram-negative susceptibility [GNS] card). The MIC results were interpreted according to CLSI categories (16).
Table 1.
Indigo-pigmented A. baumannii strain characteristics and associated clinical data of the patients
| Strain | Isolation date | Patient age (yr)/sexa | Underlying diseaseb | Diagnosis at admissionc | Nosocomial diagnosisd | Servicee | Source of infectionf | Antibiotic resistance profileg |
|---|---|---|---|---|---|---|---|---|
| 66 | 3/28/2012 | 47/F | ND | Surgical site infection | TS | Surgical wound | TMP, AMK, CAZ, FEP, CIP, PIP-TZ, IMP, MEM, GEN | |
| M30393 | 6/25/2012 | 55/M | DBT | Diabetic foot infection | Surgical site infection | TS | SSTI | TMP, AMK, CAZ, FEP, CIP, PIP-TZ, IMP, MEM, GEN |
| M30121 | 6/19/2012 | 31/F | NUD | Pyomyositis | Surgical site infection | TS | SSTI | TMP, AMK, CAZ, FEP, CIP, PIP-TZ, IMP, MEM, GEN |
| 167 | 6/25/2012 | 65/M | DBT | Diabetic foot infection | Surgical site infection | TS | SSTI | TMP, AMK, CAZ, FEP, CIP, PIP-TZ, IMP, MEM, GEN |
| 192 | 7/17/2012 | 64/M | DBT, CRF | Prosthesis infection | Surgical site infection | TS | Mini-BAL | TMP, AMK, CAZ, FEP, CIP, PIP-TZ, IMP, MEM, GEN |
| 186 | 7/12/2012 | 56/M | Diabetic foot infection | Surgical site infection | TS | SSTI | TMP, AMK, CAZ, FEP, CIP, PIP-TZ, IMP, MEM, GEN | |
| M32757 | 8/15/2012 | 64/M | DBT, CRF | Prosthesis infection | Surgical site infection | TS | SSTI | TMP, CAZ, FEP, CIP, PIP-TZ, IMP, MEM, GEN |
| M32467 | 8/8/2012 | Unknown/F | NUD | Burn infection | Surgical site infection | PSU | SSTI | TMP, AMK, CAZ, FEP, CIP, PIP-TZ, IMP, MEM, GEN |
| M32469 | 8/8/2012 | Unknown/F | NUD | Burn infection | Surgical site infection | PSU | SSTI | TMP, CAZ, FEP, CIP, PIP-TZ, IMP, MEM, GEN |
| M33405 | 8/28/2012 | 65/M | Scheduled CABG | Surgical site infection | CCU | Mini-BAL | TMP, CAZ, FEP, CIP, PIP-TZ, IMP, MEM, GEN | |
| M33614 | 9/3/2012 | 45/M | Hemorrhagic stroke | VAP | ICU | Mini-BAL | TMP, AMK, CAZ, FEP, CIP, PIP-TZ, IMP, MEM, GEN | |
| M34050 | 9/10/2012 | 60/M | Hemorrhagic stroke | VAP | ICU | Tracheal aspirate | TMP, AMK, CAZ, FEP, CIP, PIP-TZ, IMP, MEM, GEN | |
| F33943 | 9/7/2012 | 82/F | Respiratory failure, CAP | VAP | CCU | Mini-BAL | TMP, CAZ, FEP, CIP, PIP-TZ, IMP, MEM, GEN |
F, female; M, male.
DBT, diabetes; NUD, nonunderlying disease; CRF, chronic renal failure.
ND, not determined; CABG, coronary artery bypass graft; CAP, community-acquired pneumonia.
All infections were caused by Acinetobacter baumannii. VAP, ventilator-associated pneumonia.
TS, traumatology service; PSU, plastic surgery unit; CCU, coronary care unit; ICU, intensive care unit.
SSTI, skin and soft tissue infection; BAL, bronchoalveolar lavage.
TMP, trimethoprim-sulfamethoxazole; AMK, amikacin; CAZ, ceftazidime; FEP, cefepime; CIP, ciprofloxacin; PIP-TZ, piperacillin-tazobactam; IMP, imipenem; MEM, meropenem; GEN, gentamicin.
DNA techniques.
Total DNA was extracted with a MasterPure DNA purification kit (Epicentre, Madison, WI, USA) according to the manufacturer's instructions. To determine the presence of the most prevalent OXA carbapenemase genes in our region (17), such as blaOXA-23-like and blaOXA-58-like, PCRs were carried out using the primers and cycling conditions described in the literature (17). For the blaOXA amplification reactions, the strains AB3 (blaOXA-23-like) and Ab1(blaOXA-58-like) were used as positive controls (17). The presence of insertion sequences (ISs) (ISAba1, ISAba3, IS125, IS26, IS825, ISCR1, and ISCR2), class 1 and 2 integrons, and the corresponding variable regions of integrons were determined using specific primers as previously described (17–20). To further characterize the strains, the occurrence of AbaR islands using specific previously described primers (4F, 4R, 2F, and 2R) was determined (21, 22). We also searched for the presence of tetracycline-resistant genes using specific primers to amplify tet(B), tet(A), tet(M), tet(39), and tet(H) genes under the conditions described in the literature (21, 22). Amplification of the iacA gene (using the primers iacAF [5′-ATGAATAAGTTGTCTAAAATGGAG] and iacAR [5′-GCAAAACAACACGCGTAATG]), involved in indigo production, was carried out using the A. baumannii ATCC 19606 and ATCC 17978 strains as positive controls.
In addition, the relatedness of the strains was determined using two different molecular typing techniques, a PCR assay using degenerate oligonucleotide primers (DO-PCR) and the recently described pan-PCR assay, which consists of a multiplex PCR of 6 genes and allows for the identification of relevant relationships among strains (23, 24). To carry out the DO-PCR, the primer 19 (5′-GGTCGACYTTNGYNGGRTC) was employed using a low-stringency amplification protocol (5 min of denaturation at 95°C, 40 cycles for 1 min at 93°C, and 1.5 min at 36°C, 2 min at 72°C, and 10 min at 72°C) (23, 24). The pan-PCR assay was performed as described by Yang et al. using the 6 designed pairs of primers that allow amplification of a group of genes whose variable presence enables identification of the strains of interest (23, 24). The cycling conditions employed for the reaction were initial denaturation at 95°C for 5 min, followed by 20 cycles of denaturation (95°C for 30 s), annealing (60°C for 30 s), and extension (72°C for 1 min 30 s), and a single final extension at 72°C for 10 min.
Sequence analysis.
Sequencing was performed on the two DNA strands using an ABI Prism 3100 BioAnalyzer and Taq FS terminator chemistry (Taq FS; Perkin-Elmer). The sequences were examined and assembled with Sequencher 4.7 software (Gene Codes Corp.) and BLAST (version 2.0) software (http://www.ncbi.nlm.nih.gov/BLAST/).
Nucleotide sequence accession numbers.
The sequences determined in this study were submitted to GenBank under accession numbers KF410895 and KF410896.
RESULTS
All strains were identified as A. baumannii using several methods: (i) the bionumber obtained by the Vitek 2 Compact was 0201010303500352, giving identification of an A. baumannii complex with a 99% probability, (ii) the ARDRA profile obtained was 11123, which is characteristic of A. baumannii, (iii) the sequence analysis of the 16S rRNA gene (GenBank accession number KF410895) revealed 99% identity with the sequences corresponding to the 16S rRNA gene of A. baumannii (GenBank accession number CP003846), and (iv) the sequence of the rpoB gene (GenBank accession number KF410896) was 100% identical to that of the rpoB gene of A. baumannii (GenBank accession number DQ207471).
Although two antibiotic resistance profiles among the 13 indigo-pigmented A. baumannii strains were identified (Table 1), all strains were categorized as extensively drug resistant (XDR) according to the recent definitions suggested by Magiorakos et al. (25).
The clinical outcome of patients involved in the outbreak included five deaths. However, the association between A. baumannii colonization or infection and mortality could not be established because patients were medically compromised or had underlying diseases.
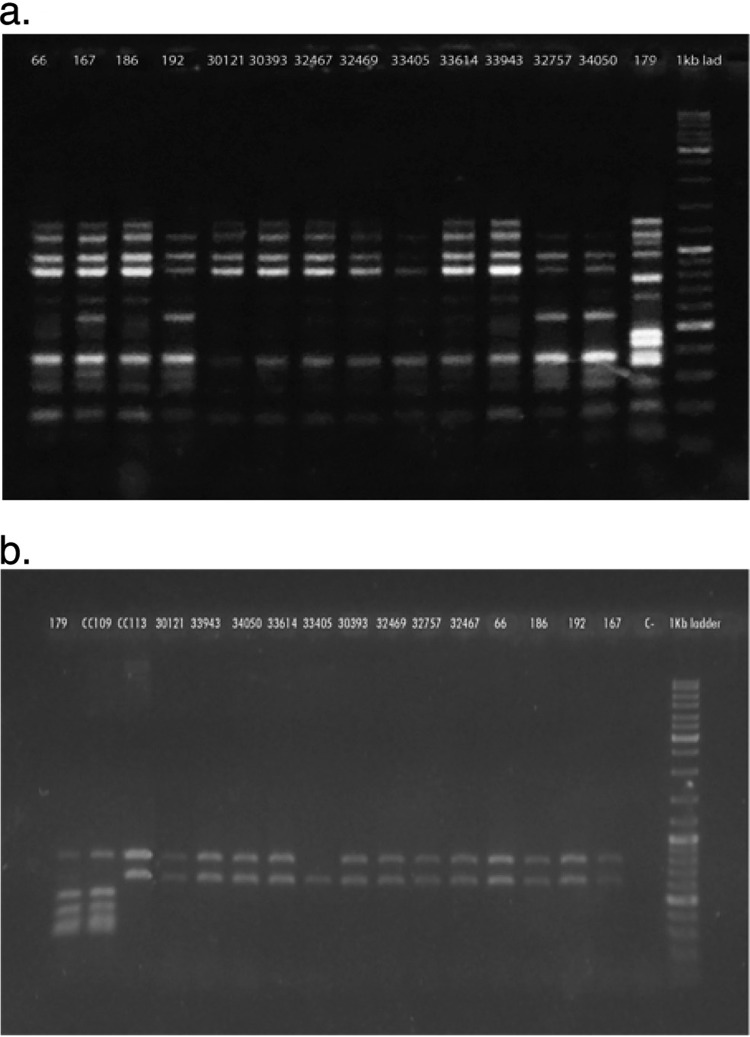
The indigo-pigmented strains showed a unique pattern by DO-PCR that clustered all of them in a single clone (Fig. 1a). We also decided to perform the new, recently described pan-PCR assay, which has been demonstrated to distinguish among strains with identical multilocus sequence types (MLSTs) (24). This technique showed unique amplification patterns, with the exception of one strain (M33405) in which one band is missing, confirming the relevant relationships among the indigo-pigmented A. baumannii strains (Fig. 1b). As this technique is defined as a highly discriminatory PCR assay, we consider that the pan-PCR assay showed a distinct variation in the gene content of this particular strain (33405).
Fig 1.
Molecular typing of the A. baumannii isolates. (a) Ethidium bromide-stained 1.5% agarose gel electrophoresis showing DO-PCR patterns obtained from the A. baumannii strains. The number at the top of each lane corresponds to a strain number. Lane 15 corresponds to a 1-kb ladder (O'GeneRuler 1 kb Plus DNA ladder; Fermentas). (b) Ethidium bromide-stained 1% agarose gel electrophoresis showing pan-PCR amplification patterns obtained in the A. baumannii strains. The number at the top of each lane corresponds to a strain number. Lanes 2 and 3 correspond to control strains of the CC109B/CC1P and CC113P/CC79P clonal complexes, respectively. Lane 17 corresponds to a 1-kb ladder (O'GeneRuler 1 kb Plus DNA ladder). The XDR nonpigmented A. baumannii strain (A179), recovered from the wound-healing chamber used in the traumatology service, was also added to the two assays.
Also, the use of pan-PCR allowed us to determine that the indigo-pigmented A. baumannii strains possessed the same amplification pattern as that obtained in the control strain for the CC113B/CC79P clonal complex, which was shown to be prevalent in clinical A. baumannii isolates from Argentina (26) (Fig. 1b). This particular clonal complex, which differs from the international clones I, II, and III, was also described for A. baumannii isolates from Brazil and Spain (26–28).
The dates when the A. baumannii strains were recovered clearly show that the outbreak began in the traumatology service and then spread to the other services (Table 1). Attempts to recover A. baumannii strains from environmental sources other than the hospitalized patients yielded no indigo-pigmented A. baumannii strains. A nonpigmented XDR A. baumannii strain (A179) was recovered from the wound-healing chamber used in the traumatology service.
The molecular characterization of the outbreak was carried out by PCRs, and sequence analyses were performed to identify the presence of the antibiotic-resistant genes and the genetic elements associated with the antibiotic resistance. We also searched for the iacA gene and found positive results in all the indigo-pigmented A. baumannii strains and also in the XDR nonpigmented A. baumannii strain A179.
While all the strains harbored the Tn2006 transposon, which carries blaOXA-23, amplification of the blaOXA-58 and blaOXA-143 carbapenemase genes gave negative results. All indigo-pigmented strains possessed IS125 and IS26 and the strA, strB, and florR genes. Class 2 integrons were also found in all the indigo-pigmented A. baumannii strains, whereas no class 1 integrons were found. These results are in accordance with our previous studies, which showed that class 2 integrons are more abundant than class 1 integrons in A. baumannii strains from Argentina (20, 29, 30). PCR cartography and sequence analyses revealed that In2-7, which is the common class 2 integron array, was present in the 13 strains studied (20, 29).
In addition, all indigo-pigmented strains were positive for the presence of not only AbaR-type genomic islands but also a cluster containing ISAba1, sul2, and ISCR2. The A. baumannii A297 strain harboring a similar cluster, which contains ISAba1, sul2, ISCR2, strB, and strA, has been described (19). However, no positive result to link the strB and strA genes with ISCR2 was obtained in our A. baumannii indigo-pigmented strains.
The PCRs we used to amplify tetracycline-resistant determinants, such as tet(A) and tet(B), gave negative results. In addition, no evidence of the aadB, aac(6′)-Ib, and aphA1 genes was found.
Our first thoughts were that the A179 strain, which was found in a wound-healing chamber in the traumatology service, may have been the source of the described outbreak and/or the source of the antimicrobial-resistant mechanisms found in the indigo-pigmented strains. To confirm our hypothesis, we characterized the A179 strain.
The nonpigmented A. baumannii A179 strain was susceptible only to minocycline, tigecycline, amikacin, and colistin. The same profile was described for nine of the indigo-pigmented strains (Table 1). As we found in the indigo-pigmented strains, the A179 strain harbored Tn2006 and the AbaR-type genomic island. The clonal relationship found by DO-PCR and pan-PCR showed that the A179 strain belongs to a different clone (Fig. 1a and b). Also, when we compared this strain with the indigo-pigmented strains, we found some differences in the contents of the genetic determinants analyzed in the present work. Instead of class 2 integrons, we found a class 1 integron, which carries the aacC1-orfP-orfQ-aadA1 array. This integron was previously described not only in the widespread international clone I but also in Argentinean isolates (30, 31). As we found in the indigo-pigmented strains, the A179 strain harbors Tn2006 and the AbaR-type genomic island. However, no evidence of the ISAba125 or ISCR2 or the strA, strB, or floR gene was detected.
DISCUSSION
To our knowledge, this is the first report of an outbreak of XDR indigo-pigmented A. baumannii strains. The molecular characterizations of the strains clearly exposed the large number of genetic elements present in these strains and thus support the general idea that A. baumannii has a particular ability to acquire different genetic elements to evolve rapidly to XDR and pandrug-resistant (PDR) strains.
In all the strains, we observed the presence of not only ISAbaI and ISAba125, which are the most prevalent ISs in this microorganism, but also IS26 and the ISCR2 elements. Our findings are in agreement with the concept that insertion sequences have a predominant role in the acquisition and dissemination of antibiotic resistance within A. baumannii. The virulence associated with indigo-pigmented isolates remains to be established.
Our study also highlights the importance of rigorous infection prevention and control measures for managing an outbreak of A. baumannii. Once the organism is identified, universal hygiene measures should be observed to avoid further spread and outbreaks.
ACKNOWLEDGMENTS
M.S.R. and D.C. are career investigators of CONICET, Argentina, and E.V. has doctoral fellowships from CONICET. This study was supported by grant PIP 11420100100152 to M.S.R. and grant UBACyTs to M.S.R. and D.C., Buenos Aires, Argentina.
We declare no conflicts of interest.
Footnotes
Published ahead of print 28 August 2013
REFERENCES
- 1. Roca I, Espinal P, Vila-Farres X, Vila J. 2012. The Acinetobacter baumannii oxymoron: commensal hospital dweller turned pan-drug-resistant menace. Front. Microbiol. 3:148. 10.3389/fmicb.2012.00148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Visca P, Seifert H, Towner KJ. 2011. Acinetobacter infection—an emerging threat to human health. IUBMB Life 63:1048–1054 [DOI] [PubMed] [Google Scholar]
- 3. Peleg AY, de Breij A, Adams MD, Cerqueira GM, Mocali S, Galardini M, Nibbering PH, Earl AM, Ward DV, Paterson DL, Seifert H, Dijkshoorn L. 2012. The success of Acinetobacter species; genetic, metabolic and virulence attributes. PLoS One 7:e46984. 10.1371/journal.pone.0046984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Peleg AY, Seifert H, Paterson DL. 2008. Acinetobacter baumannii: emergence of a successful pathogen. Clin. Microbiol. Rev. 21:538–582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Jawad A, Seifert H, Snelling AM, Heritage J, Hawkey PM. 1998. Survival of Acinetobacter baumannii on dry surfaces: comparison of outbreak and sporadic isolates. J. Clin. Microbiol. 36:1938–1941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Zarrilli R, Di Popolo A, Bagattini M, Giannouli M, Martino D, Barchitta M, Quattrocchi A, Iula VD, de Luca C, Scarcella A, Triassi M, Agodi A. 2012. Clonal spread and patient risk factors for acquisition of extensively drug-resistant Acinetobacter baumannii in a neonatal intensive care unit in Italy. J. Hosp. Infect. 82:260–265 [DOI] [PubMed] [Google Scholar]
- 7. Chmielarczyk A, Higgins PG, Wojkowska-Mach J, Synowiec E, Zander E, Romaniszyn D, Gosiewski T, Seifert H, Heczko P, Bulanda M. 2012. Control of an outbreak of Acinetobacter baumannii infections using vaporized hydrogen peroxide. J. Hosp. Infect. 81:239–245 [DOI] [PubMed] [Google Scholar]
- 8. Domenech de Cellès M, Salomon J, Marinier A, Lawrence C, Gaillard JL, Herrmann JL, Guillemot D. 2012. Identifying more epidemic clones during a hospital outbreak of multidrug-resistant Acinetobacter baumannii. PLoS One 7:e45758. 10.1371/journal.pone.0045758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Goic-Barisic I, Towner KJ, Kovacic A, Sisko-Kraljevic K, Tonkic M, Novak A, Punda-Polic V. 2011. Outbreak in Croatia caused by a new carbapenem-resistant clone of Acinetobacter baumannii producing OXA-72 carbapenemase. J. Hosp. Infect. 77:368–369 [DOI] [PubMed] [Google Scholar]
- 10. Ansaldi F, Canepa P, Bassetti M, Zancolli M, Molinari MP, Talamini A, Ginocchio F, Durando P, Mussap M, Orengo G, Viscoli C, Icardi G. 2011. Sequential outbreaks of multidrug-resistant Acinetobacter baumannii in intensive care units of a tertiary referral hospital in Italy: combined molecular approach for epidemiological investigation. J. Hosp. Infect. 79:134–140 [DOI] [PubMed] [Google Scholar]
- 11. Doukyu N, Toyoda K, Aono R. 2003. Indigo production by Escherichia coli carrying the phenol hydroxylase gene from Acinetobacter sp. strain ST-550 in a water-organic solvent two-phase system. Appl. Microbiol. Biotechnol. 60:720–725 [DOI] [PubMed] [Google Scholar]
- 12. Doukyu N, Nakano T, Okuyama Y, Aono R. 2002. Isolation of an Acinetobacter sp. ST-550 which produces a high level of indigo in a water-organic solvent two-phase system containing high levels of indole. Appl. Microbiol. Biotechnol. 58:543–546 [DOI] [PubMed] [Google Scholar]
- 13. Lin GH, Chen HP, Huang JH, Liu TT, Lin TK, Wang SJ, Tseng CH, Shu HY. 2012. Identification and characterization of an indigo-producing oxygenase involved in indole 3-acetic acid utilization by Acinetobacter baumannii. Antonie Van Leeuwenhoek 101:881–890 [DOI] [PubMed] [Google Scholar]
- 14. Vaneechoutte M, Dijkshoorn L, Tjernberg I, Elaichouni A, de Vos P, Claeys G, Verschraegen G. 1995. Identification of Acinetobacter genomic species by amplified ribosomal DNA restriction analysis. J. Clin. Microbiol. 33:11–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Weisburg WG, Barns SM, Pelletier DA, Lane DJ. 1991. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 173:697–703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.CLSI 2011. Performance standards for antimicrobial susceptibility testing; 21st informational supplement. CLSI document M100-S21. Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 17. Merkier AK, Catalano M, Ramirez MS, Quiroga C, Orman B, Ratier L, Famiglietti A, Vay C, Di Martino A, Kaufman S, Centron D. 2008. Polyclonal spread of blaOXA-23 and blaOXA-58 in Acinetobacter baumannii isolates from Argentina. J. Infect. Dev. Ctries. 2:235–240 [DOI] [PubMed] [Google Scholar]
- 18. Orman BE, Pineiro SA, Arduino S, Galas M, Melano R, Caffer MI, Sordelli DO, Centron D. 2002. Evolution of multiresistance in nontyphoid Salmonella serovars from 1984 to 1998 in Argentina. Antimicrob. Agents Chemother. 46:3963–3970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Nigro SJ, Post V, Hall RM. 2011. The multiresistant Acinetobacter baumannii European clone I type strain RUH875 (A297) carries a genomic antibiotic resistance island AbaR21, plasmid pRAY and a cluster containing ISAba1-sul2-CR2-strB-strA. J. Antimicrob. Chemother. 66:1928–1930 [DOI] [PubMed] [Google Scholar]
- 20. Ramírez MS, Pineiro S, Centron D. 2010. Novel insights about class 2 integrons from experimental and genomic epidemiology. Antimicrob. Agents Chemother. 54:699–706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Vilacoba E, Almuzara M, Gulone L, Traglia GM, Figueroa SA, Sly G, Fernandez A, Centron D, Ramirez MS. 2013. Emergence and spread of plasmid-borne tet(B)::ISCR2 in minocycline-resistant Acinetobacter baumannii isolates. Antimicrob. Agents Chemother. 57:651–654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Shaikh F, Spence RP, Levi K, Ou HY, Deng Z, Towner KJ, Rajakumar K. 2009. ATPase genes of diverse multidrug-resistant Acinetobacter baumannii isolates frequently harbour integrated DNA. J. Antimicrob. Chemother. 63:260–264 [DOI] [PubMed] [Google Scholar]
- 23. Limansky AS, Viale AM. 2002. Can composition and structural features of oligonucleotides contribute to their wide-scale applicability as random PCR primers in mapping bacterial genome diversity? J. Microbiol. Methods 50:291–297 [DOI] [PubMed] [Google Scholar]
- 24. Yang JY, Brooks S, Meyer JA, Blakesley RR, Zelazny AM, Segre JA, Snitkin ES. 2013. Pan-PCR, a computational method for designing bacterium-typing assays based on whole-genome sequence data. J. Clin. Microbiol. 51:752–758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Magiorakos AP, Srinivasan A, Carey RB, Carmeli Y, Falagas ME, Giske CG, Harbarth S, Hindler JF, Kahlmeter G, Olsson-Liljequist B, Paterson DL, Rice LB, Stelling J, Struelens MJ, Vatopoulos A, Weber JT, Monnet DL. 2012. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 18:268–281 [DOI] [PubMed] [Google Scholar]
- 26. Stietz MS, Ramirez MS, Vilacoba E, Merkier AK, Limansky AS, Centron D, Catalano M. 2013. Acinetobacter baumannii extensively drug resistant lineages in Buenos Aires hospitals differ from the international clones I–III. Infect. Genet. Evol. 14:294–301 [DOI] [PubMed] [Google Scholar]
- 27. Grosso F, Carvalho KR, Quinteira S, Ramos A, Carvalho-Assef AP, Asensi MD, Peixe L. 2011. OXA-23-producing Acinetobacter baumannii: a new hotspot of diversity in Rio de Janeiro? J. Antimicrob. Chemother. 66:62–65 [DOI] [PubMed] [Google Scholar]
- 28. Villalón P, Valdezate S, Medina-Pascual MJ, Rubio V, Vindel A, Saez-Nieto JA. 2011. Clonal diversity of nosocomial epidemic Acinetobacter baumannii strains isolated in Spain. J. Clin. Microbiol. 49:875–882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ramírez MS, Morales A, Vilacoba E, Marquez C, Centron D. 2012. Class 2 integrons dissemination among multidrug resistance (MDR) clones of Acinetobacter baumannii. Curr. Microbiol. 64:290–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ramírez MS, Stietz MS, Vilacoba E, Jeric P, Limansky AS, Catalano M, Centron D. 2011. Increasing frequency of class 1 and 2 integrons in multidrug-resistant clones of Acinetobacter baumannii reveals the need for continuous molecular surveillance. Int. J. Antimicrob. Agents 37:175–177 [DOI] [PubMed] [Google Scholar]
- 31. D'Arezzo S, Capone A, Petrosillo N, Visca P, Ballardini M, Bartolini S, Bordi E, Di Stefano A, Galie M, Minniti R, Meledandri M, Pacciani L, Parisi G, Prignano G, Santini C, Valmarin M, Venditti M, Ziantoni S. 2009. Epidemic multidrug-resistant Acinetobacter baumannii related to European clonal types I and II in Rome (Italy). Clin. Microbiol. Infect. 15:347–357 [DOI] [PubMed] [Google Scholar]