Abstract
Commutability of quantitative reference materials has proven important for reliable and accurate results in clinical chemistry. As international reference standards and commercially produced calibration material have become available to address the variability of viral load assays, the degree to which such materials are commutable and the effect of commutability on assay concordance have been questioned. To investigate this, 60 archived clinical plasma samples, which previously tested positive for cytomegalovirus (CMV), were retested by five different laboratories, each using a different quantitative CMV PCR assay. Results from each laboratory were calibrated both with lab-specific quantitative CMV standards (“lab standards”) and with common, commercially available standards (“CMV panel”). Pairwise analyses among laboratories were performed using mean results from each clinical sample, calibrated first with lab standards and then with the CMV panel. Commutability of the CMV panel was determined based on difference plots for each laboratory pair showing plotted values of standards that were within the 95% prediction intervals for the clinical specimens. Commutability was demonstrated for 6 of 10 laboratory pairs using the CMV panel. In half of these pairs, use of the CMV panel improved quantitative agreement compared to use of lab standards. Two of four laboratory pairs for which the CMV panel was noncommutable showed reduced quantitative agreement when that panel was used as a common calibrator. Commutability of calibration material varies across different quantitative PCR methods. Use of a common, commutable quantitative standard can improve agreement across different assays; use of a noncommutable calibrator can reduce agreement among laboratories.
INTRODUCTION
Quantitative viral load testing has become integral to the care of patients in a variety of clinical settings. These tests are of particular importance in the immunocompromised patient population, where measurement of cytomegalovirus (CMV), Epstein-Barr virus (EBV), BK virus, and adenovirus, among other agents, has proven utility in terms of initiating preemptive treatment strategies, determination and risk stratification of active disease states, and assessment of therapeutic response. While the methodology to support such determinations has progressed substantially in recent years, there remain significant limitations that directly hinder optimal utilization of such testing. The inability to produce results with a high degree of concordance within or across various reagents, testing platforms, and institutions is a fundamental problem. Numerous studies have now documented the wide variation of assay results seen when common specimens are tested in parallel by different laboratories (1–5). Such variation prevents the portability of results, such that patients require repeat baseline testing when transferring between institutions. Viral load results obtained in one laboratory cannot be interpreted by the same criteria as those from another laboratory. On a broader scale, this has prevented the development of consensus quantitative trigger points for intervention and treatment endpoints. It also means that studies from different centers using differing methodologies cannot be directly compared.
The variation in assay results is related to the use of a wide variety of highly complex assays without a common basis of standardization. The potential sources of such variation are many, and data support a multifactorial origin for suboptimal reproducibility (2). However, it is clear that a key issue is the use of quantitative calibrators that have typically been developed by individual laboratories, much as they have developed their own detection reagents, in the absence of a clear, international reference standard with which results might be normalized (6, 7). Studies have now shown, both for CMV and for other viruses, that the introduction of common calibration material can markedly improve agreement among laboratories using otherwise disparate detection methods (1, 8). Such data have been the basis for the recent development of new international reference standards for CMV and EBV (9–11) and have raised hopes that this material would substantially address the issue of interlaboratory agreement for these often critical tests.
However, the use of common material does not necessarily guarantee that similar results will be produced. In order for reference materials to reliably reflect patient results among different assays, they must be shown to behave like patient samples in each of those assays. This defines the concept of commutability, first gaining widespread recognition in the field of clinical chemistry, but more recently applied to quantitative virologic tests (8, 12–14). It has increasingly been recognized that the trueness of quantitative clinical tests is fundamental to their proper interpretation and therefore to appropriate patient care. That trueness, in turn, depends upon the use of reliable, well-characterized reference materials that should be stable, homogenous, and commutable. Reference materials typically are composed of either purified nucleic acid, naturally occurring virus (e.g., pooled patient samples), or cultured virus that is diluted into clinically relevant matrix or artificial matrix (7). While such analytes and matrices are designed to mimic naturally occurring measurand found in individual patient samples sent for routine testing, it cannot be assumed that such is the case. Moreover, the biologic form of analytes present within different clinical sample types has often not been clearly determined. Commutability must be demonstrated for each new reference material and should be shown for each assay in which that material is to be incorporated. This has been widely demonstrated in clinical chemistry assays, and a recent study showed that the use of a commutable CMV viral load calibrator markedly improved concordance of results from two laboratories (8). However, a broader comparison of several laboratories to demonstrate the importance of commutability for the standardization of viral load testing has yet to be published. The work presented here addresses this issue and has potential implications across a wide range of current and future applications in the clinical laboratory.
MATERIALS AND METHODS
Clinical samples and commercial calibrators.
Sixty patient EDTA plasma specimens were collected at the Emory University Hospital (Atlanta, GA) between November 2007 and September 2008. Samples were chosen for the study from material remaining after routine clinical testing. All had previously tested positive for CMV by standard diagnostic testing with a laboratory developed real-time PCR assay and represented a wide range of CMV concentrations (300 to approximately 450,000 copies/ml). Specimens were completely deidentified, and the study was considered nonhuman subject research, exempt from institutional review board (IRB) review. Samples were stored at −70°C until use in the study. They were then thawed at room temperature, aliquoted into at least five vials, and refrozen at −70°C prior to shipment to the five participating laboratories. In addition to the 60 patient plasma samples, each participating laboratory received the OptiQuant CMVtc calibration panel (referred to here as CMV panel and now branded as Acrometrix CMVtc panel [Life Technologies, Carlsbad, CA]), which consisted of four concentrations of human CMV strain AD29 (500 to 500,000 copies/ml) in addition to a negative (plasma-only) member.
Quantitative PCR testing.
Calibration panel members and clinical samples were extracted and quantified by each laboratory according to its routine testing protocol. Quantification was performed with laboratory-specific calibration materials (“lab standards”). This study design resulted in five unique combinations of sample preparation, amplification/detection reagents, and calibrators. A summary of all laboratories' methods is shown in Table 1. It may be noted that laboratory 5 utilized Luminex MultiCode CMV reagents (analyte-specific reagents), formerly produced by Eragen Biosciences. The MultiCode reagents incorporate a unique chemistry that utilizes labeled primers and results in the diminution of fluorescent signal as amplicon is produced.
Table 1.
CMV test methodsb
| Test method, component, or characteristic | Lab 1 | Lab 2 | Lab 3 | Lab 4 | Lab 5 |
|---|---|---|---|---|---|
| Assay system | Lab-developed test | Qiagen Artus CMV (ASR) | Qiagen Artus CMV (ASR) | Lab-developed test | Luminex MultiCode CMV (ASR)a |
| Laboratory calibrators | LD (synthetic) | Qiagen Quant standards | Qiagen Quant standards | LD (synthetic) | LD (synthetic) |
| Gene target | Glycoprotein B (UL55) and IE-1 (UL123) | IE-2 (UL122) | IE-2 (UL122) | Glycoprotein B (UL55) | DNA polymerase (UL54) |
| Amplicon size (bp) | 64 (UL55), 76 (UL123) | 105 | 105 | 254 | 52 |
| Extraction method | MagNA Pure TNA kit; magnetic silica particles | MagNA Pure TNA kit; magnetic silica particles | Qiagen M48; MagAttract viral RNA; magnetic silica particles | QIAamp DNA minikit; column based; QIAcube | Qiagen M48; MagAttract DNA minikit; magnetic silica particles |
| Probe chemistry | TaqMan; cleaved probe | TaqMan; cleaved probe | TaqMan; cleaved probe | FRET; probes remain intact | Labeled primers; no probes |
| Detection system | ABI StepOnePlus | ABI Prism 7500 | ABI Prism 7500 | LightCycler 2.0 | ABI Prism 7500 |
Formerly produced by Eragen Biosciences.
LD, lab developed; ASR, analyte-specific reagent; FRET, fluorescent resonance energy transfer.
The five participating laboratories each performed four PCR runs. Each run included the OptiQuant CMVtc calibration panel (CMV panel), with each panel member tested in triplicate on the first run and in duplicate on subsequent runs. A single lot of the CMV panel material was used for the entire study. The 60 clinical samples were tested once per laboratory as unknowns using lab standards included in each PCR run for calibration. At the completion of the study, each laboratory had tested nine replicates of the CMV panel (each separately extracted) and a single aliquot of each of the 60 clinical samples. The mean quantitative result for each member of the CMV panel was calculated across all four runs per laboratory and plotted against the nominal values (log10 copies/ml) using Microsoft Office Excel 2007. The ordinary least-squares regression line was plotted for each laboratory.
Calibration.
In all analyses, calibration refers to the method of constructing an ordinary least-squares regression line comparing the nominal values (log10 copies/ml) to the observed values (cycle threshold [CT]) of the calibrator (lab standards or CMV panel) and using the regression equation to calculate quantitative values for each clinical sample based on observed CT values. For all calibrations, the clinical sample data were calibrated using the results of the calibrators from the same run.
Quantitative comparisons.
To determine the performance of the common calibrator (CMV panel) between the five laboratories in relation to the clinical specimens, the means of the observed log10 concentrations of the CMV panel were calculated across all four PCR runs for each laboratory and plotted against the nominal values, and regression lines were generated using Microsoft Office Excel 2007. The observed values were compared in a pairwise fashion using all possible combinations of participating labs. Paired data were plotted against each other using JMP statistical analysis software (version 9.0.0; SAS Institute, Cary, NC). An orthogonal regression line for the clinical specimens was plotted in addition to the line of identity (y = x). Ellipses representing the 50%, 90%, and 95% confidence curves were added around the clinical sample results, assuming the bivariate normal distribution for the X and Y variables.
Commutability analysis.
For each pair of laboratories, the difference in log10 values for mean CMV panel results and clinical sample results was plotted against the mean values achieved when results of the two laboratories were averaged (Bland-Altman plots). JMP statistical analysis software was used to plot regression lines and 95% prediction intervals. The CMV panel was considered commutable between the two test methods if its plotted values were within the 95% prediction interval for the clinical specimens.
To determine whether commutability of a common calibrator correlated with improved comparability of results between laboratories, clinical specimen results were calibrated independently with both the lab standards and the CMV panel. For each clinical specimen, the absolute difference in quantitative value between each pair of laboratories was calculated and the number of specimens within various ranges was counted and plotted as a percentage of the total number of samples. This was plotted side-by-side for results calibrated using lab standards and CMV panel (recalibration plots). Data analysis was performed using Microsoft Office Excel 2007. A shift in the plotted curve to the left indicated increased agreement between the two laboratories; a shift to the right indicated diminished agreement between the laboratories. Minimal shift between the plots meant that there was no change in agreement between the quantitative values achieved by the two labs. To quantify the shifts in these plots further, the log10 difference in results between laboratory pairs when using the CMV panel for calibration was calculated for various fractions of samples (e.g., 1/60, 2/60, up to 60/60) and subtracted from the log10 difference seen when using the lab standards for calibration. A mean difference of greater than +0.25 log10 was considered to represent improved agreement, while a mean difference of less than −0.25 was considered to indicate reduced agreement.
RESULTS
Quantitative comparisons.
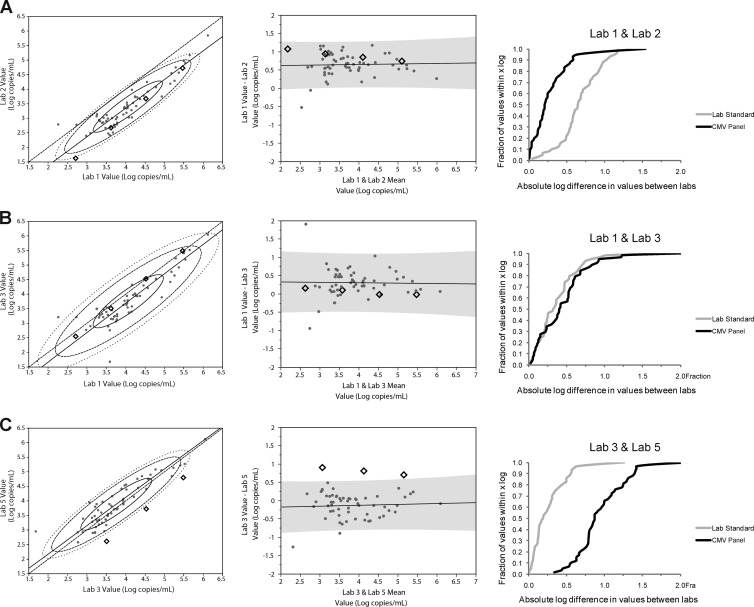
The mean of the observed log10 concentrations for each CMV panel concentration was plotted against the nominal concentrations (Fig. 1 and Table 2). This analysis demonstrated that the assays fell in two groups (labs 1 and 3 and labs 2, 4, and 5), with similar linear regression slopes but with y-intercepts that were approximately 1 log10 unit apart. The linear regression equations representing each line are shown in Table 2. An even greater divergence of results between the evaluated tests was observed when measurements of clinical specimens and mean results of calibrators were plotted in a pairwise fashion (Fig. 2, left panel; see Fig. S1 in the supplemental material). Some laboratory pairs showed good agreement, demonstrated by the intersection of the line of identity with the 50% confidence ellipse for the clinical sample results (Fig. 2B and C). For other laboratory pairs, there is poor agreement, with no intersection of the 50% confidence ellipse and the line of identity. In the extreme cases, the line of identity is outside the 90% or 95% confidence ellipse (Fig. 2A). Similarly, Bland-Altman plots (“difference plots”) illustrate the degree of agreement between laboratory pairs (Fig. 2, center panel; also Fig. S2). In the latter graphs, the closer the plot approaches zero on the y axis, the closer the agreement between quantitative results of the two laboratories.
Fig 1.
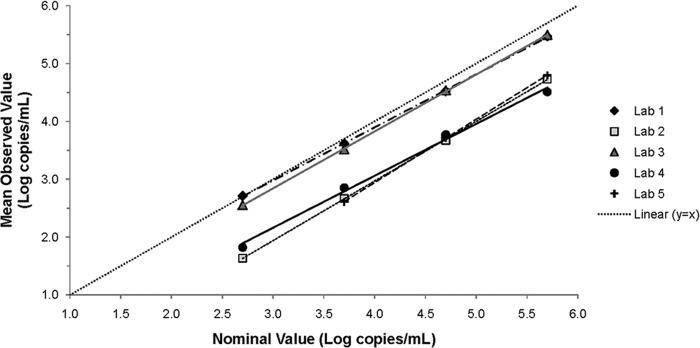
Quantitative comparison of CMV panel results among five laboratories. The mean of the observed log10 concentrations for each level of the CMV panel (n = 9 from four runs) in each laboratory was plotted against the nominal log10 concentrations along with the regression line. The line of identity (y = x) is shown as a dotted line. The 2.7-log10-copy/ml panel member was excluded from the lab 5 data set because it was not reliably detected.
Table 2.
Results of AcroMetrix CMVtc panel in five laboratories
| Nominal value | Log10 copies/ml for each lab (regression line): |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Lab 1 (y = 0.92x + 0.22) |
Lab 2 (y = 1.03x − 1.15) |
Lab 3 (y = 0.98x − 0.10) |
Lab 4 (y = 0.90x − 0.53) |
Lab 5 (y = 1.09x − 1.43) |
||||||
| Mean | SD | Mean | SD | Mean | SD | Mean | SD | Mean | SD | |
| 2.7 | 2.71 | 0.17 | 1.63 | 0.32 | 2.55 | 0.15 | 1.82 | 0.27 | ||
| 3.7 | 3.62 | 0.10 | 2.67 | 0.12 | 3.51 | 0.20 | 2.85 | 0.16 | 2.61 | 0.34 |
| 4.7 | 4.52 | 0.05 | 3.67 | 0.10 | 4.53 | 0.18 | 3.77 | 0.10 | 3.72 | 0.15 |
| 5.7 | 5.47 | 0.04 | 4.73 | 0.09 | 5.49 | 0.10 | 4.51 | 0.28 | 4.80 | 0.13 |
Fig 2.
Representative plots showing comparison of CMV panel and clinical specimen results and correlation with interlaboratory agreement. Orthogonal regressions (left column), Bland-Altman plots (middle column), and recalibration analyses (right column) for three pairs of laboratories are shown: lab 1-lab 2 results (A), lab 1-lab 3 results (B), and lab 3-lab 5 results (C). The clinical sample results are displayed as gray circles, and CMV panel results are illustrated as black diamonds.
Commutability.
Bland-Altman plots are shown in Fig. 2 (center panels) and in Fig. S2 in the supplemental material, demonstrating the difference in values achieved between pairs of labs (mean values were used from each lab for calibrators and individual values for patient samples) plotted against the mean values for each pair of labs on the x axis. Commutability of the CMV panel was demonstrated if results for individual panel members fell within the 95% prediction interval shown for the clinical samples in each plot (Fig. 2A and B). Given this definition, the CMV panel showed commutability in 6 of the 10 lab pairs (Fig. 2A and B, Bland-Altman plots; see Fig. S2 for additional Bland-Altman data; pairwise commutability findings are summarized in Table 3).
Table 3.
Lab comparison summary
| Lab pair | Commutable? | Comparability (improved, no difference, or worse) |
|---|---|---|
| Lab 1-lab 2 | Y | Improved |
| Lab 1-lab 3 | Y | No difference |
| Lab 1-lab 4 | Y | Improved |
| Lab 2-lab 4 | Y | No difference |
| Lab 2-lab 5 | Y | No difference |
| Lab 3-lab 4 | Y | Improved |
| Lab 3-lab 5 | N | Worse |
| Lab 4-lab 5 | N | No difference |
| Lab 1-lab 5 | N | Worse |
| Lab 2-lab 3 | N | No difference |
The effect of calibrator commutability on comparability of results between laboratories was determined by measurement of clinical samples using lab standards followed by recalibration using the CMV panel. The absolute difference in clinical sample viral load between each lab pair was plotted versus the fraction of values that fell within a given difference (Fig. 2, right panel; see also Fig. S3 in the supplemental material). A shift in the curve to the left when recalibrated using the CMV panel indicated increased agreement between the two laboratories; a shift to the right indicated diminished agreement. To further quantify the effect of recalibrating clinical sample results using the CMV panel versus the lab standards, the difference between the two curves on the x axis in Fig. 2 and Fig. S3 was calculated and plotted for various fractions of samples (Fig. 3). Positive mean difference values (indicated in Fig. 3 by mean diamonds above +0.25) indicated improved agreement, and negative mean differences (below −0.25 in Fig. 3) indicated worsened agreement. Table 3 summarizes the impact of adopting the CMV panel as a common calibrator. In half of the cases (3/6) where the CMV panel was shown to be commutable between two labs, its use as a calibrator improved agreement in clinical laboratory results between those two laboratories (compared to the use of lab-specific standards [Fig. 2, right panel; Fig. 3; and Table 3; see also Fig. S3 in the supplemental material]). Its use neither improved nor diminished agreement in the other three laboratory pairs. However, in half of the cases (2/4) where a lack of commutability was seen, the use of that noncommutable calibrator was associated with reduced agreement compared to the use of lab-specific calibrators. No difference in results was seen in the other two laboratory pairs that showed a lack of commutability. Agreement was never improved when the calibrator was noncommutable.
Fig 3.
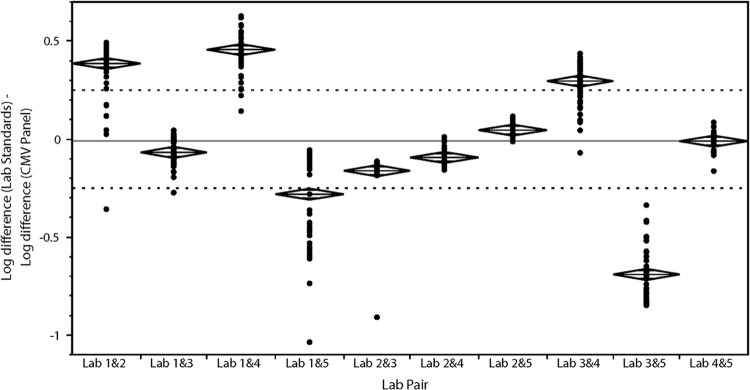
Comparison in interlaboratory agreement between lab standards and CMV panel. To quantify the closeness of recalibration curves for each lab pair as shown in the right column of Fig. 2 and in Fig. S3 in the supplemental material, the spread on the x axis in the former plots was quantified at various fractions and plotted on the y axis for each lab pair. Each dot shows the log10 difference in values for a certain fraction of samples (e.g., 1/60 or 0.017) when calibrated with the lab standards subtracted by the log10 difference in values when calibrated by the CMV panel. Positive values represent cases in which the interlaboratory difference is improved when calibrated with the CMV panel, and negative values represent cases in which the interlaboratory difference is worse. The solid line marks a difference of zero, and the two dotted lines mark ±0.25-log differences, an area considered equivalent. Mean diamonds, with the mean and 95% confidence interval of the mean, are displayed for each lab pair.
DISCUSSION
Findings here expand our understanding of the importance of commutability as we continue to strive toward standardization of viral load testing. The development of commercially available quantitative standards is necessary but not sufficient to improve agreement between laboratories and methods. Likewise, the data above show that commutability is a necessary attribute of any material that is to be used in this manner, and yet this property alone does not guarantee improved agreement. That it is essential is illustrated by the fact that none of the four laboratory pairs for which the CMV panel was noncommutable showed improved agreement, compared to half of the pairs for which commutability was demonstrated. Diminished quantitative agreement was shown in half of the noncommutable pairs, but in none of those for which commutability was shown.
These results build upon previous findings regarding the importance of commutability in the standardization of quantitative clinical laboratory testing (13, 14). This was first described in the clinical chemistry literature, initially with respect to measurements of hepatic enzyme activity and later for cardiac markers and other analytes (12, 15–18). While required in current International Organization for Standardization (ISO) guidance regarding reference material (19), such evaluations are far from universal and to date have seldom been performed even on material used as WHO international reference standards. Studies in clinical chemistry have shown that a significant proportion (for some analytes, a majority) of commercial control materials lack commutability for commonly used measurement procedures (18, 20). Quantitative testing in molecular virology is a comparatively young methodology, with a limited number of assays and reference materials currently on the market. However, the importance of intralaboratory agreement is fundamental to the usefulness of such test results. Data are building on the contribution of quantitative standardization to the improvement of interlaboratory agreement, but only recently has work been published exploring the importance of commutability in the context of viral load testing (8). That work showed that commutable standards improved comparability of viral loads between two laboratories, with a lack of improvement when standards were noncommutable for the measurement procedures used.
This expanded multicenter study is an important next step in the investigation of commutability as applied to quantitative nucleic acid amplification assays. Like the previous study, we again found that only the use of commutable reference material improved quantitative agreement between laboratories. A lack of commutability was associated with an increased disparity between results. This is supported by previous work showing clinically significant changes in the results of clinical chemistry tests when noncommutable reference materials were used (18). Not all laboratory pairs in the present study showed improvement when the calibrator was commutable, nor did all demonstrate diminished agreement with noncommutable standards. For labs 1 and 3, the lack of improvement after recalibration with the CMV panel may be explained by the observation that in these two labs, the CMV panel performed similarly (Fig. 1 and 2, left panel), and the clinical sample results were also close to the line of identity before recalibration (Fig. 2, left panel).
These observations point to the complexity of quantitative assays and the numerous factors already shown to affect viral load results, including not only selection of calibration materials but also aspects of nucleic acid extraction, assay design, and amplification target (2). As such, each assay had its own performance characteristics and limitations. For example, although all clinical samples were detected by laboratory 5, the performance of this test using reference materials indicated a lower level of sensitivity than those of other assays. Additional test verification (not shown) was performed to demonstrate its reliability for routine clinical testing. A related point is that even minor changes in such complex procedures can markedly affect commutability. Labs 2 and 3 in the present study used identical methodologies with the exception of a differing extraction method. Yet, that change alone was enough to result in a lack of commutability between the two tests.
These findings emphasize the fact that commutability must be assessed for all assays, however similar, for which a given reference material might be used, as required in current ISO guidelines. Not only must international reference standards be commutable, but secondary reference materials traceable to the international standard must also demonstrate this property for all assays in which they will be used. While relatively straightforward if only a few commercial assays are in use for a given analyte (such as for HIV or hepatitis C virus [HCV]), the challenge becomes much more burdensome if commercial assays are lacking or absent, as in the case of CMV and EBV, for which testing is performed largely with laboratory-developed tests. The difficulty in establishing commutability is compounded if multiple calibrators are on the market and is almost untenable if no commercial material is available. Even in cases such as CMV, these studies require large volumes of clinical specimens in order to simultaneously compare many tests. A primary limitation here was the ability to assess only five quantitative assays. To move beyond this to a more comprehensive evaluation of a greater number of assays would require large volumes of clinical specimens and might not be feasible with a single blood draw. Alternative solutions might be needed, such as pooling of clinical specimens from the same patient over multiple time points. While advantageous in creating a larger volume for comparative purposes, a potential drawback is dilution of matrix effects that may be present only at a single time point in a given patient. However, the progression of work in this area will continue to demand this or similar solutions. Another potential limitation could relate to the use of only a single lot of commercial CMV calibrator among all of the sites in the study. While advantageous in removing lot-to-lot variation from the assessment of commutability, this may also have produced better interlab agreement than might occur with routine use of reagents from multiple manufacturing lots.
Our findings help confirm and expand upon prior work, pointing to the critical importance of standardization in quantitative clinical lab testing. More than that is the finding that standardization and the trueness that hopefully accompanies it will not reliably be achieved through the use of common reference materials alone. This study demonstrates that commutability of such reference materials, particularly quantitative calibrators, must be proven for every assay and sample matrix to which those calibrators are applied. Future work in this area will be essential, for in its absence there is a risk of generating results that lack the accuracy and portability needed for optimal patient care.
Supplementary Material
ACKNOWLEDGMENTS
R.T.H. has a sponsored research agreement with Roche Diagnostics and research agreements with Luminex and Focus Diagnostics. A.M.C. receives clinical trial support from Roche Molecular and Qiagen and is a Roche Diagnostics Advisory Board member. M.D.S., J.B., and E.R.S. are employed by Life Technologies. A.V. has grant support from Qiagen and serves on an advisory board for Qiagen. J.K.P. had a recent sponsored research agreement with Qiagen.
This work was supported in part by the Emory Center for AIDS Research (P30 AI050409) and by the American Lebanese Syrian Associated Charities (ALSAC).
We thank Jess Ingersoll, Deborah Abdul-Ail, and Doris Igwe for their technical expertise.
Footnotes
Published ahead of print 11 September 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JCM.02036-13.
REFERENCES
- 1.Hayden RT, Hokanson KM, Pounds SB, Bankowski MJ, Belzer SW, Carr J, Diorio D, Forman MS, Joshi Y, Hillyard D, Hodinka RL, Nikiforova MN, Romain CA, Stevenson J, Valsamakis A, Balfour HH, Jr, US EBV Working Group 2008. Multicenter comparison of different real-time PCR assays for quantitative detection of Epstein-Barr virus. J. Clin. Microbiol. 46:157–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hayden RT, Yan X, Wick MT, Rodriguez AB, Xiong X, Ginocchio CC, Mitchell MJ, Caliendo AM. 2012. Factors contributing to variability of quantitative viral PCR results in proficiency testing samples: a multivariate analysis. J. Clin. Microbiol. 50:337–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pang XL, Fox JD, Fenton JM, Miller GG, Caliendo AM, Preiksaitis JK. 2009. Interlaboratory comparison of cytomegalovirus viral load assays. Am. J. Transplant. 9:258–268 [DOI] [PubMed] [Google Scholar]
- 4.Preiksaitis JK, Pang XL, Fox JD, Fenton JM, Caliendo AM, Miller GG. 2009. Interlaboratory comparison of Epstein-Barr virus viral load assays. Am. J. Transplant. 9:269–279 [DOI] [PubMed] [Google Scholar]
- 5.Wolff DJ, Heaney DL, Neuwald PD, Stellrecht KA, Press RD. 2009. Multi-site PCR-based CMV viral load assessment-assays demonstrate linearity and precision, but lack numeric standardization: a report of the association for molecular pathology. J. Mol. Diagn. 11:87–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Holden MJ, Madej RM, Minor P, Kalman LV. 2011. Molecular diagnostics: harmonization through reference materials, documentary standards and proficiency testing. Expert Rev. Mol. Diagn. 11:741–755 [DOI] [PubMed] [Google Scholar]
- 7.Madej RM, Davis J, Holden MJ, Kwang S, Labourier E, Schneider GJ. 2010. International standards and reference materials for quantitative molecular infectious disease testing. J. Mol. Diagn. 12:133–143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Caliendo AM, Shahbazian MD, Schaper C, Ingersoll J, Abdul-Ali D, Boonyaratanakornkit J, Pang XL, Fox J, Preiksaitis J, Schonbrunner ER. 2009. A commutable cytomegalovirus calibrator is required to improve the agreement of viral load values between laboratories. Clin. Chem. 55:1701–1710 [DOI] [PubMed] [Google Scholar]
- 9.National Institute for Biological Standards and Control 2013. 1st WHO international standard for Epstein-Barr virus for nucleic acid amplification techniques. NIBSC code 09/260, National Institute for Biological Standards and Control, Potters Bar, Hertfordshire, United Kingdom [Google Scholar]
- 10.Fryer JF, Heath AB, Anderson R, Minor PD, Collaborative Study Group 2010. Collaborative study to evaluate the proposed 1st WHO international standard for human cytomegalovirus (HCMV) for nucleic acid amplification (NAT)-based assays. National Institute for Biological Standards and Control, Potters Bar, Hertfordshire, United Kingdom [Google Scholar]
- 11.Haynes RJ, Kline MC, Toman B, Scott C, Wallace P, Butler JM, Holden MJ. 2013. Standard reference material 2366 for measurement of human cytomegalovirus DNA. J. Mol. Diagn. 15:177–185 [DOI] [PubMed] [Google Scholar]
- 12.Fasce CF, Jr, Rej R, Copeland WH, Vanderlinde RE. 1973. A discussion of enzyme reference materials: applications and specifications. Clin. Chem. 19:5–9 [PubMed] [Google Scholar]
- 13.Miller WG, Myers GL, Rej R. 2006. Why commutability matters. Clin. Chem. 52:553–554 [DOI] [PubMed] [Google Scholar]
- 14.Vesper HW, Miller WG, Myers GL. 2007. Reference materials and commutability. Clin. Biochem. Rev. 28:139–147 [PMC free article] [PubMed] [Google Scholar]
- 15.Cattozzo G, Franzini C, d'Eril GV. 2001. Myoglobin and creatine kinase isoenzyme MB mass assays: intermethod behaviour of patient sera and commercially available control materials. Clin. Chim. Acta 303:55–60 [DOI] [PubMed] [Google Scholar]
- 16.Cattozzo G, Franzini C, Melzi d'Eril GM. 2001. Commutability of calibration and control materials for serum lipase. Clin. Chem. 47:2108–2113 [PubMed] [Google Scholar]
- 17.Eckfeldt JH, Copeland KR. 1993. Accuracy verification and identification of matrix effects. The College of American Pathologists' protocol. Arch. Pathol. Lab. Med. 117:381–386 [PubMed] [Google Scholar]
- 18.Franzini C, Ceriotti F. 1998. Impact of reference materials on accuracy in clinical chemistry. Clin. Biochem. 31:449–457 [DOI] [PubMed] [Google Scholar]
- 19.ISO 2009. In vitro diagnostic medical devices—measurement of quantities in samples of biological origin—requirements for certified reference materials and the content of supporting documentation. ISO 15194:2009. ISO, Geneva, Switzerland [Google Scholar]
- 20.Ross JW, Miller WG, Myers GL, Praestgaard J. 1998. The accuracy of laboratory measurements in clinical chemistry: a study of 11 routine chemistry analytes in the College of American Pathologists Chemistry Survey with fresh frozen serum, definitive methods, and reference methods. Arch. Pathol. Lab. Med. 122:587–608 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.